Abstract
Background
Human sinoatrial cardiomyocytes are essential building blocks for cell therapies of conduction system disorders. However, current differentiation protocols for deriving nodal cardiomyocytes from human pluripotent stem cells (hPSCs) are very inefficient.
Methods
By employing the hPSCs to cardiomyocyte (CM) in vitro differentiation system and generating E2A-knockout hESCs using CRISPR/Cas9 gene editing technology, we analyze the functions of E2A in CM differentiation.
Findings
We found that knockout of the transcription factor E2A substantially increased the proportion of nodal-like cells in hESC-derived CMs. The E2A ablated CMs displayed smaller cell size, increased beating rates, weaker contractile force, and other functional characteristics similar to sinoatrial node (SAN) cells. Transcriptomic analyses indicated that ion channel-encoding genes were up-regulated in E2A ablated CMs. E2A directly bounded to the promoters of genes key to SAN development via conserved E-box motif, and promoted their expression. Unexpect enhanced activity of NOTCH pathway after E2A ablation could also facilate to induct ventricle workingtype CMs reprogramming into SAN-like cells.
Interpretation
Our study revealed a new role for E2A during directed cardiac differentiation of hESCs and may provide new clues for enhancing induction efficiency of SAN-like cardiomyocytes from hPSCs in the future.
Funding
This work was supported by the NSFC (No.82070391, N.S.; No.81870175 and 81922006, P.L.), the National Key R&D Program of China (2018YFC2000202, N.S.; 2017YFA0103700, P.L.), the Haiju program of National Children's Medical Center EK1125180102, and Innovative research team of high-level local universities in Shanghai and a key laboratory program of the Education Commission of Shanghai Municipality (ZDSYS14005).
Keywords: Transcription factor E2A, human embryonic stem cells; cardiomyocyte differentiation; sinoatrial node
Research in context.
Evidence before this study
As the classic bHLH protein, the role of E2A has been well demonstrated in lymphopoiesis. And a growing body of studies indicated E2A involves in regulating neural specification and differention in both embryonic mice and pluripotent cells. However, far less is known about its function in heart development. Cardiomyocytes differentiated from human pluripotent stem cells (hPSCs) provide a promising approach to reveal the regulatory mechanism in cardiac development. It is necessary to better understand the impacts of E2A in cardiac-specific differentiation of hPSCs.
Added value of this study
We found ablation of E2A expression markedly increased the ratio of nodal-like cardiomyocytes from original ∼5% to as high as 40% in our defined 2D cardiac-specific differentiation system of hESCs. E2A could bound to the promoter region of TBX5, SHOX2, and TBX3, and regulated expression of these genes key to SAN node development. Furthermore, the activity of NOTCH signaling pathway was also affected by E2A during hESC cardiac differentiation. NOTCH inhibitor partially blocked the increasing nodal-like phenotypes of E2A deficient CMs.
Implications of all the available evidence
Our data revealed a new role of the transcription factor E2A in hESC cardiac differentiation and provided new insights into promoting induction efficiency of SAN-like CMs from cardiac-specific differentiation of hPSCs.
Alt-text: Unlabelled box
1. Introduction
With prolongation of human lifespans, the overall morbidity and mortality caused by cardiac conduction system (CCS) diseases and sinoatrial node (SAN) malfunction are increasing [1]. Biological pacemaker has been proved to be promising for the treatment of CCS diseases [2,3]. However, human pacemaker cells, for the sake of future clinical application, are very difficult to obtain. Although it is now possible to derive SAN-like cardiomyocytes (CMs) from human pluripotent stem cells (hPSCs), the induction efficiency for nodal-like cells in current 2D cardiac-specific differentiation protocol is very low, usually at a ratio of less than 7-8% [4]. A recent study reported ∼40% efficiency for producing SAN-like CMs from hESCs based on 3D differentiation and flowcytometry sorting, which is a great breakthrough for the previous very low efficiency in deriving nodal-like cells [5].
E2A belongs to a member of E proteins that are known as class I helix-loop-helix (HLH) protein family. E2A functions in various developmental and disease processes by forming either homodimers or heterodimers with class II HLH proteins that further bind to the canonical or non-canonical E-box (CANNTG) motif to regulate target gene transcription [6,7]. The role of E2A in heart development and cardiac differentiation is not clear yet. Previous studies showed that E2A interacts with the transcription factor MESP1, a key regulator of cardiac progenitor cell (CPC) development [8,9]. E2A knockdown during early CMs differentiation indirectly up-regulated expression of cardiogenic mesoderm markers [10], while E2A overexpression impeded CMs maturation [11]. These data suggest that E2A plays a role in mammalian heart development and cardiac-specific differentiation of hPSCs.
Using the defined 2D cardiac-specific differentiation system of hESCs, we here tested our hypothesis on the role of E2A in CM differentiation. Surprisingly, we found that a substantially increasing number of E2A knockout hESC-derived CMs exhibited morphological and functional characteristics of SAN-like cells. E2A participated in regulating the expression of these genes key to SAN node development via direct or indirect ways and further controlled pacemaker cell committee fate. On one hand, the direct regulatory effect of E2A on TBX5, SHOX2, and TBX3 expression relied on the higy affinity with E-box motifs in the promoter region of these genes; and on the other hand, E2A knockout could lost the restraint of NOTCH signaling activity in late cardiac differentiation process which indirectly promoted the expression of vital regulators in SAN development and reprogrammed ventrical cardiomyocyte into nodal-like cells. Thus, our data revealed a new role for the transcription factor E2A in hESC cardiac differentiation and may provide new insights into promoting induction efficiency of SAN-like CMs from cardiac-specific differentiation of hPSCs in the future.
2. Methods
2.1. hESCs culture and differentiation
Human embryonic stem cells (hESCs) were maintained on culture plates coated with Matrigel (Corning, 356231) using mTeSR-1 medium (Stemcell Technologies, #85852), and passaged with Accutase (GIBCO, A1110501) at the ratio of 1:4 to 1:5 when confluence reached ∼90%. Cardiac differentiation of hESCs were induced by chronologically treating with WNT agonist CHIR-99021(Selleck, S2924) and antagonist IWR-1(Sigma, I0161). In brief, RPMI 1640 (Corning)/B-27 minus insulin (Life Technologies) containing 12μM CHIR-99021 were used to activate WNT signaling on Day 0 and 1. Medium was changed with RPMI/B-27 minus insulin on Day2. Cells were treated with fresh medium containing 5μM IWR-1 for another 48h. From Day5 onward, cells were cultured with CDM3 (RPMI1640; BSA; AscobicAcid) medium, and changed every other day. Beating CMs appeared on day 7-8 after differentiation. To further explore the possibility of E2A on regulating nodal-like cells differentiation via Notch signaling, the NOTCH inhibitor RO4929097 (Selleck, S1575) was applied to the CDM3 culture medium at a final concentration of 30 µM from day 9 to 15 of differentiation. CMs of D30 after cardiac-differentiation were utilized for downstream tests in this study.
2.2. Generation of E2A knockout hESC lines
To generate E2A null hESCs lines, we utilized CRISPR/CAS9 genome editing system. sgRNAs were designed targeting on exon 3 of the human E2A gene (http://crispr.mit.edu/) and cloned into the D10A-mutant nickase version of Cas9 (pX462 from Addgene). pX462-sgRNAa and pX462-sgRNAb were transfected into hESCs by electroporation (Neon Transfection System, ThermoFisher Scientific). E2A-null clones were picked and screened by Western blot and furtherly confirmed by genomic sequencing after selected with puromycin for one week.
2.3. Quantitative Real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen, 15596018) according to the manufacturer's instructions. 1 µg RNA was reverse-transcribed into cDNA by using ReverTra Ace qPCR RT Kit (TOYOBO). Real-time PCR was done using the Hieff qPCR SYBR Green Master Mix (YEASEN, 11202ES08) based on the manufacturers’ protocol on a Light-Cycler 96 System (Roche). Gapdh were used as an internal control. Primer sequences were listed in Supplementary Table 1.
2.4. Flow cytometry
CMs were treated with Collagenase I (Sigma) for 20 min ∼1 h, and dissociated into single cells with 0.25% Trypsin (GIBCO). CMs were fixed and permeabilized with permeabilze solution (BD Biosciences), immunotained with the anti-cTnT primary antibody (Abcam) or IgG control antibody for 2 h at 4°C, washed 2-3 times, and then stained with PE-labeled secondary antibodies for 1 h at 4°C. Stained-cells were resuspended and detected with a FACSCalibur (BD Biosciences) and analysed by the FlowJo software. Information of the used antibodies in this study are listed in Supplementary Table 2.
2.5. Alkaline phosphatase staining
hESCs were dissociated and plated on slides for 2∼3 days culture. Cells were then stained with the FRV, Naphthol and H2O (1:1:1) mixture for 15 min at room temperature avoiding light. Images were taken by a Leica Dmi8 microscope (Leica).
2.6. Immunofluorescence staining
The cells were rinsed with PBS and fixed with 4% paraformaldehyde for 5 min. The cells were permeabilized with 0.05% Triton X-100 at room temperature for 15 min, and washed three times with PBS, and then incubated in 4% goat serum for 30 min. The cells were stained by primary antibodies overnight at 4 degree.The next day, the cells were incubated with Alexa Fluor conjugated secondary antibodies at 37°C for 1 h, and then counterstained with DAPI (Abcam) at room temperature for 5 min, observed under a Leica DMi8 fluorescence microscope (Lecia). To assess pluripotency, hESC colonies were stained with antibody against SOX2 (Abcam), OCT4 (Santa cruz), SSEA4 (Abcam) and NANOG (Santa cruz). To assess differentiated cardiomyocytes, CMs were stained with antibody against cTNT2 (Abcam), α-actinin (Abcam), MLC-2v (Proteintech), TBX18(Proteintech). Secondary antibodies labeled with Alexa Fluor 488 or 594 were applied (Proteintech). Finally, cells were stained with DAPI (Abcam) and Cell area was further quantified by the ImageJ software (National Institutes of Health) as previously described [12]. Information of the used antibodies in this study are listed in Supplementary Table 2.
2.7. In vivo teratoma formation assays
4-5 × 106 WT or E2A mutant hESCs were resuspended in 100 μl matrigel (Corning, 356231) and then subcutaneously injected to six NOD-SCID mice. All mice were housed in an animal facility with a 12h light/dark cycle and free access to food and water. All procedures were performed in accordance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee of Fudan University (Protocol: 20140226-056). Eight weeks later, teratomas were dissected, and fixed with 4% PFA. Hematoxylin-eosin staining was performed to detect whether three germ layers formed.
2.8. Western blot
Total protein was extracted from cultured cells lysed in RIPA lysis buffer (Beyotime, P0013C) with protease inhibitors (Bimake, B14001). 30 μg protein was electrophoresed and separated in SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were blocked with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween-20 for 2 h. The membranes were then treated with primary antibodies of human E2A (Santa Cruz) or β-actin (Abcam) overnight at 4°C. Membranes were gently washed three times with PBS-T buffer for 10 minutes at a time,then Secondary antibodies (Proteintech) were applied for 60 min at room temperatue.Membranes were washed again in PBS-T, the method is the same as above. Immunoblots were visualized by chemiluminescent imaging system (Tanon 5200) and quantified by the ImageJ software (National Institutes of Health). Information of the used antibodies in this study are listed in Supplementary Table 3.
2.9. Cell size measurement
Cardiomyocyte size was determined in cardiomyocytes stained with cTNT antibody (Abcam) and anti-rabbit CoraLite 594 (Proteintech). At least 60 cells from randomly selected fields were analyzed using the ImageJ software (NIH) following online instruction. Briefly, scale was set to present measurement results in calibrated units. Then cells surface was measured by using the “Freehand selections” button. The measurements for each cell area were displayed in the “Results” window. To exclude observational errors and personal influences, we improved precision by averaging the test values of repeated measurement.
2.10. Cardiomyocyte contractility measurement
Single cardiomyocyte cells were seeded on matrigel-coated confocal dishes (Nalge Nunc International). After spontaneously beating, the contractility was detected by video-based motion edge detection system (Zeiss CFM500 inversion fluorescence microscope). And real-time contractile data were recorded by the FelixGX 4.2.2 software (FelixGX, PTI). Voltage of Y axes indirectly reflected the relative contractility of beating cardiomyocytes and the number of peaks in 30 s indicated the relative contracting rate indirectly.
2.11. Whole cell patch clamp recordings
Cardiomyocytes of day30 were mechanically and enzymatically dissociated to obtain single cells, and were then seeded on Matrigel-coated glass coverslips (Warner Instruments). Cells with spontaneous beatings were selected and action potentials were recorded using an EPC-10 patch clamp amplifier (HEKA) as previously described [13]. Continuous extracellular solution perfusion was achieved using a rapid solution exchanger (Bio-logic Science Instruments). Data were acquired using PatchMaster software (HEKA) and digitized at 1 kHz. Data analyses were performed using Igor Pro (Wavemetrics) and Prism (Graphpad). A TC-344B heating system (Warner Instruments) was used to maintain the temperature at 35.5-37°C. Tyrodes solution was used as the external solution containing 140 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM glucose, 1.8 mM CaCl2 and 10 mM HEPES (pH 7.4 with NaOH at 25°C). The internal solution contained 120 mM KCl, 1 mM MgCl2, 10 mM HEPES, 3 mM Mg-ATP, and 10 mM EGTA (pH 7.2 with KOH at 25°C).
2.12. Multi-electrode array (MEA) recordings
The multi-electrode array (MEA) system (Multi-Channel Systems, MEA-2100, Germany) was used to record the extracellular field potential. Beating CMs were dissociated and plated on matrigel-coated MEAs dishes for 2∼3 days to achieve attachment to the electrodes. Electric signals were collected by a MCS rack system (Multichannel Systems) and analyzed using the Spike2 7.05 software (Cambridge Electronic Design, UK) [14,15]. β-adrenergic (1 μM isoproterenol; Sigma-Aldrich) and its antagonist metoprolol (1μM carbachol; Sigma-Aldrich) were applied to assess the adrenergic response.
2.13. Dual-Luciferase reporter assays
From public ATAC-seq and Dnase-seq data, the promoters of TFs, including TBX5, TBX3, SHOX2 and TBX18 were predicted to interact with E2A. About 1Kb fragment of these TFs core promoter region were amplified by PCR and inserted into the multiple cloning site (MCS) located in the dual-luciferase reporter vector pGL3 respectively. 293T cells were co-transfected with each constructed reporter vector plus the E2A-pCMV-C-HA plasmid by DNA Transfection Reagent (Bimake) for 24 h. Cells were then harvested using a Dual-Luciferase Reporter Gene Assay Kit (Beyotime,RG027) according to the manufacturer's protocol. The relative luciferase activity was normalized to Renilla luciferase activity. The pGL3-basic and PCMV-C-HA was simultaneously transfected as an internal control.
2.14. RNA extraction, Illumina library preparation, and sequencing
Total RNA were extracted from WT and E2A knockout hESCs at day 0-15 or day 30 post directed differentiation using Trizol (Invitrogen, 15596018). The mRNAs containing Poly-A tail were purified by poly-T oligo-linked magnetic beads from 1μg total RNA and then digested into small fragments using divalent cations. The fragmented mRNAs were synthesized on the first-strand cDNA and the second strand cDNAs were then synthesized using DNA Polymerases I and RNase H (Invitrogen). After an end repair process by adding a single "A" base and ligation of the adapters, these products were then purified and enriched with PCR to create the final cDNA library. The concentration of cDNA was then determined by the Qubit® RNA Assay Kit in Qubit® 3.0. The clustering of the index-coded samples was performed on a cBot cluster generation system using HiSeq PE Cluster Kit v4-cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation, the libraries were sequenced on an Illumina sequencing platform (Illumina HiseqTM 4000, San Francisco, USA).
2.15. Statistical analysis
Data were represented as mean ± SEM. Two-tailed Student's t-test was used to compare the statistical differences between two independent groups. Analysis of variance (ANOVA) tests were used to compare statistical differences among more than two groups. Significant differences were considered when the P-value was less than 0.05.
3. Results
3.1. Expression of E2A during cardiac-specific differentiation of hESCs
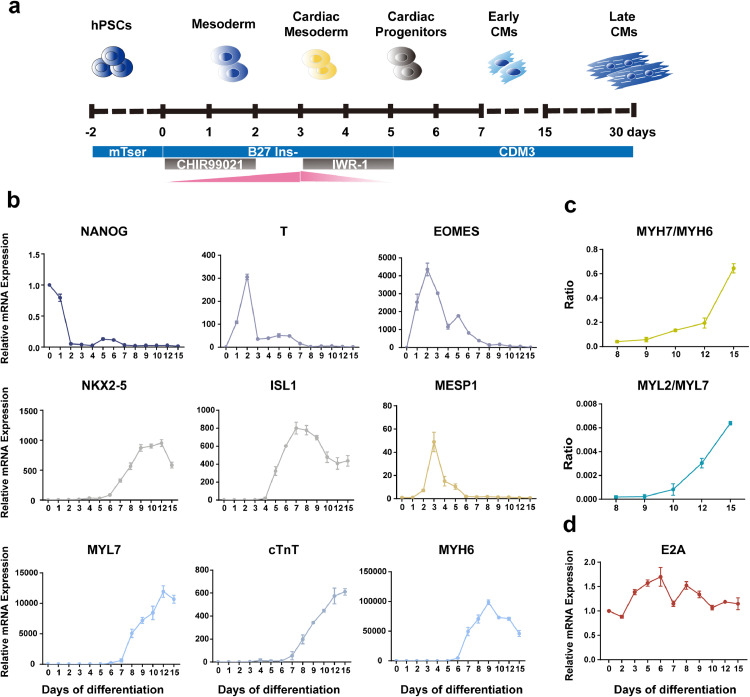
The 2D cardiac-specific differentiation protocol for hPSCs has been well-established and is a good model for studying human cardiac development and differentiation [16,17]. Here we utilized this protocol and dissected E2A expression and function during this process (Fig. 1a). We observed spontaneous beating CMs at day7 post differentiation. The expression patterns of pluripotency marker (NANOG), early mesoderm markers (T, EOMES), cardiac mesoderm markers (MESP1), cardiac progenitor cell (CPC) markers (NKX2-5, ISL1), and CMs markers (TNNT2, MYH6, MYL7) during cardiac differentiation are consistent with previous reports from other groups (Fig. 1b) [18]. Increasing ratios of MYH7/MYH6 and MYL2/MYL7 expression also suggested a time-dependent maturation for these hESC-derived CMs (Fig. 1c). These results indicated that our hESC cardiac differentiation system was successful. We next examined E2A mRNA levels during the hESC cardiac differentiation process and found that, unlike the stage-specific expression of many cardiogenic transcription factors, E2A was constantly expressed throughout the differentiation process (Fig. 1d).
Fig. 1.
Expression pattern of E2A and key markers regulating in vitro CMs directed differentiation.
(a) Directed differentiation of hESCs into CMs by modulating Wnt signaling activity. (b) Line graphs depicting the mRNA levels of key markers at different differentiation stages of cardiomyocytes differentiation. (c) The increased ratios of MYH7/MYH6 and MYL2/MYL7 demonstrated the maturation of cardiomyocytes over prolonged culture. (d) mRNA levels of E2A during CMs differentiation process. All data were normalized to the values on day 0 and presented as mean ± SEM (n=3).
3.2. E2A knockout did not affect pluripotency but altered cardiac-specific differentiation of hESCs
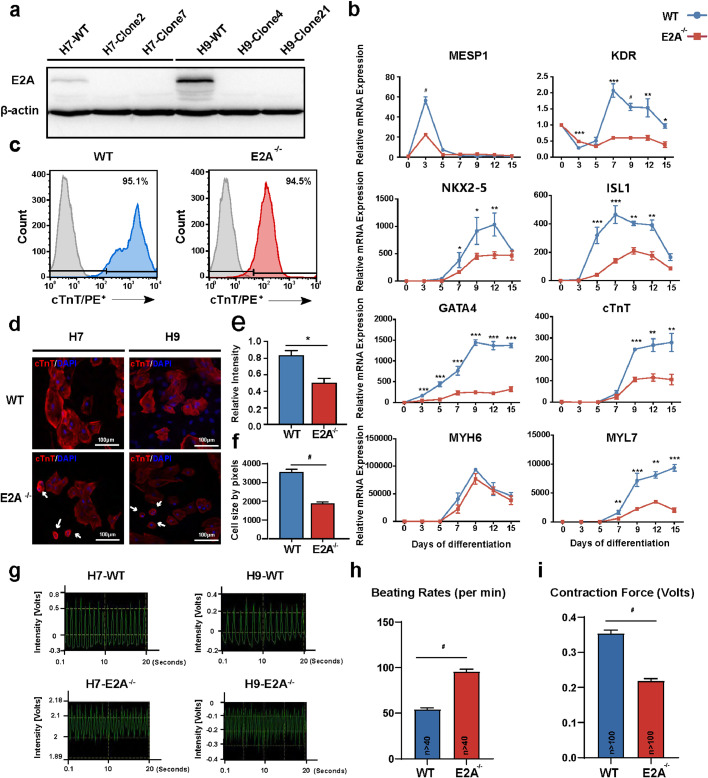
To evaluate the role of E2A in human heart development and cardiac-specific differentiation, we established E2A knockout hESC cell lines by CRISPR/Cas9 mediated genomic editing. The guide RNAs were designed to target exon 3 of the E2A gene (Supplementary Fig. 1a). Successful interruption of E2A expression by frameshifting indels in single H7 and H9 hESC clones were confirmed by sequencing and Western blotting (Supplementary Fig. 1b and Fig. 2a). The morphology and growth of E2A knockout hESCs were similar to those of the wildtype (WT) hESCs. Immunofluorescence staining of pluripotency markers OCT4 and NANOG also showed similar nuclear staining pattern (Supplementary Fig. 2a). The mRNA levels of certain pluripotency markers were slightly up-regulated in E2A knockout hESCs (Supplementary Fig. 2b). We also injected E2A knockout hESCs into NOD/SCID immunodeficient mice for teratoma development. Three embryonic germ layers were found in both E2A knockout and WT teratomas (Supplementary Fig. 2c). These data indicated that E2A deficiency did not compromise the pluripotency of hESCs.
Fig. 2.
E2A deficiency altered cardiac-specific differentiation of hESCs.
(a) Representative western blots showing E2A levels in H7 and H9 knockout homozygous clones. (b) Expression pattern of cardiogenic mesoderm markers (MESP1); cardiac progenitor cells markers (NKX2-5, ISL1) and cardiomyocytes structural genes (TNNT2 and MYL7, except for MYH6) were down-regulated in E2A knockout differentiated cells. Data are shown as the mean ± SEM from at least three independent experiments. * P value < 0.05, ** P value < 0.01, *** P value < 0.001, # P value < 0.0001(Two-tailed Student's t-test). (c) Quantification of cTnT positive cardiomyocytes in E2A knockout CMs by flow cytometry. Gray shapes represented the blank control. (d) IF analysis of differentiated cardiomyocytes generated from H7 and H9 hESCs, showing attenuated cTnT relative intensity. White arrows pointed at small size CMs. (e) Quantitation of cTnT intensity was weakened in E2A knockout CMs. (f) Quantitation of CMs cell area was reduced in E2A knockout CMs. (g) Contraction traces of wildtype and E2A knockout CMs. (h) Rising beating rates were observed in CMs derived from E2A knockout CMs. (i) E2A knockout CMs showed attenuated contractile force, n >100.* P value < 0.05,** P value < 0.01, *** P value < 0.01, # P value < 0.0001(Two-tailed Student's t-test).
To see whether E2A is important in the specification and differentiation of cardiomyocytes, we next analyzed expressions of crucial cardiac-related markers in E2A knockout hESCs during differentiation. The mRNA levels of cardiogenic mesoderm genes MESP1, CPC marker genes ISL1 and NKX2-5, as well as CM structural genes TNNT2 and MYL7 (except for MYH6) were all down-regulated at various degrees in E2A knockout hESCs compared with those in WT (Fig. 2b). Both H7 and H9 E2A knockout hESC clones were able to differentiate into monolayer beating cardiomyocyte sheets. Flowcytometry analysis showed that the percentage of cTnT-positive CMs were similar in WT and E2A knockout hESC differentiation, with E2A knockout cells showing a decreased cTnT expression intensity (Fig. 2c). Immunostaining of E2A knockout hESC-derived CMs further showed a reduced content of contractile protein cTnT (Fig. 2d and 2e). Remarkably, we also noticed a significantly increased proportion of smaller and more round shaped cardiomyocytes in E2A knockout group indicatd with white arrows in Fig. 2d. And quantification of average cell area showed a substantial decrease in E2A knockout group (Fig. 2f). To investigate the influence of E2A knockout on cardiomyocyte contractility, we next measured the relative contraction force of dissociatd single beating cardiomyocytes. From day15 post differentiation, E2A knockout hESCs derived cardiomyocytes gradually beat much faster than WT hESC-CMs (Fig. 2g and 2h; Supplementary video 1-4) and exhibited relative attenuated contraction force (Fig. 2i). These results indicated that hESC cardiomyocytes differentiation was affected in some degree by E2A gene knockout.
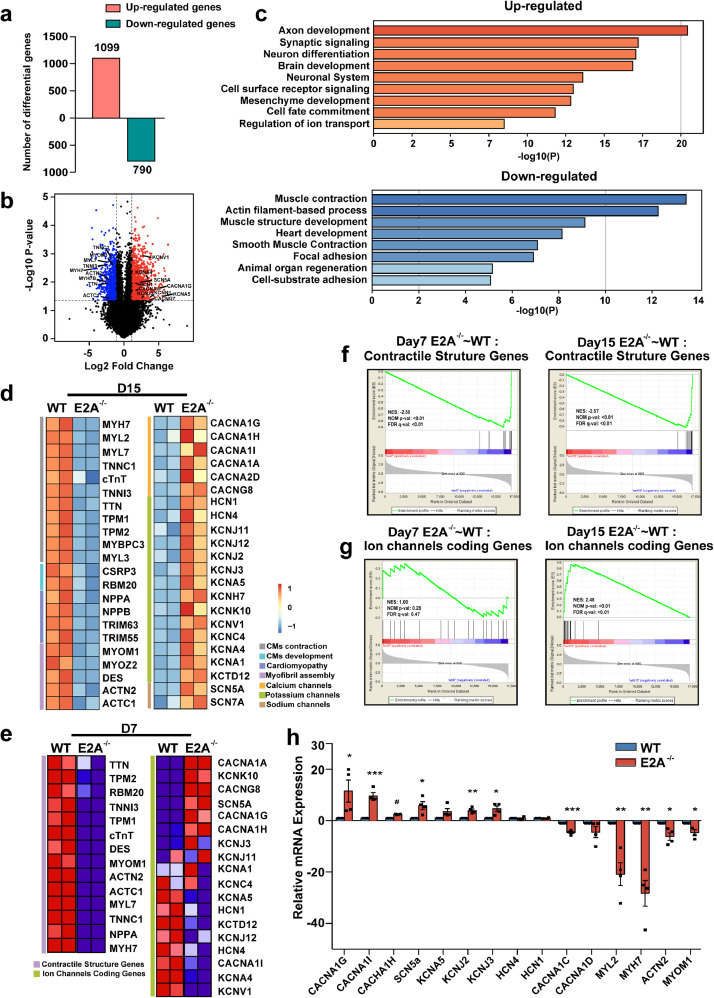
3.3. Global transcriptomic analysis of E2A knockout hESC-CMs
To further examine the phenotypes of E2A knockout cardiomyocytes derived from hESCs and better determine the genome-wide impact of E2A deficiency, we performed whole genome RNA sequencing analyses and compared WT and E2A knockout cells at day 3, 7 and 15 post differentiation. As shown in Fig. 3a and 3b, there are 1099 up-regulated genes and 790 down-regulated genes in E2A ablated hESC-CMs (cutoff = 2.0, P value < 0.05) relative to WT at day15 post differentiation. GO analysis indicated that up-regulated genes were associated with ion transport, cell fate commitment, and mesenchyme development etc. Muscle contraction, actin filament-based process, and muscle structure development were among the most affected biological processes in down-regulated genes (Fig. 3c). In detail, genes associated with cardiomyocyte contractile structure, myofibril assembly, and cardiomyocyte development, such as MYH7, TNNC1, MYOM1 and ACTN2, were down-regulated, while many ion channel encoding genes, such as CACNA1G, KCNJ3, and KCNA5, were up-regulated in E2A knockout cardiomoycytes (Fig. 3d). Of note, GO analysis also showed that up-regulated genes on day3 and day7 post differentiation, which is the time of cardiac mesoderm and the initial formation of CMs from hESCs, were enriched in neuron development and differentiation, while down-regulated genes were enriched in categories such as cardiovascular system development and muscle contration (Supplmentary Fig. 3). Gene set enrichment analysis (GSEA) confirmed the concordant down-regulated change of myofibril strucutural genes in both early cardiomyocytes (day7) and late cardiomyocytes (day15) (Fig. 3d and 3f). While ion channel encoding genes were only up-regulated in day15 cardiomyocytes (Fig. 3d and 3g). We further validated these differential gene expressions by quantitative PCR in day30 E2A knockout hESC-CMs. Expressions of T-type calcium channels CACNA1G, CACNA1I and CACNA1H, were all up-regulated, while expressions of L-type calcium channels, such as CACNA1C and CACNA1D were decreased (Fig. 3h). These results suggested that E2A deficiency drives cardiac differentiation of hESCs toward an uncommon direction, with many channel genes up-regulated after day15 post differentiation.
Fig. 3.
Bulk RNA-sequence analysis of CMs derived from hESCs at Day15.
(a) 1099 genes were up-regulated and 790 were down in day15 CMs after E2A knockout (cutoff = 2.0, P value < 0.05). (b) GO analyses discovered biological processes regulated by differential genes in day15 CMs. (c) Volcano plots showed that ion channels encoding genes were up-regulated, while CMs structural genes down-regulated after E2A deficiency. (d) Heatmap of genes expression associated with CMs contractile structure, myofibril assembly (left) and ion channels (right) in day15 wildtype and E2A knockout groups. (e) Heatmap of genes expression associated with CMs contractile structure (left) and ion channels (right) in day7 wildtype and E2A knockout groups. (f) Enrichment of genes related to contractile structure in day 7 and day15 CMs by GSEA. (g) Enrichment of ion channel encoding genes in day7 and day15 CMs by GSEA. (h) mRNA levels of representative ion channels and structure genes in day30 CMs. Data were normalized to GAPDH level and presented as mean ± SEM. * P value < 0.05, ** P value < 0.01, *** P value < 0.01, # P value < 0.0001(Two-tailed Student's t-test).
3.4. E2A knockout significantly up-regulated the ratio of nodal-like CMs in hESC cardiac-specific differentiation
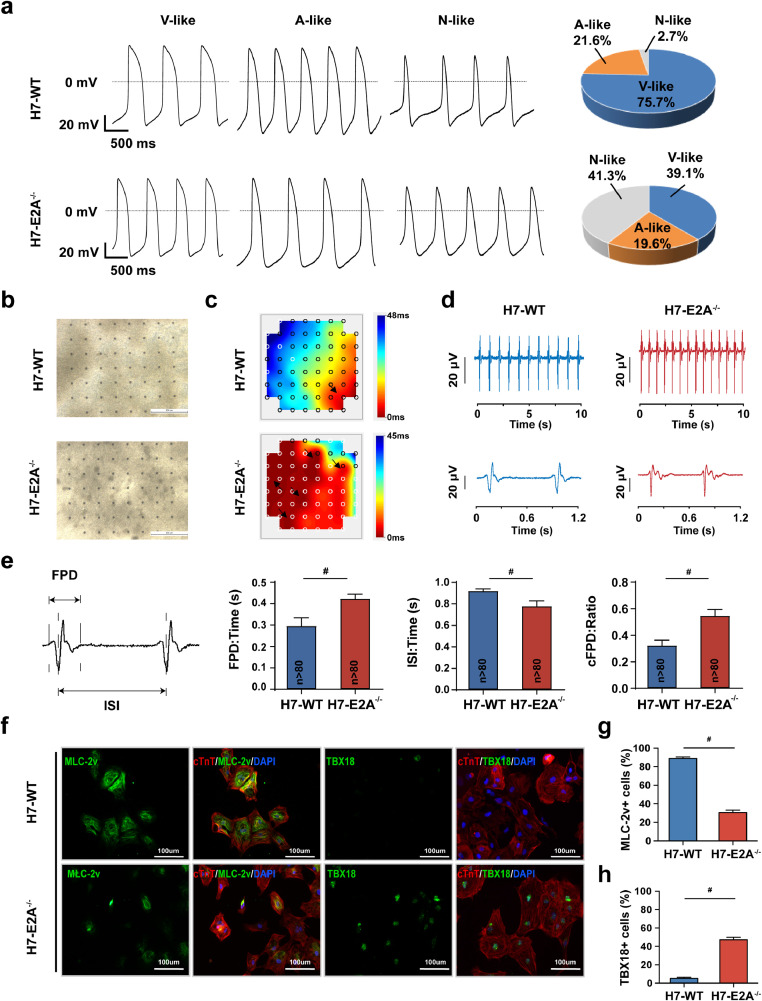
Considering the fact that T-type calcium channels are enriched in conductive cardiomyocytes and L-type calcium channels in working cardiomyocytes, the increased expression of T-type calcium channels and some other channel genes in E2A knockout cardiomoyctes further suggested that the phenotype may associate with an increase in the amount of nodal-like cardiomyocytes. Examination of action potential (AP) profiles for single cardiomyoycte is a standard approach to distinguish the heterogeneous cardiomyocyte subtypes [19]. To test our hypothesis, we next recorded APs of day30 single cardiomyocytes derived from both WT and E2A knockout hESCs using the single-cell patch clamp technique. By comparing key AP parameters, including maximum diastolic potential (MDP), overshoot, AP amplitude (APA), AP duration (APD), AP duration at 50% repolarization (APD50), AP duration at 70% repolarization (APD70), AP duration at 90% repolarization (APD90), maximal upstroke velocity (Vmax), and beating rates, we classified WT and E2A knockout cardiomyocytes into three different subtypes. We found that, after E2A knockout in H7 hESCs, the percentage of nodal-like (N-like) CMs increased from 2.7% to 41.3%, while that of ventricular-like (V-like) CMs decreased from 75.7% to 39.1% and atrial-like (A-like) CMs decreased from 21.6% to 19.6% (Fig. 4a and Table 1). Similarly, we further confirmed that the number of nodal-like cardiomyocytes also significantly increased to >25% in H9 E2A knockout hESCs (supplementary Fig. 4a and Table 2).
Fig. 4.
E2A deficiency significantly up-regulated the ratio of nodal-like CMs in hESC cardiac-specific differentiation.
(a) Whole-cell patch-clamp recording showed that nodal-like CMs increased to 41.3%, while ventricular-like CMs decreased to 39.1% after E2A knockout CMs derived from H7. (b) Low magnification bright field picture of the CMs monolayer seeded in MEA plates. (c) Representative snapshots of color map depicting electrical signal propagation of monolayer CMs. The color maps showed that the electrical signal is initiated at the bottom right corner (red) and is propagated to the upper left corner (blue) in wildtype CMs while E2A deficiency led to disruptive propagation of electrical signals with multiple initiation sites indicated by black arrows. (d) Electric pulse pattern of wildtype and E2A deficient CMs derived from H7 hESCs. (e) Quantification of FPD, ISI and cFPD (ratio of FPD/ISI) in wildtype and E2A knockout CMs. All data are presented as mean ± SEM. All data are presented as mean ± SEM, *** P value <0.001, # P value <0.0001(Two-tailed Student's t-test). (f) Expression of MLC-2v+ reduced, while TBX18+ increased after E2A knockout CMs derived from H7 hESCs. (g) Statistic of MLC-2v+ cells in wildtype and E2A knockout CMs derived from H7 hESCs. All data are presented as mean ± SEM, # P value <0.0001 (Two-tailed Student's t-test). (h) Statistic of TBX18+ cells in wildtype and E2A knockout CMs derived from H7 hESCs. All data are presented as mean ± SEM, # P value <0.0001 (Two-tailed Student's t-test).
Table 1.
H7-WT&H7-KO single cell Patch-Clamp recording Statistics
| MDP (mV) | Overshoot (mV) | APA (mV) | APD (ms) | APD90 (ms) | APD70 (ms) | APD50 (ms) | Vmax-D (V/s) | Beating rates (per minute) | SD of Interspike Interval | % Total | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ventricular-like (V-like) CMs | WT(n=28) | -56.7±0.8 | 45.9±0.8 | 102.7±1.0 | 358.3±25.8 | 290.1±22.9 | 270.0±21.9 | 251.4±20.3 | 8.0±0.4 | 87.7±7.0 | 45.2±7.6 | 75.7 |
| KO(n=18) | -59.7±0.8 | 42.6±1.2 | 102.2±1.4 | 340.1±33.9 | 256.6±30.0 | 238.8±28.9 | 220.4±27.4 | 9.8±0.5 | 95.5±8.2 | 37.3±7.5 | 39.1 | |
| Atrial-like (A-like) CMs | WT(n=8) | -56.0±1.9 | 38.8±1.7 | 94.8±1.1 | 282.6±49.5 | 219.4±39.5 | 198.0±34.8 | 175.1±28.7 | 5.7±0.3 | 121.0±12.2 | 19.5±2.2 | 21.6 |
| KO(n=9) | -58.6±1.4 | 35.5±1.0 | 94.1±1.5 | 218.8±16.2 | 142.7±9.0 | 125.3±7.7 | 110.2±6.5 | 8.2±1.1 | 118.9±12.3 | 27.6±12.1 | 19.6 | |
| Nodal-like (N-like) CMs | WT(n=1) | -46.1 | 27.5 | 73.6 | 143 | 89.5 | 75.1 | 63.7 | 3.6 | 101.5 | 54.2 | 2.7 |
| KO(n=19) | -54.3±1.2 | 27.4±1.4 | 81.7±1.9 | 192.2±10.7 | 101.7±5.9 | 81.7±5.2 | 67.4±4.4 | 6.8±0.9 | 130.2±8.5 | 22.9±3.9 | 41.3 |
Table 2.
H9-WT&H9-KO single cell Patch-Clamp recording Statistics
| MDP (mV) | Overshoot (mV) | APA (mV) | APD (ms) | APD90 (ms) | APD70 (ms) | APD50 (ms) | Vmax-D (V/s) | Beating rates (per minute) | SD of Interspike Interval | % Total | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ventricular-like (V-like) CMs | WT(n=32) | -54.2±0.9 | 47.6±0.8 | 101.9±1.3 | 317.3±17.3 | 245.6±12.7 | 229.3±12.4 | 212.4±11.8 | 13.8±0.6 | 92.5±6.6 | 72.1±16.6 | 76.2 |
| KO(n=23) | -56.8±0.9 | 47.2±1.5 | 104.0±2.1 | 316.5±23.4 | 276.4±23.2 | 262.5±23.0 | 247.6±22.6 | 12.0±0.9 | 93.3±5.5 | 64.7±16.0 | 65.7 | |
| Atrial-like (A-like) CMs | WT(n=6) | -51.2±2.6 | 43.9±1.6 | 95.1±2.5 | 293.6±47.0 | 183.3±26.1 | 162.5±23.7 | 143.6±21.1 | 14.3±2.4 | 131.0±30.2 | 29.7±11.70 | 2.4 |
| KO(n=3) | -57.1±1.1 | 43.8±3.3 | 100.9±3.6 | 277.3±62.1 | 221.4±49.8 | 199.2±44.2 | 176.5±37.7 | 10.0±3.2 | 110.6±23.0 | 36.8±19.5 | 8.6 | |
| Nodal-like (N-like) CMs | WT(n=4) | -49.6±3.2 | 35.6±1.3 | 85.2±2.2 | 275.9±56.8 | 114.2±13.9 | 86.60±12.5 | 70.60±11.2 | 9.7±0.3 | 80.5±39.2 | 84.1±25.4 | 9.5 |
| KO(n=9) | -49.8±2.5 | 32.9±1.8 | 82.7±1.9 | 208.9±15.4 | 137.4±14.6 | 113.2±13.8 | 96.1±12.0 | 7.4±0.7 | 115.2±8.2 | 43.6±9.6 | 25.7 |
We next assessed the electric activity of E2A knockout cardiomyocytes by multi-electrode arrays (MEAs). WT or E2A knockout cardiomyocytes were plated in MEA probes in a beating monolayer as shown in Fig. 4b and supplementary Fig. 4b. As it is known, the synchronized contraction of heart is controlled by the spread of electrical signal from cardiacmyocyte to cardiacmyocyte. The assessment of the conduction velocity of the cardiac impulse can be reflected by electrophysiological recordings of the 60 electrodes in the MEA probe. After determination of local activation time at each electrode, we obtained the detailed activation maps depicting the development of synchroninzed action potential propagation from earliest activition (red) towards latest activation (blue). We observed that the initiation site of electric activity was stably localized in a specific site in both H7 and H9 WT cardiomyocytes monolayer. In contrast, the electric activity of both H7 and H9 E2A knockout cardiomyocytes monolayers were highly random with multiple initiation sites (Fig. 4c and Supplementary Fig. 4c), suggesting there were increased nodal cell-generated pacing sites. E2A deficient cardiomyocytes monolayer beat faster than WT and had prolonged field potential duration (FPD, WT, 0.30 ± 0.04s; KO, 0.42 ± 0.02s; P value < 0.0001), shorter inter-spike interval (ISI, WT, 0.92 ± 0.02s; KO, 0.78 ± 0.05s; P value < 0.0001) and increased cFPD (ratio of FPD/ISI, WT, 0.32 ± 0.05; KO, 0.55 ± 0.05; P value < 0.0001) (Fig. 4d-4e, and Supplementary Fig. 4d), which indicated that E2A deficiency led to shorter periods of electric activity.
Nodal-like cardiomyocytes could be generated from TBX18+ progenitors, which is a prominent factor in SAN formation [20]. We further stained WT and E2A knockout cardiomyocytes with TBX18 to estimate the percentage of nodal-like cells. It showed that the percentage of TBX18+ cells in E2A knockout cardiomyocytes was markedly increased, while that of MLC-2v positive ventricular-like cells was significantly decreased, consistent with our whole-cell patch clamp data (Fig. 4f-4h and Supplementary Fig. 4e). Flowcytometry analyses further confirmed that the percentage of cTnT and MLC-2v double positive ventricular-like cardiomyocytes was significantly decreased from ∼90% to ∼40-45% after E2A knockout (Supplementary Fig. 5). Collectively, these data indicated that E2A ablation led to the proportional increase of nodal-like cells and decrease of ventricular-like cells in hESC-derived cardiomyocytes.
3.5. E2A directly regulates transcription factors associated with the development of conduction cells
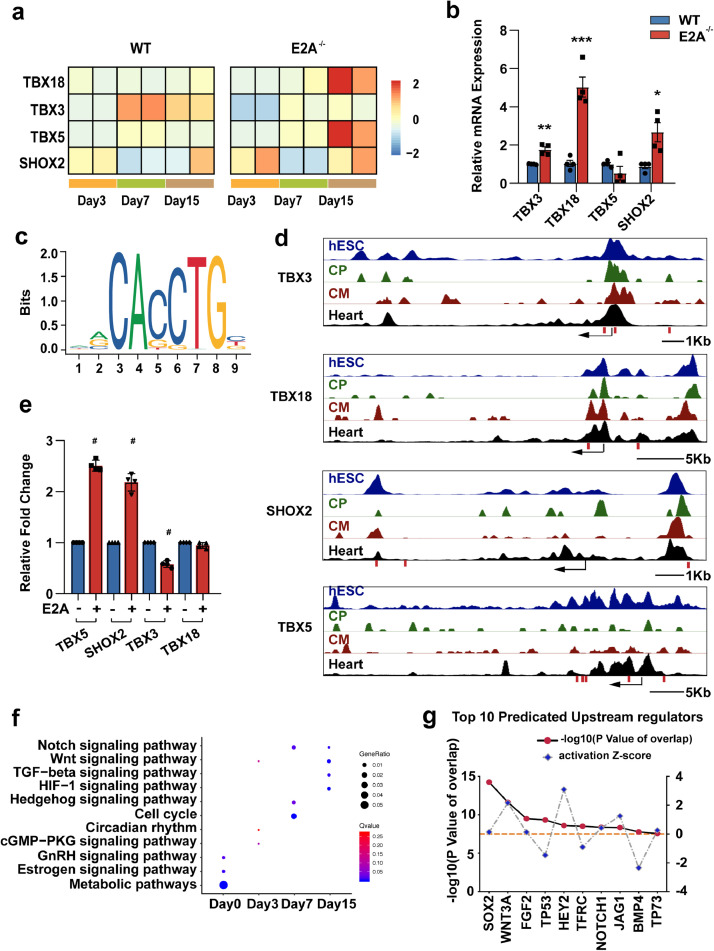
SAN is composed of a heterogeneous cell population and its development is controlled by several transcription factors, mainly including TBX18, TBX3, TBX5, and SHOX2 [21]. Our RNA-seq data showed that expression of TBX18, TBX5, and SHOX2 were all up-regulated in E2A knockout hESC-CMs at day15 after differentiation (Fig. 5a). The mRNA abundance of TBX18 and SHOX2 persisted to increase at day30 after differentiation (Fig. 5b). And we observed that the expression of the other vital transcription factor TBX3 was also significantly up-regulated (Fig. 5b). But the TBX5 mRNA level did not show statistical significant difference between WT and E2A knockout cardiomyocytes at day30 (Fig. 5b). Since TBX5 acts as the activator of TBX3 and SHOX2, there could be a fine balance between the expression level of TBX5 and TBX3 during cardiac specification. To investigate whether the above-mentioned transcription factors are directly regulated by E2A, we analyzed public ATAC-seq and DNase-seq data from hESCs, cardiac progenitors (CP), cardiomyocytes (CMs), and fetal heart tissues to locate the open chromatin regions of these genes. Considering the high affinity of E2A with conserved E-box motif (CANNTG), we discovered several E2A binding sites in the open chromatin region of these transcription factors (+324bp, -51bp and -2053bp from TSS of TBX3 gene, +1412bp and -3402bp from TSS of TBX18 gene, +6112bp, +7819bp and -4462bp from TSS of SHOX2 gene, +1792bp, +8497bp, +9062bp,+9991bp and -3418bp from TSS of TBX5 gene, respectively) (Fig. 5c-5d). Using the dual-luciferase reporter assay, we further confirmed that E2A directly bound to the promoter region and regulated the transcription of TBX5, SHOX2, and TBX3, except TBX18 (Fig. 5e). Interestingly, E2A displayed differential regulation mode of TBX3, TBX5, and SHOX2 expression. Since E2A usually form a heterodimer with another bHLH protein to regulate downstream gene expression, these results suggested that secondary factors were also involved in the regulatory activities mediated by E2A, which together realized the exquisite control of genes related to nodal cell differentiation.
Fig. 5.
E2A directly regulates transcription factors associate with the development of cardiac conduction cells and multiple signaling pathways
(a) Expression of transcription factors SHOX2, TBX3, TBX5 and TBX18 in day3, 7, and 15 CMs post differentiation. (b) mRNA levels of SHOX2, TBX3, TBX5 and TBX18 in day30 CMs. Data were normalized to GAPDH level and presented as mean ± SEM. * P value < 0.05, ** P value < 0.01, *** P value < 0.01, # P value < 0.0001(Two-tailed Student's t-test) (c) E2A binding profile from http://jaspar.genereg.net/. (d) E2A binding sites were predicted in chromatin open regions of SHOX2, TBX3, TBX5 and TBX18 in hESCs (CistromeDB:79373), human CPC (CistromeDB:80590), human CM (CistromeDB:80589), and fetal heart tissue (CistromeDB:40936), arrows indicate transcription start site (TSS) of genes, E-box elements are marked by red lines below and corresponding sequences are listed in the gray boxes. (e) Dual-Luciferase reporter assay in 293T showed that E2A directly regulated expression of SHOX2, TBX3, and TBX5, but not TBX18. Data were presented as mean ± SEM. # P value < 0.0001(Two-tailed Student's t-test) (f) KEGG analyses of up-regulated signal pathway in day0,3,7,15 E2A deficient cells. (g) Top 10 predicated upstream regulators.
Considering the increased expression level of above regulators is inconsistent with the direct effect of E2A, we next test whether E2A could manipulate these genes expression via indirect actions. Interestingly, KEGG and GSEA analysis of our RNA-seq data showed that the activities of multiple signaling pathways, including NOTCH and WNT pathways, were dramatically altered in cardiac-specific differentiation of E2A knockout hESCs compared with WT cardiomyocytes (Fig. 5f, Supplementary Fig. 6 and 7). Especially, NOTCH pathway was dramatically activated from day7 post differentiation in E2A knockout hESCs (Supplementary Fig. 6a). Evidence proved that these pathways contribute to SAN development and CCS diseases. Public ChIP-seq data in human myeloblasts showed that E2A bound to multiple binding sites of several NOTCH genes (Supplementary Fig. 6b). Upstream regulator analysis from Ingenuity Pathway Analysis (IPA) predicted several Notch signaling participators (HEY2, NOTCH1, and JAG1) and one canonical Wnt/β-catenin pathway ligand (WNT3a) as the top 10 upstream molecules leading to the observed changes in gene expression (Fig. 5g). These data suggested that E2A deficiency drives ESC-derived nodal-like cell specification and differentiation through alteration of activities of these key signaling pathways.
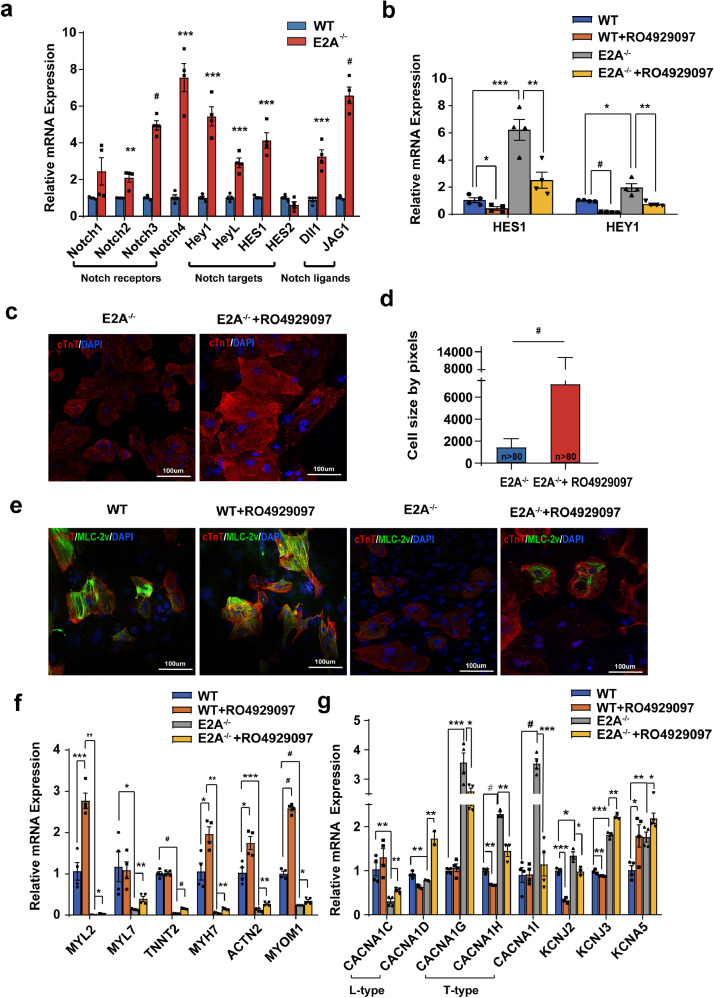
Our patch clamp and flowcytometry data showed that the number of nodal-like cardiomyocytes derived from E2A knockout hESCs increased significantly while the amount of ventricular-like cardiomyocytes reduced significantly. Since it was reported that overexpression of NOTCH extracellular domain (NICD) can directly transform ventricular cardiomyocytes into cells with conductive phenotypes in mice [22], we next examined expressions of several key genes in NOTCH signaling in day30 hESC-CMs. The result showed that expression of NOTCH receptors (NOTCH2, NOTCH3, and NOTCH4), targets (HEY1 and HES1), and ligands (DLL1 and JAG1) were all up-regulated in day30 E2A knockout cardiomyocytes relative to those in WT cardiomyocytes (Fig. 6a). To see whether blocking NOTCH signaling can reverse the cardiomyocyte phenotype after E2A knockout in hESCs, the small molecule RO4929097, an inhibitor of NOTCH signaling pathway, were applied to the WT and E2A knockout cardiomyocytes from day9 to day15 of cardiac differentiation. We observed that RO4929097 significantly reduced expression of Notch signaling targets HES1 and HEY1 in both WT and E2A knockout hESC-CMs (Fig. 6b). After treating with RO4929097, smaller cell areas of E2A knockout cardiomyocytes were reversed (Fig. 6c and 6d) and expression of cTnT and MLC-2v were partially restored (Fig. 6e and Supplementary Fig. 8). The mRNA levels of structural genes in cardiomyocytes, including MYL2, MYL7, TNNT2, were also up-regulated at different degrees both in WT and E2A knockout cardiomyocytes treated with RO4929097 (Fig. 6f). Furthermore, after RO4929097 treatment, ion channnels enriched in working cardiomyocytes, such as L-type calcium channels (CACNA1C and CACNA1D), KCNJ2, and KCNJ3, were restored to some extent, while T-type calcium channels (CACNA1G, CACNA1H and CACNA1I) enriched in conductive CMs were downregulated in E2A knockout cells (Fig. 6g). These results suggested that interference of NOTCH signaling partially switched the differentiation direction of E2A knockout cardiomyocytes back toward wildtype.
Fig. 6.
E2A partially relies on regulating the activity of NOTCH signal pathway for involving in CMs differentiation process and its inhibitor reversed the phenotypes of E2A knockout CMs.
(a) mRNA levels of representative NOTCH signal pathway genes after E2A knockout. Data were normalized to GAPDH level and presented as mean ± SEM. * P value < 0.05, ** P value < 0.01, *** P value < 0.01, # P value < 0.0001(Two-tailed Student's t-test). (b) Expression of NOTCH target genes HES1 and HEY1 in day30 CMs of WT and E2A−/- after treated with NOTCH inhibitor. Data were normalized to GAPDH level and presented as mean ± SEM. * P value < 0.05, ** P value < 0.01, *** P value < 0.01, # P value < 0.0001(ANOVA) (c) Cell areas of E2A−/- CMs increased after inhibition of NOTCH signal pathway. (d) Statistic of cell size of E2A−/- CMs after interfering NOTCH signaling. (e) Increased expression of cTnT and MLC-2v in E2A knockout CMs after treated with NOTCH inhibitor (RO4929097). All data were presented as mean ± SEM. # P value < 0.001(Two-tailed Student's t-test). (f) mRNA levels of structures genes were partially rescued in E2A−/- CMs after inhibition of NOTCH signal pathway. Data were normalized to GAPDH level and presented as mean ± SEM. * P value < 0.05, ** P value < 0.01, *** P value < 0.01, # P value < 0.0001(ANOVA). (g) mRNA expression of L-type calcium channels genes was increased, while which of T-type calcium channels and several other ion channels were decreased in E2A−/- CMs after treated with NOTCH inhibitor. All data were presented as mean ± SEM. * P value < 0.05, ** P value < 0.01, *** P value < 0.001, # P value < 0.001(ANOVA).
4. Disccusion
In this study, we found that the bHLH family transcription factor E2A participated in regulating differentiation of distinct cardiomyocyte subtypes from hESCs. E2A knockout cardiomyocytes displayed characteristic transcriptional expression pattern, increased beating rates, smaller cell size, attenuated sarcomere structure, and action potentials close to the unique features of SAN pacemaker cells and distinct from those of WT hESC-CMs. The patch clamp study also revealed E2A ablation dramatically promoted the proportion of nodal-like cells from previous <7-8% to as high as ∼40% in hESCs-derived cardiomocytes, while significantly suppressed the number of ventricular-like cardiomyocytes. We further found that spatiotemporal inhibition of Notch signaling at day 9-15 of differentiation diminished the enrichment of nodal-like cells derived from E2A knockout hESCs. Together, these results pointed to a new role of transcription factor E2A in the derivation of nodal-like cells from hESCs.
It is undeniable that identification of nodal-like cells from working type cardiomyocytes is still a big challenge since the SAN itself consists of a heterogeneous population of cardiomyocytes, let alone in an in vitro differentiation system without anatomic landmarks. Thus, it is better to use a combination of special physiological, functional, and molecular characteristics as a whole to screen nodal-like cells from working type cardiomyocytes. And, it should be noted to distinguish the role of E2A deficiency on elevating the population of the nodal-like cells rather than suppressing cardiomyocytes maturation.
There are several evidences present in our study showing that a substantial increasing number of E2A knockout hESC-CMs were nodal-like cells. First, the parameters used to idenfiy action potential of separate subtype cardiomyocytes derived from hESCs, which is a commonly accepted approach [19,23,24], showed in our study, a marked increase in the percentage of nodal-like cells in E2A knockout hESC-CMs. Second, E2A knockout hESC-CMs monolayers displayed random electric activity and increased pacing points in MEA assays, which is consistent with the property of SAN cardiomyocytes. Third, a substantial increasing number of E2A knockout cardiomyocytes displayed morphological and functional properties substantiating the nodal-like cell phenotype. Pacemaker cells derived from hESCs have been characterized by a special appearance as the round shaped and relatively smaller mononuclear cell (10-30 μm in size) [25], which is distinct from multiangular working type cardiomyocytes. A substantial increasing number of E2A knockout hESC-CMs displayed the morphology matching the special appearance of pacemaker cells derived from hESCs. Fourth, in our study, bulk RNA sequencing analysis demonstrated that the expression patterns of respresentive genes, such as ion channel encoding genes and contractile structure genes, were more close to the nodal-like cells. Moreover, expression of the key SAN development transcription factors SHOX2, TBX3 and TBX18 were all up-regulated in E2A knockout cardiomyocytes. By analyzing the public ATAC-seq and DNase-seq data from hESC, cardiac progenitors (CP), cardiomyocytes, and fetal heart tissues, we traced the conserved E-box binding motifs near the promoters of these transcription factors, which indicated the possible regulation of E2A on their expresssion. Further, we proved that E2A directly inhibited TBX3 expression by luciferase reporter assays. TBX3 is known to play a vital role in specification and development of the conduction system during vertebrate embryogensis [26,27]. TBX3 overexpression also efficiently programed mESCs or hiPSCs into sinoatrial bodies exhibiting highly beating rates and robust capability of pacing myocardium in vitro [28], [29], [30]. Vincent van Eif and et al., concluded a gene program including TBX3, SHOX2, ISL1, HOX family members, BMP and Notch signaling components are conserved between human and mouse [31]. Single cell sequencing analysis done by Liang and et al. determined TBX3 and SHOX2 are core in the gene regulatory network of pacemaker activity [32]. And functional genes encoding HCN channels, K+ channels, Ca2+ channels are all expressed in SAN across species; while the expression of voltage-gated potassium channel gene was low in SAN cells. These gene expression pattern was similar in our hESCs-derived cardiomyocyte samples from day15 after differentiation. Of note, two new SAN specific markers, Smoc2 identified by Vincent van Eif and et al., and Vsnl1 characterized by Liang in above studies, were also significantly up-regulated in our E2A knockout cardiomyocytes. All these data support our findings that E2A ablation promoted pacemaker cells formation from hESCs specific differentiation.
However we also noticed that E2A displayed a positive regulation of SHOX2 and TBX5 expression which is conflict with the alternation in the expression level of these genes after E2A ablation. Given that bHLH family proteins usually function as homodimer or heterodimer, the different regulation of E2A on TBX3 and SHOX2 expression may resulted from lacking of other bHLH family partners in our in vitro luciferase reporter assay system. It is possible that, in vivo, E2A binds to different bHLH family partners to form different heterodimers and perform precise regulation of cardiomyocyte specification in cardiac differentiation of hESCs. An insight into the dimerization brought by the shared bHLH domain may help identify the key mechanism of E2A in future study. More importantly, we assumed there may exist some indirect function of E2A to finalize its regulation on the downstream targets. Interestingly, we got a hint from the KEGG and GSEA analysis of our RNA-seq data at successive stages during the differentiation process. The NOTCH pathway was dramatically activated in E2A knockout early cardiomyocytes around day7 post differentiation (Supplementary Fig. 6a). Furthermore, the receptors, ligands, and downstream targets of NOTCH signaling were all significantly up-regulated in E2A knockout late cardiomyocytes around day30 post differentiation (Fig. 6a). With respect to the NOTCH signaling participators, including HEY2, NOTCH1 and JAG1, our data also support the previous data showing that in vitro or in vivo activation of myocardial NOTCH signaling by overexpressing the Notch intracellular domain (NICD) reprogrammed CMs to a CCS phenotype [22]. And overexpression of NOTCH extracellular domain (NICD) was proven to stimulate the expression level of TBX5 and other regulators [22,33]. By further confirming that NOTCH signaling inhibitor partially reversed the altered differentiation phenotypes of E2A knockout hESCs (Fig. 6). Considering the accordant changes of these regulators’ expression level with the activity of NOTCH signaling, we think this synergistic effect of E2A on above molecular determinants in SAN development is one of the biggest factors in high yield of nodal-like cells in E2A knockout hESCs-derived cardiomyocytes.
One limitation of our current study is not able to isolate the funny current which serves as a distinct feature of mature nodal like cells and contributes to generate the diastolic depolarization [34]. It is worth noting that the abundance of typical sinoatrial HCN4 did not change in our E2A knockout cardiomyocytes, which is responsible for the If current generation and serves as a functional marker of pacemaker cells [35], [36], [37]. One of the possible reasons is that our current small molecule-based cardiac specific differentiation protocol was performed to yield a majority of chamber cardiac myocytes from embryonic stem cells. Sartiani et al. [38] indicated that hESCs-derived cardiomyocytes at early stage express a robust If current which decreased in the late stage. Meanwhile the decreased mRNA level of HCN4 was also observed with time. But Bosman et al pointed out that although the mRNA abundance of HCN4 decreased during maturation of hESC-CMs, the HCN4 protein level actually increased [39]. And considering E2A knockout cardiomyocytes in our differentiation system may yeild the immature nodal like cells, these typical sinoatrial gene expression pattern will be more inclined to nodal-like pacemaker cells after adjusting differentiation condition. Thus, it is important to isolate If current from E2A knockout CM in our future study. And If blocker and caffeine can be applied to minimize the membrane clock or debilitate the Ca2+-clock which will provide more detailed information about Ca2+ handling in E2A knockout cardiomyocytes.
Interestingly, the electric activity recorded by MEA indicated the exceptional conductive ability of E2A knockout cardiomyocytes. Moreover, patch clamp recording revealed that E2A knockout markedly increased the proportion of nodal-like cardiomyocytes derived from hESCs, while at the same time suppressed the number of ventricular-like cardiomyocytes. This notion leads to a critical question as to in which stage E2A plays a role in the specificiation or differentiation of nodal-like cells. Previous studies provided evidences that overexpression of vital transcription factors reprogrammed ventricular cardiomyocytes into pacemaker-like cells in vitro [40], [41], [42]. In our study, we also noticed that there was a significant down-regulation of genes (e.g. MESP1, NKX2-5, ISL1) in early mesoderm development and cardiac progenitors derived from E2A knockout hESCs (Fig. 2b). Importantly, among these genes, NKX2.5 is a major regulaor for cardiomyocyte specification and differentiaiton. Previous studies also showed that pacemaker cells are recruited from NKX2.5-negative progenitors [5]. Thus, the down-regulation of NKX2.5 in E2A knockout cardiomyocytes progenitors may be key to lineage specification for nodal-like cells. Moreover, we also observed the increased expression of SHOX2, which is well-known to antagonize the expression of NKX2.5 [43,44], in E2A knockout cells. These results suggested that the changed transcriptional regulation of these genes in E2A knockout hESCs during cardiac specification may result in the altered differentiation outcome. To further investigate the mechanisms of changes in cardiac-specific differentiation regulated by E2A deficiency, we also focused our analysis on the genes encoding signaling molecules regulating early mesoderm and cardiogeneic specification. Signalling molecules of BMP, Retinoic Acid, WNT and other family served as the cardiac-inducing factors in a spatiotemporal-dependent manner to activate expression of the NKX2.5, GATA4 and T box factors [18,[45], [46], [47]]. Our current small molecule-based cardiac specific differentiation protocol was performed by adding small molecules regulating Wnt signaling to yield cardiac myocytes from embryonic stem cells [18]. As mentioned, upstream regulator analysis from IPA predicted WNT3A as one of the top 10 upstream molecules leading to the observed changes in gene expression after E2A knockout. We noticed that the activity of WNT signaling pathway were dramaticly changed compared with that in the WT hESC differentiation (Supplementary Fig. 7). The canonical WNT 11 is known to activate NKX2.5 and GATA4, while WNT3a and WNT8 were shown to suppress cardiogenesis in differentiating ES cells in vitro [48,49]. It is possible that E2A interrupted the activitiy of WNT signaling and thus affect the specification pattern of cardiac progenitors. To manipulate the activity of these signalling pathways could be an effective way to improve the yield of nodal-like cells for human cell based biopacemakers in the furture. Yechikov S and et al. recently reported they improved the yield of pacemaker-like cells up to 2.4 fold by interfering the activity of NODAL and WNT pathway in cardiac mesoderm stage [42]. Here our data indicatd that beyond its direct regulation of SAN specific genes, E2A may also play multiple actions on regulating the activity of NOTCH and other pathways to augment the proportion of nodal like cells in the heterogenous cardiomyocyte population. To address the above question, single cell sequencing could be a better solution to isolate subpopulations from the heterogenous cardiomyocytes and traces developmental fates of cardiomyocyte lineages during directed differentiation in the future.
Based on these criteria, we reasoned that E2A deficiency interfered both the specification and differentiation of nodal-like cells in our cardiac-specific differentiation system. It has drawn much attention for E2A-mediated transcriptional regulation contributed in differentiation of many cell lineages, including lymphoid, muscle, and neuronal cells [50], [51], [52], [53]. However its role in cardiac development remains unrevealed. Of note, Cunningham and others pointed out that E2A acts as the antagonist of cardiogenetic mesoderm formation from mESCs and the inhibitory activity were regulated by another bHLH protein member, Id proteins [10]. In their work, E2A knockdown by siRNA efficiently promoted the emergence of KDR positive cardiogeneic mesoderm progenitors. Their conclusion about the regulatory effect of E2A is inconsistent with our study. This may be caused by the very dissimilarity between mouse and human ESCs and in their in vitro culture/differentiation conditions. Interestingly, we also noticed that the expression level of the other two E proteins (HEB and E2-2) were up-regulated in E2A knockout hESCs and cardiomyocytes (data not shown). The complementarity or redundancy among these E proteins have been fairly confirmed in embryonic differentiation and development [54]. Whether combined or total manipulation of E protein expression benefit the differentiation efficiency of nodal-like cardiomyocytes worth further exploration. Moreover, E47 and E12, two variants of E2A generated by alternative splicing, have been identified to display different roles in stem cell pluripotency maintenance and differentiation [55], [56], [57]. Since they have biased preferences for heterodimer partners and E box motifs, it will be worth to explore the potential role of these two isoforms and their partners during cardiomyocytes differentiation in the future.
In summary, our study revealed a new role of the transcription factor E2A during directed differentiation of hESCs into cardiomyocytes that E2A ablation substantially increased the proportion of nodal-like cells. Our study may provide an important clue to develop the approach to generate distinct cardiomyocyte subtypes, and further provide SAN-like cardiomyocytes for biological cardiac pacemaker therapy.
Contributors
Xiuya Li, Fei Gao, Xiaochen Wang and Qianqian Liang: Considered equal contribution; Xiuya Li and Fei Gao carried out the majority of the experiments, performed analyses, and generated data figures and supplemental information, and Xiuya Li wrote the first draft of our manuscript; Xiaochen Wang carried out Whole cell patch clamp recordings generated data figures, and participated in writing the manuscript; Qianqian Liang and Aobing Bai contributed significant roles in RNA-seq and subsequent Bioinformation analysis; Zhuo Liu, Xinyun Chen and Ermin Li: Contributed to technique support for experiments described in the manuscript. Sifeng Chen, Chao Lu and Ruizhe Qian participated in data interpretation; Ning Sun, Ping Liang and Chen Xu: conceived the study, provided funding acquisition and finalized the manuscript.
Xiuya Li, Fei Gao and Chen Xu verified the underlying data.
Funding source
This work was supported by the National Natural Science Foundation of China (No.82070391, N.S.; No.81870175 and 81922006, P.L.), the National Key R&D Program of China 2018YFC2000202 (N.S.), the Haiju program of National Children's Medical Center EK1125180102, National Key R&D Program of China 2017YFA0103700 (P.L.); Chen Xu and Qianqian Liang are members of Innovative research team of high-level local universities in Shanghai and are supported by a key laboratory program of the Education Commission of Shanghai Municipality (ZDSYS14005).
Data sharing statement
Please contact Dr. Xu and Dr. Sun for requesting all the data and reagents described in this article.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgment
We thank all members in Sun Lab and Liang lab for critical discussion and proofreading. We apologize to people whose work was relevant to but not cited in this study due to limited space.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103575.
Contributor Information
Ning Sun, Email: sunning@fudan.edu.cn.
Ping Liang, Email: pingliang@zju.edu.cn.
Chen Xu, Email: xuchenfd@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Vedantham V. New approaches to biological pacemakers: links to sinoatrial node development. Trends Mol Med. 2015;21(12):749–761. doi: 10.1016/j.molmed.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehat I, Khimovich L, Caspi O, Gepstein A, Shofti R, Arbel G. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol. 2004;22(10):1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 3.Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol. 2018;36(7):597–605. doi: 10.1038/nbt.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blazeski A, Zhu R, Hunter DW, Weinberg SH, Boheler KR, Zambidis ET. Electrophysiological and contractile function of cardiomyocytes derived from human embryonic stem cells. Prog Biophys Mol Biol. 2012;110(2-3):178–195. doi: 10.1016/j.pbiomolbio.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat Biotechnol. 2017;35(1):56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- 6.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 7.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 8.Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014;16(9):829–840. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res. 2010;107(12):1414–1427. doi: 10.1161/CIRCRESAHA.110.227058. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham TJ, Yu MS, McKeithan WL, Spiering S, Carrette F, Huang CT. Id genes are essential for early heart formation. Genes Dev. 2017;31(13):1325–1338. doi: 10.1101/gad.300400.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda R, Gunawan F, Beisaw A, Jimenez-Amilburu V, Maischein HM, Kostin S. Proteolysis regulates cardiomyocyte maturation and tissue integration. Nat Commun. 2017;8:14495. doi: 10.1038/ncomms14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li E, Li X, Huang J, Xu C, Liang Q, Ren K. BMAL1 regulates mitochondrial fission and mitophagy through mitochondrial protein BNIP3 and is critical in the development of dilated cardiomyopathy. Protein Cell. 2020 doi: 10.1007/s13238-020-00713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo F, Sun Y, Wang X, Wang H, Wang J, Gong T. Patient-specific and gene-corrected induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of short QT syndrome. Circ Res. 2019;124(1):66–78. doi: 10.1161/CIRCRESAHA.118.313518. [DOI] [PubMed] [Google Scholar]
- 14.Kehat I, Gepstein A, Spira A, Itskovitz-Eldor J, Gepstein L. High-resolution electrophysiological assessment of human embryonic stem cell-derived cardiomyocytes: a novel in vitro model for the study of conduction. Circ Res. 2002;91(8):659–661. doi: 10.1161/01.res.0000039084.30342.9b. [DOI] [PubMed] [Google Scholar]
- 15.Feld Y, Melamed-Frank M, Kehat I, Tal D, Marom S, Gepstein L. Electrophysiological modulation of cardiomyocytic tissue by transfected fibroblasts expressing potassium channels: a novel strategy to manipulate excitability. Circulation. 2002;105(4):522–529. doi: 10.1161/hc0402.102661. [DOI] [PubMed] [Google Scholar]
- 16.Vidarsson H, Hyllner J, Sartipy P. Differentiation of human embryonic stem cells to cardiomyocytes for in vitro and in vivo applications. Stem Cell Rev Rep. 2010;6(1):108–120. doi: 10.1007/s12015-010-9113-x. [DOI] [PubMed] [Google Scholar]
- 17.Mummery CL, Zhang J, Ng ES, DA Elliott, Elefanty AG, Kamp TJ. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ Res. 2012;111(3):344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301(5):H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104(3):388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 21.Liang X, Evans SM, Sun Y. Development of the cardiac pacemaker. Cell Mol Life Sci. 2017;74(7):1247–1259. doi: 10.1007/s00018-016-2400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentschler S, Yen AH, Lu J, Petrenko NB, Lu MM, Manderfield LJ. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation. 2012;126(9):1058–1066. doi: 10.1161/CIRCULATIONAHA.112.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolossov E, Lu Z, Drobinskaya I, Gassanov N, Duan Y, Sauer H. Identification and characterization of embryonic stem cell-derived pacemaker and atrial cardiomyocytes. FASEB J. 2005;19(6):577–579. doi: 10.1096/fj.03-1451fje. [DOI] [PubMed] [Google Scholar]
- 24.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93(1):32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 25.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108(3):407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62(3):489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 28.McCann RA, Armstrong CM, Skopp NA, Edwards-Stewart A, Smolenski DJ, June JD. Virtual reality exposure therapy for the treatment of anxiety disorders: an evaluation of research quality. J Anxiety Disord. 2014;28(6):625–631. doi: 10.1016/j.janxdis.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Schweizer PA, Darche FF, Ullrich ND, Geschwill P, Greber B, Rivinius R. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res Ther. 2017;8(1):229. doi: 10.1186/s13287-017-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H, Wang F, Zhang W, Yang M, Tang Y, Wang X. Overexpression of TBX3 in human induced pluripotent stem cells (hiPSCs) increases their differentiation into cardiac pacemaker-like cells. Biomed Pharmacother. 2020;130 doi: 10.1016/j.biopha.2020.110612. [DOI] [PubMed] [Google Scholar]
- 31.van Eif VWW, Stefanovic S, van Duijvenboden K, Bakker M, Wakker V, de Gier-de Vries C. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development. 2019;146(8) doi: 10.1242/dev.173161. [DOI] [PubMed] [Google Scholar]
- 32.Liang D, Xue J, Geng L, Zhou L, Lv B, Zeng Q. Cellular and molecular landscape of mammalian sinoatrial node revealed by single-cell RNA sequencing. Nat Commun. 2021;12(1):287. doi: 10.1038/s41467-020-20448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gude N, Joyo E, Toko H, Quijada P, Villanueva M, Hariharan N. Notch activation enhances lineage commitment and protective signaling in cardiac progenitor cells. Basic Res Cardiol. 2015;110(3):29. doi: 10.1007/s00395-015-0488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bleeker WK, Mackaay AJ, Masson-Pevet M, Bouman LN, Becker AE. Functional and morphological organization of the rabbit sinus node. Circ Res. 1980;46(1):11–22. doi: 10.1161/01.res.46.1.11. [DOI] [PubMed] [Google Scholar]
- 35.DiFrancesco D. The role of the funny current in pacemaker activity. Circ Res. 2010;106(3):434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- 36.Baruscotti M, Bucchi A, Difrancesco D. Physiology and pharmacology of the cardiac pacemaker ("funny") current. Pharmacol Ther. 2005;107(1):59–79. doi: 10.1016/j.pharmthera.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 37.DiFrancesco D. Funny channels in the control of cardiac rhythm and mode of action of selective blockers. Pharmacol Res. 2006;53(5):399–406. doi: 10.1016/j.phrs.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25(5):1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 39.Bosman A, Sartiani L, Spinelli V, Del Lungo M, Stillitano F, Nosi D. Molecular and functional evidence of HCN4 and caveolin-3 interaction during cardiomyocyte differentiation from human embryonic stem cells. Stem Cells Dev. 2013;22(11):1717–1727. doi: 10.1089/scd.2012.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31(1):54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94(3):439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 42.Yechikov S, Kao HKJ, Chang CW, Pretto D, Zhang XD, Sun YH. NODAL inhibition promotes differentiation of pacemaker-like cardiomyocytes from human induced pluripotent stem cells. Stem Cell Res. 2020;49 doi: 10.1016/j.scr.2020.102043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol. 2009;327(2):376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye W, Wang J, Song Y, Yu D, Sun C, Liu C. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 2015;142(14):2521–2532. doi: 10.1242/dev.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Y, Lee MY, Schliffke S, Paavola J, Amos PJ, Ge X. Small molecule Wnt inhibitors enhance the efficiency of BMP-4-directed cardiac differentiation of human pluripotent stem cells. J Mol Cell Cardiol. 2011;51(3):280–287. doi: 10.1016/j.yjmcc.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Wobus AM, Kaomei G, Shan J, Wellner MC, Rohwedel J, Ji G. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol. 1997;29(6):1525–1539. doi: 10.1006/jmcc.1997.0433. [DOI] [PubMed] [Google Scholar]
- 48.Terami H, Hidaka K, Katsumata T, Iio A, Morisaki T. Wnt11 facilitates embryonic stem cell differentiation to Nkx2.5-positive cardiomyocytes. Biochem Biophys Res Commun. 2004;325(3):968–975. doi: 10.1016/j.bbrc.2004.10.103. [DOI] [PubMed] [Google Scholar]
- 49.Gessert S, Kuhl M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res. 2010;107(2):186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- 50.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29(2):217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6(8):1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- 52.Pfurr S, Chu YH, Bohrer C, Greulich F, Beattie R, Mammadzada K. The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development. Development. 2017;144(21):3917–3931. doi: 10.1242/dev.145698. [DOI] [PubMed] [Google Scholar]
- 53.Hashimoto Y, Tsutsumi M, Myojin R, Maruta K, Onoda F, Tashiro F. Interaction of Hand2 and E2a is important for transcription of Phox2b in sympathetic nervous system neuron differentiation. Biochem Biophys Res Commun. 2011;408(1):38–44. doi: 10.1016/j.bbrc.2011.03.113. [DOI] [PubMed] [Google Scholar]
- 54.Yoon SJ, Foley JW, Baker JC. HEB associates with PRC2 and SMAD2/3 to regulate developmental fates. Nat Commun. 2015;6:6546. doi: 10.1038/ncomms7546. [DOI] [PubMed] [Google Scholar]
- 55.Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2009;5(11) doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamazaki T, Liu L, Lazarev D, Al-Zain A, Fomin V, Yeung PL. TCF3 alternative splicing controlled by hnRNP H/F regulates E-cadherin expression and hESC pluripotency. Genes Dev. 2018;32(17-18):1161–1174. doi: 10.1101/gad.316984.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tijchon E, Havinga J, van Leeuwen FN, Scheijen B. B-lineage transcription factors and cooperating gene lesions required for leukemia development. Leukemia. 2013;27(3):541–552. doi: 10.1038/leu.2012.293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.