Abstract
Wastewater-based epidemiology (WBE) is an emerging tool that gives temporal and spatial information on a population's health status. Here, we report the epidemiological dynamics of a population of ~1.2 million residents in the metropolitan region of Mendoza province, Argentina, within the period July 2020 to January 2021. We combined the use of WBE of two wastewater treatment plants with epidemiological surveillance of the corresponding populations. We applied two viral concentration methods (polyethylene glycol precipitation and aluminum-based adsorption-flocculation) and RNA isolation methods in each wastewater sample to increase the possibility of detection and quantification of nucleocapsid markers (N1 and N2) of SARS-CoV-2 by RT-qPCR. Overall, our results allowed us to trace the rise, exponential growth, plateau, and fall of SARS-CoV-2 infections for 26 weeks. Individual analysis for each wastewater treatment plant showed a positive correlation between the viral load of SARS-CoV-2 genetic markers and COVID-19 cases that were diagnosed per week. Our findings indicate that WBE is a useful epidemiological indicator to anticipate the increase in COVID-19 cases and monitor the advance of the pandemic and different waves of infections.
Abbreviations: CE, Campo Espejo; COVID-19, Coronavirus disease-19; Cq, quantification cycle; EP, El Paramillo; LOD, limit of detection; PAC, aluminum polychloride; PEG, polyethylene glycol; SARS-CoV-2, acute respiratory syndrome-coronavirus 2; WBE, wastewater-based epidemiology; WWTP, wastewater treatment plant
Keywords: COVID-19, SARS-CoV-2, Wastewater-based epidemiology, Pandemic, Epidemiologic surveillance
Graphical abstract
1. Introduction
Coronaviruses have caused epidemic diseases in the last century (Leung et al., 2004; Lim et al., 2016), which have been managed with varying degrees of success before becoming a global health problem. Coronavirus disease-19 (COVID-19), caused by acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), is a current health problem that affects more than one hundred million people and has caused more than three million deaths (April 2021, WHO, 2021). Human-to-human transmission occurs by droplets, aerosols, and fomites that originate in the respiratory system of an infected person (Jones et al., 2020; Sharma et al., 2021). Moreover, this virus reproduces in the human gastrointestinal tract (Xiao et al., 2020) and is then expelled through feces (Guan et al., 2020; Ling et al., 2020; Wang et al., 2020).
Governments around the world have adopted different non-pharmaceutical interventions to reduce the viral spread (Amer et al., 2020; Haug et al., 2020; Hsiang et al., 2020; Kucharski et al., 2020), many of which have been economically and socially disruptive (Alzúa and Gosis, 2020; Lin and Meissner, 2020; Lustig et al., 2020). Among them, social lockdowns have been used (Alvi et al., 2020; Ferguson et al., 2020) with the goal of flattening the epidemic curve, avoiding intensive care unit saturation, and reducing the pressure exerted on the basic components of the health system. However, it is plausible that the transmission of SARS-CoV-2 do not stop, mainly due to its high contagiousness and the occurrence of asymptomatic patients who spread viruses silently without ever being detected (Kronbichler et al., 2020; Li et al., 2020).
The prevalence of COVID-19 has been assessed by implementing different clinic diagnostic tests (Chau et al., 2020; Hanson et al., 2020). The success of this strategy is variable, particularly when applied in places where the scarcity of consumables or reagents limits the epidemiological monitoring of the population health. These limitations have prompted the development of complementary epidemiological strategies to reliably demonstrate viral community circulation (Foladori et al., 2020).
Wastewater-based epidemiology (WBE) provides information about the population exposure to environmental chemicals, prevalence of pharmaceuticals (Gracia-Lor et al., 2018), and diseases that produce biomarkers (Lorenzo and Picó, 2019). Thus, WBE could give temporal and spatial information on the health status of a population regarding etiologic agents that are eliminated mainly in human's feces or urine (Ahmed et al., 2021; Barril et al., 2010; Ferreyra et al., 2015; Hasan et al., 2021; Hata et al., 2021; Hokajärvi et al., 2021; Kumar et al., 2020; La Rosa et al., 2020; Mueller et al., 2009; Prado et al., 2021; Randazzo et al., 2020; Sherchan et al., 2020; Westhaus et al., 2021; Yanez et al., 2014). Moreover, the use of WBE for the detection of SARS-CoV-2 RNA in the sewage system (Ahmed et al., 2020b; Medema et al., 2020) may anticipate the increase in clinical cases at the population level (Ahmed et al., 2021; Larsen and Wigginton, 2020; Nemudryi et al., 2020) and overcome the difficulty of detecting asymptomatic people that represent a natural reservoir of SARS-CoV-2 (Thompson et al., 2020) and escape clinical diagnostic tests (Ling et al., 2020).
A variety of SARS-CoV-2 concentration protocols in wastewater have been reported (Ahmed et al., 2020a; Jafferali et al., 2021; Lu et al., 2020), which enhance the possibility of detecting SARS-CoV-2 RNA in wastewater. Likewise, polyethylene glycol (PEG) precipitation and aluminum polychloride (PAC) flocculation are efficient methods for SARS-CoV-2 recovery from complex aqueous matrixes (Barril et al., 2021; Pérez-Cataluña et al., 2021); however, there are no reports on the joint application of both concentration protocols during an epidemiological surveillance.
In this study, we aimed to monitor the epidemic evolution of SARS-CoV-2 in Mendoza, Argentina, between July 2020 and January 2021, by analyzing sewage samples from two wastewater treatment plants, which together serve a population of ⁓1.2 million residents. We applied two combinations of viral concentration methods (PEG and PAC) and RNA isolation methods in each wastewater sample to increase the possibility of detection and quantification of nucleocapsid markers (N1 and N2) of SARS-CoV-2 by RT-qPCR. Then, we compared the temporal changes in the number of SARS-CoV-2 genetic copies per 100 mL of wastewater in each treatment plant with the weekly COVID-19 cases reported by the Mendoza Health Ministry.
2. Materials and methods
2.1. Sampling
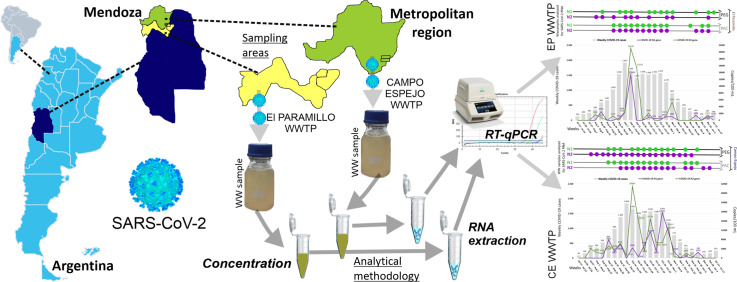
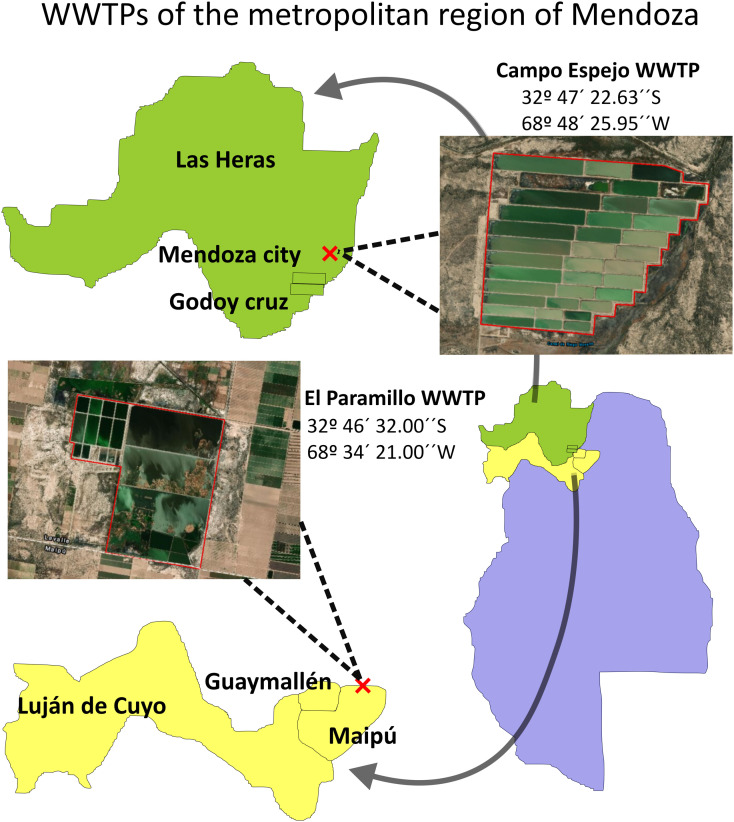
Mendoza province is located in the center-west of Argentina and has an approximate population of 1,915,759 inhabitants (Mendoza Health Ministry; https://www.mendoza.gov.ar/prensa/salud). The metropolitan region, with a population of ⁓1,191,649 inhabitants (Mendoza Health Ministry, January 2020), is integrated by six geopolitical districts, i.e., Las Heras, Mendoza city, Godoy Cruz, Guaymallén, Luján de Cuyo, and Maipú. Two wastewater treatment plants, namely El Paramillo (EP) and Campo Espejo (CE), receive and treat domestic and commercial wastewater from these districts (Fig. 1 ). EP is a secondary treatment plant (⁓350 ha) with stabilization lagoons that include four series of 3 lagoons each (anaerobic, facultative, and maturation), and two wetlands (Campo Este y Campo Norte; Irrigation General Department, http://aquabook.agua.gob.ar/). This plant receives 151,474 m3/day (GDI) and serves a population of ⁓545,000 inhabitants (from Guaymallén, Luján de Cuyo, and Maipú) (Fig. 1). CE is a secondary treatment plant (⁓321 ha) with stabilization lagoons that include twelve series of 3 lagoons each (anaerobic, facultative, and maturation) (Irrigation General Department, http://aquabook.agua.gob.ar/). This plant receives an average flow rate of 128,000 m3/day and serves a population of ⁓470,000 inhabitants (Las Heras, Mendoza city, and Godoy Cruz; Fig. 1). The reclaimed water in the secondary treatment plants is not discharged into aquatic environments (rivers or lakes) nor reused as recreational water. However, this water circulates continuously through canals and is then used for crop irrigation.
Fig. 1.
Mendoza province sampling areas, Argentina. Campo Espejo and El Paramillo treatment plants are delimited by red lines on satellite images. Rectangular spaces within the red lines are the treatment lagoons arranged to guarantee the wastewater flow between them by the unevenness of the terrain.
The operating company of both treatment plants (AYSAM S.A.) performed all wastewater samplings during 26 weeks. Sampling comprised the weekly manual collection of influent samples (obtained between 11:00 am and 1:00 pm) from each treatment plant between July 22, 2020, and November 30, 2020. In addition, samplings were obtained every two weeks during December 2020 and January 2021. No rainfall or storm events occurred on the sampling days or before them.
Immediately after being collected, samples were stored at 4 °C and shipped to the laboratory for thermal inactivation by immersing bottles in a water bath at 60 °C for 90 min (Hasan et al., 2021; Pastorino et al., 2020) to increase the safety of the laboratory personnel and work environment. Triplicate aliquots of 300 mL untreated wastewater were stored at −20 °C to maintain the integrity of genetic material (Hasan et al., 2021).
2.2. PEG and PAC concentration methods
We concentrated SARS-CoV-2 genetic material from wastewater samples following reported protocols of viral concentration (PEG precipitation and PAC flocculation methods), which have shown high viral recovery efficiency from wastewater samples (Barril et al., 2021; Randazzo et al., 2020). PEG precipitation had a mean recovery of 8.4% and PAC flocculation of 24.0% when wastewater samples were seeded with SARS-CoV-2 at a concentration higher than 4.300 genetic copies by mL (Barril et al., 2021).
The procedure of PEG precipitation was similar to that reported by Lewis and Metcalf (1988) and as “protocol 4” in Barril et al. (2021). The PAC flocculation procedure was similar to that reported by Randazzo et al. (2020), with some modifications. Briefly, 30 mL of a 8.4% PAC (pH = 4) solution was added to 300 mL of a wastewater sample and mixed on a shaker at 150 rpm for 15 min. Then, it was centrifuged at 1700 ×g for 20 min to flocculate PAC and SARS-CoV-2 genetic material from the bulk sample. After discarding the supernatant, the floc was suspended in 10 mL 3% beef extract (pH = 7.4) and mixed on a shaker at 150 rpm for 10 min. Then it was centrifuged at 1900 ×g for 30 min. Finally, the obtained precipitate was suspended in 2.5 mL PBS pH = 7.4 and stored at -20 °C for further RNA extraction and RT-qPCR assays.
2.3. RNA isolation and extraction
To assess for the presence of SARS-CoV-2 in wastewater samples (Section 2.2), we performed nucleic acid purification by using NucleoZOL® in combination with either (1) a manual method based on nucleic acid retention by silica spin columns (NucleoSpin RNA Set for NucleoZOL®, Macherey-Nagel, Germany) for PEG-processed samples, or (2) the chemical and mechanical disruption and further retention on a silica membrane column (NucleoSpin RNA Stool®, Macherey-Nagel, Germany) for PAC-processed samples.
We performed both methods of RNA isolation for each wastewater sample. Two hundred microliters of the PEG-processed sample were lysed with NucleoZOL®, and DNA and proteins were precipitated by centrifugation. Mixing the supernatant with a buffer helped bind the RNA to the silica membrane of the column. After that, we made two washing cycles with a specific buffer solution provided in the commercial kit and obtained the RNA by elution with 60 μL of RNAse-free water. On the other hand, 220 μL of the PAC-processed sample were treated with NucleoZOL® and a Lysis Buffer following the manufacturer's protocol. These samples were mixed with ceramic beads and then lysed on a shaker at room temperature for 10 min. After sample centrifugation at 10,000 ×g for 15 min, the obtained supernatant was passed through an Inhibitor Removal Column® to retain molecules that could interfere in RT-qPCR assays, and through a silica column to retain nucleic acids. The putative retained DNA was digested with DNAse, followed by three washing steps with specific buffers provided in the commercial kits. Finally, we performed the elution of RNA with 80 μL of RNAse-free water.
The total RNA was quantified using Nanodrop® equipment, and then the remaining eluate was aliquoted and stored at −20 °C for further processing by RT-qPCR assays.
2.4. SARS-CoV-2 genetic markers detection and quantification
SARS-CoV-2 genetic markers were detected by RT-qPCR using iTaq universal probe one-step kit (Bio-Rad Life Science, USA). The 2019-nCoV CDC EUA Kit (IDT#10006606, L#0000512209) targeting both N1 and N2 regions that codify for the viral nucleocapsid. A 5 μL-aliquot of the RNA-containing eluate (Section 2.3) was used as a template in a final volume of 20 μL. The RT-qPCR amplification cycles were performed according to the manufacturer's recommendations: 50 °C for 10 min and 95 °C for 3 min, followed by 45 cycles at 95 °C for 15 s and 60 °C for 50 s. We included positive (SARS-CoV-2 RNA from COVID-19 patients) and negative (nuclease-free water) reactions as analytical quality control. We considered the RT-qPCR reactions positive if the observed Cq (quantification cycle) was less than 40. We assumed the presence of inhibitors in those samples containing RNA that were negative for RT-qPCR; in these cases, a dilution of sample (1:5, v/v) was made (Peccia et al., 2020). All RT-qPCR assays were performed on a BioRad CFX 96 thermal cycler, and the run data were analyzed in CFX Maestro software (Biorad). The threshold was set at 30, and each sample was quantified in duplicate, for which we provided a mean value.
We quantified the viral load in wastewater samples using standard curves of a plasmid (2019-nCoV_N_Positive Control, IDT) that codifies for N1 and N2 regions. Linear dynamic range was evaluated among 25 and 5000 genetic copies for each marker. Standard curves had an R2 value of 0.995 (slope = -3.231; y-intercept = 39.619) for N1 and an R2 value of 0.994 (slope = -3.733; y-intercept = 40.925) for N2. We expressed the results as the number of genetic copies in 100 mL of wastewater (copies/100 mL). Results of viral loads obtained from PAC-processed samples, after RNA dilutions, are shown in Supplementary Tables 1–4.
2.5. Variables related to epidemiological surveillance
The number of both, accumulated and weekly COVID-19 cases, was provided by the Mendoza Health Ministry. In addition, the weekly number of deaths due to COVID-19 was obtained from daily communications by the Ministry (https://www.mendoza.gov.ar/prensa/salud/). The clinical test positivity (%) was calculated according to the recommendations of the Centers for Disease Control and Preventions (CDC, USA) (https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/calculating-percent-positivity.html), i.e., the average (week) daily number of positive clinical test divided by the average (week) daily number of performed clinic tests and then multiplied by 100.
2.6. Statistical analysis
Pearson's correlations analyses were performed to test the correlation between weekly COVID-19 cases and the number of genetic copies by 100 mL of wastewater samples from both treatment plants. The significance level was fixed at P < 0.05.
3. Results
3.1. Epidemiological data in the Mendoza province, Argentina
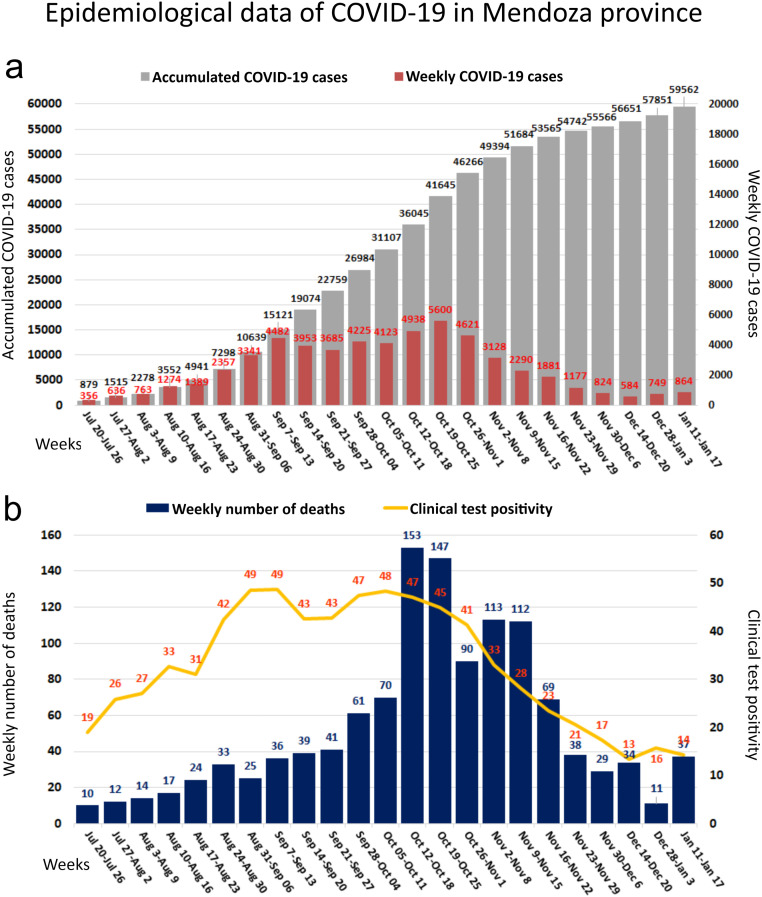
Fig. 1 shows the location and the area served by Campo Espejo and El Paramillo, with a population of ~1.2 million people. We processed the samples from these plants during the COVID-19 outbreak from July 2020 to January 2021. The Mendoza Health Ministry declared the community viral circulation by conglomerate on July 20, 2020, when weekly COVID-19 cases reached 356, and the accumulated COVID-19 cases were 879 (Fig. 2a). The COVID-19 cases increased weekly between August 10, 2020, and September 13, 2020, and this variable reached the maximum value (5600 cases) at the end of October. The weekly changes in the percentage of positive clinical tests (%) and the weekly number of deaths due to COVID-19 showed a similar temporal pattern (Fig. 2b). The percent of positive clinical tests was higher than 40% between the end of August and the end of October. On the other hand, the highest number of deaths occurred between October 12 and 25, 2020, i.e., when high numbers of weekly COVID-19 cases (4938–5600) were newly diagnosed (Fig. 2).
Fig. 2.
Epidemiological data of SARS-CoV-2 pandemic in Mendoza, Argentina, from July 2020 to January 2021. a. Temporal evolution in the accumulated and weekly COVID-19 cases. b. Temporal evolution in the weekly number of deaths by COVID-19 and percentage of positive clinical tests (%).
3.2. SARS-CoV-2 detection in wastewater samples
We detected SARS-CoV-2 RNA by RT-qPCR using two primer sets, N1 and N2 (Supplementary Tables 1 and 2), already used and validated for clinical laboratory testing. The average Cq for wastewater samples processed using PEG precipitation was 34.1 ± 0.35 (mean ± SEM) and 34.4 ± 0.38 (mean ± SEM) for N1 and N2 primers, respectively. The average Cq for wastewater samples processed using PAC flocculation was 34.3 ± 0.34 (mean ± SEM) for N1 and 34.6 ± 0.41 (mean ± SEM) for N2. Some RT-qPCR results were negative when RNA was extracted from wastewater samples processed by flocculation with PAC. This finding suggested that a material that was co-purified with total RNA inhibited the RT-qPCR. We found that the dilution (1/5; v/v) of the total RNA allowed the amplification and detection of the SARS-CoV-2 genetic markers N1 and N2 (Supplementary Tables 3 and 4).
3.3. Correlation between WBE from each treatment plant and epidemiological data
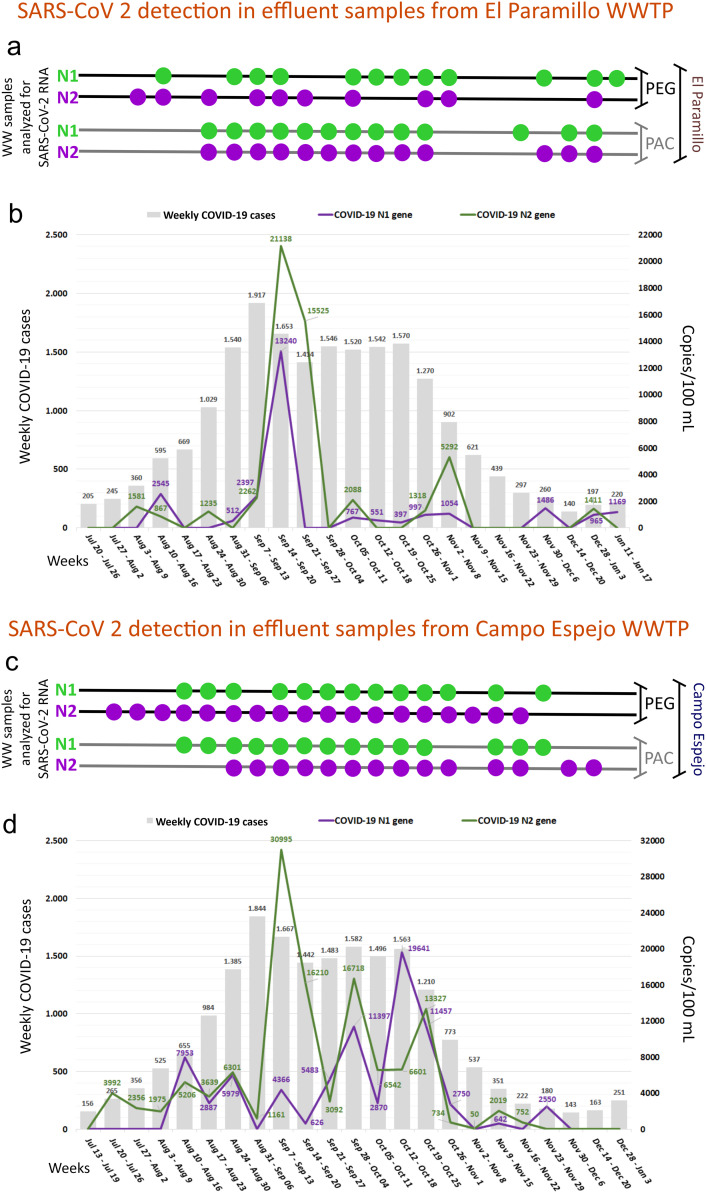
Fig. 3 shows a representative analysis of SARS-CoV-2 RNA from EP and CE, concerning the epidemiology of the places they serve. N2 of SARS-CoV-2 RNA was the first marker to become positive on August 3rd, 2020. The SARS-CoV-2 RNA detection frequencies were similar in both viral concentration procedures (PEG = 15/23 samples vs. PAC = 14/23 samples; Supplementary Table 1 and Fig. 3a). From early to mid-August, SARS-CoV-2 N1 and N2 markers became positive when low weekly COVID-19 cases (360 to 595) were reported in the places served for the EP WWTP (Fig. 3a). Wastewater samples were detectable for SARS-CoV-2 RNA genetic markers on August 3rd and 11th before the biweekly doubling of weekly COVID-19 cases. Between September 7–13, 2020 –week with the highest value of COVID-19 cases (N = 1917)– and the end of October, wastewater samples were positive for N1 and N2 of SARS-CoV-2 during eight consecutive weeks. Surprisingly, wastewater samples from EP remained detectable between late November and December, when Mendoza Health Ministry reported between 140 and 290 weekly COVID-19 cases. A correlation analysis between the weekly COVID-19 cases and a load of SARS-CoV-2 genetic markers in the influent samples was also made (N1: r = 0.3185, P = 0.1386; N2: r = 0.3890, P = 0.1514). Both genetic markers, N1 and N2, showed the highest values in the number of genetic copies per 100 mL (copies/100 mL) of wastewater sample (N1 = 13,240; N2 = 21,138) during September 14–20, 2020 (Fig. 3b and Supplementary Table 1). In addition, the N2 marker showed a second peak (5292 copies/100 mL) in the week of November 2–8, 2020 (Fig. 3b and Supplementary Table 1).
Fig. 3.
SARS-CoV-2 detection and epidemiological data from the area served by El Paramillo (a–b) and Campo Espejo (c–d) WWTPs. Wastewater sample positivity for both genetic markers, N1 (green circles) and N2 (purple circles), which codify for the viral nucleocapsid of SARS-CoV-2 (a and c). Relationship between weekly COVID-19 cases and the number of SARS-CoV-2 genetic copies per 100 mL of wastewater (b and d).
Campo Espejo WWTP receives sewage influents from people living in Mendoza city, Las Heras, and Godoy Cruz districts. CE WWTP data showed (Fig. 3c-d) a temporal pattern similar to that observed in the El Paramillo WWTP. N2 from SARS-CoV-2 RNA was the first marker to become positive on July 30, 2020. The SARS-CoV-2 RNA detection frequencies were similar in both viral concentration procedures (PEG = 19/23 samples vs. PAC = 17/23 samples; Supplementary Table 2 and Fig. 3c). Weekly COVID-19 cases were correlated with the number of genetic copies of SARS-CoV-2 (N1, r = 0.5468, P = 0.0069; N2, r = 0.6282, P = 0.3946). Wastewater samples were detectable for SARS-CoV-2 genetic markers on August 11th (PEG) and on August 19th (PEG and PAC), before the exponential growth phase of weekly COVID-19 cases. After that, the wastewater samples were positive for SARS-CoV-2 (PEG and PAC) during twelve consecutive weeks (Fig. 3c), with four synchronous peaks in the number of genetic copies of N1 and N2 (Fig. 3d). From November 9 to November 29, weekly COVID-19 cases diminished gradually while the wastewater samples remained positive. Between mid-November and December, weekly COVID-19 cases and the number of genetic copies were low (Fig. 3d).
4. Discussion
Many authors in different countries started to detect the SARS-CoV-2 virus in wastewater a few weeks after the WHO declared the COVID-19 pandemic (Ahmed et al., 2020b; Haramoto et al., 2020; La Rosa et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). Those studies highlighted the utility of viral RNA monitoring in wastewater for SARS-CoV-2 infection surveillance at a population-wide level. The present paper is part of a research program aimed to examine wastewater-based surveillance for assessing the circulation of the SARS-CoV-2 virus in a population.
The use of WBE in the surveillance of COVID-19 is based on human SARS-COV-2 excretion through the gastrointestinal tract. The viral genetic material can thus be detected in the community drainage system and wastewater treatment plants (Ahmed et al., 2021; Medema et al., 2020), even before COVID-19 patients are detected in the population and up to 21 days after the number of diagnosed cases decrease to baseline (Amirian, 2020; Yeo et al., 2020). However, the effective use of WBE depends on (a) the coverage of the sewage system of the community under study, (b) the development of standardized protocols for SARS-CoV-2 concentration from wastewater samples, and (c) the detection and quantification of molecular markers (Bogler et al., 2020; Kantor et al., 2021; Mohapatra et al., 2020; Thompson et al., 2020).
In this work, monitoring of SARS-CoV-2 was evaluated by a combination of viral concentration and RNA extraction strategies (PEG precipitation–NucleoZOL–silica membrane column; PAC flocculation–NucleoZOL–Inhibitor Removal Column–silica membrane column). These approaches have already been used to recover SARS-CoV-2 from environmental water samples (Barril et al., 2021; La Rosa et al., 2020; Pérez-Cataluña et al., 2021; Randazzo et al., 2020) because they showed adequate viral recoveries when wastewater matrices artificially were seeded with SARS-CoV-2 (Ahmed et al., 2020a; Barril et al., 2021; Lewis and Metcalf, 1988). Some comparisons between both viral concentration strategies may highlight the possible significance for a combined methodological application in WBE: (a) PEG precipitation appears more sensitive than PAC flocculation, (b) the temporal window of SARS-CoV-2 detection becomes larger when both methodologies are applied, (c) two SARS-CoV-2 RNA extraction kits were assessed to remove RT-qPCR inhibitors, (d) the viral load in samples processed by PAC are often higher than those processed by PEG, which appears to be associated with a reported high recovery in the former (Barril et al., 2021), and (e) the Ct values for N1 and N2 ranged between 30 and 40 for both vital concentration protocols as reported previously (Medema et al., 2020; Pérez-Cataluña et al., 2021).
The SARS-CoV-2 RNA detection frequencies were similar in both treatment plants using the two viral concentration procedures (Supplementary Tables 1 and 2). For this reason, we can point out that the simultaneous application of optimized and standardized protocols of viral recovery and molecular markers may facilitate the detection of SARS-CoV-2 in wastewater.
The increases in SARS-CoV-2 N1 and N2 genetic markers (copies/100 mL) in treatment plants occurred 3–6 days ahead of those in the weekly confirmed COVID-19 cases. In a similar study in New Haven, Connecticut, USA, Peccia et al. (2020) showed that SARS-CoV-2 RNA concentrations from primary sewage sludge were 1–4 days ahead of local hospital admissions and 6–8 days ahead of SARS-CoV-2 positive test results by reporting date.
The limit of detection (LOD) of the 2019-nCoV plasmid (CDC, USA) by RT-qPCR for N1 and N2 was 25 copies/reaction. In both treatment plants, SARS-CoV-2 N2 became positive one week earlier than the N1 marker, and the former showed the highest concentrations in the plateau during the first wave (Fig. 3). Medema et al. (2020) have reported discrepancies among CDC N1 trials with CDC N2, CDC N3, and E_Sarbeco for various wastewater samples in the emerging epidemic in the Netherlands. Wu et al. (2020) reported variable levels of SARS-CoV-2 RNA in wastewater samples in Massachusetts, USA, using CDC N1, N2, and N3 assays. Reciently, Ahmed et al. (2021) reported SARS-CoV-2 N1 and E but no N2 markers in wastewater samples at three WWTPs in Southeast Queensland, Australia. The virus nucleocapsid (N) gene is widely used for WBE; however, additional studies are necessary to know the kinetics of each SARS-CoV-2 molecular marker in wastewater samples.
Despite the differences in the detections of both genes, the combined analysis reflects the epidemic profile obtained by the clinical cases reported by the Health Ministry in both studied regions. Overall, our results allow us to trace the rise, exponential growth, plateau, and fall of SARS-CoV-2 infections during 26 weeks (Fig. 2, Fig. 3). A similar study in France showed an increase in SARS-CoV-2 genetic copies before that in COVID-19 cases (Wurtzer et al., 2020). These results show the usefulness of WBE to reveal the dynamics of infection before diagnostic tests and provide nearer real-time information on the prevalence of the disease (Larsen and Wigginton, 2020).
After the first detection, a constant increase in the number of genetic copies occurred at the beginning of the first epidemic wave of COVID-19 in Mendoza, Argentina. Then, the increases in viral load in both WWTPs were coincidental with an increase in reported clinical cases.
The first episodic detections of SARS-CoV-2 in wastewater –in both treatment plants in November though only in El Paramillo in December– evidenced the end of the epidemic outbreak. Finally, the decrease in the detection of positive samples for SARS-CoV-2 in wastewater coincides with the decrease in confirmed clinical cases of COVID-19, marking the end of the first epidemic COVID-19 wave.
5. Conclusions
The combined approach of WBE and clinical testing helped us report the public health status of a population of ⁓1 million residents in the metropolitan region of Mendoza, Argentina, between July 2020 and January 2021. We show that using at least two protocols to obtain RNA samples and detect different genetic markers of SARS-CoV-2 in wastewater samples is helpful to predict the increase in COVID-19 cases and to follow up the progression of the epidemic outbreak. Our findings thus indicate that WBE is a reliable tool that objectively provides a global vision and analysis of the progress of the pandemic in a community, overcoming the limitations of other epidemiological control tools that are economically costly, such as lockdown and massive testing, and is a valid alternative for detection of asymptomatic people.
CRediT authorship contribution statement
Maximiliano Giraud-Billoud: PAC precipitation, RNA extraction, RT-qPCR, Validation, Formal analysis, Investigation, Writing - review & editing, Graphical design and elaboration, Funding acquisition. Paula Cuervo: PEG precipitation, PAC precipitation validation, Writing - original draft. Jorgelina Altamirano: PEG precipitation validation, writing - original draft, Funding acquisition. Julieta Aranibar: PEG precipitation validation, writing - original draft. Marcela Pizarro: PAC precipitation, writing - original draft, Funding acquisition. Adolfo Catapano: sampling in WWTPs, Resources. Héctor Cuello: RT-qPCR validation. Gisela Masachessi: Conceptualization, Methodology, Formal analysis, Writing - original draft. Israel A. Vega: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Writing - original draft, Project administration, Funding acquisition.
Funding
This work was supported by grants from Ministerio de Ciencia, Tecnología e Innovación (Argentina) through “Programa de Articulación y Fortalecimiento Federal de las Capacidades en Ciencia y Tecnología COVID-19”. The project received financial support from the Departamento General de Irrigación de Mendoza and the AYSAM S.A. Agua y Saneamiento Mendoza.
Declaration of competing interest
The authors have declared that no competing interests exist.
Acknowledgments
We thank Dr. Sergio Carminati, Lic. Valeria Toti, and Lic. Arnaldo Sola for their technical assistance. We also thank the personnel of the operating company of both WWTPs (AYSAM S.A.), who collaborated with the samplings, and the Mendoza Health Ministry (Dirección de Planificación), who provided epidemiological data. Finally, we want to thank the selfless collaboration of Dr. Cristian Rodriguez and Dr. Alfredo Castro-Vazquez for the critical reading of the manuscript.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.148887.
Appendix A. Supplementary data
Supplementary tables
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., et al. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi M.M., Sivasankaran S., Singh M. Pharmacological and non-pharmacological efforts at prevention, mitigation, and treatment for COVID-19. J. Drug Target. 2020;28:742–754. doi: 10.1080/1061186X.2020.1793990. [DOI] [PubMed] [Google Scholar]
- Alzúa M.L., Gosis P. UNDP LAC C; 2020. Social and Economic Impact of Covid-19 and Policy Options in Argentina; p. 19. [Google Scholar]
- Amer F., Hammoud S., Farran B., Boncz I., Endrei D. Assessment of countries’ preparedness and lockdown effectiveness in fighting COVID-19. Disaster Med. Public Health Prep. 2020;217:1–8. doi: 10.1017/dmp.2020.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barril P., Giordano M., Isa M., Masachessi G., Ferreyra L., Castello A., et al. Correlation between rotavirus a genotypes detected in hospitalized children and sewage samples in 2006, Córdoba, Argentina. J. Med. Virol. 2010;82:1277–1281. doi: 10.1002/jmv.21800. [DOI] [PubMed] [Google Scholar]
- Barril P.A., Pianciola L.A., Mazzeo M., Ousset M.J., Jaureguiberry M.V., Alessandrello M., et al. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler A., Packman A., Furman A., Gross A., Kushmaro A., Ronen A., et al. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020;3:981–990. [Google Scholar]
- Chau C.H., Strope J.D., Figg W.D. COVID-19 clinical diagnostics and testing technology. Pharmacotherapy. 2020;40:857–868. doi: 10.1002/phar.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N., Laydon D., Nedjati Gilani G., Imai N., Ainslie K., Baguelin M., et al. Vol. 9. Imperial College COVID-19 Response Team; 2020. Report 9: Impact of Non-pharmaceutical Interventions (NPIs) to Reduce COVID19 Mortality and Healthcare Demand; pp. 1–20. [Google Scholar]
- Ferreyra L., Giordano M., Martinez L., Barril P.A., Masachessi G., Isa M.B., et al. Tracking novel adenovirus in environmental and human clinical samples: no evidence of endemic human adenovirus type 58 circulation in Cordoba city, Argentina. Epidemiol. Infect. 2015;143:1427–1431. doi: 10.1017/S0950268814002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., et al. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia-Lor E., Rousis N.I., Fl Hernández, Zuccato E., Castiglioni S. Wastewater-based epidemiology as a novel biomonitoring tool to evaluate human exposure to pollutants. Environ. Sci. Technol. 2018;52:10224–10226. doi: 10.1021/acs.est.8b01403. [DOI] [PubMed] [Google Scholar]
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Lee M.J., Loeb M., et al. Infectious Diseases Society of America guidelines on the diagnosis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., et al. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug N., Geyrhofer L., Londei A., Dervic E., Desvars-Larrive A., Loreto V., et al. Ranking the effectiveness of worldwide COVID-19 government interventions. Nat. Hum. Behav. 2020;4:1303–1312. doi: 10.1038/s41562-020-01009-0. [DOI] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang S., Allen D., Annan-Phan S., Bell K., Bolliger I., Chong T., et al. The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature. 2020;584:262–267. doi: 10.1038/s41586-020-2404-8. [DOI] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;55:3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kronbichler A., Kresse D., Yoon S., Lee K.H., Effenberger M., Shin J.I. Asymptomatic patients as a source of COVID-19 infections: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;98:180–186. doi: 10.1016/j.ijid.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski A.J., Klepac P., Conlan A.J., Kissler S.M., Tang M.L., Fry H., et al. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 2020;20:1151–1160. doi: 10.1016/S1473-3099(20)30457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung G.M., Hedley A.J., Ho L.M., Chau P., Wong I.O., Thach T.Q., Ghani A.C., Donnelly C.A., Fraser C., Riley S., Ferguson N.M., Anderson R.M., Tsang T., Leung P.Y., Wong V., Chan J.C., Tsui E., Lo S.V., Lam T.H. The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: an analysis of all 1755 patients. Ann. Intern. Med. 2004;141:662–673. doi: 10.7326/0003-4819-141-9-200411020-00006. [DOI] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shi J., Xia J., Duan J., Chen L., Yu X., et al. Asymptomatic and symptomatic patients with non-severe coronavirus disease (COVID-19) have similar clinical features and virological courses: a retrospective single center study. Front. Microbiol. 2020;11:1570. doi: 10.3389/fmicb.2020.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Meissner C.M. 2020. Health Vs. Wealth? Public Health Policies and the Economy During Covid-19; p. w27099. [Google Scholar]
- Ling Y., Xu S.-B., Lin Y.-X., Tian D., Zhu Z.-Q., Dai F.-H., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133:1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health. 2019;9:77–84. [Google Scholar]
- Lu D., Huang Z., Luo J., Zhang X., Sha S. Primary concentration–the critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: a mini-review. Sci. Total Environ. 2020;747 doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig N., Pabon V.M., Sanz F., Younger S.D. 2020. The Impact of COVID-19 Lockdowns and Expanded Social Assistance on Inequality, Poverty and Mobility in Argentina, Brazil, Colombia and Mexico; pp. 1–36. [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mohapatra S., Menon N.G., Mohapatra G., Pisharody L., Pattnaik A., Menon N.G., et al. The novel SARS-CoV-2 pandemic: possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci. Total Environ. 2020;765 doi: 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J.E., Bessaud M., Huang Q.S., Martinez L.C., Barril P.A., Morel V., et al. Environmental poliovirus surveillance during oral poliovirus vaccine and inactivated poliovirus vaccine use in Cordoba Province, Argentina. Appl. Environ. Microbiol. 2009;75:1395–1401. doi: 10.1128/AEM.02201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B., Touret F., Gilles M., de Lamballerie X., Charrel R.N. Heat inactivation of different types of SARS-CoV-2 samples: what protocols for biosafety, molecular detection and serological diagnostics? Viruses. 2020;12:735. doi: 10.3390/v12070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., et al. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.K., Jinadatha C., Lichtfouse E., Decroly E., van Helden J., Choi H., et al. COVID-19 epidemiologic surveillance using wastewater. Environ. Chem. Lett. 2021;19:1911–1915. doi: 10.1007/s10311-021-01188-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., et al. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO World Health Organization: Coronavirus Disease (COVID-19) Dashboard. 2021. https://covid19.who.int/ Accessed 26th Feb 2021.
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems. 2020;5:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., et al. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez L.A., Lucero N.S., Barril P.A., Díaz MdP, Tenaglia M.M., Spinsanti L.I., et al. Evidence of hepatitis A virus circulation in central Argentina: seroprevalence and environmental surveillance. J. Clin. Virol. 2014;59:38–43. doi: 10.1016/j.jcv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables