Abstract
N-glycosylation is a ubiquitous posttranslational modification that affects protein structure and function, including those of the central nervous system. N-glycans attached to cell membrane proteins play crucial roles in all aspects of biology, including embryogenesis, development, cell–cell recognition and adhesion, and cell signaling and communication. Although brain function and behavior are known to be regulated by the N-glycosylation state of numerous cell surface glycoproteins, our current understanding of brain glycosylation is limited, and glycan variations associated with functional brain regions remain largely unknown. In this work, we used a well-established cell surface glycomic nanoLC-Chip-Q-TOF platform developed in our laboratory to characterize the N-glycome of membrane fractions enriched in cell surface glycoproteins obtained from specific functional brain areas. We report the cell membrane N-glycome of two major developmental divisions of mice brain with specific and distinctive functions, namely the forebrain and hindbrain. Region-specific glycan maps were obtained with ∼120 N-glycan compositions in each region, revealing significant differences in “brain-type” glycans involving high mannose, bisecting, and core and antenna fucosylated species. Additionally, the cell membrane N-glycome of three functional regions of the forebrain and hindbrain, the cerebral cortex, hippocampus, and cerebellum, was characterized. In total, 125 N-glycan compositions were identified, and their region-specific expression profiles were characterized. Over 70 N-glycans contributed to the differentiation of the cerebral cortex, hippocampus, and cerebellum N-glycome, including bisecting and branched glycans with varying degrees of core and antenna fucosylation and sialylation. This study presents a comprehensive spatial distribution of the cell-membrane enriched N-glycomes associated with five discrete anatomical and functional brain areas, providing evidence for the presence of a previously unknown brain glyco-architecture. The region-specific molecular glyco fingerprints identified here will enable a better understanding of the critical biological roles that N-glycans play in the specialized functional brain areas in health and disease.
Keywords: glycocalyx, glycomics, mouse brain, N-glycans, N-glycome, N-glycosylation
Abbreviations: AMPA, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid; ECC, extracted compound chromatogram; EIC, extracted ion chromatograms; Fuc, fucose; GlcNAc, N-acetylglucosamine; Hex, hexose; HexNAc, N-acetylhexosamine; nanoLC-Q-TOF-MS, nano-liquid chromatography–chip-quadrupole–time of flight–mass spectrometry; NeuAc, N-acetyl neuraminic acid; NMDA, N-Methyl- d-aspartic acid or N-methyl-D-aspartate receptor; PGC, porous graphitized carbon; SPE, solid-phase extraction
Graphical Abstract
Highlights
-
•
Cell membrane N-glycome maps of functional mouse brain areas were obtained.
-
•
Highly diverse N-glycome varies in a region-specific manner.
-
•
75+ glycan motives discriminate region-specific cell membrane N-glycome.
In Brief
We have characterized the cell-membrane N-glycome of two major developmental divisions of the brain, the forebrain and hindbrain, and three functional derivatives from them, including the cerebral cortex, hippocampus, and cerebellum, revealing an extraordinary diversity of N-glycans expressed in a global and functional region-specific manner in the adult mouse. Furthermore, we identified +25 N-glycans able to differentiate the forebrain and hindbrain N-glycome. Additionally, over 35 N-glycans distinguished the cortex, hippocampus, and cerebellum N-glycomes and may serve as region-specific glycan biomarkers.
The mammalian brain harbors an enormous number of cells, predominately neurons and glial, that are intricately organized to perform diverse and complex functions at regional levels. Protein N-glycosylation is a posttranslational modification that occurs in more than 70% of mammalian proteins, directly affecting protein structure, stability, and function, including those of the central nervous system. N-glycans attached to cell membrane proteins play a pivotal role in nearly all aspects of biology, including cell signaling and communication, cell–cell recognition, adhesion, and migration, embryogenesis, differentiation, and development (1, 2).
Brain function and behavior are regulated by the N-glycosylation state of numerous cell surface glycoproteins involved in developing the nervous system and neurophysiology (3, 4, 5). N-glycosylation regulates the plasma membrane expression and function of voltage-gated ion channels and ligand-gated ion channels, thus influencing neuronal excitability, learning, memory, and behavior (6, 7, 8, 9, 10, 11, 12). N-glycans can also affect synaptic function and plasticity by altering the expression, assembly, and conductance of neurotransmitters receptors, such as serotonin and nicotinic acetylcholine receptors, γ-aminobutyric acid (GABA) and ionotropic glutamate receptors, including AMPA, kainate, and NMDA (13, 14, 15, 16, 17, 18, 19, 20).
Abnormal glycosylation is also linked to human brain cancers, including neuroblastoma, gliomas, and the most aggressive form, glioblastoma multiforme (GBM) (21, 22, 23). The critical role of N-glycosylation in brain health is revealed in over 70 human genetic disorders known as Congenital Disorders of Glycosylation (CDG). Individuals with these diseases present genetic deficiencies in the N-Glycosylation pathway, and typical manifestations include developmental delay, intellectual disability, seizures, neuropathy, and metabolic abnormalities in multiple organ systems (2).
However, despite the recognition that N-glycosylation is a critical determinant of the function of several types of cell membrane proteins expressed by neurons and glial cells, our current understanding of brain N-glycans remains limited. Early research work used inhibitors of N-glycosylation, such as tunicamycin or site-specific mutagenesis, to prevent the addition of N-glycans to the proteins. However, the glycans structures attached to the glycoproteins have not been thoroughly characterized. Mass-spectrometry-based glycomic analysis has rapidly advanced our understanding of glycans modifying glycoproteins and lipids in cells and tissues (24). Glycomics contributes to developing glycan-based biomarkers for the early diagnosis, prognosis, and therapeutic interventions of human diseases such as cancer and brain cancer (23, 24, 25). Brain glycomic studies have been reported among vertebrates and animal models, including fish (zebrafish) and mammalians (26, 27, 28, 29, 30). Human brain glycomic studies have mainly reported the characterization of brain glycosylation from patient biopsy samples, including cancerous tumor tissue and surrounding normal tissue (23). Recently, analysis of glycosaminoglycans from coronal sections of human brain tissues with Parkinson's disease has been reported (31). Several brain N-glycomic studies in rodent animal models have been carried out earlier using whole-brain tissues (32, 33, 34) and coronal sections from rodent brains (27, 29, 35). Glycoproteomic studies have also been performed on glycoproteins derived from synaptic membranes from the whole mouse brain (36, 37). Glycoproteomic analysis of the striatum and substantia nigra tissues in aging-mouse studies uncovered age-specific and region-specific changes in protein expression and N-glycans (29). Nonetheless, spatial variations in the expression of cell surface brain N-glycans and region-specific N-glycomic maps of additional functional divisions of the brain remain largely unexplored.
Here we describe the cell-membrane-enriched N-glycome of select functional mouse brain areas obtained using a well-established nano-LC-MS/MS N-glycomic platform developed in our laboratory (38). We have characterized the cell membrane fraction N-glycome of two major developmental divisions of the brain, the forebrain, and hindbrain, which revealed a high diversity of N-glycans expressed in a region-specific manner in the adult mouse. Importantly, we identified +25 N-glycans able to differentiate the forebrain and hindbrain N-glycome. Additionally, we report the cell-membrane-enriched N-glycome of three functional derivatives from the forebrain and hindbrain, the cerebral cortex, hippocampus, and cerebellum, uncovering additional unique region-specific glycan arrays. Among them, we found that 75 N-glycans can distinguish the cortex, hippocampus, and cerebellum N-glycome, which may be potential region-specific glycan-based biomarkers.
These studies show the detailed spatial distribution of cell membrane N-glycomes associated with broad regions and discrete functional brain areas, providing evidence of the presence of functional brain glyco-architectures not previously described. The application of the method will enable further research to advance our understanding of brain function and how region-specific brain N-glycomes are established, controlled, and modulated in health and disease.
Experimental Procedures
Experimental Design and Statistical Rationale
Mouse brain N-glycomic studies were performed on nine young adults (∼2 months old) male C57B6/J mice. Three mouse brains were used for studying the forebrain and hindbrain N-glycome as biological replicates (FB-M1-3 and HB-M1-3, respectively). Additionally, six mouse brains were used as biological replicates to characterize the cerebral cortex, hippocampus, and cerebrum N-glycomes (Ctx 1–6, Hippo1–6, and Cer1–6, respectively). All brain tissues isolated by surgical dissection were processed using the workflow described below. LC-MS/MS instrument performance was assessed with a quality control mixture of N-glycan obtained from human serum and RNAseB. LC-MS data acquisition was randomized. Statistical evaluation of significant glycan abundance differences was performed using one-way ANOVA and the TUKEY post-hoc test, and Student T-tests with Benjamini–Hochberg multicomparison correction. Differences were considered significant if p < 0.05. Multivariate Analysis of N-glycomes was performed using partial least squares discriminant analysis (PLS-DA) using packages pls and plsVarSel in R (3.5.1). Variable Importance in Projection (VIP) and loading scores were calculated to determine the contribution of each N-glycan to the PLS-DA model.
Chemicals and Materials
Sucrose and HEPES were acquired from Sigma-Aldrich, and protease inhibitor cocktail V was purchased from Millipore-Sigma. Glycerol-free Peptide N-glycosidase F (PNGase F) was obtained from New England Biolabs. Acetonitrile was purchased from Honeywell, and formic acid was purchased from Fisher Scientific. Porous graphitized carbon (PGC) 96-well plates were obtained from Glygen Corp. MilliQ water was used throughout the study.
Mice
C57BL/6J adult male and female mice were purchased from Jackson Lab and bred in-house at UC Davis. For the study, nine in-house bred C57BL/6J adult (8–9 weeks) mice were used. Upon euthanasia with CO2 and cervical dislocation, brains were manually collected, snap-frozen in liquid N2, and then stored at −80 °C until further analysis. Three brains were thawed and dissected into the forebrain and hindbrain regions, and from another six brains, the cerebral cortex, hippocampus, and cerebellum were surgically isolated. All brain regions were processed to obtain the cell-membrane fractions enriched in cell surface glycoproteins as described below. All procedures and protocols were reviewed and approved by the Institutional Animal Care and Use Committee at the University of California, Davis (IACUC protocol #20072).
Cell Surface Membrane Enrichment and Isolation
The extraction of region-specific brain cell membrane fractions was performed by a procedure previously described by our lab (38). Briefly, dissected brains tissues (forebrain, hindbrain, cortex, hippocampus, and cerebellum) were transferred into tubes containing homogenization buffer (0.25 M sucrose, 20 mM HEPES-KOH pH 7.4, and 1:100 protease inhibitor), and tissue lysis was performed at 4 °C using a chilled cooling rack and a probe sonicator (Qsonica) with five pulses of 5 s on, alternating with intervals of 10 s off, respectively. Brain tissue lysates were centrifuged at 9000g for 10 min to remove the nucleus, mitochondria, and cellular debris. The supernatants were transferred to 1.5 ml ultracentrifuge tubes (Beckman Coulter) for ultracentrifugation at 200,000g for 45 min at 4 °C. Pellets obtained were resuspended in 1 ml 0.2 M Na2CO3 pH 11 and pelleted again by ultracentrifugation (at 200,000g for 45 min at 4 °C), followed by a wash with 1 ml of chilled water and ultracentrifugation as described above. The resulting brain region-specific membrane fractions enriched in cell surface glycoproteins were stored at −80 °C until further processing.
N-Glycan Release and Purification
Three membrane fractions from the forebrain (FB-M1-3) and hindbrain (HB-M1-3) regions and six membrane fractions from the cortex (Ctx1–6), hippocampus (Hippo1–6), and cerebellum (Cer1–6) regions were enzymatically de-N-glycosylated. N-glycans were also prepared from a mixture of human serum and RNAseB and use as reference material (QC). Samples were resuspended with 100 to 200 μl of 100 mM ammonium bicarbonate in 5 mM dithiothreitol and heated for 5 min at 100 °C to thermally denature the proteins. To cleave N-glycans from membrane proteins, 4 μl of PNGase F was added to the samples and incubated at 37 °C in a microwave reactor (CEM Corporation) for 10 min at 20 W, followed by overnight incubation at 37 °C in a water bath. After deglycosylation, samples were ultracentrifuged to pellet residual membrane fraction and deglycosylated proteins. The supernatant containing the released N-glycans was collected, and N-glycans were purified by solid-phase extraction using a 96-well plate containing a porous graphitized carbon (PGC) matrix. The plate was conditioned with 80% Acetonitrile in water (200 μl × three times), followed by water (200 μl × three times). N-glycan samples were loaded, and the flow-through was reapplied twice, before washing with water (200 μl × 4–5 times). Elution was performed with 40% Acetonitrile in water (200 μl × two times), and eluted fractions were dried in vacuo.
Glycomic Nano-LC Mass Spectrometry Analysis
N-glycan samples were reconstituted in nanopure water for analysis, randomized, and analyzed using an Agilent nanoLC/ESI-Q-TOF-MS system (Agilent Technologies). Samples were introduced into the MS with a microfluidic chip, consisting of enrichment and analytical columns packed with PGC and a nanoelectrospray tip. A binary gradient was applied to separate and elute glycans at a flow rate of 0.3 μl/min: (A) 3% (v/v) acetonitrile and 0.1% (v/v) formic acid in water and (B) 90% (v/v) acetonitrile in 1% (v/v) formic acid in water. MS spectra were acquired at 1.5 s per spectrum over a mass range of m/z 600 to 2000 in positive ionization mode. Mass inaccuracies were corrected with reference mass m/z 1221.991. Collision-induced dissociation (CID) was performed with nitrogen gas using a series of collision energies (Vcollision) dependent on the m/z values of the N-glycans, based on the equation: Vcollision= slope (m/z) + offset; where the slope and offset were set at (1.8/100 Da) V and −2.4 V, respectively.
Data Analysis
N-glycan compounds were identified and quantified using Agilent MassHunter Qualitative Analysis B.07.01 (Agilent Technologies) in combination with an in-house retrosynthetic library of all possible glycan compositions according to accurate mass (39). Glycan signals above a signal-to-noise ratio of 5 were filtered, and data were deconvoluted using the Molecular Feature Extractor algorithm included in the MassHunter software and a 10 ppm mass error. Tandem MS analysis was performed manually to confirm glycan compositions with library matches and select structural motives. Fragment ions were assigned with ≤25 ppm mass error. All glycan structures are proposed as MS/MS data of native glycans alone are insufficient to determine complete glycan structures. Glycan relative abundances were determined by integrating ion counts for observed glycan masses and normalizing to the summed ion counts of all glycans detected. Glycan subtypes, including high mannose, complex, and hybrid, were grouped accordingly by knowledge of the mammalian N-glycan biosynthetic pathway. Statistical evaluation of significant glycan abundance changes was performed using one-way ANOVA and the TUKEY post-hoc test, and Student T-tests with Benjamini–Hochberg multicomparison correction; differences were considered significant if p < 0.05. Multivariate Analysis of N-glycomes was performed using partial least squares discriminant analysis (PLS-DA) using packages pls and plsVarSel in R (3.5.1). Variable Importance in Projection (VIP) and loading scores were calculated to determine the contribution of each N-glycan to the PLS-DA model.
Results
Regional Expression of Cell Membrane N-Glycomes in the Mouse Brain
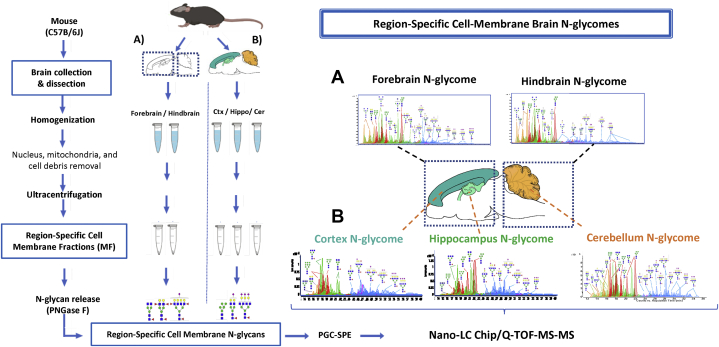
Figure 1 shows the scheme of the N-glycomic workflow used for the characterization of region-specific cell membrane enriched N-glycomes in the 8-week-old adult mouse brain. First, we interrogated the forebrain and hindbrain N-glycomes (Fig. 1A). Secondly, we characterized the cerebral cortex, hippocampus, and cerebellum N-glycomes, as functional derivates from the forebrain and hindbrain (Fig. 1B).
Fig. 1.
Scheme of the cell membrane N-glycomic workflow used for the characterization of region-specific cell surface mouse brain N-glycomes. After extraction, brains were surgically dissected either into forebrain and hindbrain (A) or the functional substructures derived from both regions, including the cortex and hippocampus (from forebrain) and cerebellum (from hindbrain) (B). Tissues dissected were homogenized, cell debris, nucleus, and mitochondria were removed by centrifugation from region-specific brain lysates. Cell membrane fractions enriched in cell surface glycoproteins were obtained by ultracentrifugation as previously described (38). Cell membrane glycans were enzymatically released, purified, and analyzed by nano-LC-Chip-Q-TOF mass spectrometry.
Forebrain and Hindbrain N-Glycomes
We first characterized the neuro-N-glycome of two major developmental divisions of the brain with specific and distinct functions, the forebrain and the hindbrain. Over 120 N-glycan compositions were identified in the forebrain and hindbrain N-glycomes with relative abundances values >0.05%, representing ∼99% of the total N-glycans identified. Supplemental Table S1 compares the relative abundance of all glycans identified in the forebrain and hindbrain regions obtained from a randomized sample set of biological triplicates (n = 3/region). To determine the degree of N-glycan processing in each brain area, and the type of decorations present on each N-glycome, individual glycan species were grouped into five categories based on their monosaccharide compositions, and the presence of fucose and/or sialic acids as decorations and their relative abundance summed. Oligo-mannose glycans were grouped under the high mannose category. Complex and hybrid glycans without fucose or sialic acids were grouped and categorized as undecorated glycans. Neutral glycans containing fucose were categorized as fucosylated glycans. Acidic glycans containing sialic acid were categorized as sialylated glycans, and glycans containing fucose and sialic acid residues were categorized as sialofucosylated glycans. Supplemental Fig. S1A and B shows the overlays of annotated extracted compound chromatograms (ECCs) obtained from three randomized biological replicates of the forebrain and hindbrain (supplemental Table S4). The comparative relative abundances of N-glycans in each category are described below. Method reproducibility was assessed from N-glycans profiles of reference material (supplemental Fig. S2). Coefficients of variance (CV) values determined for each glycan group were below 10% for glycan groups with relative abundance >5%, deeming our method highly reproducible.
The forebrain neuroglycome was dominated by fucosylated glycan species accounting for ∼45% of total N-glycans identified (supplemental Fig. S1C), containing from 1 to 5 fucose residues. High mannose and sialofucosylated glycans were also abundant, representing ∼26% and ∼19%, of the total glycans identified, respectively. Undecorated glycans represented ∼11% of the total glycans species identified, and sialylated only glycans around 1%. In contrast, the hindbrain showed lower levels of neutral fucosylated glycans (∼35%) but similar levels of high mannose glycan species (∼30%). Sialofucosylated glycans were the third most abundant group of glycans in the forebrain, with an abundance of ∼20% (supplemental Fig. S1D). Undecorated glycans were also more abundant (14%) in the forebrain, but sialylated glycan species only represented 1%. Overall, the degree of total fucosylation was higher in the forebrain (∼62%) than in the hindbrain (∼54%) (supplemental Fig. S1, C and D).
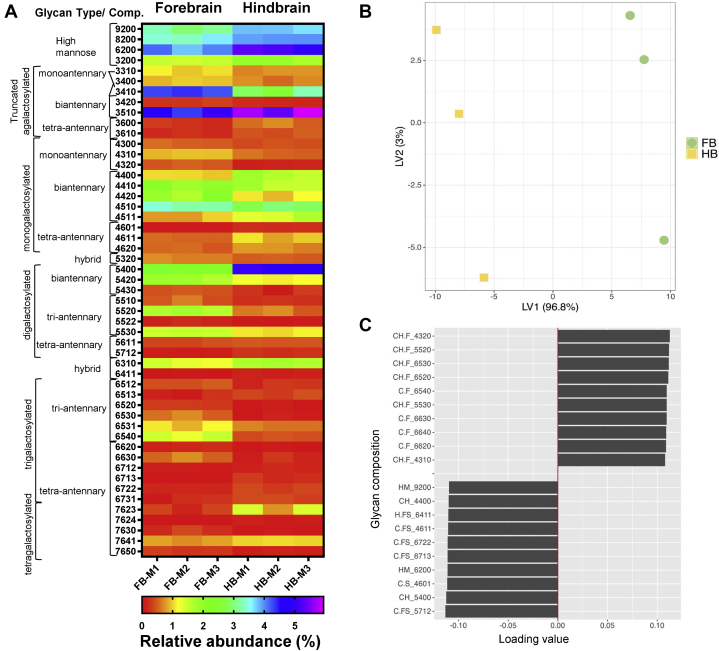
Comparing the expression of individual glycans between the forebrain and hindbrain regions showed that ∼43% of glycans among all glycan groups were differentially expressed (p < 0.05) (supplemental Table S1). Figure 2A shows a heatmap with 51 N-glycan species differentially expressed in the forebrain and hindbrain. Glycans compositions were ordered from high mannose to complex and hybrid glycans according to the biosynthetic pathway and increasing glycan mass. Major differences observed corresponded to different levels of expression of any given glycan in the forebrain or hindbrain and not the presence or absence. Among high mannose species, glycans with compositions Man3 (core), Man6, Man 8, and Man9 were significantly more abundant in the hindbrain. However, hybrid glycans with compositions 5320, 6310, and 6411 given as (HexHexNAcFucNeuAc) were more abundant in the forebrain. Similarly, 12 agalacto (truncated) and galactosylated mono- and biantennary complex species were also significantly more abundant in the forebrain. These included: a) truncated monoantennary glycans with compositions 3310 and 3410; b) monogalactosylated monoantennary glycans with compositions 4300, 4310, and 4320; c) truncated or agalacto-biantennary glycans with composition 3400, 3420; d) monogalactosylated biantennary glycans with composition 4410, 4420, and 4510; and e) digalactosylated biantennary glycans with compositions 5420 and 5430. A few mono and biantennary glycans with compositions 3510, 4400, 4511, and 5400 were more abundant in the hindbrain. Importantly, in agreement with previous reports, several of these glycans species in the forebrain and hindbrain corresponded to bisecting glycans and are further discussed later (32). Tri-antennary and tetra-antennary fucosylated and sialofucosylted glycans with compositions 5510, 5520, 5530, 6512, 6531, 6540, 6630, and 7650 were more abundant in the forebrain. Interestingly, a single tri-antennary glycan with composition 6513 was more abundant in the hindbrain. However, several undecorated, fucosylated and sialofucosylated tri- and tetra-antennary glycans were higher in the hindbrain. These corresponded to truncated glycans with compositions 3600 and 3610; agalacto and galactosylated tri and tetra-antennary glycans with compositions 4601, 4611, 4620, 5611, 5712, 6712, 6722, 6731, 7623, and 7641. Notably, among these highly abundant multifucosylated and sialylated glycans, species carrying Lewis-fucosylation and sialyl-Lewis motives were identified by tandem MS, also in agreement with previous reports (32, 40). Additionally, a few glycans expressed in a region-specific manner were also identified among less abundant glycans accounting for 0.7% to 0.3% of the total glycans identified in the forebrain and hindbrain, respectively. Di and tri-fucosylated glycans with compositions 5522, 6520, 6530, 7630 were detected almost exclusively in the forebrain, and glycans with compositions 6713 and 7624 in the hindbrain. These results show the complexity of the forebrain and hindbrain N-glycomes and reveal that these specific functional brain areas have specific N-glycomes.
Fig. 2.
Comparison of cell membrane N-glycome obtained from the forebrain and hindbrain. Heatmap of N-glycans differentially expressed in the Forebrain (FB) and Hindbrain (HB), ordered from high mannose to complex glycans approximately according to N-glycan biosynthesis (p < 0.05%, as analyzed by two-way ANOVA and the TUKEY post-hoc test) (A). Glycan mass of hybrid and complex glycans increases from top to bottom. Glycan composition is abbreviated as HexHexNAcFucNeuAc where Hex = Hexose, HexNAc = N-Acetylhexosamine, Fuc = Fucose, NeuAc = N-Acetylneuraminic Acid; and numbers indicate the number of each monosaccharide residue. The abundance for each glycan composition was normalized to the abundance of total glycans identified. Data from biological replicates (n = 3) were used (FBM1–3 and HBM1–M3). Partial least squares–discriminant analysis (PLS-DA) score plot of cell surface N-glycan profiles from FB and HB (B). Each symbol represents a mouse (n = 3). Loadings plots from the PLS-DA model showing the N-glycan species (compositions) contributing to the separation in the first dimension (x-axis of the scores plot) for the FB and HB N-glycomes (C).
Multivariant analysis using Partial Least Squares–Discriminant Analysis (PLS-DA) was performed to determine further the significance of differences observed between the forebrain and hindbrain cell membrane N-glycome. The PLS-DA scores plots show a clear separation of the forebrain-hindbrain N-glycomes (Fig. 2B). One-way ANOVA showed that 27 glycans out of the 51 described above made a significant contribution to the differentiation of the FB and HB N-glycomes (supplemental Table S1, FDR p value column). Remarkably, the top glycans contributing to the separation in the first dimension (x-axis of the scores plot) of the model are shown in the loadings plot (Fig. 2B). Neutral mono and tri-antennary fucosylated glycans containing from 1 to 4 fucose residues were the main discriminators identified between the FB and HB N-glycomes. Additionally, glycans carrying bisecting GlcNAc residues as well as sialofucosylated glycans were also significant.
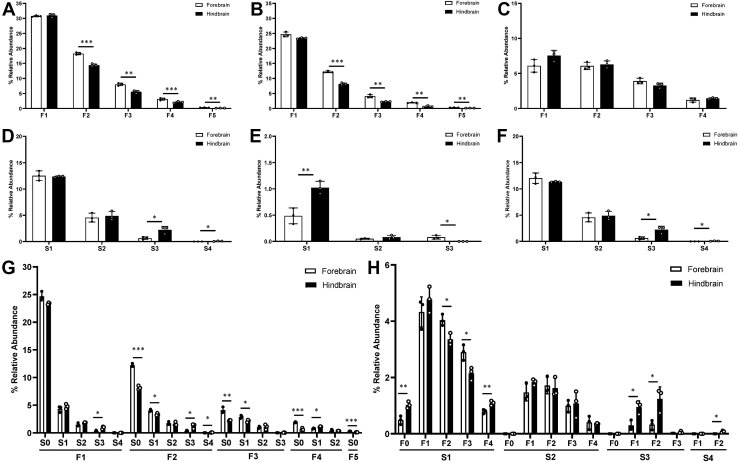
Therefore, we further investigated the distribution of fucose and sialic residues in complex and hybrid glycans in the forebrain and hindbrain (Fig. 3). All complex and hybrid glycans containing fucose, sialic acid, or both residues were separated according to the number of fucose or sialic acid residues present in mono, di, tri, tetra, and penta-fucosylated (F1–F5) or sialylated species (S1–S4) (Fig. 3, A and D). Monofucosylated species were the most abundant in the forebrain and hindbrain (∼30%). In contrast, di, tri, tetra, and pentafucosylated glycans, which indicate core and outer arm fucosylation, were significantly more abundant in the forebrain (Fig. 3A), showing a distinct distribution between the two regions, and represent an increase of fucosylation in the forebrain. This is consistent with a recent report of differential fucosylation observed in the rat brain's striatum and substantia nigra regions (30). Similarly, monosialylated species were highly expressed in both the forebrain and hindbrain, with abundances ∼12 to 14% (Fig. 3D). When neutral fucosylated glycans were evaluated, monofucosylated species were high in both regions, although species containing 2 to 5 fucose residues were significantly higher in the forebrain (Fig. 3B). Among sialofucosylated glycans, there were no significant differences between the two regions in species containing from 1 to 4 fucose residues (Fig. 3C). However, among sialylated only glycans, monosialylated species were more abundant in the hindbrain, and trisialylated species were more abundant in the forebrain (Fig. 3E). The distribution of sialic acid residues in sialofucosylated species showed an equal abundance of mono and disisalylated species in both regions, but the expression levels of tri and tetrasialylated species were significantly higher in the hindbrain (Fig. 3F). To better understand the combined distribution of fucose and sialic residues in sialofucosylated glycans in each region, glycans were grouped based on the presence and number of sialic acids and fucose residues and plotted in Figure 3, G and H. Nonsialylated mono and difucosylated species F1S0 and F2S0 were the predominant glycans in both regions (with relative abundances ∼25% and 8–14%, respectively). However, F2S0 was significantly higher in the forebrain, followed by F2S1 (∼5%). Similarly, neutral or monosialylated tri- and tetrafucosylated species (F3S0, F4S0, F3S1, and F4S1) were significantly more abundant in the forebrain (∼3%), while difucosylated, tri-, and tetrasialylated species (F2S3 and F2S4), and tetrafucosylated monosialylated glycans (F4S1) were significantly higher in the hindbrain (∼1–2%). Among sialylated species, the forebrain showed higher content of monosialylated di- and trifucosylated glycan species (S1F2 and S1F3), in contrast to the most abundant monosialylated mono and tetrafucosylated glycans (S1F1 and S1F4) in the hindbrain. Among disialylated species (S2F1–F4), no significant differences were observed between the two regions, although trisialylated species mono and difucosylated (S3F1 and S3F2) were significantly higher in the hindbrain. These results clearly show the region-specific distribution of terminal decorations patterns in brain N-glycans, including fucosylation and sialylation.
Fig. 3.
Fucosylation and sialylation in the forebrain and hindbrain N-glycome. Comparative distribution of mono (F1), di (F2), tri (F3), tetra (F4), and penta (F5) fucosylated glycans in all fucosylated glycans (A); neutral fucosylated only glycans (B); and sialofucosylated glycans (C). Comparative distribution of mono (S1), di (S2), tri (S3), and tetra-sialylated (S4) species in: all sialylated glycans (D); sialylated only glycans (E), and sialofucosylated species (F). Combined distribution of fucose and sialic acid residues (FxSy) sorted by the number of fucose residues (x) in (G) or sialic acid residues (y) in (H). Asterisks indicate the statistical significance between groups compared (∗p < 0.05%; ∗∗p < 0.01%; ∗∗∗p < 0.001%).
Although the current method does not provide a resolution regarding the linkage types between sugar residues that would allow the identification of terminal N-glycan epitopes, tandem MS analysis was performed to support the identification of relevant structural motives and their regional distribution in the brain. We used tandem MS to characterize neutral multifucosylated glycans in the forebrain. The presence of two or more fucose residues is indicative of core and outer arm fucosylation, the latter likely forming LewisX determinants that have been reported to be highly expressed among neutral glycans from whole rodent brains (32). Using CID, antenna fucosylation was identified based on characteristic and abundant trisaccharide oxonium fragment ion with m/z 512.3 (Hex1HexNAc1Fuc1) (supplemental Fig. S3). The presence of core and antenna fucosylation, likely forming Lex determinant, was confirmed in select representative tri-antennary glycans with compositions 6530 and 6540. These glycans showed a region-specific expression profile and were among the most significant glycans species able to differentiate the forebrain and hindbrain N-glycomes (supplemental Table S1). Notably, some of these tri-antennary glycans have also been shown to present a region-specific expression in the striatum and substantia nigra in rat brains and decrease with age (29).
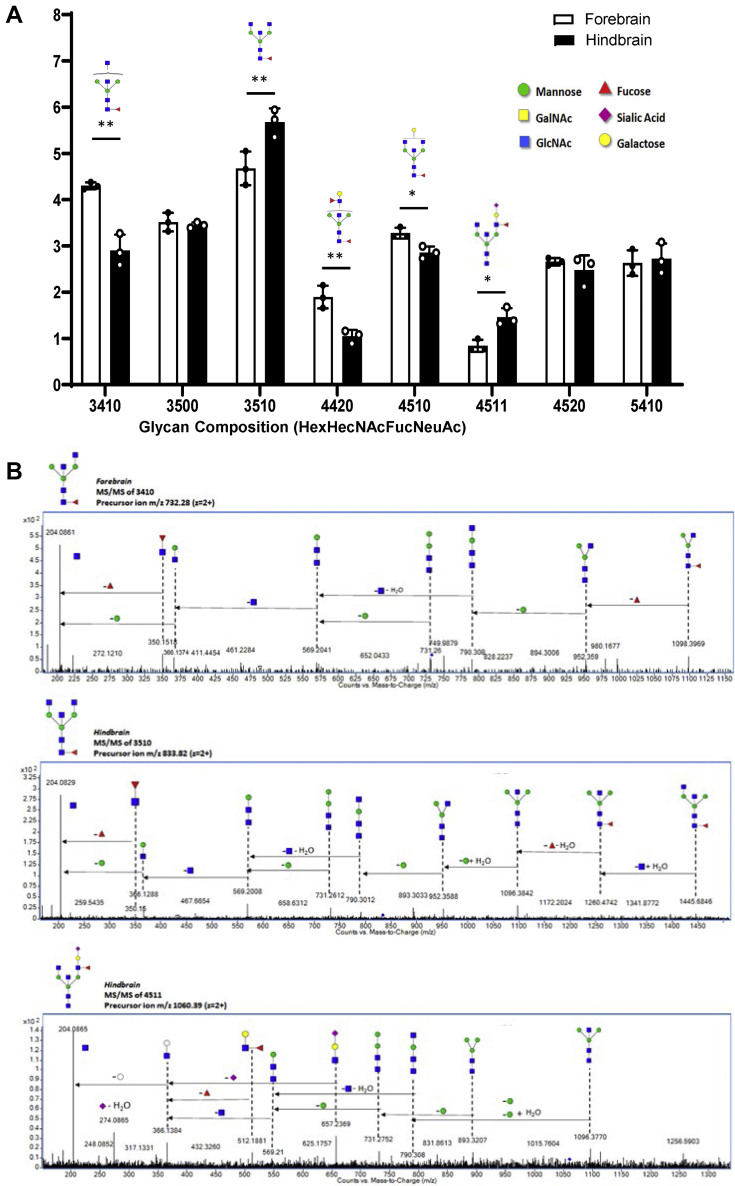
Glycans potentially carrying bisecting GlcNAc were highly expressed in the mouse forebrain and hindbrain and were among the glycans that discriminated between the forebrain and hindbrain N-glycomes. Figure 4A shows the differential spatial expression of the most abundant bisecting N-glycans confirmed by tandem MS in the forebrain and hindbrain N-glycomes, corresponding to mono and biantennary bisecting species. While monoantennary bisecting species with compositions 3410 and 4420 were significantly more abundant in the forebrain N-glycome, biantennary bisecting species with compositions 3510 and 4511 were more abundant in the hindbrain. Biantennary bisecting glycan with composition 4510 was also significantly higher in the forebrain. Additional abundant bisecting species identified with compositions 3500, 4520, and 5410 did not show significant region-specific differences. Figure 4B shows tandem MS spectra for select glycans where bisecting GlcNAc residues were confirmed using the tetra-saccharide oxonium fragment ion with m/z 790.3 (Hex1HexNAc3 + H20, C fragment type according to Domon and Costello, 1998) (41).
Fig. 4.
Region-specific expression of bisecting N-glycans in forebrain and hindbrain. Comparison of most abundant and differentially expressed glycans containing bisecting GlcNAc in forebrain and hindbrain N-glycomes (A). Glycan composition is abbreviated as HexHexNAcFucNeuAc where Hex = Hexose, HexNAc = N-Acetylhexosamine, Fuc = Fucose, NeuAc = N-Acetylneuraminic Acid; and numbers indicate the number of each monosaccharide residue. The abundance for each glycan composition was normalized to the abundance of total glycans identified. Glycan structures are presented with symbols recommended by the CFG (http://www.functionalglycomics.org/static/consortium/Nomenclature.shtml). Asterisks indicate the statistical significance between groups compared (∗p < 0.05%; ∗∗p < 0.01). Tandem MS/MS spectra of select N-glycans containing bisecting GlcNAc in the Forebrain and Hindbrain (B). The fragment ion with m/z 790.3 (C fragment of Hex1HexNAc3 + H20) was used to identify bisecting GlcNAc. Putative structures are shown with the corresponding accurate masses.
Tandem MS was also used to characterize the sialofucosylated glycans that showed a region-specific distribution in the forebrain and hindbrain. Antenna fucosylation and sialyl-Lewis epitopes were identified based on characteristics oxonium fragment ions with m/z 512.3 and m/z 803.3, corresponding to B fragment ions of the trisaccharide Hex1HexNAc1Fuc1 and tetrasaccharide Hex1HexNAc1Fuc1NeuAc1, respectively. Tandem MS will identify sialyl Lewis epitopes but cannot discriminate between Lex and Lea. Tandem MS analysis of sialofucosylated tri- and tetra-antennary glycans with compositions 6531, 6722, and 7623 is shown in supplemental Fig. S4. All sialylated glycans with or without fucose substitutions (sialofucosylated and sialylated only glycans) identified contained N-Acetyl neuraminic acid (NeuAc) residues. Glycans containing N-glycolyl neuraminic acid (NeuGc) that have been described to be present in trace amounts (<3%) in the rat brain were not found in this set of samples analyzed in agreement with a previous report in mouse brain (35).
Altogether, our results show for the first time the cell membrane N-glycan maps of the forebrain and hindbrain. The ability to assess the mouse brain's N-glycome in a region-specific manner with high-resolution and high-sensitivity allowed us to reveal region-specific expression of N-glycans with discriminant values in distinguishing these N-glycomes. Additionally, we provide evidence of a region-specific distribution of terminal N-glycans decorations in the brain, including fucosylation and sialylation, and showed a significantly increased fucosylation level in the forebrain.
Cerebral Cortex, Hippocampus, and Cerebellum N-Glycomes
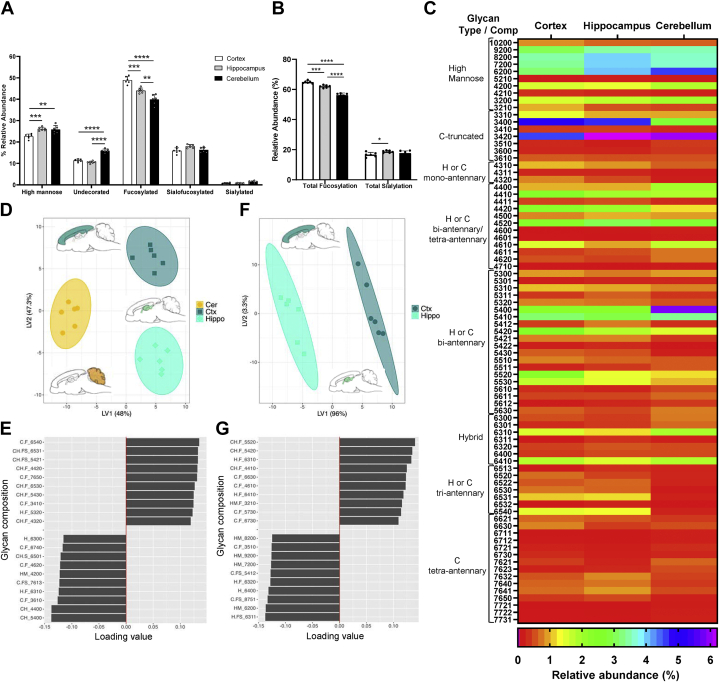
To further understand the regionalization of the brain glycome, we determined the N-glycome of three well-defined brain regions derived from the forebrain and hindbrain (cerebral cortex and the hippocampus parts of the forebrain and the cerebellum part of the hindbrain) (Fig. 1B). Over 600 glycan compound peaks, including isomers, were observed in the cerebral cortex, hippocampus, and cerebellum N-glycomes corresponding to a total of 125 N-glycan compositions on each region with normalized abundances above 0.05% (supplemental Table S2). Although the number of glycan compositions identified in the cortex, hippocampus, and cerebellum N-glycomes was comparable to those obtained for the forebrain and hindbrain, an increase in the spatial resolution on the glycans differentially expressed in these three regions was observed. Figure 5A shows the relative abundances of N-glycans in each glycan category in the cerebral cortex, hippocampus, and cerebellum. The cerebral cortex N-glycome was dominated by fucosylated N-glycans (∼50%), followed by high-mannose glycans (∼22%) and sialylofucosylated species (∼18%). Undecorated and sialylated species were less abundant, with values around 10% and 1%, respectively. Overall, this distribution pattern resembled the pattern of the parental forebrain N-glycome, but with slightly higher levels of fucosylation. The hippocampus N-glycome showed relatively lower levels of fucosylated N-glycans (45%) than the cortex but higher levels of high mannose glycans (27%). However, undecorated glycans, as well as sialofucosylated and sialylated glycans, were at the same levels in the hippocampus and the cortex. Notably, the cerebellum N-glycome presented the lowest levels of fucosylated N-glycans (39%) and similar levels of high-mannose species when compared with the hippocampus (27%), but the highest levels of undecorated species (∼18%), comparable to the parental hindbrain N-glycome. Sialofucosylated glycans in the cerebellum were similar to the levels found in the cortex and hippocampus N-glycomes. Lower levels of high mannose and undecorated glycans in the cortex N-glycome, along with higher levels of fucosylated glycans, were statistically higher compared with the hippocampus and cerebellum N-glycomes. There were also significantly higher levels of undecorated glycans in the cerebellum and increasing levels of neutral fucosylated glycans from the cerebellum to hippocampus and cortex N-glycomes (Cer<Hippo<Ctx). Total levels of fucosylation were significantly different among the three regions (p < 0.001), whereas total sialylation was significantly different between the hippocampus and cortex N-glycome (p < 0.05) (Fig. 5B). Together, these results revealed a distinct region-specific distribution of all N-glycans types in the three-region examined. Additionally, Figure 5C shows the heatmap of 82 individual N-glycan compositions from the cerebral cortex, hippocampus, and cerebellum N-glycomes, abundances of which were found to vary significantly in a region-specific manner (p < 0.05, supplemental Table S1). Remarkably, approximately ∼65% of all glycans identified in these regions were found to be differentially expressed in a region-specific manner.
Fig. 5.
Comparison of cell membrane N-glycome obtained from the cortex, hippocampus, and cerebellum. Relative abundances of N-glycan groups in the cerebral cortex, hippocampus, and cerebellum (A), and total levels of fucosylation and sialylation (B). Asterisks indicate the statistical significance between groups compared (∗p < 0.05%; ∗∗p < 0.01%; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Heatmap of N-glycans differentially expressed in the cortex, hippocampus and cerebellum, ordered from high mannose to complex glycans approximately according to N-glycan biosynthesis (p < 0.05%, as analyzed by two-way ANOVA and the TUKEY post-hoc test) (C). Glycan mass of hybrid and complex glycans increases from top to bottom. Glycan composition is abbreviated as Hex_HexNAc_Fuc_NeuAc where Hex = Hexose, HexNAc = N-Acetylhexosamine, Fuc = Fucose, NeuAc = N-Acetylneuraminic Acid; and numbers indicate the number of each monosaccharide residue. Abundance for each glycan composition was normalized to the abundance of total glycans identified. Data from six biological replicates were used. Partial least squares–discriminant analysis (PLS-DA) scores plots for cell membrane N-glycan profiles of the cortex, hippocampus, and cerebellum (D), and cortex and hippocampus (F). The 95% confidence interval is indicated as an ellipse, and each symbol represents a mouse (n = 6). The loadings plots (E and G) from the PLS-DA model show glycans contributing to the separation in the first dimension (x-axis of the scores plot) for cortex, hippocampus, and cerebellum (E), and cortex and hippocampus (G). C, complex; H, hybrid.
PLS-DA scores plots of cell surface N-glycan profiles of cortex, hippocampus, and cerebellum (Fig. 5D), and cortex and hippocampus (Fig. 5F) show they can be differentiated with a 95% confidence interval. Top glycans expressed in a region-specific manner contributing to the separation in the first dimension of these models are shown in the loading plots (Fig. 5, E and G, respectively). One-way ANOVA revealed that 39 glycan compositions with relative abundances above 0.5% were the main discriminators among cortex, hippocampus, and cerebellum N-glycome (p < 0.001). An additional 35 glycans species with relative abundances below 0.5% were also significant (p < 0.05 and p < 0.01) (supplemental Table S3). Among highly discriminating glycan species (with p < 0.001), five different expression patterns were recognized (I–V):
I- Glycans With a Distinct Region-Specific Level of Expression in the Cortex, Hippocampus, and Cerebellum N-Glycome
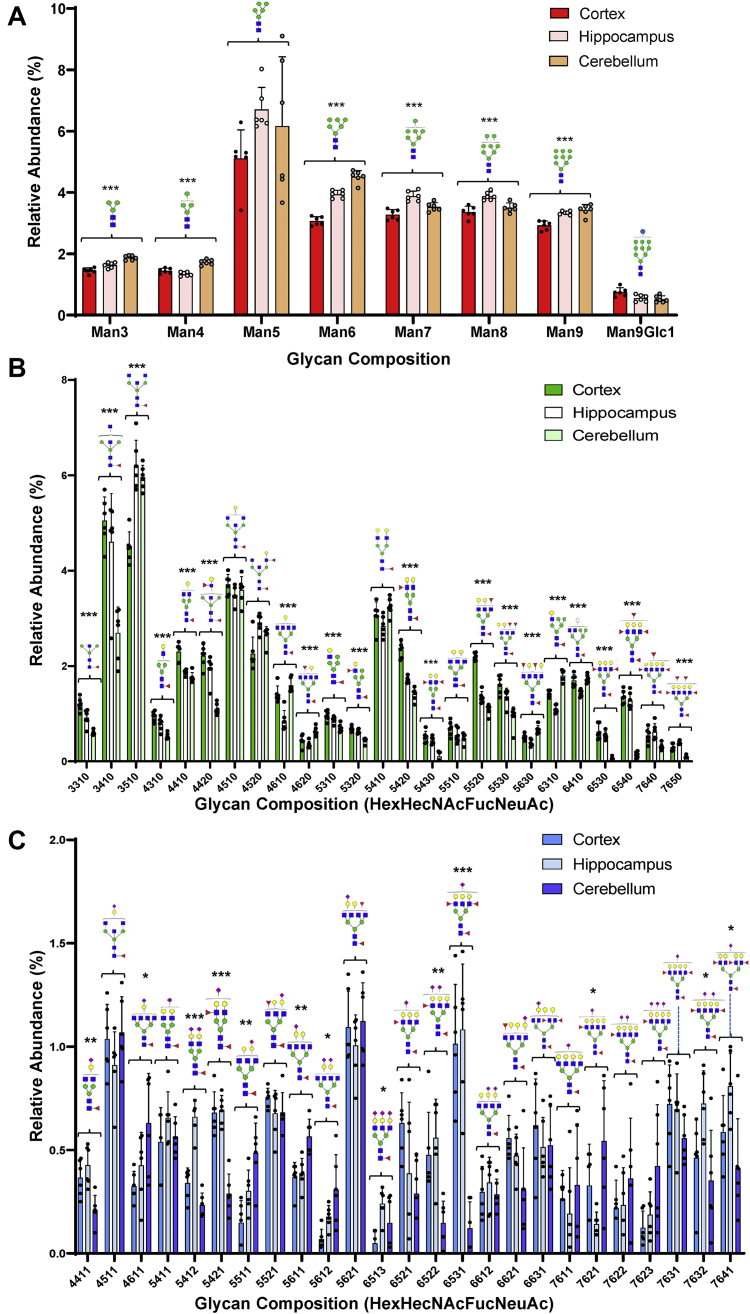
Eleven glycans were observed in this group corresponding to a) high mannose species Man3, Man6, and Man7, and b) neutral fucosylated species with compositions 3310, 4310, 4320, 5420, 5520, 5630, 6310, and 7650, given as (HexHexNacFucNeuAc) (Fig. 6, A and B and supplemental Table S3, bolded letters).
Fig. 6.
Region-specific distribution of most abundant glycans in the cortex, hippocampus, and cerebellum N-Glycomes. Comparative distribution of high mannose glycans (A), neutral fucosylated glycans (B), and sialofucosylated glycans (C) in the cerebral cortex, hippocampus, and cerebellum N-Glycomes. Glycan composition is abbreviated as follow HexHexNAcFucNeuAc where Hex = Hexose, HexNAc = N-Acetylhexosamine, Fuc = Fucose, NeuAc = N-Acetylneuraminic Acid; and numbers indicate the number of each monosaccharide residue. Values are reported as the average relative abundance of six biological replicates (n = 6). Asterisks indicate the statistical significance between groups compared (∗p < 0.05%; ∗∗p < 0.01%; ∗∗∗p < 0.001).
II- Glycans With Higher Expression in the Cortex and Hippocampus N-Glycome
Ten glycans were identified in this group corresponding to a) eight fucosylated glycans with compositions 3410, 4420, 5310, 5320, 5430, 5530, 6530, and 6540, and b) two sialofucosylated species with compositions 5421 and 6531 (Fig. 6, B and C and supplemental Table S3).
III- Glycans With Higher Expression in the Cerebellum N-Glycome
In this group, seven glycans were found corresponding to a) high mannose Man4, b) undecorated glycans with compositions 4400, 5400, and 6300, c) fucosylated species with compositions 3610 and 4620, and d) sialylated glycan with composition 6301.
IV- Glycans With Higher Expression in the Hippocampus N-Glycome
In this group, five glycans were observed corresponding to a) high mannose Man8 and b) sialofucosylated species with compositions 5311, 5412, 5422, and 6311 (Fig. 6, B and C and supplemental Table S3).
Also, among less abundant discriminating species (with p > 0.001, supplemental Table S3), similar expression profiles were observed (I–V). A majority of these species corresponded to sialofucosylated glycans. Figure 6C shows sialofucosylated glycans with abundances above 0.15% and compositions 5511, 5611, 5612, 6513, 6522, 6531, 7621, 7632, and 7640.
Additionally, glycan species able to distinguish between the cerebral cortex and hippocampus were neutral fucosylated species, with compositions 3210, 4410, 4610, 5420, 5520, and 6310 (with p < 0.001), and glycans with compositions 6630, 6730 (p < 0.05) (Fig. 6B).
Tandem MS analysis was also performed to support the presence of structural motives that show a differential spatial distribution among the cortex, hippocampus, and cerebellum N-glycomes. Comparably, to our discoveries in the parental N-glycomes, numerous glycans carrying bisecting GlcNAc were identified among highly significant glycans also able to discriminate the cerebral cortex, hippocampus, and cerebellum N-glycomes. Supplemental Fig. S5 shows the differential spatial expression of the most abundant bisecting N-glycans in the cortex, hippocampus, and cerebellum N-glycome. Monoantennary bisecting species with compositions 3410 and 4420 were significantly higher in both the cerebral cortex and hippocampus N-glycome. This was similar to the pattern displayed in the parental forebrain N-glycome shown in Figure 4. Likewise, biantennary bisecting species with composition 3510 showed higher levels in the cerebellum and comparable to levels observed in the parental hindbrain N-glycome. However, the hippocampus also showed equally high levels of this glycan but was significantly lower in the cortex N-glycome, the latter resembling the parental forebrain N-glycome. Bisecting monogalactosylated biantennary glycan with composition 4520 also showed a higher level in the hippocampus and cerebellum N-glycomes. However, no spatial differences were observed between the parental forebrain and hindbrain N-glycomes. Interestingly, bisecting glycan with composition 4511 did not show significant differences among the cortex, hippocampus, and cerebellum N-Glycomes, though it was significantly higher in the hindbrain N-glycome. Additional abundant bisecting species with composition 3500 and 5410 showed no significant spatial differences neither in the parental N-glycomes nor in the cortex, hippocampus, and cerebellum N-glycomes. Confirmatory tandem MS analysis of select mono and biantennary bisecting glycans with and without fucose expressed in the cortex, hippocampus, and cerebellum N-glycomes are shown in supplemental Fig. S6 (Bisecting species with composition 4510 (in cortex), 3510 and 4420 (in hippocampus), and 4520 (in cerebellum).
Additionally, tandem MS analysis of core and Le-fucosylated glycans expressed in a region-specific in cortex, hippocampus, and cerebellum N-glycomes is shown in supplemental Fig. S7 (glycans with compositions 4420 and 4520 (in cortex), and 7650 (in hippocampus). Additional MS spectra of Le-fucosylated glycans also containing bisecting GlcNAc are shown in supplemental Fig. S6 (glycan 4420 (in hippocampus) and 4520 (in cerebellum). Lastly, Tandem MS analysis of tri- and tetra-antennary sialofucosylated glycans with compositions 7641 (in cortex), 6531 and 7623 (in hippocampus), and 5621 (in cerebellum) is shown in supplemental Fig. S8. All sialylated glycans with or without fucose substitutions (sialofucosylated and sialylated only glycans) identified in the cortex, hippocampus, and cerebellum contained N-Acetyl neuraminic acid (NeuAc) residues. Glycans containing N-glycolyl neuraminic acid (NeuGc) were not found in this set of samples analyzed in agreement with our results in the parental forebrain and hindbrain N-glycomes.
In summary, we show the cell membrane N-glycan maps of the cortex, hippocampus, and cerebellum regions and reveal an increased resolution of region-specific expression of N-glycans. We also identified more than 70 glycan compositions with statistically significant discriminant values in distinguishing these N-glycomes and categorized them into five different expression patterns. We confirmed by tandem MS important structural motives, including bisecting, core and outer-arm fucosylation (Le-fucosylation), and sialyl-Le species that show region-specific expression profiles.
Altogether, our results reveal that the cerebral cortex, hippocampus, and cerebellum also present a unique region-specific cell surface N-glycome (glyco-phenotype), suggesting that the expression of glyco-related genes in the brain may be tightly regulated.
Discussion
N-glycosylation regulates brain development, function, and behavior (3, 4, 5, 42, 43, 44). Numerous brain glycomic studies have been reported in vertebrate animal models, from fish to rodents, and in human brain tissues (26, 27, 29, 30, 31, 32, 33). Despite significant advances, the cell surface brain N-glycome and the extent to which the N-glycan array and N-glycosylation pattern correspond to functional neuroanatomic areas in the mammalian brain remain largely unknown. In this work, we used a well-established methodology developed in our laboratory (38) to comprehensively map the cell surface membrane N-glycome of global and discrete functional areas in the 8-week-old mouse brain (Fig. 1). Here, we report for the first time the cell surface N-glycomes of the forebrain and hindbrain, revealing an impressive diversity of N-glycan arrays expressed in a region-specific manner. Furthermore, we also report the N-glycomes of three distinctive functional derivatives from the forebrain and hindbrain, specifically the cerebral cortex, hippocampus, and cerebellum, revealing additional unique region-specific glycan arrays. Our results show in detail the spatial distribution of cell surface N-glycome associated with highly specialized functional brain areas, demonstrating the presence of a previously unknown functional brain glyco-architecture.
Over 120 individual glycan compositions are identified in the forebrain and hindbrain N-glycomes with high sensitivity and reproducibility (supplemental Table S1). In agreement with earlier glycomic studies (32, 33, 34, 40, 45), our results show the presence of “brain-type” glycans represented by high levels of high mannose glycans, neutral species containing bisecting GlcNAc and branched di-, tri-, and tetra-antennary glycans with varying levels of galactosylation, fucosylation, and sialylation. However, we observed an increased level of fucosylated glycans, containing up to five fucose residues, sialofucosylated glycans containing up to four sialic acid residues, and overall a higher degree of fucosylation and sialofucosylation in the forebrain. These quantitative differences in the abundance of glycans subgroups are likely due to differences in methodological aspects in sample preparation, as we report the cell surface membrane N-glycomes derived from plasma-membrane-enriched proteins derived from region-specific brain tissues, and earlier reports profiled glycans from total protein extracts derived from whole or coronal section brain homogenates (32, 33, 40, 45). Interestingly, in a recent spatiotemporal comparative glycomic study of human and mouse brains, the N-glycome of the forebrain and hindbrain regions were not addressed (46). We observed that nearly 43% of N-glycans identified in the forebrain and hindbrain were differentially expressed (Fig. 2) and 27 of these N-glycans expressed in region-specific-manner can serve as discriminants of the forebrain and hindbrain N-glycomes (Figs. 2 and 3). Mono- and tri-antennary fucosylated glycans containing 1 to 4 fucose residues were the main discriminators between the forebrain and hindbrain N-glycomes. Similarly, high mannose and sialylated species were also differentially expressed. These results are consistent with recent reports of differential content in oligomannose, fucosylation, and sialylation determined in the striatum and substantia nigra regions in the rat brain (29, 30). Notably, several sialofucosylated glycans species observed in the FB and HB N-glycome were shown to decrease in a region-specific manner during aging in the nigrostriatal pathway in rat brains (29). This study also reports the differential expression of bisecting glycans in the forebrain and hindbrain (Fig. 4). We present tandem MS analysis confirming the presence of bisecting, core and antenna fucosylation (Le-fucosylation), and sialyl-Le-fucosylation motives in glycans expressed in a region-specific manner and that are in agreement with previous reports in rodent brains (32, 40, 45). Bisecting glycans are important in normal and pathological neural physiology, as they regulate the biosynthesis of other terminal modifications in N-glycans and are upregulated in the brain with Alzheimer's disease (AD) (44). A recent report shows that posttranscriptional regulations may play a role in modulating terminal N-glycans fucosylation and sialylation levels (45). Additionally, earlier reports correlated the relative abundance of bisecting glycans and fucosylated species carrying core and antennary fucose residues (forming LewisX (LeX) epitopes) in the brain tissues with high levels of transcripts encoding the respective enzymes responsible for the addition of those residues in N-glycans structures (Mgat3, Fut8, and Fut9) (33). Thus, the distinct region-specific abundance of bisecting and fucosylated species here identified may reflect in part a complex and dynamic interplay of transcriptional and posttranscriptional regulations of glycosyltransferases that deserves further studies.
To further understand the glyco-regionalization of the brain architecture, we characterized the cell-membrane N-glycome of the cerebral cortex and the hippocampus derived from the forebrain and the cerebellum derived from the hindbrain. We report a highly diverse array of 125 glycans compositions in the cortex, hippocampus, and cerebellum N-glycomes (supplemental Table S2), demonstrating a suborgan specific expression of N-glycans in the brain. This spatial location and distribution of N-glycan agree with previous brain glycomic studies using MALDI–imaging mass spectrometry (MALDI-IMS) on-tissue of coronal sections of mouse brains (27). However, we achieve a full region-specific N-glycome as we performed the analysis of entire subregions surgically isolated from the mouse brain. Our results obtained from 8-week-old adult mice are also in good agreement with Lee and colleague's work reporting the spatiotemporal variations in the brain of 6-week-old and 10-week-old mice (46). We achieved similar numbers of glycan species identified (∼120–130) on the cortex, hippocampus, and cerebellum N-glycome, although we report a higher number of glycans with a relative abundance ≥ of 0.05%. Furthermore, we also agree on numerous glycans species identified by PLS-DA analysis with discriminatory power to distinguish N-glycomes from different brain regions, including neutral multifucosylated glycans with core and antenna fucosylation, and a sialofucosylated glycan with compositions 4320, 4420, 5430, 5520, and 6531, respectively.
The region-specific resolution of the cell surface membrane N-glycome could reflect at least in part the underlying regional heterogeneities found at cellular and subcellular levels in the brain. For instance, at a cellular level, neuronal and nonneuronal components (i.e., glial cells) vary in a region-specific manner in the cortex, hippocampus, and cerebellum in function, origin, density, morphology, turnover, and metabolism (47). At the molecular level, N-glycan biosynthesis occurs in a protein-specific manner, and transcriptomic studies have revealed that distinct spatial protein expression patterns are associated with different classes of neurons and cell density across the brain (48). Furthermore, transcriptional and protein regulation of glyco-related genes, including glycosyltransferases as discussed before, sugar transporters, and trafficking proteins, are also accepted mechanisms that give rise to glycan heterogeneities.
We compared the N-glycosylation profile in each region and identified ∼75 glycan compositions being expressed in a region-specific manner that can serve as discriminants of the cerebral cortex, hippocampus, and cerebellum cell membrane N-glycomes (supplemental Table S3 and Fig. 6). Furthermore, we grouped these glycans into five expression pattern groups (I–V) to provide unique glyco-fingerprints that distinguish the spatial resolution of N-glycans in the cortex, hippocampus, and cerebellum that are in good agreement with Lee et al., 2020 (46). We monitored the presence of other acidic glycans carrying sulfated epitopes (including the HNK-1 epitope). Signals potentially compatible with these glycans were identified; however, their low intensity precluded identity confirmation by MS/MS. This potential discrepancy with Lee’s work is likely due to differences in methodological aspects of sample preparation. However, it is not surprising as this modification is expressed in trace amounts on specific glycoproteins and cell types only in areas of active neurogenesis in the adult mouse brain (5).
In this study we observed a ubiquitous but distinctive spatial distribution of high mannose glycans, with higher abundances in the hindbrain and cerebellum N-glycomes, and lower abundance in the forebrain and cerebral cortex N-glycome. Intriguingly, the hippocampus N-glycome showed high mannose glycans levels as high as those detected in the cerebellum. High mannose glycans have been reported to modify essential postsynaptic proteins such as GABA, AMPA, and NMDA receptors, and cell surface adhesion molecules such as the neural cell adhesion molecule L1 (3, 5). High mannose glycans in these glycoproteins participate in fundamental cell–cell adhesion interactions modulating synaptic transmission (3, 5, 36). Thus, the ubiquitous expression of high mannose glycans in all areas analyzed agrees with the abundant expression of cell surface neurotransmitter receptors and synaptic proteins expressed across the brain that controls basic brain electrophysiology and synaptic transmission. Interestingly, altered levels of proteins carrying high mannose glycans were recently reported in a mouse model of AD (49). These glycoproteomic studies were performed using whole-brain protein extracts, and thus the spatial resolution of this alteration was not resolved. Since AD progression occurs in a region-specific manner and affects the hippocampus and prefrontal cortex preferentially, mapping the glycan alterations in a region-specific manner would be of great significance to better understand the pathophysiology of the disease.
In addition to differences in the forebrain and hindbrain N-glycome, in this study, we also report a region-specific expression of bisected glycans in the cortex, hippocampus, and cerebellum N-glycomes. In addition to modulating the biosynthesis of higher branched N-glycan structures as previously discussed, bisected glycans also serve as protein trafficking tags, modulating the expression of crucial brain glycoproteins at the cell surface level (45, 50). Moreover, site-specific occupancy of bisecting GlcNAc glycan has also been reported, indicating that specific glycoproteins at specific glyco-sites are modified by bisecting GlcNAc residues (45). The highly regulatory functions of bisecting glycans highlight the value of the region-specific expression patterns of bisecting moieties here identified as a potential mechanism involved in the establishment or modulation of regional-brain-N-glycomes.
We also report the complexity and region-specific variations of fucosylated N-glycans in the cortex, hippocampus, and cerebellum N-glycomes (Figs. 5 and 6). We show that a majority of glycans (∼50%) able to discriminate the three N-glycomes are fucosylated species (supplemental Table S3, p < 0.001). Higher levels of fucosylation and increased core fucosylated glycans were detected in the cerebral cortex and hippocampus N-glycomes. Importantly, core fucosylation is a crucial modification involved in neuronal plasticity and is required for learning and memory (4). Specifically, loss of core fucosylation impairs Long-Term-Potentiation (LTP) and the ability to consolidate memory formation (51). The cerebral cortex and hippocampus are heavily involved in learning and memory; thus, our findings may be indicative of a glycan structure-region function correlation previously unknown. Our results are also in agreement with a report of differential core fucosylation levels in the stratum and substantia nigra regions in the rat brain (30).
We also describe prominent region-specific differences in the abundances of multifucosylated N-glycans containing up to five fucose residues (Fig. 6 and supplemental Table S3) in agreement with early and latest reports (46). Tandem MS confirmed the presence of core and antenna fucosylation (Le X) in agreement with several brain glycomic studies in adult rodents (32, 33, 40, 45). LeX epitopes are developmentally regulated, and in the adult brain, LeX occurs in neurogenic zones and is expressed at high levels in different populations of glial cells (i.e., astrocytes and oligodendrocytes) (5). This is in support of the significant heterogeneity and differential distribution of LeX reported here. Interestingly, numerous proteins carry this modification, including neural cell adhesion molecules (NCAM and L1), synaptic proteins (synapsin I), and myelin proteins (contactin-1) are involved in axon and neurite outgrowth, synaptic transition, and myeline sheath stabilization highlighting the broad functional relevance of LeX in numerous key neurophysiological processes.
Likewise, we report the complexity of cell membrane highly branched sialofucosylated glycans, containing up to four sialic acids and up to five fucose residues. Our results are in good agreement with earlier N-glycomic reports obtained using whole mouse brain tissues (33, 45), fixed and fresh frozen coronal brain sections (34, 35), and cell membrane fractions (46). We provide with high resolution the region-specific occurrence of sialofucosylated glycans and their values in differentiating the N-glycome from the cortex, hippocampus, and cerebellum that is unknown thus far. All species identified presented NeuAc residues, and the expression of trace amounts of NeuGc reported in brain tissue (52) was not detected in these regions, in accordance with our results from the forebrain and hindbrain. Sialic acids (NeuAc) are essential functional regulators of brain electrophysiology by modulating cell surface expression, gating, and kinetics of numerous ion channels (Na+2, Ca+2, and K+) and neurotransmitter transporters (Acetylcholine and GABA receptors) (3, 10, 12, 13, 14, 16). The forebrain as a unit and the cortex and hippocampus as specialized derived structures centralize the processing of information related to cognitive activities and behavior. Dysregulation of brain sialylation levels has been reported in CDG, psychiatric diseases such as schizophrenia, during aging, and in neurodegenerative disorders such as Alzheimer's and Parkinson's diseases (2, 29, 31, 49, 53). Thus, additional research to unveil the functional relevance of regional spatial distribution of sialic acid in the brain will shed light to understand better the modulatory role of N-glycosylation in brain function and cognition at a suborgan and region-specific level.
In conclusion, this study reports the cell-membrane enriched N-glycome maps obtained from global functional regions of the brain: the forebrain, hindbrain, and three discrete functional brain areas cerebral cortex, hippocampus, and cerebellum. These unique cell membrane N-glycome maps reveal the extraordinary complexity and diversity of N-glycans distribution in a functional and region-specific manner in the adult mouse brain. To the best of our knowledge, this is the first report of the forebrain and hindbrain N-glycome. Region-specific glycomic profiles identified in this work are groundbreaking discoveries as they reveal the resolution of cell surface N-glycosylation in the mammalian brain. The data provide new insights into specific glycans structures and their expression patterns in the brain, suggesting tight controls over the expression of enzymes involved in the biosynthesis of brain N-glycans. The application of the method, as described here, will enable further research to advance our understanding of brain function and how it can be regionally modulated by N-glycosylation in normal and pathophysiological conditions.
Data Availability
Data that support the findings of this study are available from the corresponding author on reasonable request. Glycomic data was deposited in MassIVE (https://massive.ucsd.edu) with the dataset identifier MSV000086642.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Kavi Morgan Rude for their help with graphic brain illustrations.
Author contributions
M. B., H. E. R., and C. B. L. conceptualization; K. S. and T. A. K. formal analysis; M. B., M. G. G., H. E. R., and C. B. L. funding acquisition; M. B. and J. L. investigation; M. G. G., H. E. R., and C. B. L. resources; M. B. supervision; K. S. and T. A. K. visualization; M. B., H. E. R., and C. B. L. writing—original draft; M. B. writing—reviewing and editing.
Funding and additional information
This work was supported by the National Institutes of Health (NIH) grants, from the National Institute of Diabetes and Digestive and Kidney Diseases R21DK118379 to M. B., the National Center for Complementary and Integrative Health R01AT009365 to M. G. G., and R01AT007079 to H. E. R., and the National Institute of General Medical Sciences R01GM049077 to C. B. L. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplemental Data
References
- 1.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng B.G., Freeze H.H. Perspectives on glycosylation and its congenital disorders. Trends Genet. 2018;34:466–476. doi: 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott H., Panin V.M. The role of protein N-glycosylation in neural transmission. Glycobiology. 2014;24:407–417. doi: 10.1093/glycob/cwu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inaba H., Kai D. N-glycosylation in the hippocampus is required for the consolidation and reconsolidation of contextual fear memory. Neurobiol. Learn. Mem. 2016;135:57–65. doi: 10.1016/j.nlm.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 5.Sytnyk V., Leshchyns'ka I., Schachner M. Neural glycomics: The sweet side of nervous system functions. Cell. Mol. Life Sci. 2021;78:93–116. doi: 10.1007/s00018-020-03578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson D., Montpetit M.L., Stocker P.J., Bennett E.S. The sialic acid component of the beta1 subunit modulates voltage-gated sodium channel function. J. Biol. Chem. 2004;279:44303–44310. doi: 10.1074/jbc.M408900200. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe I., Zhu J., Sutachan J.J., Gottschalk A., Recio-Pinto E., Thornhill W.B. The glycosylation state of Kv1.2 potassium channels affects trafficking, gating, and simulated action potentials. Brain Res. 2007;1144:1–18. doi: 10.1016/j.brainres.2007.01.092. [DOI] [PubMed] [Google Scholar]
- 8.Hall M.K., Reutter W., Lindhorst T., Schwalbe R.A. Biochemical engineering of the N -acyl side chain of sialic acids alters the kinetics of a glycosylated potassium channel Kv3.1. FEBS Lett. 2011;585:3322–3327. doi: 10.1016/j.febslet.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Weiss N., Black S.A.G., Bladen C., Chen L., Zamponi G.W. Surface expression and function of Cav3.2 T-type calcium channels are controlled by asparagine-linked glycosylation. Pflügers Arch. 2013;465:1159–1170. doi: 10.1007/s00424-013-1259-3. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe I., Zhu J., Recio-Pinto E., Thornhill W.B. The degree of N-glycosylation affects the trafficking and cell surface expression levels of Kv1.4 potassium channels. J. Membr. Biol. 2015;248:187–196. doi: 10.1007/s00232-014-9756-7. [DOI] [PubMed] [Google Scholar]
- 11.Nowycky M.C., Wu G., Ledeen R.W. Springer; New York, NY: 2014. Glycobiology of Ion Transport in the Nervous System; pp. 321–342. [DOI] [PubMed] [Google Scholar]
- 12.Ednie A.R., Bennett E.S. Vol 2. John Wiley & Sons, Inc; Hoboken, NJ: 2012. Modulation of voltage-gated ion channels by sialylation; pp. 1269–1301. (Comprehensive Physiology). [DOI] [PubMed] [Google Scholar]
- 13.Chen L. In pursuit of the high-resolution structure of nicotinic acetylcholine receptors. J. Physiol. 2010;588:557–564. doi: 10.1113/jphysiol.2009.184085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.daCosta C.J.B., Kaiser D.E.E., Baenziger J.E. Role of glycosylation and membrane environment in nicotinic acetylcholine receptor stability. Biophys. J. 2005;88:1755–1764. doi: 10.1529/biophysj.104.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishizaki T. N-glycosylation sites on the nicotinic ACh receptor subunits regulate receptor channel desensitization and conductance. Mol. Brain Res. 2003;114:172–176. doi: 10.1016/s0169-328x(03)00171-2. [DOI] [PubMed] [Google Scholar]
- 16.Lo W.-Y., Lagrange A.H., Hernandez C.C., Harrison R., Dell A., Haslam S.M., Sheehan J.H., Macdonald R.L. Glycosylation of {beta}2 subunits regulates GABAA receptor biogenesis and channel gating. J. Biol. Chem. 2010;285:31348–31361. doi: 10.1074/jbc.M110.151449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandel M.B., Yamamoto S., Midorikawa R., Morise J., Wakazono Y., Oka S., Takamiya K. N-glycosylation of the AMPA-type glutamate receptor regulates cell surface expression and tetramer formation affecting channel function. J. Neurochem. 2018;147:730–747. doi: 10.1111/jnc.14565. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi Y., Morise J., Morita I., Takematsu H., Oka S. Role of site-specific N-glycans expressed on GluA2 in the regulation of cell surface expression of AMPA-type glutamate receptors. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tucholski J., Simmons M.S., Pinner A.L., McMillan L.D., Haroutunian V., Meador-Woodruff J.H. N-linked glycosylation of cortical N-methyl-D-aspartate and kainate receptor subunits in schizophrenia. Neuroreport. 2013;24:688–691. doi: 10.1097/WNR.0b013e328363bd8a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichnerova K., Kaniakova M., Park S.P., Skrenkova K., Wang Y.-X., Petralia R.S., Suh Y.H., Horak M. Two N-glycosylation sites in the GluN1 subunit are essential for releasing N-methyl-d-aspartate (NMDA) receptors from the endoplasmic reticulum. J. Biol. Chem. 2015;290:18379–18390. doi: 10.1074/jbc.M115.656546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheray M., Petit D., Forestier L., Karayan-Tapon L., Maftah A., Jauberteau M.-O., Battu S., Gallet F.P., Lalloué F. Glycosylation-related gene expression is linked to differentiation status in glioblastomas undifferentiated cells. Cancer Lett. 2011;312:24–32. doi: 10.1016/j.canlet.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y., Mayampurath A., Khan S., Cohen J.K., Mechref Y., Volchenboum S.L. N-linked glycan profiling in neuroblastoma cell lines. J. Proteome Res. 2015;14:2074–2081. doi: 10.1021/pr5011718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veillon L., Fakih C., Abou-El-Hassan H., Kobeissy F., Mechref Y. Glycosylation changes in brain cancer. ACS Chem. Neurosci. 2017;9:51–72. doi: 10.1021/acschemneuro.7b00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhaak L.R., Xu G., Li Q., Goonatilleke E., Lebrilla C.B. Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev. 2018;118:7886–7930. doi: 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues J.G., Balmaña M., Macedo J.A., Poças J., Fernandes Â., de-Freitas-Junior J.C.M., Pinho S.S., Gomes J., Magalhães A., Gomes C., Mereiter S., Reis C.A. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell. Immunol. 2018;333:46–57. doi: 10.1016/j.cellimm.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Yamakawa N., Vanbeselaere J., Chang L.Y., Yu S.Y., Ducrocq L., Harduin-Lepers A., Kurata J., Aoki-Kinoshita K.F., Sato C., Khoo K.H., Kitajima K., Guerardel Y. Systems glycomics of adult zebrafish identifies organ-specific sialylation and glycosylation patterns. Nat. Commun. 2018;9:4647. doi: 10.1038/s41467-018-06950-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers T.W., Jones E.E., Betesh L.R., Romano P.R., Gao P., Copland J.A., Mehta A.S., Drake R.R. Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal. Chem. 2013;85:9799–9806. doi: 10.1021/ac402108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torii T., Yoshimura T., Narumi M., Hitoshi S., Takaki Y., Tsuji S., Ikenaka K. Determination of major sialylated N-glycans and identification of branched sialylated N-glycans that dynamically change their content during development in the mouse cerebral cortex. Glycoconj. J. 2014;31:671–683. doi: 10.1007/s10719-014-9566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raghunathan R., Polinski N.K., Klein J., Hogan J.D., Shao C., Khatri K., Leon D., McComb M.E., Manfredsson F.P., Sortwell C.E., Zaia J. Glycomic and proteomic changes in aging brain nigrostriatal pathway. Mol. Cell. Proteomics. 2018;17:1778–1787. doi: 10.1074/mcp.RA118.000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samal J., Saldova R., Rudd P.M., Pandit A., O’Flaherty R. Region-specific characterization of N-glycans in the striatum and substantia nigra of an adult rodent brain. Anal. Chem. 2020;92:12842–12851. doi: 10.1021/acs.analchem.0c01206. [DOI] [PubMed] [Google Scholar]
- 31.Raghunathan R., Hogan J.D., Labadorf A., Myers R.H., Zaia J. A glycomics and proteomics study of aging and Parkinson’s disease in human brain. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-69480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y.J., Wing D.R., Guile G.R., Dwek R.A., Harvey D.J., Zamze S. Neutral N-glycans in adult rat brain tissue - complete characterisation reveals fucosylated hybrid and complex structures. Eur. J. Biochem. 1998;251:691–703. doi: 10.1046/j.1432-1327.1998.2510691.x. [DOI] [PubMed] [Google Scholar]
- 33.Nairn A.V., York W.S., Harris K., Hall E.M., Pierce J.M., Moremen K.W. Regulation of glycan structures in animal tissues: Transcript profiling of glycan-related genes. J. Biol. Chem. 2008;283:17298–17313. doi: 10.1074/jbc.M801964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turiák L.T., Shao C., Meng L., Khatri K., Leymarie N., Wang Q., Pantazopoulos H., Leon D.R., Zaia J. Workflow for combined proteomics and glycomics profiling from histological tissues. Anal. Chem. 2014;86:9670–9678. doi: 10.1021/ac5022216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji I.J., Hua S., Shin D.H., Seo N., Hwang J.Y., Jang I.-S., Kang M.-G., Choi J.-S., An H.J. Spatially-resolved exploration of the mouse brain glycome by tissue glyco-capture (TGC) and nano-LC/MS. Anal. Chem. 2015;87:2869–2877. doi: 10.1021/ac504339t. [DOI] [PubMed] [Google Scholar]
- 36.Trinidad J.C., Schoepfer R., Burlingame A.L., Medzihradszky K.F. N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics. 2013;12:3474–3488. doi: 10.1074/mcp.M113.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M.-Q., Zeng W.-F., Fang P., Cao W.-Q., Liu C., Yan G.-Q., Zhang Y., Peng C., Wu J.-Q., Zhang X.-J., Tu H.-J., Chi H., Sun R.-X., Cao Y., Dong M.-Q. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017;8:438. doi: 10.1038/s41467-017-00535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q., Xie Y., Wong M., Barboza M., Lebrilla C.B. Comprehensive structural glycomic characterization of the glycocalyxes of cells and tissues. Nat. Protoc. 2020;15:2668–2704. doi: 10.1038/s41596-020-0350-4. [DOI] [PubMed] [Google Scholar]
- 39.Kronewitter S.R., An H.J., de Leoz M.L., Lebrilla C.B., Miyamoto S., Leiserowitz G.S. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamze S., Harvey D.J., Chen Y.J., Guile G.R., Dwek R.A., Wing D.R. Sialylated N-glycans in adult rat brain tissue - a widespread distribution of disialylated antennae in complex and hybrid structures. Eur. J. Biochem. 1998;258:243–270. doi: 10.1046/j.1432-1327.1998.2580243.x. [DOI] [PubMed] [Google Scholar]
- 41.Domon B., Costello C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988;5:397–409. [Google Scholar]
- 42.Kleene R., Schachner M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004;5:195–208. doi: 10.1038/nrn1349. [DOI] [PubMed] [Google Scholar]
- 43.Ye Z., Marth J.D. N-glycan branching requirement in neuronal and postnatal viability. Glycobiology. 2004;14:547–558. doi: 10.1093/glycob/cwh069. [DOI] [PubMed] [Google Scholar]
- 44.Kizuka Y., Taniguchi N. Neural functions of bisecting GlcNAc. Glycoconj. J. 2018;35:345–351. doi: 10.1007/s10719-018-9829-4. [DOI] [PubMed] [Google Scholar]
- 45.Nakano M., Mishra S.K., Tokoro Y., Sato K., Nakajima K., Yamaguchi Y., Taniguchi N., Kizuka Y. Bisecting GlcNAc is a general suppressor of terminal modification of N-glycan. Mol. Cell. Proteomics. 2019;18:2044–2057. doi: 10.1074/mcp.RA119.001534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J., Ha S., Kim M., Kim S.W., Yun J., Ozcan S., Hwang H., Ji I.J., Yin D., Webster M.J., Weickert C.S., Kim J.H., Yoo J.S., Grimm R., Bahn S. Spatial and temporal diversity of glycome expression in mammalian brain. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28743–28753. doi: 10.1073/pnas.2014207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan Y.L., Yuan Y., Tian L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry. 2020;25:351–367. doi: 10.1038/s41380-019-0609-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko Y., Ament S.A., Eddy J.A., Caballero J., Earls J.C., Hood L., Price N.D. Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3095–3100. doi: 10.1073/pnas.1222897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang P., Xie J., Sang S., Zhang L., Liu M., Yang L., Xu Y., Yan G., Yao J., Gao X., Qian W., Wang Z., Zhang Y., Yang P., Shen H. Multilayered N-glycoproteome profiling reveals highly heterogeneous and dysregulated protein N-glycosylation related to Alzheimer’s disease. Anal. Chem. 2020;92:867–874. doi: 10.1021/acs.analchem.9b03555. [DOI] [PubMed] [Google Scholar]
- 50.Kizuka Y., Kitazume S., Fujinawa R., Saito T., Iwata N., Saido T.C., Nakano M., Yamaguchi Y., Hashimoto Y., Staufenbiel M., Hatsuta H., Murayama S., Manya H., Endo T., Taniguchi N. An aberrant sugar modification of BACE 1 blocks its lysosomal targeting in A lzheimer’s disease. EMBO Mol. Med. 2015;7:175–189. doi: 10.15252/emmm.201404438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu W., Fukuda T., Isaji T., Hang Q., Lee H.H., Sakai S., Morise J., Mitoma J., Higashi H., Taniguchi N., Yawo H., Oka S., Gu J. Loss of α1,6-fucosyltransferase decreases hippocampal long term potentiation: Implications for core fucosylation in the regulation of AMPA receptor heteromerization and cellular signaling. J. Biol. Chem. 2015;290:17566–17575. doi: 10.1074/jbc.M114.579938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies L.R.L., Pearce O.M.T., Tessier M.B., Assar S., Smutova V., Pajunen M., Sumida M., Sato C., Kitajima K., Finne J., Gagneux P., Pshezhetsky A., Woodss R., Varki A. Metabolism of vertebrate amino sugars with N-glycolyl groups: Resistance of α2-8-linked N-glycolylneuraminic acid to enzymatic cleavage. J. Biol. Chem. 2012;287:28917–28931. doi: 10.1074/jbc.M112.365056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams S.E., Mealer R.G., Scolnick E.M., Smoller J.W., Cummings R.D. Aberrant glycosylation in schizophrenia: A review of 25 years of post-mortem brain studies. Mol. Psychiatry. 2020;25:3198–3207. doi: 10.1038/s41380-020-0761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available from the corresponding author on reasonable request. Glycomic data was deposited in MassIVE (https://massive.ucsd.edu) with the dataset identifier MSV000086642.