Abstract
Abstract
Bioprospecting for innovative basidiomycete cytochrome P450 enzymes (P450s) is highly desirable due to the fungi’s enormous enzymatic repertoire and outstanding ability to degrade lignin and detoxify various xenobiotics. While fungal metagenomics is progressing rapidly, the biocatalytic potential of the majority of these annotated P450 sequences usually remains concealed, although functional profiling identified several P450 families with versatile substrate scopes towards various natural products. Functional knowledge about the CYP5035 family, for example, is largely insufficient. In this study, the families of the putative P450 sequences of the four white-rot fungi Polyporus arcularius, Polyporus brumalis, Polyporus squamosus and Lentinus tigrinus were assigned, and the CYPomes revealed an unusual enrichment of CYP5035, CYP5136 and CYP5150. By computational analysis of the phylogeny of the former two P450 families, the evolution of their enrichment could be traced back to the Ganoderma macrofungus, indicating their evolutionary benefit. In order to address the knowledge gap on CYP5035 functionality, a representative subgroup of this P450 family of P. arcularius was expressed and screened against a test set of substrates. Thereby, the multifunctional enzyme CYP5035S7 converting several plant natural product classes was discovered. Aligning CYP5035S7 to 102,000 putative P450 sequences of 36 fungal species from Joint Genome Institute-provided genomes located hundreds of further CYP5035 family members, which subfamilies were classified if possible. Exemplified by these specific enzyme analyses, this study gives valuable hints for future bioprospecting of such xenobiotic-detoxifying P450s and for the identification of their biocatalytic potential.
Graphical abstract
Key points
• The P450 families CYP5035 and CYP5136 are unusually enriched in P. arcularius.
• Functional screening shows CYP5035 assisting in the fungal detoxification mechanism.
• Some Polyporales encompass an unusually large repertoire of detoxification P450s.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-021-11444-2.
Keywords: P. arcularius, Enzyme discovery, CYPome, CYP5035, Detoxification
Introduction
The genomes of white-rot Basidiomycota code for the highest percentage of CYPome compared to the total proteome of all organisms (Chen et al. 2014). Such high density of cytochrome P450 enzymes (P450s/CYPs) helps these fungi to fully degrade the most recalcitrant aromatic polymer lignin or its low molecular weight degradation products (Peralta et al. 2017) and to survive in harsh conditions by detoxifying a vast variety of plant-based xenobiotics and other environmental hazards (Kües 2015). In order to investigate these fascinating features of fungi and their enzyme toolbox for their application in bioenergy processing or bioremediation (Yadav et al. 2019; Mäkelä et al. 2020), programmes such as the 1000 Fungal Genome project were initiated and already sequenced numerous genomes of white-rot fungi (Grigoriev et al. 2011; Grigoriev et al. 2014), providing free access to an incredible amount of P450 sequences.
Computational efforts were mobilised to annotate such enzyme sequences with unknown functions (Gerlt et al. 2011; McKay et al. 2015). With the model white-rot fungus Phanerochaete chrysosporium being the first (Syed and Yadav 2012), the P450s of several other wood-degrading basidiomycetes were analysed computationally (Suzuki et al. 2012; Syed et al. 2013a; Syed et al. 2014; Hori et al. 2014; Kües et al. 2015; Zhu et al. 2015; Mgbeahuruike et al. 2017) and grouped into (sub)families according to the 40% (family) and 55% (subfamily) sequence identity rules of the International P450 Nomenclature Committee (Nelson 2006). Yet, such low sequence identity, usually high selectivity of P450 enzymes and a scarce number of functionally characterised basidiomycete P450s render simulated predictions of biochemical capacities extremely difficult (Ichinose 2013). Even the closest homologues may have divergent reactivity (Gerlt 2007). Therefore, some studies attempted to express and functionally analyse the entire CYPome of P. chrysosporium and model brown-rot fungus Postia placenta (Hirosue et al. 2011; Ide et al. 2012; Ichinose 2013). A few individual P450s with interesting activities were also looked at more closely (Kasai et al. 2009; Syed et al. 2010; Kasai et al. 2010a; Kasai et al. 2010b; Chigu et al. 2010; Syed et al. 2011; Ichinose and Wariishi 2012; Syed et al. 2013b; Syed et al. 2013c; Hatakeyama et al. 2016; Sakai et al. 2018; Yang et al. 2018; Wang et al. 2019). However, the research about fungal P450s is still in its early stages and often shares ideas and limited information rather than comprehensive details on the P450 function. Generally, the biocatalytic repertoire of basidiomycetes as a whole remains greatly understudied (Schmidt-Dannert 2016). Compared to bacterial P450 enzymes, one reason for this might be the more challenging recombinant expression and scarce availability in general, as well as the lack of access to isolated enzymes. In part, this is due to the generally low expression of mostly membrane-bound enzymes and their need for several co-expressed interacting peptide chains to become enzymatically fully functional.
A comparison of the CYPomes of six model wood-degrading fungi revealed that 11 out of 68 P450 families were enriched, including CYP5035, CYP5136 and CYP5150 (Syed et al. 2014). While a few studies analysed enzymes of the latter two families (Syed et al. 2011; Ichinose and Wariishi 2012; Syed et al. 2013c; Hatakeyama et al. 2016), the function of CYP5035 is still inconclusive (Syed et al. 2014). Four members of its subfamilies A and B from P. chrysosporium were expressed in yeast and accepted naproxen, flavone or dehydroabietic acid to form yet unresolved products (Hirosue et al. 2011). However, no activity could be observed for the 13 other expressed CYP5035 enzymes of subfamilies A–E from the same fungus (Hirosue et al. 2011) or of subfamily F of P. placenta (Supplementary Table S1) (Ide et al. 2012). Hence, the knowledge about this enriched P450 family is largely insufficient and calls for further investigation.
In this study, putative P450 sequences from publicly available genomes of phylogenetically closely related (Krüger and Gargas 2004; Sotome et al. 2008; Seelan et al. 2015) white-rot fungi Polyporus arcularius, Polyporus brumalis, Polyporus squamosus and Lentinus tigrinus were extracted and their P450 families assigned. Thereby, it was noticed that the CYP5035 and CYP5136 families were enhanced even more than in any of the model white-rot fungi already analysed in the literature, except in Ganoderma species. By sketching their phylogeny in contrast to P. chrysosporium and other model basidiomycetes, the evolution of these enzyme families was studied in the systematic order of Polyporales. To fill part of the described functional gap, a representative group of nine CYP5035 sequences dispersed over the available CYP5035 subfamilies in P. arcularius were heterologously expressed in Komagataella phaffii (Pichia pastoris) and screened for activity towards a test set of structurally diverse substrates representing several different natural product classes. Thereby, a promiscuous CYP5035 was discovered.
Materials and methods
All solvents and chemicals were purchased from Sigma-Aldrich/Merck (Steinheim/Darmstadt, Germany), VWR International (Fontenay-sous-Bois, France), Carl Roth GmbH (Karlsruhe, Germany) or Fisher Scientific (Loughborough, UK) in best available purity and were used as received without further purification. HPLC tubes were bought from Macherey-Nagel (Düren, Germany) and the corresponding caps and inserts from Bruckner Analysentechnik (Linz, Austria). An Agilent Technologies 1100 Series executed the HPLC analysis, and a Shimadzu GCMS-QP2010 SE instrument equipped with an AOC-20i/s autosampler and injector unit together with a Zebron ZB-5MSi capillary column (30 m × 0.25 mm × 0.25 μm, Phenomenex) performed the GC-MS measurements. OD values were determined with an Eppendorf BioPhotometer plus. The CYP5035 coding regions, identified from the publicly available databases, were ordered as double-stranded DNA fragments from TWIST Bioscience. Cells of P. pastoris with expressed and versatile P450 3A4 were obtained from bisy GmbH (Hofstaetten, Austria) and used as a positive control for biotransformations. These cells had been cultivated, then stored as frozen pellets at −80°C. Figures were generated in the programmes GraphPad Prism 8 and CS ChemDraw Ultra.
Fungal CYPome determination
Publicly available protein sequences of the genomes of the following species were downloaded from the Joint Genome Institute (JGI) Genome Portal website (https://genome.jgi.doe.gov/portal/) of the US Department of Energy (Grigoriev et al. 2014):
-
Polyporus arcularius (Varga et al. 2019) HHB13444 v1.0: Project: 1006899
-
Polyporus brumalis (Miyauchi et al. 2018) BRFM 1820 v1.0: Project: 1051563
-
Polyporus squamosus CCBS676 v1.0: Project 1108915
(https://genome.jgi.doe.gov/portal/Polsqu1/Polsqu1.download.html);
-
Lentinus tigrinus (Wu et al. 2018) ALCF2SS1-6 v1.0: Project 1020066
(https://genome.jgi.doe.gov/portal/Lenti6_1/Lenti6_1.download.html).
In each case, the Files -> Annotation -> Filtered Models (‘best’) -> Proteins -> ‘Species’_GeneCatalog_proteins.aa.fasta.gz files were used.
Additionally, the necessary genome P450s sequences (Martinez et al. 2004; Martinez et al. 2009; Eastwood et al. 2011; Suzuki et al. 2012; Floudas et al. 2012; Morin et al. 2012; Binder et al. 2013) or CYPome statistics of the fungi Phanerochaete chrysosporium (Syed and Yadav 2012; Syed et al. 2014), Phanerochaete carnosa (Suzuki et al. 2012; Syed et al. 2014), Agaricus bisporus (Syed et al. 2014), Postia placenta (Ide et al. 2012; Syed et al. 2014), Ganoderma sp. (Syed et al. 2013a; Syed et al. 2014; Kües et al. 2015), Serpula lacrymans (Syed et al. 2014), Trametes versicolor (Syed and Mashele 2014), Bjerkandera adusta (Syed et al. 2013a), Phlebia brevispora (Syed et al. 2013a), Heterobasidion irregulare (Mgbeahuruike et al. 2017), Phlebiopsis gigantea (Hori et al. 2014), Lignosus rhinoceritis (Yap et al. 2014), Ganoderma lucidum (Chen et al. 2012; Kües et al. 2015) and Ganoderma sinense (Zhu et al. 2015) were obtained from the cited literature or accessed according to their instructions (Online Resource 1).
The CYPomes of P. arcularius, P. brumalis, P. squamosus and L. tigrinus (Online Resource 2) were determined according to the P450 identification and annotation strategy described by Syed and Mashele (2014) with a slight adjustment as the old BLAST server on the Cytochrome P450 Homepage (Nelson 2009) did not work and the new P450 BLAST page (http://www.p450.unizulu.ac.za/?page_id=21) had not been installed at the time:
Superfamily annotation of the protein sequences by the Batch CD-Search Tool of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi).
Verification of the P450 signature motifs ‘E-x-x-R’ and ‘C-x-G’ in the putative P450 sequences using the ScanProsite tool (https://prosite.expasy.org/scanprosite/).
P450 family assignment by applying the FCPB BLAST Search of the verified P450 protein sequence (http://p450.riceblast.snu.ac.kr/blast.php) (Moktali et al. 2012).
Verification of the P450 family assignment by alignment against already assigned P450 sequences of previous publications using the Protein BLAST option of the NCBI. P450 families were verified according to the 40% (family) and 55% (subfamily) sequence identity rules of the International P450 Nomenclature Committee (Nelson 2006).
Step 4 was of utter importance to assign less common P450 families correctly because the FCPB did not consider enzyme families of the Ganoderma macrofungus. P450 sequences that did not get a good match were left unassigned.
Evolutionary P450 sequence analysis
Following the same strategy as Syed et al. (2014), evolutionary analyses of the desired P450 protein sequences were conducted in MEGA X (Kumar et al. 2018; Stecher et al. 2020). The evolutionary history was inferred using the minimum evolution method (Rzhetsky and Nei 1992). The evolutionary distances were computed using the Poisson correction method (Zuckerhandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. The minimum evolution tree was searched using the close-neighbour-interchange algorithm (Nei and Kumar 2000) at a search level of 1. The neighbour-joining algorithm (Saitou and Nei 1987) was used to generate the initial tree. All ambiguous positions were removed for each sequence pair (pairwise deletion option).
CYP5035 enzyme expression in Pichia pastoris
Due to the advantages for the functional overexpression of membrane-bound eukaryotic P450 enzymes and success in co-expression experiments of foreign P450 genes with the P450 reductase of P. pastoris (PpCPR) described previously (Geier et al. 2012), representative P. arcularius P450 genes were expressed by P. pastoris. Nine CYP5035 sequences of P. arcularius were ordered as synthetic double-stranded DNA fragments from TWIST Bioscience, amplified and cloned into the expression vector (Supplementary Fig. S1) equipped with Zeocin resistance and a bidirectional promoter for co-expression of the PpCPR gene by Gibson assembly. The P. pastoris strain BSYBG11 (aox1Δ, MUTS), a derivative of P. pastoris strain BG08 of BioGrammatics Inc. (Carlsbad, USA) (Sturmberger et al. 2016), was transformed with the resulting linearised plasmids for genomic integration of the expression cassettes. Small-scale cultivations were done for the 7-methoxy-4-(trifluoromethyl)coumarin (MFC) demethylation assay and carried out following the deep-well plate (DWP) and induction protocols reported previously (Weis et al. 2004). For the substrate screening, cultivations were scaled up to 250-mL shake-flasks inoculating 45 mL BMD1 (pH 7.4), adding 5 mL BMM10 (pH 7.4) after 60 h (Weis et al. 2004) and three times further feeding with 0.5 mL methanol every 12 h. Having harvested and washed cells twice in 50 mM potassium phosphate buffer (pH 7.4), cells were resuspended in the same phosphate buffer until an OD600 of 100 was obtained. A cell broth volume of 400 μL was filled into each well of a DWP and 4 μL of the 100 mM compound stock solutions added to get a final substrate concentration of 1 mM. The biotransformations were carried out for 17 h at 28°C, 80% humidity and 320 rpm in a tilted orientation on the shaker to ensure maximal oxygen availability. After stopping the reaction with the addition of 300 μL of an acetonitrile/methanol (1:1; v/v) solution, the resulting mixture was vortexed and centrifuged and 200 μL of supernatant of each well were transferred into 96-well GreinerV plates for HPLC analysis. The biotransformations of all compounds were analysed by HPLC, except for squalene by GC-MS. Percentage conversions were calculated by peak area integration of new peaks that were not present in the negative control (Supplementary Fig. S2). Separation was carried out via a Kinetex C18 (100 Å; 50 × 4.6 mm; 2.6 μm) reverse-phase column. Water containing 0.1% acetic acid (A) and acetonitrile (B) was used for elution at 25°C in the following ratios: 0 min: A/B 80/20; 1 min: A/B 80/20; 1.01 min: A/B 50/50; 4 min: A/B 0/100; 5.50 min: A/B 0/100; 5.51 min: A/B 80/20; and 6.50 min: A/B 80/20. GC-MS analysis; equal volumes of dichloromethane containing 0.01% undecane were added to the biotransformations and after phase separation, the organic layer was dried with anhydrous Na2SO4. The following method (linear velocity of 39.5 cm s−1 using He carrier gas; total and column flow of 15.2 mL min−1 and 1.21 mL min−1, respectively; injection temperature of 250°C; split ratio of 9.1) was applied: 35°C for 5 min, 20°C min−1 to 300°C and 300°C for 5 min in a total run time of 23.25 min.
Results
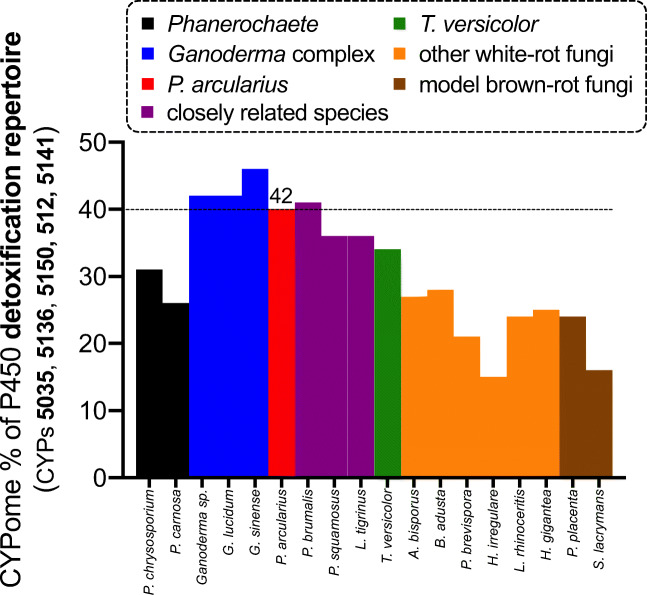
Using the publicly available genome and translated protein sequences from the JGI Genome Portal website (Grigoriev et al. 2014), a genome-wide search for putative P450s in P. arcularius (Supplementary Fig. S3) was carried out following the identification and annotation strategy of Syed and Mashele (2014) (Online Resource 2) with a few adjustments as outlined in the ‘Materials and methods’ section. The CYPome of P. arcularius showed a similar collection of P450s to that of Ganoderma sp., primarily owing to the presence of numerous CYP5359 and a few CYP5144 (Online Resource 1). Likewise, a CYPome comparison between P. arcularius and well-known model white- (e.g. P. chrysosporium) and brown-rot (e.g. P. placenta) fungi analysed extensively by previous studies (Syed et al. 2013a; Syed et al. 2014; Syed and Mashele 2014) revealed an unusually large number of CYP5035 (23) and CYP5136 (12) in this fungus: approximately 2-fold more (13; 5) than in P. chrysosporium (Fig. 1). In fact, even when extending the comparison to a total of 17 wood-degrading fungi, P. arcularius had the highest percentages of these two P450 families in its CYPome (Supplementary Figs. S4 and S5). Only the absolute number of CYP5035 and CYP5136 was surpassed slightly by Ganoderma sinense and P. brumalis, respectively (Supplementary Figs. S6 and S7) (Zhu et al. 2015). Additionally, CYP5150 coding gene sequences were frequent in the genome (Supplementary Fig. S8), with the percentage of this family in the genome higher in Ganoderma species and Trametes versicolor (Supplementary Fig. S9).
Fig. 1.
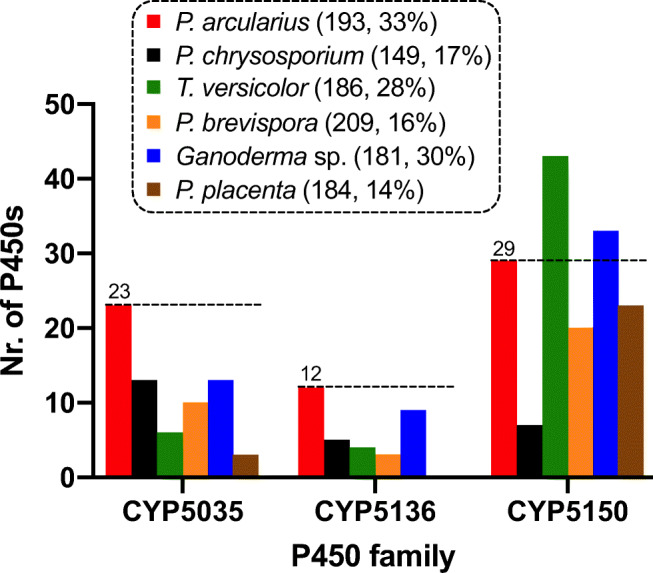
A comparison of the number of P450 genes of the families CYP5035, 5136 and 5150 identified in the genome of P. arcularius (Parc) with several model white- (P. chrysosporium, T. versicolor, Phlebia brevispora, Ganoderma sp.) and brown-rot fungi (P. placenta, Ppla) is shown. The total number of P450s and the percentage of these three families in comparison to all P450s in the translated genome of each species are given in parenthesis. A more detailed CYPome comparison is provided in Online Resource 1
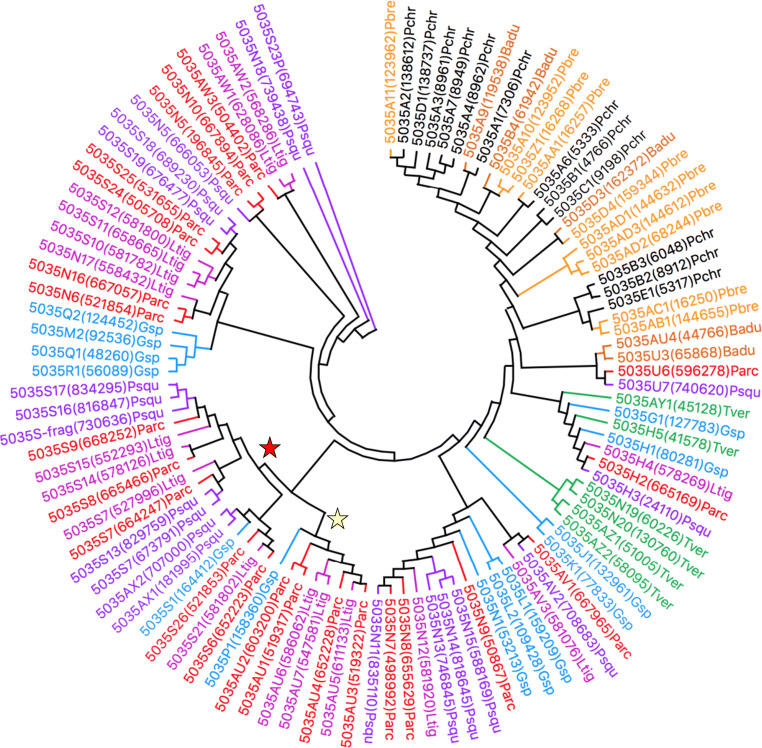
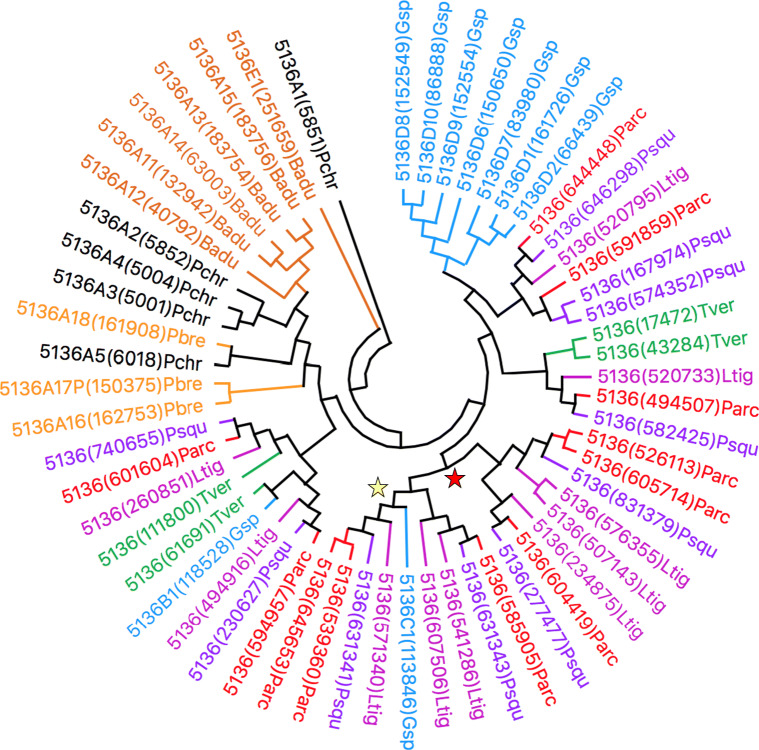
When consulting literature articles for the relative phylogeny of P. arcularius to that of white-rot fungal species for which CYPomes had been analysed previously (Justo and Hibbett 2011; Floudas et al. 2012; Binder et al. 2013), the Ganoderma macrofungus indeed turned out to be the fungus most closely related (Supplementary Fig. S10). Perhaps the evolved enrichment of CYP5035 and CYP5136 had its origin at approximately this branching point in fungal diversification and continued downstream to P. arcularius. In order to answer this hypothesis, the CYPomes of P. brumalis (Supplementary Fig. S11) (Miyauchi et al. 2018), P. squamosus (Supplementary Fig. S12) and L. tigrinus (Supplementary Fig. S13) (Wu et al. 2018) were determined according to the aforementioned genome-wide P450 identification strategy. The selected fungal genomes unveiled a similarly high number of CYP5035 and CYP5136 (Online Resource 1). The computation of minimum evolution trees of both P450 families shown in Figs. 2 and 3, respectively, further supported the proposed evolutionary theory. Due to its close relationship to P. arcularius (Supplementary Fig. S14), P. brumalis was only included together with further white- and brown-rot fungi in extended phylogenetic trees (Supplementary Figs. S15 and S16), which contributed towards the same conclusions.
Fig. 2.
A minimum evolution tree of the CYP5035 family involving 103 amino acid sequences from eight different organisms. The phylogeny of CYP5035 enzymes of the fungus P. arcularius (Parc; red) compared to related species L. tigrinus (Ltig; violet) and P. squamosus (Psqu; purple) and the other model white-rot fungi Ganoderma sp. (Gsp; blue), T. versicolor (Tver; green), P. chrysosporium (Pchr; black), B. adusta (Badu; dark orange) and P. brevispora (Pbre; orange) in order to get an insight into the evolution of this P450 family. The yellow or red stars indicate a diversification process and new branch of the P450 families compared to Ganoderma sp., respectively. The tree was constructed using the close-neighbour-interchange algorithm in MEGA X. An extended tree can be found in Supplementary Fig. S15
Fig. 3.
Displayed is a minimum evolution tree of the CYP5136 family involving 58 amino acid sequences from eight different organisms. Phylogeny of CYP5136 enzymes of fungus P. arcularius (Parc; red) compared to related species L. tigrinus (Ltig; violet) and P. squamosus (Psqu; purple) and the other model white-rot fungi Ganoderma sp. (Gsp; blue), T. versicolor (Tver; green), P. chrysosporium (Pchr; black), B. adusta (Badu; dark orange) and P. brevispora (Pbre; orange) in order to get an insight into the evolution of this P450 family. The yellow or red stars indicate a diversification process and new branch of the P450 families compared to Ganoderma sp., respectively. The tree was constructed using the close-neighbour-interchange algorithm in MEGA X. An extended tree can be found in Supplementary Fig. S16
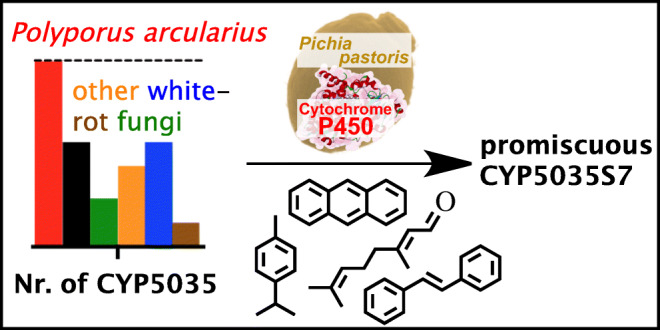
The presence of the large number of CYP5035 sequences in the genome of P. arcularius awoke our interest in studying this P450 family in more detail. It was thus decided to pick a small, representative selection of nine CYP5035 sequences of P. arcularius distributed among the available subfamilies (Supplementary Table S1, Online Resource 2) to express them in P. pastoris and to test their activities and substrate scope employing recombinant whole-cell biotransformation. These nine CYP5035 were cloned into bidirectional co-expression plasmids (Vogl et al. 2018) together with P. pastoris’ native P450 reductase (Supplementary Figure S1). Fourteen transformants of each CYP5035 variant were screened for activity using the MFC demethylation assay (Donato et al. 2004) to select the best clonal variants. Although only CYP5035S7 was active when employing the simple MFC screening assay, the formation of a blue colour upon conversion of indole to indigo (Çelik et al. 2005) by both CYP5035S7 and CYP5035H2 indicated successful expression and oxidative activity also for other expressed P450 genes, which encouraged us to move forward with randomly selected individual transformants for each of the enzymes other than CYP5035S7 and test alternative substrates by chromatographic analysis. For this substrate specificity screening, the selected transformants were cultivated and applied in whole-cell biotransformation experiments with over 40 structurally diverse and complementary compounds of eight different natural product classes (terpenes, steroids, alkaloids, stilbenoid and flavonoid backbones, phenylpropanoids, fatty acid derivative and coumarins) and also pharmaceuticals, (nitrogen-containing) polycyclic aromatic hydrocarbons ((N)PAHs) and other chemicals. Due to its known broad substrate acceptance, the human P450 3A4 co-expressed with its human CPR in P. pastoris was used as a positive control for all biotransformations (Fessner et al. 2020). As a negative control, an empty vector strain only overexpressing the yeast’s intrinsic P450 reductase was used to identify a possible involvement of intrinsic P. pastoris enzymes in the substrate conversions.
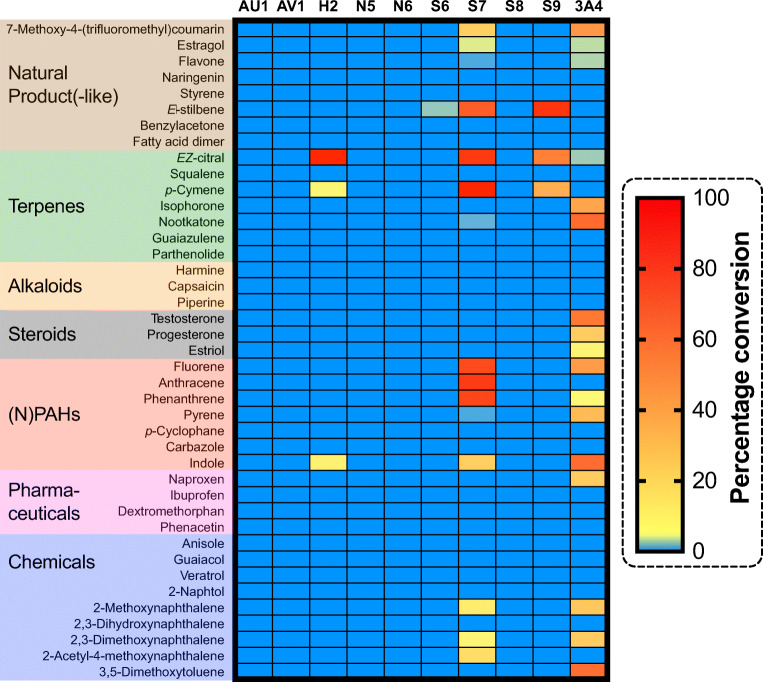
The heat map in Fig. 4 illustrates the activity pattern that was obtained from the HPLC or GC analysis. Notably, the observed activities imply different substrate scopes even among P450s of the same subfamily CYP5035S. While the three enzymes CYP5035H2, -S6 and -S9 only converted at least one of (E)-stilbene, (E/Z)-citral, p-cymene and indole, the fourth variant CYP5035S7 demonstrated a much larger substrate scope being active especially on PAHs and terpenes but also across other natural product classes such as the phenylpropanoid estragol and the stilbenoid-backbone (E)-stilbene (Fig. 5). In contrast, cells with expression constructs for CYP5035S8 and four other individual CYP5035 genes did not show any activity whatsoever, and their expression constructs were not further studied.
Fig. 4.
This heat map illustrates the result of the recombinant whole-cell activity screening of P. pastoris transformants expressing nine different CYP5035 enzymes of P. arcularius and the positive control co-expressing human P450 3A4 together with human CPR. An empty vector strain overexpressing the yeast’s intrinsic P450 reductase only was used as a negative control to identify possible background conversions caused by intrinsic P. pastoris enzymes. The percentage conversions of the initial 1 mM substrate concentrations were calculated from peak integration of newly appeared peaks in HPLC profiles (Supplementary Fig. S2). Only the biotransformation of squalene was analysed by GC-MS. Reaction conditions were as follows: OD600 of 100, 28°C, 320 rpm and 1 mM substrate concentration
Fig. 5.
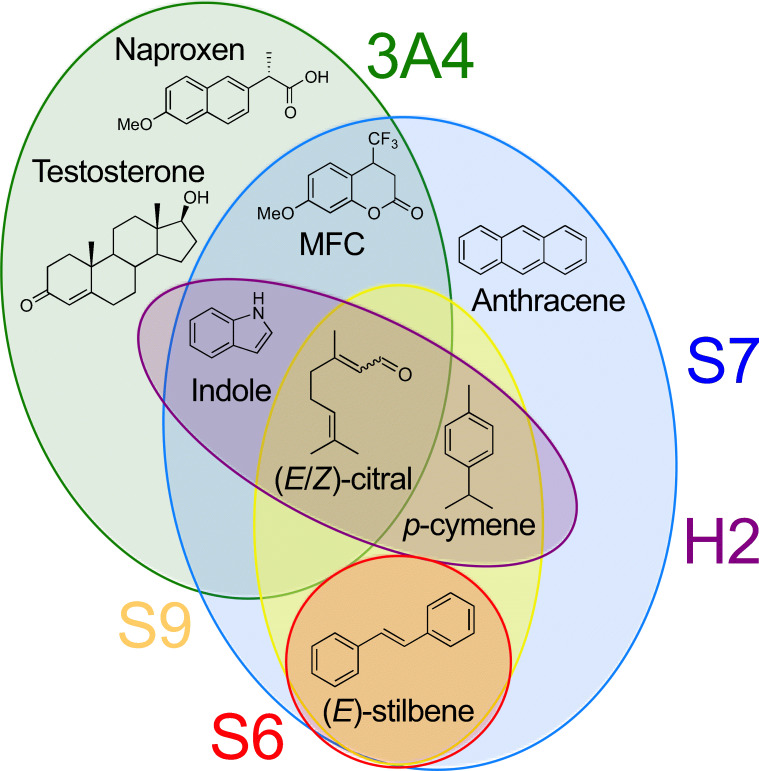
Venn diagram illustrating the catalytic promiscuity of several accepted substrates of the CYP5035 candidates and human P450 3A4 used for the whole-cell activity screening displayed in Fig. 4. Only a fraction of the substrates tolerated by only one or both of CYP5035S7 and P450 3A4 is shown
Having identified the promiscuity of CYP5035S7, a BLAST search for hits with this enzyme sequence against the personal P450 collection of David Nelson encompassing 102,000 sequences from JGI was performed to locate orthologous CYP5035 or similar sequences in other fungi (Online Resource 3). A total of 314 sequences in 36 different fungal species were longer than 450 amino acids and aligned with >40% identity, indicating an allocation to the same CYP5035 family, though only sequences of fungi used in this study were found to belong to the same subfamily with >55% identity. Hence, better allocation was achieved by blasting the 314 sequences each against all CYP5035 sequences named so far (Online Resource 3). The results were sorted by subfamily, percentage identity and species.
Discussion
P. arcularius was selected for this work because in 2019, a study took the fungus into closer consideration as one of the candidates with high potential for organic pollutant degradation (Dao et al. 2019), and in the same year, its genome was sequenced (Varga et al. 2019). The observed enrichment of CYP5035, CYP5136 and CYP5150 in the investigated white-rot Polyporales pointed towards a higher diversity of P450s compared to other white-rot fungi. Most likely, such enrichments happened in order to adapt to harsher conditions in new ecological niches and to detoxify the high diversity of degradation products in such a world of different available carbon sources. The increase in functional P450s of these two families must have secured some evolutionary advantage (Syed et al. 2014; Kües et al. 2015). Former studies had clearly classified CYP5136 and CYP5150 as enzymes participating in the fungal defence mechanism and metabolic diversity degrading both plant material and xenobiotics. Equipped with broad substrate scopes, members of these P450 families were able to oxidise hydrocarbons, plant chemicals, steroids and pharmaceuticals (Kasai et al. 2010a; Syed et al. 2011; Ichinose and Wariishi 2012; Syed et al. 2013c; Syed et al. 2014; Hatakeyama et al. 2016; Lu et al. 2019). However, more limited activity information was available for CYP5035 (Supplementary Table S1). The fact that nonwood-degrading basidiomycetes conversely only possessed a few P450 genes, and these three enriched P450 families were completely absent in their genomes, strengthened their results (Syed et al. 2014). On the other hand, the heterologous expression of CYP5150L8 from G. lucidum was recently found to enable the biosynthesis of a ganoderic acid in S. cerevisiae (Wang et al. 2018). Therefore, the observed diversity of P450s in these studied organisms also suggests the essential roles of those enzymes in the synthesis of natural products.
This study aimed at investigating the functional potential of the enriched CYP5035 genes that might harbour an evolutionary advantage for some fungal species, keeping in mind the possibility of successful bioprospecting for innovative P450 enzymes useful for synthetic and industrial purposes. For efficient heterologous expression, the yeast P. pastoris was used as the host organism due to previous success with similar enzymes (Syed et al. 2010; Syed et al. 2011; Geier et al. 2012; Syed et al. 2013c) and a broad toolbox for complex protein expressions such as stepwise inducible strong promoters and expression strategies established in our laboratory (Vogl and Glieder 2013; Vogl et al. 2014; Weninger et al. 2015; Vogl et al. 2016; Vogl et al. 2018) and the advantage of the presence of an intrinsic, fungal P450 reductase (CPR) at a low abundance of intracellular P450s. Overexpression of intrinsic yeast CPR demonstrated beneficial effects for heterologous eukaryotic P450 expression before (Braun et al. 2012). Based on literature results undertaking CYPome functionality studies, also the native CPR of the yeast Saccharomyces cerevisiae generally seemed to couple well with basidiomycete P450s across different families (Kasai et al. 2009; Kasai et al. 2010a; Kasai et al. 2010b; Hirosue et al. 2011; Nazir et al. 2011; Ide et al. 2012). It was thus presumed that the same would apply to the related yeast Pichia pastoris, which had proven to be an excellent host for P450 expression with the potential for upscaling in a bioreactor (Martinez and Rupashinghe 2013; Byrne 2015; Fessner et al. 2020). Indeed, the functional screening confirmed the expected functional interaction with the co-expressed PpCPR.
Only a small selection of nine representative CYP5035 monooxygenases of P. arcularius was picked in order to balance efforts to cover the high enzymatic diversity representatively on the one hand and the screening efforts faced for the many substrates per enzyme planned on the other. In the end, this limited CYP5035 selection was sufficient to give a representative idea of the enzyme family’s capabilities, which revealed a much broader substrate acceptance than anticipated so far. Especially, CYP5035S7 showed a broad substrate scope encompassing PAHs and several natural product classes, offering itself as an attractive P450 for natural product modification. The high conversion of PAHs (82–87%; Fig 4) is in line with several articles that identified white-rot fungal P450s of different families with remarkable PAH conversion abilities towards various ring sizes (Syed et al. 2010; Syed et al. 2011; Syed et al. 2013b). Particularly, striking was the mutual conversion of p-cymene, (E)-stilbene and (E/Z)-citral by three different CYP5035 enzymes each (Fig. 5) because carvacrol and resveratrol, which are derivatives of the former two compounds, and (E/Z)-citral itself are known fungicidal agents (Yoneyama and Natsume 2010; Jian et al. 2016).
Since only CYP5035S7 was active in the fluorescence assay employing substrate MFC for activity screening, transformants of the other tested CYP5035 expression strains were picked randomly encouraged by the visible blue colour upon indigo formation also by CYP5035H2, which suggested possible activity of the enzymes beyond the MFC demethylation assay. Especially for the highly selective families of P450 enzymes, surrogate screening substrates only provide insufficient information about the real biocatalytic potential. This subset of nine new and different P450 genes was expected to provide also some information about which fraction of new fungal P450 genes can be expected to be functionally expressed in such blind discovery approach. Previously published studies demonstrated that functional expression of basidiomycete P450s is challenging and commonly based on trial and error (Schmidt-Dannert 2016). For five of the nine individual CYP5035 genes, which did not show any activity, it remains unclear if the lack of activity was due to the lack of a suitable substrate in the employed test set or if functional overexpression using the P. pastoris/PpCPR system failed. Due to the usually low expression of recombinant membrane-bound P450s, the lack of suitable antibodies and the limited information about P450 functionality, which can be obtained from P450 quantification by carbon monoxide spectroscopy in the case of multicomponent membrane-bound P450 systems, those expression constructs were not further studied in this functional approach for P450 screening. Ultimately, authentic activity information and the efficient bioprospecting for fungal enzymes towards industrial application rely on efficient heterologous expression (Mitrovic and Glieder 2015).
Interestingly, none of the active enzymes tested here accepted any of the tested active pharmaceutical ingredients (APIs), although three of four active CYP5035 of P. chrysosporium converted naproxen in a previous study (Hirosue et al. 2011). These observations highlight the deviating nature of P450s of phylogenetically similar species or even the same subfamily with sequence identities of >60% but different chemoselectivity. However, this study identified with CYP5035S7, an interesting new P450 that showed in this study a similarly versatile substrate acceptance as the broadly active human P450 3A4, which initiates detoxification pathways for many compounds in the human liver. The high conversion rates (82–100% for red squares; Fig. 4) for the test substrates in the applied substrate screening procedure suggest sufficient expression and activity for preparative applications just as demonstrated for human P450 3A4 whole-cell biocatalyst before (Fessner et al. 2020). At the same time, different candidates of CYP5035 variants have now been shown to be multifunctional with a diverse catalytic activity oxidising PAHs, pharmaceuticals and various plant materials encompassing fungicidal agents, which strongly suggests that CYP5035 are part of the fungal detoxification mechanism just like CYP5136 and CYP5150.
In addition to the other two enriched families CYP5136 and CYP5150 mentioned earlier, this further increases the percentage of those enriched P450s with a detoxifying function to a third of the CYPome in the genome of P. arcularius (Supplementary Fig. S17). Apparently, P. arcularius and other species following the phylogenetic ladder up to the Ganoderma complex have an extensive repertoire of such xenobiotic-biodegrading P450s. As shown in Fig. 6, this phenomenon is preserved even when including P450 families CYP512 and CYP5141, which have also been flagged with xenobiotic-degrading functions (Syed et al. 2014). Interestingly, T. versicolor as the phylogenetic parent of the Ganoderma macrofungus also possesses an enhanced collection of such P450s in its genome compared to the other white-rot fungi. However, this is mainly due to the unmatched number of CYP5150 (Fig. 1, Supplementary Figs. S8 and S9).
Fig. 6.
A comparison of the number of P450s considered to be part of the detoxification repertoire (CYP5035, CYP5136, CYP5150, CYP512 and CYP5141) of various white-rot fungi as a percentage of the total number of P450s in the fungus’s genome. The white-rot fungi focused on by this study possess an unusually high number of such detoxifying P450s compared to model white- and brown-rot fungi
Computing the phylogeny of both P450 families with minimum evolution trees made it possible to follow the evolutionary tree of this enrichment up to the Ganoderma complex, where the diversification of CYP5035 members might have branched off. The CYP5136 family expansion was found to have occurred separately for Ganoderma species and was intensified later. These conclusions were drawn due to the following observations: (1) the closer phylogenetic P450 relation within the group of P. arcularius, P. squamosus and L. tigrinus to Ganoderma sp. than to white-rot fungi P. chrysosporium, P. brevispora, T. versicolor or Bjerkandera adusta became clearly visible. (2) An intensified diversification process of the P450 families can be noticed especially in Fig. 2 for CYP5035 in parallel to almost all of Ganoderma sp. enzymes, often yielding higher numbers of homologous P450s (examples marked by yellow stars). (3) Additionally, from some common nodes, new branches diverged, which do not contain an enzyme member of Ganoderma sp. (examples marked by red stars). (4) Therefore, the CYP5035 family expansion likely happened before speciation with Ganoderma as the starting point and continued further downstream. (5) In contrast, diversification of CYP5136 occurred separately in the case of Ganoderma as indicated by the lack of ortholog pairs and started only later in time.
It remains unclear whether the Ganoderma macrofungus itself really is the starting point or merely one of the species in the row of the evolution of CYP5035 enrichment. However, the range of 13–26 members of CYP5035 and 7–9 members of CYP5136 within Ganoderma sp., G. sinense and Ganoderma lucidum is one argument for the former. The starting point of the diversification of CYP5136 is also unknown. Syed et al. suggested P450 gene duplication due to environmental adaptation as the origin for such family expansions (Syed et al. 2014). For example, the ortholog P450 of CYP5035C1 of P. chrysosporium was duplicated several times in P. carnosa, indicating some evolutionary advantage.
The sequence alignment search of CYP5035S7 against 102,000 P450 sequences from JGI only revealed orthologous enzymes (>80% ID) within the fungal species used in this study and generally indicated relatively small amounts of CYP5035 in each of the >30 other fungal species (Online Resource 3). Only Dichomitus squalens and Earliella scabrosa possessed a considerable number of CYP5035 sequences, suggesting that large numbers of this P450 family are rather rare. In addition, 38 of the 314 blasted sequences had <55% identity and will belong to yet undefined CYP5035 subfamilies.
In combination with the observed reasonable success rate for functional recombinant expression of those membrane-bound multicomponent P450 complexes and the phylogenetic analysis of CYP5035 and CYP5136, this BLAST result provides a valuable starting point for future bioprospecting for xenobiotic-degrading P450s with activity towards plant compounds similar or complementary to P. arcularius. The large repertoire of such detoxification monooxygenases within the white-rot fungal genomes shown in Fig. 6 incites further interest because it may provide a versatile toolbox of white-rot fungal P450 enzymes for natural product modification. Information about the activities of only three subfamilies were uncovered to date, although already >50 different CYP5035 subfamilies were categorised and will increase further. Hence, despite the efforts of this study, there remains a large functional gap and an unhidden catalytic potential for CYP5035.
Despite their biosynthetic potential (Fessner 2019), the majority of P450s remains the so-called functionally uncharacterised ‘orphan P450s’ (Kelly and Kelly 2013) due to the shortage of studies investigating them (Durairaj et al. 2016) and the expression difficulties (Schmidt-Dannert 2016) also observed in this study and the sheer amount of sequences available in the sequenced genomes (Ferrer et al. 2016). Therefore, this study aimed at and substantially helped to obtain more information about the function of the hardly studied CYP5035. A surprising multifunctional enzyme CYP5035S7 from P. arcularius was identified holding a versatile synthetic potential that remains to be investigated further in lab- and pilot-scale preparative application experiments.
Supplementary Information
(PDF 5011 kb)
(XLSX 44 kb)
(XLSX 172 kb)
(XLSX 148 kb)
Acknowledgements
The authors thank Prof. Margit Winkler, Prof. Harald Pichler, Prof. Rolf Breinbauer and Prof. Wolfgang Kroutil for providing substrates for the screening. The authors also acknowledge the JGI Mycocosm Fungal Genomics Resource database for the publicly accessible genomes. The authors are grateful to Prof. Francis Martin and the CSP 1974 ‘1KFG: Deep sequencing of ecologically-relevant Dikaria’ consortium for access to the unpublished genome data of P. squamosus. The genome sequence data were produced by the US Department of Energy Joint Genome Institute in collaboration with the user community.
Author contribution
N.D.F. and A.G. conceived and designed the research. N.D.F. conducted experiments, analysed data and wrote the original manuscript. D.R.N. annotated the P450s and performed the alignment search. N.D.F. and A.G. jointly reviewed and edited the manuscript. A.G. supervised and managed scientific and financial project reporting. All authors read and approved the manuscript.
Funding
Open access funding provided by Graz University of Technology. This project has received funding from the European Union’s Horizon 2020 research and innovation programme, OXYtrain MSCA-ITN, under grant agreement no. 722390.
Data availability
The data supporting the findings of this study are available within this article and the supplementary materials. The fungal genome data, on which this study is based, are publicly available online as outlined in the ‘Materials and methods’ section.
Code availability
Any software used in this study for genomic data processing is publicly available online via the links provided in the ‘Materials and methods’ section.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
This article does not contain any studies with human participants.
Consent for publication
The authors give formal consent for the publication of this study.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Binder M, Justo A, Riley R, Salamov A, Lopez-Giraldez F, Sjökvist E, Copeland A, Foster B, Sun H, Larsson E, Larsson K-H, Townsend J, Grigoriev IV, Hibbett DS. Phylogenetic and phylogenomic overview of the Polyporales. Mycologia. 2013;105:1350–1373. doi: 10.3852/13-003. [DOI] [PubMed] [Google Scholar]
- Braun A, Geier M, Bühler B, Schmid A, Mauersberger S, Glieder A. Steroid biotransformations in biphasic systems with Yarrowia lipolytica expressing human liver cytochrome P450 genes. Microb Cell Factories. 2012;11:106. doi: 10.1186/1475-2859-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B. Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol. 2015;32:9–17. doi: 10.1016/j.sbi.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Çelik A, Speight RE, Turner NJ. Identification of broad specificity P450CAM variants by primary screening against indole as substrate. Chem Commun. 2005;3652:3652–3654. doi: 10.1039/b506156c. [DOI] [PubMed] [Google Scholar]
- Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y, Luo H, Li Y, Song J, Henrissat B, Levasseur A, Qian J, Li J, Luo X, Shi L, He L, Xiang L, Xu X, Niu Y, Li Q, Han MV, Yan H, Zhang J, Chen H, Lv A, Wang Z, Liu M, Schwartz DC, Sun C. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun. 2012;3:913. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lee M-K, Jefcoate C, Kim S-C, Chen F, Yu J-H. Fungal cytochrome P450 monooxygenases: their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol Evol. 2014;6:1620–1634. doi: 10.1093/gbe/evu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigu NL, Hirosue S, Nakamura C, Teramoto H, Ichinose H, Wariishi H. Cytochrome P450 monooxygenases involved in anthracene metabolism by the white-rot basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 2010;87:1907–1916. doi: 10.1007/s00253-010-2616-1. [DOI] [PubMed] [Google Scholar]
- Dao ATN, Vonck J, Janssens TKS, Dang HTC, Brouwer A, de Boer TE. Screening white-rot fungi for bioremediation potential of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Ind Crop Prod. 2019;128:153–161. doi: 10.1016/j.indcrop.2018.10.059. [DOI] [Google Scholar]
- Donato MT, Jiménez N, Castell JV, Gómez-Lechón MJ. Fluorescence-based assays for screening nine cytochrome P450 (P450) activities in intact cells expressing individual human P450 enzymes. Drug Metab Dispos. 2004;32:699–706. doi: 10.1124/dmd.32.7.699. [DOI] [PubMed] [Google Scholar]
- Durairaj P, Hur J-S, Yun H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb Cell Factories. 2016;15:125. doi: 10.1186/s12934-016-0523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee Y-H, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie X, Kues U, Hibbett DS, Hoffmeister D, Hogberg N, Martin F, Grigoriev IV, Watkinson SC. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Martínez-Martínez M, Bargiela R, Streit WR, Golyshina OV, Golyshin PN. Estimating the success of enzyme bioprospecting through metagenomics: current status and future trends. Microb Biotechnol. 2016;9:22–34. doi: 10.1111/1751-7915.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessner ND. P450 Monooxygenases enable rapid late-stage diversification of natural products via C−H bond activation. ChemCatChem. 2019;11:2226–2242. doi: 10.1002/cctc.201801829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessner ND, Srdic M, Weber H, Schmid C, Schoenauer D, Schwaneberg U, Glieder A. Preparative-scale production of testosterone metabolites by human liver cytochrome P450 3A4. Adv Synth Catal. 2020;362:2725–2738. doi: 10.1002/adsc.202000251. [DOI] [Google Scholar]
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St. John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- Geier M, Braun A, Emmerstorfer A, Pichler H, Glieder A. Production of human cytochrome P450 2D6 drug metabolites with recombinant microbes - a comparative study. Biotechnol J. 2012;7:1346–1358. doi: 10.1002/biot.201200187. [DOI] [PubMed] [Google Scholar]
- Gerlt JA. A protein structure (or function ?) initiative. Structure. 2007;15:1353–1356. doi: 10.1016/j.str.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Allen KN, Almo SC, Armstrong RN, Babbitt PC, Cronan JE, Dunaway-Mariano D, Imker HJ, Jacobson MP, Minor W, Poulter CD, Raushel FM, Sali A, Shoichet BK, Sweedler JV. The enzyme function initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, Kuske C, Magnuson JK, Martin F, Spatafora JW, Tsang A, Baker SE. Fueling the future with fungal genomics. Mycology. 2011;2:192–209. doi: 10.1080/21501203.2011.584577. [DOI] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:699–704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M, Kitaoka T, Ichinose H. Heterologous expression of fungal cytochromes P450 (CYP5136A1 and CYP5136A3) from the white-rot basidiomycete Phanerochaete chrysosporium: functionalization with cytochrome b5 in Escherichia coli. Enzym Microb Technol. 2016;89:7–14. doi: 10.1016/j.enzmictec.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Hirosue S, Tazaki M, Hiratsuka N, Yanai S, Kabumoto H, Shinkyo R, Arisawa A, Sakaki T, Tsunekawa H, Johdo O, Ichinose H, Wariishi H. Insight into functional diversity of cytochrome P450 in the white-rot basidiomycete Phanerochaete chrysosporium: involvement of versatile monooxygenase. Biochem Biophys Res Commun. 2011;407:118–123. doi: 10.1016/j.bbrc.2011.02.121. [DOI] [PubMed] [Google Scholar]
- Hori C, Ishida T, Igarashi K, Samejima M, Suzuki H, Master E, Ferreira P, Ruiz-Dueñas FJ, Held B, Canessa P, Larrondo LF, Schmoll M, Druzhinina IS, Kubicek CP, Gaskell JA, Kersten P, St. John F, Glasner J, Sabat G, Splinter BonDurant S, Syed K, Yadav J, Mgbeahuruike AC, Kovalchuk A, Asiegbu FO, Lackner G, Hoffmeister D, Rencoret J, Gutiérrez A, Sun H, Lindquist E, Barry K, Riley R, Grigoriev IV, Henrissat B, Kües U, Berka RM, Martínez AT, Covert SF, Blanchette RA, Cullen D. Analysis of the Phlebiopsis gigantea genome, transcriptome and secretome provides insight into its pioneer colonization strategies of wood. PLoS Genet. 2014;10:e1004759. doi: 10.1371/journal.pgen.1004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H. Cytochrome P450 of wood-rotting basidiomycetes and biotechnological applications. Biotechnol Appl Biochem. 2013;60:71–81. doi: 10.1002/bab.1061. [DOI] [PubMed] [Google Scholar]
- Ichinose H, Wariishi H. Heterologous expression and mechanistic investigation of a fungal cytochrome P450 (CYP5150A2): Involvement of alternative redox partners. Arch Biochem Biophys. 2012;518:8–15. doi: 10.1016/j.abb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Ide M, Ichinose H, Wariishi H. Molecular identification and functional characterization of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch Microbiol. 2012;194:243–253. doi: 10.1007/s00203-011-0753-2. [DOI] [PubMed] [Google Scholar]
- Jian W, He D, Song S. Synthesis, biological evaluation and molecular modeling studies of new oxadiazole-stilbene hybrids against phytopathogenic fungi. Sci Rep. 2016;6:31045. doi: 10.1038/srep31045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justo A, Hibbett DS. Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset. Taxon. 2011;60:1567–1583. doi: 10.1002/tax.606003. [DOI] [Google Scholar]
- Kasai N, Ikushiro S, Hirosue S, Arisawa A, Ichinose H, Uchida Y, Wariishi H, Ohta M, Sakaki T. Atypical kinetics of cytochromes P450 catalysing 3’-hydroxylation of flavone from the white-rot fungus Phanerochaete chrysosporium. J Biochem. 2010;147:117–125. doi: 10.1093/jb/mvp155. [DOI] [PubMed] [Google Scholar]
- Kasai N, Ikushiro S, Hirosue S, Arisawa A, Ichinose H, Wariishi H, Ohta M, Sakaki T. Enzymatic properties of cytochrome P450 catalyzing 3′-hydroxylation of naringenin from the white-rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun. 2009;387:103–108. doi: 10.1016/j.bbrc.2009.06.134. [DOI] [PubMed] [Google Scholar]
- Kasai N, Ikushiro S, Shinkyo R, Yasuda K, Hirosue S, Arisawa A, Ichinose H, Wariishi H, Sakaki T. Metabolism of mono- and dichloro-dibenzo-p-dioxins by Phanerochaete chrysosporium cytochromes P450. Appl Microbiol Biotechnol. 2010;86:773–780. doi: 10.1007/s00253-009-2413-x. [DOI] [PubMed] [Google Scholar]
- Kelly SL, Kelly DE. Microbial cytochromes P450: biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos Trans R Soc B Biol Sci. 2013;368:20120476. doi: 10.1098/rstb.2012.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D, Gargas A. The basidiomycete genus Polyporus - an emendation based on phylogeny and putative secondary structure of ribosomal RNA molecules. Feddes Repert. 2004;115:530–546. doi: 10.1002/fedr.200311052. [DOI] [Google Scholar]
- Kües U. Fungal enzymes for environmental management. Curr Opin Biotechnol. 2015;33:268–278. doi: 10.1016/j.copbio.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Kües U, Nelson DR, Liu C, Yu G-J, Zhang J, Li J, Wang X-C, Sun H. Genome analysis of medicinal Ganoderma spp. with plant-pathogenic and saprotrophic life-styles. Phytochemistry. 2015;114:18–37. doi: 10.1016/j.phytochem.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Feng J, Chen X, Bao Y-J, Wang Y, Wu Q, Ma Y, Zhu D. Distinct regioselectivity of fungal P450 enzymes for steroidal hydroxylation. Appl Environ Microbiol. 2019;85:1–13. doi: 10.1128/AEM.01182-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä MR, Hildén K, Kowalczyk JE, Hatakka A. Progress and research needs of plant biomass degradation by basidiomycete fungi. In: Nevalainen H, editor. Grand Challenges in Fungal Biotechnology. Cham: Grand Challenges in Biology and Biotechnology. Springer Nature Switzerland; 2020. pp. 405–438. [Google Scholar]
- Martinez C, Rupashinghe S. Cytochrome P450 bioreactors in the pharmaceutical industry: challenges and opportunities. Curr Top Med Chem. 2013;13:1470–1490. doi: 10.2174/15680266113139990111. [DOI] [PubMed] [Google Scholar]
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, Splinter BonDurant S, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavín JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kües U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, Rokhsar D, Berka R, Cullen D. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- McKay T, Hart K, Horn A, Kessler H, Dodge G, Bardhi K, Bardhi K, Mills JL, Bernstein HJ, Craig PA. Annotation of proteins of unknown function: initial enzyme results. J Struct Funct Genom. 2015;16:43–54. doi: 10.1007/s10969-015-9194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mgbeahuruike AC, Kovalchuk A, Ubhayasekera W, Nelson DR, Yadav JS. CYPome of the conifer pathogen Heterobasidion irregulare: inventory, phylogeny, and transcriptional analysis of the response to biocontrol. Fungal Biol. 2017;121:158–171. doi: 10.1016/j.funbio.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Mitrovic A, Glieder A. Discovery of fungal enzymes and pathways. In: Gupta VK, Mach RL, Sreenivasaprasad S, editors. Fungal biomolecules: sources, applications and recent developments. Ltd, Chichester, UK: John Wiley & Sons; 2015. pp. 51–62. [Google Scholar]
- Miyauchi S, Rancon A, Drula E, Hage H, Chaduli D, Favel A, Grisel S, Henrissat B, Herpoël-Gimbert I, Ruiz-Dueñas FJ, Chevret D, Hainaut M, Lin J, Wang M, Pangilinan J, Lipzen A, Lesage-Meessen L, Navarro D, Riley R, Grigoriev IV, Zhou S, Raouche S, Rosso M-N. Integrative visual omics of the white-rot fungus Polyporus brumalis exposes the biotechnological potential of its oxidative enzymes for delignifying raw plant biomass. Biotechnol Biofuels. 2018;11:201. doi: 10.1186/s13068-018-1198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moktali V, Park J, Fedorova-Abrams ND, Park B, Choi J, Lee Y-H, Kang S. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics. 2012;13:1–13. doi: 10.1186/1471-2164-13-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagye LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Deakin G, Doddapaneni H, Floudas D, Grimwood J, Hilden K, Kues U, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wosten HAB, Xu J, Eastwood DC, Foster GD, Sonnenberg ASM, Cullen D, de Vries RP, Lundell T, Hibbett DS, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci. 2012;109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir KHMNH, Ichinose H, Wariishi H. Construction and application of a functional library of cytochrome P450 monooxygenases from the filamentous fungus Aspergillus oryzae. Appl Environ Microbiol. 2011;77:3147–3150. doi: 10.1128/AEM.02491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Nelson DR. Cytochrome P450 nomenclature, 2004. In: Philips IR, Shephard EA, editors. Cytochrome P450 Protocols. New Jersey: Humana Press; 2006. pp. 1–10. [Google Scholar]
- Nelson DR. The cytochrome P450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta RM, da Silva BP, Gomes Côrrea RC, Kato CG, Vicente Seixas FA, Bracht A (2017) Enzymes from Basidiomycetes—peculiar and efficient tools for biotechnology. In: Brahmachari G (ed) Biotechnology of Microbial Enzymes. Elsevier Inc., pp 119–149
- Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol. 1992;9:945–967. doi: 10.1093/oxfordjournals.molbev.a040771. [DOI] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai K, Matsuzaki F, Wise L, Sakai Y, Jindou S, Ichinose H, Takaya N, Kato M, Wariishi H, Shimizu M. Biochemical characterization of CYP505D6, a self-sufficient cytochrome P450 from the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 2018;84:1–15. doi: 10.1128/AEM.01091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Dannert C. Biocatalytic portfolio of Basidiomycota. Curr Opin Chem Biol. 2016;31:40–49. doi: 10.1016/j.cbpa.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelan JSS, Justo A, Nagy LG, Grand EA, Redhead SA, Hibbett D. Phylogenetic relationships and morphological evolution in Lentinus, Polyporellus and Neofavolus , emphasizing southeastern Asian taxa. Mycologia. 2015;107:460–474. doi: 10.3852/14-084. [DOI] [PubMed] [Google Scholar]
- Sotome K, Hattori T, Ota Y, To-anun C. Salleh B, Kakishima M. Phylogenetic relationships of Polyporus and morphologically allied genera. Mycologia. 2008;100:603–615. doi: 10.3852/07-191R. [DOI] [PubMed] [Google Scholar]
- Stecher G, Tamura K, Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmberger L, Chappell T, Geier M, Krainer F, Day KJ, Vide U, Trstenjak S, Schiefer A, Richardson T, Soriaga L, Darnhofer B, Birner-Gruenberger R, Glick BS, Tolstorukov I, Cregg J, Madden K, Glieder A. Refined Pichia pastoris reference genome sequence. J Biotechnol. 2016;235:121–131. doi: 10.1016/j.jbiotec.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, MacDonald J, Syed K, Salamov A, Hori C, Aerts A, Henrissat B, Wiebenga A, VanKuyk PA, Barry K, Lindquist E, LaButti K, Lapidus A, Lucas S, Coutinho P, Gong Y, Samejima M, Mahadevan R, Abou-Zaid M, de Vries RP, Igarashi K, Yadav JS, Grigoriev IV, Master ER. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genomics. 2012;13:444. doi: 10.1186/1471-2164-13-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS. Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs) Biochem Biophys Res Commun. 2010;399:492–497. doi: 10.1016/j.bbrc.2010.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Mashele SS. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS One. 2014;9:e95616. doi: 10.1371/journal.pone.0095616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Nelson DR, Riley R, Yadav JS. Genomewide annotation and comparative genomics of cytochrome P450 monooxygenases (P450s) in the polypore species Bjerkandera adusta, Ganoderma sp. and Phlebia brevispora. Mycologia. 2013;105:1445–1455. doi: 10.3852/13-002. [DOI] [PubMed] [Google Scholar]
- Syed K, Porollo A, Lam YW, Grimmett PE, Yadav JS. CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbons, alkylphenols, and alkanes. Appl Environ Microbiol. 2013;79:2692–2702. doi: 10.1128/AEM.03767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Porollo A, Lam YW, Yadav JS. A fungal P450 (CYP5136A3) capable of oxidizing polycyclic aromatic hydrocarbons and endocrine disrupting alkylphenols: role of Trp129 and Leu324. PLoS One. 2011;6:e28286. doi: 10.1371/journal.pone.0028286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Porollo A, Miller D, Yadav JS. Rational engineering of the fungal P450 monooxygenase CYP5136A3 to improve its oxidizing activity toward polycyclic aromatic hydrocarbons. Protein Eng Des Sel. 2013;26:553–557. doi: 10.1093/protein/gzt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Shale K, Pagadala NS, Tuszynski J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model Basidiomycete fungi. PLoS One. 2014;9:e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Yadav JS. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol. 2012;38:339–363. doi: 10.3109/1040841X.2012.682050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga T, Krizsán K, Földi C, Dima B, Sánchez-García M, Sánchez-Ramírez S, Szöllősi GJ, Szarkándi JG, Papp V, Albert L, Andreopoulos W, Angelini C, Antonín V, Barry KW, Bougher NL, Buchanan P, Buyck B, Bense V, Catcheside P, Chovatia M, Cooper J, Dämon W, Desjardin D, Finy P, Geml J, Haridas S, Hughes K, Justo A, Karasiński D, Kautmanova I, Kiss B, Kocsubé S, Kotiranta H, LaButti KM, Lechner BE, Liimatainen K, Lipzen A, Lukács Z, Mihaltcheva S, Morgado LN, Niskanen T, Noordeloos ME, Ohm RA, Ortiz-Santana B, Ovrebo C, Rácz N, Riley R, Savchenko A, Shiryaev A, Soop K, Spirin V, Szebenyi C, Tomšovský M, Tulloss RE, Uehling J, Grigoriev IV, Vágvölgyi C, Papp T, Martin FM, Miettinen O, Hibbett DS, Nagy LG. Megaphylogeny resolves global patterns of mushroom evolution. Nat Ecol Evol. 2019;3:668–678. doi: 10.1038/s41559-019-0834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol. 2013;30:385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Vogl T, Kickenweiz T, Pitzer J, Sturmberger L, Weninger A, Biggs BW, Köhler E-M, Baumschlager A, Fischer JE, Hyden P, Wagner M, Baumann M, Borth N, Geier M, Ajikumar PK, Glieder A. Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nat Commun. 2018;9:3589. doi: 10.1038/s41467-018-05915-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Ruth C, Pitzer J, Kickenweiz T, Glieder A. Synthetic core promoters for Pichia pastoris. ACS Synth Biol. 2014;3:188–191. doi: 10.1021/sb400091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Sturmberger L, Kickenweiz T, Wasmayer R, Schmid C, Hatzl AM, Gerstmann MA, Pitzer J, Wagner M, Thallinger GG, Geier M, Glieder A. A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth Biol. 2016;5:172–186. doi: 10.1021/acssynbio.5b00199. [DOI] [PubMed] [Google Scholar]
- Wang J, Ohno H, Ide Y, Ichinose H, Mori T, Kawagishi H, Hirai H. Identification of the cytochrome P450 involved in the degradation of neonicotinoid insecticide acetamiprid in Phanerochaete chrysosporium. J Hazard Mater. 2019;371:494–498. doi: 10.1016/j.jhazmat.2019.03.042. [DOI] [PubMed] [Google Scholar]
- Wang W-F, Xiao H, Zhong J-J. Biosynthesis of a ganoderic acid in Saccharomyces cerevisiae by expressing a cytochrome P450 gene from Ganoderma lucidum. Biotechnol Bioeng. 2018;115:1842–1854. doi: 10.1002/bit.26583. [DOI] [PubMed] [Google Scholar]
- Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A. Reliable high-throughput screening with by limiting yeast cell death phenomena. FEMS Yeast Res. 2004;5:179–189. doi: 10.1016/j.femsyr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Weninger A, Glieder A, Vogl T. A toolbox of endogenous and heterologous nuclear localization sequences for the methylotrophic yeast Pichia pastoris. FEMS Yeast Res. 2015;15:fov082. doi: 10.1093/femsyr/fov082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Xu Z, Knudson A, Carlson A, Chen N, Kovaka S, LaButti K, Lipzen A, Pennachio C, Riley R, Schakwitz W, Umezawa K, Ohm RA, Grigoriev IV, Nagy LG, Gibbons J, Hibbett D. Genomics and development of Lentinus tigrinus: a white-rot wood-decaying mushroom with dimorphic fruiting bodies. Genome Biol Evol. 2018;10:3250–3261. doi: 10.1093/gbe/evy246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav AN, Gupta A, Mishra S, Singh S. Recent advancement in white biotechnology through fungi. Cham: Springer International Publishing; 2019. [Google Scholar]
- Yang C, Li W, Li C, Zhou Z, Xiao Y, Yan X. Metabolism of ganoderic acids by a Ganoderma lucidum cytochrome P450 and the 3-keto sterol reductase ERG27 from yeast. Phytochemistry. 2018;155:83–92. doi: 10.1016/j.phytochem.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Yap H-YY, Chooi Y-H, Firdaus-Raih M, Fung S-Y, Ng S-T, Tan C-S, Tan N-H. The genome of the tiger milk mushroom, Lignosus rhinocerotis, provides insights into the genetic basis of its medicinal properties. BMC Genomics. 2014;15:635. doi: 10.1186/1471-2164-15-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Natsume M. Allelochemicals for plant–plant and plant–microbe interactions. In: Liu H-W, Mander L, editors. Comprehensive natural products II: Chemistry and Biology. Amsterdam: Elsevier Ltd.; 2010. pp. 539–561. [Google Scholar]
- Zhu Y, Xu J, Sun C, Zhou S, Xu H, Nelson DR, Qian J, Song J, Luo H, Xiang L, Li Y, Xu Z, Ji A, Wang L, Lu S, Hayward A, Sun W, Li X, Schwartz DC, Wang Y, Chen S. Chromosome-level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense. Sci Rep. 2015;5:11087. doi: 10.1038/srep11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerhandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 5011 kb)
(XLSX 44 kb)
(XLSX 172 kb)
(XLSX 148 kb)
Data Availability Statement
The data supporting the findings of this study are available within this article and the supplementary materials. The fungal genome data, on which this study is based, are publicly available online as outlined in the ‘Materials and methods’ section.
Any software used in this study for genomic data processing is publicly available online via the links provided in the ‘Materials and methods’ section.