Abstract
Background
Shortening antibiotic-treatment durations is a key recommendation of antibiotic-stewardship programmes, yet it is based on weak evidence. We investigated whether halving antibiotic courses would reduce antibiotic-resistance genes (ARG) in the intestinal microbiomes of patients treated for gram-negative bacteraemia.
Methods
This nested prospective cohort study included adult patients hospitalized at Geneva University Hospitals (Switzerland) participating in the PIRATE randomized trial assessing non-inferiority of shorter antibiotic courses (7 versus 14 days) for gram-negative bacteraemia (‘cases’) and, simultaneously, hospitalized patients with similar demography and comorbidity yet no antibiotic therapy (‘controls’). Stool was collected from case and control patients on days 7, 14, 30 and 90 after antibiotic initiation (day 1) and days 7 and 14 after admission, respectively, and analysed by whole-metagenome shotgun sequencing. The primary outcome was ARG abundance at day 30; secondary outcomes included microbiota-species composition and clustering over time.
Findings
Forty-five patients and 11 controls were included and evaluable; ARG analyses were conducted on the 29 per-protocol patients receiving 7 (±2) days or 14 (±3) days of antibiotic therapy. At day 30, ARGs were not detected at similar abundance in patients receiving 7 and 14 days (median counts/million [mCPM]: 96 versus [vs] 71; p=.38). By day 30, total ARG content between both groups was not significantly different from that of controls at D7 (362 and 370 mCPM vs 314 mCPM, p=.24 and 0.19). There were no significant differences amongst antibiotic-treated patients at any timepoint in bacterial diversity or clustering, but Shannon species diversity was significantly reduced compared to controls through day 14 (median 3.12 and 3.24 in the 7-day and 14-day groups vs 3.61 [controls]; p=.04 and 0.012). Patients treated for 14 days had reduced faecal phage content during and after therapy compared to other patient groups.
Interpretation
Reducing antibiotic durations by half did not result in decreased abundance of ARGs in patients treated for gram-negative bacteraemia, nor did it improve microbiota species diversity.
Funding
The study was funded by the University of Geneva's Louis-Jeantet Foundation (grant no. S04_12) and the Swiss National Science Foundation (NRP Smarter Healthcare, grant no. 407,440_167359).
Keywords: Resistome, Antibiotics, Faecal microbiota, Whole-metagenome shotgun sequencing, Treatment duration
Research in context.
Evidence before this study
We searched PubMed with the terms “antibiotics” AND “duration” AND “microbiota” AND “resistome” in ‘All Fields’ for previously published reviews and articles from database creation to December 15, 2020. Ten studies on the effects of large- and narrow-spectrum antibiotics administration on infant or adult microbiota have been documented in a systematic review. Antibiotic course is known to have dramatic effect on microbiota composition as well as in the selection of antibiotic-resistance genes (ARG). In particular, microbiota species diversity can be heavily decreased also after short-term therapy with narrow-spectrum antibiotics. However, whether shorter antibiotic treatments result in a reduction of genetic resistance in the intestinal microbiota as well as to an earlier recovery of microbiota diversity remains an open question. No study has examined the comparative effects of antibiotic therapy on the intestinal microbiota of patients randomized to different antibiotic durations.
Added value of this study
Our metagenomic study shows that short-term (≤7-day) antibiotic therapy for gram-negative bacteraemia did not prompt reduced ARG abundance nor improved microbiota diversity compared to longer (≥14-day) therapy.
We provide insights on how microbiota and resistome change upon short-term versus long-term antibiotic courses. In particular, we showed that one week of antibiotic therapy alone is already sufficient to drastically impact faecal microbiota and resistome composition, and does not result in faster recovery than that seen with two weeks of antibiotics.
Implication of all the available evidence
Antibiotic resistance has increased and antimicrobial drug development is slowing. One major factor driving resistance is antibiotic usage in clinical practice. Here we document that antibiotic therapy increases ARG abundance and compromises microbiota species diversity irrespective of its duration. Our findings highlight the importance of reinforcing surveillance on antibiotic administration.
Alt-text: Unlabelled box
1. Introduction
Antibiotic overuse is one of the key drivers of antibiotic resistance [1]. Epidemiologic data demonstrate that long antibiotic courses select for collateral resistance in the microbiota [2], putting patients at increased risk for later infections by antibiotic-resistant pathogens [3,4]. Metagenomic data are beginning to capture the immediate changes in gut microbiota composition and antibiotic-resistance genes (ARG) after administration of commonly used antibiotics such as ciprofloxacin [5] and imipenem [6]. Yet data are lacking as to whether patients randomized to significantly shorter antibiotic courses will see a proportionate reduction in detectable ARG in the intestinal microbiome. It is also as yet unknown whether those patients retain more microbiota diversity and experience earlier recovery.
To answer these questions, we analysed faecal samples collected from hospitalized patients at the time of their inclusion in the ‘PIRATE’ study [7,8], a multicentre, point-of-care-randomized trial assessing clinical non-inferiority of 7-day and individualized (C-reactive-protein [CRP]-guided) antibiotic courses to 14-day courses for gram-negative bacteraemia, and in the three months thereafter. The PIRATE study confirmed non-inferiority of shorter antibiotic durations, with similar clinical cure rates in all arms in the three months following randomization.
2. Methods
2.1. The ‘PIRATE RESISTANCE project’: study design and participants
Between July 2018 and April 2019, this nested prospective observational cohort study included adult (≥18 years) patients hospitalized at the Geneva University Hospitals and enroled in the PIRATE trial (‘cases’) and, simultaneously, hospitalized patients with similar demography and comorbidity but without antibiotic therapy (‘controls’). Entry criteria for the PIRATE trial are described elsewhere [7,8]; briefly, immunocompetent adults with gram-negative bacteraemia without complications (e.g., abscess) or source requiring long-term therapy (e.g., endocarditis) were eligible.
Given the lack of data allowing for hypothesized effect sizes, the sample size was not calculated; a convenience sample of 51 cases and 11 controls was chosen, with its size guided chiefly by budgetary and logistic feasibility. Stool samples were collected from case patients on days 7 (±2) (D7), 14 (±3) (D14), 30 (±7) (D30) and 90 (±14) (D90) after antibiotic initiation and from control patients on days 7 (±2) and 14 (±3) after hospital admission. Characteristics of the study cohort are reported in Table S1. Importantly, control patients were not treated with antibiotics in the three months preceding inclusion into PIRATE RESISTANCE.
2.2. Antibiotic therapy duration and patient group assignment
For each patient, antibiotic duration was tabulated in days; a gap of ≤24 h mid-course (e.g., missed dose) was accepted as one day of therapy. Patients were classified by treatment duration and analysis population (Fig. S1, Table S2).
Three analysis populations were created. All enroled case patients but one were included in the ‘baseline population’: one patient (number 159) was excluded from all analyses because he had received vancomycin for 22 days prior to enrolment. Patients in the baseline population were subclassified into ‘group S’ (for “short”; antibiotic duration ≤10 days) or ‘group L’ (for “long”; duration >10 days).
The ‘per-protocol’ population consisted of case patients with the following criteria:
-
(1)
≥2 faecal samples, one being a baseline sample (D7 or D14) and the other a D30 sample (primary outcome),
-
(2)
treatment duration of either 7 (±2) (‘D7’) or 14 (±3) days (‘D14’), and
-
(3)
no further receipt of antibiotics after D14.
The third population consisted of control patients. Results of the per-protocol analyses are presented here; results from baseline-population analyses can be found in the supplement.
2.3. Stool sampling
Stools were collected from a Commode Specimen Container (Covidien Medtronic, Dublin, Ireland) placed over the toilet-seat opening. A few nut-sized sections of faecal material were added to Sarstedt feces Tube 76 × 20 mm (Nümbrecht, Germany) and then refrigerated at 4 °C. Samples were delivered within 24 h to the Genomic Research Laboratory for storage at −80 °C until DNA extraction.
2.4. DNA extraction and whole-metagenome shotgun sequencing
DNA was extracted from about 150 mg of stool (95–300 mg) using Quick-DNA Fecal/Soil Microbe Miniprep Kit (Zymo) and eluted in 55 µL of water. Bacterial and human DNA were quantified by qPCR and stored at –20 °C.
The concentration of human DNA was determined by qPCR experiments targeting human β-actin genes on a Bio-Rad CFX96 qPCR system. The assay was performed in 20 µL of ABsolute qPCR Mix (Thermo Scientific) containing 300 nM each forward and reverse β-actin primers, 200 nM β-actin probe and 1 µL of non-diluted DNA extract. The amplification parameters were as follows: 95 °C/15 min, followed by 95 °C/15 s and 60 °C/60 s for 42 cycles. Human genomic DNA used at known concentrations to generate the reference curve and β-actin primers and probe were from the Applied Biosystems TaqMan β-Actin Control Reagents kit (Thermo Fisher Scientific).
The concentration of bacterial DNA was determined using primers targeting the V3 region of bacterial 16S rRNA genes (Escherichia coli positions 338–534) on an Mx3005P qPCR system (Agilent) as previously described [9]. One microlitre of the 1/10 diluted DNA extract was added to the qPCR reaction mix. The cycling conditions included an initial step of 10 min at 95 °C followed by 40 cycles of 95 °C for 5 s and 68 °C for 20 s. All reactions were carried out in duplicate and the reference curves for DNA absolute quantification were obtained using known concentrations of genomic DNA of E. coli strain DH5α.
Four no-sample controls were performed by extracting DNA using the same extraction procedure but omitting the addition of stool sample. After DNA fragmentation by sonication, the sequencing library was constructed using TruSeq Nano DNA sample preparation kit (Illumina) under conditions aimed at producing the insert size of 250 bp. Library was sequenced (2 × 150) on a NovaSeq 6000 System (Illumina) at Fasteris (Plan-les-Ouates, Switzerland). In total we sequenced 161 samples: 136 were from 46 case patients, 21 were from 11 control patients, and the remaining four were from no-sample sequencing controls.
2.5. Quality filtering, dereplication and taxonomic assignment
The quality of fastq files was inspected with FastQC v0.11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Raw reads were scanned with Trimmomatic v0.36[10] with a sliding window of 10 nt. Reads were trimmed when the average quality within the window fell below a Phred score of 30 and filtered out if their length before or after trimming step was <120 nt. Quality filtered reads were dereplicated with in-house Perl script (no publicly available). Reads which were classified by CLARK v1.2.5 [11] to the species Homo sapiens using a confidence score of 0.5, were removed. For this step, the human genome reference assembly vGRCh38.p7 was used. Dereplicated reads classified as non-human were then assigned by CLARK to bacterial, viral and fungal species when a confidence score of 0.8 was obtained (for genome reference database, see below). The relative abundance of bacterial species was expressed as percentage of total number of reads mapping to bacteria.
Since phages can influence the abundance of commensal species [12] and may play a role in the dissemination of ARG in the microbiome [13,14], we also aimed to capture the dynamics of changes in intestinal phage content according to therapy duration. We did so by selecting reads that remained unclassified in mapping to human, bacterial, viral and fungal species. Then, we queried those reads against phage genomes by CLARK with a confidence score of 0.8.
The abundance of phages was computed by dividing the total number of reads mapping to phages with the total number of dereplicated non-human reads; such ratio was then multiplied by one million (counts per million; CPM).
2.6. Filtering of bacterial species
We filtered out species for which the ratio between mean relative abundance in negative extraction controls and mean relative abundance in test samples was superior to 1. We retained 3020 of 5373 (56%) bacterial species for further analyses. On average, 5.6 million read pairs per sample were mapped to those selected species.
2.7. Identification of genes encoding antibiotic resistance
Dereplicated and quality-filtered forward non-human reads were mapped against Resfinder database [15] by USEARCH v11.0.667 [16] using the following settings: -id 0.9 -strand both -top_hits_only -mincols 100 -maxaccepts 20.
Generally, one read was associated with one best hit (single best hit). In case one read mapped to more than one best hit (multiple best hits), we chose the hit that was the most abundant in the sample. When two or more best hits had the same abundance for a given read, their names were concatenated and considered as a new hit. Best hits were labelled with the acronym combining the gene name(s) and corresponding accession number(s). Roughly 5800 forward reads per sample were classified to 904 different ARG hits. Importantly, we used forward reads for two reasons: first the sequencing quality of forward reads was better than reverse reads thereby allowing a more reliable detection of ARGs; second, mapping both forward and reverse reads might lead to spurious hits. Of 904 identified ARGs, 456 [50.4%] resulted from combining equally abundant multiple best hits.
In the present study, the ARG class takes the name of the antibiotic class against which a given gene encodes resistance. We assigned each hit to the corresponding ARG class by querying the gene name against the table reported in the ResFinder file “phenotype.txt”. When the hit was not assigned to any class in “phenotype.txt”, we queried the gene name of the hit against the table reported in the ResFinder “notes.txt” file. If thereafter no association was identified, antibiotic class was manually assigned. Importantly, one read could be assigned to more than one ARG class because it mapped to different genes from different ARG classes. When this happened, the classes’ names were combined and treated as a new class.
The abundance of ARGs and classes was normalized (per sample) to bacteria-assigned reads: read counts assigned to an ARG or to an ARG class in a given sample were divided by the total number of reads classified to bacteria. The ratio was then multiplied by one million and ARG relative abundance expressed as counts per million (CPM) of bacterial reads.
For each individual, we investigated resistome composition according to the type of antibiotic administered to case patients (beta-lactam, quinolone, folate-pathway antagonist [FPA], aminoglycoside, macrolide and metronidazole).
To gain a temporal resolution of resistome changes, we analysed the relative abundance of ARGs encoding resistance against the antibiotic classes used in the study, except for metronidazole, by treatment duration and distance from last treatment day. For each antibiotic class, we considered only patients receiving antibiotics of that class.
2.8. Databases
The two databases used to classify reads to bacteria, archaea, viruses and fungi, and to phages, respectively, were downloaded from NCBI on 13 December 2019. The first contained 5550 bacterial, 257 archaeal, 1000 viral and 288 fungal genomic sequences, the second 2450 and 23 genomes belonging to the orders of Caudovirales and Ligamenvirales, respectively. The ResFinder database was downloaded on 7 January 2020.
2.9. Ethics
The present study was approved by the Cantonal Ethics Committee of Geneva (no. 2017-0018); all participants provided written informed consent before enrolment.
2.10. Statistics
Only complete data on the variables accounted in a given analyses were considered. Principal coordinates analysis (PCoA) was performed to visually explore differences between microbiota species profiles (so-called ‘beta diversity’) amongst case and control patients at a given time point. PCoA analysis reduces the number of the variables determining the microbiota profile (e.g. all microbiota species) to two abstract dimensions that capture the highest variance present in the dataset. These two dimensions are then used to draw a plot where microbiota profile from each sample is represented as a dot. The closer two dots are on the plot, the more similar the corresponding microbiota profiles will be. PCoA were performed in R v3.2.3 using the vegan v2.3–5 package [17]. Bray-Curtis dissimilarity was computed on square-rooted species relative abundance with vegdist R function (vegan). Samples coordinates were computed with betadisper R function (vegan).
Shannon diversity was computed with diversity R function (vegan). The formula used for the diversity index computation is here pi represents the relative abundance of the ith species (the number of reads mapping to the ith species, divided by the total number of reads). For species richness we considered the number of species with read counts > 0 in a given sample. Counts of reads classified to bacterial species were rarefied to 500,000 with rarefy R function (vegan) prior to computation of Shannon and richness indices.
To investigate differences in microbiota composition between groups of patients, we applied permutational analysis of variance (pairwise PERMANOVA, with 9999 permutations, unrestricted permutation of raw data and Type III sums of squares) and distance-based linear models (DistLM, with 9999 permutations) based on the Bray-Curtis similarity matrices of square-root-transformed species’ relative abundance. PERMANOVA and DistLM were performed in PRIMER v7 (PRIMER-E Ltd, Plymouth, UK). Importantly, when PERMANOVA was performed in presence of covariables (e.g., age and comorbidity indices), we used the following settings: permutation of residuals under a reduced model; fixed effects sum to zero for mixed term; 9999 permutations.
A result was considered significant when its associated p was <0.05 (two-sided).
We selected species whose changes in relative abundance between two conditions were associated with a Wilcoxon signed-rank and/or Wilcoxon rank-sum test p value <0.05. Importantly, the conclusions from these analyses were based on the results of statistical tests applied.
For graphical representation, we took into account mean relative abundance and the number of samples in which the species was detected as additional parameters to select species along with fold change between conditions, thus avoiding taxa associated with significant p values yet of low prevalence in samples.
Data visualization was performed in R with ggplot2 v3.3.2 package.
2.11. Role of the funding source
This study was initiated by the investigators. Funders had no role in project design, during sample collection and project execution, analyses, results interpretation, report writing and in any other aspects concerning the study (e.g. journal submission).
3. Results
Forty-six case patients and 11 control patients were included; amongst case patients, 45 and 29 were included in the baseline and per-protocol populations, respectively (Fig. S1, Table S2). In the baseline population, 23 patients received ≤10 days of antibiotic therapy (group S) and 22 >10 days (group L). In the per-protocol population, 15 patients received 7 days of therapy and 14 received 14 days. Patient demographics and individual antibiotic regimens are described in Tables S1 and S2, respectively. We did not find significant associations between microbiota species composition of faecal samples with age or comorbidity index (Table S3).
3.1. The resistome
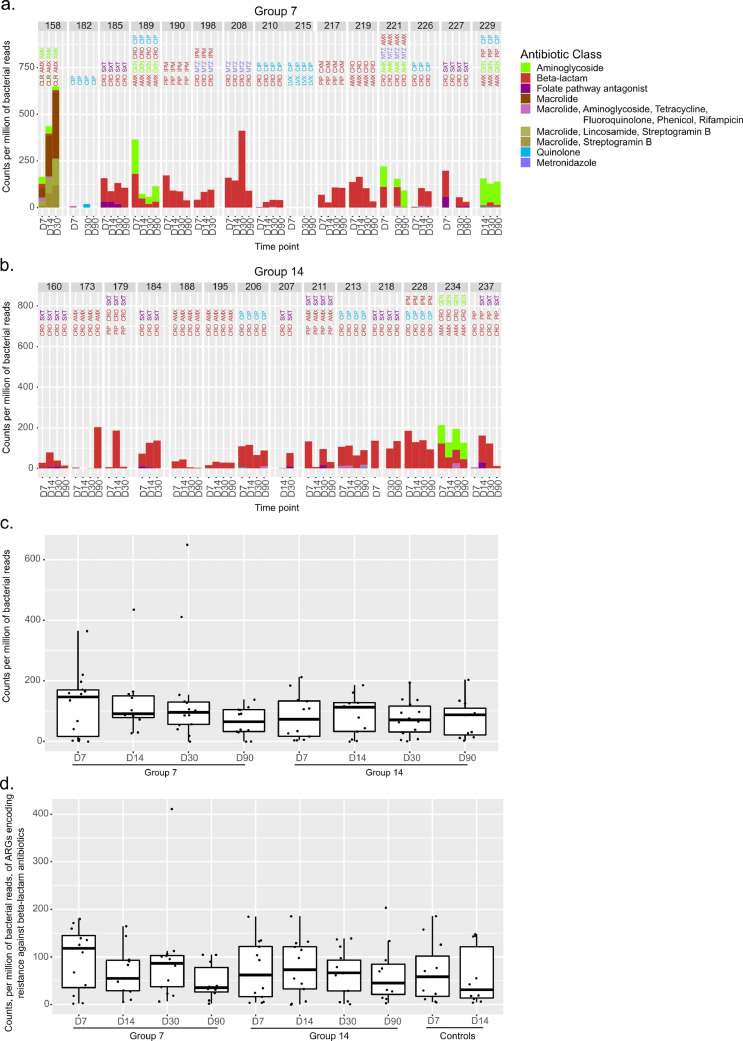
We identified ARGs encoding resistance against 14 unique antibiotic classes and 10 combinations of ≥2 antibiotic classes. In per-protocol patients, ARGs conferring resistance to the antibiotic administered were identified (Fig. 1a-b). Yet abundance of ARGs at day 30 did not differ significantly between patients receiving 7 days and those receiving 14 days (median counts/million (mCPM): 96 versus 71, Wilcoxon rank sum test p=.38; Fig. 1c). Indeed, when assessing only ARGs conferring resistance to antibiotics given to the patient, the fractions of patients with decreased ARG abundance at D30 were similar in the 7-day and 14-day groups (6/13 [0.46] and 7/13 [0.54], Fisher's exact test p = 1; Table 1).
Fig. 1.
Resistome abundance compared to antibiotic therapy. (a.-b.) Bar plots representing the abundance of antibiotic-resistance-encoding genes in faecal samples at time points (D7, D14, D30 and D90) of patients assigned to 7 days (a.) or 14 days of antibiotic therapy (b.). For each patient, we selected genes encoding resistance against the antibiotics used in treatments till a given time point. When a time point is not included, faeces were not sampled. Antibiotics’ abbreviations are reported above bars and coloured according to the corresponding antibiotic class. Reads mapping to genes conferring resistance to the same class of antibiotic were summed and normalized to the number of reads mapping to bacteria. The y-axis thus reports the number of reads mapping to a given class per million of bacterial reads. Like antibiotics’ abbreviations, bars are coloured according to antibiotic classes (see Abbreviations section). (c.) Boxplots reporting the changes of selected components of the resistome and summarizing data shown in panels a. and b. Dots represent single values. (d.) Boxplots reporting the changes of ARGs conferring resistance to beta-lactam antibiotics in patients treated with this class of antibiotic and in controls.
Table 1.
Summary table reporting the number of patients receiving 7- or 14-day antibiotic courses for whom a decrease or increase in resistome abundance was observed when comparing time points. The number of the patients for each comparison refers to Fig. 1.
| Antibiotic duration | Comparison | Decrease in ARGs abundance at time point 1 | Increase in ARGs abundance at time point 1 | ARGsnot detected | Total no. of samples with detected ARGs | Total no. of samples collected at both time points 1 and 2 | Total no. of patients per group | |
|---|---|---|---|---|---|---|---|---|
| Time point 1 | Time point 2 | |||||||
| 7 days | D30 | D7 | 6 /13 [0.46] | 7/13[0.54] | 1 | 13 | 14 | 15 |
| D30 | D14 | 5/11 [0.45] | 6/11[0.55] | 11 | 11 | |||
| D14 | D7 | 5/10 [0.5] | 5/10 [0.5] | 10 | 10 | |||
| 14 days | D30 | D7 | 7 /13 [0.54] | 6/13 [0.46] | 13 | 13 | 14 | |
| D30 | D14 | 7/13 [0.54] | 6/13 [0.46] | 13 | 13 | |||
| D14 | D7 | 4/12 [0.33] | 8/12 [0.67] | 12 | 12 | |||
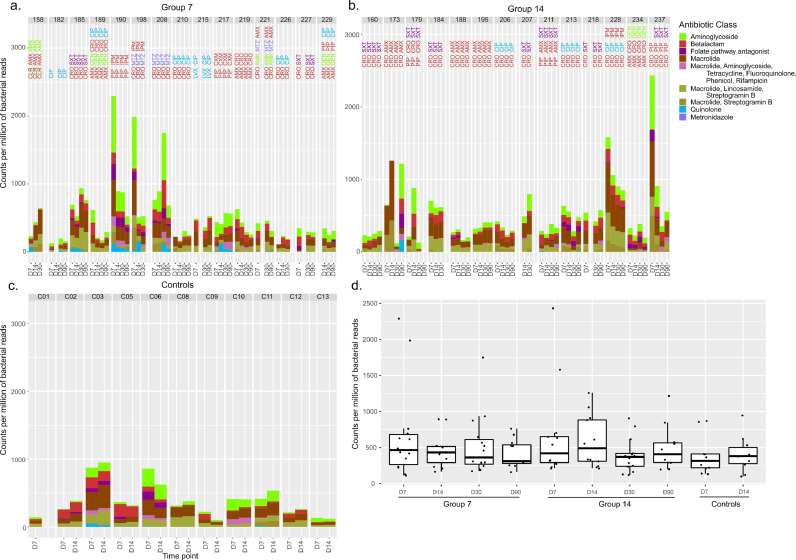
Further, ARGs conferring resistance to a given antibiotic class were also found in control patients and in case patients not receiving the antibiotic class (Fig. 2c-d). When analysing all ARG content at D30 in the 7- and 14-day groups, we found a median relative abundance of 362 mCPM (interquartile range [IQR] 270–611) and 370 mCPM (IQR 239–419; Wilcoxon rank sum test p=.425), respectively. Relative abundance of ARGs of controls did not differ significantly from that of case patients (median 314 mCPM [IQR 218–412] at D7 and 380 mCPM [IQR 278–502] at D14; Fig. 2d).
Fig. 2.
Resistome abundance in treated and control patients. (a.-b.-c.) Bar plots as in Fig. 1, except that the temporal changes in abundance are reported for genes encoding resistance against all the antibiotics used to treat bacteraemia. Values are reported for the cohort of 29 treated patients and for 11 untreated controls (see Abbreviations section). (d) Boxplots reporting the changes of the resistome and summarizing data shown in panels a., b. and c. Dots represent single values.
Median values of ARG normalised counts peaked at the end of treatment for both case-patient groups (D7 and D14, respectively; Figs. 1c and 2d). Since all patients except two from the 7-day group (patients 182 and 215) were treated with beta-lactam antibiotics (Table S2), we examined the temporal changes specifically of beta-lactam-resistance-encoding ARG (Fig. 1d): medians were lower at D30 (end of therapy) than on-treatment time points in both case-patient groups (Fig. 1d); such differences were not found to be statistically significant.
3.2. ARG abundance by treatment duration and distance from last treatment day
Increases in ARG targeting metronidazole were not detected in patients treated with it (Fig. S2). Only folate-pathway-antagonist (FPA) use was associated with FPA-targeting ARGs, with their abundance decreasing after FPA use was discontinued. Resistance against antibiotics persisted days after the end of antibiotic treatment irrespective of treatment duration (Fig. S2). More specifically, we detected ARGs 23 (IQR 10-77) versus 18 (IQR 11-74) days after therapy discontinuation in the 7- and 14-day groups.
3.3. Microbiota species composition
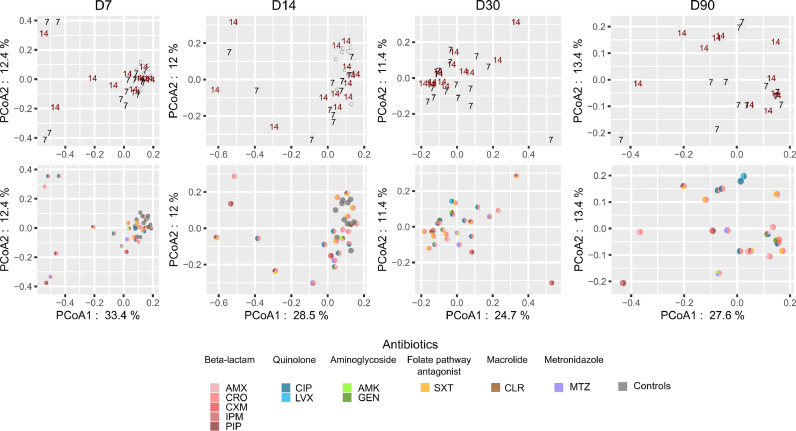
When all species were analysed together, microbiota composition at D30 between patients receiving 7 and 14 days did not differ significantly (PERMANOVA test, t = 1.03, p=.3).
In PCoA plots, D30 samples tended to cluster by treatment duration and by antibiotic treatment (Fig. 3). The sample size did not allow for assessment of clustering at the antibiotic level.
Fig. 3.
Global species microbiota profiles in the 7-day and 14-day groups and in controls. Principal coordinates analysis (PCoA) plots representing global differences between bacterial species communities of faecal samples at a given sampling time point. Top row: PCoA plots where samples are labelled according to treatment duration: 7 = 7-day group; 14= 14-day group. C= control patients. Bottom row: dots are coloured according to antibiotic treatment. Antibiotics belonging to the same class have similar colours. For time points D7 and D14, samples from control patients are also shown (grey dots). The percentage of total data variance is reported for PCoA1 and PCoA2.
Microbiota species composition amongst case patients at D7 and D14 differed significantly from that of control patients (Table S4). When age and comorbidity index were included as covariables in such analyses, alone or in combination, several differences in microbiota composition remained significant (PERMANOVA test p<.05), (Table S4). Species richness and diversity were significantly reduced in per-protocol patients receiving 7 days of antibiotic therapy at D7 (Wilcoxon rank sum test p<.02) and D14 (p<.05) compared to controls (Fig. S3; Table S5). For patients receiving 14 days, only Shannon diversity was significantly decreased at D7 and at D14 compared to controls (Table S5). In addition, in both treated patient groups these ecological indices increased at D30 and D90 compared to D7 and D14 (Fig. S3).
We analysed the temporal changes of median values of the Firmicutes to Bacteroidetes (F:B) ratio over time and observed that medians increased in later time points in both groups (Fig. S4a). However, the F:B ratio increased in 8/14 and decreased in 8/13 patients from group 7 and 14, respectively, at D30 compared to the end-of-treatment time points (D7, D14 for group 7 and 14, respectively). These differences were not statistically significant. The temporal changes observed for ratio reflect the dynamics of changes in relative abundance observed for Firmicutes (Fig. S4c).
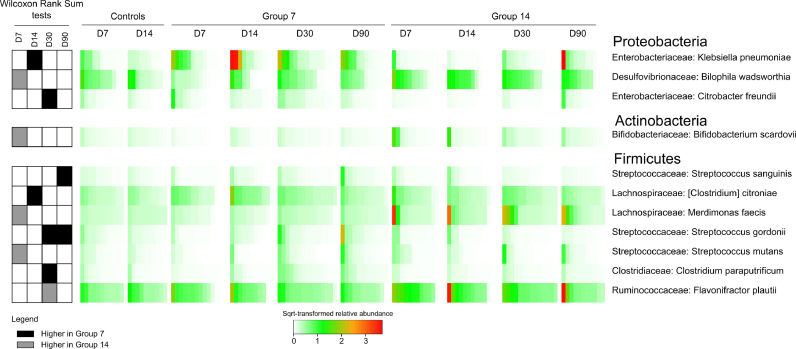
In per-protocol patients, Klebsiella pneumoniae and Citrobacter freundii abundance decreased significantly at D14 and D30, respectively, in patients receiving 14 days versus 7 days (Fig. 4). Conversely, Flavonifractor plautii (Ruminococcaceae) abundance significantly increased at D30 in patients receiving 14 days versus 7 days (Fig. 4). Members of the Bifidobacteriaceae (Actinobacteria), Clostridiaceae, Lachnospiraceae, Peptostreptococcaceae and Ruminococcaceae (Firmicutes) families were significantly more abundant at D30 and D90 than they were at D7 in per-protocol patients receiving 7 days of antibiotics (Fig. S5). Alistipes obesi, Anaerobutyricum hallii, Coprococcus comes, Romboutsia timonensis, Roseburia intestinalis and Veillonella parvula became more abundant by D30 in comparison to the end-of-treatment time point in all per-protocol patients (Figs. S5 and S6).
Fig. 4.
Differentially abundant species between patients treated for 7 days and 14 days. For each of the represented species, we report the significance of differences in the relative abundance between the 7-day and 14-day groups at a given time point (heat map on the left), the relative abundance in the groups (heat map in the middle) and the corresponding phylum and family (on the right). We reported species with differences in relative abundance between the 7- and 14-day groups associated with a significant Wilcoxon rank sum test (p < .05), a fold change ≥ 1.5 in the relative abundance and a mean relative abundance ≥ 0.05% in at least one of the compared groups. Finally, the species had to be detectable (relative abundance higher than 0) in at least 3 samples of one of the compared groups. Heat map on the left: coloured cells report significant differences (p < .05) associated with a ≥ 1.5 fold change in the relative abundance whereas white cells correspond to other cases. In particular, black represents the species significantly more abundant in the 7-day group whereas grey those significantly more abundant in the 14-day group. Heat map in the middle: square-root-transformed relative abundance is colour-scaled as indicated in the legend.
3.4. Faecal phage composition
On average 0.14% of human-depleted reads mapped to known phage sequences. We detected 875 different phages amongst which the five most abundant were specific to Escherichia (11.7%), Pseudomonas and Salmonella (7.5%), Staphylococcus (7.4%) and Lactococcus (6.4%) hosts. Overall phage content was ∼4-fold and ∼3-fold decreased in the 14-day group (221 mCPM per sample) compared to the 7-day group (809 mCPM per sample, Wilcoxon rank sum test p=.012) and controls (626 mCPM per sample, p=.105), respectively. The difference in phage content was less marked between the 7-day group and control samples (p=.859).
We investigated the temporal dynamics of detected phages in both case and control patients. We observed that the median values of overall phage content in the 7-day group decreased over time and dropped at D30 (344 mCPM) compared to D7 (1021 mCPM, Wilcoxon signed-rank test p = 1) and D14 (622 mCPM, p= .577; Fig. S7a). On the other hand, median values of phage content remained relatively stable over time for the 14-day group and controls (Wilcoxon signed-rank test p>.1 and = 0.049, respectively; Fig. S7a).
Observations made at the population level were accompanied by high variability amongst individual patients of the same group (Fig. S7b). Roughly half of the 7-day patients had decreased phage content at D30 compared to D7 (7/14) and D14 (6/11).
The number of ≥2-fold increase in CPM D30 vs D7 and D30 vs D14 were comparable between group 7 and group 14 (D30 vs D7: 5/14 and 3/13; D30 vs D14: 2/11 and 2/13, respectively). For ≥2-fold decrease in CPM, we observed similar trends (D30 vs D7: 5/14 and 3/13; D30 vs D14: 4/11 and 4/13).
4. Discussion
In this metagenomic study of patients randomized to different durations of antibiotic therapy for gram-negative bacteraemia, we did not find reduced ARG abundance or improved microbiota diversity in patients receiving shorter antibiotic courses. Patients treated for only seven days had higher median amounts of ARGs on day 30, but not on day 90, as those treated for 14 days. These results suggest that one week of antibiotic therapy alone suffices to set in motion changes to the resistome and microbiota that cannot easily be reversed in the short-term.
Despite individual patient variability, we observed differences in phage abundance: patients treated for 14 days had consistently suppressed phage content at all time points, except when treated with anti-FPA drugs. The significance of this finding is difficult to determine. First, the observed differences in phage content were not significant between patient groups; second, the identification of most phage-host pairs in the intestinal microbiota remains uncharacterized, and we are currently able to work only with known phage sequences, leaving much viral ‘dark matter’ undefined [18].
When comparing antibiotic-treated patients to untreated control patients, significant differences in both microbiota profiles and species diversity were apparent, as observed in other metagenomic studies [19,20]. Once antibiotic therapy was discontinued, case patients saw a gradual recovery in microbiota species composition, with their microbiota profiles approaching those of control patients at later time points. Yet inter-individual variability was high and depended on the antibiotic class received and, likely, other factors (e.g., age and comorbidities). Indeed, when analysing other parameters such as the Firmicutes-to-Bacteroidetes ratio and ecological indices, there was no shared pattern of changes.
Given this variability, we performed pairwise comparisons between groups of treated patients to analyse which species were affected by treatment duration. Longer antibiotic treatment seemed to be accompanied by decreased abundance of K. pneumoniae and C. freundii, both Enterobacteriaceae. Therefore, a prolonged antibiotic regimen would affect more the abundance of these species in the intestine than a short-term treatment, at least in a setting where multidrug-resistant strains of these bacteria are still relatively rare. Yet, in all case patients, antibiotic discontinuation resulted in a significant increase of the butyrate-producing species A. hallii, C. comes, and R. intestinalis; butyrate plays an important role in preserving intestinal epithelium from inflammation [21], in opposing pathogen expansion and in enhancing pathogen clearance from the gut [22].
This study has limitations. Patients were randomized to antibiotic duration only and could receive different antibiotics for their bacteraemia, a variability that reduces the nested study's statistical power given its limited sample size. We were unable to collect samples from patients before their antibiotic therapy began, as bacteremia cannot be confirmed until day 2 or 3 of antibiotic therapy. However, the inclusion of several untreated hospitalized patients as controls mitigated this limitation. Detection of ARG was limited to acquired genetic antibiotic resistance; chromosomal mutations, which might lead to similar resistance phenotypes, were not investigated. Selection of differentially abundant species was based on arbitrary cut-offs on relative abundance and fold change, potentially missing information on other important species. Furthermore, we were unable to study how antibiotic duration impacts transmission of multidrug-resistant organisms (MDRO). Most clinically relevant MDRO such as extended-spectrum beta-lactamase or carbapenemase producing Enterobacterales or methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus species are not selected de novo through antibiotic exposure but rather acquired through transmission from other patients or the environment. It is likely that the impact of antibiotic duration on transmission of MDRO shows a different dynamic than the impact on the intestinal microbiota.
Bacteraemic patients randomized to shorter antibiotic courses did not have fewer antibiotic-resistance genes or improved intestinal microbiota diversity in the weeks after therapy, suggesting that one week of antibiotic therapy alone is enough to significantly impact the intestinal microbiota and resistome. The sample size of this cohort, however, allows a certain degree of uncertainty surrounding the results. Thus, larger studies are needed to better characterize these changes and better determine how their impact might be minimized.
Abbreviations
| Acronym | Full name | Antibiotic class |
| AMK | Amikacin | Aminoglycoside |
| GEN | Gentamicin | Aminoglycoside |
| AMX | Amoxicillin | Beta-lactam |
| CRO | Ceftriaxone | Beta-lactam |
| CXM | Cefuroxime | Beta-lactam |
| FEP | Cefepime | Beta-lactam |
| IPM | Imipenem | Beta-lactam |
| PIP | Piperacillin | Beta-lactam |
| CIP | Ciprofloxacin | Quinolone |
| LVX | Levofloxacin | Quinolone |
| SXT | Sulfamethoxazole (with trimethoprim) | Folate-pathway antagonist |
| CLR | Clarithromycin | Macrolide |
| MTZ | Metronidazole | Metronidazole |
Contributors
BH, VL and AH conceptualized and supervised the study. EVD, VP and AH collected the data. SL, VL and AH contributed to the analyses and interpretation of data, and drafting and revision of the manuscript. SL, VL and AH verified the underlying data. EVD, LK, VP, JS and BH contributed to the interpretation of data and revision of the manuscript. All authors read and approved the final version of the manuscript.
Funding
The PIRATE RESISTANCE project was funded by the University of Geneva's Louis-Jeantet Foundation (grant no. S04_12) and the Swiss National Science Foundation (NRP 74 Smarter Healthcare, grant no. 407440_167359).
Data Sharing
Quality-filtered dereplicated non-human reads are available on European Nucleotide Archive Database under the study accession number PRJEB40995 (https://www.ebi.ac.uk/ena/browser/view/PRJEB40995). Patient-sample (time point) identifier of each FASTQ file can be visualized on the ENA study webpage by clicking “Sample title” option in “Read files” tab. Codes for metagenomic analyses are not publicly available as they do not contain further information (e.g. values of parameters for sequencing read processing and data mining) beyond those already provided in the present work. Supplementary Figures and Tables are available in Supplementary appendix.
Declaration of Competing Interest
All authors declare no competing interests.
Acknowledgments
We thank Ms. Myriam Girard for wet-lab work and Ms. Nadia Gaïa for bioinformatics support. We thank Ms. Donatienne Wynar of the Clinical Research Centre of the Geneva University Hospitals and Faculty of Medicine for logistic support and Dr Shawna McCallin for assistance with data exportation and compilation.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103566.
Appendix. Supplementary materials
References
- 1.Holmes A.H., Moore L.S., Sundsfjord A., Steinbakk M., Regmi S., Karkey A. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2015 doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 2.Bell B.G., Schellevis F., Stobberingh E., Goossens H., Pringle M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect Dis. 2014;14:13. doi: 10.1186/1471-2334-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zarkotou O., Pournaras S., Tselioti P., Dragoumanos V., Pitiriga V., Ranellou K. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect. 2011;17(12):1798–1803. doi: 10.1111/j.1469-0691.2011.03514.x. [DOI] [PubMed] [Google Scholar]
- 4.Chastre J., Wolff M., Fagon J.Y., Chevret S., Thomas F., Wermert D. Comparison of 8 vs 15 days of antibiotic therapy for ventilator-associated pneumonia in adults: a randomized trial. JAMA. 2003;290(19):2588–2598. doi: 10.1001/jama.290.19.2588. [DOI] [PubMed] [Google Scholar]
- 5.Stewardson A.J., Gaïa N., François P., Malhotra-Kumar S., Delémont C., Martinez de Tejada B. Collateral damage from oral ciprofloxacin versus nitrofurantoin in outpatients with urinary tract infections: a culture-free analysis of gut microbiota. Clin Microbiol Infect. 2015;21(4):344. doi: 10.1016/j.cmi.2014.11.016. e1-.e11. [DOI] [PubMed] [Google Scholar]
- 6.Grall N., Lazarevic V., Gaïa N., Couffignal C., Laouénan C., Ilic-Habensus E. Unexpected persistence of extended-spectrum β-lactamase-producing Enterobacteriaceae in the faecal microbiota of hospitalised patients treated with imipenem. Int J Antimicrob Agent. 2017;50(1):81–87. doi: 10.1016/j.ijantimicag.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Huttner A., Albrich W.C., Bochud P.-.Y., Gayet-Ageron A., Rossel A., Ev Dach. PIRATE project: point-of-care, informatics-based randomised controlled trial for decreasing overuse of antibiotic therapy in Gram-negative bacteraemia. BMJ Open. 2017;7(7) doi: 10.1136/bmjopen-2017-017996. -e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Dach E., Albrich W.C., Brunel A.-.S., Prendki V., Cuvelier C., Flury D. Effect of C-reactive Protein–guided antibiotic treatment duration, 7-day treatment, or 14-day treatment on 30-day clinical failure rate in patients with uncomplicated Gram-negative bacteremia: a randomized clinical trial. JAMA. 2020;323(21):2160–2169. doi: 10.1001/jama.2020.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazarevic V., Gaïa N., Girard M., Schrenzel J. Decontamination of 16S rRNA gene amplicon sequence datasets based on bacterial load assessment by qPCR. BMC Microbiol. 2016;16:73. doi: 10.1186/s12866-016-0689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ounit R., Wanamaker S., Close T.J., Lonardi S.C. fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 2015;16(1):236. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornuault J.K., Petit M.-.A., Mariadassou M., Benevides L., Moncaut E., Langella P. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018;6(1):65. doi: 10.1186/s40168-018-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Schaik W. The human gut resistome. Philos Trans R Soc Lond B Biol Sci. 2015;370(1670) doi: 10.1098/rstb.2014.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerin E., Hill C. Shining Light on Human Gut Bacteriophages. Front Cell Infect Microbiol. 2020;10(481) doi: 10.3389/fcimb.2020.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 17.J. Oksanen, F.G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P.R. Minchin, R.B. O'Hara, G.L. Simpson, P. Solymos, M. Henry, H. Stevens, E. Szoecs, H. Wagner. vegan: community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan. 2019.
- 18.Sutton T.D.S., Hill C. Gut bacteriophage: current understanding and challenges. Front Endocrinol (Lausanne) 2019;10(784) doi: 10.3389/fendo.2019.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leo S., Lazarevic V., Girard M., Gaïa N., Schrenzel J., de Lastours V. Metagenomic characterization of gut microbiota of carriers of extended-spectrum beta-lactamase or carbapenemase-producing Enterobacteriaceae following treatment with oral antibiotics and fecal microbiota transplantation: results from a multicenter randomized trial. Microorganisms. 2020;8(6) doi: 10.3390/microorganisms8060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willmann M., Vehreschild M.J.G.T., Biehl L.M., Vogel W., Dörfel D., Hamprecht A. Distinct impact of antibiotics on the gut microbiome and resistome: a longitudinal multicenter cohort study. BMC Biol. 2019;17(1):76. doi: 10.1186/s12915-019-0692-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tedelind S., Westberg F., Kjerrulf M., Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13(20):2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorbara M.T., Dubin K., Littmann E.R., Moody T.U., Fontana E., Seok R. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216(1):84–98. doi: 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.