Abstract
Bats have a primeval evolutionary origin and have adopted various survival methods. They have played a central role in the emergence of various viral diseases. The sustenance of a plethora of virus species inside them has been an earnest area of study. This review explains how the evolution of viruses in bats has been linked to their metabolic pathways, flight abilities, reproductive abilities and colonization behaviors. The utilization of host immune response by DNA and RNA viruses is a commencement of the understanding of differences in the impact of viral infection in bats from other mammals. Rabies virus and other lyssa viruses have had long documented history as bat viruses. While many others like Ebola virus, Nipah virus, Hantavirus, SARS-CoV, MERS-CoV and other new emerging viruses like Sosuga virus, Menangle and Tioman virus are now being studied extensively for their transmission in new hosts. The ongoing pandemic SARS-CoV-2 virus has also been implicated to be originated from bats. Certain factors have been linked to spillover events while the scope of entitlement of other conditions in the spread of diseases from bats still exists. However, certain physiological and ecological parameters have been linked to specific transmission patterns, and more definite proofs are awaited for establishing these connections.
Keywords: Bat, Emerging viruses, Nipah virus, Rabies virus, Sosuga virus, Menangle virus, Tioman virus, Ebola virus, Hanta virus, Corona viruses
Introduction
Bats are diverse, abundant mammalian species that have co-evolved with humans and serve as a reservoir to numerous infectious agents, remarkably viruses. Bats act as a reservoir for sundry viral agents, ranging from flaviviruses and rabies to novel coronaviruses (CoVs) [1, 2]. Bats are the second most gargantuan mammalian species, constituting 20% of mammals on earth, second only to rodents, with more than 1400 species [3]. Bats are momentous since they act as reservoirs for emerging and re-emerging viruses, in addition to their role in pollination, restraining the population of insects and act as an ancillary for reseeding of forests. Moreover, guano of bats can be utilized as fertilizer, ingredient in soaps and antibiotics [4–6]. In bats, more than 130 types of viruses have been recognized, including around 60 types of zoonotic viruses which are extremely pathogenic in humans [7], for example, SARS-like coronavirus (SL-CoV), Ebola virus, Nipah virus, and Hendra virus [8–11] (Fig. 1). The recent outbreaks of novel viruses have fascinated enormous inquisitiveness to delineate the exclusivity of bats in harboring these high-profile viruses, which contribute to a high rate of mortality owing to the snowballing spillage events. It appears to be likely that bats may act as natural hosts, which play a critical role in viral inter-host transmission. Furthermore, the innovations in molecular and phylogenetic techniques provide an opportunity to delimit the evolutionary dynamics of bats, pertaining to the origin of new viral diseases. Study on the habitat of bat-associated viruses can help in preventing the new emerging viruses and re-emerging zoonosis. Here, we explain the viral spectrum found in different bat species and the ecological and genetic characteristics of the few novel bat viruses.
Fig. 1.
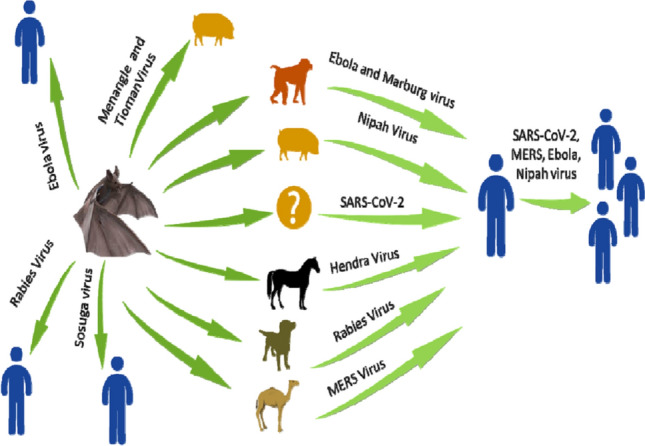
The emergence of superfluous viruses harbored by bats and the generalized route of transmission to humans
Evolution of bats
Bats are known for more than 52.5 million years ago and have a primeval evolutionary origin, when they commenced attaining unique evolutionary determinants for their survival. They are inimitable in enduring all the climatic conditions and are contemporaneous across the globe except Antarctica. Being mammals, they are distinctive in having the ability of flight, providing those with an opportunity to migrate to far-off places, which escalates the ventures of inter and intra-species transmission globally [12]. Bats have an ability to hang upside down owing to the presence of single valves in arteries to prevent the backflow and due to the unique attachment of tendons directly to upper body, without utilizing muscle as a connector between the two. Bats belong to the order Chiroptera, meaning ‘hand-wings’ in Greek [13] and based on phylogenetic and molecular basis have two sub-divisions, Yinpterochiroptera and Yangochiroptera. At the same time, Yangochiroptera that includes microbats is stated as Microchiroptera, with 16 families comprising 135 genera and 759 species [14]. The annexation of enormous species in this order makes it a hefty breeding ground for spillover and dispersion of novel viruses.
Bats have miscellany feeding habits, varying from blood, insects to fish, fruits and pollens. However, due to dearth of the fossil records of bats, it has been hypothesized that they emerged later but evolved precipitously in the cretaceous era, diversifying the population dynamics of bats [15, 16]. The evolution of bats can be ascribed to ‘survival of the fittest theory’ since it was following the fifth mass extinction. During this K-T extinction bats started to evolve that occurs 65 million years ago. The K-T extinction was a consequence of a collision of huge bolide that created a crater in Yucatan in Mexico subsequently leading to extinction of 70% of vivacity worldwide, including 50% of marine life at genus level and 70–80% at the species level [17, 18]. This annihilation affected all forms of life, from viruses to mammals as a consequence of thermal pulse released by ballistic eruption that paved the path for future evolutionary instances.
Consequently, birds and animals coped up with this thermal change by hiding in burrows, soil, sand etc. While bats being the small mammals, adopted distinctive survival strategies for their persistence in the environment. They act as the perseverance of infectious agents without overt infectious complications, ability to transmit the infectious agent both horizontally and vertically, with a broad host range and lesser impact on population size and survival of the host [19]. The inimitable physiognomies progressively acquired by bats embrace the flight proficiency, high metabolic rate, and prolonged longevity, despite being flying mammals with high metabolic rate. The most elite trait adopted by bats is ‘echolocation’ that enables them to wander in the dark [20, 21]. Moreover, aerosols generated during echolocation alongside ultrasonic waves facilitate the transmission of virus between bats. The co-existence of viruses with bats invoked inquisitiveness to outline a symbiotic relationship between the two, enhancing the extant viral biodiversity. In contrast to viral diversity of 10–50% in fecal samples of bats, the viral diversity is lesser in the most abundant mammalian genera of rodents [22]. However, the prevalence of hantavirus and arenavirus outnumber in rodents than bats, thereby highlighting the dearth of existing information pertaining to viral reservoirs, their pathogenesis and transmission dynamics.
Co-evolution of viruses and bats
A total of 66 genera from 15 families of RNA with 5 families of DNA viruses have been isolated from 75 bat genera [23]. After the K-T evolution, the viral diversity was ostentatious owing to the non-availability of host to maintain a minimum threshold required for sustaining the transmission of viruses. The co-existence of viruses with diverse bat species is remarkable, as viruses constitute the treasured earthbound biotic communes. RNA viruses are more commonly associated with bats than DNA viruses owing to a higher error-prone rate of enzymes, subsequently, leading to higher antigenic variability and the surfs of pandemics. RNA viruses have an effective role in blocking innate arm of defense, modulation of signaling and disturbances in normal metabolic pathways [24]. The transmission of viruses from bats to humans is credited to the disturbance in taciturn interaction of viral proteins with host receptors, in addition to dampened immune response. Toward the start of the SARS epidemic, practically all the recorded patients before developing infection had animal exposure. After the identification of causal agent of SARS, SARS-CoV or its antibodies were detected in palm civets and people handling animals [25–27]. However, later investigations revealed that SARS-CoV found in palm civets were transmitted from other animals [26, 27]. Later in 2005, two groups separately discovered that novel coronavirus is related to human SARS-CoV in horseshoe bats [8, 28]. These researches suggested that civets were intermediate hosts for SARS-CoV and bats may be the natural hosts. Later numerous CoVs were discovered in bats from China, Europe, Africa and Southeast Asia, which were phylogenetically related to SARS-CoVs [8, 28, 29].
Bats are ground to many emerging viral illnesses like rabies, Ebola, Hendra, Nipah, SARS-CoV, SARS CoV-2, Influenza and Marburg virus [13]. There have been shreds of evidence of other vertebrate viruses like Hepatitis B virus [30]. Bat’s harbor most of the virus species than any other mammalian host species, which can be linked to their ecological and phylogeny richness. The sharing of mammalian cell receptors with evolution would make them harbor viruses that can be transmitted to mammals including humans [7, 31]. There are approximately 70 virus families found associated with bats, which include 5 DNA virus families, 2 dsRNA families, 2 reverse transcribing virus families, 4 ssDNA families, 7 positive ssRNA families, 9 negative ssRNA families. Microcheroptera hosts the majority of viruses (8527) in the database [32]. The transmission from bats to humans can be either direct or indirect. Direct transmission to humans is after either bat bites or exposure to aerosols during cave trips. Another implicated risk factor is ingestion of bat meat, soup or other delicacies made from the bat. Bats come in contact with humans due to various push and pull factors. A ‘push’ refers to demand for increased space and resources due to overpopulation, which destroys bat habitats and leads to shortage of food [33]. Natural environmental changes, such as typhoons, droughts, forest fires can also place stresses on bats. A ‘pull’ involves the living environments built by humans, due to increased urbanization, intensive agriculture and food animal breeding; these attract bats and bring them close to human habitat. Since fruit bats cannot consume large feed in a single instance, they chew part of fruit to devour the desired sugar and discard the rest of the fruit, which if eaten by animals and humans, can transmit the infection. The spillover leading to emergence of zoonotic infections in new populations requires coordination of certain ecological factors [34]. The episodic shedding due to nutritional deficient times in bats and in epochs of pregnancy and lactation, in proximity with protracted environmental load of virus and intersection of host might augment for spillover [35, 36]. The spillover is often through an indirect recipient. The indirect transmission is via an intermediate host like pigs, civets, camels or pangolins. Only those animals can serve as intermediate host, which are in close proximity to humans and can amplify the virus for efficient transmission [33, 35].
Rabies
Rabies is an ancient disease that was discovered 400 years ago, and later, bats were identified as one of the potential reservoirs of the disease. Three varied species of bats have been implicated in the causation, namely Desmodus rotundus, Diphylla ecaudata (hairy-legged vampire bat) and Diaemus youngi (white-winged vampire bat). Of these, vampire bat is the main reservoir of rabies, implicated in human transmission. Another astounding factor noted in the transmission of rabies is the high prevalence of virus in colonial genera rather than the solitary genera [37]. Lately, it has been found that bats were incriminated in 90% of the cases in United States. However, the number is understated, as most of the cases could not recall the history of bat bites [38]. There are reports of difference in clinical characteristics of bat acquired lyssavirus infections in humans with average incubation period of 51 days while in carnivores acquired infection it was 65 days, even clinical period was found shorter in bat lyssa viruses [39]. Studies from India have revealed that the rabies virus strains circulating in India are mainly of canine origin. However, bats are increasingly being implicated as causative agent of rabies in some parts of the country [39, 40]. Bats have been found to be seropositive (5.1%) for lyssavirus in Nagaland. However, the virus couldn’t be detected from their brain tissues by RT-PCR and immunofluorescence [40].
Other lyssa viruses
Other lyssaviruses transmitted by bats include Irkut virus (IRKV), Shimoni bat virus (SHIBV), Duvenhage virus (DUVV), West Caucasian bat virus (WCBV), European bat lyssavirus 1 (EBLV-1), European bat lyssavirus 2 (EBLV-2) and Australian bat lyssavirus (ABLV) [41–44]. Of these viruses, WCBV is the most divergent species of lyssavirus, which has been isolated from Southeast Europe. The bat species implicated in the same involves Miniopterus schreibersii. Astoundingly, antivirals were unable to protect the mice leading to death of the mice [45]. IRKV has been reported after bite from bats in Russia and subsequently, cases were reported from China and Ukraine. The same virus has been isolated from Murina leucogaster in China, underlining the bat-to-human transmission [46]. Virus, when introduced into mice, could establish same symptomatology, and no symptoms were noted when the mice was given pre-exposure prophylaxis. Analogously, DUVV was first isolated from a Dutch patient after bat bite (Miniopterus) in Kenya. Inspite of the medical assistance, the patient died. SHIBV is known to transmit via Hipposideros commersoni, which when introduced into mice eventually led to fatality, even after administration of antivirals [47, 48]. ABLV is the species of lyssavirus that has been isolated from the ‘rabies-free Australia’ in 1996 [49] and subsequently, all the bats in Australia were found to harbor the virus. The species of bats implicated in the same included Saccolaimus flaviventris [50, 51]. The co-existence of varied lyssaviruses in diverse bat species connotes the specificity of virus tropism to certain receptors, minutiae of which are still cryptic [13, 52].
Hendra virus
Hendra virus was first isolated from Hendra, Australia in 1994, following an outbreak of acute respiratory illness in humans and animals [53]. The bat species implicated in illness embrace fruit bats (flying foxes) of the genus Pteropus, including black flying fox, P. alecto; little red flying fox, P. scapulatus; gray headed flying fox, Pteropus poliocephalus; and spectacled flying fox, P. conspicillatus. Of these diverse species, P. scapulatus is the chief incriminated species involve in interspecies transmission [13]. Multiple spillover events from bats form the cornerstone in transmission dynamics of virus, abetting in the intersection of interspecies interfaces. Similarly, Nipah virus has caused encephalitis outbreaks in Malaysia, Bangladesh and India [54–56]. Pteropus bat species is associated with such spillover events that facilitate the transmission. In the Malaysian outbreak, contact with pigs has been implicated as the immediate event initiating the outbreak. Whereas in the Bangladesh outbreak, the human-to-human transmission was the keystone of the event, with no association with pigs [54, 57, 58]. A study conducted in Haryana; North India detected 48% of bats to be seropositive for henipavirus. The bats were captured using mist nets, and blood was drawn from cephalic or brachial artery [59]. Recently, bats were incriminated in an outbreak in Kerela, India, without involvement of any intermediate host. The fruit bat (Pteropus medius; n = 52) and Leschenault’s rousette (Rousettus leschenaulti; n = 12) from an area near the index case were euthanized; throat and rectal swabs were collected and tested for Nipah virus. The euthanized animals were transported to ICMR-NIV in a liquid nitrogen transport chamber and different organs were dissected like liver, spleen, lung, kidney, brain and intestines. The virus was detected in 25% of throat and rectal swabs and the same could be retrieved from liver, spleen or both organs. A high level of homology, 99.7–100% was reported between Pteropus spp and human NiV sequences of N gene, whereas the homology decreased (85.14–96.15%) when compared to strains from Bangladesh, Malaysia and Cambodia [60]. The ephrin-B2/B3 receptors in Henipavirus at as attachment for virus entry in the host by binding their glycoprotein G [61].
Other viruses
Meanangle virus was discovered during the outbreak investigation of stillborn piglets in Australia in 1997 [62]. The litters were characterized by autolysis, mummification and stillbirths, with few of them exhibiting the teratogenic effects in the form of brachygnathia, kyphosis and arthrogryposis. The studies have revealed tissue tropism of virus for lymphoid organs in animals and human gastrointestinal epithelium. Though bats were not directly implicated in causation, research has divulged the presence of neutralizing antibodies in little red flying foxes [62, 63]. Tioman virus is distinct from Meanangle virus and has been isolated from Pteropus bats in Australia [64]. The same was noted in Malaysia and Singapore during an outbreak investigation of suspected Nipah virus. Tioman virus was recently characterized from North-eastern region of India in 2016, from the tissue of Pteropus giganteus by RT-PCR and a homology of 99.87% and 99.99% was found with TioV strains of Malaysia [65]. Tioman virus has also been found to infect the lymphoid tissue of pigs and due to its closeness with other paramyxoviruses affecting humans the study indicated pigs as amplifying host which could lead to future outbreaks of disease in pig farmers [66].
Nipah virus
Nipah virus (NiV) is an emerging virus having zoonotic potential with a wide distribution of reservoir bat species. The World Health Organization considered as a global health priority that may have pandemic potential because of human-to-human transfer, the high case fatality rate in humans, and unavailability of vaccine or therapeutic agents [67]. The NiV or related viruses have been detected by serological surveillance or PCR in different bat species, but NiV viruses have not been successfully isolated. The NiV has been reported in Pteropus hypomelanus bats [68, 69] and P. vapmyrus bats in Malaysia [10] and P. lylei bats in Cambodia [70]. An outbreak in Malaysia was reported between September 1998 and April 1999 in pigs and humans. The outbreak infects 265 people and causing 105 deaths and the slaughter of around 1 million pigs. Initial reports, suggested Japanese encephalitis virus as the cause of the outbreak, but later culturing in Vero cells and electron microscopy confirmed the Nipah virus [71]. The virus is transmitted from bats to pigs and pigs to humans. Bangladesh reported NiV outbreak in 2001, and India reported two outbreaks in West Bengal in 2007 and Kerala during May 2018. These infections in Bangladesh and India were found in the bat reservoir in P. medius [72, 73]. Chua et al. [68] reported that wildlife species showed neutralizing antibodies against NiV during the outbreak in P. hypomelanus. They reported the syncytial cytopathic effect in Vero cells in urine and swabs of eaten fruits of Island flying foxes.
Japanese encephalitis virus
Japanese encephalitis virus (JEV) is a zoonotic arbovirus of the family Flaviviridae and the genus Flavivirus with a tick or mosquito-borne characteristics. The genome segment of JEV (~ 11 kb) encodes three structural proteins and seven non-structural proteins. The serologically Flavivirus group into eight species and two subtype viruses, West Nile virus (WNV); Kunjin virus (a subtype of WNV); Murray Valley encephalitis virus (MVEV); Alfuy virus (a subtype of MVEV); St. Louis encephalitis virus (SLEV); Usutu virus (USUV); Koutango virus; Yaounde virus; and Cacipacore virus [74, 75]. The JEV has been isolated from mosquitoes, pigs, birds and horses [76, 77]. The JEV has been isolated from bats since 1963; however, the role of bats in JEV epidemiology is fully defined [78, 79]. Wang et al. [80] reported the molecular and virulence characterization of two bat JEV isolates from China. In Asia, around 30,000 to 50,000 cases of Japanese encephalitis occur, and are mostly related Culex mosquitos. JEV antibodies are identified in several bat species like Hipposideros armiger, H. speoris, H. cineraceus, H. pomona, H. bicolor, Rhinolophus comutus, R. ferrumequinum, R. rouxi, Myotis macrodactylus, Vespertilio superans, and Miniopterus schreibersii [13]. The bats (R. leschenaulti) having anti-JEV antibodies are prevalent in Southern China. These bats are cave dwelling, highly gregarious and migratory, which makes them suitable for carrying, circulation and spillover of the virus. They also feed on fruits like litchi or longan shared by humans which facilitates the viral cross-infection [81].
Alpha virus
The equine encephalitis virus (EEV) is an RNA virus of the family Togaviridae and the genus Alphavirus. It comprises of Eastern Equine Encephalitis Virus, Western Equine Encephalitis Virus, Venezuelan Equine Encephalitis Virus, and Highland J virus, which are transmitted by arthropods and flies. However, reservoir animals like birds and bats have an important role in the transmission of the virus. The distribution of these viruses is limited according to the climatic conditions and dispersal is dependent on the geographical distribution of hosts, vectors and reservoirs. The bats of the genus Desmodus rotundus are considered as the most vulnerable of contracting EEV [82, 83]. However, the most common route for EEV transmission is via mosquito bites. The virus-infected equines show high fever, hyporexia, restlessness, ataxia, bruxism, and imbalance towards the environment. Humans were also found to be infected with the EEV and currently only preventing vaccines are available. In a study from Mexico, analyzed pulmonary, cardiac and hepatic tissue of bats (Carollia perspicillata, Artibeus turpis and Glossophaga soricina) and found EEV in a male species of Artibeus turpis [84]. Other studies in the departments of Sucre and Córdoba reported EEV in bat species Artibeus planirostris and Sturnira lilium. These reports suggest that fruit bats from the Caribbean region in Columbia are may play a role in the enzootic cycle of equine encephalitis virus [82].
Ebola virus
The four Ebola virus strains; Ebola Zaire virus, Ebola Reston virus, Ebola Sudan virus, Ebola Ivory Coast virus and Marburg virus are known to cause severe viral hemorrhagic fever in humans except for Reston. RESTV the only subtype reported outside African continent causes mild disease in pigs, severe disease in non-human primates with no reports of disease in human [85–88]. The bat species implicated involves little free-tailed bat (Chaerephon pumilus), Angola free tailed bat (Mopscondylurus) and Wahlberg’s epauletted fruit bat species (Epomophorus wahlbergi) [9]. Moreover, the serological evidence of the same has been noted in 4% of the bat population. The presence of Niemann-Pick type C disease/NPC1 receptor in bats determines the susceptibility of filoviruses in bats; small changes in these genes could reduce their susceptibility to Ebola virus [89]. The presence of high viral load in bats facilitates human-to-human transmission.
Marburg virus
Marburg was reported from Frankfurt, Germany and Belgrade of Yugoslavia. The disease has a high case fatality rate. It is spread either via contact with infected blood or body fluids of the infected individual or via monkeys and bats of Rousettus aegyptiacus species. Though the molecular methods had unearthed virus in bats, the same isn’t adequate to ascertain bats as the perpetrator, since the intermediate host might be involved in the cryptic transmission dynamics [90, 91]. Studies from North-eastern part of India have revealed the presence of neutralizing antibodies against varied filoviruses in humans (5.1%) and the same has been noted in R. leschenaultii (13.3%) and E. spelaea (6.2%), underscoring the spillover events from bats. The basis could be co-roosting of different species of bats and the rolling parturition pattern of bats in caves [92]. This highlights the significance of surveillance and testing of both humans and bats at high-risk interfaces.
Sosuga virus
Sosuga is a species of Paramyxovirus and was identified for the first time in a female wildlife biologist after a short expedition for collection of bats in South Sudan and Uganda in 2012. Though the reservoir has not been established yet, but Egyptian Rousette bats (Rousettus aegyptiacus) have been found to have some evidence of virus serologically [93]. The symptoms of the disease were high fever, generalized myalgia and arthralgia, sore throat, neck stiffness, and maculopapular rash. The deep sequencing and metagenomic analysis confirmed the Sosuga virus closely related to Rubula like viruses found in several species of African and Asian fruit bats [93].
Coronaviruses (SARS-CoV and MERS-CoV)
The emergence of novel coronavirus in 2002 was an upheaval, having origin in live wet markets of China, subsequently spreading across the globe, affecting ~ 8000 cases. Apart from humans, raccoon dogs and palm civets were found to harbor the virus and the Himalyan palm civets were eventually identified as the intermediate host for SARS-CoV. The reservoir for the infection was illustrated to be Rhinolophus species of Chinese horseshoe bats, which spilled over crossing the inter-species barrier, owing to the gradual adaptive mutations, making it suitable for human-to-human transmission. No immediate ancestor of SARS-CoV was found in bat population during the 15 years of research, regardless of frequent RNA recombination in coronaviruses [94, 95]. Therefore, it is highly considered that SARS-CoV recently rose through recombination of bat SARSr-CoVs in the yet to be recognized bat caves. This speculation is predictable with past information indicating that an immediate forebear of SARS-CoV developed before 2002 [96, 97]. Recombination examination additionally firmly supported the speculation that the civet SARS-CoV strain SZ3 emerged through the recombination of two WIV16 and Rf 4092 bat strains. Moreover, WIV16, the nearest relative of SARS-CoV found in bats, likely emerged through recombination of two other pervasive bat SARSr-CoV strains [98].
Another novel coronavirus was Middle East Respiratory syndrome virus that originated in Saudi Arabia in 2012. The outbreak had a high case fatality rate of 45%, and the reservoir was identified as Egyptian tomb bat, Taphozous with dromedary camels as the intermediate host [99, 100]. As the development of SARS is related to palm civets, the majority of the early MERS cases involved the dromedary camels. MERS-CoV strains from dromedary camels were identical to the strains isolated from humans [101–103]. Furthermore, MERS-CoV-specific antibodies were prevalent in camels from the Middle East, Africa and Asia [104, 105]. Besides, the MERS-CoV infection was detected in the serum of camels collected in 1983 [106], which shows that MERS-CoV was present 30 years ago in camels. Sequence analysis demonstrated that MERS-CoV species like HKU4 (Tylonycteris bat coronavirus) and HKU5 (Pipistrellus bat coronavirus) are phylogenetically related. MERSr-CoVs are found in two bat families (Vespertilionidae and Nycteridae) having 14 bat species. In any case, none of these MERSr-CoVs is an immediate ancestor of MERS-CoV, as their S proteins differ from that of MERS-CoV [104, 107]. DPP4 receptor responsible for tissue tropism of MERS-CoVs is found evidently in respiratory and intestinal epithelium of most of the frugivorous bat species. While in insectivorous bats the tropism is more limited to intestinal epithelium expressing this receptor [108].
SARS-CoV 2
The recent outbreak of SARS-CoV 2 has been associated with live, wet markets of Wuhan, China [109] and pangolins have been identified as the intermediate host with ~ 99% of homology between the sequences of SARS-CoV-2 and virus isolated from pangolins [110]. The reservoir has been illustrated to be bats, and it appears plausible that the virus originated in RaTG13 strain of coronavirus, isolated from bat caves in Yunnan province of China in 2013, through natural selection. The entry of SARS coronavirus into the host is mediated via its spike glycoprotein (amino acids residues at positions 479 and 487 of receptor-binding motif, RBM), which interacts with the host ACE-2 receptor (major hotspot regions being 31 and 353). The similar receptor has been implicated in SARS-CoV-2 (corresponding to amino acids residues at 493 and 501 of RBM). The strength of interaction between spike protein RBM and ACE2 receptors determines the propensity for human-to-human transmission, predilection being more definite if asparagine (N) and threonine (T) are present at 479 and 487 positions respectively [111].
Few changes occurred in SARS CoV was identified in Himalyan palm civets, the intermediate host in SARS. The changes in SARS CoV had lysine (K) and serine (S) at these positions, however over the years, K and S mutated to N and T respectively, allowing the virus to dock in human host. While, bat coronavirus had asparagine (N) at both these places, emphasizing the requisite of an intermediate host for transmission to humans, as they lack of heterogeneity required for direct human transfer. Since the sequence, similarity between RaTG13 and human SARS-CoV-2 strains throughout the genome is 96.2% and protein-sequence identity is approximately 98%, suggesting a need for an intermediate host. Moreover, in SARS-CoV-2 there is single variation merely at position 479, where glutamine is present in human SARS-CoV-2 and tyrosine in RaTG13, endowing the hypothesis of direct transmission from bats to humans without any intermediate host [111].
This is further substantiated by the fact that the homology between spike glycoproteins of SARS bat and human coronavirus was merely 78% and this upsurge to 98% in civets, thus favoring the transmission from civets to humans. Though a difference of 1100 nucleotides has been comprehended between bat and human SARS-CoV-2, in contrast to mere 7 nucleotide disparity noted in Himalyan palm civet and humans SARS virus [112]. This might be attributed either to lack of variation in the amino-acid encoded, or the presence of this variation in a region other than the one encoding the spike glycoprotein [111]. A study has delineated the circulation of different bat coronaviruses from eight Rousettus spp. and 21 Pteropus spp. from seven varied states of India. RdRp region was extensively studied and the phylogenetic analysis revealed distinct sub-clustering of all these isolates [113].
Bats as a preferred choice for viruses
The inquisitiveness about why bats are preferred by viruses as their abode is the scorching theme of the hour, in view of the emerging and re-emerging viruses of pandemic potential. The origin and subsequent dispersion of these novel viruses have led to snooping within researchers to unearth out the bases behind this friendship. Amongst thousands of viruses, only a few cross the human-animal interface to have an efficient human-to-human transmission. Several hypotheses have been propositioned to rationalize this propinquity between the persistence of viruses in bats without impairing the homeostasis of bats. The most putative explanations for the same incorporate the inimitable innate and adaptive immune system, along with lymphoid development affecting B and T lymphocytes, macrophages and immunoglobulins [13, 114]. The model of bats harboring such high-profile viruses originated in the 1920’s, when rabies virus was isolated from bats in South and Central America, and the bats were confirmed to harbor a wide array of RNA and DNA viruses in 1974. The hypothesis based on constitutive expression of type 1 interferon in bat cells suggested that this could lead to low level intermittent viral expression that would lead to reduced immune stimulation of weak adaptive response and poor antibody stimulation [115]. Inflammation in bats is controlled, which aids in more prolonged survival of viruses. NLRP3 inflammasome, which plays an important role in controlling RNA virus infections in mammals, was found down regulated in bats preventing downstream activation of inflammatory mediators [116, 117]. Response to DNA virus infection in bats is controlled by dampened STING response. STING is an adaptor protein involved in DNA sensing pathways; it was found to have mutation in serine residue leading to loss in its activity. As high metabolic rate during flight leads to damage in DNA and self-DNA is released into the cytoplasm so a dampened STING response could also help viruses to conserve their DNA in bats [118].
Bat cell line persistently infected with MERS-CoV shows selection of mutations in different viral genes, which could further help in adaptation in bats as well as other hosts [117]. The symbiotic relationship between bats and viruses is beneficial in view of habitat and dispersal for the virus; and for bats to enhance the immune system by cross-protection. The cross-protection is indebted to the perseverance of viruses that further inhibits other pathogens to establish disease in bats, by enhancing the immune response. Viral persistence in the bat population could be probably due to consistent introduction of naïve individuals or due to persistent infection in infected individuals [119]. Owing to the protection from predators, viruses are referred as ‘biological weapons’ for bats, which are emancipated into the target population in events of imminent threat to their existence, but remain innocuous in peace with the host otherwise [120–122]. Another hypothesis to support the co-existence of viruses with bats is owed to the low reproductive ability of the bats, with one or two offspring per year. The high-profile viruses in turn support the low reproducibility, in order to have a robust defense phenomenon in place to combat the survival threats in the alleyway of evolution [31].
Flight ability
The exceptional flight ability owned by bats is speculated to be the foremost keystones pertained to viral symbiosis. The migratory ability can be solitary as seen in silver-haired bats or colonial as noted in Mexican free-tailed bats, which further expedites the exchange of viruses among non-migratory species as well. The migration is consequential to complex interplay of multiple factors as sex-specific mating; circumvention of inbreeding and the major advantage is subsequent gene flow. Majority of species migrate owing to the unfavorable conditions, while migration in temperate bats is connoted with hibernation. The ability to fly and migrate to far places with a range of 130 mm to 2 m is due to the presence of finger-like hollow bones, without bone marrow that curtails the immune response in the absence of B cell production. The flight mechanism in bats raises the metabolic rate in body, producing a variety of metabolic by-products. These by-products in turn, generate an oxidative burst in the body that damages mitochondrial DNA and other cellular mechanisms. This high metabolic rate in turn can prove detrimental, as creatures with high metabolic rate are usually short-lived. However, bats have a long-life span, underlining the connotation of mitochondrial DNA in defying the damage of oxidative outburst [123, 124]. Moreover, the less prevalence of tumors in bats unveils the presence of such resistant mechanisms in bats, as high level of oxidative outburst promotes tumorigenesis [125]. The high metabolic rate generated in the body is 15-folds as compared to sevenfold rise in rodents and twofold rise in other flying mammals. Moreover, this high metabolic rate in bats mimics the exceptional febrile response, which helps in foresee of viral replication inside the body, subsequently, snowballing the efficiency of immune response. The “flight as fever” hypothesis suggests that increased body temperature while flight could provide a metabolic mechanism for virus survival in them without causing any disease [126]. The cost of immune response is subsidized owing to the proportionate rise in metabolic rate on encountering the pathogen as compared to the rate during flight [125, 127, 128].
Consequently, the high metabolic rate assists in facilitation and subvention of immune cost in bats. However, bats hibernate during winter leading to a decline in metabolic rate, but it has been noted that bats arouse multiple times during their hibernating period. Long hibernation periods have been linked to persistent coronavirus infection in bats [129]. The torpor has also been associated with inhibition in viral replication and has inverse correlation with viral richness, but this needs to be extensively studied [78, 130]. Few studies have revealed the role of diversified receptors that facilitate this multitude of viruses in bats, but very limited research has been accomplished in this aspect.
Conclusion and future prospects
Bats serve as the abode to many emerging viruses owing to the unique characteristics attained during evolution. The recent rise in emergence of novel viral species is attributed to significant changes in the behavioral patterns and habitats of bats that arouse stress due to encroachment of their habitat by humans, which further leads to altered foraging and behavior patterns, leading to viral niche expansion and closer proximity to humans. The temptation to speculate the reasons have led to the most accepted hypothesis of tendency to flight with migratory capacity and high metabolic rate as the mainly incriminated dynamic elements, which acts as the intensive selective force driving the evolution. The available data on receptor binding, pathogenesis and genetic evolution suggest that SARS-CoV evolved in bats from bat SARSr-CoV through sequential recombination.
However, further extensive studies are anticipated to delineate the bat-ecology interaction that drives the outbreaks. Very little is known about the pathogenesis and replication of these viruses. So, future work should be planned for studying virus isolation, reverse genetics and in-vivo and in-vitro infection assays. These results would help in the prevention of SARS-like or MERS-like diseases in the future. The bat physiological studies ought to be employed to analyze the immune response in captive bats trained in a restricted environment in tunnels which are then, verified for high metabolic rates and the consequent immune responses in distinctive scenarios. Currently, there is no medication or vaccine available for any of the human coronavirus. As the RBD is conserved in SARS-CoV and bat SARSr-CoVs, some strategies like anti-RBD antibodies or RBD-based vaccines should be developed. There is an urgent need for establishing a systematic surveillance network, using the metagenomics approach, to quest for the viruses in varied wildlife to delimit the ‘virosphere’ and myriad of elements impelling the cross-species barrier. It is well established that viruses exist in their natural environment for a long time. Moreover, the spillover of the viruses from the natural environment/host to humans is due to human activities like agriculture and urbanization. Thus, the most effective procedure to prevent viral zoonosis is to maintain a barrier between human society and natural reservoirs of viruses, keeping in mind the ‘one health’ concept. The evolution of immune regulation in assisting the prolonged existence of a legion of virus species in bats needs to be studied to establish a connection of new emergences with ancestral viral species. In many of the emerging diseases host shifts occur and molecular evolutionary dynamics has shown the role of selection, viral and host genetic backgrounds in such shifts. Further, the role of selection pressure inside the reservoir host that could aid in diversification of genetic elements that could lead to outbreaks in a new population also could help to predict outbreaks.
Declarations
Conflict of interest
The author declared that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, et al. Bats worldwide carry hepatitis e virus-related viruses that form a putative novel genus within the family hepeviridae. J Virol. 2012;86(17):9134–9147. doi: 10.1128/jvi.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiwari R, Dhama K, Sharun K, Iqbal Yatoo M, Malik YS, Singh R, et al. COVID-19: animals, veterinary and zoonotic links. Vet Q. 2020;40(1):169–182. doi: 10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turmelle AS, Olival KJ. Correlates of viral richness in bats (Order Chiroptera) EcoHealth. 2009;6(4):522–539. doi: 10.1007/s10393-009-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill JE, Smith JD. Bats: a natural history. Austin: University of Texas Press; 1984. [Google Scholar]
- 5.Morrison DW. Bat Ecology. American Association for the Advancement of Science; 1983.
- 6.Bird B. Bats, mosquitoes and dollars. Am J Pub Health. 1926;16(9):929–930. doi: 10.2105/AJPH.16.9.929. [DOI] [Google Scholar]
- 7.Luis AD, Hayman DTS, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JRC, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proceed Royal Soc B: Biol Sci. 2013;280(1756):20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 9.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 10.Rahman SA, Hassan SS, Olival KJ, Mohamed M, Chang L-Y, Hassan L, et al. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats. Malaysia Emerg Infect Dis. 2010;16(12):1990–1993. doi: 10.3201/eid1612.091790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81(8):1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- 12.Kunz TH, Fenton MB. Bat ecology. USA: University of Chicago Press; 2005. [Google Scholar]
- 13.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19(3):531. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solari S, Baker RJ. Mammal Species of the world: a taxonomic and geographic reference by DE Wilson; DM Reeder. American Society of Mammalogists 810 East 10th Street, PO Box 1897, Lawrence; 2007.
- 15.Teeling EC, Springer MS, Madsen O, Bates P, Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307(5709):580. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 16.Jones KE, Purvis A, MacLarnon ANN, Bininda-Emonds ORP, Simmons NB. A phylogenetic supertree of the bats (Mammalia: Chiroptera) Biol Rev. 2002;77(2):223–259. doi: 10.1017/S1464793101005899. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez LW, Alvarez W, Asaro F, Michel HV. Extraterrestrial cause for the cretaceous-tertiary extinction. Science. 1980;208(4448):1095. doi: 10.1126/science.208.4448.1095. [DOI] [PubMed] [Google Scholar]
- 18.Sharpton VL, Brent Dalrymple G, Marín LE, Ryder G, Schuraytz BC, Urrutia-Fucugauchi J. New links between the Chicxulub impact structure and the Cretaceous/Tertiary boundary. Nature. 1992;359(6398):819–821. doi: 10.1038/359819a0. [DOI] [Google Scholar]
- 19.Robertson DS, McKenna MC, Toon OB, Hope S, Lillegraven JA. Survival in the first hours of the Cenozoic. GSA Bull. 2004;116(5–6):760–768. doi: 10.1130/B25402.1. [DOI] [Google Scholar]
- 20.Speakman J. A first for bats. Nature. 2008;451(7180):774–775. doi: 10.1038/451774a. [DOI] [PubMed] [Google Scholar]
- 21.Simmons NB, Seymour KL, Habersetzer J, Gunnell GF. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature. 2008;451(7180):818–821. doi: 10.1038/nature06549. [DOI] [PubMed] [Google Scholar]
- 22.Chu DKW, Chin AWH, Smith GJ, Chan K-H, Guan Y, Peiris JSM, et al. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J Gen Virol. 2010;91(Pt 10):2457–2462. doi: 10.1099/vir.0.022764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Openshaw JJ, Hegde S, Sazzad HMS, Khan SU, Hossain MJ, Epstein JH, et al. Bat hunting and bat-human interactions in Bangladeshi villages: implications for zoonotic disease transmission and bat conservation. Transbound Emerg Dis. 2017;64(4):1287–1293. doi: 10.1111/tbed.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. Metagenomic analysis of the viromes of three north American bat species: viral diversity among different bat species that share a common habitat. J Virol. 2010;84(24):13004. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J et al. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. bioRxiv : the preprint server for biology. 2020. 10.1101/2020.03.02.972935.
- 26.Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79(18):11892. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu C, Crameri G, Kong X, Chen J, Sun Y, Yu M, et al. Antibodies to SARS coronavirus in civets. Emerg Infect Dis. 2004;10(12):2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi H-W, Wong BHL, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102(39):14040. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balboni A, Gallina L, Palladini A, Prosperi S, Battilani M. A real-time pcr assay for bat sars-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci World J. 2012;2012:989514. doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett AJ, Bushmaker T, Cameron K, Ondzie A, Niama FR, Parra H-J, et al. Diverse RNA viruses of arthropod origin in the blood of fruit bats suggest a link between bat and arthropod viromes. Virology. 2019;528:64–72. doi: 10.1016/j.virol.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L-F, Walker PJ, Poon LLM. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol. 2011;1(6):649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Liu B, Yang J, Jin Q. DBatVir: the database of bat-associated viruses. Database. 2014 doi: 10.1093/database/bau021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han HJ, Hl W, Zhou CM, Chen FF, Luo LM, Liu JW, et al. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. doi: 10.1016/j.virusres.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu T. The socioeconomic and environmental drivers of the COVID-19 pandemic: a review. Ambio. 2021;50(4):822–833. doi: 10.1007/s13280-020-01497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, et al. Ecological dynamics of emerging bat virus spillover. Proceed Royal Soc B Biol Sci. 2015;282(1798):20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webber QMR, Brigham RM, Park AD, Gillam EH, O’Shea TJ, Willis CKR. Social network characteristics and predicted pathogen transmission in summer colonies of female big brown bats (Eptesicus fuscus) Behav Ecol Sociobiol. 2016;70(5):701–712. doi: 10.1007/s00265-016-2093-3. [DOI] [Google Scholar]
- 37.Turner DC. The vampire bat; a field study in behavior and Ecology. 1975.
- 38.Rupprecht CE, Gibbons RV. Prophylaxis against rabies. N Engl J Med. 2004;351(25):2626–2635. doi: 10.1056/NEJMcp042140. [DOI] [PubMed] [Google Scholar]
- 39.Begeman L, GeurtsvanKessel C, Finke S, Freuling CM, Koopmans M, Müller T, et al. Comparative pathogenesis of rabies in bats and carnivores, and implications for spillover to humans. Lancet Infect Dis. 2018;18(4):e147–e159. doi: 10.1016/S1473-3099(17)30574-1. [DOI] [PubMed] [Google Scholar]
- 40.Reddy RVC, Mohana Subramanian B, Surendra KSNL, Babu RPA, Rana SK, Manjari KS, et al. Rabies virus isolates of India—Simultaneous existence of two distinct evolutionary lineages. Infect Genet Evol. 2014;27:163–172. doi: 10.1016/j.meegid.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Mani RS, Dovih DP, Ashwini MA, Chattopadhyay B, Harsha PK, Garg KM, et al. Serological evidence of lyssavirus infection among Bats in Nagaland, a North-Eastern state in India. Epidemiol Infect. 2017;145(8):1635–1641. doi: 10.1017/S0950268817000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nel LH, Rupprecht CE. Emergence of Lyssaviruses in the old world: The case of Africa. In: Childs JE, Mackenzie JS, Richt JA, editors. Wildlife and emerging zoonotic diseases: The biology, circumstances and consequences of cross-species transmission. Berlin, Heidelberg; 2007. p. 161–93.
- 43.Swanepoel R, Barnard BJ, Meredith CD, Bishop GC, Brückner GK, Foggin CM, et al. Rabies in southern Africa. Onderstepoort J Vet Res. 1993;60(4):325–346. [PubMed] [Google Scholar]
- 44.Speare R, Skerratt L, Foster R, Berger L, Hooper P, Lunt R, et al. Australian bat lyssavirus infection in three fruit bats from north Queensland. Commun Dis Intell. 1997;21(9):117–120. [PubMed] [Google Scholar]
- 45.Hanlon CA, Kuzmin IV, Blanton JD, Weldon WC, Manangan JS, Rupprecht CE. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 2005;111(1):44–54. doi: 10.1016/j.virusres.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Botvinkin AD, Poleschuk EM, Kuzmin IV, Borisova TI, Gazaryan SV, Yager P, et al. Novel lyssaviruses isolated from bats in Russia. Emerg Infect Dis. 2003;9(12):1623–1625. doi: 10.3201/eid0912.030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markotter W, Kuzmin IV, Rupprecht CE, Nel LH. Lagos bat virus virulence in mice inoculated by the peripheral route. Epidemiol Infect. 2009;137(8):1155–1162. doi: 10.1017/S0950268808001945. [DOI] [PubMed] [Google Scholar]
- 48.van Thiel P-PAM, de Bie RMA, Eftimov F, Tepaske R, Zaaijer HL, van Doornum GJJ, et al. Fatal human rabies due to duvenhage virus from a bat in kenya: Failure of treatment with coma-induction, ketamine, and antiviral drugs. PLoS Negl Trop Dis. 2009;3(7):e428. doi: 10.1371/journal.pntd.0000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser GC, Hooper PT, Lunt RA, Gould AR, Gleeson LJ, Hyatt AD, et al. Encephalitis caused by a Lyssavirus in fruit bats in Australia. Emerg Infect Dis. 1996;2(4):327–331. doi: 10.3201/eid0204.960408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gould AR, Kattenbelt JA, Gumley SG, Lunt RA. Characterisation of an Australian bat lyssavirus variant isolated from an insectivorous bat. Virus Res. 2002;89(1):1–28. doi: 10.1016/S0168-1702(02)00056-4. [DOI] [PubMed] [Google Scholar]
- 51.Warrilow D, Smith IL, Harrower B, Smith GA. Sequence analysis of an isolate from a fatal human infection of Australian Bat Lyssavirus. Virology. 2002;297(1):109–119. doi: 10.1006/viro.2002.1417. [DOI] [PubMed] [Google Scholar]
- 52.Shipley R, Wright E, Selden D, Wu G, Aegerter J, Fooks AR, et al. Bats and viruses: emergence of novel lyssaviruses and association of bats with viral zoonoses in the EU. Tropical Med Infect Disease. 2019;4(1):31. doi: 10.3390/tropicalmed4010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268(5207):94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 54.Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PSK, Ksiazek TG, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. The Lancet. 1999;354(9186):1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- 55.CDC Outbreak of Hendra-like virus—Malaysia and Singapore, 1998–1999. MMWR Morb Mortal Wkly Rep. 1999;48(13):265–269. [PubMed] [Google Scholar]
- 56.CDC Update: outbreak of Nipah virus—Malaysia and Singapore, 1999. MMWR Morb Mortal Wkly Rep. 1999;48(16):335–337. [PubMed] [Google Scholar]
- 57.Chua KB, Chua BH, Wang CW. Anthropogenic deforestation, El Niño and the emergence of Nipah virus in Malaysia. Malays J Pathol. 2002;24(1):15–21. [PubMed] [Google Scholar]
- 58.Goh KJ, Tan CT, Chew NK, Tan PSK, Kamarulzaman A, Sarji SA, et al. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342(17):1229–1235. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- 59.Epstein JH, Prakash V, Smith CS, Daszak P, McLaughlin AB, Meehan G, et al. Henipavirus infection in fruit bats (Pteropus giganteus) India Emerg Infect Dis. 2008;14(8):1309–1311. doi: 10.3201/eid1408.071492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yadav PD, Shete AM, Kumar GA, Sarkale P, Sahay RR, Radhakrishnan C, et al. Nipah virus sequences from humans and bats during Nipah outbreak, Kerala, India, 2018. Emerg Infect Dis. 2019;25(5):1003–1006. doi: 10.3201/eid2505.181076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci USA. 2005;102(30):10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, et al. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg Infect Dis. 1998;4(2):269–271. doi: 10.3201/eid0402.980214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chant K, Chan R, Smith M, Dwyer DE, Kirkland P. Probable human infection with a newly described virus in the family Paramyxoviridae. The NSW Expert Group. Emerg Infect Dis. 1998;4(2):273–5. doi: 10.3201/eid0402.980215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chua KB, Wang L-F, Lam SK, Crameri G, Yu M, Wise T, et al. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology. 2001;283(2):215–229. doi: 10.1006/viro.2000.0882. [DOI] [PubMed] [Google Scholar]
- 65.Yadav P, Sarkale P, Patil D, Shete A, Kokate P, Kumar V, et al. Isolation of Tioman virus from Pteropus giganteus bat in North-East region of India. Infect Genet Evol. 2016;45:224–229. doi: 10.1016/j.meegid.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yaiw KC, Bingham J, Crameri G, Mungall B, Hyatt A, Yu M, et al. Tioman virus, a paramyxovirus of bat origin, causes mild disease in pigs and has a predilection for lymphoid tissues. J Virol. 2008;82(1):565–568. doi: 10.1128/jvi.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sweileh WM. Global research trends of World Health Organization’s top eight emerging pathogens. Glob Health. 2017;13(1):9. doi: 10.1186/s12992-017-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chua KB, Lek Koh C, Hooi PS, Wee KF, Khong JH, Chua BH, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microb Infect. 2002;4(2):145–151. doi: 10.1016/S1286-4579(01)01522-2. [DOI] [PubMed] [Google Scholar]
- 69.Singh RK, Dhama K, Chakraborty S, Tiwari R, Natesan S, Khandia R, et al. Nipah virus: epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—a comprehensive review. Vet Q. 2019;39(1):26–55. doi: 10.1080/01652176.2019.1580827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reynes J-M, Counor D, Ong S, Faure C, Seng V, Molia S, et al. Nipah virus in Lyle's flying foxes. Cambodia Emerg Infect Dis. 2005;11(7):1042–1047. doi: 10.3201/eid1107.041350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288(5470):1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 72.Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, et al. Nipah virus encephalitis reemergence. Bangladesh Emerg Infect Dis. 2004;10(12):2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yadav PD, Raut CG, Shete AM, Mishra AC, Towner JS, Nichol ST, et al. Detection of Nipah Virus RNA in Fruit Bat (Pteropus giganteus) from India. Am J Trop Med Hyg. 2012;87(3):576–578. doi: 10.4269/ajtmh.2012.11-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mackenzie JS, Barrett ADT, Deubel V. The Japanese encephalitis serological group of flaviviruses: A brief introduction to the group. In: Mackenzie JS, Barrett ADT, Deubel V, editors. Japanese encephalitis and west nile viruses. Berlin, Heidelberg: Springer; 2002. pp. 1–10. [DOI] [PubMed] [Google Scholar]
- 75.Singh RK, Dhama K, Malik YS, Ramakrishnan MA, Karthik K, Tiwari R, et al. Zika virus—emergence, evolution, pathology, diagnosis, and control: current global scenario and future perspectives—a comprehensive review. Vet Q. 2016;36(3):150–175. doi: 10.1080/01652176.2016.1188333. [DOI] [PubMed] [Google Scholar]
- 76.Solomon T, Ni H, Beasley DWC, Ekkelenkamp M, Cardosa MJ, Barrett ADT. Origin and evolution of japanese encephalitis virus in southeast Asia. J Virol. 2003;77(5):3091–3098. doi: 10.1128/jvi.77.5.3091-3098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang HY, Takasaki T, Fu SH, Sun XH, Zhang HL, Wang ZX, et al. Molecular epidemiological analysis of Japanese encephalitis virus in China. J Gen Virol. 2007;88(3):885–894. doi: 10.1099/vir.0.82185-0. [DOI] [PubMed] [Google Scholar]
- 78.Sulkin SE, Allen R. Virus infections in bats. Monogr Virol. 1974;8:1–103. doi: 10.1159/000395511. [DOI] [PubMed] [Google Scholar]
- 79.Karthikeyan A, Shanmuganathan S, Pavulraj S, Prabakar G, Pavithra S, Porteen K, et al. Japanese encephalitis, recent perspectives on virus genome, transmission, epidemiology, diagnosis and prophylactic interventions. J Exp Biol Agri Sci. 2017;5(6):730–748. [Google Scholar]
- 80.Wang J-L, Pan X-L, Zhang H-L, Fu S-H, Wang H-Y, Tang Q, et al. Japanese encephalitis viruses from bats in Yunnan. China Emerg Infect Dis. 2009;15(6):939–942. doi: 10.3201/eid1506.081525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cui J, Counor D, Shen D, Sun G, He H, Deubel V, et al. Detection of Japanese Encephalitis Virus Antibodies in Bats in Southern China. Am J Trop Med Hyg. 2008;78(6):1007–1011. doi: 10.4269/ajtmh.2008.78.1007. [DOI] [PubMed] [Google Scholar]
- 82.Guzmán C, Calderón A, Martinez C, Oviedo M, Mattar S. Eco-epidemiology of the Venezuelan equine encephalitis virus in bats of Córdoba and Sucre. Colombia Acta Trop. 2019;191:178–184. doi: 10.1016/j.actatropica.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guzmán C, Calderón A, Oviedo T, Mattar S, Castañeda J, Rodriguez V, et al. Molecular and cellular evidence of natural Venezuelan equine encephalitis virus infection in frugivorous bats in Colombia. Veter World. 2020;13(3):495–501. doi: 10.14202/vetworld.2020.495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong-Chia C, Scherer WF. Isolation of the Venezuelan encephalitis virus from a frugivorous bat (Artibeus turpis) in Mexico. Bol Oficina Sanit Panam. 1971;70(4):339–343. [PubMed] [Google Scholar]
- 85.Cantoni D, Hamlet A, Michaelis M, Wass MN, Rossman JS. Risks posed by Reston, the forgotten Ebolavirus. mSphere. 2016;1(6):e00322–16. doi: 10.1128/mSphere.00322-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peñas JA, Miranda ME, de Los Reyes VC, Sucaldito MNL, Magpantay RL. Risk assessment of Ebola Reston virus in humans in the Philippines. Western Pacific Surv Res J WPSAR. 2019;10(3):1–8. doi: 10.5365/wpsar.2017.3.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh RK, Dhama K, Malik YS, Ramakrishnan MA, Karthik K, Khandia R, et al. Ebola virus—epidemiology, diagnosis, and control: threat to humans, lessons learnt, and preparedness plans—an update on its 40 year's journey. Vet Q. 2017;37(1):98–135. doi: 10.1080/01652176.2017.1309474. [DOI] [PubMed] [Google Scholar]
- 88.Kuldeep D, Yashpal Singh M, Satya Veer Singh M, Raj KS. Ebola from emergence to epidemic: the virus and the disease, global preparedness and perspectives. J Inf Develop Count. 2015 doi: 10.3855/jidc.6197. [DOI] [PubMed] [Google Scholar]
- 89.Ng M, Ndungo E, Kaczmarek ME, Herbert AS, Binger T, Kuehne AI, et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. eLife. 2015 doi: 10.7554/eLife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monath TP. Ecology of Marburg and Ebola Viruses: Speculations and directions for future research. J Infect Dis. 1999;179(Supplement_1):S127–S38. doi: 10.1086/514281. [DOI] [PubMed] [Google Scholar]
- 91.Morvan JM, Deubel V, Gounon P, Nakouné E, Barrière P, Murri S, et al. Identification of Ebola virus sequences present as RNA or DNA in organs of terrestrial small mammals of the Central African Republic. Microb Infect. 1999;1(14):1193–1201. doi: 10.1016/S1286-4579(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 92.Dovih P, Laing ED, Chen Y, Low DHW, Ansil BR, Yang X, et al. Filovirus-reactive antibodies in humans and bats in Northeast India imply zoonotic spillover. PLoS Negl Trop Dis. 2019;13(10):e0007733. doi: 10.1371/journal.pntd.0007733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Albariño CG, Foltzer M, Towner JS, Rowe LA, Campbell S, Jaramillo CM, et al. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerg Infect Dis. 2014;20(2):211–216. doi: 10.3201/eid2002.131620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masters PS. The molecular biology of coronaviruses. Advances in Virus Research. Academic Press; 2006. p. 193–292. [DOI] [PMC free article] [PubMed]
- 95.Malik YS, Sircar S, Bhat S, Sharun K, Dhama K, Dadar M, et al. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40(1):68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao Z, Li H, Wu X, Zhong Y, Zhang K, Zhang Y-P, et al. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol Biol. 2004;4(1):21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hon C-C, Lam T-Y, Shi Z-L, Drummond AJ, Yip C-W, Zeng F, et al. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-Like coronavirus and its implications on the direct ancestor of SARS coronavirus. J Virol. 2008;82(4):1819–1826. doi: 10.1128/jvi.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu B, Zeng L-P, Yang X-L, Ge X-Y, Zhang W, Li B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11):e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eurosurveillance. 2012;17(40):20290. doi: 10.2807/ese.17.40.20290-en. [DOI] [PubMed] [Google Scholar]
- 100.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20(6):1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14(2):140–145. doi: 10.1016/s1473-3099(13)70690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sabir JSM, Lam TT-Y, Ahmed MMM, Li L, Shen Y, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016 doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 103.Chu DKW, Hui KPY, Perera RAPM, Miguel E, Niemeyer D, Zhao J, et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci. 2018;115(12):3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng H-Y, et al. MERS coronavirus in dromedary camel herd. Saudi Arabia Emerg Infect Dis. 2014;20(7):1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harcourt JL, Rudoler N, Tamin A, Leshem E, Rasis M, Giladi M, et al. The prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) antibodies in dromedary camels in Israel. Zoonoses Public Health. 2018;65(6):749–754. doi: 10.1111/zph.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Müller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg Infect Dis. 2014;20(12):2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, et al. Rooting the phylogenetic tree of middle east respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88(19):11297–11303. doi: 10.1128/jvi.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Widagdo W, Begeman L, Schipper D, Run PRv, Cunningham AA, Kley N, , et al. Tissue Distribution of the MERS-Coronavirus Receptor in Bats. Sci Rep. 2017;7(1):1193. doi: 10.1038/s41598-017-01290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.WHO. WHO Statement regarding cluster of pneumonia cases in Wuhan, China. 2020. https://www.who.int/china/news/detail/09-01-2020-who-statement-regarding-cluster-of-pneumonia-cases-in-wuhan-china. Accessed 18 Jan 2020.
- 110.News S. Coronavirus: Pangolins identified as “most likely” intermediate host. 2020. https://news.sky.com/story/coronavirus-illegal-trade-of-pangolins-could-have-helped-spread-deadly-virus-11928443. Accessed 10 Feb 2020.
- 111.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–e220. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malik YS, Sircar S, Bhat S, Vinodhkumar OR, Tiwari R, Sah R, et al. Emerging Coronavirus Disease (COVID-19), a pandemic public health emergency with animal linkages: current status update. Preprint. 2020 doi: 10.20944/preprints202003.0343.v1. [DOI] [Google Scholar]
- 113.Yadav P, Shete-Aich A, Nyayanit D, Pardeshi P, Majumdar T, Balasubramanian R, et al. Detection of coronaviruses in Pteropus & Rousettus species of bats from different states of India. Indian J Med Res. 2020;151(2):226–235. doi: 10.4103/ijmr.IJMR_795_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Davenport BJ, Willis DG, Prescott J, Farrell RM, Coons TA, Schountz T. Generation of competent bone marrow-derived antigen presenting cells from the deer mouse (Peromyscus maniculatus) BMC Immunol. 2004;5(1):23. doi: 10.1186/1471-2172-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schountz T, Baker ML, Butler J, Munster V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front Immunol. 2017 doi: 10.3389/fimmu.2017.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahn M, Anderson DE, Zhang Q, Tan CW, Lim BL, Luko K, et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol. 2019;4(5):789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Banerjee A, Subudhi S, Rapin N, Lew J, Jain R, Falzarano D, et al. Selection of viral variants during persistent infection of insectivorous bat cells with Middle East respiratory syndrome coronavirus. Sci Rep. 2020;10(1):7257. doi: 10.1038/s41598-020-64264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, et al. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23(3):297–301.e4. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Subudhi S, Rapin N, Misra V. Immune system modulation and viral persistence in bats: understanding viral spillover. Viruses. 2019;11(2):192. doi: 10.3390/v11020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roossinck MJ. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol. 2011;9(2):99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 121.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447(7142):326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 122.Fontana JM, Bankamp B, Rota PA. Inhibition of interferon induction and signaling by paramyxoviruses. Immunol Rev. 2008;225(1):46–67. doi: 10.1111/j.1600-065X.2008.00669.x. [DOI] [PubMed] [Google Scholar]
- 123.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32(4):157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 124.Indran IR, Tufo G, Pervaiz S, Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim Biophys Acta. 2011;1807(6):735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 125.Koopal S, Furuhjelm JH, Järviluoma A, Jäämaa S, Pyakurel P, Pussinen C, et al. Viral oncogene-induced DNA Damage Response Is Activated in Kaposi Sarcoma Tumorigenesis. PLoS Pathog. 2007;3(9):e140. doi: 10.1371/journal.ppat.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brook CE, Dobson AP. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23(3):172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Adelman R, Saul RL, Ames BN. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc Natl Acad Sci. 1988;85(8):2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Munshi-South J, Wilkinson GS. Bats and birds: exceptional longevity despite high metabolic rates. Ageing Res Rev. 2010;9(1):12–19. doi: 10.1016/j.arr.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 129.Subudhi S, Rapin N, Bollinger TK, Hill JE, Donaldson ME, Davy CM, et al. A persistently infecting coronavirus in hibernating Myotis lucifugus, the North American little brown bat. J Gen Virol. 2017;98(9):2297–2309. doi: 10.1099/jgv.0.000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.George DB, Webb CT, Farnsworth ML, O'Shea TJ, Bowen RA, Smith DL, et al. Host and viral ecology determine bat rabies seasonality and maintenance. Proc Natl Acad Sci. 2011;108(25):10208–10213. doi: 10.1073/pnas.1010875108. [DOI] [PMC free article] [PubMed] [Google Scholar]


