Abstract
Endothelial cell senescence is one of the most important causes of vascular dysfunction and atherosclerosis. Circular RNAs (circRNAs) are endogenous RNA molecules with covalently closed-loop structures, which have been reported to be abnormally expressed in many human diseases. However, the potential role of circRNAs in endothelial cell senescence and atherosclerosis remains largely unknown. Here, we compared the expression patterns of circRNAs in young and senescent human endothelial cells with RNA sequencing. Among the differentially expressed circRNAs, circGNAQ, a circRNA enriched in vascular endothelium, was significantly downregulated in senescent endothelial cells. circGNAQ silencing triggered endothelial cell senescence, as determined by a rise in senescence-associated β-galactosidase activity, reduced cell proliferation, and suppressed angiogenesis; circGNAQ overexpression showed the opposite effects. Mechanistic studies revealed that circGNAQ acted as an endogenous miR-146a-5p sponge to increase the expression of its target gene PLK2 by decoying the miR-146a-5p, thereby delaying endothelial cell senescence. In vivo studies showed that circGNAQ overexpression in the endothelium inhibited endothelial cell senescence and atherosclerosis progression. These results suggest that circGNAQ plays critical roles in endothelial cell senescence and consequently the pathogenesis of atherosclerosis, implying that the management of circGNAQ provides a potential therapeutic approach for limiting the progression of atherosclerosis.
Keywords: circGNAQ, circular RNA, vascular endothelium, endothelial cell senescence, atherosclerosis
Graphical abstract

Endothelial cell senescence is one of the most important causes of vascular dysfunction and atherosclerosis. We found that circGNAQ, a circular RNA enriched in vascular endothelium, inhibited endothelial cell senescence and atherosclerosis progression, implying that circGNAQ-based gene therapy could serve as a novel therapeutic strategy to protect against atherosclerosis development.
Introduction
Aging is an important risk factor for cardiovascular diseases, and the incidence of cardiovascular events increases with age.1,2 Vascular endothelial cells (ECs) are critically involved in the maintenance of vascular homeostasis by regulating vascular tone and integrity.3,4 A dysfunctional endothelium has lost this tightly regulated balance and displays oxidant, vasoconstrictor, proinflammatory, and prothrombotic properties.4 Recent studies have shown that the integrity of EC function is compromised with aging, and EC senescence plays pivotal roles in the evolution of age-related vascular disorders, such as atherosclerosis.5,6 Therefore, it is critical to explore the mechanisms underlying EC senescence so as to better predict and prevent endothelial contributions to the pathogenesis of cardiovascular diseases associated with aging.
Circular RNAs (circRNAs) are a large, novel type of endogenous transcripts that comprise a closed continuous loop whereby the 3′ and 5′ ends are joined together.7,8 In the 1990s, only a handful of circRNAs were identified in humans and rodents and found to be produced in the back-splicing of precursor mRNA (pre-mRNA).9 However, these circRNAs were present at low levels and considered to be functionless by-products of splicing.10 Recently, high-throughput transcriptome sequencing technology and novel computational approaches have shown that the expression of circRNAs is widespread.8,11 Furthermore, in biological experiments many circRNAs have been demonstrated to be highly expressed in a tissue-specific or cell type-specific manner.12,13 Salient features of circRNAs also include remarkable stability, high abundance, and evolutionary conservation.13, 14, 15 Studies have shown that circRNAs can function as microRNA (miRNA) sponges,8 binding partners of proteins,16 and regulators of gene expression17 or can be translated into proteins.18 With accumulated knowledge of characteristics and functions of circRNAs, it has been described that circRNAs play critical roles in human diseases, such as cancer,19,20 neurological disorders,21,22 and heart disease.16,23,24 However, the overall pathophysiological contributions of circRNAs to age-related vascular diseases remain largely unknown.
In this study, we analyzed the expression profile of circRNAs in young and senescent ECs by using RNA sequencing (RNA-seq). We further identified that circGNAQ (hsa_circ_0006459), a circRNA enriched in the vascular endothelium, was significantly downregulated in senescent ECs. Further functional and mechanistic investigations revealed that circGNAQ may act as a competing endogenous RNA (ceRNA) to regulate the expression of Polo-like kinase 2 (PLK2) by decoying the miR-146a-5p, thereby delaying EC senescence. Additionally, circGNAQ overexpression in the endothelium delayed vascular EC senescence and prevented atherosclerosis progression in vivo. In this work, we shed light on the roles of circGNAQ in EC senescence and atherosclerosis, which may represent a promising strategy for vascular aging and age-related vascular disease interventions.
Results
Identification of endothelial cell senescence-associated circRNAs
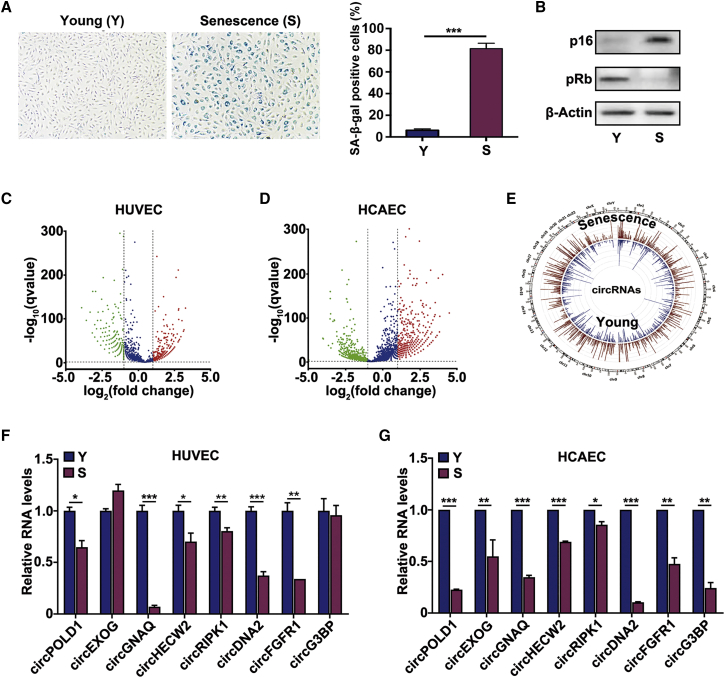
Compared with the young ECs, senescent ECs displayed a flattened and enlarged morphology and increased senescence-associated β-galactosidase (SA-β-gal) activity, a widely used senescence marker (Figure 1A). The expression of senescent marker p16 was increased, whereas that of pRb was decreased, in senescent ECs (Figure 1B). RNA-seq was performed on young and senescent ECs (Figures 1C and 1D). We mapped the RNA-seq data to the human reference genome (GRCH38/hg38) by HISAT2. Counts of reads mapping across an identified back-splice were normalized by transcripts per million (TPM), which could be used to quantitatively compare the back-splices between different samples. With a cutoff criterion of log2 fold change > 1.0 and p < 0.01, differentially expressed circRNAs were identified by RNA-seq in human umbilical vein endothelial cells (HUVECs) and human coronary artery endothelial cells (HCAECs). The majority of the identified circRNAs were generated from exons, and the others originated from introns (Figure S1A). Furthermore, most of the identified circRNAs were <5,000 nucleotides in length (Figure S1B). Interestingly, differentially expressed circRNAs were enriched in senescent ECs compared with young ECs (Figure 1E). These differentially expressed circRNAs may have potential function in EC senescence and need to be further explored.
Figure 1.
Identification of endothelial cell senescence-associated circRNAs
(A) Representative images of SA-β-gal staining in young and senescent HUVECs. Y, young; S, senescent. (B) Western blot analysis of the levels of p16 and pRb in young and senescent HUVECs. (C and D) Volcano plot showing circRNA expression in HUVECs and HCAECs (senescent versus young). The red and green dots represent circRNAs with statistically significant differences in expression. (E) Circos plots showing all circRNAs from young and senescent HUVECs. (F) qRT-PCR analysis of changes in circRNA expression in young and senescent HUVECs. (G) qRT-PCR analysis of changes in circRNA expression in young and senescent HCAECs. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by Student’s t test.
Characterization of circGNAQ in endothelial cells
To verify the RNA-seq results, we examined differentially expressed circRNAs between young and senescent ECs by quantitative real-time PCR (qRT-PCR) using circRNA-specific divergent primers (Table S1). Among them, circGNAQ expression was consistently and significantly downregulated in senescent ECs compared with young ECs (Figures 1F and 1G). However, the expression of GNAQ mRNA had no apparent change (Figure S1C). We confirmed the head-to-tail splicing in the RT-PCR product of circGNAQ with expected size by Sanger sequencing (Figure 2A). The sequence is consistent with circBase database annotation. Subsequently, we treated RNAs extracted from HUVECs with or without RNase R. By northern blotting and qRT-PCR, we confirmed that although RNase R treatment decreased GNAQ linear mRNA levels, it did not affect circGNAQ levels (Figures 2B and 2C). These results confirmed the characteristics of circGNAQ as a circRNA and implied that its function may benefit from the biological stability of this molecule.
Figure 2.
Characterization of circGNAQ in endothelial cells
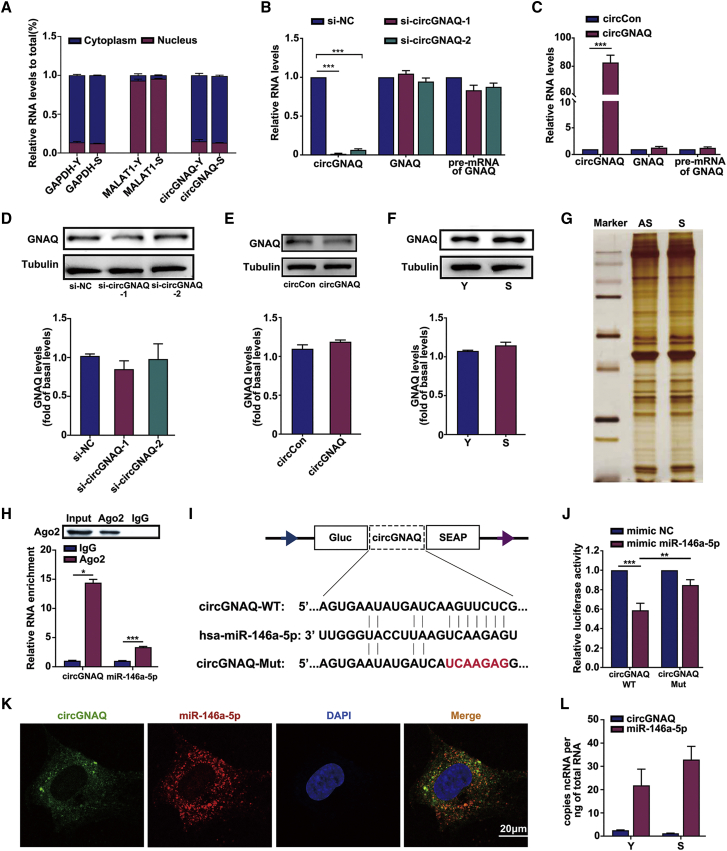
(A) Genomic loci of the circGNAQ gene. circGNAQ is produced at the GNAQ gene locus containing exons 4–5. The back-splice junction of circGNAQ was identified by Sanger sequencing. (B) Northern blot analysis showed the expression of circGNAQ after infection with lentiviruses expressing circGNAQ. RNase R digestion did not affect circGNAQ levels (lower) but degraded linear RNAs (upper). (C) qRT-PCR results show the abundance of circRNAs and linear RNAs in HUVECs treated with RNase R. The levels of circGNAQ and GNAQ were normalized to the values measured after mock treatments (n = 3). (D) qRT-PCR analysis of circGNAQ levels in the aorta tissue of young (n = 10, 1.5 months old) and old (n = 11, 24 months old) C57BL/6 mice. (E) qRT-PCR analysis of circGNAQ levels in various tissues of C57BL/6 mice. (F) circGNAQ and GNAQ expression was detected by qRT-PCR in the aortic intima from 8-week-old C57BL/6 mice (n = 4; each sample represents RNA pooled from two mice). As a control, total RNAs obtained from the leftover samples after endothelium-enriched RNA preparation (media + adventitial regions: M+A) were used. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by Student’s t test.
Next, the endogenous circGNAQ expression was assessed in humans and mice. Consistent with the RNA-seq and validation results, circGNAQ was downregulated in the aorta tissue of aged C57BL/6 mice (Figure 2D) and the whole blood of older adults (Figure S1D). We found that circGNAQ was commonly expressed in various tissues and relatively more highly expressed in the aorta tissue (Figure 2E). Of note, circGNAQ expression in the vascular endothelium evidently exceeded that in the aortic media and adventitia (M+A) (Figure 2F), which implied its important role in endothelium function. Since EC senescence plays pivotal roles in atherosclerosis, we further evaluated whether circGNAQ is associated with atherosclerotic progression. As shown in Figure 2G, circGNAQ expression was reduced in the aortic intima of Ldlr−/− mice after high-fat diet (HFD) for 4, 8, or 12 weeks. These data demonstrate that the aortic intimal expression of circGNAQ fell with atherosclerotic progression, suggesting that it may be involved in the pathogenesis of atherosclerosis.
circGNAQ suppresses cellular senescence in endothelial cells
To explore the biological function of circGNAQ in EC senescence, the circGNAQ lentivirus and short interfering RNAs (siRNAs) targeting the junction of circGNAQ were established and transfected into ECs. The results showed that circGNAQ siRNAs significantly decreased the expression of circGNAQ but had no effect on GNAQ mRNA (Figure 3A; Figure S2A). Similarly, circGNAQ was successfully overexpressed in HUVECs and HCAECs, whereas GNAQ mRNA expression had no obvious change (Figure 3B; Figure S2B). These data indicated that the expression of GNAQ was unaffected by circGNAQ.
Figure 3.
circGNAQ suppresses cellular senescence in endothelial cells
(A) qRT-PCR analysis of the levels of circGNAQ and GNAQ in young HUVECs after transfection of negative control (si-NC) or si-circGNAQ-1/2. (B) qRT-PCR analysis of the levels of circGNAQ and GNAQ in senescent HUVECs after infection with lentiviruses expressing circCon or circGNAQ. (C, E, and G) BrdU assays, SA-β-gal staining, and tube formation in young HUVECs after transfection with si-NC or si-circGNAQ-1/2. (D, F, and H) BrdU assays, SA-β-gal staining, and tube formation in senescent HUVECs after infection with lentiviruses expressing circCon or circGNAQ. (I) circGNAQ and angiogenesis in vivo. Left, Representative photomicrographs of Matrigel plugs. VEGF(−), negative control; VEGF(+), positive control. Right, Quantification of hemoglobin in the homogenized Matrigel plugs; results are expressed as hemoglobin content in the Matrigel plug (mg/dL). (J) Western blot analysis of p16 in young HUVECs after transfection of si-NC or si-circGNAQ-1/2. (K) Western blot analysis of p16 in senescent HUVECs after infection with lentiviruses expressing circCon or circGNAQ. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by Student’s t test.
In this study, silencing of circGNAQ led to reduced percentage of 5-bromo-2′-deoxyuridine (BrdU)-positive cells and increased SA-β-gal activity in HUVECs and HCAECs (Figures 3C and 3E; Figures S2C and S2E). Conversely, stably overexpressing circGNAQ exhibited the opposite effect (Figures 3D and 3F; Figures S2D and S2F). Additionally, knockdown of circGNAQ significantly impaired the tube formation of HUVECs and HCAECs, whereas overexpression of circGNAQ promoted tube formation (Figures 3G and 3H; Figures S2G and S2H). As circGNAQ had an effect on the tube formation ability of ECs, we then directly assessed whether circGNAQ induced angiogenesis in vivo. The results showed that the angiogenesis was decreased in the plugs supplemented with HUVECs after circGNAQ silencing compared with the si-NC group (Figure 3I). Moreover, silencing of circGNAQ led to an upregulation of senescent marker p16, whereas overexpression of circGNAQ reduced p16 expression (Figures 3J and 3K). Taken together, our results showed that circGNAQ could play a suppressive role in EC senescence. Given this evidence, we set out to investigate the underlying mechanism of circGNAQ regulation of the senescent phenotype in ECs.
circGNAQ acts as a sponge for miR-146a-5p in endothelial cells
To observe the cellular localization of circGNAQ, we conducted qRT-PCR analysis for nuclear and cytoplasmic circGNAQ RNA. The results showed that the circGNAQ transcript was preferentially located in the cytoplasm, and there was no significant change in the localization of circGNAQ between young and senescent ECs (Figure 4A). RNA fluorescence in situ hybridization (FISH) also showed that circGNAQ was predominantly localized in the cytoplasm (Figure 4K).
Figure 4.
circGNAQ acts as a sponge for miR-146a-5p in endothelial cells
(A) Levels of circGNAQ in the nuclear and cytoplasmic fractions of HUVECs. GAPDH and MALAT1 were used as positive controls in the cytoplasm and nucleus, respectively. (B) qRT-PCR analysis of the levels of circGNAQ, GNAQ, and GNAQ pre-mRNA in young HUVECs after transfection of si-NC or si-circGNAQ-1/2. (C) qRT-PCR analysis of the levels of circGNAQ, GNAQ, and GNAQ pre-mRNA in young HUVECs after infection with lentiviruses expressing circCon or circGNAQ. (D) Western blot analysis of GNAQ in HUVECs after transfection of si-NC or si-circGNAQ-1/2. (E) Western blot analysis of GNAQ in HUVECs after infection with lentiviruses expressing circCon or circGNAQ. (F) Western blot analysis of GNAQ in young and senescent HUVECs. (G) RNA pull-down assay by biotin-labeled circGNAQ and its antisense RNA followed by silver staining of protein extract from HUVECs. S, sense strand of circGNAQ; AS, anti-sense strand of circGNAQ. (H) RIP was performed with AGO2 antibody in HUVECs, and then the enrichment of circGNAQ and miR-146a-5p was detected. (I) Schematic illustration demonstrates complementarity to the miR-146a-5p seed sequence with circGNAQ. Red letters indicate mutated nucleotides. (J) The luciferase activity of circGNAQ-WT or circGNAQ-Mut in HEK293T cells after co-transfection with miR-146a-5p. (K) Co-localization between miR-146a-5p and circGNAQ was observed by RNA FISH in HUVECs. Nuclei were stained with DAPI. Scale bar, 20 μm. (L) Absolute quantification for circGNAQ and miR-146a-5p in young and senescent HUVECs. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by Student’s t test.
To elucidate the mechanisms responsible for the roles of circGNAQ in EC senescence, we investigated its effects on the expression of its host gene GNAQ. However, our results indicated that silencing or overexpression of circGNAQ did not affect the pre-mRNA, mRNA, and protein levels of its parental gene GNAQ (Figures 4B–4E). Similarly, there was no significant change in the protein level of GNAQ between young and senescent HUVECs (Figure 4F). Alternatively, circRNAs could serve as essential regulators of RNA-binding proteins.16,25 Unfortunately, the RNA pull-down assay showed that there were no specific bands significantly enriched by the circGNAQ probe (Figure 4G).
Given that circRNAs can reportedly function as sponges for miRNAs and that circGNAQ is stable and located in the cytoplasm, we tried to explore whether circGNAQ could bind to miRNAs. By circRNA in vivo precipitation (circRIP) experiments, we purified the circGNAQ-associated miRNAs and analyzed the 188 candidate miRNAs in the complex. Interestingly, we found a specific enrichment of circGNAQ and miR-146a-5p compared with the controls (Table S2), indicating that miR-146a-5p could be the circGNAQ-associated miRNA in ECs. Next, we predicted the miR-146a-5p binding sites of circGNAQ, using the bioinformatics database RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/), and circGNAQ contains one site that is complementary to miR-146a-5p. To further confirm this result, we performed a luciferase assay using miR-146a-5p. Luciferase reporters were constructed by inserting either the wild-type circGNAQ-miR-146a-5p sequence (WT) or the sequence with mutated binding sites of miR-146a-5p (Mut) into the 3′ UTR of Gaussia luciferase (GLuc) (Figure 4I). Upregulation of miR-146a-5p remarkably reduced the luciferase activities of the WT reporter but not the mutant (Figure 4J), suggesting that miR-146a-5p could interact with circGNAQ via the complementary seed region. In this study, we also performed RNA immunoprecipitation (RIP) assay to observe the enrichment of argonaute 2 (AGO2) on circGNAQ and miR-146a-5p in vascular ECs. Our data revealed that circGANQ and miR-146a-5p were both enriched by AGO2 through RIP assay (Figure 4H). Using an RNA FISH assay, we demonstrated that circGNAQ and miR-146a-5p were co-localized in the cytoplasm (Figure 4K). To support that circGNAQ could function as a ceRNA to sponge miR-146a-5p, copy number analysis revealed that the observed miR-146-5p-to-circGNAQ ratio seemed to be compatible with a sponge activity (Figure 4L). In addition, miR-146a-5p mimic or anti-miR-146a-5p transfection did not affect the expression level of circGNAQ (Figure S3), indicating that miR-146a-5p had no effect on the degradation of circGNAQ. Together, these data revealed that circGNAQ could act as a sponge for miR-146a-5p in ECs. Moreover, our results revealed that miR-146a-5p overexpression significantly increased senescence of ECs, whereas miR-146a-5p knockdown dramatically decreased EC senescence (Figures S4A and S4B). Collectively, these results suggest that circGNAQ functions as a negative regulator of miR-146a-5p, and thus suppresses EC senescence.
circGNAQ/miR-146a-5p/PLK2 axis regulates endothelial cell senescence
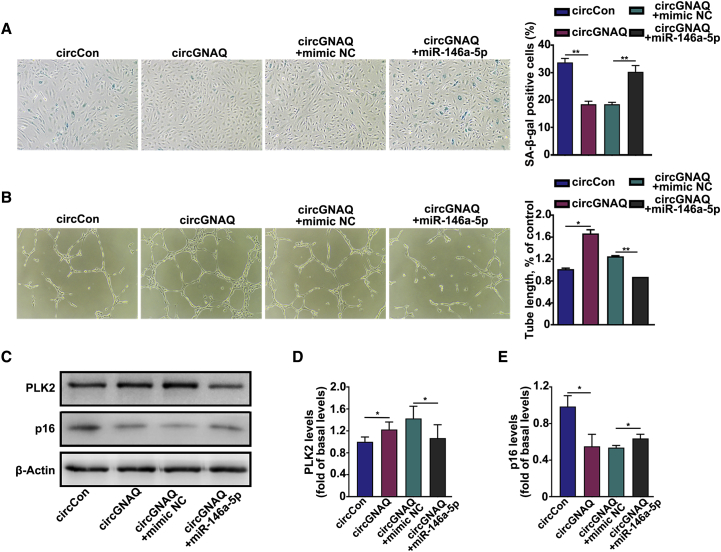
To address whether circGNAQ inhibits cellular senescence via interacting with miR-146a-5p, we transfected miR-146a-5p mimics into circGNAQ lentivirus-infected ECs. SA-β-gal staining displayed that overexpression of circGNAQ resulted in a significant decrease of SA-β-gal-positive cells and the effect could be abrogated by miR-146a-5p mimics (Figure 5A). In addition, the circGNAQ lentivirus-infected ECs transfected with miR-146a-5p mimics showed impairment in tube formation compared with circGNAQ overexpression alone (Figure 5B). PLK2 is a validated target of miR-146a.26 We confirmed that miR-146a-5p negatively regulates PLK2 expression in ECs (Figures S4C and S4D). In this study, downregulation of circGNAQ reduced the protein levels of PLK2 (Figure S5). Upregulation of circGNAQ enhanced the protein levels of PLK2 and decreased the levels of p16, and the effect could be reversed by miR-146a-5p mimics (Figures 5C–5E). Collectively, co-transfection of miR-146a-5p mimics could reverse circGNAQ overexpression-mediated effects in ECs.
Figure 5.
circGNAQ/miR-146a-5p/PLK2 axis regulates endothelial cell senescence
(A) SA-β-gal staining in circGNAQ lentivirus-infected endothelial cells after transfection with mimic NC or miR-146a-5p. (B) Angiogenesis assay in circGNAQ lentivirus-infected endothelial cells after transfection with mimic NC or miR-146a-5p. (C–E) Western blot analysis of PLK2 and p16 in circGNAQ lentivirus-infected endothelial cells after transfection with mimic NC or miR-146a-5p. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05; ∗∗p < 0.01 by Student’s t test.
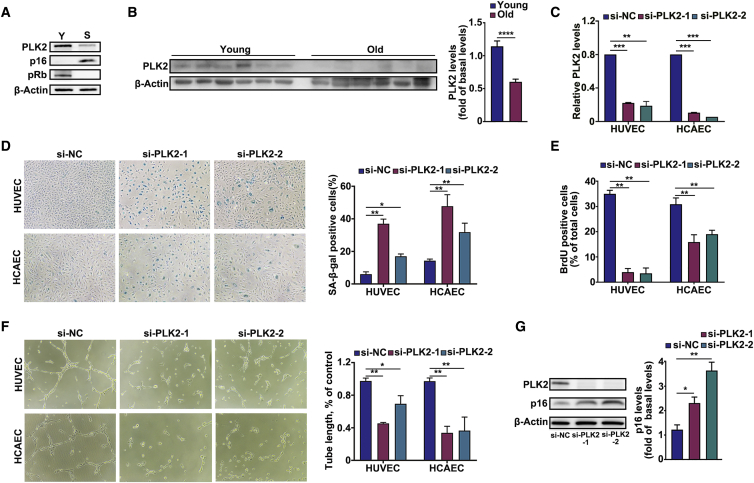
PLK2 protein apparently decreased in the senescent HUVECs and was downregulated in the aorta of old mice (Figures 6A and 6B). Furthermore, we specifically knocked down PLK2 expression using designed si-PLK2-1/2. The inhibition of PLK2 in ECs was confirmed (Figure 6C), and silencing of PLK2 promoted cellular senescence and inhibited cell proliferation and tube formation (Figures 6D–6F). Moreover, western blot assays confirmed that knockdown of PLK2 led to an upregulation of p16 (Figure 6G). These data indicated that PLK2 could inhibit EC senescence. Here we further investigated whether circGNAQ played a role in EC senescence through PLK2 regulation. As shown in Figure S6, PLK2 siRNAs significantly attenuated the effects of circGNAQ on PLK2. In addition, silencing of PLK2 could diminish the effect of circGNAQ overexpression on proliferation and tube formation and attenuated the effects of circGNAQ on the inhibition of EC senescence (Figure S6). Our data showed that the phenotype could be reversed by knockdown of PLK2 in the circGNAQ overexpression cells, suggesting that the role of circGNAQ in EC senescence is dependent on PLK2 expression. Collectively, circGNAQ served as a sponge for miR-146a-5p to regulate PLK2 and suppress cellular senescence via the ceRNA mechanism in ECs.
Figure 6.
PLK2 regulates endothelial cell senescence
(A) Young and senescent HUVECs were extracted, and levels of PLK2, p16, and pRb in the extracts were determined by western blotting. (B) Levels of PLK2 protein measured by western blotting in the aortas of 1.5- and 24-month-old mice (n = 6 per group). (C) qRT-PCR analysis of the levels of PLK2 in young HUVECs and HCAECs after transfection of si-NC or si-PLK2-1/2. (D) SA-β-gal staining in young HUVECs and HCAECs after transfection with si-NC or si-PLK2-1/2. (E) BrdU assay in young HUVECs and HCAECs after transfection with si-NC or si-PLK2-1/2, respectively. (F) Angiogenesis assay in young HUVECs and HCAECs after transfection with si-NC or si-PLK2-1/2. (G) Western blot analysis of p16 in young HUVECs after transfection of si-NC or si-PLK2-1/2. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by Student’s t test.
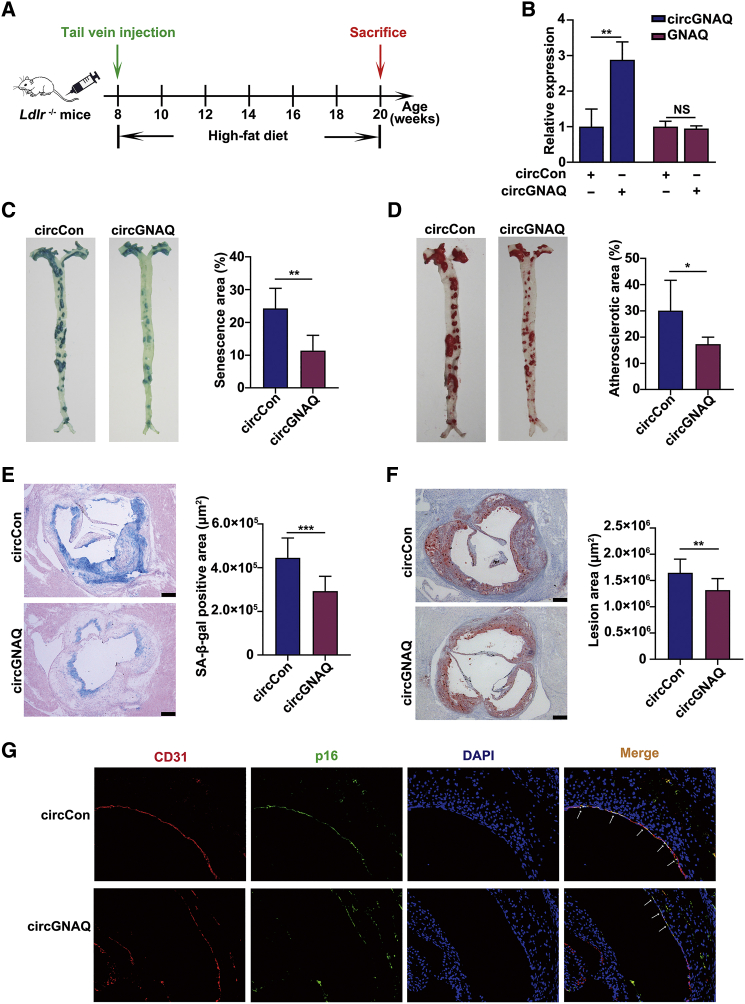
circGNAQ inhibits endothelial cell senescence and reduces atherosclerosis in Ldlr−/− mice
Since circGNAQ levels decrease in senescent ECs and aged aortas, we hypothesized that EC-specific overexpression of circGNAQ may prevent the progression of atherosclerosis. Therefore, we generated a recombinant adeno-associated virus (AAV) cassette with circGNAQ expression driven by a mouse Tie2 promoter, which ensures endothelium-specific expression.27, 28, 29 Ldlr−/− mice were injected with the AAV particles and fed a HFD for 12 weeks (Figure 7A). As shown in Figure 7B, the expression of circGNAQ in the aortic intima from mice injected with AAV-circGNAQ was 2.8-fold higher than that in mice injected with the AAV-circCon (mock vector with no circGNAQ sequence), whereas no overexpression of GNAQ was observed. Consistent with in vitro data, overexpression of circGNAQ decreased SA-β-gal activity in the aortas and the aortic roots (ARs) (Figures 7C and 7D) of Ldlr−/− mice. When we examined additional measures of cellular senescence, we found that the abundance of p16 in the intimal layer of the aorta was lower in circGNAQ-treated mice than in circCon-treated mice (Figure 7E). These findings were in agreement with our SA-β-gal staining results. To explore the potential therapeutic benefit of circGNAQ in atherosclerosis, we next evaluated its ability to retard the progression of atherosclerosis. As expected, overexpression of circGNAQ attenuated plaque lesion formation in the aortas and in the ARs (Figures 7F and 7G). Taken together, these results suggested that circGNAQ could inhibit EC senescence and have protective effects against atherosclerotic lesion formation.
Figure 7.
circGNAQ reduces atherosclerosis in Ldlr−/− mice
Ldlr−/− mice received tail vein injection of AAV-circCon or AAV-circGNAQ and were fed a high-fat diet for 12 weeks as described in Materials and methods. (A) Schema of experimental procedure. (B) Quantification of the circGNAQ expression in the aortic intima from AAV-circCon and AAV-circGNAQ groups (n = 4; each sample represents RNA pooled from two mice). (C and D) SA-β-gal activity assays of whole aorta and aortic root sections. Scale bars, 250 μm. (E) Immunofluorescence staining of p16 and CD31 in the atherosclerotic lesions. Arrows indicate p16 positive staining in the endothelium. Scale bars, 100 μm. (F and G) Lesion areas were detected by oil red O (ORO) staining of whole aorta and aortic root sections. Scale bars, 250 μm. n = 4–6 per group. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by Student’s t test. Error bars indicate SD.
Discussion
In this study, we screened for differentially expressed circRNAs in young and senescent ECs by RNA-seq. A novel circ RNA, circGNAQ (hsa_circ_0006459), was found to be decreased in senescent ECs and aging blood vessels. Interestingly, circGNAQ was enriched in vascular endothelium, suggesting that circGNAQ could be involved in regulating vascular function. Moreover, circGNAQ delayed cellular senescence by harboring miR-146a-5p to abolish the suppressive effect on the target gene PLK2 in ECs. Additionally, circGNAQ overexpression inhibited EC senescence and prevented atherosclerosis progression in Ldlr−/− mice. Thus, our data suggest that circGNAQ could play an important role in EC senescence and atherosclerosis.
It has been demonstrated that circRNAs execute their function through several molecular mechanisms, such as acting as miRNA sponges,30, 31, 32 binding partners of protein,16 and regulators of parental gene expression.17,33 In this study, the pre-mRNA, mRNA, and protein levels of GNAQ were not significantly changed after circGNAQ silencing or overexpression in ECs, suggesting that circGNAQ has no effect on the expression of its parental gene GNAQ. The RNA pull-down assay revealed that specific proteins did not significantly enrich by the circGNAQ probe. These results limit the possibility that circGNAQ functions as a regulator of parental gene expression and/or a partner of specific proteins.
Growing evidence has indicated that some circRNAs could serve as sponges for miRNAs, regulating the expression of miRNA target genes in multiple human diseases.8,11 Here we showed that circGNAQ was primarily expressed in the cytoplasm of ECs and contained a conserved miR-146a-5p target site that was validated by circRIP, luciferase assay, RIP, and FISH analyses. Therefore, we inferred that circGNAQ might protect ECs against senescence via sponging miR-146a-5p. It is reported that miR-146a expression is upregulated not only in HUVECs but also in human aortic endothelial cells (HAECs) and in senescent HCAECs, confirming that its upregulation is associated with the senescent phenotype in different vascular cell types.34 Another study showed that significant miR-146a modulation was observed in senescent versus young ECs.35 miR-146a is also found to be upregulated in senescent HCA2 fibroblasts and exhibited a role in the cell non-autonomous effects of cellular senescence, a phenomenon linked to both cancer and aging.36 These findings are consistent with our observation that miR-146a-5p is associated with promoted EC senescence. Together, our findings demonstrated that circGNAQ served as a senescence suppressor by sponging miR-146a-5p in ECs and revealed the significance of interaction between circGNAQ and miR-146a-5p in EC senescence.
PLK2 is a serine-threonine kinase that acts as a critical regulator of cell cycle progression.37 Deng et al. reported that miR-146a regulates lineage-negative bone marrow cell (lin-BMC) senescence by suppressing its target PLK2 expression.26 A report by Yang et al. found that PLK2 can control angiogenesis during vascular development by specifically regulating EC lamellipodia.38 Additionally, PLK2 has been observed to mediate angiogenesis through regulating RAP1 activity and localization during vascular development.38 In this study, PLK2 knockdown could mimic the effects of circGNAQ silencing, which promoted EC senescence and inhibited tube formation. During EC dysfunction, circGNAQ overexpression becomes a sink for miR-146a-5p and releases the repressive effect of miR-146a-5p on PLK2 expression. Therefore, the circGNAQ/miR-146a-5p/PLK2 axis is responsible for regulating EC senescence.
Several circRNAs have already been identified as important for cardiovascular biology.23,39 For example, circANRIL, circACTA2, and circLRP6 have recently been identified in smooth muscle cells (SMCs) and are involved in the regulation of SMC migration, proliferation, and phenotypic modulation.23,39,40 A recent study showed that circMAP3K5 contributed to TET2-mediated SMC differentiation by sequestering miR-22-3p and attenuated intimal hyperplasia.41 circRNAs have also been identified as regulators of various pathways that are involved in aging and cellular senescence.30,42,43,44 Du et al. found that circ-Foxo3 promotes cardiac senescence by interacting with multiple factors associated with stress and senescence responses.16 Here, we have shown that the expression of circGNAQ was significantly decreased during replicative senescence of ECs. There is emerging evidence that EC senescence contributes to the pathogenesis of human atherosclerosis.45,46 Therefore, we anticipate that circGNAQ overexpression in the endothelium may play a protective role against arteriosclerosis. In this study, we showed that AAV-Tie2-circGNAQ, targeting ECs, could inhibit vascular EC senescence and reduce aortic atherosclerosis in mice. Our results may reveal an important role of circGNAQ in delaying EC senescence and provide a fresh perspective on circRNAs in atherosclerosis.
In summary, our study provided a portrayal of circRNAs in young and senescent ECs. We further characterized and functionally evaluated a novel circRNA, circGNAQ, that is enriched in vascular endothelium. Our findings revealed that circGNAQ expression was significantly decreased in senescent ECs. Functionally and mechanistically, circGNAQ prevented EC senescence by sponging miR-146a-5p and upregulating PLK2 expression, indicating its role as a senescence suppressor in the vascular aging process. Furthermore, the in vivo overexpression of circGNAQ delayed EC senescence and prevented atherosclerosis progression. Hence, circGNAQ-based gene therapy could serve as a novel therapeutic strategy to protect against atherosclerosis development.
Materials and methods
Cell culture
HUVECs and HCAECs were purchased from ScienCell (Carlsbad, CA, USA). The cells were cultured in endothelial cell medium (ECM) supplemented with 5% fetal bovine serum and 1% endothelial cell growth supplement (ECGS, ScienCell) at 37°C in 5% CO2 and 95% humidity. Replicative senescent ECs were prepared by culturing HUVECs for an extended period until passages 20–22. HUVECs in passages 4–5 were used as proliferating young control cells.
RNA sequencing, identification, and quantification of circRNAs
The total RNAs extracted from young and senescent ECs (HUVECs and HCAECs) were treated with the Epicenter Ribo-Zero rRNA Removal Kit (Epicenter, USA) for deleting rRNA, according to the manufacturer’s instructions. Next, the rRNA-depleted and RNase R-digested RNA samples were fragmented and cDNA synthesized with random primer. The PCR amplification products of cDNA were purified, and then the libraries were quality controlled and sequenced by HiSeq 2500 (Illumina, USA).
qRT-PCR analysis
Total RNA was used for gene expression analysis by reverse transcription followed by qRT-PCR analysis. Reverse transcription was performed with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). qPCR was carried out with gene-specific primers and the TB Green Premix Ex Taq Kit (Takara, Japan) in a Light Cycler 96 instrument (Roche). β-Actin was used as an internal reference for the quantification of circRNA and mRNA, and U6 was used for miRNA. Each reaction was performed in triplicate, and analysis was performed with the 2−ΔΔCt method as described previously.47 All primers used are listed in Table S3. Whole blood collection for RNA isolation was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical University.
Digital PCR analysis
A chip-based digital PCR (dPCR) platform (QuantStudio 3D Digital PCR System; Life Technologies, USA) was used for quantification of the copy number of circGNAQ and miR-146a-5p using cDNA synthesized from RNA isolated from young and senescent HUVECs, as described above. The reaction conditions were as follows: hot start at 96°C for 10 min, denaturation at 98°C for 30 s, annealing/extension at 64°C for 2 min for a total of 39 cycles, followed by a final extension step at 64°C for 2 min. The data analysis was performed with QuantStudio 3D AnalysisSuite Cloud Software version 3.1.2.
RNase R treatment
For the RNA digestion assay, 10 μg of RNA isolated from HUVECs was either untreated (mock) or treated with 1 μL of RNase R (Epicenter, USA) in the presence of 1× RNase R buffer and 1 μL of RiboLock RNase Inhibitor (Thermo Scientific, USA) and incubated for 15 min at 37°C, followed by qRT-PCR analysis of circRNA that was resistant to RNase R treatment. Forward and reverse primers (Table S3) were used to sequence the amplified PCR products and identify the junction sequence.
Northern blots
Total RNA with or without RNase R treatment and digoxigenin (DIG)-labeled RNA molecular weight marker (Roche) were loaded on a 1% agarose gel containing 1% formaldehyde and were run in 1× MOPS buffer. The RNA was then transferred onto Amersham Hybond-N1 membranes (GE Healthcare, Pittsburgh, PA) by capillary transfer. Membranes were then crosslinked, pre-hybridized (Roche), and hybridized with a 3′-DIG-labeled probe versus circGNAQ. The membranes were then washed twice in 2× saline sodium citrate (SSC) with 0.1% SDS at room temperature and washed two additional times at 68°C. After washing, the membranes were exposed and analyzed. The prober sequence is listed in Table S4.
Preparation of nuclear and cytoplasmic fractions
The nuclear and cytoplasmic fractions were extracted with the PARIS Kit (Invitrogen, USA) according to the manufacturer’s instructions. Briefly, cells (2 × 106) were washed with PBS and immediately lysed on ice with 300 μL of cell fractionation buffer for 10 min. The nuclear and cytoplasmic portions were separated by centrifuging at 500 × g for 5 min. Finally, RNA extraction was carried out according to the manufacturer’s instructions.
Transfection and viral infection
Transfection of miRNA mimics/inhibitors or siRNAs was performed with Lipofectamine RNAiMAX (Life Technologies, USA) according to the manufacturer’s instructions. To knock down circGNAQ, two siRNAs targeting the back-splice junction site of circGNAQ and a siRNA-NC were synthesized by RiboBio (Guangzhou, China), after efficiency examination by qRT-PCR. miRNA mimics/inhibitors or siRNAs were purchased from RiboBio (Guangzhou, China).
To overexpress circGNAQ, the full-length cDNA of circGNAQ was amplified in HEK293T cells. To construct lentivirus-circGNAQ (LV-circGNAQ), circGNAQ cDNA was inserted into pLCDH-ciR (Geneseed, China), which was reconstructed by inserting the front circular frame and back circular frame to promote RNA circularization. The front circular frame contains the endogenous flanking genomic sequences with EcoRI restriction enzyme site and an AG splice acceptor, and the back circular frame contains part of the inverted upstream sequence with BamHI site and a GT splice donor. A mock vector with no circGNAQ sequence served as a control, called circCon. Cell transfections of circGNAQ and circCon were conducted by lentiviral infection. Briefly, cells were infected with 1 × 106 recombinant lentivirus-transducing units and 8 μg/mL polybrene (Sigma), followed by 2 μg/mL puromycin treatment for 1 week.
Senescence-associated β-galactosidase staining
Cellular senescence was assessed through detecting the activity of β-galactosidase with a SA-β-gal staining kit (Beyotime) according to the manufacturer’s instructions. Briefly, cells were fixed with a fixative solution for 15 min at room temperature. After rinsing with PBS, cells were incubated with freshly prepared staining solution overnight at 37°C. The percentage of positively stained cells (blue cells) versus the total number of cells was determined for six randomly selected microscopic fields. The images were captured at 100× magnification with a Nikon Eclipse TS100 microscope (Nikon, Japan). SA-β-gal activity was also examined in tissue. Briefly, sections from the AR were fixed for 10 min, stained for 1 week, and observed by light microscopy. Whole aortas were stained for SA-β-gal as described previously.48
BrdU incorporation
Cell proliferation was analyzed by the BrdU incorporation assay. Briefly, cells were exposed to 40 μM BrdU for 1 h before fixation. The formaldehyde-fixed cells were permeabilized with 0.05% trypsin, followed by blocking for 1 h in 3% bovine serum albumin. Incorporated BrdU was detected by incubating for 30 min to 1 h with anti-BrdU mouse monoclonal antibody (CST, USA). Detection was performed with an anti-mouse immunoglobulin G (IgG) antibody conjugated with Alexa Fluor 488 (CST, USA). BrdU-positive cells were visualized and images were captured at 100× magnification with a fluorescence microscope (Olympus, Japan) and presented as the percentage of BrdU-positive nuclei over the total number of nuclei counted.
In vitro angiogenesis assay
The in vitro angiogenic activity of endothelial cells was determined by the Matrigel tube formation assay. Matrigel matrix basement membrane (BD Biosciences, USA) was thawed at 4°C, pipetted into pre-cooled 48-well plates, and incubated at 37°C for 1 h. After Matrigel polymerization, treated cells were suspended in ECM, added to the Matrigel-coated wells at a density of 3 × 104 cells/well, and incubated at 37°C for 4–6 h. Quantification of angiogenic activity was calculated by measuring the length of tube walls formed between discrete ECs in each well relative to the control. The tube length was quantified with ImageJ software.
In vivo Matrigel plug assay
Athymic nude mice received a 400-μL subcutaneous injection of Matrigel plugs supplemented with saline (VEGF−, negative control) or VEGF (250 ng/mL, positive control) or mixed with HUVECs after transfection of si-NC or si-circGNAQ-1/2. Seven days after implantation, animals were sacrificed by cervical dislocation and the Matrigel plugs were removed and photographed. To evaluate angiogenesis, the hemoglobin content of the removed Matrigel plug was measured with the Quanti Chrom Hemoglobin Assay Kit (BioAssay Systems, DIHB-250).
Western blot analysis
Cell lysates were prepared in RIPA buffer (Beyotime) containing protease inhibitors and were separated by SDS polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Incubation with primary antibodies recognizing p16 (CST, USA), p21 (CST, USA), pRb (CST, USA), GNAQ (Abcam, USA), PLK2 (CST, USA), tubulin, and β-actin (Santa Cruz, USA) was followed by incubation with the appropriate secondary antibodies conjugated to horseradish peroxidase (HRP) (Millipore, Billerica, MA, USA). Signals were developed with enhanced chemiluminescence.
circRNA in vivo precipitation
Biotin-labeled circGNAQ and control probes were synthesized by Invitrogen. The circRIP assay was performed according to the reported literature with minor alterations.19,49 circGNAQ-overexpressing HUVECs were seeded in a 10-cm dish at a density that allowed them to grow for 48 h without reaching complete confluence. Then, the cells were washed with ice-cold PBS, fixed by 1% formaldehyde for 10 min, lysed, and sonicated. After centrifugation, 50 μL of the supernatant was retained as input and the remaining part was incubated with a circGNAQ-specific probe-streptavidin Dynabeads mixture for 3 h at 37°C. Subsequently, the probe-Dynabeads-circRNA mixture was washed and incubated with 200 mL of lysis buffer and proteinase K to reverse the formaldehyde crosslinking. Finally, RNA was extracted from the mixture with TRIzol reagent (Invitrogen). Purified mRNA and miRNA were detected by qRT-PCR assay using the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, USA).
Biotin-labeled RNA pull-down
The RNA pull-down assay was conducted as previously described.50,51 Briefly, the circGNAQ probe was synthesized and labeled with Biotin RNA Labeling Mix (Roche) by in vitro transcription. The biotin-labeled RNA was first folded in RNA structure buffer and then incubated with HUVEC whole-cell lysate at 4°C for 1 h with rotation. After incubation, RNA-protein complexes were retrieved by streptavidin-coupled T1 beads (Dynabeads), washed five times in immunoprecipitation (IP) buffer, and eluted in Laemmli buffer. The binding proteins were separated by SDS-PAGE and visualized by silver staining.
Dual-luciferase reporter assay
The sequences of circGNAQ and the corresponding mutant versions without miR-146a-5p binding sites were synthesized and subcloned into luciferase reporter vector pEZX-GA02 (GeneCopoeia, USA), termed circGNAQ-WT and circGNAQ-Mut, respectively. At 24 h before transfection, HEK293T cells were seeded in 24-well plates at a density of 1 × 105. The cells were co-transfected with 500 ng of reporter vectors and 50 nmol of miRNA mimic. After 48 h, the luciferase activity was measured with the Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia, USA). For each assay, one internal control (secreted alkaline phosphatase, SEAP) and one negative control (miRNA negative control) were used. To take into account the transfection efficiency variability, the GLuc activity was first normalized to the SEAP activity. The fold change was determined by comparing the activity of the miRNA mimic to that of the miRNA negative control.
RNA immunoprecipitation
The Magna RIP Kit (Millipore, USA) was used according to the manufacturer’s protocol. HUVECs were lysed in complete RIP lysis buffer, and the cell extract was incubated with magnetic beads conjugated with anti-Argonaute 2 (Ago2, Abcam, USA) or control anti-IgG antibody (Millipore, USA) for 6 h at 4°C. The beads were washed and incubated with proteinase K to remove proteins. Finally, purified RNA was subjected to qRT-PCR analysis.
RNA fluorescent in situ hybridization
Cy3-labeled locked nucleic acid miR-146a-5p probe and FAM-labeled circGNAQ probe were designed and synthesized by GenePharma (Suzhou, China). The signals of the probes were detected with a Fluorescent In Situ Hybridization Kit (GenePharma, China) according to the manufacturer’s instructions. The images were acquired with a Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments, Japan).
Mouse experiments
Animal studies were approved by the Animal Care Committee of Guangdong Medical University and conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, 8th Edition, 2011). C57BL/6 mice (male, 6–8 weeks) were purchased from the Laboratory Animal Centre of Southern Medical University (Guangzhou, China). Six-week-old male low-density lipoprotein receptor knockout (Ldlr−/−) mice, which have been widely used to investigate the mechanisms of atherogenesis,52 were purchased from GemPharmatech (Nanjing, Jiangsu, China) and then randomized into 2 groups to form groups with equivalent mean body weights. Recombinant AAV carrying circGNAQ and empty vector with the mouse endothelial specific promoter Tie2 (AAV-Tie2-circGNAQ and AAV-Tie2-empty) were manufactured by Hanbio Biotechnology (Shanghai, China). AAV-Tie2-empty served as negative control. AAV-Tie2-circGNAQ/empty vectors (1.6 × 1011 vector genomes [vg]/mice) were delivered by intravenous injection. Mice were subsequently maintained on a HFD (Guangdong Medical Lab Animal Center, China) for 12 weeks. Aortas were carefully excised from mice and examined for immunohistology and characterization of atherosclerotic lesions.
Intimal RNA isolation from aorta tissue
Isolation of intimal RNA from aorta was modified from a previous study.53 In brief, the mice were sacrificed and aortas were isolated and cleaned of peri-adventitial tissue. After being flushed with PBS, the lumen of aorta was flushed with TRIzol by a 29-gauge insulin syringe into a microfuge tube. The TRIzol elute was used for RNA extraction to determine the gene expression in vascular ECs. The aorta left over after flushing with TRIzol was used to prepare RNA from M+A.
Atherosclerotic lesion analysis
The hearts and aortas were then perfused with PBS through the left ventricle. The hearts were embedded in OCT compound (Sakura Finetek, 4583). The aortas were dissected from the proximal ascending aorta to the bifurcation of the iliac artery, and adventitial fat was removed. For en face analysis, the aortas were split longitudinally, pinned onto flat plates, and fixed overnight in 10% formaldehyde in PBS. Fixed aortas were stained with oil red O solution for 15 min and briefly washed with PBS. Images of the aortas were captured with a digital camera. The percentage of the luminal surface area stained by oil red O was determined and analyzed with ImageJ software. For analyzing AR plaque lesions, cryosectioning was performed. Briefly, 10-μm-thick cryostat sections, selected 80 μm apart and covering 320 μm of the sinus, were stained with oil red O and counterstained in Mayer’s hematoxylin for quantification of atherosclerosis. Images were quantitated with ImageJ software. For each mouse, lesion size (μm2) was determined from the average of 5 cross sections, and the results of each group are expressed as lesion size (μm2) ± SEM.
Immunofluorescence
Frozen tissue sections of atherosclerotic lesions were incubated with antibodies against CD31 as a marker for ECs and against p16 as a senescence marker overnight at 4°C and then stained with fluorophore-conjugated secondary antibodies, after which DAPI staining was performed and cells were observed with a confocal microscope.
Statistical analysis
Statistical analyses were performed with SPSS 19.0 (IBM, SPSS, Chicago, IL, USA) and GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA). Data are shown as mean ± standard deviation (SD) or SEM. The differences between groups were assessed with the Student’s t test. A p value < 0.05 was considered statistically significant.
Data availability
Raw and normalized data files for the RNA-seq analysis have been deposited in the NCBI Gene Expression Omnibus under accession number: GSE151475.
Acknowledgments
The study was supported by grants from the National Natural Science Foundation of China (81871120, 82071576), the Natural Science Foundation of Guangdong Province (2019A1515010334, 2019KZDXM059), the program for Training High-level Talents of Dongguan (201901019), and the discipline construction project of Guangdong Medical University (4SG21008G).
Author contributions
X.-D.X. conceived and designed the project. W.-P.W., M.-Y.Z., D.-L.L., and X.M. planned and performed the experiments. T.S., Z.-Y.X., X.J., and M.-Y.C. helped with the experiments. W.-P.W., M.-Y.Z., D.-L.L., and X.M. wrote the manuscript. S.X., X. Liang, M.M., and X. Liu helped with the revision. X.-D.X. supervised the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.020.
Supplemental information
References
- 1.Ungvari Z., Tarantini S., Donato A.J., Galvan V., Csiszar A. Mechanisms of Vascular Aging. Circ. Res. 2018;123:849–867. doi: 10.1161/CIRCRESAHA.118.311378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izzo C., Carrizzo A., Alfano A., Virtuoso N., Capunzo M., Calabrese M., De Simone E., Sciarretta S., Frati G., Oliveti M. The Impact of Aging on Cardio and Cerebrovascular Diseases. Int. J. Mol. Sci. 2018;19:481. doi: 10.3390/ijms19020481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donato A.J., Machin D.R., Lesniewski L.A. Mechanisms of Dysfunction in the Aging Vasculature and Role in Age-Related Disease. Circ. Res. 2018;123:825–848. doi: 10.1161/CIRCRESAHA.118.312563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gimbrone M.A., Jr., García-Cardeña G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossman M.J., Kaplon R.E., Hill S.D., McNamara M.N., Santos-Parker J.R., Pierce G.L., Seals D.R., Donato A.J. Endothelial cell senescence with aging in healthy humans: prevention by habitual exercise and relation to vascular endothelial function. Am. J. Physiol. Heart Circ. Physiol. 2017;313:H890–H895. doi: 10.1152/ajpheart.00416.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bierhansl L., Conradi L.C., Treps L., Dewerchin M., Carmeliet P. Central Role of Metabolism in Endothelial Cell Function and Vascular Disease. Physiology (Bethesda) 2017;32:126–140. doi: 10.1152/physiol.00031.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 8.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 9.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 11.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 12.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L., Zhang R.Y., Wang X.Q., Liu Z.H., Shen Y., Ding F.H., Meng H., Wang L.J., Yan X.X., Yang K. C1q/TNF-related protein-1: an adipokine marking and promoting atherosclerosis. Eur. Heart J. 2016;37:1762–1771. doi: 10.1093/eurheartj/ehv649. [DOI] [PubMed] [Google Scholar]
- 15.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 16.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 20.Liu G., Huang K., Jie Z., Wu Y., Chen J., Chen Z., Fang X., Shen S. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol. Cancer. 2018;17:170. doi: 10.1186/s12943-018-0917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han B., Zhang Y., Zhang Y., Bai Y., Chen X., Huang R., Wu F., Leng S., Chao J., Zhang J.H. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Du L., Bai Y., Han B., He C., Gong L., Huang R., Shen L., Chao J., Liu P. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry. 2020;25:1175–1190. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating miRNA-145 Function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 24.Piao L., Zhao G., Zhu E., Inoue A., Shibata R., Lei Y., Hu L., Yu C., Yang G., Wu H. Chronic Psychological Stress Accelerates Vascular Senescence and Impairs Ischemia-Induced Neovascularization: The Role of Dipeptidyl Peptidase-4/Glucagon-Like Peptide-1-Adiponectin Axis. J. Am. Heart Assoc. 2017;6:e006421. doi: 10.1161/JAHA.117.006421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng S., Wang H., Jia C., Zhu S., Chu X., Ma Q., Wei J., Chen E., Zhu W., Macon C.J. MicroRNA-146a Induces Lineage-Negative Bone Marrow Cell Apoptosis and Senescence by Targeting Polo-Like Kinase 2 Expression. Arterioscler. Thromb. Vasc. Biol. 2017;37:280–290. doi: 10.1161/ATVBAHA.116.308378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T.N., Tozawa Y., Deutsch U., Wolburg-Buchholz K., Fujiwara Y., Gendron-Maguire M., Gridley T., Wolburg H., Risau W., Qin Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 28.Korhonen J., Lahtinen I., Halmekytö M., Alhonen L., Jänne J., Dumont D., Alitalo K. Endothelial-specific gene expression directed by the tie gene promoter in vivo. Blood. 1995;86:1828–1835. [PubMed] [Google Scholar]
- 29.Sun S., Qin W., Tang X., Meng Y., Hu W., Zhang S., Qian M., Liu Z., Cao X., Pang Q. Vascular endothelium-targeted Sirt7 gene therapy rejuvenates blood vessels and extends life span in a Hutchinson-Gilford progeria model. Sci. Adv. 2020;6:eaay5556. doi: 10.1126/sciadv.aay5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panda A.C., Grammatikakis I., Kim K.M., De S., Martindale J.L., Munk R., Yang X., Abdelmohsen K., Gorospe M. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017;45:4021–4035. doi: 10.1093/nar/gkw1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C., Ge H.M., Liu B.H., Dong R., Shan K., Chen X., Yao M.D., Li X.M., Yao J., Zhou R.M. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc. Natl. Acad. Sci. USA. 2019;116:7455–7464. doi: 10.1073/pnas.1814874116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z.G., Awan F.M., Du W.W., Zeng Y., Lyu J., Wu D., Gupta S., Yang W., Yang B.B. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol. Ther. 2017;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao T., Pan Y.H., Xiong X.D. Circular RNA: an important player with multiple facets to regulate its parental gene expression. Mol. Ther. Nucleic Acids. 2020;23:369–376. doi: 10.1016/j.omtn.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivieri F., Lazzarini R., Recchioni R., Marcheselli F., Rippo M.R., Di Nuzzo S., Albertini M.C., Graciotti L., Babini L., Mariotti S. MiR-146a as marker of senescence-associated pro-inflammatory status in cells involved in vascular remodelling. Age (Dordr.) 2013;35:1157–1172. doi: 10.1007/s11357-012-9440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olivieri F., Lazzarini R., Babini L., Prattichizzo F., Rippo M.R., Tiano L., Di Nuzzo S., Graciotti L., Festa R., Brugè F. Anti-inflammatory effect of ubiquinol-10 on young and senescent endothelial cells via miR-146a modulation. Free Radic. Biol. Med. 2013;63:410–420. doi: 10.1016/j.freeradbiomed.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 36.Bhaumik D., Scott G.K., Schokrpur S., Patil C.K., Orjalo A.V., Rodier F., Lithgow G.J., Campisi J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 2009;1:402–411. doi: 10.18632/aging.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnke S., Kemmler S., Hames R.S., Tsai H.L., Hoffmann-Rohrer U., Fry A.M., Hoffmann I. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2004;14:1200–1207. doi: 10.1016/j.cub.2004.06.059. [DOI] [PubMed] [Google Scholar]
- 38.Yang H., Fang L., Zhan R., Hegarty J.M., Ren J., Hsiai T.K., Gleeson J.G., Miller Y.I., Trejo J., Chi N.C. Polo-like kinase 2 regulates angiogenic sprouting and blood vessel development. Dev. Biol. 2015;404:49–60. doi: 10.1016/j.ydbio.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y., Yang Z., Zheng B., Zhang X.H., Zhang M.L., Zhao X.S., Zhao H.Y., Suzuki T., Wen J.K. A Novel Regulatory Mechanism of Smooth Muscle α-Actin Expression by NRG-1/circACTA2/miR-548f-5p Axis. Circ. Res. 2017;121:628–635. doi: 10.1161/CIRCRESAHA.117.311441. [DOI] [PubMed] [Google Scholar]
- 41.Zeng Z., Xia L., Fan S., Zheng J., Qin J., Fan X., Liu Y., Tao J., Liu Y., Li K. Circular RNA CircMAP3K5 Acts as a MicroRNA-22-3p Sponge to Promote Resolution of Intimal Hyperplasia Via TET2-Mediated Smooth Muscle Cell Differentiation. Circulation. 2021;143:354–371. doi: 10.1161/CIRCULATIONAHA.120.049715. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S., Zhu D., Li H., Li H., Feng C., Zhang W. Characterization of circRNA-Associated-ceRNA Networks in a Senescence-Accelerated Mouse Prone 8 Brain. Mol. Ther. 2017;25:2053–2061. doi: 10.1016/j.ymthe.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K., Zhang Y., Xiong W., Zhang Z., Wang Z., Lv L., Liu C., Hu Z., Zheng Y.T., Lu L. CircGRIA1 shows an age-related increase in male macaque brain and regulates synaptic plasticity and synaptogenesis. Nat. Commun. 2020;11:3594. doi: 10.1038/s41467-020-17435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muniz L., Lazorthes S., Delmas M., Ouvrard J., Aguirrebengoa M., Trouche D., Nicolas E. Circular ANRIL isoforms switch from repressors to activators of p15/CDKN2B expression during RAF1 oncogene-induced senescence. RNA Biol. 2021;18:404–420. doi: 10.1080/15476286.2020.1812910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minamino T., Komuro I. Vascular cell senescence: contribution to atherosclerosis. Circ. Res. 2007;100:15–26. doi: 10.1161/01.RES.0000256837.40544.4a. [DOI] [PubMed] [Google Scholar]
- 46.Wang J.C., Bennett M. Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ. Res. 2012;111:245–259. doi: 10.1161/CIRCRESAHA.111.261388. [DOI] [PubMed] [Google Scholar]
- 47.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Itahana K., Campisi J., Dimri G.P. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol. Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- 49.Su X., Wang H., Ge W., Yang M., Hou J., Chen T., Li N., Cao X. An In Vivo Method to Identify microRNA Targets Not Predicted by Computation Algorithms: p21 Targeting by miR-92a in Cancer. Cancer Res. 2015;75:2875–2885. doi: 10.1158/0008-5472.CAN-14-2218. [DOI] [PubMed] [Google Scholar]
- 50.Lan Y., Xiao X., He Z., Luo Y., Wu C., Li L., Song X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46:5809–5821. doi: 10.1093/nar/gky214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong C.H., Lou U.K., Li Y., Chan S.L., Tong J.H., To K.F., Chen Y. CircFOXK2 Promotes Growth and Metastasis of Pancreatic Ductal Adenocarcinoma by Complexing with RNA-Binding Proteins and Sponging MiR-942. Cancer Res. 2020;80:2138–2149. doi: 10.1158/0008-5472.CAN-19-3268. [DOI] [PubMed] [Google Scholar]
- 52.Loyer X., Potteaux S., Vion A.C., Guérin C.L., Boulkroun S., Rautou P.E., Ramkhelawon B., Esposito B., Dalloz M., Paul J.L. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ. Res. 2014;114:434–443. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 53.Nam D., Ni C.W., Rezvan A., Suo J., Budzyn K., Llanos A., Harrison D., Giddens D., Jo H. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H1535–H1543. doi: 10.1152/ajpheart.00510.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and normalized data files for the RNA-seq analysis have been deposited in the NCBI Gene Expression Omnibus under accession number: GSE151475.