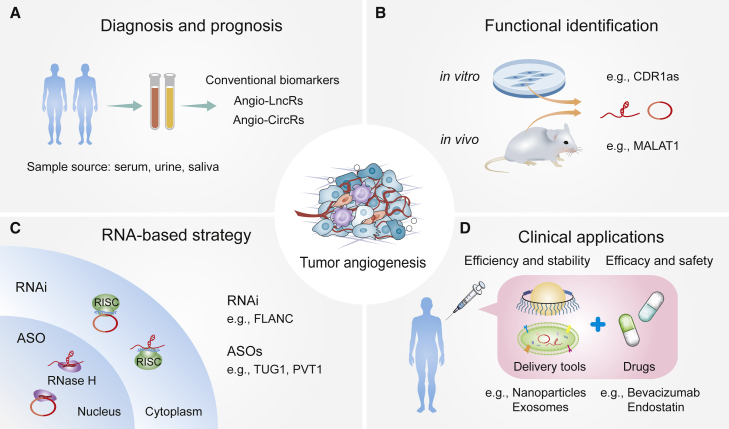
Abstract
Long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) execute a wide array of functions in physiological and pathological processes, including tumor progression. Angiogenesis, an elaborate multistep process driving new blood vessel formation, accelerates cancer progression by supplying nutrients and energy. Dysregulated lncRNAs and circRNAs can reportedly impact cancer progression by influencing angiogenesis. However, the expanding landscape of lncRNAs and circRNAs in tumor progression-dependent angiogenesis remains largely unknown. This review summarizes the major functions of angiogenic lncRNAs (Angio-LncRs) and angiogenic circRNAs (termed Angio-CircRs) and their cancer mechanisms. Moreover, we highlight the commonalities of lncRNAs and circRNAs in epigenetic, transcriptional, and post-transcriptional regulation as well as illustrate how Angio-LncRs and Angio-CircRs induce cancer onset and progression. We also discuss their potential clinical applications in diagnosis, prognosis, and anti-angiogenic therapies.
Keywords: long non-coding RNAs, circular RNAs, tumor angiogenesis, cancer progression, anti-angiogenic therapy
Graphical abstract
This review summarizes the functions and mechanisms of lncRNAs and circRNAs in tumor angiogenesis, and it discusses their potential clinical applications in diagnosis, prognosis, and anti-angiogenic therapies.
Introduction
Non-coding RNAs (ncRNAs) are transcribed from the genome but generally lack protein-coding potential. Long ncRNAs (lncRNAs) and circular RNAs (circRNAs) are two major groups of large ncRNAs that play key roles in various pathophysiological processes, especially cancers.1, 2, 3 lncRNAs are widely defined as a large and heterogeneous class of regulatory transcripts that are at least 200 nt long.4,5 circRNAs are also a subtype of endogenous ncRNAs with tissue- and cell-specific expression patterns, whose biogenesis is regulated by a particular form of alternative splicing, termed backsplicing.6 With the development of high-throughput technologies and extensive research reports, lncRNAs and circRNAs have gained wide attention for their roles in the development of many human diseases.5,7 lncRNAs and circRNAs are generally low in abundance; however, they are significantly differentially expressed in specific cell types, tissues, developmental stages, and disease states.8, 9, 10 Recently, accumulating independent studies have shown that the dysregulation of lncRNAs and circRNAs plays multifunctional roles in cancer progression, including tumor angiogenesis.11,12 However, the commonalities of their functions and mechanisms in tumor progression-dependent angiogenesis remain largely obscure.
Angiogenesis is a multistep process that involves the formation of new blood vessels from pre-existing vessels, contributing to cancer onset and progression.13 Within tumors, the angiogenic switch is activated, thereby inducing the continuous formation of new blood vessels.14 Targeting angiogenesis is an effective therapeutic strategy against cancer that has been applied in a plethora of cancers.15 Of note, diverse arrays of molecules play multifaceted roles in the mechanisms underlying tumor angiogenesis.16,17 To date, the role of ncRNAs in manipulating cancer phenotypes across various tumor types has been studied, and lncRNAs and circRNAs are emerging as part of a widespread regulatory mechanism that orchestrates gene expression in tumor angiogenesis.
Recent reviews have mainly focused on the respective roles of lncRNAs and circRNAs in angiogenesis;18, 19, 20, 21, 22 however, their molecular mechanisms and commonalities have not been deeply explored, especially in tumor angiogenesis. This review summarizes the expanding landscape of lncRNAs and circRNAs ranging from epigenetic to transcriptional to post-transcriptional regulation in tumor angiogenesis. This work is the first review on the advance of circRNAs in tumor angiogenesis and the functional commonalities of lncRNAs and circRNAs. Furthermore, we illustrate the roles of angiogenesis-related lncRNAs and circRNAs in multistep tumor development and explore their great potential in tumor angiogenesis as new biomarkers and tumor therapeutic targets.
lncRNAs and circRNAs: Functional commonalities in tumor angiogenesis
Functional commonalities between lncRNAs and circRNAs
Increasing independent research has indicated that lncRNAs and circRNAs can play a broad and diverse spectrum of pathophysiological roles, including tumor angiogenesis.23, 24, 25 Mechanistically, lncRNAs and circRNAs can directly interact with diverse partners in the form of cis or trans interactions to regulate gene expression and signal transduction.5,26,27 Based on their action mechanism, lncRNAs are generally divided into four functional archetypes: signal, decoy, guide, and scaffold.24 Interestingly, similar complex mechanisms of circRNAs have emerged in a wide variety of cell types and diseases. Moreover, both lncRNAs and circRNAs exhibit common functional features that have not been previously appreciated.
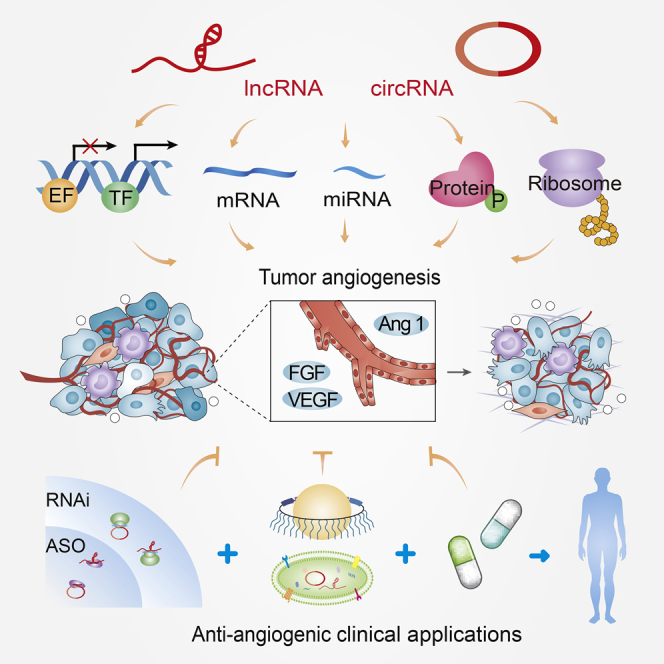
The expanding mechanisms of lncRNAs and circRNAs orchestrating gene expression can be summarized into the following aspects (Figure 1): (1) at the epigenetic layer, lncRNAs and circRNAs recruit diverse epigenetic factors to orchestrate gene transcription and signal transduction;28, 29, 30 (2) at the transcriptional level, gene transcription is regulated by lncRNAs and circRNAs via interaction with transcriptional factors and co-factors or target gene promoters;31, 32, 33, 34 (3) at the post-transcriptional level, lncRNAs and circRNAs modulate precursor (pre-)mRNAs and mRNAs of many genes by interacting with microRNAs (miRNAs), splicing factors, and diverse RNA-binding proteins (RBPs);35, 36, 37, 38, 39 (4) lncRNAs and circRNAs orchestrate a broad repertoire of RNA or protein modifications to affect their activation and stability;40, 41, 42, 43 and (5) notably, lncRNAs and circRNAs can encode functional peptides that exert crucial roles in distinct biological processes.44,45 Taken together, gene regulation by lncRNAs and circRNAs is an extensive and complex process that exists in a plethora of human diseases. The emerging commonalities of lncRNAs and circRNAs show expanding mechanisms that provide hope for new therapeutic interventions in diverse diseases. However, the detailed functions of most lncRNAs and circRNAs are still unclear and require detailed, comprehensive research in preclinical models.
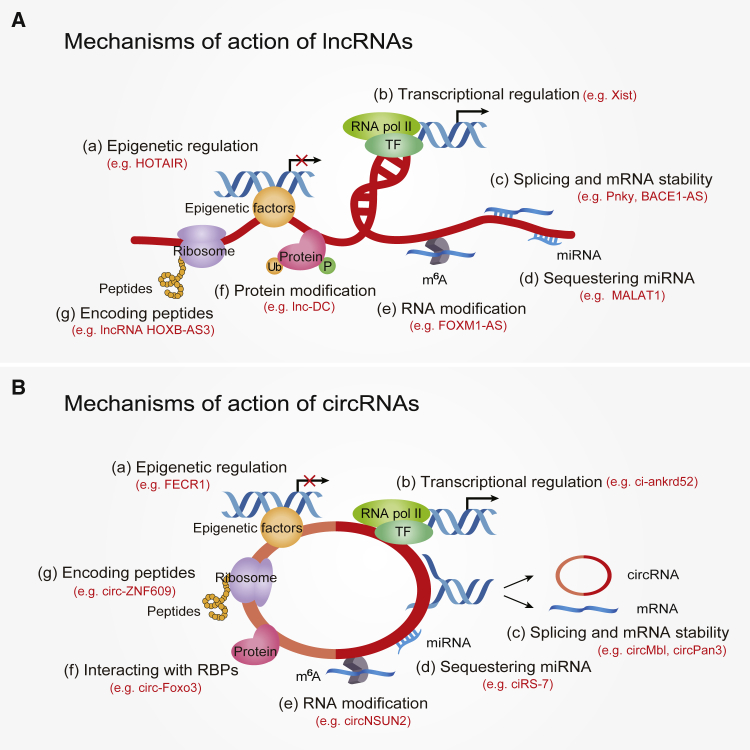
Figure 1.
The functional commonalities of lncRNAs and circRNAs
(A and B) Diverse mechanisms have been described for lncRNA regulation (A) and circRNA regulation (B), including (a) epigenetic regulation, (b) transcriptional regulation, (c) splicing and mRNA stability, (d) sequestering miRNA, (e) RNA modification, (f) protein modification or interaction with RBPs, and (g) encoding peptides.
lncRNAs and circRNAs: Novel players in tumor angiogenesis
Angiogenesis is a complicated multistep process stimulated by various pro-angiogenic factors (such as vascular endothelial growth factor [VEGF]). The vascular network’s original dynamic balance is compromised, the capillary basement membrane degrades, and endothelial cells begin to migrate and proliferate, forming new primary capillary networks. Capillaries undergo arteriovenous differentiation and remodeling in the form of sprouting and intussusception, and this primitive network expands widely to form a network of vascular systems with complex functions.13,46 Generally speaking, angiogenesis occurs in response to physiological and pathological processes, including tumor growth, inflammation, tissue regeneration, and reproduction.18 During tumor growth, a large number of new vascular systems are needed to provide adequate nutrients and remove metabolic waste. Therefore, blocking tumor blood vessel formation is equivalent to inhibiting tumor growth and progression; however, tumor-induced blood vessels often present morphological disorders characterized by precocious vascular structure, tortuosity, poor permeability, and dysfunction.47 These abnormal vascular networks create a hypoxic microenvironment within the tumor, triggering the expression of multitudinous oncogenes and inducing immunosuppression and metastasis.19 Therefore, some scholars think that anti-angiogenic therapy may facilitate the metastasis and invasion of cancer cells by activating a cancer cell hypoxic response.48, 49, 50 However, this is a controversial view and may not occur during clinical therapy.51 In essence, anti-angiogenesis has always been the focus of anticancer drug development due to the critical role of angiogenesis in tumor growth and progression.15
VEGF and its receptors are one of the well-documented signaling pathways in tumor angiogenesis.16 In most solid tumors, hypoxia is one of the key drivers of angiogenesis and induces the expression of angiogenic factors via hypoxia-inducible factors (HIFs).52 In tumor hypoxia, HIF-1α induces the expression of VEGF, which activates VEGF receptor 2 (VEGFR2), thereby stimulating tip cell migration from the arteries to initiate angiogenesis.52 Notch regulates the proliferation and migration of the tip cells and stalk cells, and Neuropilin-1 accelerates the separation of arteries and veins,53,54 thus continuously promoting the establishment of the vascular system network. Currently, lncRNAs and circRNAs have been identified as novel and versatile players involved in tumor angiogenesis via manipulating angiogenic factors. Specific well-known lncRNAs (such as H19 and MALAT1) have been identified as vital modulators of angiogenic factors.55,56 Similarly, circRNA circRhoC enhances ovarian cancer (OC) angiogenesis by regulating the expression of VEGFA.57 Increasing evidence indicates that lncRNAs and circRNAs are intricately involved in tumor angiogenesis via diverse mechanisms.
Recently, Yu and Wang18 named lncRNAs regulating angiogenesis and vascular diseases as Angio-LncRs. Herein, we have termed the circRNAs that modulate angiogenesis and vascular disease as Angio-CircRs. In reality, Angio-LncRs and Angio-CircRs play extensive regulatory roles in distinct vascular diseases, such as cancer, atherosclerosis, aneurysm, and diabetic retinopathy, among others.18,22 In the field of oncology, the study of ncRNAs has attracted great attention and made rapid progress. Reviewing the current reports of Angio-LncRs and Angio-CircRs in tumor angiogenesis could help further explore the novel potential targets of ncRNA-related antitumor strategies and provide clinical enlightenment for cancer diagnosis and treatment.
Angio-LncRs: Mechanisms of action in cancers
Current studies have identified that the dysregulation of lncRNAs contributes to tumorigenesis and tumor progression. Generally, lncRNAs exert tumor-promotive and -suppressive roles to regulate gene expression via diverse mechanisms of action. Accumulating evidence shows that lncRNA-modulated cancer progression, especially tumor angiogenesis, remains to be further explored. In the subsequent sections, we comprehensively summarize the expanding functions and mechanisms of lncRNAs in tumor angiogenesis (Table 1; Figure 2), especially inducing the expression of several key angiogenic mediators, including VEGF and VEGFR.
Table 1.
The expanding roles of Angio-LncRs in various cancers
| Mechanism/level | lncRNA | Expression | Function | Molecular mechanism | Cancer type | Refs. |
|---|---|---|---|---|---|---|
| Epigenetic regulation | PVT1 | up | pro-angiogenesis | interacts with PRC2 and inhibits transcription of ANGPTL4 | CCA | 58 |
| H19 | up | pro-angiogenesis | increases VASH1 expression in HAMSCs via binding to EZH2 | multiple tumors | 56 | |
| LINC00313 | down | anti-angiogenesis | binds PRC2 and deceases transcription of cell migration-regulating genes | Kaposi sarcoma | 59 | |
| BZRAP1-AS1 | up | pro-angiogenesis | inhibits THBS1 transcription by interacting with DNMT3b to induce THBS1 promoter methylation | HCC | 60 | |
| AK001058 | up | pro-angiogenesis | promotes the methylation level of ADAMTS12 to decrease the expression of ADAMTS12 | CRC cell lines | 61 | |
| CRNDE | up | pro-angiogenesis | regulates the mTOR signaling pathway through epigenetic mechanisms | hepatoblastoma | 62 | |
| Transcriptional regulation | RAB11B-AS1 | up | pro-angiogenesis | enhances the expression of VEGFA and ANGPTL4 by increasing the recruitment of RNA Pol II | breast cancer cell lines | 11 |
| LINC00312 | up | pro-angiogenesis | promotes the expression of VEGFA by binding to YBX1 | lung adenocarcinoma | 63 | |
| Linc00665 | up | pro-angiogenesis | interacts with YB-1 to promote transcription of ANGPT4, ANGPTL3, and VEGFA | lung adenocarcinoma | 64 | |
| PVT1 | up | pro-angiogenesis | activates transcription of VEGFA and PVT1 by interacting with STAT3 | gastric cancer | 65 | |
| CPS1-IT1 | down | anti-angiogenesis | competitively combines with BRG1 to inhibit the expression of Cyr61 and its downstream targets VEGF and MMP9 | melanoma | 66 | |
| HOTAIR | up | pro-angiogenesis | activates VEGFA transcription by directly targeting the VEGFA promoter, or upregulates GRP78 expression to mediate VEGFA and Ang2 expression | nasopharyngeal carcinoma | 67 | |
| LINC00284 | up | pro-angiogenesis | recruits NF-κB1 and downregulates MEST expression | ovarian cancer | 68 | |
| LINC00858 | up | pro-angiogenesis | upregulates HNF4α and downregulates WNK2 expression | colon cancer | 69 | |
| HNF1A-AS1 | up | pro-angiogenesis | increases the OTX1 expression via interacting with transcription factor PBX3 to activate the ERK/MAPK pathway | colon cancer | 70 | |
| Sequestering miRNAs | MALAT1 | up | pro-angiogenesis | sponges miR-126-5p to increase expression of VEGFA, SLUG, and TWIST | CRC | 71 |
| MALAT1 | up | pro-angiogenesis | increases VEGFA expression by sponging miR-140, re-directing the M2 polarization of macrophages | HCC | 72 | |
| MALAT1 | up | pro-angiogenesis | promotes VEGF expression via sponging miR-145 | breast cancer | 73 | |
| MALAT1 | up | pro-angiogenesis | competitively binds miR145-5p and elevates the NEDD9 protein expression | NSCLC | 74 | |
| TUG1 | up | pro-angiogenesis | sponges miR-143-5p to mediate HIF-1α expression | osteosarcoma | 75 | |
| TUG1 | up | pro-angiogenesis | sequesters miR-299 to induce VEGF expression | glioblastoma | 76 | |
| TUG1 | up | pro-angiogenesis | sponges miR-34a-5p and increases VEGFA expression | hepatoblastoma | 77 | |
| ZFAS1 | up | pro-angiogenesis | SP1-induced ZFAS1 upregulates VEGFA via competitively binding miR-150-5p, thereby activating the Akt/mTOR signaling pathway | CRC | 78 | |
| HULC | up | pro-angiogenesis | upregulates the TF E2F1 by sponge miR-107 to increase SPHK1 expression | HCC | 79 | |
| mRNA Stability | lncRNA-APC1 | down | anti-angiogenesis | inhibits exosome production via directly interacting with Rab5b mRNA and decreasing its stability | CRC | 80 |
| lncRNA MVIH | up | pro-angiogenesis | interacts with RPS24c mRNA to enhance the stability of each other, inhibiting the secretion of PGK1 | CRC | 81 | |
| TPT1-AS1 | up | pro-angiogenesis | interacts with NF90 and enhances VEGFA mRNA stability | CRC | 82 | |
| LINC00346 | up | pro-angiogenesis | promotes the degradation of ZNF655 mRNA, further creating an ANKHD1/LINC00346/ZNF655 feedback loop | glioma | 83 | |
| HITT | down | anti-angiogenesis | titrates away YB-1 from the 5′ UTR of HIF-1α mRNA via a high-stringency YB-1-binding motif, thereby repressing HIF-1α expression | colon cancer | 84 | |
| Protein modification | CamK-A | up | pro-angiogenesis | triggers PNCK to phosphorylate IκBα and activate NF-κB | multiple tumors | 85 |
| FLANC | up | pro-angiogenesis | increases the half-life of phosphorylated STAT3 to induce VEGFA expression | CRC | 86 | |
| lncRNA-MM2P | up | pro-angiogenesis | regulates M2 polarization of macrophages by increasing phosphorylation on STAT6 | osteosarcoma cell lines | 87 | |
| lnc-CCDST | down | anti-angiogenesis | facilitates DHX9 degradation through acting as a scaffold to promote the interplay between MDM2 and DHX9 | cervical cancer | 88 | |
| NBAT1 | down | anti-angiogenesis | interacts with Sox9 and reduces its protein stability | gastric cancer | 89 | |
| TNK2-AS1 | up | pro-angiogenesis | interacts with STAT3 and elevate VEGFA expression | NSCLC | 90 | |
| Encoding peptides | LINC00908 | down | anti-angiogenesis | encodes ASRPS, which interacts with STAT3 and diminishes STAT3 phosphorylation, inhibiting VEGF expression | TNBC | 91 |
Expression is in relation to normal tissue. Cell lines means cell samples, and the rest are human tissue samples. All lncRNAs were measured with qRT-PCR in these studies. CCA, cholangiocarcinoma; HCC, hepatocellular carcinoma; CRC, colorectal cancer; RNA Pol II, RNA polymerase II; NSCLC, non-small cell lung cancer; TNBC, triple-negative breast cancer.
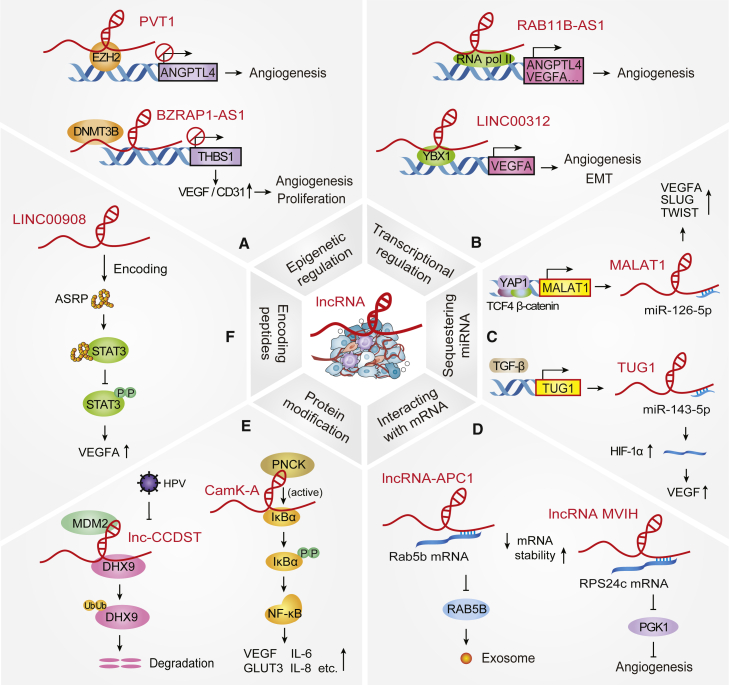
Figure 2.
Mechanisms of action of Angio-LncRs in cancer progression
(A) lncRNAs epigenetically regulate histone or DNA methylation, e.g., PVT1 and BZRAP1-AS1. (B) lncRNAs mediate transcription of angiogenesis-related genes, e.g., RAB11B-AS1 and LINC00312. (C) lncRNAs act as molecular sponges of miRNAs, e.g., MALAT1 and TUG1. (D) lncRNAs interact with mRNAs to affect the stability of mRNA, e.g., lncRNA-APC1 and lncRNA MVIH. (E) lncRNAs regulate protein modification (phosphorylation and ubiquitination), e.g., CamK-A and lnc-CCDST. (F) lncRNAs also encode bioactive peptides, e.g., LINC00908.
Angio-LncRs influence chromatin modification
Multiple lines of studies have revealed that many lncRNAs influence epigenetic changes by recruiting or interacting with chromatin remodeling complexes to specific genomic loci, thus leading to epigenetic activation or silencing of gene expression. Enhancer of zeste homolog 2 (EZH2), a catalytic subunit of the polycomb repressive complex 2 (PRC2), can catalyze the methylation of histone H3 lysine-27 for epigenetic regulation.92,93 Emerging studies have shown that numerous lncRNAs interact with PRC2 for the epigenetic regulation of tumor angiogenesis. For instance, lncRNA PVT1 is significantly elevated in cholangiocarcinoma (CCA). Mechanistically, by binding to PRC2, PVT1 could mediate the histone methylation of the promoter of angiopoietin-like 4 (ANGPTL4) via reducing the interaction between EZH2 and H3K27 trimethylation sites throughout the promoters of ANGPTL4, resulting in the promotion of cell angiogenesis, cell proliferation, migration, and apoptosis (Figure 2A).58 Similarly, H19 is closely related to a plethora of tumor types via manipulating cell proliferation, invasion, and angiogenesis. Silencing of H19 inhibited the capability of EZH2 to recruit methyl groups to the promoter region of the angiogenesis inhibitor vasohibin-1 (VASH1), thereby elevating VASH1 expression and secretion of human amniotic mesenchymal stem cells (HAMSCs) and restraining angiogenesis by interacting with EZH2.56 A novel Kaposi’s sarcoma-associated herpesvirus (KSHV) reactivation-activated lncRNA, LINC00313, suppresses endothelial cell migration and tube formation via interaction with HIV Tat. Mechanistically, LINC00313 blocks endothelial cell angiogenesis-related properties by binding to PRC2 and decreasing migration-regulating gene expression.59 In addition to the interaction with PRC2, certain lncRNAs also bind to other epigenetic factors. Another hypoxia-mediated lncRNA, GATA6-AS, is upregulated in endothelial cells under hypoxic conditions. Silencing of GATA6-AS inhibits transforming growth factor (TGF)-β-induced epithelial-to-mesenchymal transition (EMT) and tumor angiogenesis. Mechanistically, GATA6-AS interacts with the epigenetic mediator LOXL2 to regulate endothelial gene transcription by inducing histone methylation.94 Taken together, increasing evidence shows that dysregulation of lncRNAs contributes to tumor angiogenesis and tumor progression via the recruitment of chromatin-modifying enzymes and their subunits.
In addition to histone methylation, DNA methylation is an important epigenetic modification involved in the expression of many cancer-related genes.95 For example, BZRAP1-AS1 suppresses the transcription of thrombospondin-1 (THBS1) via interaction with the DNA methyltransferase 3B and induction of methylation of the THBS1 promoter; this promotes the proliferation, migration, angiogenesis, and tumor growth of hepatocellular carcinoma (HCC) (Figure 2A).60 On the contrary, lncRNA AK001058 promotes tumor growth, accounting for increased cell apoptosis and tumor angiogenesis in colorectal cancer (CRC), which partly depends on the methylation of ADAMTS12, a potential anti-oncogene located on chromosome 5, and resulting in increased expression of VEGFA and angiopoietin II.61 Additionally, various lncRNAs are also associated with chromatin-modifying complexes to manipulate tumor growth and angiogenesis in distinct signaling pathways, such as the mammalian target of rapamycin (mTOR) signaling pathway.62 Collectively, a growing number of studies have shown that lncRNAs epigenetically regulate transcription of tumor angiogenesis-related genes by either directly or indirectly affecting chromatin-modifying factors. This broadens our understanding of angiogenesis and provides a positive direction for the potential application of therapeutic intervention in regulating tumor progression at the epigenetic level.
Angio-LncRs in transcriptional regulation
Studies have shown that lncRNAs can manipulate gene transcription by binding specific transcription factors (TFs) to promoters of adjacent or distant genes, thus modulating tumor angiogenesis. For instance, in hypoxic breast cancer cells, RAB11B-AS1 is induced by HIF-2, promoting tumor angiogenesis and distant metastasis in response to hypoxia. Mechanistically, RAB11B-AS1 facilitates the transcription of VEGFA and ANGPTL4 by promoting RNA polymerase II recruitment (Figure 2B).11 The levels of LINC00312 were high in lung adenocarcinoma (LUAD) patients, which positively correlated with tumor node metastasis. LINC00312 promotes the transcription of VEGFA by directly binding to the TF Y-box binding protein 1 (YB-1, YBX1) and inducing migration and vasculogenic mimicry (VM) (Figure 2B).63 Similarly, in LUAD cells, linc00665 suppresses ubiquitination-dependent proteolysis and induces YB-1 nuclear translocation by directly binding to YB-1. The accumulated nuclear YB-1 promoted transcription of VEGFA, ANGPTL4, and ANGPTL3 by binding to their promoters, thus facilitating tumor angiogenesis.64 The lncRNA PVT1 level was elevated and markedly correlated with poor prognosis in gastric cancer (GC). Mechanistically, PVT1 directly interacted with phospho-STAT3 in the nucleus, thereby promoting VEGFA expression by activating the VEGFA promoter to stimulate tumor angiogenesis. Moreover, STAT3 could bind to the PVT1 promoter and facilitate its transcription, which established a positive feedback loop of PVT1 and STAT3 to strengthen their oncogenic effects.65 A well-known lncRNA, HOTAIR, was extremely abundant in nasopharyngeal carcinoma (NPC); it promoted tumor cell growth and angiogenesis by directly activating VEGFA transcription via targeting VEGFA promoter and upregulating the GRP78/VEGFA/Ang2 axis.67 lncRNA CPS1-IT1 acts as a tumor suppressor and suppresses EMT, cell migration, and angiogenesis in melanoma. Mechanistic studies uncovered that CPS1-IT1 impedes the transcription of the angiogenic factor cysteine-rich 61 (Cyr61) by blocking the binding of BRG1 to the Cyr61 promoter.66
In addition to VEGF/VEGFA, other angiogenic factors are mediated by different lncRNAs to induce cancer angiogenesis and development. For example, Ruan and Zhao68 identified that LINC00284 is associated with angiogenesis during OC development. Silencing LINC00284 inhibits tumor angiogenesis via recruitment of nuclear factor κB1 (NF-κB1), thereby upregulating mesoderm-specific transcript (MEST) in OC. Another example, LINC00858, is mainly located in the nucleus and exerts a promotive role in colon cancer growth. Functional assays uncovered that LINC00858 elevates the transcription of HNF4α, thereby promoting angiogenesis by the HNF4α/WNK2 axis.69 Additionally, lncRNA HNF1A-AS1 is highly expressed in colon cancer. HNF1A-AS1 promotes OTX1 transcription via interacting with the TF PBX3, thereby activating the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway in colon cancer.70 Taken together, lncRNAs may act as transcriptional activators or inhibitors, which can directly or indirectly regulate the transcription of angiogenesis-related genes, thereby affecting the progression of various malignant tumors.
Sequestering miRNAs
lncRNAs can also function as competing endogenous RNAs (ceRNAs) to compete together with miRNAs and dampen the gene expression of miRNA targets, thus participating in post-transcriptional regulation.96 This section provides an overview of the direct and indirect mechanisms of lncRNAs by sponging diverse miRNAs to affect tumor angiogenesis. For instance, MALAT1, a widely reported lncRNA, is highly expressed in a variety of tumors. Functional assays indicated that YAP1-mediated MALAT1 could sponge miR-126-5p to facilitate the expression of VEGFA, SLUG, and TWIST, which could promote CRC angiogenesis and EMT (Figure 2C).71 Similarly, MALAT1 exerts a promotive role in regulating tumorigenesis by sponging miR-140 or miR-145, thereby increasing VEGF and VEGFA expression in HCC and breast cancer.72,73 Additionally, estrogen receptor β promotes the progression of lung cancer by upregulating MALAT1 to alter miR-145-5p/NEDD9 signaling, resulting in the facilitation of VM formation and cell invasion.74 Similar to MALAT1, the lncRNA TUG1 is also significantly highly expressed in various tumors. Cancer-associated fibroblast (CAF)-derived TGF-β promotes TUG1 expression, and the interplay between CAFs and osteosarcoma (OS) cells induces TUG1 to facilitate OS angiogenesis, proliferation, and metastasis. TUG1 acts as a sponge for miR-143-5p, thereby elevating HIF-1α levels to promote VEGF expression (Figure 2C).75 Similarly, TUG1 knockdown suppresses angiogenesis and tumor growth by decreasing VEGFA expression via upregulation of miR-299 and miR-34a-5p in glioblastoma and hepatoblastoma, respectively.76,77 Several groups have shown that many lncRNAs (PVT1, DANCR, LINC01116, linc01105, and SNHG15) contribute to tumor angiogenesis and progression, upregulating VEGF/VEGFA expression by competitively binding to distinct miRNAs in a wide variety of cancers types.97, 98, 99, 100, 101
Recently, lncRNAs were reported to be involved in cellular signaling pathways such as the NF-κB, phosphatidylinositol 3-kinase (PI3K)/AKT, mTOR, and Wnt/β-catenin pathways. As relatively upstream regulatory molecules, lncRNAs act by impacting downstream molecules via competitively sponging miRNAs. For example, LINC01410 facilitates GC angiogenesis and metastasis via interaction with miR-532-5p, increasing NCF2 expression and activating the NF-κB pathway.102 RP11-79H23.3 inhibits the angiogenesis and pathogenesis of bladder cancer (BCa) by inducing the miR-107/PTEN axis and inactivating the PI3K/Akt signaling pathway.103 Similarly, PTENP1, a pseudogene of PTEN, inhibits the oncogenic PI3K/AKT pathway by sponging miR-17, miR-19b, and miR-20a, which results in eliciting pro-death autophagy and dampening angiogenesis.104 Additionally, MCM3AP-AS1 and HOXA-AS2 regulate the PI3K/AKT pathway and contribute to glioblastoma multiforme (GBM) angiogenesis by influencing miRNA targets.105,106 The SP1-induced lncRNA ZFAS1 contributes to CRC progression by elevating VEGFA levels via sequestering miR-150-5p, thereby activating the Akt/mTOR signaling pathway.78
Furthermore, lncRNAs indirectly regulate the expression of angiogenesis-related genes through miRNA-mediated TFs. For example, XIST and LINC00339 can act as ceRNAs to sponge miR-137 and miR-539-5p, elevating the expression of the miRNA targets (FOXC1, ZO-2, TWIST1) by increasing promoter activity and downstream gene expression, thereby promoting glioma angiogenesis.107,108 lncRNA HULC sequesters miR-107 via a ceRNA model, which increases the transcription of miR-107 target E2F1. As a result, the TF E2F1 binds to the SPHK1 promoter and enhances SPHK1 transcription to induce HCC angiogenesis.79 In addition, lncRNAs modulate the expression of other angiogenic factors via sequestering different miRNAs to mediate tumor angiogenesis. For example, H19, SNHG16, and MCM3AP-AS were significantly upregulated in diverse cancer types; they regulated the expression of vasohibin 2, ALDH1A1, and KPNA4, respectively, by sponging certain miRNAs to promote tumor growth and angiogenesis.109, 110, 111 Taken together, current progress indicates that lncRNAs often act as ceRNAs in tumor angiogenesis and may serve as promising targets for cancer therapy.
Angio-LncRs interact with mRNAs
Many studies report that lncRNAs can bind mRNAs to affect their stability and the translation process, thus regulating the expression and secretion of related molecules for tumor progression and tumor angiogenesis. For instance, lncRNA-APC1, as a downstream factor of adenomatous polyposis coli (APC) in CRC, suppresses CRC angiogenesis and metastasis by inhibiting exosome production via directly interacting with the Rab5b mRNA, which results in decreased Rab5b mRNA stability. This study revealed that CRC-derived exosomes regulated the lncRNA-APC1/Rab5b axis to inhibit the angiogenesis of CRC, linking APC signaling to the canonical Wnt pathway (Figure 2D).80 Another lncRNA, MVIH, activates CRC angiogenesis via interacting with the RPS24c isoform and enhances each other’s stability, thereby inhibiting the secretion of PGK1 (Figure 2D).81 lncRNA TPT1-AS1 directly binds nuclear factor 90 (NF90) and induces the interplay between NF90 and the VEGFA mRNA, thus enhancing VEGFA mRNA stability and facilitating CRC angiogenesis and metastasis.82 Moreover, LINC00346 can bind to the ZNF655 mRNA by its Alu elements, thereby promoting the degradation of the ZNF655 mRNA in a Staufen1-mediated mRNA decay manner. Furthermore, ANKHD1 targets LINC00346 and elevates its stability. ZNF655 targets the promoter region of ANKHD1 and forms a positive feedback loop that contributes to glioma angiogenesis.83
Hypoxia plays an oncogenic role in angiogenesis, metabolism, tumor invasion, and metastasis. A novel lncRNA termed HIF-1α inhibitor at translation level (HITT) is downregulated in colon cancer. Mechanistically, HITT inhibits HIF-1α translation by directly interacting with YB-1 and blocking the interplay between YB-1 and the 5′ UTR of HIF-1α. Furthermore, HITT and HIF-1α form an autoregulatory feedback loop, where HIF-1α destabilizes HITT by inducing miR-205, which directly targets HITT for degradation.84 In summary, lncRNAs play a diverse spectrum of roles by regulating mRNA stability; thus, pharmacological targeting to modulate mRNA stability could serve as an antitumor angiogenesis strategy.
Angio-LncRs induce protein modifications
Accumulating studies have revealed that multifaceted functions of lncRNAs are achieved by protein modification to manipulate the activation of proteins. The post-translational protein modifications, such as phosphorylation and ubiquitination, have been modulated by distinct ncRNAs. lncRNAs also manipulate oncogenic and tumor-suppressive protein activation and stability, thereby manipulating tumor phenotypes involved in angiogenesis. For example, the lncRNA CamK-A is highly expressed in many human cancer types and activates the Ca2+/calmodulin-dependent kinase PNCK, which phosphorylates IκBα and triggers calcium-dependent NF-κB activation. This results in the active expression of the NF-κB target genes, such as VEGF, GLUT3, IL-6, and IL-8, promoting cancer microenvironment remodeling, including macrophage recruitment, angiogenesis, and tumor progression (Figure 2E).85 Furthermore, the expression of FLANC was increased and associated with poor survival. Mechanistically, elevated FLANC prolonged the half-life of phosphorylated STAT3, promoting VEGFA transcription and inducing CRC angiogenesis. Moreover, pharmacological targeting of FLANC significantly suppressed the metastases of CRC.86 lncRNA-MM2P levels are elevated during M2 polarization but decreased in M1 macrophages. Silencing of lncRNA-MM2P significantly inhibited M2 polarization and macrophage-induced tumorigenesis and tumor angiogenesis by regulating STAT6 phosphorylation.87
In addition to modulating protein phosphorylation, lncRNAs mediate ubiquitination to regulate the activation and ubiquitination-dependent degradation of distinct proteins. As an example, lnc-CCDST is significantly downregulated in cervical cancer (CC) tissues. Mechanistically, lnc-CCDST interacts with DHX9 and enhances its degradation via the ubiquitin-proteasome pathway by recruiting the E3 ubiquitin ligase MDM2. In HPV-positive cell lines, HPV oncogenes E6 and E7 can abolish the negative effects of lnc-CCDST by increasing DHX9 expression, which facilitates CC cell motility and angiogenesis (Figure 2E).88 Another lncRNA, NBAT1, inhibits GC progression and capillary tube formation. Further analysis uncovered that NBAT1 interacted with SRY-related high-mobility-group box 9 (Sox9) and reduced its protein stability by promoting polyubiquitination and proteasome-dependent degradation. Furthermore, Sox9 acts as a TF and interacts with the NBAT1 promoter to diminish its transcription, forming a negative feedback loop of NBAT1 and Sox9.89 Similarly, TNK2-AS1 levels were markedly increased in non-small cell lung cancer (NSCLC) and promoted oncogenesis, angiogenesis, and metastasis. Mechanistically, TNK2-AS1 directly binds STAT3 and enhances its protein stability by weakening its proteasome-mediated degradation. In return, STAT3 occupies the TNK2-AS1 promoter to promote its transcription. The positive feedback loop between TNK2-AS1 and STAT3 enhances STAT3 signaling by elevating VEGFA expression.90 Taken together, lncRNAs directly or indirectly interact with key proteins through different mechanisms of action and orchestrate tumor angiogenesis by impacting protein modifications such as phosphorylation and ubiquitination.
Angio-LncRs encode functional peptides
Although lncRNAs are not widely translated by definition, emerging studies have shown that lncRNAs can encode functional peptides to regulate various biological processes, including tumor progression.26 As an example, Huang et al.112 identified that the lncRNA HOXB-AS3 encodes a conserved 53-aa peptide that suppresses aerobic glycolysis and inhibits colon cancer growth. The biological effect of lncRNA-encoded peptides has attracted considerable attention. Moreover, this novel mode of action has been found to regulate tumor angiogenesis and progression. LINC00908, which encodes a 60-aa functional polypeptide termed automatic speech recognition and processing server (ASRPS; a small regulatory peptide of STAT3), is downregulated in triple-negative breast cancer (TNBC). ASRPS directly interacts with STAT3 through the coiled-coil domain and diminishes STAT3 phosphorylation, resulting in VEGF expression inhibition. In a mouse xenograft breast cancer model, silencing of ASRPS promoted tumor growth and angiogenesis, which suggested that LINC00908 and its encoded ASRPS are potential prognostic markers and therapeutic targets for TNBC (Figure 2F).91 However, circRNAs may be evolutionarily conserved in a wide range of species compared with most peptides encoded by lncRNAs,113 in which the expression pattern of angiogenesis-mediating circRNAs plays a fundamental role in transcriptional regulation for tumor progression. To date, the functional peptides encoded by lncRNAs have been applied in cancer diagnostics, prognostics, and therapy; this makes them potential new targets for drug development.
Angio-CircRs: Mechanisms of action in cancers
circRNAs may act as multifunctional regulators to modulate gene expression and signal transduction at multidimensional levels and influence the fate of their targets. Notably, emerging studies disclosed that circRNAs influence cellular functions in tumor progression-dependent angiogenesis. These functions of Angio-CircRs are described in detail below (Table 2; Figure 3).
Table 2.
The emerging roles of Angio-CircRs in various cancers
| Mechanism/level | circRNA | Expression | Function | Molecular mechanism | Cancer type | Refs. |
|---|---|---|---|---|---|---|
| Epigenetic and transcriptional regulation | circ-CCAC1 | up | pro-angiogenesis | Increases SH3GL2 expression by sequestering EZH2 in the cytoplasm | CCA | 12 |
| Splicing and mRNA stability | circSMARCA5 | down | anti-angiogenesis | interacts with SRSF1 to regulate VEGFA mRNA Splicing | GBM | 114 |
| circPOK | up | pro-angiogenesis | promotes the ILF2/3 complex to bind and stabilize interleukin (IL)-6 and VEGF mRNA | mesenchymal tumor | 115 | |
| Interacting with RBPs | circRhoC | up | pro-angiogenesis | directly binds and modulates VEGFA expression, sponges miR-302e to regulate VEGFA expression | ovarian cancer | 57 |
| circ-GLI1 | up | pro-angiogenesis | interacts with p70S6K2 to upregulate Cyr61 via inducing activation of Hedgehog/GLI1 and Wnt/β-catenin pathways | melanoma | 116 | |
| Sequestering miRNAs | circ-ATXN1 | up | pro-angiogenesis | sponges miR-526b-3p to upregulate the expression of MMP2/VEGFA | glioma | 117 |
| circSCAF11 | up | pro-angiogenesis | sponges miR-421 to promote SP1 expression, which activates the transcription of VEGFA | glioma | 118 | |
| circRNA-MYLK | up | pro-angiogenesis | sponges miR-29a to trigger VEGFA/VEGFR2 and the downstream Ras/ERK signaling pathway | bladder carcinoma | 119 | |
| circ0001429 | up | pro-angiogenesis | sponges miR-205-5p to increase VEGFA expression | bladder cancer | 120 | |
| circCCT3 | up | pro-angiogenesis | sponges miR-613 to upregulate VEGFA and WNT3 expression | CRC | 121 | |
| circ_0056618 | up | pro-angiogenesis | sponges miR-206 to upregulate CXCR4 and VEGFA | CRC | 122 | |
| circHIPK3 | down | anti-angiogenesis | sponges miR-558 to inhibit the expression of HPSE | bladder cancer | 123 |
Expression is in relation to normal tissue. These studies used human tissue samples. All circRNAs were measured with qRT-PCR in these studies. GBM, glioblastoma multiforme; PDAC, pancreatic ductal adenocarcinoma.
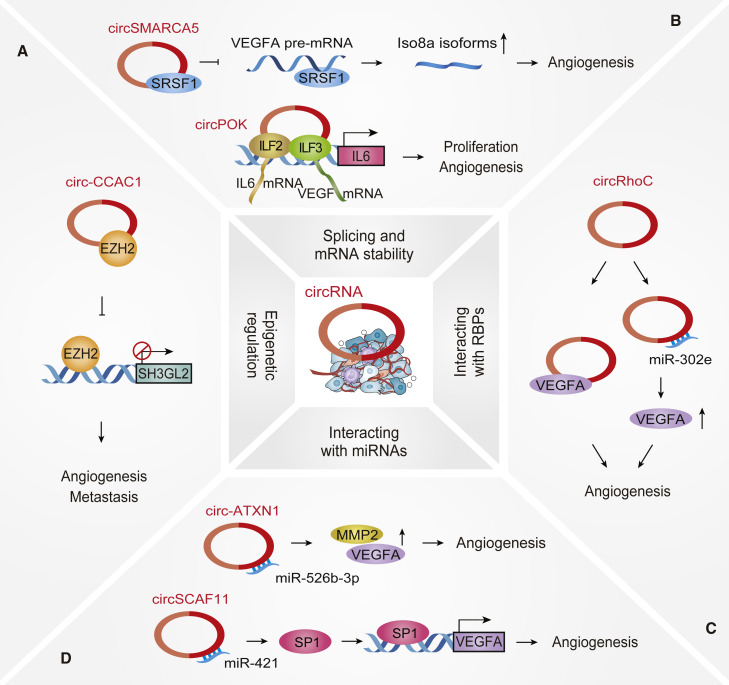
Figure 3.
Mechanisms of action of Angio-CircRs in cancer progression
(A) circRNAs epigenetically mediate transcriptional regulation, e.g., circ-CCAC1. (B) circRNAs participate in splicing regulation and mRNA stability, e.g., circSMARCA5 and circPOK. (C) circRNAs directly interact with RBPs, e.g., circRhoC. (D) circRNAs act as molecular sponges to sequester miRNAs, e.g., circ-ATXN1 and circSCAF11.
Angio-CircRs regulate gene transcription
Emerging evidence shows that circRNAs function as epigenetic transcriptional regulators to manipulate distinct gene expression. A few circRNAs recruit specific proteins to certain cellular locations; for example, the circRNA FECR1 recruits TET1 to the promoter region of its host gene FLI1, thereby demethylating the CpG sites and triggering transcription.6 Silencing of the cerebellar degeneration-related protein 1 transcript (CDR1as) promotes IGF2BP3-mediated melanoma invasion and metastasis via epigenetic silencing of LINC00632 in a miR-7-independent manner.124 Notably, CCA-associated circRNA 1 (circ-CCAC1) is elevated in cancerous bile-resident extracellular vesicles (EVs) and tissues and epigenetically accelerates CCA tumorigenesis and metastasis. Furthermore, CCA-derived EVs transfer circ-CCAC1 to endothelial monolayer cells and promote angiogenesis by disrupting endothelial barrier integrity. Functional assays uncovered that circ-CCAC1 strengthens cell leakiness by sequestering EZH2 in the cytoplasm, thus promoting SH3GL2 transcription to decrease intercellular junction protein levels. Additionally, circ-CCAC1 facilitates CCA progression via sponging miR-514a-5p to enhance YY1 translation. YY1 acts as a TF to activate CAMLG transcription by directly binding to its promoter (Figure 3A).12 Overall, current studies have shown that circRNAs influence angiogenesis by epigenetically inducing gene transcription, establishing a favorable microenvironment for tumor progression.
Angio-CircRs mediate splicing and mRNA stability
In addition to regulating gene transcription, circRNAs appear to orchestrate tumor angiogenesis by impacting post-transcriptional processes such as alternative splicing and mRNA stability. For example, circSMARCA5 regulates the VEGFA mRNA splicing by interacting with the splicing factor serine and arginine-rich splicing factor 1 (SRSF1), resulting in the production of pro-angiogenic (Iso8a) and anti-angiogenic (Iso8b) mRNA isoforms of VEGFA in GBM. SRSF1, a proximal splice site of VEGF, promotes the expression of pro-angiogenic isoforms (VEGF-Axxxa), contributing to tumor angiogenesis. This study shows that circSMARCA5 is an upstream regulator of the ratio of pro-angiogenic to anti-angiogenic VEGFA isoforms within GBM cells, which suggests that circSMARCA5 acts as a promising prognostic anti-angiogenic biomarker (Figure 3B).114 Another novel circRNA, circPOK, is encoded by the Zbtb7a gene in mesenchymal tumor progression. circPOK interacts with the ILF2/3 protein complex and enhances its ability to stabilize the VEGF mRNA, thereby inducing pro-angiogenic and pro-proliferative roles in tumor growth. circPOK binds to the promoter of Il6 as a transcriptional co-activator of ILF2/3, resulting in enhanced Il6 transcription. These findings indicate that circPOK exerts diverse functions in regulating mRNA transcription and stability (Figure 3B).115 Therefore, targeting angiogenesis-related circRNAs during cancer development and metastasis may be a promising RNA-based therapeutic approach.
Angio-CircRs interacting with RBPs
RBPs exert fundamental roles in orchestrating protein expression, RNA metabolism, and transport and localization of distinct transcripts.125 The binding of circRNAs to RBPs has a wide range of functions, such as affecting epigenetic regulation, transcriptional regulation, splicing regulation, and the formation of circRNAs themselves.34,39,114 Recent studies have shown that circRNAs directly bind to VEGFs and thus regulate tumor angiogenesis. For example, circRhoC, derived from the RhoC mRNA, was significantly highly expressed in advanced-stage OC and contributed to OC angiogenesis and metastasis. Mechanistic analysis indicated that circRhoC functioned not only as a miR-302e sponge to promote VEGFA translation but also directly bound and regulated VEGFA expression (Figure 3C).57 Additionally, circ-GLI1 interacted with p70S6K2 to induce GSK3β phosphorylation at Ser9, thereby blocking the binding of GSK3β with GLI1 and β-catenin, elevating their protein expression. This study uncovered that circ-GLI1 facilitates melanoma angiogenesis and metastasis by elevating Cyr61 via p70S6K2-dependent activation of the Hedgehog/GLI1 and Wnt/β-catenin pathways.116 In summary, the interplay between circRNAs and RBPs could be a common mechanism that is deeply involved in diverse physiological processes.
Angio-CircRs act as miRNA sponges
It is well established that circRNAs sponge diverse miRNAs, which is the most extensive mechanism to regulate tumor angiogenesis. Earlier studies identified many miRNA-binding sites on circRNAs.35,36 In tumor cells, circRNAs can also sponge miRNAs and elevate the expression of angiogenic mediators such as VEGF and VEGFA.126
A typical example is circ-ATXN1 that can facilitate cell viability, migration, and angiogenesis of glioma-exposed endothelial cells (GECs) by sponging miR-526b-3p to upregulate the expression of MMP2 and VEGFA (Figure 3D).117 Additionally, in gliomas, circSCAF11 activates VEGFA expression by targeting miR-421 to upregulate the TF SP1 (Figure 3D).118 As another example, circRNA-MYLK could sponge miR-29a in BCa, further triggering VEGFA/VEGFR2 and Ras/ERK pathways, thereby promoting EMT, angiogenesis, and metastasis.119 In addition, circRNA-MYLK can sponge miR-513a-5p and upregulate the expression of VEGFC, thereby promoting cell proliferation, metastasis, and angiogenesis in renal cell carcinoma.127 Similarly, in BCa tissues, circ0001429 can increase the expression of VEGFA to promote cell proliferation, migration, and invasion by sponging miR-205-5p.120 In CRC, the expression of circ-001971, circCCT3, and circ0056618 was significantly increased, contributing to VEGFA expression. They function as ceRNAs through the adsorption of diverse miRNAs and promote cell proliferation, migration, invasion, and angiogenesis.121,122,128
In addition to VEGF, circRNAs regulate the expression of other angiogenic regulators by modulating miRNAs to control tumor angiogenesis. A study uncovered that circ0020710 acts as a molecular sponge to adsorb miR-370-3p, promoting CXCL12 expression in melanoma. Its downstream CXCL12/CXCR4/CXCR7 axis mediates angiogenesis, tumor development, and recruitment of immunosuppressive cells.129 circPRRC2A sponges miR-6776-5p and miR-514a-5p to prevent the degradation of the TRPM3 mRNA, thereby promoting angiogenesis and metastasis of renal cell carcinoma.130 Another circRNA, SHKBP1, upregulates FOXP1 and FOXP2 expression by sponging miR-544a and miR-379, respectively, in glioma, while FOXP1/FOXP2 transcriptionally promote AGGF1 expression to facilitate viability, migration, and tube formation of GECs through the PI3K/AKT/ERK pathway.131 Similarly, circ-DICER1 and circ002136 promote the tube formation of GECs by the sponging of miRNAs and the resulting upregulation of ZIC4 and SOX13.132,133 Additionally, in pancreatic ductal adenocarcinoma (PDAC), hsa_circ_001653 promotes the restoration of cell cycle progression, angiogenesis, and invasiveness, sequestering miR-377 and elevating HOXC6 expression.134 circ-ASH2L regulates Notch 1 expression by sequestering miR-34a, promoting tumor invasion, proliferation, and angiogenesis.135 hsa_circ_0000092 sponges miR-338-3p to upregulate HN1 in HCC, and hsa_circRNA_002178 sponges miR-338-3p to upregulate COL1A1 in breast cancer, thus promoting angiogenesis.136,137 On the contrary, circHIPK3 is decreased in BCa and inversely correlates with cancer grade and lymph node metastasis. Functional assays uncovered that circHIPK3 inhibited the expression of heparanase (HPSE) by sponging miR-558.123 Altogether, these studies indicate that circRNAs extensively modulate miRNAs to regulate angiogenesis, which provides a novel insight to guide tumor therapy through targeting circRNAs.
Emerging paradigms of lncRNAs and circRNAs in angiogenesis
Angio-LncRs and Angio-CircRs induce cancer onset and progression
Increasing numbers of studies have shown that lncRNAs and circRNAs orchestrate tumor proliferation, migration, invasion, metastasis, and the local microenvironment for tumor cell colonization.138,139 Herein, we provide examples of studies on Angio-LncRs and Angio-CircRs from the perspective of immune escape, EMT and migration, intravasation and extravasation, and the pre-metastasis niche (Figure 4).
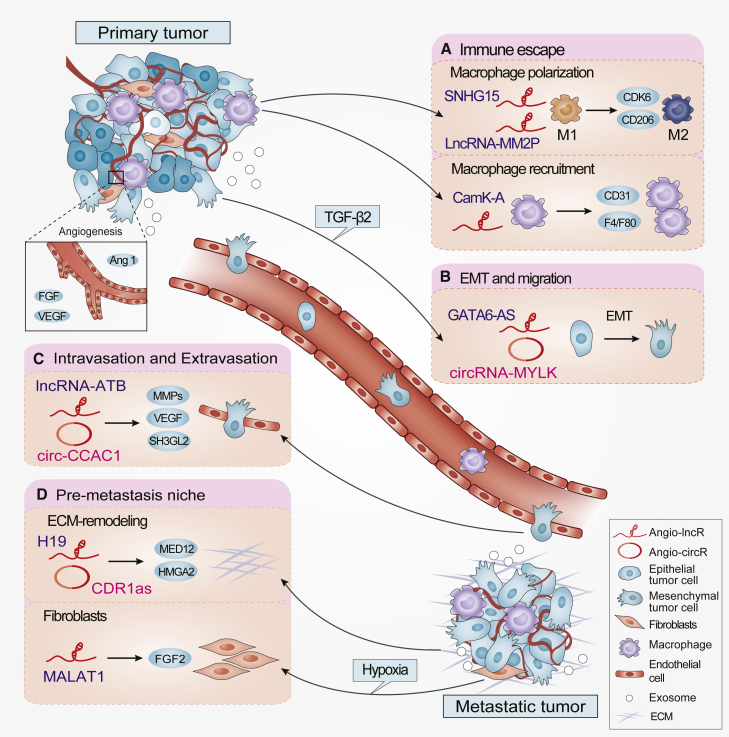
Figure 4.
Angio-LncRs and Angio-CircRs in the multistep tumor progression
(A–D) Angio-LncRs and Angio-CircRs exert diverse regulatory roles in tumor progression and metastasis, including (A) immune escape (e.g., SNHG15, LncRNA-MM2P, and CamK-A), (B) EMT and migration (e.g., GATA6-AS and circRNA-MYLK), (C) intravasation and extravasation (e.g., lncRNA-ATB and circ-CCAC1), and (D) the pre-metastatic niche (e.g., H19, CDR1as, and MALAT1).
Immune escape has always been considered a crucial step in tumor development and progression, including the recruitment of immune cells and macrophage M1/M2 (classically activated/ alternatively activated) polarization.140,141 lncRNAs can serve as key regulators of macrophages to impact innate immunity. For example, lncRNA-MM2P and SNHG15 promoted M2 polarization of macrophages, and CamK-A was involved in macrophage recruitment.85,87,142 EMT is a cellular process that causes cancer cells to attenuate epithelial characteristics and acquire mesenchymal characteristics, separating them from neighboring cells and allowing them to migrate and invade more efficiently.143, 144, 145 GATA6-AS increases TGF-β2-induced EMT and promotes the formation of blood vessels in mice.94 MALAT1 promotes angiogenesis and EMT in CRC by elevating VEGFA, SLUG, and TWIST expression.71 The circRNA-MYLK contributes to EMT and BCa development by activating VEGFA/VEGFR2 and its downstream Ras/ERK signaling pathways.119 Another circRNA, circ-CSPP1, promotes EMT and OC development by upregulating EMT-related markers.146 Intravasation and extravasation are the biological processes by which invasive cancer cells pass in and out of circulation through the walls of blood vessels and spread to the site of metastasis.147 A recent study uncovered that circ-CCAC1 could destroy the endothelial barrier integrity and promote angiogenesis by interacting with EZH2.12 lncRNA-ATB can mediate TGF-β to promote EMT, invasion, intravasation, and colonization of tumor cells.148,149 Furthermore, the pre-metastatic niche is the changed supportive microenvironment of distant organs and tissues before the tumor cells reach the site of metastasis, including the changes in cellular constituents, immune status, blood supply, and extracellular matrix (ECM). In particular, fibroblasts and ECM dynamics are involved in the adhesion, growth, and metastasis of tumor cells.150,151 H19 promotes the expression of key ECM-remodeling genes through a variety of mechanisms.152 Additionally, functional enrichment analysis suggested that CDR1as was associated with angiogenesis, ECM organization, and especially interaction with ECM receptor to modulate the tumor microenvironment.153 MALAT1 and the lncRNA XIST can mediate the expression and secretion of fibroblast growth factor (FGF).154,155 Current studies indicate that lncRNAs and circRNAs manipulate the progression of diverse cancers via modulating the tumor microenvironment. The new links between Angio-LncRs/Angio-CircRs and multistep tumor progression will open up a new perspective for ncRNA-induced angiogenesis, thus accelerating rational development of antitumor treatment strategies from an anti-angiogenic perspective.
The potential clinical application of lncRNAs and circRNAs
As discussed above, Angio-LncRs and Angio-CircRs contribute to tumor progression by manipulating a plethora of phenotypes, including tumor angiogenesis, cell proliferation, EMT, apoptosis, and metastasis. Therefore, Angio-LncRs and Angio-CircRs could act as promising biomarkers and efficacious therapeutic targets for tumor therapy. To date, only a small portion of lncRNAs and circRNAs have been used as promising tools for diagnostic, prognostic, and therapeutic biomarkers in the clinical application for diverse tumors (Figure 5).156,157 A few tumor Angio-LncRs, such as H19, MALAT1 and HULC, can be used as plasma biomarkers in a plethora of cancers.158, 159, 160, 161 In the case of tumor Angio-CircRs, circHIPK3 can be used as a biomarker for the prognosis of various tumors and new therapies in patients with NPC.157 hsa-circRNA-002178 could be used as a potential non-invasive biomarker for the early detection of LUAD and could act as a potential target of immune therapy.162 However, diverse functional identifications need to be explored in vivo and in vitro to target Angio-LncRs/Angio-CircRs as novel and promising therapeutic options in the clinical setting.
Figure 5.
The potential clinical application of Angio-LncRs and Angio-CircRs
(A) These Angio-LncRs and Angio-CircRs can be detected from tumor patient samples and are potential diagnostic and prognostic biomarkers. (B) The functional identifications of Angio-LncRs and Angio-CircRs have been elucidated in preclinical models. (C) RNA-based strategy can effectively target Angio-LncRs and Angio-CircRs in the cytoplasm and nucleus. (D) Combination of targeting ncRNAs and using conventional anti-angiogenic agents enhance therapeutic efficacy through diverse delivery tools in vivo.
Targeting Angio-LncRs and Angio-CircRs might represent a promising strategy for inhibiting tumor development and progression. Currently, RNA-based therapeutic approaches mainly include RNA interference (RNAi) and antisense oligonucleotides (ASOs), designed to target diverse RNAs and specific regions. The first RNAi-based drug patisiran was approved by the US Food and Drug Administration to treat hereditary transthyretin amyloidosis.163 ASOs have also displayed a promising prospect in targeting lncRNAs in a study on patients with Angelman syndrome.164 Similarly, intravenous treatment with ASOs targeting lncRNAs TUG1 and PVT1 can inhibit tumor proliferation and differentiation by combining with the drug delivery system.165,166 In vivo RNAi and pharmacological therapeutic strategy targeting FLANC significantly suppressed tumor angiogenesis and CRC metastasis, while specific small interfering RNAs (siRNAs) significantly suppress the metastases.86 Using a double-stranded DNA plasmid, DTA-H19-targeting cancer cells that overexpress H19 induced diphtheria toxin-A expression, thereby reducing the size of multiple tumor types.167 In addition, emerging research suggests that the peptides encoded by Angio-CircRs have increasing potential for tumor therapy. As in TNBC, the peptide ASRPS encoded by LINC00908 can inhibit tumor angiogenesis, and the peptide CIP2A-BP encoded by LINC00665 can significantly reduce invasion and metastasis, indicating that they are effective anti-TNBC peptides.91,168 In the future, the combinatorial application of conventional therapy with targeting Angio-LncRs or Angio-CircRs adds synergy in anti-angiogenic cancer therapy, which broadens our understanding of the progress made in cancer therapy.
Accumulating studies have uncovered that a variety of Angio-LncRs and Angio-CircRs regulate chemotherapy resistance, radiotherapy sensitivity, and immunotherapeutic tolerance. For example, some Angio-LncRs, such as MALAT1, MEG3, and CRNDE, are involved in resistance to 5-fluorouracil, adriamycin, mitomycin, and oxaliplatin in distinct cancers.169, 170, 171 Similarly, Angio-CircRs also influence chemotherapy resistance in different cancers. For example, the upregulation of circ-SMARCA5 can enhance the sensitivity of intrahepatic CCA cells to cisplatin and gemcitabine.172 hsa_circ_0023404 was upregulated in cervical tumor cells to promote cisplatin resistance.173 In addition, cZNF292 increased the hypoxic tumor cell radiosensitivity of radiotherapy in HCC.174 hsa-circRNA-002178 can enhance the expression of programmed cell death-1 (PD-1) and programmed cell death ligand-1 (PD-L1) in tumor cells and T cells and enhance immunotherapy tolerance.162 To date, several anti-angiogenic drugs such as bevacizumab have been approved in targeting angiogenesis for tumor therapy.16 However, there are no clinical applications for targeting Angio-LncRs/Angio-CircRs in tumor therapy. Therefore, the detection of lncRNA and circRNA levels in various patients could predict the efficacy and safety of various drugs and suggest implementing different therapeutic doses or methods to promote individualized treatment regimens. Altogether, the development of new biomarkers and treatment strategies for Angio-LncRs and Angio-CircRs could develop potential clinical significance for tumor treatment.
Conclusions and future perspectives
This review emphasizes the emerging functions and mechanisms of angiogenic lncRNAs and circRNAs in tumor progression and their potential diagnostic and therapeutic applications. Importantly, Angio-LncRs and Angio-CircRs orchestrate the carcinogenesis and development of diverse cancers via a broad repertoire of molecular mechanisms. We highlight the common dysregulation of Angio-LncRs and Angio-CircRs in tumor progression to manipulate multiple signaling pathways associated with angiogenesis, cell proliferation, EMT, apoptosis, and metastasis. Additionally, exploring the functional commonalities between lncRNAs and circRNAs may provide a vital hint to understand the biological functions of ncRNAs and suggest clinical values of Angio-LncRs and Angio-CircRs in tumor angiogenesis.
Since the exact mechanisms of action of lncRNAs and circRNAs have not been fully elucidated, their known roles in cancer biology may represent only the tip of the iceberg. Some Angio-LncRs and Angio-CircRs may have multiple levels of mechanical functions in tumor angiogenesis. For instance, PVT1 can not only regulate chromatin remodeling, transcriptional activation, and protein modification, but it also can adsorb a variety of miRNAs regulating downstream target genes.175 Besides affecting angiogenesis, Angio-LncRs and Angio-CircRs also influence multiple different functional phenotypes. For example, MALAT1 regulates EMT, migration, invasion, metastasis, and ECM remodeling.176 In that this review focuses on the roles of Angio-LncRs and Angio-CircRs in tumor angiogenesis, we did not elaborate on other functional mechanisms.
Although lncRNAs and circRNAs have shown attractive breakthroughs and prospects, the field is yet to conquer certain challenges and limitations. Utilizing or developing novel technologies, such as CRISPR-mediated gene editing and genome-wide chromatin interrogation, will continue to accelerate advances in preclinical studies for antitumor treatment.18 The development of RNA sequencing (RNA-seq), global run-on sequencing (GRO-seq), and quantitative RT-PCR (qRT-PCR) techniques has greatly improved the ability to determine the transcription and expression of genes and to elucidate the function of ncRNAs. Moreover, siRNA or short hairpin RNA (shRNA) is often used to silence ncRNAs, and the RNA pull-down assays or immunoprecipitation (RIP) has helped elucidate the interaction between ncRNAs and proteins.18,177 Emerging advancements in new technologies and approaches provide exciting opportunities for ncRNA research and its translation to the clinic. However, in vivo treatment strategies related to lncRNAs and circRNAs are still limited because of their poor intracellular uptake and stability. Currently, liposomes, lentiviruses, adenoviruses, exosomes, and nanoparticles are commonly used in vivo delivery systems for lncRNAs and circRNAs.178, 179, 180, 181 Future studies, combined with novel technologies, should focus on delivering lncRNAs and circRNAs to target cells in a tissue-specific manner.
Despite current challenges and limitations, significant advances in the field of Angio-LncR and Angio-CircR research are nonetheless being made. Recent advances describing the functions and mechanisms of angiogenic lncRNAs and circRNAs facilitate cancer diagnosis. However, there are a series of key questions to be answered. Are a series of angiogenic ncRNAs more promising as diagnostic markers in angiogenic diseases? Does targeting angiogenic ncRNAs have any impact on the expression of essential genes in normal and tumor tissues? Could the combination of targeting ncRNAs and using conventional anti-angiogenic agents enhance therapeutic efficacy in heterogeneous tumors? In essence, exploring the functional commonalities between lncRNAs and circRNAs may provide a vital hint to understand the biological functions of ncRNAs. Understanding the mechanism of interaction between lncRNAs or circRNAs and other biomolecules may identify certain lncRNAs and circRNAs that have the potential to be used as biomarkers or therapeutic targets in tumor angiogenesis; this would be a major advance in the beneficial treatment of cancer patients.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81872412 to H.-W.X.), the Natural Science Foundation of Hubei Province, China (2019CFB591 to Z.M.), the Bureau of Science and Technology of Jingzhou Municipality, China (2020CB21-35 to Z.M.), and Guangdong Provincial-level Special Funds for Promoting High-quality Economic Development, China (2020032 to Q.Z.).
Author contributions
Z.M. conceived the review. Z.M., H.-W.X., and Q.Z. wrote and revised the manuscript. C.-G.L., J.L., and Y.X. drafted the manuscript and prepared the figures. W.L. and S.-X.F. helped to modify the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Qing Zhang, Email: lsszq@mail.sysu.edu.cn.
Hong-Wu Xin, Email: hongwu_xin@126.com.
Zhaowu Ma, Email: zhaowu823@126.com.
References
- 1.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 2.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 3.Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47(D1):D1034–D1037. doi: 10.1093/nar/gky905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 7.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 8.Batista P.J., Chang H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J., Meng J., Zhu L., Peng Y. Exosomal noncoding RNAs in glioma: Biological functions and potential clinical applications. Mol. Cancer. 2020;19:66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu Y., Bao L., Chen Y., Wang C., Luo M., Zhang B., Zhou M., Wang J.E., Fang Y.V., Kumar A. HIF2-induced long noncoding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res. 2020;80:964–975. doi: 10.1158/0008-5472.CAN-19-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y., Leng K., Yao Y., Kang P., Liao G., Han Y., Shi G., Ji D., Huang P., Zheng W. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73:1419–1435. doi: 10.1002/hep.31493. [DOI] [PubMed] [Google Scholar]
- 13.Herbert S.P., Stainier D.Y. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 2011;12:551–564. doi: 10.1038/nrm3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D., Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 15.Jayson G.C., Kerbel R., Ellis L.M., Harris A.L. Antiangiogenic therapy in oncology: Current status and future directions. Lancet. 2016;388:518–529. doi: 10.1016/S0140-6736(15)01088-0. [DOI] [PubMed] [Google Scholar]
- 16.Ferrara N., Kerbel R.S. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Wang L., Chen C., Chu X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol. Cancer. 2018;17:22. doi: 10.1186/s12943-018-0766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu B., Wang S. Angio-LncRs: lncRNAs that regulate angiogenesis and vascular disease. Theranostics. 2018;8:3654–3675. doi: 10.7150/thno.26024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J., Li L., Han Z.Y., Wang Z.X., Qin L.X. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am. J. Cancer Res. 2019;9:1367–1381. [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar M.M., Goyal R. lncRNA as a therapeutic target for angiogenesis. Curr. Top. Med. Chem. 2017;17:1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng S.R., Wu J.S., Tang Y.L., Liang X.H. Long noncoding RNAs: Emerging regulators of tumor angiogenesis. Future Oncol. 2017;13:1551–1562. doi: 10.2217/fon-2017-0149. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Yang Y., Wang Z., Fu X., Chu X.M., Li Y., Wang Q., He X., Li M., Wang K. Insights into the regulatory role of circRNA in angiogenesis and clinical implications. Atherosclerosis. 2020;298:14–26. doi: 10.1016/j.atherosclerosis.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 23.Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M., Date S., Sugimoto S., Kanai T., Sato T. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. 2017;545:187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 24.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu P., Mo Y., Peng M., Tang T., Zhong Y., Deng X., Xiong F., Guo C., Wu X., Li Y. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 30.Chen N., Zhao G., Yan X., Lv Z., Yin H., Zhang S., Song W., Li X., Li L., Du Z. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHugh C.A., Chen C.K., Chow A., Surka C.F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I. lncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 35.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 36.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 37.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., 3rd, Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bögler O. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon J.H., Abdelmohsen K., Kim J., Yang X., Martindale J.L., Tominaga-Yamanaka K., White E.J., Orjalo A.V., Rinn J.L., Kreft S.G. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 44.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho W.C., Jour G., Aung P.P. Role of angiogenesis in melanoma progression: Update on key angiogenic mechanisms and other associated components. Semin. Cancer Biol. 2019;59:175–186. doi: 10.1016/j.semcancer.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Bottaro D.P., Liotta L.A. Cancer: Out of air is not out of action. Nature. 2003;423:593–595. doi: 10.1038/423593a. [DOI] [PubMed] [Google Scholar]
- 49.Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 50.Ebos J.M., Kerbel R.S. Antiangiogenic therapy: Impact on invasion, disease progression, and metastasis. Nat. Rev. Clin. Oncol. 2011;8:210–221. doi: 10.1038/nrclinonc.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blagosklonny M.V. Antiangiogenic therapy and tumor progression. Cancer Cell. 2004;5:13–17. doi: 10.1016/s1535-6108(03)00336-2. [DOI] [PubMed] [Google Scholar]
- 52.Chatterjee S., Heukamp L.C., Siobal M., Schöttle J., Wieczorek C., Peifer M., Frasca D., Koker M., König K., Meder L. Tumor VEGF:VEGFR2 autocrine feed-forward loop triggers angiogenesis in lung cancer. J. Clin. Invest. 2013;123:1732–1740. doi: 10.1172/JCI65385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siekmann A.F., Lawson N.D. Notch signalling and the regulation of angiogenesis. Cell Adhes. Migr. 2007;1:104–106. doi: 10.4161/cam.1.2.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fantin A., Schwarz Q., Davidson K., Normando E.M., Denti L., Ruhrberg C. The cytoplasmic domain of neuropilin 1 is dispensable for angiogenesis, but promotes the spatial separation of retinal arteries and veins. Development. 2011;138:4185–4191. doi: 10.1242/dev.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Wu Z., Yuan J., Sun L., Lin L., Huang N., Bin J., Liao Y., Liao W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 56.Yuan Z., Bian Y., Ma X., Tang Z., Chen N., Shen M. lncRNA H19 knockdown in human amniotic mesenchymal stem cells suppresses angiogenesis by associating with EZH2 and activating vasohibin-1. Stem Cells Dev. 2019;28:781–790. doi: 10.1089/scd.2019.0014. [DOI] [PubMed] [Google Scholar]
- 57.Wang L.L., Zong Z.H., Liu Y., Guan X., Chen S., Zhao Y. circRhoC promotes tumorigenicity and progression in ovarian cancer by functioning as a miR-302e sponge to positively regulate VEGFA. J. Cell. Mol. Med. 2019;23:8472–8481. doi: 10.1111/jcmm.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Y., Zhang M., Liu J., Xu B., Yang J., Wang N., Yan S., Wang F., He X., Ji G. Long non-coding RNA PVT1 promotes cell proliferation and migration by silencing ANGPTL4 expression in cholangiocarcinoma. Mol. Ther. Nucleic Acids. 2018;13:503–513. doi: 10.1016/j.omtn.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W.S., Lin T.Y., Chang L., Yeh W.W., Huang S.C., Chen T.Y., Hsieh Y.T., Chen S.T., Li W.C., Pan C.C. HIV-1 Tat interacts with a Kaposi’s sarcoma-associated herpesvirus reactivation-upregulated antiangiogenic long noncoding RNA, LINC00313, and antagonizes its function. J. Virol. 2020;94:e01280-19. doi: 10.1128/JVI.01280-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W., Chen G., Wang B., Yuan Z., Liu G., Niu B., Chen Y., Zhou S., He J., Xue H. Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation. J. Transl. Med. 2019;17:421. doi: 10.1186/s12967-019-02145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng S., Lin F., Zhang M., Mu N., Ge X., Fu J. Long non-coding RNA AK001058 regulates tumor growth and angiogenesis in colorectal cancer via methylation of ADAMTS12. Am. J. Transl. Res. 2019;11:6117–6123. [PMC free article] [PubMed] [Google Scholar]
- 62.Dong R., Liu X.Q., Zhang B.B., Liu B.H., Zheng S., Dong K.R. Long non-coding RNA-CRNDE: A novel regulator of tumor growth and angiogenesis in hepatoblastoma. Oncotarget. 2017;8:42087–42097. doi: 10.18632/oncotarget.14992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng Z., Wang J., Shan B., Li B., Peng W., Dong Y., Shi W., Zhao W., He D., Duan M. The long noncoding RNA LINC00312 induces lung adenocarcinoma migration and vasculogenic mimicry through directly binding YBX1. Mol. Cancer. 2018;17:167. doi: 10.1186/s12943-018-0920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cong Z., Diao Y., Li X., Jiang Z., Xu Y., Zhou H., Qiang Y., Wu H., Shen Y. Long non-coding RNA linc00665 interacts with YB-1 and promotes angiogenesis in lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2020;527:545–552. doi: 10.1016/j.bbrc.2020.04.108. [DOI] [PubMed] [Google Scholar]
- 65.Zhao J., Du P., Cui P., Qin Y., Hu C., Wu J., Zhou Z., Zhang W., Qin L., Huang G. lncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–4109. doi: 10.1038/s41388-018-0250-z. [DOI] [PubMed] [Google Scholar]
- 66.Zhou X., Rao Y., Sun Q., Liu Y., Chen J., Bu W. Long noncoding RNA CPS1-IT1 suppresses melanoma cell metastasis through inhibiting Cyr61 via competitively binding to BRG1. J. Cell. Physiol. 2019;234:22017–22027. doi: 10.1002/jcp.28764. [DOI] [PubMed] [Google Scholar]
- 67.Fu W.M., Lu Y.F., Hu B.G., Liang W.C., Zhu X., Yang H.D., Li G., Zhang J.F. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7:4712–4723. doi: 10.18632/oncotarget.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruan Z., Zhao D. Long intergenic noncoding RNA LINC00284 knockdown reduces angiogenesis in ovarian cancer cells via up-regulation of MEST through NF-κB1. FASEB J. 2019;33:12047–12059. doi: 10.1096/fj.201900101RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu T., Wu K., Zhang L., Zheng S., Wang X., Zuo H., Wu X., Tao G., Jiang B., Zhang L. Long non-coding RNA LINC00858 exerts a tumor-promoting role in colon cancer via HNF4α and WNK2 regulation. Cell Oncol. (Dordr.) 2020;43:297–310. doi: 10.1007/s13402-019-00490-8. [DOI] [PubMed] [Google Scholar]
- 70.Wu J., Meng X., Jia Y., Chai J., Wang J., Xue X., Dang T. Long non-coding RNA HNF1A-AS1 upregulates OTX1 to enhance angiogenesis in colon cancer via the binding of transcription factor PBX3. Exp. Cell Res. 2020;393:112025. doi: 10.1016/j.yexcr.2020.112025. [DOI] [PubMed] [Google Scholar]
- 71.Sun Z., Ou C., Liu J., Chen C., Zhou Q., Yang S., Li G., Wang G., Song J., Li Z. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627–2644. doi: 10.1038/s41388-018-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Hou Z.H., Xu X.W., Fu X.Y., Zhou L.D., Liu S.P., Tan D.M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am. J. Physiol. Cell Physiol. 2020;318:C649–C663. doi: 10.1152/ajpcell.00510.2018. [DOI] [PubMed] [Google Scholar]
- 73.Huang X.J., Xia Y., He G.F., Zheng L.L., Cai Y.P., Yin Y., Wu Q. MALAT1 promotes angiogenesis of breast cancer. Oncol. Rep. 2018;40:2683–2689. doi: 10.3892/or.2018.6705. [DOI] [PubMed] [Google Scholar]
- 74.Yu W., Ding J., He M., Chen Y., Wang R., Han Z., Xing E.Z., Zhang C., Yeh S. Estrogen receptor β promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene. 2019;38:1225–1238. doi: 10.1038/s41388-018-0463-1. [DOI] [PubMed] [Google Scholar]
- 75.Yu X., Hu L., Li S., Shen J., Wang D., Xu R., Yang H. Long non-coding RNA taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF-1α via miR-143-5p. Cell Death Dis. 2019;10:280. doi: 10.1038/s41419-019-1509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cai H., Liu X., Zheng J., Xue Y., Ma J., Li Z., Xi Z., Li Z., Bao M., Liu Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 77.Dong R., Liu G.B., Liu B.H., Chen G., Li K., Zheng S., Dong K.R. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016;7:e2278. doi: 10.1038/cddis.2016.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X., Zeng K., Xu M., Hu X., Liu X., Xu T., He B., Pan Y., Sun H., Wang S. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9:982. doi: 10.1038/s41419-018-0962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu Z., Xiao Z., Liu F., Cui M., Li W., Yang Z., Li J., Ye L., Zhang X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1) Oncotarget. 2016;7:241–254. doi: 10.18632/oncotarget.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang F.W., Cao C.H., Han K., Zhao Y.X., Cai M.Y., Xiang Z.C., Zhang J.X., Chen J.W., Zhong L.P., Huang Y. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J. Clin. Invest. 2019;129:727–743. doi: 10.1172/JCI122478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y., Wu Y., Xiao K., Zhao Y., Lv G., Xu S., Wu F. RPS24c isoform facilitates tumor angiogenesis via promoting the stability of MVIH in colorectal cancer. Curr. Mol. Med. 2020;20:388–395. doi: 10.2174/1566524019666191203123943. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y., Sun J., Qi Y., Wang Y., Ding Y., Wang K., Zhou Q., Wang J., Ma F., Zhang J., Guo B. Long non-coding RNA TPT1-AS1 promotes angiogenesis and metastasis of colorectal cancer through TPT1-AS1/NF90/VEGFA signaling pathway. Aging (Albany NY) 2020;12:6191–6205. doi: 10.18632/aging.103016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang C., Zheng J., Liu X., Xue Y., He Q., Dong Y., Wang D., Li Z., Liu L., Ma J. Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via Staufen1-mediated mRNA decay. Mol. Ther. Nucleic Acids. 2020;20:866–878. doi: 10.1016/j.omtn.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X., Li L., Zhao K., Lin Q., Li H., Xue X., Ge W., He H., Liu D., Xie H. A novel lncRNA HITT forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. Cell Death Differ. 2020;27:1431–1446. doi: 10.1038/s41418-019-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sang L.J., Ju H.Q., Liu G.P., Tian T., Ma G.L., Lu Y.X., Liu Z.X., Pan R.L., Li R.H., Piao H.L. lncRNA CamK-A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Mol. Cell. 2018;72:71–83.e7. doi: 10.1016/j.molcel.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 86.Pichler M., Rodriguez-Aguayo C., Nam S.Y., Dragomir M.P., Bayraktar R., Anfossi S., Knutsen E., Ivan C., Fuentes-Mattei E., Lee S.K. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut. 2020;69:1818–1831. doi: 10.1136/gutjnl-2019-318903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao J., Dong R., Jiang L., Gong Y., Yuan M., You J., Meng W., Chen Z., Zhang N., Weng Q. lncRNA-MM2P identified as a modulator of macrophage M2 polarization. Cancer Immunol. Res. 2019;7:292–305. doi: 10.1158/2326-6066.CIR-18-0145. [DOI] [PubMed] [Google Scholar]
- 88.Ding X., Jia X., Wang C., Xu J., Gao S.J., Lu C. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell Death Differ. 2019;26:1750–1765. doi: 10.1038/s41418-018-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan J., Huang W., Huang X., Xiang W., Ye C., Liu J. A negative feedback loop between long noncoding RNA NBAT1 and Sox9 inhibits the malignant progression of gastric cancer cells. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180882. BSR20180882. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Wang Y., Han D., Pan L., Sun J. The positive feedback between lncRNA TNK2-AS1 and STAT3 enhances angiogenesis in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018;507:185–192. doi: 10.1016/j.bbrc.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y., Wu S., Zhu X., Zhang L., Deng J., Li F., Guo B., Zhang S., Wu R., Zhang Z. lncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190950. jem.20190950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Comet I., Riising E.M., Leblanc B., Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 93.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neumann P., Jaé N., Knau A., Glaser S.F., Fouani Y., Rossbach O., Krüger M., John D., Bindereif A., Grote P. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 2018;9:237. doi: 10.1038/s41467-017-02431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jung G., Hernández-Illán E., Moreira L., Balaguer F., Goel A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020;17:111–130. doi: 10.1038/s41575-019-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mao Z., Xu B., He L., Zhang G. PVT1 promotes angiogenesis by regulating miR-29c/vascular endothelial growth factor (VEGF) signaling pathway in non-small-cell lung cancer (NSCLC) Med. Sci. Monit. 2019;25:5418–5425. doi: 10.12659/MSM.917601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin X., Yang F., Qi X., Li Q., Wang D., Yi T., Yin R., Zhao X., Zhong X., Bian C. lncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol. Carcinog. 2019;58:2286–2296. doi: 10.1002/mc.23117. [DOI] [PubMed] [Google Scholar]
- 99.Ye J., Zhu J., Chen H., Qian J., Zhang L., Wan Z., Chen F., Sun S., Li W., Luo C. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int. J. Cancer. 2020;146:248–261. doi: 10.1002/ijc.32483. [DOI] [PubMed] [Google Scholar]
- 100.Zhao X., Liu Y., Li Z., Zheng S., Wang Z., Li W., Bi Z., Li L., Jiang Y., Luo Y. linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J. Cell. Mol. Med. 2018;22:655–667. doi: 10.1111/jcmm.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma Y., Xue Y., Liu X., Qu C., Cai H., Wang P., Li Z., Li Z., Liu Y. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol. Rep. 2017;38:3265–3277. doi: 10.3892/or.2017.5985. [DOI] [PubMed] [Google Scholar]
- 102.Zhang J.X., Chen Z.H., Chen D.L., Tian X.P., Wang C.Y., Zhou Z.W., Gao Y., Xu Y., Chen C., Zheng Z.S. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660–2675. doi: 10.1038/s41388-018-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Chi H., Yang R., Zheng X., Zhang L., Jiang R., Chen J. lncRNA RP11-79H23.3 functions as a competing endogenous RNA to regulate PTEN expression through sponging hsa-miR-107 in the development of bladder cancer. Int. J. Mol. Sci. 2018;19:2531. doi: 10.3390/ijms19092531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen C.L., Tseng Y.W., Wu J.C., Chen G.Y., Lin K.C., Hwang S.M., Hu Y.C. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and microRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 105.Gao Y., Yu H., Liu Y., Liu X., Zheng J., Ma J., Gong W., Chen J., Zhao L., Tian Y., Xue Y. Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the miR-373/EGFR axis. Cell. Physiol. Biochem. 2018;45:131–147. doi: 10.1159/000486253. [DOI] [PubMed] [Google Scholar]
- 106.Yang C., Zheng J., Xue Y., Yu H., Liu X., Ma J., Liu L., Wang P., Li Z., Cai H., Liu Y. The effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front. Mol. Neurosci. 2018;10:437. doi: 10.3389/fnmol.2017.00437. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 107.Yu H., Xue Y., Wang P., Liu X., Ma J., Zheng J., Li Z., Li Z., Cai H., Liu Y. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6:e303. doi: 10.1038/oncsis.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo J., Cai H., Liu X., Zheng J., Liu Y., Gong W., Chen J., Xi Z., Xue Y. Long non-coding RNA LINC00339 stimulates glioma vasculogenic mimicry formation by regulating the miR-539-5p/TWIST1/MMPs axis. Mol. Ther. Nucleic Acids. 2018;10:170–186. doi: 10.1016/j.omtn.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jia P., Cai H., Liu X., Chen J., Ma J., Wang P., Liu Y., Zheng J., Xue Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381:359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 110.Wang D., Zheng J., Liu X., Xue Y., Liu L., Ma J., He Q., Li Z., Cai H., Liu Y. Knockdown of USF1 inhibits the vasculogenic mimicry of glioma cells via stimulating SNHG16/miR-212-3p and linc00667/miR-429 axis. Mol. Ther. Nucleic Acids. 2019;14:465–482. doi: 10.1016/j.omtn.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Li X., Yu M., Yang C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J. Cell. Biochem. 2020;121:2258–2267. doi: 10.1002/jcb.29448. [DOI] [PubMed] [Google Scholar]
- 112.Huang J.Z., Chen M., Chen D., Gao X.C., Zhu S., Huang H., Hu M., Zhu H., Yan G.R. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell. 2017;68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 113.Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.R. ncRNA-encoded peptides or proteins and cancer. Mol. Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barbagallo D., Caponnetto A., Brex D., Mirabella F., Barbagallo C., Lauretta G., Morrone A., Certo F., Broggi G., Caltabiano R. circSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (Basel) 2019;11:194. doi: 10.3390/cancers11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guarnerio J., Zhang Y., Cheloni G., Panella R., Mae Katon J., Simpson M., Matsumoto A., Papa A., Loretelli C., Petri A. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29:628–640. doi: 10.1038/s41422-019-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen J., Zhou X., Yang J., Sun Q., Liu Y., Li N., Zhang Z., Xu H. circ-GLI1 promotes metastasis in melanoma through interacting with p70S6K2 to activate Hedgehog/GLI1 and Wnt/β-catenin pathways and upregulate Cyr61. Cell Death Dis. 2020;11:596. doi: 10.1038/s41419-020-02799-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu X., Shen S., Zhu L., Su R., Zheng J., Ruan X., Shao L., Wang D., Yang C., Liu Y. SRSF10 inhibits biogenesis of circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2 pathway. J. Exp. Clin. Cancer Res. 2020;39:121. doi: 10.1186/s13046-020-01625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meng Q., Li S., Liu Y., Zhang S., Jin J., Zhang Y., Guo C., Liu B., Sun Y. Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol. Ther. Nucleic Acids. 2019;17:669–677. doi: 10.1016/j.omtn.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L., Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 120.Cao W., Zhao Y., Wang L., Huang X. circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. 2019;25:101–113. doi: 10.3233/CBM-182380. [DOI] [PubMed] [Google Scholar]
- 121.Li W., Xu Y., Wang X., Cao G., Bu W., Wang X., Fang Z., Xu Y., Dong M., Tao Q. circCCT3 modulates vascular endothelial growth factor A and Wnt signaling to enhance colorectal cancer metastasis through sponging miR-613. DNA Cell Biol. 2020;39:118–125. doi: 10.1089/dna.2019.5139. [DOI] [PubMed] [Google Scholar]
- 122.Zheng X., Ma Y.F., Zhang X.R., Li Y., Zhao H.H., Han S.G. circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4190–4202. doi: 10.26355/eurrev_202004_20999. [DOI] [PubMed] [Google Scholar]
- 123.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., Jiang G. circHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hanniford D., Ulloa-Morales A., Karz A., Berzoti-Coelho M.G., Moubarak R.S., Sánchez-Sendra B., Kloetgen A., Davalos V., Imig J., Wu P. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70.e15. doi: 10.1016/j.ccell.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vos P.D., Leedman P.J., Filipovska A., Rackham O. Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cell. Mol. Life Sci. 2019;76:3745–3752. doi: 10.1007/s00018-019-03163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., Ren D., Ye X., Li C., Wang Y. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li J., Huang C., Zou Y., Yu J., Gui Y. Circular RNA MYLK promotes tumour growth and metastasis via modulating miR-513a-5p/VEGFC signalling in renal cell carcinoma. J. Cell. Mol. Med. 2020;24:6609–6621. doi: 10.1111/jcmm.15308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen C., Huang Z., Mo X., Song Y., Li X., Li X., Zhang M. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J. Exp. Clin. Cancer Res. 2020;39:91. doi: 10.1186/s13046-020-01594-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 129.Wei C.Y., Zhu M.X., Lu N.H., Liu J.Q., Yang Y.W., Zhang Y., Shi Y.D., Feng Z.H., Li J.X., Qi F.Z., Gu J.Y. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol. Cancer. 2020;19:84. doi: 10.1186/s12943-020-01191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li W., Yang F.Q., Sun C.M., Huang J.H., Zhang H.M., Li X., Wang G.C., Zhang N., Che J.P., Zhang W.T. circPRRC2A promotes angiogenesis and metastasis through epithelial-mesenchymal transition and upregulates TRPM3 in renal cell carcinoma. Theranostics. 2020;10:4395–4409. doi: 10.7150/thno.43239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.He Q., Zhao L., Liu Y., Liu X., Zheng J., Yu H., Cai H., Ma J., Liu L., Wang P. circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol. Ther. Nucleic Acids. 2018;10:331–348. doi: 10.1016/j.omtn.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He Q., Zhao L., Liu X., Zheng J., Liu Y., Liu L., Ma J., Cai H., Li Z., Xue Y. MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4 expression change. J. Exp. Clin. Cancer Res. 2019;38:9. doi: 10.1186/s13046-018-0990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.He Z., Ruan X., Liu X., Zheng J., Liu Y., Liu L., Ma J., Shao L., Wang D., Shen S. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in glioma. J. Exp. Clin. Cancer Res. 2019;38:65. doi: 10.1186/s13046-019-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi H., Li H., Zhen T., Dong Y., Pei X., Zhang X. hsa_circ_001653 implicates in the development of pancreatic ductal adenocarcinoma by regulating microRNA-377-mediated HOXC6 axis. Mol. Ther. Nucleic Acids. 2020;20:252–264. doi: 10.1016/j.omtn.2019.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 135.Chen Y., Li Z., Zhang M., Wang B., Ye J., Zhang Y., Tang D., Ma D., Jin W., Li X., Wang S. circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2019;38:466. doi: 10.1186/s13046-019-1436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pu J., Wang J., Li W., Lu Y., Wu X., Long X., Luo C., Wei H. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J. Cell. Mol. Med. 2020 doi: 10.1111/jcmm.15010. Published online February 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu T., Ye P., Ye Y., Lu S., Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J. Cell. Mol. Med. 2020;24:2189–2201. doi: 10.1111/jcmm.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 139.Zhang Q., Wang W., Zhou Q., Chen C., Yuan W., Liu J., Li X., Sun Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer. 2020;19:14. doi: 10.1186/s12943-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mohme M., Riethdorf S., Pantel K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017;14:155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 141.Guo X., Zhao Y., Yan H., Yang Y., Shen S., Dai X., Ji X., Ji F., Gong X.G., Li L. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 2017;31:247–259. doi: 10.1101/gad.294348.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Z., Zhang J., Zheng H., Li C., Xiong J., Wang W., Bao H., Jin H., Liang P. Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2019;38:380. doi: 10.1186/s13046-019-1371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 144.Cano A., Nieto M.A. Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition. Trends Cell Biol. 2008;18:357–359. doi: 10.1016/j.tcb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 145.Cheng J.T., Wang L., Wang H., Tang F.R., Cai W.Q., Sethi G., Xin H.W., Ma Z. Insights into biological role of lncRNAs in epithelial-mesenchymal transition. Cells. 2019;8:1178. doi: 10.3390/cells8101178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li Q.H., Liu Y., Chen S., Zong Z.H., Du Y.P., Sheng X.J., Zhao Y. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed. Pharmacother. 2019;114:108832. doi: 10.1016/j.biopha.2019.108832. [DOI] [PubMed] [Google Scholar]
- 147.Reymond N., d’Água B.B., Ridley A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 148.Li W., Kang Y. A new Lnc in metastasis: Long noncoding RNA mediates the prometastatic functions of TGF-β. Cancer Cell. 2014;25:557–559. doi: 10.1016/j.ccr.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhu A.D., Sun Y.Y., Ma Q.J., Xu F. Retraction. J. Cell. Biochem. 2019;120:14360–14371. doi: 10.1002/jcb.30000. Published online June 4, 2021. [DOI] [PubMed] [Google Scholar]
- 150.Steeg P.S. Targeting metastasis. Nat. Rev. Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu Y., Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell. 2016;30:668–681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 152.Cao T., Jiang Y., Wang Z., Zhang N., Al-Hendy A., Mamillapalli R., Kallen A.N., Kodaman P., Taylor H.S., Li D., Huang Y. H19 lncRNA identified as a master regulator of genes that drive uterine leiomyomas. Oncogene. 2019;38:5356–5366. doi: 10.1038/s41388-019-0808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zou Y., Zheng S., Deng X., Yang A., Xie X., Tang H., Xie X. The role of circular RNA CDR1as/ciRS-7 in regulating tumor microenvironment: a pan-cancer analysis. Biomolecules. 2019;9:429. doi: 10.3390/biom9090429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang J.K., Ma L., Song W.H., Lu B.Y., Huang Y.B., Dong H.M., Ma X.K., Zhu Z.Z., Zhou R. lncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J. Cell. Biochem. 2017;118:4821–4830. doi: 10.1002/jcb.26153. [DOI] [PubMed] [Google Scholar]
- 155.Zhou K., Li S., Du G., Fan Y., Wu P., Sun H., Zhang T. lncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. OncoTargets Ther. 2019;12:7095–7109. doi: 10.2147/OTT.S208329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Chandra Gupta S., Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int. J. Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 157.Guo Y., Yang J., Huang Q., Hsueh C., Zheng J., Wu C., Chen H., Zhou L. Circular RNAs and their roles in head and neck cancers. Mol. Cancer. 2019;18:44. doi: 10.1186/s12943-019-1003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou X., Yin C., Dang Y., Ye F., Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zidan H.E., Karam R.A., El-Seifi O.S., Abd Elrahman T.M. Circulating long non-coding RNA MALAT1 expression as molecular biomarker in Egyptian patients with breast cancer. Cancer Genet. 2018;220:32–37. doi: 10.1016/j.cancergen.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 160.Liu M., Yang P., Mao G., Deng J., Peng G., Ning X., Yang H., Sun H. Long non-coding RNA MALAT1 as a valuable biomarker for prognosis in osteosarcoma: A systematic review and meta-analysis. Int. J. Surg. 2019;72:206–213. doi: 10.1016/j.ijsu.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 161.Li J., Wang X., Tang J., Jiang R., Zhang W., Ji J., Sun B. HULC and linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell. Physiol. Biochem. 2015;37:687–696. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 162.Wang J., Zhao X., Wang Y., Ren F., Sun D., Yan Y., Kong X., Bu J., Liu M., Xu S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 164.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Katsushima K., Natsume A., Ohka F., Shinjo K., Hatanaka A., Ichimura N., Sato S., Takahashi S., Kimura H., Totoki Y. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xu Y., Li Y., Jin J., Han G., Sun C., Pizzi M.P., Huo L., Scott A., Wang Y., Ma L. lncRNA PVT1 up-regulation is a poor prognosticator and serves as a therapeutic target in esophageal adenocarcinoma. Mol. Cancer. 2019;18:141. doi: 10.1186/s12943-019-1064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mizrahi A., Czerniak A., Levy T., Amiur S., Gallula J., Matouk I., Abu-lail R., Sorin V., Birman T., de Groot N. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guo B., Wu S., Zhu X., Zhang L., Deng J., Li F., Wang Y., Zhang S., Wu R., Lu J., Zhou Y. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020;39:e102190. doi: 10.15252/embj.2019102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yuan P., Cao W., Zang Q., Li G., Guo X., Fan J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem. Biophys. Res. Commun. 2016;478:1067–1073. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 170.Wang H., Li H., Zhang L., Yang D. Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis. Biomed. Pharmacother. 2018;106:1607–1615. doi: 10.1016/j.biopha.2018.07.131. [DOI] [PubMed] [Google Scholar]
- 171.Han P., Li J.W., Zhang B.M., Lv J.C., Li Y.M., Gu X.Y., Yu Z.W., Jia Y.H., Bai X.F., Li L. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lu Q., Fang T. Circular RNA SMARCA5 correlates with favorable clinical tumor features and prognosis, and increases chemotherapy sensitivity in intrahepatic cholangiocarcinoma. J. Clin. Lab. Anal. 2020;34:e23138. doi: 10.1002/jcla.23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guo J., Chen M., Ai G., Mao W., Li H., Zhou J. hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed. Pharmacother. 2019;115:108957. doi: 10.1016/j.biopha.2019.108957. [DOI] [PubMed] [Google Scholar]
- 174.Yang W., Liu Y., Gao R., Xiu Z., Sun T. Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell. Signal. 2019;60:122–135. doi: 10.1016/j.cellsig.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 175.Ghafouri-Fard S., Omrani M.D., Taheri M. Long noncoding RNA PVT1: A highly dysregulated gene in malignancy. J. Cell. Physiol. 2020;235:818–835. doi: 10.1002/jcp.29060. [DOI] [PubMed] [Google Scholar]
- 176.Lei L., Chen J., Huang J., Lu J., Pei S., Ding S., Kang L., Xiao R., Zeng Q. Functions and regulatory mechanisms of metastasis-associated lung adenocarcinoma transcript 1. J. Cell. Physiol. 2018;234:134–151. doi: 10.1002/jcp.26759. [DOI] [PubMed] [Google Scholar]
- 177.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dong Y., Siegwart D.J., Anderson D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019;144:133–147. doi: 10.1016/j.addr.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang D., Zhang F., Gao G. CRISPR-based therapeutic genome editing: Strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu C.F., Chen G.J., Luo Y.L., Zhang Y., Zhao G., Lu Z.D., Czarna A., Gu Z., Wang J. Rational designs of in vivo CRISPR-Cas delivery systems. Adv. Drug Deliv. Rev. 2021;168:3–29. doi: 10.1016/j.addr.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 181.Tay A., Melosh N. Nanostructured materials for intracellular cargo delivery. Acc. Chem. Res. 2019;52:2462–2471. doi: 10.1021/acs.accounts.9b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]