Abstract
Background and objective
Given that the novel coronavirus disease 2019 (COVID-19) has become a pandemic, a method to accurately distinguish COVID-19 from community-acquired pneumonia (CAP) is urgently needed. However, the spatial uncertainty and morphological diversity of COVID-19 lesions in the lungs, and subtle differences with respect to CAP, make differential diagnosis non-trivial.
Methods
We propose a deep represented multiple instance learning (DR-MIL) method to fulfill this task. A 3D volumetric CT scan of one patient is treated as one bag and ten CT slices are selected as the initial instances. For each instance, deep features are extracted from the pre-trained ResNet-50 with fine-tuning and represented as one deep represented instance score (DRIS). Each bag with a DRIS for each initial instance is then input into a citation k-nearest neighbor search to generate the final prediction. A total of 141 COVID-19 and 100 CAP CT scans were used. The performance of DR-MIL is compared with other potential strategies and state-of-the-art models.
Results
DR-MIL displayed an accuracy of 95% and an area under curve of 0.943, which were superior to those observed for comparable methods. COVID-19 and CAP exhibited significant differences in both the DRIS and the spatial pattern of lesions (p<0.001). As a means of content-based image retrieval, DR-MIL can identify images used as key instances, references, and citers for visual interpretation.
Conclusions
DR-MIL can effectively represent the deep characteristics of COVID-19 lesions in CT images and accurately distinguish COVID-19 from CAP in a weakly supervised manner. The resulting DRIS is a useful supplement to visual interpretation of the spatial pattern of lesions when screening for COVID-19.
Keywords: COVID-19, Community-acquired pneumonia, Lung CT image, Convolutional neural network, Deep learning, Multiple instance learning
1. Introduction
The novel coronavirus disease 2019 (COVID-19) has become a continuing pandemic. According to data from the World Health Organization (WHO), the number of confirmed cases of COVID-19 had surpassed 202 million and the total number of deaths had exceeded 4 million by early of August 2021 [1]. Thus, there is an urgent need to accurately and automatically differentiate COVID-19 from community-acquired pneumonia (CAP) by large-scale screening.
As a non-invasive imaging modality, chest CT has proved an effective tool in the clinical screening and diagnosis of COVID-19, although the real-time reverse-transcriptase polymerase chain reaction (RT-PCR) is the gold standard. A WHO rapid advice guide suggests using chest imaging (CT or X-ray radiography) for the diagnosis of COVID-19 when RT-PCR testing is unavailable, RT-PCR testing is available but results are delayed, or initial RT-PCR testing is negative but there is high clinical suspicion of COVID-19 [2]. Chest CT has shown high sensitivity and reproducibility during the diagnosis of COVID-19 and can be considered an important and reliable complement to RT-PCR testing [3], [4], [5].
However, the spatial uncertainty and diversity in the intensity and morphology of COVID-19 lesions in lung CT images, and their subtle differences from CAP lesions, make differential diagnosis non-trivial. COVID-19 lesions may appear as consolidative and/or ground-glass pulmonary opacities [6]. In the early stages, lesions are often atypical and primarily distributed in the lateral zones of the lungs or the subpleural areas. As the disease advances, lesions such as ground-glass opacities progress from subpleural to central. In severe and critical cases, patients may exhibit lung consolidation [7,8]. In some cases of CAP, lesions can appear similar to those of COVID-19. Even experienced radiologists may have only moderate ability to distinguish COVID-19 from CAP by visual inspection of CT images [9].
To assess and report the pulmonary involvement of COVID-19 in CT images in a standardized manner, several initiatives have been proposed [10], [11], [12]. As a typical example, the COVID-19 Reporting and Data System (CO-RADS) developed by the Dutch Radiological Society has demonstrated good interobserver agreement and performance while discriminating cases with different levels of suspicion for pulmonary involvement of COVID-19 [10].
Alongside the evaluation of CT images by radiologists, the use of artificial intelligence (AI), especially deep learning, has been proposed for the rapid and accurate differentiation of COVID-19 from other pulmonary diseases via CT images [13]. For example, Li et al. built a deep learning model (COVNet) for detecting COVID-19 that uses ResNet-50 as the backbone to generate features of each slice and max pooling to combine the extracted features [14]. Wang et al. created COVID-19Net with a DenseNet-like structure for the diagnostic and prognostic analysis of COVID-19 patients [15]. Ouyang et al. developed a dual-sampling attention network that uses online attention and 3D ResNet-34 as the backbone [16]. Zhang et al. used DeepLabv3 as the backbone to segment lung lesions and developed an AI system for diagnosing and characterizing COVID-19 [17]. It has been reported that augmentation with deep learning can allow radiologists to achieve higher accuracy, sensitivity, and specificity in their evaluations [18]. The reader is referred to two comprehensive reviews for more details regarding the role of deep learning with respect to COVID-19 [19,20].
At least three points are worth noting from the deep learning studies discussed above. First, although more than 1000 CT images were used, the data were still insufficient for training one deep convolutional neural network (CNN) model, and transfer learning was typically adopted [14,15]. Second, the segmentation of infected lesions is required, although it is worthwhile to mention that infected lesions may be falsely excluded and more annotations are frequently necessary. Third, although a post-hoc analysis of class activation maps (CAM) can coarsely identify the infected lesions in 2D slices, it is unknown which slice makes the most important contribution.
With the above considerations in mind, machine learning or a combination of machine and deep learning is a reasonable approach. One example is the representation learning model developed by Kang et al. [21]. In addition, Han et al. neatly proposed a 3D deep multiple instance learning (MIL) model with an attention mechanism for screening suspected COVID-19 cases [22]. This study demonstrated the power of MIL and improved the interpretability by visualizing key instances. The network reported by Han et al. was inspired by the attention-based MIL pooling operator [23]. In addition, mean, max, and Noisy-AND pooling are commonly applied for MIL [24,25].
In the current study, with the aim of accurately and automatically distinguishing COVID-19 from CAP using CT images, we propose a deep represented multiple instance learning (DR-MIL) method. DR-MIL is a weakly supervised learning method that only requires a patient-level label. In this method, 3D volumetric CT images of one subject are treated as one bag and ten CT slices are selected randomly as the initial instances. Each instance is transformed into deep features extracted from the pre-trained ResNet-50 with fine-tuning and subsequently represented as a score, i.e., the deep represented instance score (DRIS), using a dimension reduction approach. Each bag with 10 DRISs is then input into one MIL classifier to generate the final prediction. Using a variety of performance measures, DR-MIL is compared with 2D CNN with voting, 2D CNN of a montage of 10 CT slices, 3D MedicalNet, and other state-of-the-art models. The spatial distribution patterns generated by Grad-CAM for COVID-19 are compared to those for CAP.
The contributions and novelties of this paper are fourfold. First, we have developed the DR-MIL method for accurately distinguishing COVID-19 from CAP, which has several methodological advantages, including good applicability to small datasets of hundreds of CT examinations, the weakly supervised nature of the learning, and no requirement for lesion segmentation. Second, we have generated the score referred to as DRIS, which is significantly different between COVID-19 and CAP and can serve as a discriminative imaging biomarker for COVID-19. Third, we have shown that the spatial distribution pattern of infected lesions highlighted by Grad-CAM in COVID-19 is significantly different from that in CAP. Fourth, as a means of content-based image retrieval (CBIR), DR-MIL can identify images used as key instances, references, and citers for visual interpretation.
2. Materials and methods
2.1. Participants and datasets
A total of 241 participants (141 [58.5%] COVID-19 and 100 [41.5%] CAP cases) were included in this retrospective study, and lung CT images and related clinical information were collected. All of the subjects in the COVID-19 group were confirmed by RT-PCR to have contracted COVID-19 during the period from December 29, 2019, to February 16, 2020. The CT scans and RT-PCR sampling were conducted on the same day. Subjects with CAP were selected from datasets of the participating hospitals and they were enrolled between December 8, 2019, and February 26, 2020.
Some CAP patients received etiological confirmation from a specialized laboratory; bacterial cultures were positive for 57 patients and negative for 12 patients (9 viral pneumonia and 3 mycoplasma). For the remaining 31 patients, the etiology was uncertain, but the possibility of false-negative COVID-19 test results was excluded through strict epidemiological investigations, several RT-PCR tests, and final clinical outcomes. This study was approved by the medical ethics committees of the participating hospitals, and no informed consent was required after review by the committees.
The demographic information of the participants and the acquisition parameters of the CT images are summarized in Table 1 . The tube voltage for all CT examinations was 120 kVp and the slice thickness ranged from 0.625 to 5.0 mm. CT examinations using standard imaging protocols were conducted using scanners from various manufacturers. Typically, 50–505 CT slices were acquired in each volumetric CT examination. All CT images had the same matrix size of 512×512 with 16 bits and were collected from hospitals in the DICOM format.
Table 1.
Demographic information of the participants and acquisition parameters of the CT images.
| Information | COVID-19 | CAP | p value |
|---|---|---|---|
| Clinical type | Mild (n=2)Moderate (n=38)Severe (n=20)Critical (n=9)Uncertain (n=72) | Bacterial (n=57)Viral (n=9)Mycoplasma (n=3)Uncertain (n=31) | - |
| Gender (male/female) | 60/81 | 52/48 | 0.1474a |
| Age (years) (mean ± S.D.) | 55.16±17.71 | 40.92±20.41 | 3.76×10−8b |
| Tube voltage (kVp) | 120 | 120 | - |
| Slice thickness (mm) | 0.625 (n=39)1.0 (n=14)1.25 (n=17)1.50 (n=1)2.0 (n=25)5.0 (n=45) | 1.0 (n=30)1.50 (n=4)2.0 (n=48)3.0 (n=16)5.0 (n=2) | - |
| Pixel size (mm)(mean ± S.D.) | 0.763±0.063 | 0.697±0.103 | 1.61×10−6b |
| Tube current (mA) (mean ± S.D.) | 236.723±69.534 | 206.8±89.485 | 0.0016b |
| CT scanner manufacturer | Siemens (n=26), Toshiba (n=25),GE Medical Systems (n=90) | Siemens (n=31), Toshiba (n=69) | - |
| Manufacturer model name | Sensation 16 (n=26), Optima CT660 (n=39), Aquilion ONE (n=25), LightSpeed16 (n=51) | Aquilion (n=69),SOMATOM Definition AS+ (n=6),SOMATOM Scope (n=25) | - |
| Institution name | General Hospital of the Yangtze River Shipping (n=75),Wuhan Puren Hospital (n=66); | The Affiliated Hospital of Guizhou Medical University (n=100) | - |
p value is for the chi-square test;
p value is for the two-sample t-test.
In addition to our own dataset, we also included open-access data downloaded from the China Consortium of Chest CT Image Investigation (CC-CCII) [17]. After excluding those with fewer than 60 CT slices, with segmented images only, with incomplete lung area, and from COVID-19 patients without typical image manifestations, the final dataset contained CT images in JPEG or PNG format from 620 COVID-19 patients and 610 CAP patients. For these patients, the CT scans with the largest number of CT slices were selected. The aim of using the CC-CCII dataset was to evaluate the generalization ability of our proposed method. It should be noted that the CC-CCII database only provides images in JPEG format, although it is known that the compression from DICOM to JPEG may affect the performance during clinical diagnosis. Moreover, our own dataset and the CC-CCII dataset were not combined but separately used.
2.2. Outline of proposed method
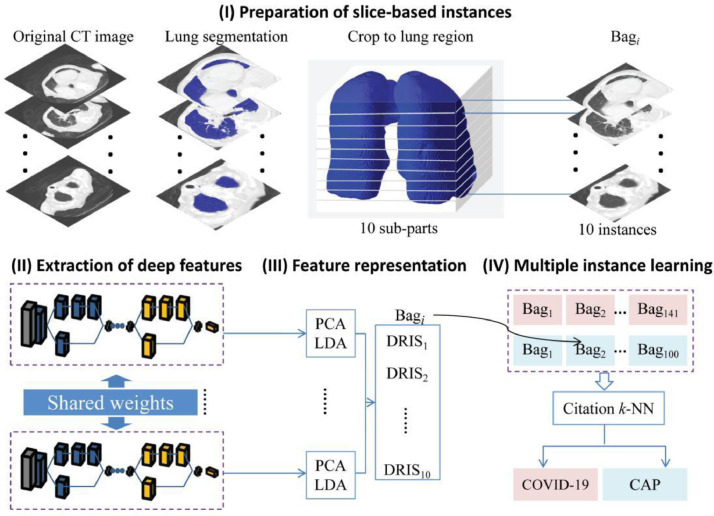
The DR-MIL method proposed in the current study consists of four key steps (Fig. 1 ): (I) preparation of slice-based instances, (II) extraction of deep features, (III) feature representation, and (IV) multiple instance learning. Descriptions of each step are provided below.
Fig. 1.
Overview of procedures used in this study.
2.3. Preparation of slice-based instances
Given that the CT images originated from different sources, preprocessing of the images was important. The CT images were first input into the Pulmonary Toolkit software (https://www.tomdoel.com/software/) and the 3D lung volume was extracted using an embedded region-growing algorithm. Threshold segmentation (CT number, 400 HU, −1500 HU) was initially used to binarize the images and the largest connected region was then identified, excluding any region that started from the image edge and grew inward to the chest region with a pixel value of 0. It should be noted that the corresponding Pulmonary Toolkit code has been embedded into our proposed software tool, such that no explicit input from or output to Pulmonary Toolkit is required.
A bounding box containing the extracted lung field was obtained. We then cropped a cuboid from the volume of the original CT images by using the bounding box (not the segmented mask of the lung field). This cuboid was further split into ten sub-parts evenly along the longitudinal direction. One random slice was selected from each sub-part, resized to the same dimensions of 224×224, and used as one instance in the subsequent processes. One bag with ten slice-based instances was obtained for each subject. It should be noted that accurate segmentation of the lung field is not required because one bounding box is needed. Even so, the quality of segmentation was confirmed by visual inspection.
After obtaining the instances, two further steps were performed: (1) the window location and level were set to 1600 and −600 HU, respectively, to clearly display the lung area, and (2) the data were normalized. Specifically, the pixel intensity was first adjusted to between 0 and 1, and the data were then normalized by the channel mean and standard deviation of the images in ImageNet.
2.4. Extraction of deep features
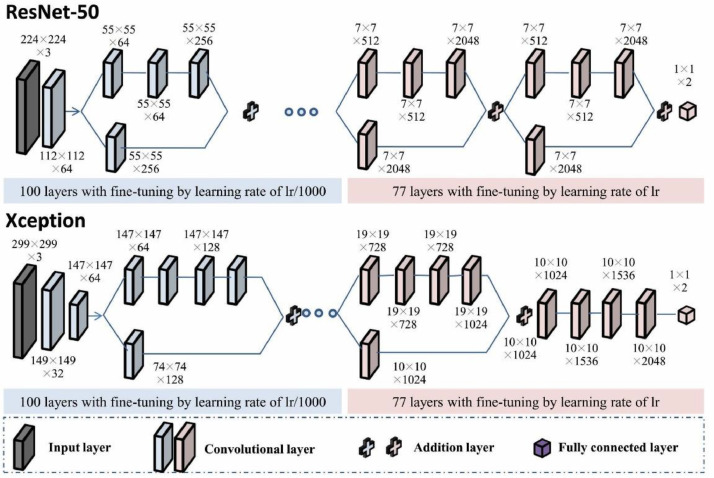
Pre-trained ResNet-50 and Xception with fine-tuning were employed as the backbone networks for deep feature extraction (Fig. 2 ) [26,27]. Two neurons were kept in the last fully connected layer of the network to adapt to the binary classification problem. The parameters of the last three layers were initialized in Glorot mode and pre-trained parameters from ImageNet were used for weight initialization of the other layers. The first 100 layers and the other 77 layers in the pre-trained model were fine-tuned using initial learning rates (lr) of lr/1000 and lr, respectively. Here, lr was 0.03. The learning rate was reduced by a factor of 0.5 for every five epochs. In shallow layers, image features are at a low abstraction level and highly transferable; they are therefore capable of being applied to most tasks of computer vision [28]. At increased depths, the quality of feature expression is more dependent on the data in the training set. Stochastic gradient descent with momentum (SGDM) was adopted for the optimization algorithm. Meanwhile, the number of epochs for training was set to 15, and the batch size was 32.
Fig. 2.
Pre-trained backbone networks (ResNet-50 and Xception) with fine-tuning for feature extraction.
Data augmentation techniques (image rotation, reflection, and translation) and L1 regularization were used to fine-tune the backbone network to avoid overfitting. Specifically, each image was translated by up to 50 pixels horizontally and vertically, rotated with an angle of up to 360°, and reflected in the left-to-right direction, with 50% probability during each epoch of training, such that each epoch used a different data set but the number of training images did not change. Finally, 100,352 deep features were obtained from the last addition layer of ResNet-50. To ensure that no information leakage occurred, the fine-tuning was controlled within each fold of the 10-fold cross-validation procedure. We also evaluated other CNN models as the feature extractor, including InceptionV3 [29], DenseNet-201 [30], GoogleNet [31], MobileNetV2 [32], ShuffleNet [33], VGG-19 [34], and AlexNet [35].
2.5. Feature representation
As the deep feature dimensionality (100,352) was too high, we adopted a combination of principal component analysis (PCA) and linear discriminant analysis (LDA) to reduce the dimensionality. PCA was applied first and the top 1200 principal components were retained. LDA was then used to project the data onto one-dimensional space, where the distance between the centers of the two categories of data (COVID-19 and CAP) was as large as possible and their covariance was as small as possible. Therefore, the deep features of each instance were represented as the score that we herein refer to as the DRIS.
2.6. Multiple instance learning and visual interpretability
Citation k-nearest neighbor (k-NN) has been used as one classifier in multiple instance learning [36]. Citation k-NN seeks to search references and citers for a particular unknown bag Uk. Specifically, in the training set, a bag Bi and its C-th nearest bag BC are first determined and their distance is denoted DiC. If the distance between the bag Uj in the test set and Bi is closer than or equal to DiC, Bi is a citer of Uj. Similarly, R nearest neighbors of Uj in the training set are considered as references of Uj. Finally, the decision for a particular unknown bag Uj is based on the majority voting of the labels of bags regarded as references and citers. The Hausdorff distance is exploited to measure the similarity between any two bags:
| (1) |
| (2) |
| (3) |
where a and b are the instances in A and B, respectively, and ‖a − b‖ and ‖b − a‖ are the Euclidean distances.
In this study, a grid search method was employed to determine R and C. The search range of the two parameters was [1,15] and the interval was 1. R=3 and C=13 represent the highest accuracy. The ratio of the number of references and citers with the label COVID-19 to the number of all references and citers was calculated to represent the probability of COVID-19. A threshold of 0.5 was used to determine the measures of accuracy, specificity, sensitivity, and F1 score.
Gradient-weighted class activation mapping (Grad-CAM) was used to interpret the results obtained from ResNet-50 and highlight important regions in the CT slices for predicting COVID-19 and CAP [37]. Details of the principles and implementation of Grad-CAM are provided in the Supplementary Material.
2.7. Comparative experiments
In the DR-MIL method, the feature extractors for ResNet-50 and Xception are denoted MResNet-50-MIL and MXception-MIL, respectively. Moreover, to assess the role of fine-tuning, we also used the strategy of freezing all previous layers before fully connecting them; the resulting model is denoted MResNet-50-MIL-frozen-layers.
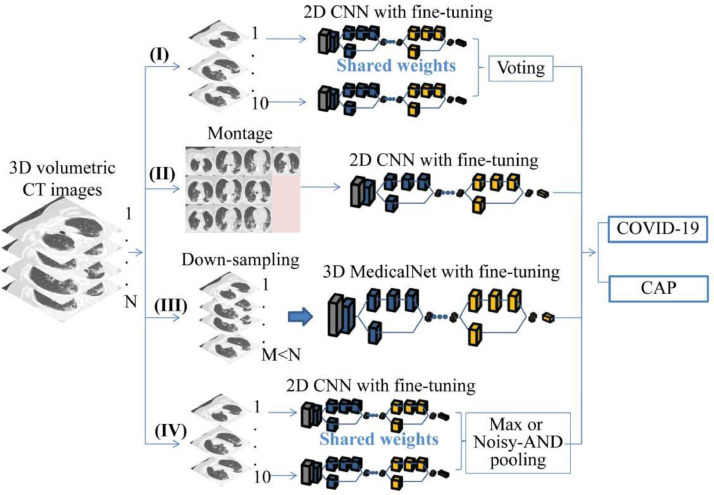
Another four categories of comparable experiments were performed as depicted in Fig. 3 . The pseudo code representation for each method is presented in Fig. 4 , with the exception of the second category, where the procedure is rather simple. The first category was 2D CNN with voting. Pre-trained ResNet-50 and Xception were initially fine-tuned by our CT slices. Ten instances (slices) were sequentially input into the fine-tuned models to generate ten slice-based predictions, and the final patient-level prediction was made on the basis of the majority voting of these predictions. These two models are denoted MXception-Voting and MResNet-50-Voting, respectively.
Fig. 3.
Four categories of comparable methods (2D voting, 2D montage, 3D down-sampling, and MIL with max or Noisy-AND pooling).
Fig. 4.
Pseudo code representations for three comparable methods.
The second category was a 2D CNN of a montage of ten CT slices. Ten instances from each patient were initially combined into a montage, and these montages were used to fine-tune pre-trained ResNet-50 and Xception. The resulting models are denoted MResNet-50-Montages and MXception-Montages, respectively.
For the third category, 241 preprocessed CT scans were used to fine-tune and test the 3D network that had been pre-trained on the 3DSeg-8 dataset, which covers various organs/tissues of interest with either CT or MR scans [38]. A fully connected layer was employed to take over from the last convolutional layer in the backbone network (ResNet-50), changing the network from segmentation into a classification architecture. The initial learning rate was lr (lr = 0.001) for the fully connected layer and lr/100 for the remaining layers. SGDM was adopted for the optimization algorithm. The learning rate was reduced by a factor of 0.99 for every epoch, and the number of epochs for training was 60. Owing to the limitations of the GPU memory, each 3D matrix was resized to 224×224×50 and the batch size was 2. The resulting model is denoted MMedicalNet.
Finally, two end-to-end deep CNNs for MIL were compared with our DR-MIL method. As shown in Fig. 3, a special MIL pooling layer combined all of the feature maps of the pre-trained ResNet-50 with fine-tuning from all 10 instances. A fully connected layer was linked to the MIL layer to afford the final prediction. We have so far applied max pooling [25] and Noisy-AND pooling [26]. The trained models are denoted MResNet-50-MIL-max-pooling and MResNet-50-MIL-Noisy-AND-pooling, respectively.
2.8. Training, testing, and evaluation
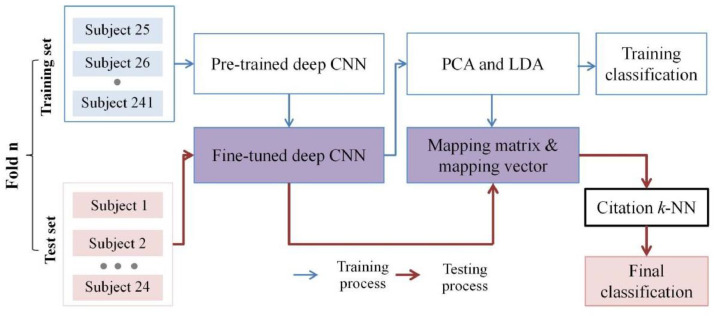
The training and testing procedures of the proposed models were performed using 10-fold cross-validation (Fig. 5 ). The entire dataset of the chest CT scans of 241 patients was split into training and test sets in a ratio of 9:1. During the training process, 8/9 of the training set was used to fine-tune the pre-trained CNN and the remaining 1/9 was used to validate it. The deep features were extracted from the fine-tuned CNN and represented as a DRIS that combines PCA and LDA. In the testing process, the deep features were extracted from the slice-based instances by the fine-tuned deep CNN and incorporated into the DRIS by the mapping matrix and mapping vector that resulted from the training process. The bag of DRISs was then input into citation k-NN to yield the final classification.
Fig. 5.
Training and testing procedures of the proposed models by 10-fold cross-validation.
MMedicalNet was implemented in the PyTorch library, and the other experiments were performed in Matlab 2019b (Deep Learning Toolbox) on a Windows 10 system. The workstation used for the implementation had an Intel Core i7-9700 3.00 GHz CPU with an NVIDIA GeForce RTX 2080 Ti GPU. Citation k-NN was executed using the Multiple Instance Learning Library (MILL) (http://www.cs.cmu.edu/∼juny/MILL).
Five performance measures, namely, the area under the curve (AUC), accuracy (ACC), sensitivity (SEN), specificity (SPE), and F1 score, were used to evaluate the different models:
| (4) |
| (5) |
| (6) |
| (7) |
where TP, TN, FP, and FN denote the true positive, true negative, false positive, and false negative, respectively. Precision = TP/(TP + FP) and Recall is equal to SEN.
3. Results
3.1. DR-MIL method and its performance
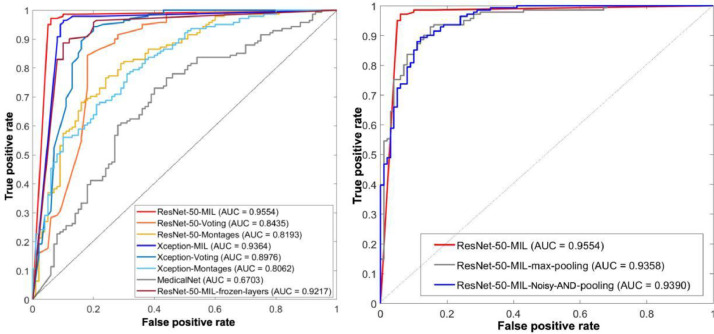
MResNet-50-MIL displayed the best patient-level performance, with an ACC of 0.959, SEN of 0.972, SPE of 0.940, AUC of 0.955, and F1 score of 0.965 (Fig. 6 and Table 2 ). For the cohort of 241 participants in our study, we asked one radiologist with 16 years of experience to differentiate COVID-19 from CAP via CT images. The accuracy of this radiologist was 66.80%, which is within the range of 60–83% reported by Bai et al. [9]. The accuracy of 95.9% observed for MResNet-50-MIL thus demonstrates the potential of this method for improving the differentiation of COVID-19 from CAP.
Fig. 6.
ROC curves and AUC values for the proposed and comparable models.
Table 2.
Performance comparison of the various models for identifying COVID-19.
| Model | ACC | SEN | SPE | AUC | F1 score |
|---|---|---|---|---|---|
| MResNet-50-MIL | 0.959 | 0.972 | 0.940 | 0.955 | 0.965 |
| MResNet-50-MIL-frozen-layers | 0.888 | 0.887 | 0.890 | 0.922 | 0.903 |
| MXception-MIL | 0.938 | 0.965 | 0.900 | 0.936 | 0.948 |
| MResNet-50-Voting | 0.900 | 0.957 | 0.820 | 0.844 | 0.918 |
| MXception-Voting | 0.920 | 0.936 | 0.900 | 0.898 | 0.933 |
| MResNet-50-Montages | 0.730 | 0.965 | 0.400 | 0.819 | 0.807 |
| MXception-Montages | 0.734 | 0.760 | 0.690 | 0.806 | 0.772 |
| MMedicalNet | 0.681 | 0.908 | 0.360 | 0.670 | 0.769 |
| MResNet-50-MIL-max-pooling | 0.896 | 0.936 | 0.840 | 0.936 | 0.914 |
| MResNet-50-MIL-Noisy-AND-pooling | 0.880 | 0.929 | 0.810 | 0.939 | 0.900 |
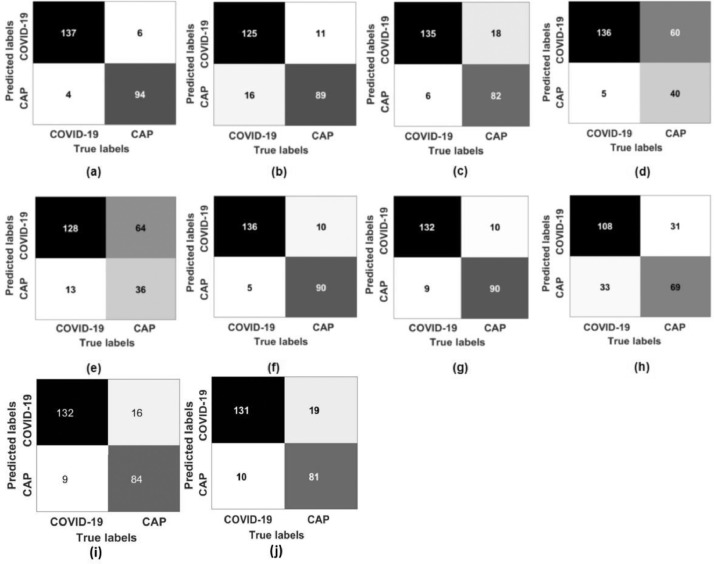
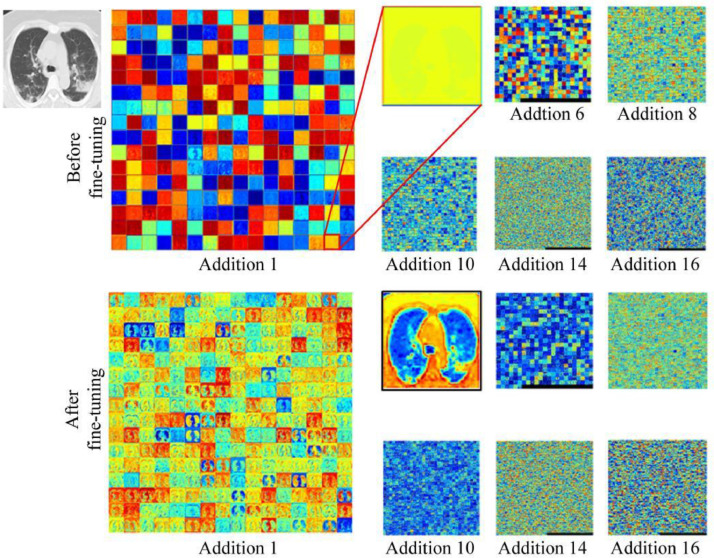
As shown in Fig. 7 , among the 141 patients with COVID-19, four were wrongly predicted as non-COVID-19, including one severe case, one moderate case, and two unknown cases. Among the 100 CAP patients, six were wrongly predicted as COVID-19 (two cases of bacterial pneumonia, four cases of unknown etiology). For one case, MXception-MIL was not as good as MResNet-50-MIL, and the former displayed an ACC of 0.938, SPE of 0.900, and AUC of 0.936. Moreover, as expected, the performance of MResNet-50-MIL-frozen-layers was inferior to that of MResNet-50-MIL, indicating that the fine-tuning helped with the extraction of deep features. Fig. 8 confirms this point, where the contrast in the feature maps (especially in the layer of Addition 1) was significantly enhanced after fine-tuning. The lung field is clearly indicated and the lesions have been highlighted.
Fig. 7.
Confusion matrices for the proposed and comparable models: (a) MResNet-50-MIL, (b) MResNet-50-MIL-frozen-layers, (c) MResNet-50-Voting, (d) MResNet-50-Montages, (e) MMedicalNet, (f) MXception-MIL, (g) MXception-Voting, (h) MXception-Montages, (i) MResNet-50-MIL-max-pooling, and (j) MResNet-50-MIL-Noisy-AND-pooling.
Fig. 8.
Feature maps for ResNet-50 before and after fine-tuning.

The results obtained using ResNet-50 and seven other CNN models as the feature extractor are presented in Table 3 . The parameter k indicates the number of the last layer with the initial learning rate, after which the learning rate was set as the initial learningrate multiplied by 10−3. The parameter k, initial learning rate, and learning rate drop factor were determined by a Bayesian optimization algorithm. It can be seen that the highest ACC for COVID-19 identification by DR-MIL was obtained when ResNet-50 was used for feature extraction. Moreover, the training time was 2440 seconds, which was shorter than those for the other CNN models with the exception of AlexNet.
Table 3.
Comparison of various feature extractors for COVID-19 identification by DR-MIL.
| Feature extractor | Size of model(MB) | k* | Initial learning rate | Learning rate drop factor | Output layer | Feature map | ACC | Training time (s) |
|---|---|---|---|---|---|---|---|---|
| ResNet-50 | 96 | 101 | 0.03 | 0.500 | 172 | 7×7×2048 | 0.959 | 2440 |
| InceptionV3 [29] | 89 | 178 | 0.031 | 0.400 | 311 | 8×8×2048 | 0.876 | 3504 |
| DenseNet-201 [30] | 77 | 400 | 0.031 | 0.400 | 703 | 7×7×1920 | 0.834 | 13050 |
| GoogleNet [31] | 27 | 94 | 0.014 | 0.286 | 139 | 7×7×1024 | 0.710 | 2748 |
| MobileNetV2 [32] | 13 | 88 | 0.027 | 0.372 | 148 | 7×7×1280 | 0.834 | 3438 |
| ShuffleNet [33] | 5.4 | 98 | 0.021 | 0.162 | 167 | 7×7×544 | 0.793 | 3246 |
| VGG-19 [34] | 535 | 28 | 0.014 | 0.293 | 36 | 14×14×512 | 0.710 | 4872 |
| AlexNet [35] | 227 | 14 | 0.009 | 0.0126 | 14 | 13×13×256 | 0.710 | 2398 |
Layer(1:k): learning rate was set as the initial learning rate; Layer(k+1:end): learning rate was set as the initial learning rate multiplied by 10−3.
The running and testing times for the various methods are compared in Table 4 . MResNet-50-MIL required 2586 seconds for running one fold and 1.17 seconds for testing one case. This testing time is considered acceptable, although it was longer than those for the other models.
Table 4.
Running and testing times for the various models.
| Model | Running time for one fold (s) | Testing time for each case (s) |
|---|---|---|
| MResNet-50-MIL | 2586 | 1.17 |
| MMedicalNet | 5370 | 0.25 |
| MResNet-50-MIL-max-pooling | 2331 | 0.39 |
| MResNet-50-MIL-Noisy-AND-pooling | 2354 | 0.39 |
| MResNet-50-Voting | 2453 | 0.49 |
| MResNet-50-Montages | 2547 | 0.02 |
3.2. Comparison between DR-MIL and other methods
MResNet-50-MIL outperformed the models based on the strategy of 2D CNN with voting (Figs. 6, 7, and Table 2), and MXception-Voting outperformed MResNet-50-Voting. These results indicate that using MIL to integrate the predictions of slice-based instances may be superior to using simple voting.
MResNet-50-MIL also performed better than the 2D CNN models based on montages. The AUC values for MResNet-50-Montages and MXception-Montages were 0.819 and 0.806, respectively, which were even lower than those for MResNet-50-Voting and MXception-Voting. There are two plausible explanations for this outcome: (1) the dataset using montages was smaller than that using voting, such that the former could not fully train the model despite the adoption of transfer learning, and (2) extracting discriminative features from the montage of ten slices was more difficult than from each slice separately.
The pre-trained 3D MedicalNet with fine-tuning (MMedicalNet) displayed the least satisfactory performance, with an ACC of 0.681, SEN of 0.908, SPE of 0.360, AUC of 0.670, and F1 score of 0.769. In this regard, MMedicalNet had more severe challenges of limited data and increased feature extraction difficulty than MResNet-50-Montages and MXception-Montages.
In our current study, MResNet-50-MIL outperformed the two end-to-end deep MIL CNNs, MResNet-50-MIL-max-pooling and MResNet-50-MIL-Noisy-AND-pooling, which displayed AUC values of 0.936 and 0.939, respectively.
3.3. Comparison between DR-MIL and current state-of-the-art studies
The performance of our model was also compared with those of current state-of-the-art studies (Table 5 ). It can be seen that our method is comparable to the other strategies, including machine learning [21], pre-trained 2D CNN and merged slice-based predictions [14,18], 3D ResNet [39], and 3D deep MIL [22]. However, it must be noted that the results were not obtained from the same datasets (not all of which have been publicly disclosed), thus limiting the significance of this comparison.
Table 5.
Performance of our DR-MIL method and current state-of-the-art studies.
| Study | Key aspects | Performance |
|---|---|---|
| Our study (MResNet-50-MIL) | - Deep features extracted by ResNet-50- DRIS represented by PCA and LDA and multiple instance learning- 241 patients (COVID-19: 141, CAP: 100)- Binary classification (COVID-19 or CAP) | ACC = 0.959, AUC = 0.955,SEN = 0.972,SPE = 0.941 |
| Kang et al., 2020 [21] | -V-Net for lung segmentation and 189 handcrafted features extracted from lesions- Complete and structured representation learning- Fully connected neural network for classification- 2522 CT images (COVID-19: 1495, CAP: 1027) | ACC = 0.955,SEN = 0.966,SPE = 0.932 |
| Li et al., 2020 [14] | - COVNet using ResNet-50 as the backbone- 4356 chest CT examinations from six hospitals (COVID-19: 1296, CAP: 1735, non-pneumonia: 1325)- Three-class classification (non-pneumonia, CAP, or COVID-19) | AUC = 0.96,SEN = 0.90,SPE = 0.96 |
| Bai et al., 2020 [18] | - Lung segmentation by 3D Slicer software and manual modification- 1186 patients (COVID-19: 521, non-COVID-19 pneumonia: 665)- 2D pre-trained EfficientNet with fine-tuning- Slice predictions concatenated using two fully connected layers | AUC = 0.95,ACC = 0.96,SEN = 0.95,SPE = 0.96 |
| Ouyang et al., 2020 [16] | - VB-Net toolkit for lung segmentation- Two 3D ResNet-34 networks- Online attention module and ensemble learning- Multi-center dataset: 2186 CT scans for training and validation, 2776 CT scans for test set- Binary classification (COVID-19 or CAP) | ACC = 0.875,AUC = 0.944,SEN = 0.869,SPE = 0.901 |
| Han et al., 2020 [22] | - Generate 3D deep instance automatically- Attention-based MIL pooling- CT examinations of 79 COVID-19 patients, 100 CAP, 130 without pneumonia- Three-class classification (non-pneumonia, CAP, or COVID-19) | ACC = 0.943,AUC = 0.988 |
| Zhang et al., 2020 [17] | - Seven-class segmentation- 3D ResNet-18 for classification- 2246 patients (COVID-19: 752, CAP: 797, non-pneumonia: 697) for training; six validation datasets for testing- Three-class classification (non-pneumonia, CAP, or COVID-19) | ACC = 85.26–92.49% |
| Jin et al., 2020 [42] | - Dataset of 10,000 CT volumes from COVID-19, influenza A/B, non-viral (CAP), and non-pneumonia subjects.- 2D pre-trained ResNet-152 for slice-level prediction- Task-specific fusion block for volume/case-level prediction | AUC = 0.978 fora test cohort of 3199 scans |
| Harmon et al., 2020 [13] | - Lung segmentation- Full 3D model and hybrid 3D model- Multinational datasets of 2617 patients | ACC = 0.908,SEN = 0.840,SPE = 0.930 |
| Di et al., 2021 [43] | - 2148 COVID-19 and 1182 CAP- Uncertainty vertex-weighted hypergraph learning (UVHL) method | ACC = 0.898 |
| Javaheri et al., 2021 [44] | - COVID-19: 111, CAP: 115, healthy control: 70- Bi-directional ConvLSTM U-Net with densely connected convolutions (BCDU-Net) | ACC = 0.95 |
The MResNet-50-MIL method was also applied to the CC-CCII dataset. Specifically, the deep features were extracted for each image instance using the same pre-trained ResNet-50 with fine-tuning using our own dataset. The deep features of each instance were further represented as DRISs by PCA and LDA. Using 10-fold cross-validation for citation k-NN yielded an ACC of 0.957, SEN of 0.925, SPE of 0.989, AUC of 0.952, and F1 score of 0.989. In this 10-fold cross-validation, one-tenth was used for training and nine-tenths for testing. The performance of our method was comparable to that reported in a previous study [17], where the accuracy ranged from 85.26% to 92.49% for one internal validation dataset, one retrospective study, three prospective pilot studies, and one additional dataset from Ecuador.
Our method is a combination of deep learning and machine learning and has the advantage of being applicable to small datasets of only a few hundred CT examinations. Moreover, with the exception of the previous study that used 3D deep MIL [22], our approach has additional benefits over comparable methods, including the weakly supervised nature of the learning and no requirement for lesion segmentation.
3.4. Deep features and DRIS
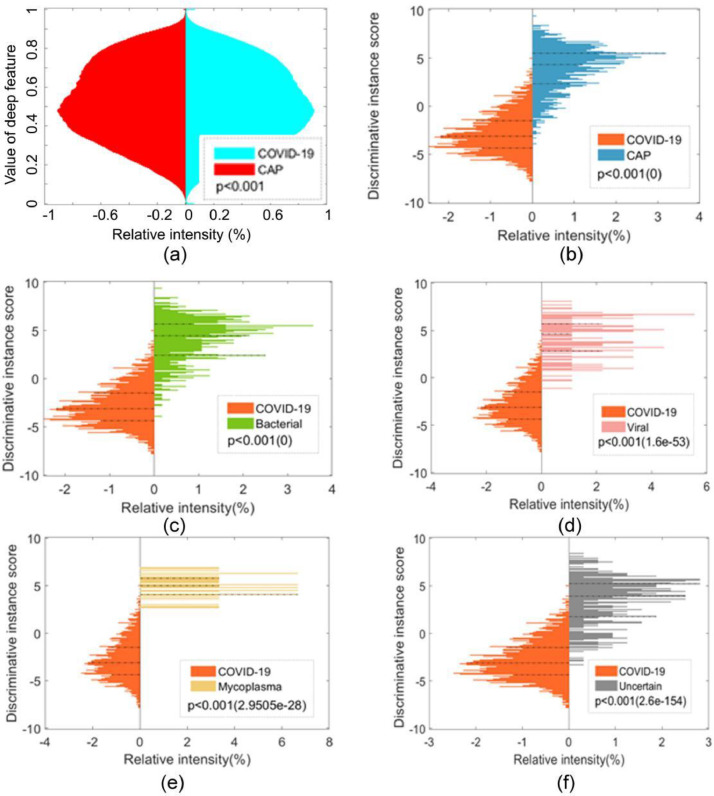
As shown in Fig. 9 (a), the distribution of deep features (100,352 features for each instance or slice) was plotted for the COVID-19 and CAP groups. A two-sided Wilcoxon rank sum test was performed to determine whether the distribution was significantly different between the two groups, which indicated a significant difference (p<0.001). As shown in Fig. 9(b), a significant difference was also observed in terms of the DRISs between the COVID-19 and CAP groups (p<0.001). Similar differences were also found between COVID-19 and the various subcategories of CAP (bacterial, viral, mycoplasma, and uncertain etiology) (Figs. 9(c)–(f)). These results demonstrate that the DRIS is highly discriminative for COVID-19 and CAP and accounts for the exceptional performance observed for MIL using the DRIS. Therefore, the DRIS can potentially be applied as a slice-based imaging biomarker to differentiate between COVID-19 and CAP.
Fig. 9.
Distribution of deep features and DRIS for COVID-19 and CAP: (a) deep features for COVID-19 versus CAP, (b) DRIS for COVID-19 versus CAP, (c) DRIS for COVID-19 versus bacterial CAP, (d) DRIS for COVID-19 versus viral CAP, (e) DRIS for COVID-19 versus mycoplasma CAP, and (f) DRIS for COVID-19 versus CAP of uncertain etiology.
3.5. Spatial patterns of COVID-19 and CAP lesions
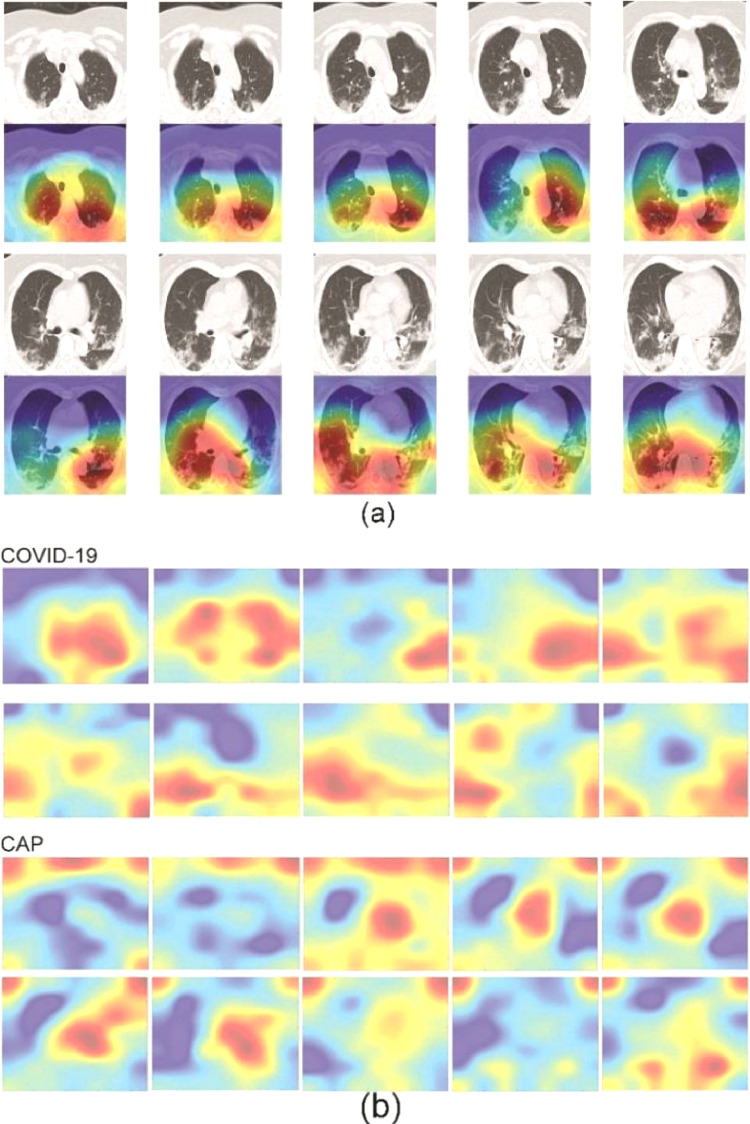
The Grad-CAM results demonstrated that our pre-trained ResNet-50 with fine-tuning could focus on the lesions in COVID-19 CT images (Fig. 10 (a)). Therefore, we were able to sum the Grad-CAM results for all COVID-19 patients to obtain the spatial pattern of lesions. Fig. 10(b) presents a comparison of the spatial patterns of lesions for COVID-19 and CAP. It can be seen that the main lesions in COVID-19 were located in the lower parts of the lung in the axial view and distributed in the subpleural areas. In contrast, the lesions in CAP were more likely to be found around the central area.
Fig. 10.
Grad-CAM results and spatial patterns of COVID-19 and CAP lesions: (a) example of Grad-CAM for one case of COVID-19 and (b) comparison of spatial patterns of lesions between COVID-19 and CAP.
These findings are in accordance with previous clinical reports. Salehi et al. reported the distribution of lesions in COVID-19 as a mixture of peripheral (76.0%), multilobar (78.8%), and bilateral (87.5%) [39]. Lesions are predominantly distributed in the posterior segments of the subpleural areas and may progress to central areas as the condition of the patient worsens [40,41,45]. Although this distribution of lesions is partially shared with that in CAP patients, there are also significant differences: a central location was observed in only 10.65% of the COVID-19 group but in 47.13% of the non-COVID-19 group.
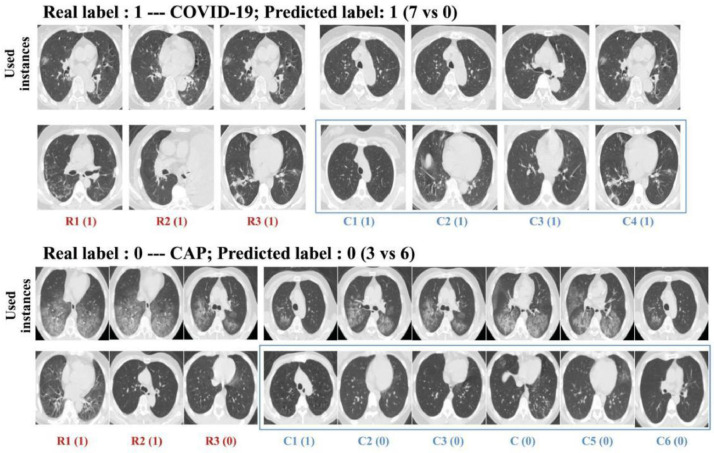
3.6. Visual interpretation for CBIR
As shown in Fig. 11 , one advantage of citation k-NN is that the key instances determining the distance between unknown bags and the corresponding citers or references can be presented as CBIR. From left to right, the distance between a particular instance in a bag is called a reference, and the corresponding instance used in the unknown bag increases as the visual similarity between them diminishes. For the example of COVID-19, all of the labels for the three references and four citers are “1” (i.e., COVID-19; 7 versus 0); hence, the predicted label is also “1”. For CAP, two references and one citer have the incorrect prediction of “1” while the others have the correct prediction. After voting, the final prediction is the correct label, i.e., “0” (3 versus 6).
Fig. 11.
Visual interpretation of used instances and the corresponding instances in the bags regarded as references and citers.
4. Discussion
In the current study, we have evaluated the suitability of the DR-MIL method for distinguishing between COVID-19 and CAP in CT images. Deep features were drawn from each slice-based instance by using the pre-trained ResNet-50 with fine-tuning and transformed into a DRIS. Each patient was treated as a bag of DRISs and citation k-NN was employed as the MIL classifier to generate the final patient-level prediction.
The key findings of this study are as follows: (1) DR-MIL afforded an ACC of 95% and an AUC of 0.943, which are superior to comparable methods and evaluation by a radiologist (66.80%). The reading time was approximately 1.17 seconds. (2) COVID-19 and CAP exhibited significant differences in terms of both the DRIS and the spatial pattern of lesions (p<0.001). (3) As a means of CBIR, DR-MIL can identify images used as key instances, references, and citers for visual interpretation. Therefore, similar to the previously published model, DR-MIL could serve as an independent reader to provide useful suggestions to radiologists. Furthermore, the short reading time of DR-MIL may help improve the productivity of radiologists for COVID-19 diagnosis. According to Jin et al. [42], the average reading time of radiologists is approximately 6.5 minutes per CT scan, while their AI system required only 2.73 seconds. Thus, DR-MIL could be used to screen candidates for confirmation by a radiologist by setting a high SEN and/or provide an error warning by setting a high ACC [42], allowing human and artificial intelligence to be combined. The visual interpretation afforded by DR-MIL could facilitate this combination and help train inexperienced radiologists. The advantages of our method and implications of our findings are discussed in the following sections.
4.1. MIL versus slice-based voting
In previous studies, MIL has yielded excellent performance while tackling the problem of uncertain lesion locations in chronic obstructive pulmonary disease [46,47]. In this study, the specificity and accuracy of MResNet-50-Voting were lower than those of MResNet-50-MIL. According to the MIL definition, a positive bag consists of “true positive instances” and “false positive instances”, while all instances in a negative bag are negative [48]. Here, the bag or instance that is labeled COVID-19 is defined as positive; otherwise, it is negative. Therefore, under the assumption that CNN can achieve accurate classification, if an instance is predicted to be positive, this bag should be regarded as positive. Obviously, the method of slice-based majority voting used in MResNet-50-Voting lacks interpretability, although it can achieve high accuracy. However, it is more reasonable to conduct majority voting on the bag level because a negative bag can be similar to a positive bag with “false positive instances”. This is why an unknown bag cannot be directly judged as positive when any of its citers or references is positive [36].
4.2. Feature representation and DRIS
It is unrealistic to directly feed features with a dimensionality that is much larger than the number of samples into the classifier because of the over-fitting problem. Therefore, a suitable feature reduction strategy is needed. A combination of PCA and LDA is one such approach to obtaining inputs for citation k-NN. LDA has been used in numerous fields, such as meal detection and face recognition [49,50], and the combination of PCA and LDA has been applied to disease diagnosis [51,52]. In our study, the combination of PCA and LDA was adopted to incorporate deep features into a single DRIS, and our experiments demonstrated that this combination was superior to PCA or LDA alone. We thus introduce the DRIS as a potential imaging biomarker to distinguish COVID-19 from CAP. This parameter can be obtained from CT images and used as a discriminative feature to construct more accurate classifiers using clinical information.
Besides these 100,352 deep features, we have tried more features from the other layers. The classification performance obtained using more features from the other layers was inferior to that achieved for the 100,352 deep features.
4.3. Visual interpretation of citation k-NN
Citation k-NN is similar to CBIR [53]. The training set is a large repository containing medical images from patients with various diseases. Given a particular image, several similar images in the repository could be retrieved by using the similarity measured by the Hausdorff distance. In this manner, radiologists and clinicians could be helped to understand why the DR-MIL method affords a given prediction. Meanwhile, CBIR is also useful for training inexperienced clinicians and developing “doctor-in-the-loop” or AI-augmented diagnosis of COVID-19 [18,54].
4.4. Irrelevant variables and standardization
In our study, the COVID-19 patients and CAP patients were examined in different hospitals using different CT protocols. There were also significant demographic differences, e.g., age. Thus, one may raise the objection that the model may just learn the differences between different protocols and ages. We discuss this issue below.
First, it should be noted that a number of the COVID-19 patients (n=25) and CAP patients (n=69) were scanned using the same CT instrument model (Aquilion), although they were used at different hospitals. Moreover, even within a single hospital, several different CT scanners and protocols were used. This is typical of most Grade-A tertiary hospitals, which often operate more than one CT scanner.
Second, to strictly control for the influence of irrelevant variables on the classification or prediction, the patients must be scanned using the same scanner and the same protocol. However, this would reduce the sample size and the generalization ability of the trained model. Therefore, in most studies [14,18], CT images from different hospitals, scanners, and protocols have been used in conjunction after proper preprocessing and standardization. In our study, we also applied preprocessing and standardization methods similar to those of Bai et al. [18].
Third, although it is known that differences in CT images can result from different hospitals, scanners, and protocols, and that these may influence the performance of trained classification models such as those that use handcrafted features in radiomics [55], we have found the robustness of models using deep learning or deep features to be significantly improved by preprocessing and standardization. As shown in Table 6 , no apparent differences in accuracy were observed among the various slice thicknesses. Harmon et al. trained their model by using a highly diverse multinational dataset with different CT scanners and acquisition protocols [13]. The developed model exhibited acceptable performance when applied to new data, compared with models trained using data from only one center, one CT scanner, or one scanning protocol. Moreover, a recent study reported a method of applying deep learning to convert reconstructed CT images by using different kernels to improve the reproducibility of radiomics models [56].
Table 6.
Performance of our proposed DR-MIL method for various slice thicknesses.
| Slice thickness (mm) | Accuracy (number of correctly predicted samples/total number of samples) |
Average accuracy | |
|---|---|---|---|
| CAP | COVID-19 | ||
| 1.0 | 0.900 (27/30) | 0.929 (13/14) | 0.910 (40/44) |
| 1.5 | 0.750 (3/4) | 1.000 (1/1) | 0.800 (4/5) |
| 2.0 | 1.000 (48/48) | 0.920 (23/25) | 0.973 (71/73) |
| 3.0 | 1.000 (16/16) | — (0/0) | 1.000 (16/16) |
| 5.0 | 0.000 (0/2) | 0.978 (44/45) | 0.936 (44/47) |
| 0.625 | — (0/0) | 1.000 (39/39) | 1.000 (39/39) |
| 1.25 | — (0/0) | 1.000 (17/17) | 1.000 (17/17) |
Fourth, we have also used two independent external datasets to evaluate our method. The promising performance observed for both sets of images suggests that the proposed method is largely insensitive to differences associated with irrelevant variables.
Fifth, it is known that COVID-19 outcomes are negatively influenced by higher age and the presence of comorbidities. To clarify whether age affects the performance of our current model, we performed further analysis. The COVID-19 group was divided into two sub-groups of COVID-19-Sub-Group 1 (age ≤ 33 years) and COVID-19-Sub-Group 2 (age > 33 years), while the CAP group was divided into CAP-Sub-Group 1 (age ≤ 31 years) and CAP-Sub-Group 2 (age > 31 years). No significant difference in age was found between CAP-Sub-Group 2 and COVID-19-Sub-Group 2 (two-sample t-test, t>0.05). Analysis by the chi-square test revealed no significant differences in accuracy for our proposed model between COVID-19-Sub-Group 1 and COVID-19-Sub-Group 2 (p>0.05) or between CAP-Sub-Group 1 and CAP-Sub-Group 2 (p>0.05). These results indicate that age does not significantly influence the performance of our proposed model.
4.5. Limitations and potential future work
The current study has several limitations. First, the size of the current dataset was small. Although citation k-NN is a kind of machine learning and not as data-hungry as deep learning, overfitting may still occur owing to a small dataset size, which could limit the general applicability of the method and the DRIS. Second, only the classification of COVID-19 and CAP was examined, while the clinical types of CAP and the severity of COVID-19 were not taken into account. This is because there were only nine (3.7%) cases of viral pneumonia and three (1.2%) cases of mycoplasma pneumonia in our dataset. The numbers of patients belonging to the various clinical types of COVID-19 were also unequally distributed. Third, some healthy controls may also present with some opacities in the lung field in CT images, and these opacities may influence the classification performance. The severity of this kind of influence is unknown because no healthy control was included in our study.
The collection of datasets with more patients and a more balanced structure of the various CAP sub-types and COVID-19 severities should lead to models with more generalization ability and clinical significance. More advanced methods, such as deep convolutional generative adversary networks (DCGAN), agile CNN, and ensemble learning, may help increase the accuracy and generalization ability of models developed for COVID-19 management [57], [58], [59]. The automatic segmentation of COVID-19 lesions and prediction of prognosis are also promising research directions [60,61].
5. Conclusions
Our DR-MIL method can effectively represent the deep characteristics of COVID-19 lesions in CT images and accurately distinguish COVID-19 from CAP in a weakly supervised manner. The combination of deep learning as feature extractor and MIL as classifier make DR-MIL suitable for small datasets containing only a few hundred patients. The resulting DRIS can potentially serve as an imaging biomarker for COVID-19 diagnosis. The finding of distinct spatial distributions of lesions in COVID-19 and CAP is in line with previous studies. Citation k-NN can provide visual interpretation as a means of CBIR, which may help train inexperienced clinicians and contribute to AI-augmented COVID-19 diagnosis.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Nos. 82072008, 81671773 and 61672146), the Fundamental Research Funds for the Central Universities (N2124006-3), and the Key R&D Program Guidance Projects in Liaoning Province (2019JH8/10300051).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cmpb.2021.106406.
Appendix. Supplementary materials
References
- 1.https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 2.Akl E.A., Blažić I., Yaacoub S., Frija G., Chou R., Appiah J.A., et al. Use of chest imaging in the diagnosis and management of COVID-19: a WHO rapid advice guide. Radiology. 2021;298(2):E63–E69. doi: 10.1148/radiol.2020203173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ai T., Yang Z., Hou H., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dangis A., Gieraerts C., Bruecker Y.D., et al. Accuracy and reproducibility of low-dose submillisievert chest CT for the diagnosis of COVID-19. Radiology. 2020;2(2) doi: 10.1148/ryct.2020200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majidi H., Niksolat F. Chest CT in patients suspected of COVID-19 infection: a reliable alternative for RT-PCR. Am. J. Emerg. Med. 2020;38:2730–2732. doi: 10.1016/j.ajem.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung M., Bernheim A., Mei X., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y., Gao Z.H., Liu Y.L., et al. Clinical and CT imaging features of 2019 novel coronavirus disease (COVID-19) J. Infect. 2020;81(1):147–178. doi: 10.1016/j.jinf.2020.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caruso D., Zerunian M., Polici M., et al. Chest CT features of COVID-19 in Rome, Italy. Radiology. 2020;296(2):E79–E85. doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai H.X., Hsieh B., Xiong Z., et al. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology. 2020;296(2):E46–E54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prokop M., van Everdingen W., van Rees Vellinga T., van Ufford H.Q., Stöger L., Beenen L., et al. CO-RADS: a categorical CT assessment scheme for patients suspected of having COVID-19 - definition and evaluation. Radiology. 2020;296(2):E97–E104. doi: 10.1148/radiol.2020201473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson S., Kay F.U., Abbara S., Bhalla S., Chung J.H., Chung M., et al. Radiological society of North America expert consensus statement on reporting chest CT findings related to COVID-19. Endorsed by the society of thoracic radiology, the American college of radiology, and RSNA-secondary publication. J. Thorac. Imaging. 2020;35:219–227. doi: 10.1097/RTI.0000000000000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues J.C.L., Hare S.S., Edey A., Devaraj A., Jacob J., Johnstone A., et al. An update on COVID-19 for the radiologist-A British society of thoracic imaging statement. Clin Radiol. 2020;75:323–325. doi: 10.1016/j.crad.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11:4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Qin L., Xu Z., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E72. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Zha Y., Li W., et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.00775-2020. 2000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang X., Huo J., Xia L., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 17.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;(6):181. doi: 10.1016/j.cell.2020.04.045. 1423–1433.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai H.X., Wang R., Xiong Z., et al. AI Augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong D., Tang Z., Wang S., et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 20.Shi F., Wang J., Shi J., et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 21.Kang H., Xia L., Yan F., et al. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging. 2020;39(8):2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 22.Han Z., Wei B., Hong Y., et al. Accurate screening of COVID-19 using attention based deep 3D multiple instance learning. IEEE Trans. Med. Imaging. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 23.M. Ilse, J.M. Tomczak, M. Welling, Attention-based deep multiple instance learning, 2018, arXiv:1802.04712.
- 24.Kraus O.Z., Ba J.L., Frey B.J. Classifying and segmenting microscopy images with deep multiple instance learning. Bioinformatics. 2016;32(12):i52–i59. doi: 10.1093/bioinformatics/btw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu W., Lou Q., Vang Y.S., et al. MICCAI. 2017. Deep multi-instance networks with sparse label assignment for whole mammogram classification; pp. 603–611. [Google Scholar]
- 26.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, NV; 2016. pp. 770–778. [DOI] [Google Scholar]
- 27.F. Chollet, Xception: deep learning with depthwise separable convolutions, 2016, arXiv:1610.02357.
- 28.Tajbakhsh N., Shin J.Y., Gurudu S.R., et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 29.Christian S., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 30.Gao H., Liu Z., Van Der Maaten L., Weinberger K.Q. Densely connected convolutional networks. CVPR. 2017;1(2):3. [Google Scholar]
- 31.Christian S., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., et al. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 32.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. IEEE/CVF Conference on Computer Vision and Pattern Recognition. IEEE; 2018. MobileNetV2: inverted residuals and linear bottlenecks; pp. 4510–4520. [Google Scholar]
- 33.X. Zhang, X. Zhou, M. Lin, J. Sun. ShuffleNet: an extremely efficient convolutional neural network for mobile devices. arXiv preprint arXiv:1707.01083v2 (2017).
- 34.S. Karen, A. Zisserman. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014).
- 35.Alex K., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012:1097–1105. [Google Scholar]
- 36.Wang J., Zucker J.D. Proceedings of the International Conference on Machine Learning. 2000. Solving multiple-instance problem: a lazy learning approach; pp. 1119–1126. [Google Scholar]
- 37.Selvaraju R.R., et al. 2017 IEEE International Conference on Computer Vision (ICCV) Vol. 1. 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 38.S. Chen, K. Ma, Y. Zheng. Med3d: transfer learning for 3D medical image analysis. arXiv preprint arXiv:1904.00625, 2019.
- 39.Salehi S., Abedi A., Balakrishnan S., et al. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am. J. Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y.C., Luo H., Liu S., et al. Dynamic evolution of COVID-19 on chest computed tomography: experience from Jiangsu Province of China. Eur. Radiol. 2020;30:6194–6203. doi: 10.1007/s00330-020-06976-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao W., Zhong Z., Xie X., et al. CT scans of patients with 2019 novel coronavirus (COVID-19) pneumonia. Theranostics. 2020;10(10):4606. doi: 10.7150/thno.45016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun. 2020;11(1):5088. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di D., Shi F., Yan F., Xia L., Mo Z., Ding Z., et al. Hypergraph learning for identification of COVID-19 with CT imaging. Med. Image Anal. 2021;68 doi: 10.1016/j.media.2020.101910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javaheri T., Homayounfar M., Amoozgar Z., Reiazi R., Homayounieh F., Abbas E., et al. CovidCTNet: an open-source deep learning approach to diagnose covid-19 using small cohort of CT images. npj Digit. Med. 2021;4:29. doi: 10.1038/s41746-021-00399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ardakani A.A., Acharya U.R., Habibollahi S., et al. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur. Radiol. 2021;31:121–130. doi: 10.1007/s00330-020-07087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheplygina V., Pena I.P., Pedersen J.H., et al. Transfer learning for multicenter classification of chronic obstructive pulmonary disease. IEEE J. Biomed. Health Inform. 2017;22(5):1486–1496. doi: 10.1109/JBHI.2017.2769800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu C., Qi S., Feng J., et al. DCT-MIL: deep CNN transferred multiple instance learning for COPD identification using CT images. Phys. Med. Biol. 2020;65 doi: 10.1088/1361-6560/ab857d. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Yan Y., Tang P., et al. Revisiting multiple instance neural networks. Pattern Recognit. 2018;74:15–24. [Google Scholar]
- 49.Kölle K., Biester T., Christiansen S., et al. Pattern recognition reveals characteristic postprandial glucose changes: non-individualized meal detection in diabetes mellitus type 1. IEEE J. Biomed. Health Inform. 2019;24(2):594–602. doi: 10.1109/JBHI.2019.2908897. [DOI] [PubMed] [Google Scholar]
- 50.Ouyang A., Liu Y., Pei S., et al. A hybrid improved kernel LDA and PNN algorithm for efficient face recognition. Neurocomputing. 2020;393:214–222. [Google Scholar]
- 51.Nayak D.R., Dash R., Majhi B. An improved pathological brain detection system based on two-dimensional PCA and evolutionary extreme learning machine. J. Med. Syst. 2018;42:19. doi: 10.1007/s10916-017-0867-4. Jan. [DOI] [PubMed] [Google Scholar]
- 52.Jeng M.J., Sharma M., Sharma L., et al. Raman spectroscopy analysis for optical diagnosis of oral cancer detection. J. Clin. Med. 2019;8(9):1313. doi: 10.3390/jcm8091313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y.Y. Multiple-instance learning based decision neural networks for image retrieval and classification. Neurocomputing. 2016;171:826–836. [Google Scholar]
- 54.Ma L., Liu X., Gao Y., et al. A new method of content based medical image retrieval and its applications to CT imaging sign retrieval. J. Biomed. Inform. 2017;66:148–158. doi: 10.1016/j.jbi.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Lambin P., Leijenaar R.T.H., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 56.Choe J., Lee S.M., Do K.H., Lee G., Lee J.G., Lee S.M., Seo J.B. Deep learning–based image conversion of CT reconstruction kernels improves for pulmonary nodules or masses. Radiology. 2019;92(2):365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
- 57.Monkam P., Qi S., Ma H., Gao W., Yao Y., Qian W. Detection and classification of pulmonary nodules using convolutional neural networks: a survey. IEEE Access. 2019;7:78075–78091. [Google Scholar]
- 58.Zhao X., Liu L., Qi S., Teng Y., Li J., Qian W. Agile convolutional neural network for pulmonary nodule classification using CT images. Int. J. Comput. Assist. Radiol. Surg. 2018;43(4):585–595. doi: 10.1007/s11548-017-1696-0. [DOI] [PubMed] [Google Scholar]
- 59.Zhang B., Qi S., Monkam P., Li C., Yang F., Yao Y., Qian W. Ensemble learners of multiple deep CNNs for pulmonary nodules classification using CT images. IEEE Access. 2019;7:110358–110371. [Google Scholar]
- 60.Fan D., Zhou T., Ji G., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 61.Wu Q., Wang S., Li L., Wu Q., Qian W., Hu Y., Li L., Zhou X., Ma H., Li H., Wang M., Qiu X., Zha Y., Tian J. Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19. Theranostics. 2020;10(6):7231–7244. doi: 10.7150/thno.46428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.