Abstract
Germ line mutations in the breast cancer susceptibility gene BRCA2 predispose to early-onset breast cancer, but the function of the nuclear protein encoded by the gene is ill defined. Using the yeast two-hybrid system with fragments of human BRCA2, we identified an interaction with the human DSS1 (deleted in split hand/split foot) gene. Yeast and mammalian two-hybrid assays showed that DSS1 can associate with BRCA2 in the region of amino acids 2472 to 2957 in the C terminus of the protein. Using coimmunoprecipitation of epitope-tagged BRCA2 and DSS1 cDNA constructs transiently expressed in COS cells, we were able to demonstrate an association. Furthermore, endogenous BRCA2 could be coimmunoprecipitated with endogenous DSS1 in MCF7 cells, demonstrating an in vivo association. Apparent orthologues of the mammalian DSS1 gene were identified in the genome of the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae. Yeast strains in which these DSS1-like genes were deleted showed a temperature-sensitive growth phenotype, which was analyzed by flow cytometry. This provides evidence for a link between the BRCA2 tumor suppressor gene and a gene required for completion of the cell cycle.
Hereditary predisposition to early-onset breast cancer is associated with germ line mutations in the breast cancer susceptibility genes BRCA1 and BRCA2, and BRCA2 mutations are found in up to half of patients with familial incidence of the disease (29, 48, 53). Both genes are thought to be tumor suppressors, since tumors in heterozygous mutation carriers have been shown to have lost the wild-type allele (3, 20, 23). Inheritance of germ line mutations in these genes is also associated with other cancers, including those of the ovary, pancreas, and prostate (17, 45, 50).
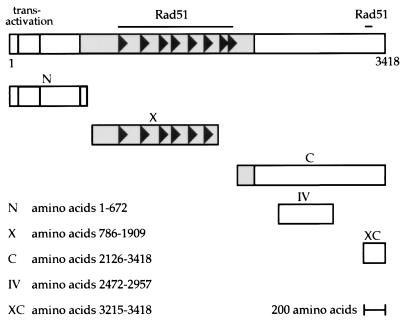
The BRCA2 gene is composed of 27 exons and encodes a 3,418-amino-acid protein with no obvious sequence similarity to other known proteins. BRCA2 appears to be a nuclear protein whose expression is regulated through the cell cycle (4). A potential role for the BRCA2 protein is suggested by the demonstration that it can bind to RAD51, the human equivalent of yeast RAD51, which is involved in recombination and DNA double-strand break repair (31, 42). This interaction has been shown to be mediated both by the C terminus of BRCA2 and by the BRC repeats, eight repeated amino acid sequences in the central, exon 11-encoded part of the protein (Fig. 1) (5, 6, 52). BRCA1 is also reported to interact with RAD51, which suggests a role for both proteins in DNA repair or maintenance of genomic integrity (41). Recently it has been suggested that BRCA2 might associate with BRCA1, suggesting a link between the two known breast cancer susceptibility genes, although the significance of this interaction is unclear (8). Further evidence for a role for BRCA2 in genomic integrity includes the finding of multiple chromosomal rearrangements in BRCA2 mutant cells (19). In addition, BRCA2 has also been implicated in the regulation of gene expression, with studies that have identified regions of the protein with transcriptional activity and histone acetylase activity (30, 44).
FIG. 1.
Diagrammatic representation of the human BRCA2 protein showing the regions used in this study. The section of the protein encoded by exon 11 is shaded, and the BRC repeats (6) are represented by filled arrows. The location of the N-terminal region reported to have transcriptional activity (30) and the Rad51-binding sites are indicated. The positions of the protein fragments expressed by the various constructs are shown beneath the diagram, with the extent indicated in amino acids.
BRCA2 is essential during mouse development, with homozygous null mutations resulting in early embryonic lethality (26, 42, 47). Reduced cell proliferation may explain the failure of the BRCA2-null embryos to develop, but heterozygous mice do not show an elevated susceptibility to cancer (26, 47). Mice with less extensive truncations of BRCA2 which retain portions of exon 11 have been developed (10, 16, 35) and used to demonstrate a hypersensitivity to γ irradiation and a defect in DNA double-strand break repair (10, 33).
Given the large size of BRCA2, it seems possible that it can interact with many proteins. To characterize further the function of BRCA2, the yeast two-hybrid system was used to identify such proteins. We have demonstrated an in vivo interaction between BRCA2 and the DSS1 (deleted in split hand/split foot) protein in mammalian cells. The DSS1 gene was originally identified on chromosome 7q21.3-q22.1 as a gene deleted in patients with the developmental disorder split hand/split foot malformation (SHFM) and codes for a 70-amino-acid, highly acidic peptide (13). Analysis of the function of the DSS1 gene in the yeast strains suggests that it is required for normal cell growth.
MATERIALS AND METHODS
Yeast two-hybrid screens.
Bait plasmids were generated by inserting selected cDNA fragments of human BRCA2 from sequenced constructs into pAS1-CYH2 (21), to produce in-frame fusions with GAL4 DNA-binding domain. These included a bait construct consisting of a C-terminal fragment encoding amino acids 2472 to 2957 of BRCA2 (BRCA2.IV). Plasmids were transformed into the Saccharomyces cerevisiae reporter strain Y190 (containing GAL4-lacZ and GAL4-HIS3 reporters), and selectants were checked for expression of the fusion proteins by immunoblotting yeast lysates with an anti-DNA-binding-domain antibody (Santa Cruz) or 12CA5 (Boehringer Mannheim), which recognizes the hemagglutinin (HA) tag epitope included in the construct. These yeast cultures were then transfected with a two-hybrid cDNA library, either from HeLa cells (Clontech) or from mammary tissue (Stratagene), and selected on triple-dropout medium supplemented with 25 mM 3-aminotriazole. Between 2.5 × 106 and 6.5 × 106 transformants were analyzed in each library screen. LacZ+ clones were identified by β-galactosidase assay using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate. Filter lifts were grown overnight at 30°C, snap frozen in liquid nitrogen, then incubated at 37°C in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 [pH 7.0]) with 40 mM β-mercaptoethanol and 0.06% X-Gal. Interacting cDNA clones were retrieved by transformation of Escherichia coli with DNA extracted from yeast clones and identified by sequencing with GAL4 activation domain primers. Library clones were checked for interaction by reintroduction into strain Y190 expressing the original bait plasmid and for specificity by transformation of Y190 with various combinations of constructs in pAS1 and pACTII (21).
Mammalian two-hybrid assay.
BRCA2.IV and DSS1 were cloned into vectors of the Mammalian Matchmaker Two-Hybrid system (Clontech) and transiently cotransfected with a GAL4-luciferase reporter construct, pFR-LUC (Stratagene), into 293T cells by using Lipofectamine (Gibco). Cells were harvested and assayed for luciferase activity after 48 h, using a luciferase assay system (Promega).
Anti-DSS1 antibody production.
Polyclonal rabbit anti-DSS1 antisera were raised by immunization with a C-terminal peptide encoding the last 13 amino acids of DSS1, AELEKHGYKMETS (Severn Biotech), or with full-length recombinant human DSS1 prepared from a thrombin-cleaved glutathione S-transferase (GST)–DSS1 fusion protein from a pGEX-KG construct expressed in E. coli BL21/DE3(pLysS). Antisera were checked for specific recognition of GST-DSS1 by immunoblotting and immunoprecipitation.
Coimmunoprecipitation.
Human DSS1 was cloned into pEFm6, a vector derived from pEF (27) which expresses an N-terminal Myc epitope-specific 9E10 tag (14). This construct was transiently cotransfected into COS cells by using Lipofectamine (Gibco) with various portions of human BRCA2 cloned into constructs derived from pMT-SM (39) which had been modified to encode N-terminal FLAG-tagged fusions. Constructs used were BRCA2.N (amino acids 1 to 672 of human BRCA2), BRCA2.X (amino acids 786 to 1909), and BRCA2.C (amino acids 2126 to 3148). Cells were harvested after 48 h by lysis in NETN buffer (0.1 M NaCl, 20 mM Tris [pH 8.0], 1 mM EDTA, 1% Nonidet P-40), and lysates were pooled and divided for parallel immunoprecipitation with 9E10 (mouse monoclonal anti-Myc epitope tag antibody), 290 (rabbit polyclonal anti-DSS1 antiserum), and a nonspecific control antibody 12CA5 (mouse monoclonal anti-HA tag antibody). Immunoprecipitates on protein G-Sepharose (Sigma) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% gel) and Western blotting with anti-FLAG antibody M2 (Kodak).
Coimmunoprecipitation of endogenous BRCA2 and DSS1 was performed with lysates in NETN buffer from asynchronously cycling MCF7 cells. Immunoprecipitations were performed in parallel with polyclonal anti-DSS1 antiserum 290, polyclonal anti-BRCA2 antiserum B (4), or their preimmune sera. The products were analyzed by SDS-PAGE (6% gel) and Western blotting with an anti-BRCA2 rat monoclonal antibody (4).
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline, permeabilized with 0.2% Triton X-100, and blocked with 10% fetal calf serum prior to antibody staining. Staining by anti-DSS1 polyclonal antiserum 290 was visualized with fluorescein isothiocyanate-labeled donkey anti-rabbit immunoglobulin G (Jackson ImmunoResearch).
S. pombe DSS1 deletion.
A Schizosaccharomyces pombe DSS1-like open reading frame located on chromosome I was identified in the database (cosmid c3G6; GenBank accession no. Z99167). We have designated this gene dss1+. A deletion cassette was constructed by cloning PCR fragments from genomic S. pombe DNA encoding 430 bp from immediately 5′ and 470 bp from immediately 3′ of the dss1+ open reading frame on either side of ura4+ in pBS.URA4 (from K. Willison). The cassette was transformed into a wild-type diploid strain (made by crossing two haploid strains, JY741 and JY742, from M. Yamamoto), Ura+ transformants were sporulated, tetrads were dissected, and the dss1/Δdss1 status of haploid clones was established by PCR. Haploid strains were maintained in YES medium (32) and routinely grown at 30°C. Phenotypic rescue was assessed by transformation of wild-type and Δdss1 strains with pREP41 (2) constructs encoding human DSS1 cDNA, S. pombe genomic dss1+, or empty vector.
S. cerevisiae DSS1 deletion.
An S. cerevisiae DSS1-like open reading frame was identified on chromosome IV (cosmid 9476; GenBank accession no. U28372). A deletion cassette was constructed by cloning PCR-amplified fragments of genomic S. cerevisiae DNA encoding 1.02 kb from immediately 5′ and 320 bp from immediately 3′ of the open reading frame on either side of URA3 in pBS.KS (38). The cassette was transformed into YPH499 strain (from M. Gustin) (7), Ura+-selected transformants were sporulated, tetrads were dissected, and haploid strains in which the DSS1/Δdss1 status was confirmed by PCR were established and maintained in YPD medium.
Yeast growth rates.
Comparative growth rates of wild-type and Δdss1 clones of S. pombe and S. cerevisiae were obtained by dilution of equivalent cultures to the same density (5 × 105 cells/ml), followed by 1:10 serial dilutions, plating on respective media, and incubation at 25, 30, or 35°C.
Flow cytometry.
The DNA content of S. pombe grown under the indicated conditions was analyzed by fluorescence-activated cell sorting (FACS) on a flow cytometer after fixation of cells with 70% ethanol and staining with propidium iodide (PI; 4 μg/ml in 50 mM sodium citrate) (15).
RESULTS
BRCA2 binds DSS1 in the yeast two-hybrid system.
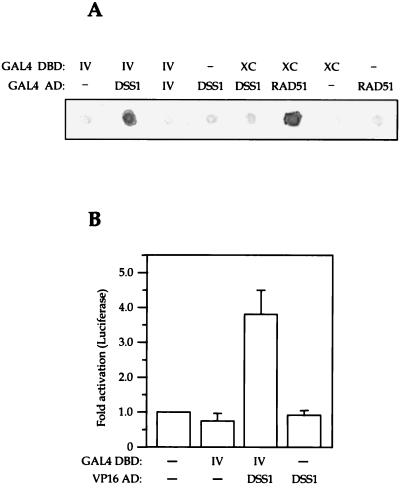
Yeast two-hybrid screens were carried out with various sections of human BRCA2 expressed as in-frame fusions with GAL4 DNA-binding domain in S. cerevisiae reporter strain Y190. Screens of two different libraries, derived from HeLa cells and human mammary tissue with BRCA2.IV (Fig. 1), repeatedly selected cDNA fragments derived from the same gene. Sequence database searches revealed that these cDNAs were derived from the previously identified DSS1 gene (13). This gene had been identified in a screen for genes on human chromosome 7 deleted in the inherited developmental disorder SHFM. Full-length cDNA clones from the DSS1 gene (GenBank accession no. U41515) and also a 5′-truncated version lacking the first nine amino acids of the open reading frame were isolated. When subcloned and reintroduced into strain Y190, DSS1 retained the ability to activate His and LacZ expression in a BRCA2.IV fragment-dependent manner, but not with other fragments of BRCA2, supporting the existence of a specific association with this region (Fig. 2A). Comparing the strength of the β-galactosidase activation, the affinity of the BRCA2.IV-DSS1 interaction in yeast was similar in magnitude to that of the extreme C terminus of human BRCA2 with human RAD51 (Fig. 2A), which has been previously demonstrated for mouse BRCA2 (31, 42).
FIG. 2.
Demonstration of an interaction between BRCA2 and DSS1 in yeast and mammalian two-hybrid systems. (A) Yeast two-hybrid interaction between human BRCA2.IV and DSS1, determined by β-Galactosidase filter assay of Y190 strains transformed with combinations of two-hybrid vectors encoding GAL4 DNA-binding domain (DBD) fusion partner and GAL4 activation domain (AD) fusion partner (human BRCA2 aa 2472 to 2957 [IV], BRCA2 aa 3215 to 3418 [XC], full-length human DSS1 cDNA clone [DSS1], full-length RAD51 cDNA clone [RAD51], or empty vector [−]). (B) Two-hybrid assay in human 293T cells. The indicated combinations of GAL4 DNA-binding domain (DBD) and VP16 activation domain (AD) fusions with BRCA2.IV and full-length human DSS1 or empty vector were transiently expressed in 293T cells with a GAL4-luciferase reporter, and activation was measured by luciferase assay on cell lysates. Data are presented as mean fold activation of the reporter above background from four separate experiments.
BRCA2 and DSS1 can interact in mammalian cells.
To support the yeast two-hybrid data, the BRCA2.IV fragment and the full-length DSS1 cDNA clone were expressed as GAL4 DNA-binding domain and VP16 activation domain fusions, respectively, for use in a mammalian two-hybrid system. The interaction was analyzed by activation of a GAL4-luciferase reporter construct in transiently transfected 293T cells (Fig. 2B). A reproducible activation, with a mean of almost fourfold, was seen in this system, suggesting that the interaction was physiologically significant.
Detection of mammalian DSS1.
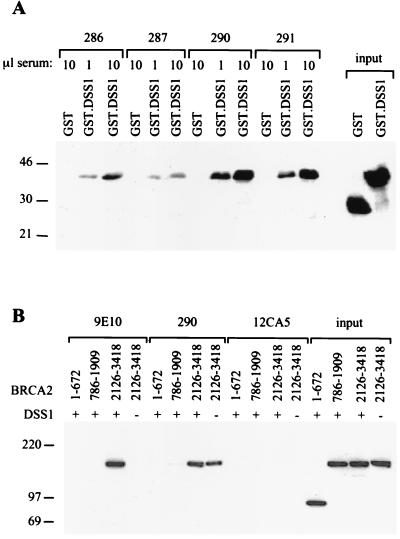
To enable the investigation of DSS1 in mammalian cells, specific rabbit antisera for human DSS1 were raised against recombinant protein purified as a cleavage product from GST-DSS1 expressed in E. coli and also against a synthetic peptide corresponding to the C-terminal 13 amino acids of DSS1. Both antisera specifically recognized GST-DSS1 on Western blots (not shown) and could also be used to specifically immunoprecipitate GST-DSS1 (Fig. 3A). Antisera 290 and 291, raised against the C-terminal DSS1 peptide, appeared to be more efficient in this assay than antisera 286 and 287, which were raised against recombinant full-length DSS1.
FIG. 3.
Coimmunoprecipitation of BRCA2 and DSS1 in mammalian cells. (A) DSS1 antisera can be used for specific immunoprecipitation. Preparations of GST or GST-DSS1 were immunoprecipitated in parallel with 1 or 10 μl of polyclonal antisera generated in this study. Antisera 286 and 287 were raised against the full-length recombinant DSS1, and antisera 290 and 291 were raised against a C-terminal peptide of DSS1. Immunoprecipitates were analyzed by immunoblotting with anti-GST antibody. The input proteins are shown. Sizes in both panels are indicated in kilodaltons. (B) Interaction of DSS1 with the C terminus of BRCA2 in transiently transfected COS cells. Lysates from COS cells transiently transfected with 9E10-tagged DSS1 (+) and various FLAG-tagged BRCA2 fragments (indicated by amino acid numbers) were immunoprecipitated with anti-Myc tag antibody 9E10, anti-DSS1 antiserum 290, or control anti-HA tag antibody 12CA5. Immunoprecipitates were analyzed by SDS-PAGE (6% polyacrylamide gel), together with samples of the input lysates representing half the protein used in the immunoprecipitations, followed by Western blot analysis with anti-FLAG tag antibody M2.
Cotransfected BRCA2 and DSS1 coimmunoprecipitate in mammalian cells.
To confirm the region of BRCA2 involved in the DSS1 interaction, we transiently transfected COS cells with Myc-tagged DSS1 and a panel of FLAG epitope-tagged BRCA2 constructs expressing various regions of the protein and performed immunoprecipitations using 9E10 antibody followed by Western blotting with FLAG antibody. These studies showed that a C-terminal fragment of BRCA2 comprising amino acids 2126 to 3148, which includes the BRCA2.IV region used in the yeast two-hybrid assay, could be coprecipitated with Myc-tagged DSS1 (Fig. 3B). This BRCA2 C-terminal fragment was efficiently coimmunoprecipitated with both 9E10 and anti-DSS1 antiserum 290 from cells expressing 9E10-tagged DSS1, but was also reproducibly coimmunoprecipitated with antiserum 290 from cells not transfected with the tagged DSS1 construct. This result suggests that COS cell (monkey) DSS1 is sufficiently conserved to be recognized by the anti-human DSS1 antiserum and to form a complex with the exogenous human BRCA2 fragment. An unrelated antibody (12CA5) gave no signal.
In vivo association of BRCA2 and DSS1.
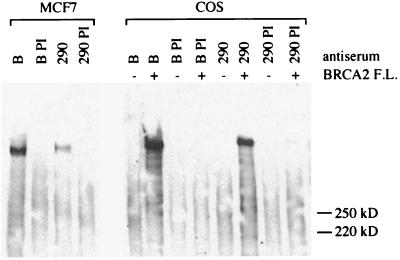
To determine whether a complex between endogenous DSS1 and endogenous BRCA2 could be detected in mammalian cells, parallel immunoprecipitations were performed with polyclonal anti-BRCA2 antiserum B and anti-DSS1 antiserum 290 on lysates from asynchronously cycling cells of the MCF7 breast cancer cell line. Subsequent detection of BRCA2 was carried out by Western blotting with a rat monoclonal anti-human BRCA2 antibody (Fig. 4). Preimmune sera did not pull down any BRCA2, but antiserum 290 coprecipitated full-length BRCA2 from MCF7 cells, suggesting the presence of an endogenous complex including BRCA2 and DSS1 proteins. Immunoprecipitations performed in parallel on lysates from COS cells transiently transfected with a full-length human BRCA2 construct demonstrate the coimmunoprecipitation of full-length BRCA2 with COS cell DSS1 (Fig. 4). We found that it was not possible to detect DSS1 from cell lysates by Western blotting with the antisera generated in this study, probably because of failure of the small, highly charged protein to bind to the nitrocellulose or polyvinylidene difluoride membrane. This limitation therefore precludes the use of this method to illustrate the converse coprecipitation of DSS1 with BRCA2.
FIG. 4.
Coimmunoprecipitation of endogenous BRCA2 with DSS1 in MCF7 cells. Lysates of MCF7 cells cultured below confluency in 10% fetal calf serum, and lysates of COS cells transiently transfected with (+) or without (−) a construct expressing full-length (F.L.) BRCA2 were immunoprecipitated, in parallel, with anti-BRCA2 antiserum B, anti-DSS1 antiserum 290, or their preimmune sera (B PI and 290 PI). Immunoprecipitates were analyzed by SDS-PAGE (6% polyacrylamide gel) and Western blotting with an anti-BRCA2 rat monoclonal antibody.
DSS1 expression is serum stimulated.
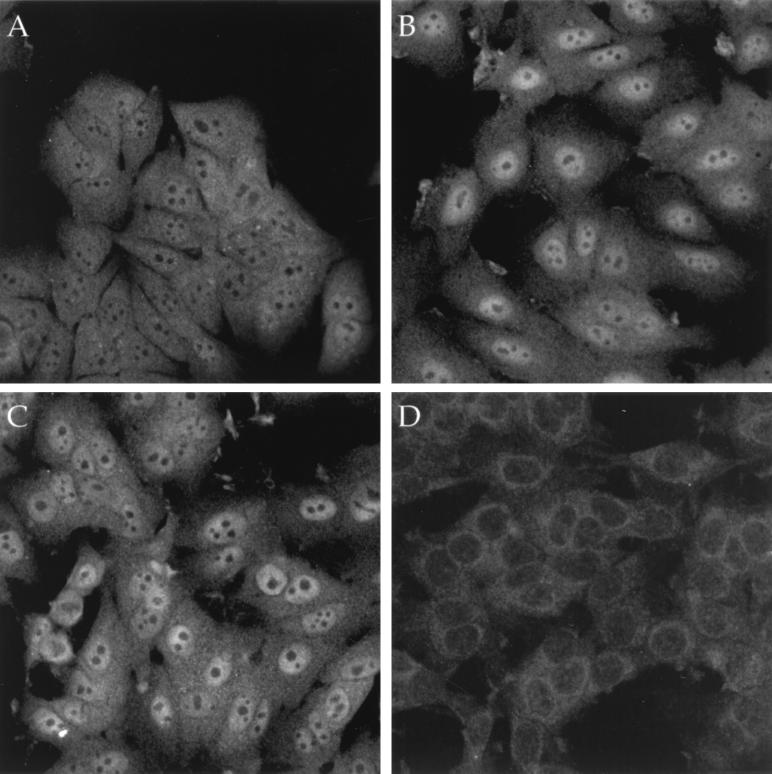
To determine whether DSS1 expression patterns are similar to those of BRCA2 in mammalian cells, we subjected various cell lines to immunofluorescence analysis using the anti-DSS1 antiserum 290. A clear nuclear staining pattern, excluding nucleoli, was seen with 290 in MCF7 cells grown in the presence of serum (Fig. 5C). The specificity of the nuclear staining with the polyclonal antiserum 290 was confirmed by preincubation with the DSS1 C-terminal immunizing peptide which blocked the nuclear immunofluorescence signal of antiserum 290, leaving only a residual, nonspecific cytoplasmic stain (Fig. 5D). A similar pattern was also seen in asynchronously cycling COS cells, confirming the recognition of COS cell DSS1 by antiserum 290 that had been suggested by coimmunoprecipitation studies (Fig. 3B and 4); in COS cells transiently transfected with 9E10-tagged DSS1, 290 gave overlapping signals with a strong nuclear staining similar to that seen with 9E10 antibody (not shown). Since previous studies have shown that serum stimulation elevates the expression of BRCA2 protein (4), we investigated whether DSS1 was regulated in a similar manner. The expression of endogenous DSS1 protein was found to be stimulated by serum in MCF7 cells (Fig. 5). Cycling MCF7 cells grown in medium supplemented with 10% fetal calf serum and insulin had strong specific nuclear staining (Fig. 5C), but cells starved of serum for 24 h showed negligible DSS1 nuclear signal (Fig. 5A). On restimulation of starved cells with 10% serum and insulin, DSS1 nuclear signal was visible after only 2 h (not shown) and continued to intensify after 6 and 24 h of incubation in the presence of serum (Fig. 5B and C). The nuclear expression of BRCA2 protein is also very low in serum-starved cells and is induced after serum stimulation, though not until late G1/S phase, which is about 12 h after serum addition in MCF7 cells (4). DSS1 expression is maintained during this phase, which suggests that the two proteins are expressed at the same point of the cell cycle.
FIG. 5.
Expression of DSS1 is serum stimulated. Immunofluorescence detection of DSS1 in MCF7 cells which had been starved of serum for 24 h (A) or subsequently refed with 10% fetal calf serum for 6 h (B) or 24 h (C and D). The staining pattern of the antiserum is specific, as shown by preblocking the antibody with the immunizing peptide (D), as opposed to a nonspecific peptide (A to C). Visualization was with fluorescein isothiocyanate conjugated goat anti-rabbit immunoglobulin G.
Identification of genes related to mammalian DSS1 in S. pombe and S. cerevisiae.
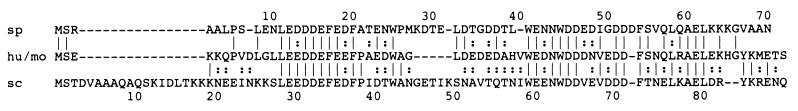
The function of DSS1 is not known; therefore, in an attempt to address its importance and aid understanding of BRCA2 function, sequence database searches were performed. An open reading frame encoding a product with significant homology to human DSS1 (49% amino acid identity) was identified in the fission yeast sequence database (GenBank accession no. Z99167; nucleotides 5154 to 5515). The 71-amino-acid peptide is encoded by three exons and shows two regions of high similarity to the human peptide sequence (Fig. 6). Also, a previously unidentified open reading frame coding for a putative 89-residue peptide (GenBank accession no. U28372; nucleotides 30070 to 30339) was found by searching translations of the S. cerevisiae sequence database with the human DSS1 peptide sequence. This gene is located on chromosome IV in the region of the CDC40 gene (51) (GenBank accession no. U17018) but had not been included in the annotation of the complete genome sequence of S. cerevisiae, as it falls below the cutoff length for open reading frames. This gene encodes a protein with 46% identity with the human DSS1 gene product and also shows considerable similarity with the S. pombe product (Fig. 6).
FIG. 6.
Yeast orthologues of DSS1. Alignment of human/mouse DSS1 protein sequence (hu/mo) (13) with amino acid sequences of putative products of DSS1-like open reading frames in S. pombe (sp) and S. cerevisiae (sc). Identical and conserved residues are indicated by solid and broken bars, respectively.
S. pombe and S. cerevisiae DSS1 deletion strains are temperature sensitive.
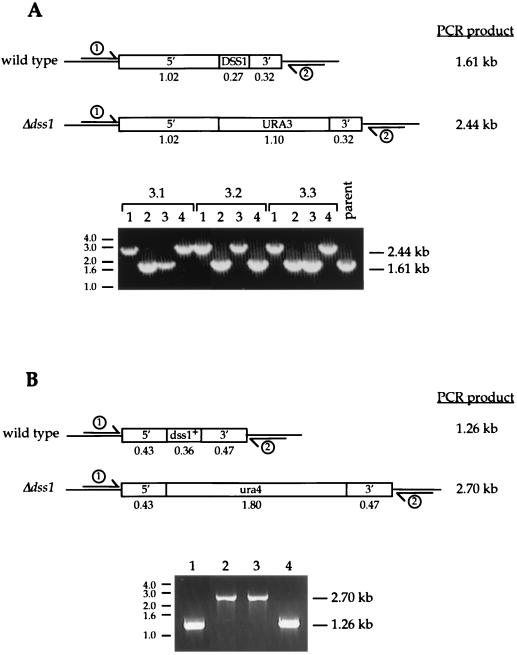
Since one can gain information about the function of a gene through analysis of the phenotype of yeast cells in which it has been deleted, the DSS1-like open reading frames from both S. pombe and S. cerevisiae were targeted for deletion. Constructs were designed by using ura4 and URA3 genes located between genomic sequence fragments immediately flanking the identified genes derived from the database sequences (Fig. 7). Δdss1 strains were obtained by transforming wild-type fission and budding yeast strains with the appropriate deletion cassettes, sporulating selected transformants, and dissecting tetrad spores (38). The DSS1/Δdss1 status of haploid clones was established by PCR (Fig. 7).
FIG. 7.
Construction of deletions of yeast DSS1 genes. (A) S. cerevisiae DSS1 deletion. The 5′ and 3′ genomic fragments flanking the DSS1 gene which were used in the deletion construct, shown in the wild-type and Δdss1 arrangements, with the extent of the deletion in nucleotides indicated beneath in kilobases. Haploid clones derived by sporulation of cells transformed with the deletion cassette were analyzed for DSS1 status by PCR on genomic DNA. Positions of forward and reverse primers 1 and 2 in the genomic sequences flanking those used in the deletion cassette are indicated in the upper panel with the expected size of the PCR product for wild-type or deleted clones. PCR results for three tetrads (3.1, 3.2, and 3.3) and the parent strain are shown at the bottom. (B) S. pombe dss1 deletion cassette, illustrating the 5′ and 3′ genomic fragments used to construct the deletion cassette, in both wild-type and Δdss1 configurations. Confirmation of the deletion by PCR is shown at the bottom.
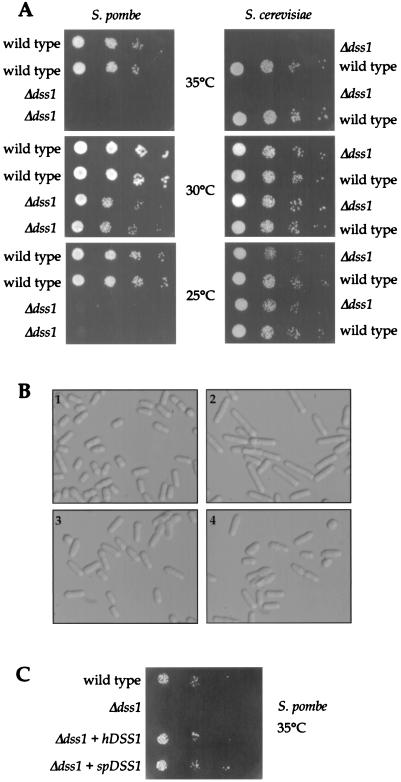
In neither S. pombe nor S. cerevisiae was the haploid DSS1 deletion found to be lethal, but a reduced growth rate of the Δdss1 clones compared to wild-type was observed, particularly at raised or lowered temperatures (Fig. 8A). This was more pronounced in S. pombe, where slower growth of the deleted clones was seen even at 30°C and negligible growth was seen at 25 or 35°C. Wild-type and Δdss1 S. cerevisiae did not show significant difference in growth rate phenotypes at 30°C, but the deleted clones grew more slowly at a higher temperature. Examination of the morphological phenotype of the S. pombe Δdss1 clones showed that many of the cells were considerably elongated compared to wild-type counterparts, which suggests a cell cycle delay (Fig. 8B). Subsequent transformation of an S. pombe Δdss1 strain with constructs expressing either the human DSS1 or the S. pombe genomic dss1+ resulted in significant rescue of both the morphology of the cells (Fig. 8B) and the growth rate at different temperatures (results at 35°C shown in Fig. 8C), indicating conservation of DSS1 protein function across species and that the S. pombe gene identified probably encodes the orthologue of human DSS1. Initial investigations of the sensitivity of Δdss1 strains of S. pombe to ionizing radiation showed a consistent but only marginal increase in sensitivity in the mutant strain compared to the wild type (not shown).
FIG. 8.
Yeast Δdss1 phenotypes. (A) Temperature-sensitive growth of S. pombe and S. cerevisiae deleted for DSS1. Equivalent cultures of wild-type and Δdss1 yeast clones were diluted to the same concentration and serially diluted 1:10. These dilutions of each yeast were plated in series, and the plates were incubated at 25, 30, or 35°C, as indicated. (B) Morphological phenotypes of cells from exponentially growing cultures of the following S. pombe strains: 1, wild type; 2, Δdss1; 3, Δdss1 expressing human DSS1; 4, Δdss1 expressing S. pombe dss1+. (C) Rescue of the temperature-sensitive growth of Δdss1 S. pombe at 35°C by expression of human DSS1 (hDSS1) or S. pombe dss1+ (spdss1).
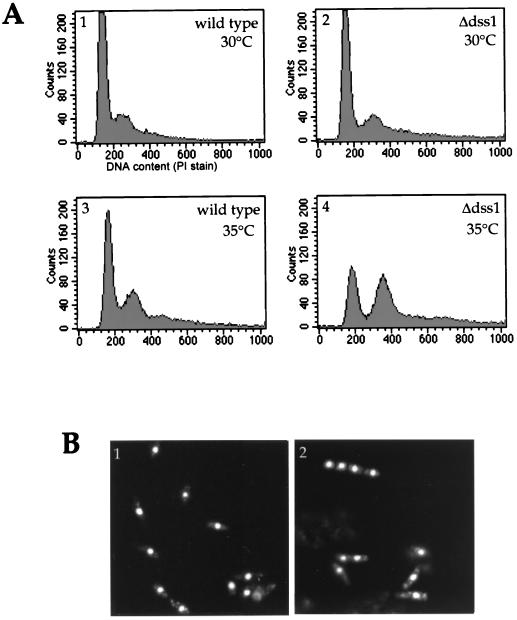
Analysis of the DNA content of S. pombe cells by staining with PI and flow cytometry (18) revealed that on incubation at 35°C instead of 30°C for 6 h, cells of the Δdss1 strain, unlike their wild-type counterparts, shift from a predominantly 2C DNA population (main peak in Fig. 9A, graph 2) toward a polyploid state with greater than 2C DNA content (Fig. 9A, graph 4). Microscopic examination of the PI-stained cells of Δdss1 S. pombe showed a heterogeneous population including cells with several septa and nuclei (Fig. 9B), which appear not to have completed division before entering the next cycle of DNA synthesis. Consistent with this defect in completion of the cell cycle, growth in nitrogen-depleted medium, which causes wild-type cells to accumulate with a 1C DNA content, was repeatedly less effective on the Δdss1 strain (not shown).
FIG. 9.
DNA content of Δdss1 S. pombe. (A) The cell cycle distributions of wild-type and Δdss1 S. pombe strains were examined by flow cytometry using PI staining of fixed cells from cultures grown at 30 and 35°C for 6 h. The data for 2 × 104 ungated events are presented as relative DNA content on the horizontal axis and cell number on the vertical axis. (B) Microscopic images of PI-stained cells of wild-type (image 1) and Δdss1 (image 2) strains of S. pombe, prepared as for FACS analysis.
DISCUSSION
The frequently observed loss of the wild-type copy of BRCA2 in breast tumors in individuals heterozygous for a loss-of-function mutation suggests that the gene encodes a tumor suppressor (3, 9). Potential functions that have been attributed to BRCA2 include a role in transcriptional regulation and an involvement in DNA repair via interaction with RAD51 (30, 42). However, further information is needed to clarify such roles and to identify additional functions of this very large protein. Using the yeast two-hybrid system with fragments of human BRCA2 as bait, we have found an interaction between a C-terminal portion of BRCA2 and the human DSS1 protein. The validity of this association was supported by demonstration of an association using a mammalian two-hybrid system, and this interaction was reproduced in mammalian cells transiently transfected with cDNA constructs. Furthermore, an endogenous complex including the two proteins was identified in breast cancer cell line MCF7. The C-terminal section of BRCA2 (amino acids 2472 to 2957) which interacts with DSS1 in yeast is distinct from regions of the protein involved in the association with RAD51 (42, 52). This fragment of BRCA2 represents a region of higher conservation between human and mouse sequences than the open reading frame as a whole, with 77% identity at the amino acid level compared with 59% overall (11). This suggests an important function of this region which is conserved. Attempts to further localize the BRCA2 residues involved in the interaction with DSS1 by using sections of amino acids 2472 to 2957 of BRCA2 in yeast two-hybrid analyses with full-length DSS1 failed to identify any shorter fragments which retained the association (data not shown), suggesting that there may be multiple independent determinants in this region for the interaction. The interaction between BRCA2 and DSS1 is likely to be direct, due to the demonstration in the yeast two-hybrid system, but the requirement of a cofactor cannot be ruled out in the absence of the use of purified proteins in vitro.
Numerous tumor-associated frameshift mutations in BRCA2 are located in or upstream of the sequence encoding the region which interacts with DSS1, resulting in truncations which delete all or part of this region of BRCA2 (12, 17, 48, 53). Such mutations include the 6174delT mutation associated with Ashkenazi Jewish lineages which results in a truncation of the BRCA2 protein prior to the eighth BRC repeat (34, 49). The frequency of reported missense polymorphisms within amino acids 2472 to 2957 of BRCA2 is very low (12, 48). However, a few frameshift mutations which would result in truncation of BRCA2 between the DSS1-interacting region and the site of the C-terminal polymorphic stop codon 3326 suggest that loss of a more C-terminal function is critical to loss of tumor suppression activity (12, 17, 28, 36). The human DSS1 gene is located at chromosome 7q21, and frequent loss of heterozygosity at 7q is seen in a range of cancers, suggesting the presence of a tumor suppressor gene in this region. However, although 7q21 rearrangements have been identified in tumors, the minimum commonly deleted region appears to be around 7q31 (1, 25).
DSS1 is one of three candidates identified on 7q21.3-q22.1 in a search for genes involved in an autosomal dominant form of the heterogeneous limb developmental disorder SHFM, which is characterized by missing or fused digits (13, 22, 40). Chromosomal rearrangement breakpoints at the SHFM1 locus on 7q from a subset of affected patients defined a critical region containing the DSS1 gene and two Distal-less homeobox genes, DLX5 and DLX6, all of which are expressed in the mouse in patterns which could implicate these genes in limb and facial development (13, 46). The human and mouse DSS1 genes encode identical 70-amino-acid highly acidic proteins of unknown function (13). The absolute conservation of human and mouse DSS1 and the identification here of highly conserved yeast orthologues, both in S. pombe and in S. cerevisiae, suggests that DSS1 has an important cellular function in eukaryotes.
To understand the significance of the interaction of BRCA2 with DSS1, we wished to gain some information as to the normal cellular role of DSS1. The yeasts S. cerevisiae and S. pombe were used as model systems. Deletions of the DSS1-like open reading frames in the fission and budding yeasts were made in order to investigate the role of DSS1 in these organisms. The deletion of DSS1 was not lethal to either yeast, but the gene appeared to be required for normal growth control. S. pombe Δdss1 strains grew more slowly than wild-type cells, particularly at reduced or elevated temperatures, exhibiting a cell cycle delay with cells gaining a considerably increased length before undergoing division. Analyses of S. pombe Δdss1 cells by flow cytometry and confocal microscopy revealed that the mutant cells have a defect in completion of cell division which is more pronounced at elevated temperature, leading to an accumulation of cells with greater than 2C DNA content. S. cerevisiae Δdss1 strains were also sensitive to elevated temperature. No BRCA2 orthologue has been identified in either S. pombe or S. cerevisiae. However this does not preclude the possibility that some other gene serves a similar function in yeast. The conservation of function of DSS1 is strongly suggested by the rescue of S. pombe Δdss1 phenotypes by overexpression of human DSS1. Given the potential role of BRCA2 in DNA repair (35, 42), we have measured the sensitivity of Δdss1 yeast to ionizing radiation. Preliminary experiments suggest that although deleted S. pombe cells appear to be marginally more sensitive than wild-type strains, the effect is much less than would be expected for a gene directly involved in DNA repair (43). The identification of yeast DSS1 should provide a useful system to analyze the function of the gene and should help elucidate its relationship to the cellular role of BRCA2.
Both DSS1 and BRCA2 genes appear to be ubiquitously expressed in mouse tissues, as assayed by reverse transcription-PCR and Northern blotting (11, 13, 27a, 37). The cellular localization was investigated; expression of DSS1 protein in MCF7 cells appears to be growth regulated, with very low amounts detected in serum-starved cells and expression elevated rapidly after refeeding with serum. BRCA2 expression is also very low in serum-starved MCF7 cells but accumulates only as the cells reach the G1/S boundary after serum stimulation (4). The maintenance of high levels of DSS1 after addition of serum to culture medium means that both proteins may be expressed together in the nucleus, particularly during S phase. DSS1 has no obvious nuclear localization signal and so may be transported and anchored in the nucleus by interaction with another protein, conceivably BRCA2.
Since submission of this report, the S. cerevisiae DSS1 gene has been described elsewhere (24). The gene is implicated in exocytosis and pseudohyphal differentiation in S. cerevisiae. The relationship between these two processes in S. cerevisiae and the phenomena that we observe in Δdss1 S. pombe is not clear but may suggest that DSS1 is involved in multiple cellular processes.
The functions of the BRCA2 gene are unclear, although there is evidence for roles in DNA repair and transcriptional regulation (3). Here we show that BRCA2 associates in yeast and mammalian cells with the product of the previously described gene DSS1. This protein appears to be required in yeast for proper completion of cell division, providing a link between BRCA2 and the cell cycle. Whether DSS1 is involved in the etiology of tumors in women carrying mutations in BRCA2 is under investigation.
ACKNOWLEDGMENTS
This work was funded by the Cancer Research Campaign.
We thank Richard Marais for plasmid pEFm6, Ian Titley for help with FACS analyses, Janet Valentine for critically reading the manuscript, and Steve Hooper and Amanda Smith for technical assistance.
REFERENCES
- 1.Achille A, Biasi M O, Zamboni G, Bogina G, Magalini A R, Pederzoli P, Perucho M, Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- 2.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 3.Bertwistle D, Ashworth A. Functions of the BRCA1 and BRCA2 genes. Curr Opin Genet Dev. 1998;8:14–20. doi: 10.1016/s0959-437x(98)80056-7. [DOI] [PubMed] [Google Scholar]
- 4.Bertwistle D, Swift S, Marston N J, Jackson L E, Crossland S, Crompton M R, Marshall C J, Ashworth A. Nuclear location and cell cycle regulation of the BRCA2 protein. Cancer Res. 1997;57:5485–5488. [PubMed] [Google Scholar]
- 5.Bignell G, Micklem G, Stratton M R, Ashworth A, Wooster R. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum Mol Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 7.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 9.Collins N, McManus R, Wooster R, Mangion J, Seal S, Lakhani S R, Ormiston W, Daly P A, Ford D, Easton D F, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- 10.Connor F, Bertwistle D, Mee P J, Ross G M, Swift S, Grigorieva E, Tybulewicz V L, Ashworth A. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 11.Connor F, Smith A, Wooster R, Stratton M, Dixon A, Campbell E, Tait T M, Freeman T, Ashworth A. Cloning, chromosomal mapping and expression pattern of the mouse Brca2 gene. Hum Mol Genet. 1997;6:291–300. doi: 10.1093/hmg/6.2.291. [DOI] [PubMed] [Google Scholar]
- 12.Couch F J, Farid L M, DeShano M L, Tavtigian S V, Calzone K, Campeau L, Peng Y, Bogden B, Chen Q, Neuhausen S, Shattuck-Eidens D, Godwin A K, Daly M, Radford D M, Sedlacek S, Rommens J, Simard J, Garber J, Merajver S, Weber B L. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet. 1996;13:123–125. doi: 10.1038/ng0596-123. [DOI] [PubMed] [Google Scholar]
- 13.Crackower M A, Scherer S W, Rommens J M, Hui C C, Poorkaj P, Soder S, Cobben J M, Hudgins L, Evans J P, Tsui L C. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 14.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsburg, S. 11 February 1999, revision date. Yeast flow cytometry protocol. [Online.] http://pingu.salk.edu/fcm/protocols/ycc.html. [11 February 1999, last date accessed.]
- 16.Friedman L S, Thistlethwaite F C, Patel K J, Yu V P, Lee H, Venkitaraman A R, Abel K J, Carlton M B, Hunter S M, Colledge W H, Evans M J, Ponder B A. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 1998;58:1338–1343. [PubMed] [Google Scholar]
- 17.Gayther S A, Mangion J, Russell P, Seal S, Barfoot R, Ponder B A, Stratton M R, Easton D. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet. 1997;15:103–105. doi: 10.1038/ng0197-103. [DOI] [PubMed] [Google Scholar]
- 18.Gould K L, Moreno S, Owen D J, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gretarsdottir S, Thorlacius S, Valgardsdottir R, Gudlaugsdottir S, Sigurdsson S, Steinarsdottir M, Jonasson J G, Anamthawat-Jonsson K, Eyfjord J E. BRCA2 and p53 mutations in primary breast cancer in relation to genetic instability. Cancer Res. 1998;58:859–862. [PubMed] [Google Scholar]
- 20.Gudmundsson J, Johannesdottir G, Bergthorsson J T, Arason A, Ingvarsson S, Egilsson V, Barkardottir R B. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res. 1995;55:4830–4832. [PubMed] [Google Scholar]
- 21.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 22.Ignatius J, Knuutila S, Scherer S W, Trask B, Kere J. Split hand/split foot malformation, deafness, and mental retardation with a complex cytogenetic rearrangement involving 7q21.3. J Med Genet. 1996;33:507–510. doi: 10.1136/jmg.33.6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingvarsson S, Geirsdottir E K, Johannesdottir G, Sigbjornsdottir B I, Eiriksdottir G, Ragnarsson G, Agnarsson B A, Gudmundsson J, Jonasson J G, Sigurdsson A, Egilsson V, Barkardottir R B. High incidence of loss of heterozygosity in breast tumors from carriers of the BRCA2 999del5 mutation. Cancer Res. 1998;58:4421–4425. [PubMed] [Google Scholar]
- 24.Jantti J, Lahdenranta J, Olkkonen V M, Soderlund H, Keranen S. SEM1, a homologue of the split hand/split food malformation candidate gene Dss1, regulates exocytosis and pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1999;96:909–914. doi: 10.1073/pnas.96.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristjansson A K, Eiriksdottir G, Ragnarsson G, Sigurdsson A, Gudmundsson J, Barkardottir R B, Jonasson J G, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 7q in human breast cancer: association with clinical variables. Anticancer Res. 1997;17:93–98. [PubMed] [Google Scholar]
- 26.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 27.Marais R, Light Y, Paterson H F, Marshall C J. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Marston, N. Unpublished data.
- 28.Mazoyer S, Dunning A M, Serova O, Dearden J, Puget N, Healey C S, Gayther S A, Mangion J, Stratton M R, Lynch H T, Goldgar D E, Ponder B A, Lenoir G M. A polymorphic stop codon in BRCA2. Nat Genet. 1996;14:253–254. doi: 10.1038/ng1196-253. [DOI] [PubMed] [Google Scholar]
- 29.Miki Y, Katagiri T, Kasumi F, Yoshimoto T, Nakamura Y. Mutation analysis in the BRCA2 gene in primary breast cancers. Nat Genet. 1996;13:245–247. doi: 10.1038/ng0696-245. [DOI] [PubMed] [Google Scholar]
- 30.Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T. Transcriptional activation functions in BRCA2. Nature. 1997;386:772–773. doi: 10.1038/386772a0. [DOI] [PubMed] [Google Scholar]
- 31.Mizuta R, LaSalle J M, Cheng H L, Shinohara A, Ogawa H, Copeland N, Jenkins N A, Lalande M, Alt F W. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci USA. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 33.Morimatsu M, Donoho G, Hasty P. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to gamma-radiation and premature senescence. Cancer Res. 1998;58:3441–3447. [PubMed] [Google Scholar]
- 34.Neuhausen S, Gilewski T, Norton L, Tran T, McGuire P, Swensen J, Hampel H, Borgen P, Brown K, Skolnick M, Shattuck-Eidens D, Jhanwar S, Goldgar D, Offit K. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet. 1996;13:126–128. doi: 10.1038/ng0596-126. [DOI] [PubMed] [Google Scholar]
- 35.Patel K J, Vu V P, Lee H, Corcoran A, Thistlethwaite F C, Evans M J, Colledge W H, Friedman L S, Ponder B A, Venkitaraman A R. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 36.Phelan C M, Lancaster J M, Tonin P, Gumbs C, Cochran C, Carter R, Ghadirian P, Perret C, Moslehi R, Dion F, Faucher M C, Dole K, Karimi S, Foulkes W, Lounis H, Warner E, Goss P, Anderson D, Larsson C, Narod S A, Futreal P A. Mutation analysis of the BRCA2 gene in 49 site-specific breast cancer families. Nat Genet. 1996;13:120–122. doi: 10.1038/ng0596-120. [DOI] [PubMed] [Google Scholar]
- 37.Rajan J V, Marquis S T, Gardner H P, Chodosh L A. Developmental expression of Brca2 colocalizes with Brca1 and is associated with proliferation and differentiation in multiple tissues. Dev Biol. 1997;184:385–401. doi: 10.1006/dbio.1997.8526. [DOI] [PubMed] [Google Scholar]
- 38.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 39.Schaap D, van der Wal J, Howe L R, Marshall C J, van Blitterswijk W J. A dominant-negative mutant of raf blocks mitogen-activated protein kinase activation by growth factors and oncogenic p21ras. J Biol Chem. 1993;268:20232–20236. [PubMed] [Google Scholar]
- 40.Scherer S W, Poorkaj P, Allen T, Kim J, Geshuri D, Nunes M, Soder S, Stephens K, Pagon R A, Patton M A, et al. Fine mapping of the autosomal dominant split hand/split foot locus on chromosome 7, band q21.3-q22.1. Am J Hum Genet. 1994;55:12–20. [PMC free article] [PubMed] [Google Scholar]
- 41.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 42.Sharan S K, Morimatsu M, Albrecht U, Lim D S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 44.Siddique H, Zou J P, Rao V N, Reddy E S. The BRCA2 is a histone acetyltransferase. Oncogene. 1998;16:2283–2285. doi: 10.1038/sj.onc.1202003. [DOI] [PubMed] [Google Scholar]
- 45.Sigurdsson S, Thorlacius S, Tomasson J, Tryggvadottir L, Benediktsdottir K, Eyfjord J E, Jonsson E. BRCA2 mutation in Icelandic prostate cancer patients. J Mol Med. 1997;75:758–761. doi: 10.1007/s001090050162. [DOI] [PubMed] [Google Scholar]
- 46.Simeone A, Acampora D, Pannese M, D’Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K, et al. Cloning and characterization of two members of the vertebrate Dlx gene family. Proc Natl Acad Sci USA. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki A, de la Pompa J L, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, Koo W, Billia P, Ho A, Fukumoto M, Hui C C, Mak T W. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 48.Tavtigian S V, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, Belanger C, Bell R, Berry S, Bogden R, Chen Q, Davis T, Dumont M, Frye C, Hattier T, Jammulapati S, Janecki T, Jiang P, Kehrer R, Leblanc J F, Goldgar D E, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 49.Teng D H, Bogden R, Mitchell J, Baumgard M, Bell R, Berry S, Davis T, Ha P C, Kehrer R, Jammulapati S, Chen Q, Offit K, Skolnick M H, Tavtigian S V, Jhanwar S, Swedlund B, Wong A K, Kamb A. Low incidence of BRCA2 mutations in breast carcinoma and other cancers. Nat Genet. 1996;13:241–244. doi: 10.1038/ng0696-241. [DOI] [PubMed] [Google Scholar]
- 50.Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson J G, Tavtigian S V, Tulinius H, Ogmundsdottir H M, Eyfjord J E. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 51.Vaisman N, Tsouladze A, Robzyk K, Ben-Yehuda S, Kupiec M, Kassir Y. The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol Gen Genet. 1995;247:123–136. doi: 10.1007/BF00705642. [DOI] [PubMed] [Google Scholar]
- 52.Wong A K C, Pero R, Ormonde P A, Tavtigian S V, Bartel P L. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31948. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 53.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]