Abstract
Non-renewable fossil fuels such as bitumen, coal, natural gas, oil shale, and petroleum are depleting over the world owing to unrestricted consumption. Biofuels such as biodiesel, biobutanol, bioethanol, and biogas are considered an eco-friendly and cost-effective alternatives of fossil fuels. For energy sustainability, the production of advanced biofuels is required. The advancement of genetic and metabolic engineering in microbial cells played a significant contribution to biofuels overproduction. Essential approaches such as next-generation sequencing technologies and CRISPR/Cas9-mediated genome editing of microbial cells are required for the mass manufacture of biofuels globally. Advanced “omics” approaches are used to construct effective microorganisms for biofuels manufacturing. A new investigation is required to augment the production of lignocellulosic-based biofuels with minimal use of energy. Advanced areas of metabolic engineering are introduced in the manufacture of biofuels by the use of engineered microbial strains. Genetically modified microorganisms are used for the production of biofuels in large quantities at a low-cost.
Keywords: Biofuels, Biogas, Microbiome, Omics approaches, Metabolic engineering, Genetic engineering
Introduction
Bitumen, coal, natural gas, oil shale, and petroleum are non-renewable fossil fuels that are key sources of energy around the globe. Because of the unrestricted use of fossil fuels, these resources become unsustainable (Westbrook et al. 2019). Bioethanol which is synthesized from corn and biodiesel which is prepared by esterification of vegetable oils are considered first-generation biofuels (Sheng and Feng 2015). Second-generation biofuels are prepared from cellulose, lignin, and hemicellulose of agricultural wastes, the wood of forestry, and municipal wastages (Yadav and Vivekanand 2019). Biofuels such as biobutanol, biodiesel, bioethanol, and biogas are considered an eco-friendly and fruitful alternative to fossil fuels. Essential approaches are required for the mass production of biofuels globally (Xing et al. 2012). Ineffective carbon uptake, the creation of growth inhibitors during biomass pre-treatment, residual chemicals in saccharification, inhibitory metabolites, and other industrial growth conditions that limit microbial growth are bottlenecks in biofuel production (Mukhopadhyay 2015). To overcome the technological bottlenecks in the successful conversion of biomass into biofuels, the industry is required to develop novel enzyme technology for the efficient production of biofuels through identifying microbial genes. Most of the microorganisms from natural environments are not culturable. Cultivation of microbes requires specific growth conditions, trophic dependencies, and syntrophic relationships which are the limitations for biofuel and biogas production. Metagenomics approaches (Fig. 1) are used to overcome traditional culture techniques and also used to identify enzymes such as amylolytic enzymes, β-glucosidase, endoglucanase, lignases, and xylanase for biofuels production (Xing et al. 2012). Production of biogas [methane (CH4), carbon dioxide (CO2), hydrogen sulphide (H2S)] from sewage and organic waste digester may be considered as an alternative source of renewable bioenergy in the rural area of the world (Awe et al. 2017). In developed countries, bioethanol is utilized as a gasoline additive, and biobutanol is used as a petroleum substitute (Shanmugam et al. 2019).
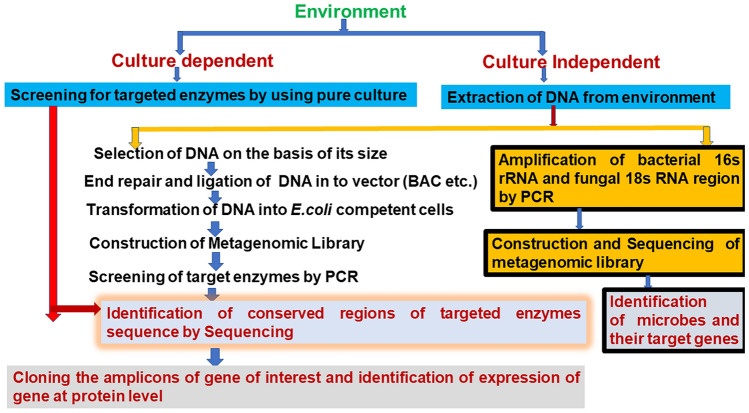
Fig. 1.
Schematic representation of culture dependent and independent methods for identifying microbial enzymes for biofuel production from environment. Hypervariable V1–V9 regions of the 16S ribosomal RNA gene are used to identify bacterial communities; whereas, 18S rRNA genes are used to identify fungal communities by next generation sequencing. Bacterial artificial chromosomes (BAC), cosmid, and fosmid are used as a vector and E. coli as a host for cloning of DNA fragment to construct metagenomic library. Sanger sequence-based screening approaches are used to identify microbial enzymes from metagenomic library of environmental samples
Biomass is considered a renewable source of energy. Biomass conversion produces biofuels such as biogas and liquid biofuels. Resources of biomass include forest products such as bark, shrubs, sawdust, trees, wood, and wood residues wastes from agricultural production, energy crops such as alfalfa, grasses, soybean, sunflower, and starch crops such as corn, barley, and wheat (Sarsekeyeva et al. 2015). Anaerobic digestion (AD) of plant material by microbes is the main source of biogas such as methane (CH4) and carbon dioxide (CO2) production (Fig. 2). In biogas plants (BGPs), renewable resources of agriculture such as grass, maize, and sugar beet, and biodegradable organic wastes are used to generate biogas by anaerobic digestion (Zhang et al. 2016). Methane production is divided into several phases such as hydrolysis of organic compounds such as carbohydrates, proteins, and lipids, production of butyrate, propionate, other short-chain volatile fatty acids (VFA), and alcohols, production of acetic acid, and finally production of CH4. Bacteria such as Clostridium bornimense, Fermentimonas caenicola, Herbinix hemicellulosilytica, Herbinix luporum, Herbivorax saccincola, Proteiniphilum saccharofermentans, Petrimonas mucosa, and Proteiniborus indolifex are involved in the process of hydrolysis and acidogenesis (Hahnke et al. 2016; Hassa et al. 2018). Clostridium acetobutylicum, C. beijerinckii, C. saccharobutylicum, and C. saccharoperbutylacetonicum are used to generate butanol (Jiang et al. 2017).
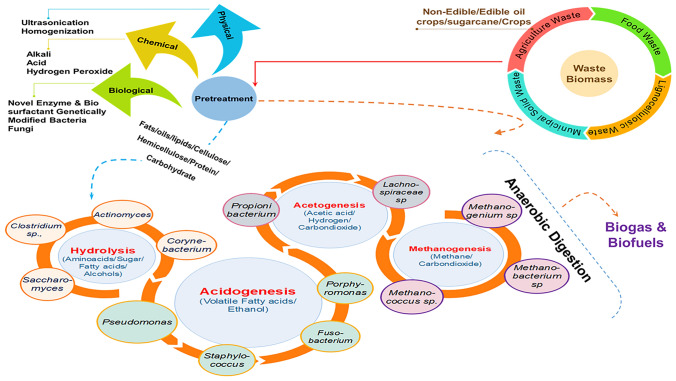
Fig. 2.
Schematic representation of biogas and biofuels production from the lignocellulosic biomass, oil crops, sugarcane/food waste through pre-treatment, and anaerobic digestion. Sugar, fat, and proteins are converted into organic acids and alcohols by acidogenesis; organic acids and alcohols are converted into hydrogen (H2), carbon dioxide (CO2), and acetic acid (CH3COOH) by acetogenic bacteria. Methanogenic bacteria are involved in the synthesis of biogas from CH3COOH, CO2, and H2
New generation sequencing technologies (NGS) are used to identify the complex microbial environments and genetic constituents of various species of microorganisms that contribute significantly to biofuel production (Hassa et al. 2018). Metagenomic characterization of biomass degrading microorganisms from diverse ecological environments may able to identify the enzymes in bioengineering (Bilal et al. 2018) (Fig. 1). Genome editing of microbial cells by CRISPR/Cas9 has been efficiently employed for the production of biofuels (Javed et al. 2019). Advanced areas of metabolic engineering and synthetic biology are introduced in the production of biofuels through the use of engineered microbial strains (Cheon et al.2016). Recently, CRISPR-Cas has been utilized in biofuel (biobutanol) producing microorganisms such as Saccharomyces cerevisiae and Clostridium (Joseph et al. 2018; Raschmanová et al. 2018). Our goal is to provide a complete overview of metagenomics and CRISPR-Cas-based genome editing approaches for better biofuels and biogas production. In this review, we discussed the management of genome editing approaches to regulating microbial strains in biofuels production and also microbial diversity involved in biogas production using metagenomics approaches.
Types of biofuels
Biofuels are classified into primary and secondary biofuels. Secondary biofuels are categorized into four generations (Rodionova et al. 2017).
Sources and application of biofuels
Biofuels are produced from biological materials such as materials from livestock and waste plants. Primary biofuels such as crop remains, fuel-wood, wood chips, and landfill gas are mainly utilized in cooking, heating, and manufacturing electricity whereas secondary biofuels such as biodiesel, bioethanol, and biogas are used in vehicles and industry (Rodionova et al. 2017; Doshi et al. 2016) (Fig. 2). Liquid biofuels are ethanol, butanol, methanol, and biodiesel. In 2011, ten billion liters of bioethanol were produced from corn and sugar beans which will be increased to 281.5 billion liters by 2020 (Sarsekeyeva et al. 2015). The global annual growth rate of biofuel production is currently around 9%, although this is expected to drop to 7% by 2022 (https://www.iea.org/reports/renewable-energy-market-update-2021/transport-biofuels). Esterification of oils from vegetables, microalgae, or other microorganisms produces biodiesel. Gasoline is produced from triacylglycerides (TAGs) and diacylglycerides (DAGs) (Georgianna and Mayfield 2012).
Different generations of biofuels
Bioethanol and butanol are the first-generation biofuels that are produced due to the fermentation of starch of barley, potato, wheat, and corn and sugars of sugar beet and sugarcane. Bioethanol is produced from crop plants having a high carbohydrate concentration by using enzymes of S. cerevisiae. Biodiesel is produced through the trans-esterification of vegetable oils from sebaceous plants such as coconut, palm, rapeseed, sunflower, and soybeans (Nigam and Singh 2011). Second generation biofuels such as biobutanol are produced from lignocellulosic materials of wood, leaves, and straw (Havlík et al. 2011). High oil content (around 60–70%) of green algae such as Chlorella vulgaris, Chamydomonas reinhardtii, and Dunaliella salina are used for biodiesel production. Microalgae become able to produce more biodiesel than conventional crops (Azad et al. 2014). Microalgal TAGs are involved in the manufacturing of third-generation biofuels (Prˇibyl et al. 2014). Hydrogen, acetone, and methane are considered to be third generation biofuels. First‐generation biofuels have several disadvantages such as low yield and high cost whereas second‐generation biofuels have several advantages such as economical, sustainable, and eco-friendly. Due to their low cost, high energy, and renewability, algae are considered to be the major source of third‐generation fuels. Due to its high energy, low cost, less unhealthy, and better solubility, biobutanol is considered to be the best alternative to non-renewable fossil fuels (Rathour et al. 2018). Fourth generation biofuels are derived from genetically engineered cyanobacteria (Sarsekeyeva et al. 2015). Algal species such as Botryococcus braunni, Chaetocero scalcitrans, Chlorella species, Isochrysisgal bana, Nanochloropsis sp., Schizochytrium limacinum, and Scenedesmus as well as other microbes such as Acinetobacter calcoacetius, Arthrobacter sp. Bacillus anthracis, and B. subtilis are identified as a source for fourth generation biofuels synthesis (Dutta et al. 2014).
Microbiome involved in biofuel production
Microbes from different ecological habitats are involved in the conversion of lignocellulosic biomass to biofuels (Pabbathi et al. 2021). The majority of cellulolytic microorganisms belong to the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria, and their cellulolytic enzymes are important in the conversion of biomass to biofuels (Prasad et al. 2019). Fungi such as Ceriporiopsis subvermispora, Pleurotus florida, Phanerochaete chrysosporium, and Trichoderma reesei have enzymes such as lignin peroxidases, laccases, and manganese peroxidases which are used for the disintegration of biomass (Zabed et al. 2016; Zhao et al. 2011). Bacterial species such as Acetovibrio, Bacillus, Bacteroides, Clostridium, Cellulomonas, Erwinia, Ruminococcus, Thermomonospora, Streptomyces, and Microbispora, and fungal genera such as Aspergillus, Fusarium, Humicola, Schizophyllum, Sclerotium, Schizophillum, Trichoderma, and Penicillium are involved in the production of hydrolytic enzymes such as cellulases, and hemicellulases which are involved in the production of monomeric sugars from biomass (Javed et al. 2019). Gram-negative bacterium Zymomonas mobilis, anaerobic thermophilic bacteria such as Clostridium thermohydrosulfuricum, Clostridium thermosaccharolyticum, Thermoanaerobacter mathranii, Thermoanaerobacter ethanolicus, and Thermoanaerobium brockii are engaged in the conversion of monomeric sugars into ethanol under anaerobic environment (Balat 2011). Bacterial species belonging to the genus Clostridium have been found to be involved in the production of 1-butanol through acetone-butanol-ethanol (ABE) pathway (Cai et al. 2016). Bacteria such as Escherichia coli and Klebsiella oxytoca and the yeast Pichia stipites are used in ethanol production from pentoses through genetic engineering approaches (Dellomonaco et al. 2010). Ethanologenic bacterium Z. mobilis is used for the synthesis of ethanol from glucose and sucrose. Expression of such as xylose isomerase, xylulokinase, transketolase, and transaldolase genes from E. coli in Z. mobilis CP4 (pZB5) enhanced ethanol production from xylose (Colin et al. 2011; Xia et al. 2019; Yang et al. 2016). In Z. mobilis ATCC39676 (pZB206), expression of E. coli genes such as araABD (l-arabinose isomerase, l-ribulokinase, l-ribulose-5-P-4-epimerase), genes encoding for transketolase and transaldolase increased ethanol production from arabinose (Colin et al. 2011; Xia et al. 2019). PDC and ADH genes (pdc and adhB) from Z. mobilis were expressed in E. coli to enhance ethanol production. E. coli and S. cerevisiae strains have been used for the production of biofuel isoprenoids. Two genes of Bacillus subtilis are involved in the conversion of isoprenyl diphosphate (IPP) into isopentenol. The expression of the Bacillus subtilis pyrophosphatase gene in E. coli increased the isopentenol production (Dellomonaco et al. 2010) (Table 1). Clostridium acetobutylicum and C. pasteurianum are involved in the production of biodiesel from glycerol (Yazdani and Gonzalez 2008).
Table 1.
Genetically modified microbial strains for biofuels production
| Strain | Genetic modifications | Biofuels | References |
|---|---|---|---|
| Zymomonas mobilis CP4 (pZB5) | Expression of Escherichia coli genes such as xylose isomerase, xylulokinase, transketolase, and transaldolase | Ethanol from xylose | Colin et al. (2011), Xia et al. (2019) and Yang et al. (2016) |
| Zymomonas mobilis ATCC39676 (pZB206) | Expression of Escherichia coli genes such as araABD (L-arabinose isomerase, L-ribulokinase, L-ribulose-5-P-4-epimerase), genes encoding for transketolase and transaldolase | Ethanol production from arabinose | Colin et al. (2011) and Xia et al. (2019) |
| Escherichia coli | Expression of pyrophosphatase gene from Bacillus subtilis | Isopentenol | Dellomonaco et al. (2010) |
| Clostridium saccharoperbutylacetonicum N1-4 | Overexpression of genes such as aldehyde/alcohol dehydrogenase genes and thiolase genes | Butanol | Wang et al. (2017a, b) |
| Clostridium beijerinckii CC101-SV6 | Overexpression of adhE2 and ctfAB genes | Butanol | Lu et al. (2017) |
| Clostridium tyrobutyricum | Overexpression of genes such as (scrB, scrA, scrK) and adhE2 | Butanol production from sucrose and sugarcane | Zhang et al. (2017) |
| Escherichia coli | Expression of genes such as thl, hbd, crt, bcd, etfAB, and adhE2 which are involved in the conversion of Acetyl-CoA to butanol | Butanol from glucose | Atsumi et al. (2008) |
| Escherichia coli | Overexpression of ilvIHCD operon | Isobutanol | Dellomonaco et al. (2010) |
| Synechococcus elongatus PCC 7942 | Expression of two genes such as pyruvate decarboxylase and alcohol dehydrogenase II from Zymomonas mobilis | Ethanol | Song et al. (2014) |
| Escherichia coli | Expression of genes such as 2-keto acid decarboxylase (aro10) and alcohol dehydrogenase (adh2) from Saccharomyces cerevisiae and acyltransferases (ws/dgat) from Acinetobacter baylyi | Isobutanol and 3-methyl-1-butanol | Guo et al. (2014) |
Monomeric sugars are converted by a simultaneous process of saccharification and fermentation (SSF) into ethanol. Bioethanol is produced due to the fermentation of hexose sugars at a lower temperature (30–32 °C) in which amylase converts starch into dextrin whereas glucoamylase converts dextrin into glucose. High-temperature tolerant microorganisms e.g., Kluyveromyces marxianus is used in the saccharification process (Javed et al. 2019). Both C6-fermenting and C5-fermenting microbes such as S. cerevisiae and Candida shehatae are used for biofuel production from monomeric sugars (das Neves et al. 2007). In consolidated bioprocessing (CBP), bacterial species such as Clostridium thermocellum and several fungal species such as Fusarium oxysporum, Neurospora crassa, and Paecilomyces sp. are involved in enzymatic hydrolysis and fermentation (Kang et al. 2014). In S. cerevisiae, fatty alcohol was produced by the expression of the Mycobacterium marinum gene carboxylic acid reductase, which is involved in the conversion of fatty acids to fatty aldehydes (Zhou et al. 2016). In E. coli, expression of genes such as 2-keto acid decarboxylase (aro10) and alcohol dehydrogenase (adh2) from S. cerevisiae and acyltransferases (ws/dgat) from Acinetobacter baylyi enhanced the production of fatty acids with short chain alcohols such as isobutanol and 3-methyl-1-butanol (Guo et al. 2014). Cell-wall degrading enzymes are involved in the conversion of lignocellulosic biomass into bio-alcohol (Tiwari et al. 2018). The extremophilic bacterium such as Caldicelluloseruptor bescii becomes more efficient to synthesize cellulolytic enzyme as compared to Trichoderma reesei (Kanafusa-Shinkai et al. 2013).
Biobutanol (or butyl alcohol) is synthesized by the fermentation of sugars from biomass such as corn stover, sugarcane bagasse, algal biomass, and food waste by using anaerobic as well as aerobic bacteria through acetone–butanol–ethanol (ABE) fermentation. Anaerobe bacteria Clostridia are involved in butanol production through acidogenesis where organic acids, carbon dioxide, and hydrogen are produced as a by-product and solventogenesis where acetone, butanol, and ethanol are produced. Clostridia is a butanol-producing Gram-positive, and strictly anaerobic bacterium (Karimi et al. 2015). ABE-producing Clostridia are involved in the fermentation of carbon sources, including fructose, glucose, galactose, sucrose, xylose, mannose, inulin, and glycerol (Visioli et al. 2014). Overexpression of genes such as aldehyde/alcohol dehydrogenase genes and thiolase genes in Clostridium saccharoperbutylacetonicum N1-4 are used in the production of butanol (Wang et al. 2017a, b). Overexpression of adhE2 and ctfAB genes in C. beijerinckii CC101-SV6 are used in the higher production of butanol (Lu et al. 2017). Overexpression of genes such as (scrB, scrA, scrK) and adhE2 in C. tyrobutyricum are used in butanol production from sucrose and sugarcane (Zhang et al. 2017) (Table 1). Escherichia coli, Saccharomyces cerevisiae, and Synechococcus elongatus are also involved in production of 1-butanol (Cheon et al. 2016). Expression of genes involved in the conversion of Acetyl-CoA to butanol such as thl, hbd, crt, bcd, etfAB, and adhE2 in E. coli enhanced the production of butanol from glucose (Atsumi et al. 2008) (Table 1). Butanol manufacture in E. coli was performed by a keto-acid-mediated pathway that utilized the leucine biosynthesis operon (leuABCD) and norvaline biosynthesis (Shen and Liao 2008). Overexpression of ilvIHCD operon in E. coli was used for isobutanol production through the conversion of pyruvate to 2-ketoisovalerate. Clostridium acetobutylicum and Clostridium beijerinckii are involved in the production of butanol from glucose (Dellomonaco et al. 2010) (Table 1).
Role of Cyanobacteria in biofuel production
Cyanobacteria is a key resource of renewable biofuels (Angermayr et al. 2009). Transgenic Synechococcus elongatus PCC 7942 is used to synthesize ethanol by expressing two genes such as pyruvate decarboxylase and alcohol dehydrogenase II from Z. mobilis. The amount of ethanol production was increased when the above mentioned genes were expressed in Synechocystis sp. PCC 6803. (Song et al. 2014) (Table 1). Isobutanol may be used in the place of gasoline. Synechococcus elongatus PCC 7942 is used to produce 1-butanol. Five genes such as aldehyde/alcohol dehydrogenase (adhE2), crotonase (crt), hydroxybutyryl-CoA dehydrogenase (hbd) from Clostridium acetobutylicum, modified trans-enoyl-CoA-reductase (ter) from Treponema denticola, and acetyl-CoA acetyltransferase or thiolase (atoB) from E. coli were inserted into the genome of Synechococcus elongatus to synthesize 1-butanol (Lan and Liao 2011). Transgenic S. elongatus was used to synthesize isobutyraldehyde (Atsumi et al. 2009). Because of their rapid development, high productivity, resilience to genetic alterations, cyanobacteria are a viable source of biomass for the generation of biofuel (Sarsekeyeva et al. 2015).
Role of ‘Knallgas’ bacteria in biofuels production
Gram negative betaproteobacterium Ralstonia eutropha is considered Knallgas bacteria. It is well-known for its ability to produce polyhydroxyalkanoate (PHA) biopolymers from a number of carbon sources such as sugars, lipids, and CO2. Genetically engineered Ralstonia eutropha is used to synthesize n-butanol, isobutanol, and terpene molecules under chemolithoautotrophic conditions. Bio-based fuels are produced from CO2. Knallgas bacteria serve as a biocatalyst for the use of CO2 and are involved in the synthesis of high-density biofuels (Brigham 2019). Rhodobacter capsulatus is involved in hydrogen production (Silva et al. 2016) whereas Rhodococcus opacus, is involved in the synthesis of triacylglycerol (Kurosawa et al. 2015).
Role of thermophilic bacteria in biofuels production from lignocellulose
Clostridium thermocellum is used to produce ethanol. To improve the production of ethanol, Clostridium thermocellum has been used as one of the co-cultured microbes with other thermophilic bacteria such as Thermoanaerobacterium thermosaccharolyticum, C. thermohydrosulfuricum, Thermoanaerobacter ethanolicus, Geobacillus stearothermophilus, and Thermoanaerobacter brockii (Taylor et al. 2009). Thermoanaerobacterium saccharolyticum is used for the hydrolysis of cellulose to produce ethanol (Boonsayompoo and Reungsang 2013). Geobacillus is used to produce ethanol through the metabolism of pentose and hexose sugars (Raita et al. 2016). The presence of more carbon sources, thermostable genes, and less risk of contamination make thermophiles more suitable for the production of lignocellulosic biofuels Jiang et al. 2017)
Application of metagenomic approaches to identify novel enzymes for biofuels and biogas production.
Metagenomics has been used to find enzymes such as lignases, xylanases, endoglucanases, amylolytic enzymes, glucosidase for bioethanol, and lipolytic enzymes for biodiesel production (Wang et al. 2019).
Microorganisms involved in hydrolysis of lignocellulose
Lignocellulose which is made up of cellulose, hemicellulose, and lignin, is the world’s most abundant renewable biofuel resource. Plant cell walls include cellulose and hemicellulose which are digested to produce glucose and galactose, which serve as a carbon source for microbes engaged in biofuel synthesis (Adegboye et al. 2021). Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria are the most common cellulolytic bacterial phyla (de Gonzalo et al. 2016). Actinobacteria, Proteobacteria (classes: Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Gammaproteobacteria), Firmicutes, Bacteroides, and Archaea were found to degrade lignin (Bugg and Rahmanpour 2015). Trichoderma longibrachiatum, Aspergillus niger, and Ustilago maydis have hemicellulose digesting enzyme endo β-1,4 xylanase. Several strains such as T. terrestris, Neurospora crassa, Podospora anserine, Aspergillus nidulans, Myceliophthora thermophila, and Sporotrichum pulyverolentum have lytic polysaccharide monooxygenases (LPMOs) which are involved in the cleavage of chitin and cellulose. Streptomyces strains such as NWU339 and NWU49 which are identified from the rhizosphere of maize use starch, xylan, and cellulose as potential substrates for biofuel synthesis. Amylase, cellulases, pectinase, and xylanase of Streptomyces fulvissimus CKS7 showed the highest production of bioethanol (Adegboye et al. 2021). By employing plant biomass as substrate, Aspergillus, Trichoderma, and Bacillus sp. have been employed to synthesize enzymes (Sakhuja et al. 2021).
Metagenomic approaches
To isolate novel enzymes from the environmental microorganisms by using metagenomic approaches, different approaches are applied such as isolation of DNA from the environment, identification of vector for gene cloning, construction of a metagenomic library, and functional characterization of genes encoding novel enzymes from the metagenomic library (Fig. 1) (Wang et al. 2019). Highly purified and large molecular weight DNA is an important factor for the preparation of metagenomic libraries (Gunawardana et al. 2014). Bacterial artificial chromosomes (BACs), cosmids, fosmids, or yeast artificial chromosomes (YACs) are used for cloning large DNA fragments in metagenomic library preparations. Escherichia coli may be preferred for gene cloning but it may not be applicable for cloning of enzymes of microbes in extreme environments (Jung et al. 2012). Agrobacterium tumefaciens, Bacillus subtilis, Burkholderia graminis, Caulobacter vibrioides, Pseudomonas putida, Ralstonia metallidurans, Sulfolobus solfataricus, and Thermus thermophilus are considered as an alternative host for library preparation (Wang et al. 2019). T. thermophilus has been considered as a better host for metagenomic library preparation for the discovery of genes having esterases and xylanases as compared to E. coli (Leis et al. 2015).
Functional isolation of positive clones having β-glucosidases has been isolated from plates containing esculin hydrate. Functional screenings have lower efficiency and several limitations (Fang et al. 2010). Carboxymethyl cellulose, 4-methylumbelliferyl β-D-celloside (MUC), AZCL (Azurine-Crosslinked Polysaccharides)-HE-cellulose, and AZCL-β-glucan are used as a substrate for screening of metagenomic library of endoglucanase. esculin, arbutin, p-nitrophenylb-d-glucopyranoside, p-nitrophenyl-β-d-cellobioside, and 4-methylumbelliferyl β-d-glucoside (MUG) are used as substrates for screening of a metagenomic library of β-glucosidase (Tiwari et al. 2018).
PCR primers are designed for DNA sequencing based on DNA sequences of a gene of interest having enzymatic properties under extreme environmental conditions. Other DNA sequence based approaches such as microarray, fluorescence in situ hybridization (FISH), and RT-PCR are used to identify the clones of genes interests (Shang et al. 2018). Environmental metagenomic communities are identified using Next generation sequencing technologies such as Illumina metagenome sequencing, and Oxford Nanopore Technologies (ONT) based MinION (Wang et al. 2019). Metagenomics approaches are used to identify the diversity of microbial communities at different taxa and functional classification of microbial genes in an environment (Simon and Daniel 2011). 16S ribosomal RNA (V1–V9 hypervariable regions) is used for the identification of bacterial taxa; whereas, internal transcribed spacer (ITS), and 18S rRNA are used for fungal identification (Morgan and Huttenhower 2012).
Metagenome-derived enzymes for biofuel synthesis
Metagenomics studies are responsible for the isolation of lignocellulose-degrading enzymes. β-Glucosidase which is used in the transformation of cellulosic biomass into glucose has been isolated from different environmental conditions such as alkaline polluted soil, hot spring, wastewater, marine water, mangrove soil, and agricultural soil (Tiwari et al. 2018). The marine microbial metagenomic library was utilized for the identification of β-glucosidase (Bgl1A) (Fang et al. 2010). Shotgun metagenomic analysis revealed that Bacillus thermozeamaize, Caldibacillus debilis, and Geobacillus thermoglucosidasius have carboxymethylcellulose and glycosyl hydrolases (GHs) which are involved in the degradation of lignocellulose (Lemos et al. 2017). Microbes having lignocellulolytic enzymes have been isolated from corn stover, rice straw, sugarcane bagasse, and wheat straw by using functional metagenomic approaches (Rattanachomsri et al. 2011).
Enzymes played a crucial role in industrial biofuel production. Metagenomics approaches are used for screening the biomass-degrading microbial novel enzymes such as carboxyl- hydrolases (esterases, lipases), polysaccharide-modifying enzymes (α-amylases, cellulases, xylanases,1,4-α-glucan branching enzymes), oxidoreductases, dehydrogenases, and oxygenases from environmental samples. These novel enzymes are capable of producing biofuels under a wide range of pH, temperature, or ionic conditions (Xing et al. 2012).
Metagenomic approaches are used to isolate the enzymes such as lignin peroxidase, manganese peroxidase, xylanase, and versatile peroxidase from environmental DNA libraries (Fang et al. 2011). Xylanase of soil derived metagenomic library showed enhanced activity in lower temperatures and reduced alkaline environment (Hu et al. 2008). The alkaline xylanase from a metagenomic library of microbiome derived fungus-growing termites showed stability over a wide range of pH. (Liu et al. 2011). Cellulose which is used for the production of biofuel is usually degraded by enzymes such as endoglucanases, exoglucanases, and β-glucosidases. Cellulases are used in the production of bioethanol (Ilmberger and Streit 2010). Cellulases such as 2β-Glucosidase, 3 Endoglucanase, 5 endoglucanases, 2 β-glucosidases, and 1 β-Cellobiohydrolase are obtained from metagenomic libraries from the guts of earthworms (Beloqui et al. 2010; Wang et al. 2009). Endoglucanase is derived from the metagenome of the compost soils (Pang et al. 2009). Exo-glucanase which is used in the degradation of xyloglucan and oligoxyloglucan is obtained from ruminal microbial metagenomes (Wong et al. 2010). The enzyme glycosyl hydrolase was obtained from the metagenomic library of cow rumen (Palackal et al. 2007). Thermophilic bacteria such as Caldicellulosiruptor danielii, Caldicellulosiruptor morganii, and Caldicellulosiruptor naganoensis NA10 use lignocellulose as a substrate (Lee et al. 2018). Xylose isomerase, xylulokinase, xylose transporter, ribulose-phosphate 3-epimerase genes in Herbivorax saccincola A7 are involved in xylose metabolism (Aikawa et al. 2018). Clostridium clariflavum possesses polysaccharide-degrading enzymes. Without pre-treatments, Caldicellulosiruptorbescii demonstrated disintegration of plant biomass (Brunecky et al. 2018). Algoriphagus ratkowskyi, Flavobacterium beibuense, Halomonas meridian, Joostella marina, and Pseudomonas putida showed degradation of lignocellulose in a saline environment (Cortes-Tolalpa et al. 2018).
Microbial communities in biogas production
Bacteria are involved in several phases of biogas formation including hydrolysis, acidogenesis, and acetogenesis, whereas archaea are involved in methanogenesis (Fig. 2). Clostridium belong to Firmicutes is involved in the hydrolysis of biopolymers such as cellulose, starch, proteins, and lipids into oligo- and monomers which are converted into volatile fatty acids such as acetate, propionate, and butyrate by acidogenic bacteria of phyla Firmicutes, Chloroflexi, Bacteroidetes, and Proteobacteria. During acetogenesis, volatile fatty acids are converted into acetate, CO2, and H2 by Syntrophomonas, Syntrophobacter, Pelotomaculum, and Thermoanaerobacter. Methanomicrobiales, Methanosarcinales, and Methanobacteriales which belong to phylum Euryarchaeota under Archaea are involved in hydrogenotrophic methanogenesis (CO2 and H2), acetoclastic pathway, and methylotrophic pathway. Thermophilic BGPs are predominated by Methanothermobacter and Methanosarcina as well as Thermotogae and Synergistetes. Mesophilic BGPs are predominated by class Clostridia, Bacteroidetes, Bacilli, Spirochaetes of the bacterial community as well as Methanomicrobiales, Methanosarcinales, and Methanobacteriales of an archaeal community (Jünemann et al.2017) (Table 2).
Table 2.
List of predominant microbial community in biogas plants
| Type of biogas plant | Predominant microbial community | References |
|---|---|---|
| Mesophilic | Acidobacteria, Firmicutes, Proteobacteria, Spirochaetes, Tenericutes, Verrucomicrobia, Chloroflexi | Stolze et al. (2016) |
| Bacteroidetes, Porphyromonadaceae, Marinilabiaceae | Moset et al. (2015) | |
| Clostridia, Bacteroidetes, Bacilli, Spirochaetes, (Bacteria) Methanomicrobiales, Methanosarcinales, Methanobacteriales (Archaea) | Jünemann et al. (2017) | |
| Thermophilic | Methanothermobacter, Methanosarcina, Thermotogae, Synergistetes | Jünemann et al.2017 |
| Firmicutes, Thermotogae, Bacteroidetes, Clostridium thermocellum, | Akinosho et al. (2014) | |
| Lachnospiraceae, Halanaerobiaceae | Stolze et al. (2016) | |
| Defluviitoga, Clostridium cluster III, Tepidanaerobacter | Maus et al. (2016a, b) | |
| Agricultural | Methanoculleus, Methanosarcina, Methanobrevibacter, Methanomicrobiales spp. Methanobacteriales spp. Methanosarcina sp. | Ziganshin et al. (2013) |
Omics approaches to understand the composition and functional relationship of bacteria and archaea in biogas plant
Bacteria and archaea are the predominant microbial communities in the biogas plant (Table 2). In biogas plants (BGP), the richness and abundance of the microbial population are influenced by temperature, fed substrates, pH, and reactor and fermentation pattern (Abendroth et al. 2015; Yu et al. 2014). The composition of microbial communities in biogas plants is determined by 16S rRNA gene amplicon sequencing. This BGP is operated under mesophilic (35–45 °C) or thermophilic (45–60 °C) conditions by using sewage sludge, manure, and agricultural wastage (Maus et al. 2017).
Composition of bacterial communities in mesophilic BGP
Firmicutes are the predominant phyla in BGP. Among phylum Firmicutes, the classes Clostridia and Bacilli are highly abundant in the mesophilic condition of BGP. Phyla such as Acidobacteria, Proteobacteria, Spirochaetes, Tenericutes, Verrucomicrobia, and Chloroflexi are also reported in BGP which is operated under mesophilic condition (Stolze et al. 2016). Degradation of complex carbohydrates such as amylose, amylopectin, cellulose, and xylan is carried out by mainly bacterial species belonging to the genus Clostridium. Acetoanaerobium sticklandii and Butyrivibrio proteoclasticus belong to Clostridia are also involved in the degradation of proteins (Hassa et al. 2018). Bacteria belong to Thermoanaerobacteriaceae, Costridiaceae, and Syntrophomonadaceae families are considered as a syntroph which are involved in the breakdown of syntrophic fatty acid (Schnürer 2016). Bacterial genera such as Syntrophus, Pelobacter, Smithella, Syntrophorhabdus, and Syntrophobacter belong to phylum Proteobacteria are associated with methanogenic Archaea (Qiu et al. 2008). Bacteroidetes, Porphyromonadaceae, and Marinilabiaceae are predominant in mesophilic BGP. Bacteroidetes are involved in the production of acetate and propionate from sugars (Moset et al. 2015).
Composition of bacterial communities in thermophilic BGP
Bacterial phyla such as Firmicutes, Thermotogae, and Bacteroidetes are predominant in thermophilic BGP. Bacteria belong to class Clostridia showed higher abundance in thermophilic BGPs as compared to mesophilic BGP. Clostridium thermocellum is considered a key cellulose degrader of thermophilic BGPs (Akinosho et al. 2014). In thermophilic conditions of BGP, Lachnospiraceae and Halanaerobiaceae showed higher abundance (Stolze et al. 2016). The species belong to Lachnospiraceae family are involved in the degradation of complex plant content. Herbinix hemicellulosilytica T3/55T and Halocella cellulolytica which are isolated thermophilic BGP are involved in the degradation of cellulose. They engage in the manufacture of end products such as acetate, ethanol, lactate, H2, and CO2 (Koeck et al. 2015). In thermophilic biogas plants, bacterial genera such as Defluviitoga, Clostridium cluster III, and Tepidanaerobacter were found to be more abundant (Maus et al. 2016a, b).
Composition of bacterial and archaeal communities in agricultural BGP
Methanoculleus, Methanosarcina, and Methanobrevibacter showed higher abundance in agricultural BGP. They can grow in both mesophilic and thermophilic conditions. Hydrogenotrophic methanogenic Archaea such as Methanomicrobiales spp. and Methanobacteriales spp. can survive in toxic ammonia containing the environment. Methanosarcina sp. are able to survive in high ammonia or salt concentrations (Ziganshin et al. 2013).
Bacterial species in biogas reactors are involved in the digestion of carbohydrates and energy conversion. Bacterial genes involved in cellulose degradation such as cellobiose phosphorylase, glucosidase, and cellulase/cellobiase are predominant in agricultural BGPs (Maus et al. 2016b). Bacterial genes involved in the metabolism of nitrogen, phosphorus, and aromatic compound were found in higher abundances in wastewater sludge with higher amounts of aromatic compounds, nitrite, organic contaminants, and phosphate (Luo et al. 2016). Bacterial genes involved in acetoclastic methanogenesis were more abundant in biogas reactors fed sewage sludge, whereas, bacterial genes involved in the hydrogenotrophic pathway were overexpressed in biogas reactors given agricultural leftovers and manure (Stolze et al. 2015). In the acetoclastic pathway, the predominant genes are acetyl-CoA synthetase and acetyl-CoA decarbonylase/synthase complex, whereas, formate dehydrogenase and formylmethanofuran dehydrogenase are highly abundant in the hydrogenotrophic pathway (Luo et al. 2016).
In agricultural BGP fed with cow and chicken manure and maize silage, methyl-coenzyme-M-reductase of hydrogenotrophic archaeal species showed higher abundance (Güllert et al. 2016). Whole metatranscriptome sequencing analyses of BGP digesting maize silage, barley, and cattle manure were revealed that taxa Anaerobaculum (Synergistetes), Clostridium cluster III (Firmicutes), Cellulosibacter (Firmicutes), Defluviitoga (Thermotogae), Methanoculleus (Euryarchaeota), and Tepidanaerobacter (Firmicutes) showed transcriptional activities (Maus et al. 2016b). In agricultural BGPs, Methanobacterium, Methanosaeta, and Methanoculleus showed higher expression of methanogenesis enzymes. Firmicutes are involved in the degradation of cellulose, whereas, Bacteroidetes are mostly involved in the digestion of polysaccharides. Firmicutes are involved in the synthesis of enzymes such as xylanases, xylosidases, and cellulases (Hassa et al. 2018).
Application of genome editing of microbial cells mediated by CRISPR/Cas9-in biofuel synthesis
Several genome editing technologies such as Zinc Finger Nucleases (ZFNs), Transcription Activator-Like (TAL) Effector Nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR) system are available in the advanced field of genetic engineering. There are various drawbacks to ZFNs and TALENs, including the lack of an effective cloning vector for delivery mechanisms, off targeted effects, cytotoxicity, and low efficiency for multiple-gene targeting. To overcome the drawbacks of ZFNs and TALENs, the CRISPR/Cas9 system has evolved (Javed et al. 2019; Shanmugam et al. 2019).
An overview of CRISPR/Cas9 system
Several bacterial and archaeal strains have the CRISPR-Cas system (Grissa et al. 2007). CRISPR-Cas genome editing tools consist of Cas protein (Cas9, Cpf1/Cas12a) having enzymatically active nuclease domains, and single guide RNA (sgRNA) which is the chimera of endogenous bacterial CRISPR (crRNA) and trans-activating crRNA (tracrRNA). Cas protein induces double-strand break (DSB) in DNA strands which are located upstream of the protospacer adjacent motif (PAM). This DSB has been repaired by non-homologous end joining (NHEJ), and the homology directed repair (HDR) pathway (Jiang et al. 2013). The CRISPR consists of tiny repeated DNA sequences flanked by small spacer DNA segments from bacteriophage or plasmid. CRISPR associated genes such as Cas9 have been involved in the unwinding of DNA double helix through its nuclease or helicase activity (Rodriguez 2016). Cas9 was originally obtained from the bacterium Streptococcus pyogenes. Using Cas9 nuclease, single guide RNA (sgRNA) along with crRNAs and trans-activating RNA (tracrRNA) are involved in breaking unique DNA sequences (Louwen et al.2014). For the improved production of biofuels, CRISPR/Cas9-mediated site directed mutagenesis in the genome of microbial cells is used (Ulaganathan et al. 2017) (Table 3). Genome editing enhances the microorganisms to survive in a highly toxic environment such as fermentation process inhibitors and specific growth inhibitors such as phenolic compounds and furan derivatives (Tkalec et al. 2014).
Table 3.
CRISPR/Cas9-based genome editing in microbial strains for biofuel production
| Microbial strains | CRISPR-Cas9 machinery | Target genes | End products | References |
|---|---|---|---|---|
| Clostridium saccharoperbutylacetonicum | Mutation created by using (ORF) of Cas9 from Streptococcus pyogenes under the influence of lactose inducible promoter (bgaL) and transcribed sgRNA from small RNA promoter of Clostridium beijerinckii | Phosphotransacetylase (pta) and butyrate kinase (buk) genes | Butanol | Wang et al. (2016, 2017a, b) |
| Escherichia coli EMJ50 strain) | Over expression | Acetoacetyl-CoA thiolase (thl), and alcohol dehydrogenase (adhE2) genes of Clostridium acetobutylicum, and formate dehydrogenase (fdh1) gene of Candida boidinii | Butanol | Shanmugam et al. (2019) |
| Clostridium acetobutylicum DSM792 and Clostridium pasteurianum ATCC6013 | SpCRISPR-dCas9 used to suppress carbon catabolite repression | Kinase/phosphorylase (hprK) gene | Butanol | Bruder et al. (2016) |
| Zymomonas mobilis | E145G, G14C, K219E, L18 P, L361 F, L606, P195 T, P511T, V355 M,V522 G, Q649 L | RpoD protein | Ethanol | Ulaganathan et al. (2017) |
| Saccharomyces cerevisiae | Single amino acid alteration in ADH3 (G416 T, T966 G, T201 A), ASG1 (G1248 A, G1248 T, A1979 G, G2881C), GIS4 (G1322C, G295 A), and SKS1 (G821 T, C617 A) | ADH3 and SKS1 | Ethanol | González-Ramos et al. (2016) |
| Escherichia coli strain BW25113 | CRISPRi system with l-rhamnose inducible dCas9 expression cassette and sgRNA block the synthesis of acetate, succinate, lactate, and ethanol | Inhibit the expression of pta, frdA, ldhA, and adhE genes | n-butanol | Kim et al. (2017) |
Genome editing of target genes in microorganisms for biofuels production
CRISPR-Cas9 genome editing has been utilized in C. saccharoperbutylacetonicum N1-4 strain for the production of butanol (Wang et al. 2017a, b). The genome editing technology has been applied in phosphotransacetylase (pta) and butyrate kinase (buk) genes of C. saccharoperbutylacetonicum for the production of acetate and butyrate. The mutation in the above mentioned genes was created by using an open reading frame (ORF) of Cas9 from S. pyogenes under the influence of lactose inducible promoter (bgaL) and transcribed sgRNA from a small RNA promoter of C. beijerinckii (Wang et al. 2016). Overexpression of acetoacetyl-CoA thiolase (thl), and alcohol dehydrogenase (adhE2) genes of C. acetobutylicum, and formate dehydrogenase (fdh1) gene of Candida boidinii in E. coli EMJ50 strain induce biobutanol synthesis (Shanmugam et al. 2019). SpCRISPR-dCas9 is used to synthesize the biobutanol from lignocellulose through carbon catabolite repression (CCR) of C. acetobutylicum DSM792 and C. pasteurianum ATCC6013 via suppression of kinase/phosphorylase (hprK) gene (Bruder et al. 2016).
A frameshift mutation at codon 41 in the NADH dehydrogenase enzyme of Z. mobilis results in enhanced ethanol production which favored growth in aerobic conditions (Ulaganathan et al. 2017). Point mutations in the alcohol dehydrogenase enzyme of C. thermocellum, are used for the enhancement of ethanol tolerance (Brown et al. 2011). Single amino acid alteration in ADH3 (G416 T, T966 G, T201 A), ASG1 (G1248 A, G1248 T, A1979 G, G2881C), GIS4 (G1322C, G295 A), and SKS1 (G821 T, C617 A) enhanced tolerance of S. cerevisiae in an acetic acid environment (González-Ramos et al. 2016). The enhanced production of bioethanol by S. cerevisiae depends on its tolerance towards inhibitors. Single amino acid alterations (E145G, G14C, K219E, L18 P, L361 F, L606, P195 T, P511T, V355 M,V522 G, Q649 L) in RpoD protein of Z. mobilis enhanced its furfural tolerance (Ulaganathan et al. 2017). The production of bioethanol generates high temperature that inhibits the development of ethanol-producing microorganisms. Enhanced thermotolerance and increased production of ethanol by Z. mobilis and S. cerevisiae may be due to its single amino acid alterations in the pyruvate kinase and NADH dehydrogenase genes (Caspeta et al. 2014). S. cerevisiae developed thermostability due to the deletion of Dfg5 glycosyl phosphatidylinositol-anchored membrane protein (Nasution et al. 2015). Replacement of amino acids of cellulases in Bacillus sp. strain KSM-64 (N179K and D194K), C. cellulovorans (E116D and V192 A), C. phytofermentans (E158 V, N144I, N291 K, andV245 G), C. thermocellum (S329 G), Humicola insolens (C313S), and Melanocarpus albomyces (S290 T, G4C/M70C/S290 T) enhanced their heat tolerance (Chokhawala et al. 2015). Hemicellulases such as Endo-1, 4-β-xylanase is involved in the production of ethanol through a breakdown of β-1, 4-xylosidic bond in xylan processing xylo-oligosaccharides (Dumon et al. 2012). Thermotolerance of Aspergillus usamii A. niger, B. subtilis, and Yarrowia lipolytica is enhanced through substitutions of amino acid in xylanases. Xylanases activity of Thermomyces lanuginosus and Geobacillus stearothermophilus enhanced through amino acid alterations (Ulaganathan et al. 2017).
Multiplex automated genome engineering (MAGE) method has been applied to create different mutations in a specific gene without alterations of other genes (Bao et al. 2016). Combinatorial approaches of CRISPR Enabled Trackable genome Engineering (CREATE) along with MAGE are used to synthesize isopropanol in E. coli (Liang et al. 2017). CRISPRi system which consists of L-rhamnose inducible dCas9 expression cassette and sgRNA has been applied for the production of n-butanol in E. coli strain BW25113 through blocking the synthesis of acetate, succinate, lactate, and ethanol by supressing the expression of endogenous genes such as pta, frdA, ldhA, and adhE (Kim et al. 2017) (Table 3).
Conclusion
Biofuels are considered a renewable energy source in the world due to the decline of fossil fuels. Microbial enzymes are utilized in the production of bioethanol, biofuels, and biodiesel from biomass. For the production of biofuels, metagenomic technology is employed to identify the novel enzymes from environmental sources. Advanced “omics “ approaches such as metagenomics, metatranscriptomics, metaproteomics, and metabolomics are used to identify the microbial communities in biogas. Each technique has its own set of constraints when it comes to producing biofuels and biogas. A new investigation is required to strengthen the production of lignocellulosic-based biofuels with the least use of energy. Microorganisms that have been genetically modified using the CRISPR/Cas9 genome editing technique could be utilized to produce large amounts of biofuels. Synthetic biology and metabolic engineering approaches in Cyanobacteria, Clostridia, Escherichia coli, S. cerevisiae, and Zymomonas mobilis are used for the synthesis of biofuels. Genetic and metabolic engineering advancements in microbial cells contributed greatly to the overproduction of biofuels for long-term energy conversion sustainability.
Declarations
Conflict of interest
None.
Ethical statement
This manuscript does not include any human participants or animals studies.
Contributor Information
J. Rajesh Banu, Email: rajeshces@gmail.com.
Gopalakrishnan Kumar, Email: gopalakrishnanchml@gmail.com.
Indranil Chattopadhyay, Email: indranil_ch@yahoo.com.
References
- Abendroth C, Vilanova C, Günther T, Luschnig O, Porcar M. Eubacteria and archaea communities in seven mesophile anaerobic digester plants in Germany. Biotechnol Biofuels. 2015;8:87. doi: 10.1186/s13068-015-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adegboye MF, Ojuederie OB, Talia PM, Babalola OO. Bioprospecting of microbial strains for biofuel production: metabolic engineering, applications, and challenges. Biotechnol Biofuels. 2021;14:5. doi: 10.1186/s13068-020-01853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa S, Baramee S, Sermsathanaswadi J, et al. Characterization and high-quality draft genome sequence of Herbivorax saccincola A7, an anaerobic, alkaliphilic, thermophilic, cellulolytic, and xylanolytic bacterium. Syst Appl Microbiol. 2018;41:261–269. doi: 10.1016/j.syapm.2018.01.010. [DOI] [PubMed] [Google Scholar]
- Akinosho H, Yee K, Close D, Ragauskas A. The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications. Front Chem. 2014;2:66. doi: 10.3389/fchem.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr SA, Hellingwerf KJ, Lindblad P, de Mattos MJ. Energy biotechnology with cyanobacteria. Curr Opin Biotechnol. 2009;20:257–263. doi: 10.1016/j.copbio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJY, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- Awe OW, Zhao Y, Nzihou A, Minh DP, Lyczko N. A review of biogas utilization, purification and upgrading technologies. Waste Biomass Valor. 2017;8:267–283. doi: 10.1007/s12649-016-9826-4. [DOI] [Google Scholar]
- Azad AK, Rasul M, Khan MMK, Sharma SC. Review of biodiesel production from microalgae: a novel source of green energy. Int Green Energy Conf. 2014 doi: 10.13140/2.1.3013.0244. [DOI] [Google Scholar]
- Balat M. Production of bioethanol from lignocellulosic materials via the biochemical pathway: a review. Energy Convers Manag. 2011;52:858–875. doi: 10.1016/j.enconman.2010.08.013. [DOI] [Google Scholar]
- Bao Z, Cobb RE, Zhao H. Accelerated genome engineering through multiplexing. Wiley Interdiscip Rev Syst Biol Med. 2016;8:5–21. doi: 10.1002/wsbm.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloqui A, Nechitaylo TY, Lopez-Cortes N, Ghazi A, Guazzaroni ME, Polaina J, et al. Diversity of glycosyl hydrolases from cellulose-depleting communities enriched from casts of two earthworm species. Appl Environ Microbiol. 2010;76:5934–5946. doi: 10.1128/AEM.00902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilal T, Malik B, Hakeem KR. Metagenomic analysis of uncultured microorganisms and their enzymatic attributes. J Microbiol Methods. 2018;155:65–69. doi: 10.1016/j.mimet.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Boonsayompoo O, Reungsang A. Thermophilic biohydrogen production from the enzymatic hydrolysate of cellulose fraction of sweet sorghum bagasse by Thermoanaerobacterium thermosaccharolyticum KKU19: optimization of media composition. Int J Hydrogen Energy. 2013;38:15777–15786. doi: 10.1016/j.ijhydene.2013.04.129. [DOI] [Google Scholar]
- Brigham C. Perspectives for the biotechnological production of biofuels from CO2 and H2 using Ralstonia eutropha and other 'Knallgas' bacteria. Appl Microbiol Biotechnol. 2019;103:2113–2120. doi: 10.1007/s00253-019-09636-y. [DOI] [PubMed] [Google Scholar]
- Brown SD, Guss AM, Karpinets TV, Parks JM, et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci. 2011;108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder MR, Pyne ME, Moo-Young M, Chung DA, Chou CP. Extending CRISPR-Cas9 technology from genome editing to transcriptional engineering in the genus Clostridium. Appl Environ Microbiol. 2016;82:6109–6119. doi: 10.1128/AEM.02128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunecky R, Chung D, Sarai NS, et al. High activity CAZyme cassette for improving biomass degradation in thermophiles. Biotechnol Biofuels. 2018;11:22. doi: 10.1186/s13068-018-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TD, Rahmanpour R. Enzymatic conversion of lignin into renewable chemicals. Curr Opin Chem Biol. 2015;29:10–17. doi: 10.1016/j.cbpa.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Cai D, Chen H, Chen C, et al. Gas stripping–pervaporation hybrid process for energy-saving product recovery from acetone–butanol–ethanol (ABE) fermentation broth. Chem Eng J. 2016;287:1–10. doi: 10.1016/j.cej.2015.11.024. [DOI] [Google Scholar]
- Caspeta L, Chen Y, Ghiaci P, et al. Altered sterol composition renders yeast thermotolerant. Science. 2014;346:75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- Cheon S, Kim HM, Gustavsson M, Lee SY. Recent trends in metabolic engineering of microorganisms for the production of advanced biofuels. Curr Opin Chem Biol. 2016;35:10–21. doi: 10.1016/j.cbpa.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Chokhawala HA, Roche CM, Kim T-W, et al. Mutagenesis of Trichoderma reesei endoglucanase I: impact of expression host on activity and stability at elevated temperatures. BMC Biotechnol. 2015 doi: 10.1186/s12896-015-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin VL, Rodríguez A, Cristóbal HA. The role of synthetic biology in the design of microbial cell factories for biofuel production. J Biomed Biotechnol. 2011;2011:601834. doi: 10.1155/2011/601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Tolalpa L, Norder J, van Elsas JD, Falcao Salles J. Halotolerant microbial consortia able to degrade highly recalcitrant plant biomass substrate. Appl Microbiol Biotechnol. 2018;102:2913–2927. doi: 10.1007/s00253-017-8714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves MA, Kimura T, Shimizu N, Nakajima M. State of the art and future trends of bioethanol production. Dyn Biochem Process Biotechnol Mol Biol. 2007;1:1–14. [Google Scholar]
- de Gonzalo G, Colpa DI, Habib MH, Fraaije MW. Bacterial enzymes involved in lignin degradation. J Biotechnol. 2016;236:110–119. doi: 10.1016/j.jbiotec.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Dellomonaco C, Fava F, Gonzalez R. The path to next generation biofuels: successes and challenges in the era of synthetic biology. Microb Cell Fact. 2010;9:3. doi: 10.1186/1475-2859-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi A, Pascoe S, Coglan L, Rainey TJ. Economic and policy issues in the production of algae-based biofuels: a review. Renew Sustain Energy Rev. 2016;64:329–337. doi: 10.1016/j.rser.2016.06.027. [DOI] [Google Scholar]
- Dumon C, Song L, Bozonnet S, Fauré R, O’Donohue MJ. Progress and future prospects for pentose-specific biocatalysts in biorefining. Process Biochem. 2012;47:346–357. doi: 10.1016/j.procbio.2011.06.017. [DOI] [Google Scholar]
- Dutta K, Daverey A, Lin JG. Evolution retrospective for alternative fuels: first to fourth generation. Renew Energy. 2014;69:114–122. doi: 10.1016/j.renene.2014.02.044. [DOI] [Google Scholar]
- Fang Z, Fang W, Liu J, et al. Cloning and characterization of a beta-glucosidase from marine microbial metagenome with excellent glucose tolerance. J Microbiol Biotechnol. 2010;20:1351–1358. doi: 10.4014/jmb.1003.03011. [DOI] [PubMed] [Google Scholar]
- Fang Z, Li T, Wang Q, Zhang X, Peng H, Fang W, et al. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl Microbiol Biotechnol. 2011;89:1103–1110. doi: 10.1007/s00253-010-2934-3. [DOI] [PubMed] [Google Scholar]
- Georgianna DR, Mayfield SP. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature. 2012;488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- González-Ramos D, de Vries ARG, Grijseels SS, et al. A new laboratory evolution approach to select for constitutive acetic acid tolerance in Saccharomyces cerevisiae and identification of causal mutations. Biotechnol Biofuels. 2016;9:173. doi: 10.1186/s13068-016-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güllert S, Fischer MA, Turaev D, et al. Deep metagenome and metatranscriptome analyses of microbial communities affiliated with an industrial biogas fermenter, a cow rumen, and elephant feces reveal major differences in carbohydrate hydrolysis strategies. Biotechnol Biofuels. 2016;9:121. doi: 10.1186/s13068-016-0534-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardana M, Chang S, Jimenez A, et al. Isolation of PCR quality microbial community DNA from heavily contaminated environments. J Microbiol Methods. 2014;102:1–7. doi: 10.1016/j.mimet.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Guo D, Zhu J, Deng Z, Liu T. Metabolic engineering of Escherichia coli for production of fatty acid short-chain esters through combination of the fatty acid and 2-keto acid pathways. Metab Eng. 2014;22:69–75. doi: 10.1016/j.ymben.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Hahnke S, Langer T, Koeck DE, Klocke M. Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosasp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int J Syst Evol Microbiol. 2016;66:1466–1475. doi: 10.1099/ijsem.0.000902. [DOI] [PubMed] [Google Scholar]
- Hassa J, Maus I, Off S, et al. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl Microbiol Biotechnol. 2018;102:5045–5063. doi: 10.1007/s00253-018-8976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlík P, Schneider UA, Schmid E, et al. Global land-use implications of first and second generation biofuel targets. Energy Policy. 2011;39:5690–5702. doi: 10.1016/j.enpol.2010.03.030. [DOI] [Google Scholar]
- Hu Y, Zhang G, Li A, Chen J, Ma L. Cloning and enzymatic characterization of a xylanase gene from a soil-derived metagenomic library with an efficient approach. Appl Microbiol Biotechnol. 2008;80:823–830. doi: 10.1007/s00253-008-1636-6. [DOI] [PubMed] [Google Scholar]
- Ilmberger N, Streit WR. Screening for cellulase-encoding clones in metagenomic libraries. Methods Mol Biol. 2010;668:177–188. doi: 10.1007/978-1-4939-6691-2_12. [DOI] [PubMed] [Google Scholar]
- Javed MR, Noman M, Shahid M, et al. Current situation of biofuel production and its enhancement by CRISPR/Cas9-mediated genome engineering of microbial cells. Microbiol Res. 2019;219:1–11. doi: 10.1016/j.micres.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Xin F, Lu J, Dong W, et al. State of the art review of biofuels production from lignocellulose by thermophilic bacteria. Bioresour Technol. 2017;245(Pt B):1498–1506. doi: 10.1016/j.biortech.2017.05.142. [DOI] [PubMed] [Google Scholar]
- Joseph RC, Kim NM, Sandoval NR. Recent Developments of the Synthetic Biology Toolkit for Clostridium. Front Microbiol. 2018;9:154. doi: 10.3389/fmicb.2018.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jünemann S, Kleinbölting N, Jaenicke S, Henke C, et al. Bioinformatics for NGS-based metagenomics and the application to biogas research. J Biotechnol. 2017;261:10–23. doi: 10.1016/j.jbiotec.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Jung SK, Parisutham V, Jeong SH, Lee SK. Heterologous expression of plant cell wall degrading enzymes for effective production of cellulosic biofuels. J Biomed Biotechnol. 2012;2012:405842. doi: 10.1155/2012/405842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanafusa-Shinkai S, Wakayama J, Tsukamoto K, et al. Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulose/hemicellulose system from the extreme thermophilic bacterium Caldicellulosiruptor bescii. J Biosci Bioeng. 2013;115:64–70. doi: 10.1016/j.jbiosc.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Kang Q, Appels L, Tan T, Dewil R. Bioethanol from lignocellulosic biomass: current findings determine research priorities. Sci World J. 2014;2014:298153. doi: 10.1155/2014/298153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi K, Tabatabaei M, Horváth IS, Kumar R. Recent trends in acetone, butanol, and ethanol (ABE) production. Biofuel Res J. 2015;2:301–308. doi: 10.18331/BRJ2015.2.4.4. [DOI] [Google Scholar]
- Kim SK, Seong W, Han GH, Lee DH, Lee SG. CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Fact. 2017;16:188. doi: 10.1186/s12934-017-0802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck DE, Ludwig W, Wanner G, et al. Herbinix hemicellulosilytica gen. nov., sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reactor. Int J Syst Evol Microbiol. 2015;65:2365–2371. doi: 10.1099/ijs.0.000264. [DOI] [PubMed] [Google Scholar]
- Kurosawa K, Laser J, Sinskey AJ. Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels. 2015;8:76. doi: 10.1186/s13068-015-0258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng. 2011;13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Lee LL, Blumer-Schuette SE, Izquierdo JA, et al. Genus-wide assessment of lignocellulose utilization in the extremely thermophilic genus Caldicellulosiruptor by genomic, pangenomic, and metagenomic analyses. Appl Environ Microbiol. 2018;84:e02694–e2717. doi: 10.1128/AEM.02694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis B, Angelov A, Mientus M, et al. Identification of novel esterase-active enzymes from hot environments by use of the host bacterium Thermus thermophilus. Front Microbiol. 2015;6:275. doi: 10.3389/fmicb.2015.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos LN, Pereira RV, Quaggio RB, et al. Genome-centric analysis of a thermophilic and cellulolytic bacterial consortium derived from composting. Front Microbiol. 2017;8:644. doi: 10.3389/fmicb.2017.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Liu R, Garst AD, et al. CRISPR Enabled Trackable genome Engineering for isopropanol production in Escherichia coli. Metab Eng. 2017;41:1–10. doi: 10.1016/j.ymben.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Liu N, Yan X, Zhang M, et al. Microbiome of fungus-growing termites: a new reservoir for lignocellulase genes. Appl Environ Microbiol. 2011;77:48–56. doi: 10.1128/AEM.01521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwen R, Staals RH, Endtz HP, et al. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev. 2014;78:74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Yu L, Varghese S, Yu M, Yang ST. Enhanced robustness in acetone-butanol-ethanol fermentation with engineered Clostridium beijerinckii overexpressing adhE2 and ctfAB. Bioresour Technol. 2017;243:1000–1008. doi: 10.1016/j.biortech.2017.07.043. [DOI] [PubMed] [Google Scholar]
- Luo G, Fotidis IA, Angelidaki I. Comparative analysis of taxonomic, functional, and metabolic patterns of microbiomes from 14 full-scale biogas reactors by metagenomic sequencing and radioisotopic analysis. Biotechnol Biofuels. 2016;9:51. doi: 10.1186/s13068-016-0465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus I, Koeck DE, Cibis KG, et al. Unravelling the microbiome of a thermophilic biogas plant by metagenome and metatranscriptome analysis complemented by characterization of bacterial and archaeal isolates. Biotechnol Biofuels. 2016;9:171. doi: 10.1186/s13068-016-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus I, Cibis KG, Bremges A, et al. Genomic characterization of Defluviitoga tunisiensis L3, a key hydrolytic bacterium in a thermophilic biogas plant and its abundance as determined by metagenome fragment recruitment. J Biotechnol. 2016;232:50–60. doi: 10.1016/j.jbiotec.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Maus I, Kim YS, Wibberg D, et al. Biphasic study to characterize agricultural biogas plants by high-throughput 16S rRNA gene amplicon sequencing and microscopic analysis. J Microbiol Biotechnol. 2017;27:321–334. doi: 10.4014/jmb.1605.05083. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol. 2012 doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moset V, PoulsenM WR, et al. Mesophilic versus thermophilic anaerobic digestion of cattle manure. Methane productivity and microbial ecology. Microb Biotechnol. 2015;8:787–800. doi: 10.1111/1751-7915.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A. Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 2015;23:498–508. doi: 10.1016/j.tim.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Nasution O, Lee J, Srinivasa K, et al. Loss of Dfg5 glycosylphosphatidylinositol-anchored membrane protein confers enhanced heat tolerance in Saccharomyces cerevisiae. Environ Microbiol. 2015;17:2721–2734. doi: 10.1111/1462-2920.12649. [DOI] [PubMed] [Google Scholar]
- Nigam PS, Singh A. Production of liquid biofuels from renewable resources. Prog Energy Combust Sci. 2011;37:52–68. doi: 10.1016/j.pecs.2010.01.003. [DOI] [Google Scholar]
- Pabbathi NPP, Velidandi A, Tavarna T, et al. Role of metagenomics in prospecting novel endoglucanases, accentuating functional metagenomics approach in second-generation biofuel production: a review. Biomass Convers Biorefin. 2021;7:1–28. doi: 10.1007/s13399-020-01186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palackal N, Lyon CS, Zaidi S, et al. A multifunctional hybrid glycosyl hydrolase discovered in an uncultured microbial consortium from ruminant gut. Appl Microbiol Biotechnol. 2007;74:113–124. doi: 10.1007/s00253-006-0645-6. [DOI] [PubMed] [Google Scholar]
- Pang H, Zhang P, Duan CJ, Mo XC, Tang JL, Feng JX. Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase. Curr Microbiol. 2009;58:404–408. doi: 10.1007/s00284-008-9346-y. [DOI] [PubMed] [Google Scholar]
- Přibyl P, Cepák V, Zachleder V. Oil overproduction by means of microalgae. In: Bajpai R, Prokop A, Zappi M, editors. Cultivation of cells and products. Dordrecht: Springer; 2014. pp. 241–273. [Google Scholar]
- Prasad RK, Chatterjee S, Mazumder PB, et al. Bioethanol production from waste lignocelluloses: a review on microbial degradation potential. Chemosphere. 2019;231:588–606. doi: 10.1016/j.chemosphere.2019.05.142. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Hanada S, Ohashi A, Harada H, Kamagata Y, Sekiguchi Y. Syntrophorhabdus aromaticivorans gen. Nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl Environ Microbiol. 2008;74:2051–2058. doi: 10.1128/AEM.02378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raita M, Ibenegbu C, Champreda V, Leak DJ. Production of ethanol by thermophilic oligosaccharide utilising Geobacillus thermoglucosidasius TM242 using palm kernel cake as a renewable feedstock. Biomass Bioenergy. 2016;95:45–54. doi: 10.1016/j.biombioe.2016.08.015. [DOI] [Google Scholar]
- Raschmanová H, Weninger A, Glieder A, Kovar K, Vogl T. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: current state and future prospects. Biotechnol Adv. 2018;36:641–665. doi: 10.1016/j.biotechadv.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Rathour RK, Ahuja V, Bhatia RK, Bhatt AK. Biobutanol: new era of biofuels. Int J Energy Res. 2018 doi: 10.1002/er.4180. [DOI] [Google Scholar]
- Rattanachomsri U, Kanokratana P, Eurwilaichitr L, Igarashi Y, Champreda V. Culture-independent phylogenetic analysis of the microbial community in industrial sugarcane bagasse feedstock piles. Biosci Biotechnol Biochem. 2011;75:232–239. doi: 10.1271/bbb.100429. [DOI] [PubMed] [Google Scholar]
- Rodionova M, Poudyal R, Tiwari I, et al. Biofuel production: challenges and opportunities. Int J Hydrogen Energy. 2017;42:8450–8461. doi: 10.1016/j.ijhydene.2016.11.125. [DOI] [Google Scholar]
- Rodriguez E. Ethical issues in genome editing using Crispr/Cas9 system. J Clin Res Bioeth. 2016 doi: 10.4172/2155-9627.1000266. [DOI] [Google Scholar]
- Sakhuja D, Ghai H, Rathour RK, Kumar P, Bhatt AK, Bhatia RK. Cost-effective production of biocatalysts using inexpensive plant biomass: a review. 3 Biotech. 2021;11(6):280. doi: 10.1007/s13205-021-02847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsekeyeva F, Zayadan BK, Usserbaeva A, Bedbenov VS, Sinetova MA, Los DA. Cyanofuels: biofuels from cyanobacteria. Reality and perspectives. Photosynth Res. 2015;125:329–340. doi: 10.1007/s11120-015-0103-3. [DOI] [PubMed] [Google Scholar]
- Schnürer A. Biogas production. Microbiology and technology. Adv Biochem Eng Biotechnol. 2016;156:195–234. doi: 10.1007/10_2016_5. [DOI] [PubMed] [Google Scholar]
- Shang M, Chan VJ, Wong DWS, Liao H. A novel method for rapid and sensitive metagenomic activity screening. MethodsX. 2018;5:669–675. doi: 10.1016/j.mex.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam S, Ngo H-H, Wu Y-R. Advanced CRISPR/Cas-based genome editing tools for microbial biofuels production: a review. Renew Energy. 2019 doi: 10.1016/j.renene.2019.10.107. [DOI] [Google Scholar]
- Shen CR, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng. 2008;10:312–320. doi: 10.1016/j.ymben.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Sheng J, Feng X. Metabolic engineering of yeast to produce fatty acid-derived biofuels: bottlenecks and solutions. Front Microbiol. 2015;6:554. doi: 10.3389/fmicb.2015.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva FT, Moreira LR, de Souza FJ, Batista FR, Cardoso VL. Replacement of sugars to hydrogen production by Rhodobacter capsulatus using dark fermentation effluent as substrate. Bioresour Technol. 2016;200:72–80. doi: 10.1016/j.biortech.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77:1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Chen L, Wang J, Lu Y, Jiang W, Zhang W. A transcriptional regulator Sll0794 regulates tolerance to biofuel ethanol in photosynthetic Synechocystis sp. PCC 6803. Mol Cell Proteomics. 2014;13:3519–3532. doi: 10.1074/mcp.M113.035675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolze Y, Zakrzewski M, Maus I, et al. Comparative metagenomics of biogas-producing microbial communities from production-scale biogas plants operating under wet or dry fermentation conditions. Biotechnol Biofuels. 2015;8:14. doi: 10.1186/s13068-014-0193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolze Y, Bremges A, Rumming M, et al. Identification and genome reconstruction of abundant distinct taxa in microbiomes from one thermophilic and three mesophilic production-scale biogas plants. Biotechnol Biofuels. 2016;9:156. doi: 10.1186/s13068-016-0565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27:398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Tiwari R, Nain L, Labrou NE, Shukla P. Bioprospecting of functional cellulases from metagenome for second generation biofuel production: a review. Crit Rev Microbiol. 2018;44:244–257. doi: 10.1080/1040841X.2017.1337713. [DOI] [PubMed] [Google Scholar]
- Tkalec M, Štefanić PP, Cvjetko P, Šikić S, Pavlica M, Balen B. The effects of cadmium-zinc interactions on biochemical responses in tobacco seedlings and adult plants. PLoS ONE. 2014;9:e87582. doi: 10.1371/journal.pone.0087582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaganathan K, Goud S, Reddy M, Kayalvili U. Genome engineering for breaking barriers in lignocellulosic bioethanol production. Renew Sustain Energy Rev. 2017;74:1080–1107. doi: 10.1016/j.rser.2017.01.028. [DOI] [Google Scholar]
- Visioli LJ, Enzweiler H, Kuhn RC, Schwaab M, Mazutti MA. Recent advances on biobutanol production. Sustain Chem Process. 2014;2:15. doi: 10.1186/2043-7129-2-15. [DOI] [Google Scholar]
- Wang F, Li F, Chen G, Liu W. Isolation and characterization of novel cellulase genes from uncultured microorganisms in different environmental niches. Microbiol Res. 2009;164:650–657. doi: 10.1016/j.micres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang ZT, Seo SO, Lynn P, Lu T, Jin YS, Blaschek HP. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable "Clean" mutant selection in Clostridium beijerinckii as an example. ACS Synth Biol. 2016;5:721–732. doi: 10.1021/acssynbio.6b00060. [DOI] [PubMed] [Google Scholar]
- Wang S, Dong S, Wang Y. Enhancement of solvent production by overexpressing key genes of the acetone-butanol-ethanol fermentation pathway in Clostridium saccharoperbutylacetonicum N1–4. Bioresour Technol. 2017;245:426–433. doi: 10.1016/j.biortech.2017.09.024. [DOI] [PubMed] [Google Scholar]
- Wang S, Dong S, Wang P, Tao Y, Wang Y. Genome Editing in Clostridium saccharoperbutylacetonicum N1–4 with the CRISPR-Cas9 System. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.00233-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hart DJ, An Y. Functional metagenomic technologies for the discovery of novel enzymes for biomass degradation and biofuel production. Bioenerg Res. 2019;12:457–470. doi: 10.1007/s12155-019-10005-w. [DOI] [Google Scholar]
- Westbrook AW, Miscevic D, Kilpatrick S, Bruder MR, Moo-Young M, Chou CP. Strain engineering for microbial production of value-added chemicals and fuels from glycerol. Biotechnol Adv. 2019;37:538–568. doi: 10.1016/j.biotechadv.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Wong DD, Chan VJ, McCormack AA, Batt SB. Cloning and characterization of an exoxylogucanase from rumenal microbial metagenome. Protein Pept Lett. 2010;17:803–808. doi: 10.2174/092986610791190381. [DOI] [PubMed] [Google Scholar]
- Xia J, Yang Y, Liu CG, Yang S, Bai FW. Engineering Zymomonas mobilis for Robust Cellulosic Ethanol Production. Trends Biotechnol. 2019;37:960–972. doi: 10.1016/j.tibtech.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Xing MN, Zhang XZ, Huang H. Application of metagenomic techniques in mining enzymes from microbial communities for biofuel synthesis. Biotechnol Adv. 2012;30:920–929. doi: 10.1016/j.biotechadv.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Yadav M, Vivekanand V. Chaetomium globosporum: a novel laccase producing fungus for improving the hydrolyzability of lignocellulosic biomass. Heliyon. 2019 doi: 10.1016/j.heliyon.2019.e01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Fei Q, Zhang Y, Contreras LM, Utturkar SM, Brown SD, Himmel ME, Zhang M. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol. 2016;9:699–717. doi: 10.1111/1751-7915.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani SS, Gonzalez R. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab Eng. 2008;10:340–351. doi: 10.1016/j.ymben.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Yu D, Kurola JM, Lähde K, Kymäläinen M, Sinkkonen A, Romantschuk M. Biogas production and methanogenic archaeal community in mesophilic and thermophilic anaerobic co-digestion processes. J Environ Manag. 2014;143:54–60. doi: 10.1016/j.jenvman.2014.04.025. [DOI] [PubMed] [Google Scholar]
- Zabed H, Sahu J, Boyce A, Faruq G. Fuel ethanol production from lignocellulosic biomass: an overview on feedstocks and technological approaches. Renew Sust Energy Rev. 2016;66:751–774. doi: 10.1016/j.rser.2016.08.038. [DOI] [Google Scholar]
- Zhang Q, Hu J, Lee D-J. Biogas from anaerobic digestion processes. Research updates. Renew Energy. 2016;98:108–119. doi: 10.1016/j.renene.2016.02.029. [DOI] [Google Scholar]
- Zhang J, Yu L, Lin M, Yan Q, Yang ST. n-Butanol production from sucrose and sugarcane juice by engineered Clostridium tyrobutyricum overexpressing sucrose catabolism genes and adhE2. Bioresour Technol. 2017;233:51–57. doi: 10.1016/j.biortech.2017.02.079. [DOI] [PubMed] [Google Scholar]
- Zhao X-Q, Zi L-H, Bai F-W, Lin H-L, Hao X-M, Yue G-J, Ho NW (2011) Bioethanol from lignocellulosic biomass. Biotechnology in China III: biofuels and Bioenergy. Springer, Berlin, pp 25–51
- Zhou YJ, Buijs NA, Zhu Z, Qin J, et al. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziganshin AM, Liebetrau J, Pröter J, Kleinsteuber S. Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl Microbiol Biotechnol. 2013;97:5161–5174. doi: 10.1007/s00253-013-4867-0. [DOI] [PubMed] [Google Scholar]