Abstract
MicroRNAs (miRNAs) are promising drug targets for obesity and metabolic disorders. Recently, miRNA mimics are providing a unique mechanism of action that guides the process for drug development and sets out the context of their therapeutic application. miRNA (miR)-21 expression in white adipose tissue (WAT) has been associated with obesity. We aimed to analyze miR-21 expression levels in relation to diabetes and obesity to determine the effect that miR-21 mimic has on processes involved in WAT functionality, to dissect the underlying molecular mechanisms, and to study the potential therapeutic application of the miR-21 mimic against obesity. We found higher miR-21 levels in WAT from non-diabetic obese compared to normoweight humans and mice. Moreover, in 3T3-L1 adipocytes, miR-21 mimic affect genes involved in WAT functionality regulation and significantly increase the expression of genes involved in browning and thermogenesis. Interestingly, in vivo treatment with the miR-21 mimic blocked weight gain induced by a high-fat diet in obese mice, without modifying food intake or physical activity. This was associated with metabolic enhancement, WAT browning, and brown adipose tissue (AT) thermogenic programming through vascular endothelial growth factor A (VEGF-A), p53, and transforming growth factor β1 (TGF-β1) signaling pathways. Our findings suggest that miR-21 mimic-based therapy may provide a new opportunity to therapeutically manage obesity and consequently, its associated alterations.
Keywords: obesity, miR-21, adipose tissue, thermogenesis, browning, brown adipose tissue, diabetes, metabolism, uncoupling protein 1, TMEM26
Graphical abstract
miR-21 mimic could serve as a potential therapeutic tool to control obesity by delaying energy storage and promoting energy expenditure in adipose tissue.
Introduction
Obesity is gradually becoming a global health epidemic. It is commonly associated with different diseases, including diabetes, cardiovascular diseases, and cancer, compromising the quality of life and putting an enormous economic burden on society.1 To date, there are no efficient therapies for the treatment of obesity.2,3
microRNAs (miRNAs) have arisen as potential therapeutic targets due to their regulatory role in many biological processes, including the transcriptional regulation of metabolism.4 In fact, dysregulation of many miRNAs in metabolic tissues such as liver, muscle, and adipose tissue (AT) has been described to contribute to the development of obesity comorbidities.4, 5, 6 miRNAs are single-stranded, small non-coding RNAs that bind to mRNAs to repress gene expression of target genes (TGs). In brief, miRNAs are synthetized as double-stranded precursors that are loaded into the RNA-induced silencing complex (RISC), which removes the passenger strand and uses the guide strand to target mRNAs.7 miRNA mimics, artificial miRNA duplexes that mimic endogenous miRNAs, are used as a strategy to gain the function of a specific miRNA, which is also known as miRNA replacement therapy.8 miRNA mimics directly join the RISC to interact with their respective mRNA targets and produce an effect on the expression of the genes targeted by the original miRNA that it is mimicking.9 miRNA mimics are being explored as potential therapeutic tools. In fact, some miRNA mimics have already entered clinical testing for cancer treatment.10,11 Thus, the administration of miRNA mimics might also be a new avenue for the management of obesity and metabolic disorders.
It is well established that white AT (WAT) dysfunction leads to the development of obesity-related metabolic disturbances.12,13 Given the existence of obese subjects without metabolic disturbances and diabetic lean individuals, it has been suggested that when the expansion capacity of AT required to store excess energy is exceeded, metabolic disorders occur.14 In this case, WAT dysfunction results in the accumulation of fat in non-fatty organs, such as muscles, pancreas, liver, and the heart, leading to lipotoxicity and impaired insulin signaling.13,15
It is well documented that obesity is driven by a systemic energy excess stored as lipids in white adipocytes, causing ectopic and excessive deposition of WAT. To shift the energy balance for obesity treatment, strategies pointing at decreasing energy intake manifest modest efficacy as well as harmful side effects.2,3 In contrast, strategies pointing at energy expenditure have sparked enormous interest, mainly due to the newly discovered brown and beige adipocytes in adult humans.16
In addition to WAT, brown AT (BAT) and beige AT, mitochondria-rich tissues, are involved in weight management and metabolic homeostasis due to their high energy-utilizing capacity through thermogenesis.16,17 Beige thermogenic adipocytes arise in WAT depots after continuous exposure to cold, β3-adrenergic stimulation or by genetic manipulation of certain specific pathways. This process is known as browning,18 which can be induced pharmacologically by activation of the β3-adrenergic receptor and thiazolidinediones.18 Interestingly, rodent models have shown that WAT browning was able to promote metabolic improvement and resistance to diet-induced obesity (DIO).19
Many miRNAs have been defined as regulators of the differentiation and function of beige AT and BAT.20 Understanding the role of miRNAs in the thermogenic activation of BAT and the browning of WAT can provide new therapeutic targets against obesity and associated metabolic diseases.
miRNA (miR)-21 was found to be frequently upregulated in many chronic diseases, such as obesity.21,22 miR-21 is upregulated in epididymal WAT from obese mice compared to normoweights (NWs)22 and in type 2 diabetic (T2D) obese compared to non-diabetic obese subjects.23 miR-21 enhances adipogenic differentiation through the modulation of transforming growth factor β (TGF-β) signaling24,25 and plays a pivotal role in angiogenesis through the regulation of vascular endothelial growth factor A (VEGF-A), which is in turn known to be a regulator of thermogenesis.26 However, the precise mechanisms underlying its relationship with obesity and T2D or insulin resistance (IR) are yet to be described.
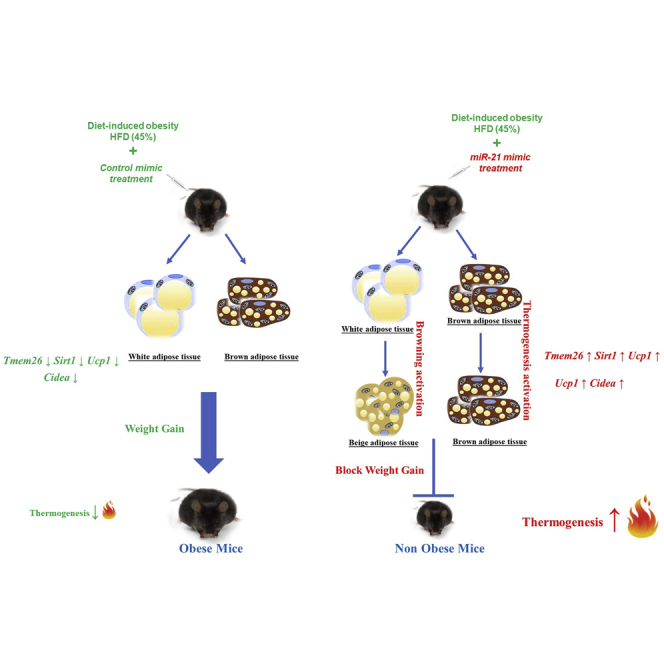
In the present work, we aimed at analyzing the expression levels of miR-21 in relation to obesity and the glycemic status in both humans and mice. Once determined that miR-21 is exclusively overexpressed in non-diabetic obese (but not in diabetic obese) compared to NW subjects and mice, we performed in vitro and in vivo experimental approaches with the miR-21 mimic to elucidate the molecular pathways related to adipose functionality that can be triggered by miR-21 upregulation. Our results indicate that the miR-21 mimic regulates processes related to WAT functionality, including angiogenesis and apoptosis, and activates the thermogenic program in BAT and browning in different WAT depots, which suggests an increase in adipose energy expenditure. Concordantly, miR-21 mimic administration also delayed weight gain in miR-21 mimic-treated mice under a high-fat diet (HFD) compared to their control littermates fed the same diet. All of this evidence places miR-21 mimic as a potential therapeutic tool to modulate adipose function and to control obesity.
Results
Visceral and subcutaneous WAT miR-21 expression is enhanced in obese patients and mice without T2D or IR
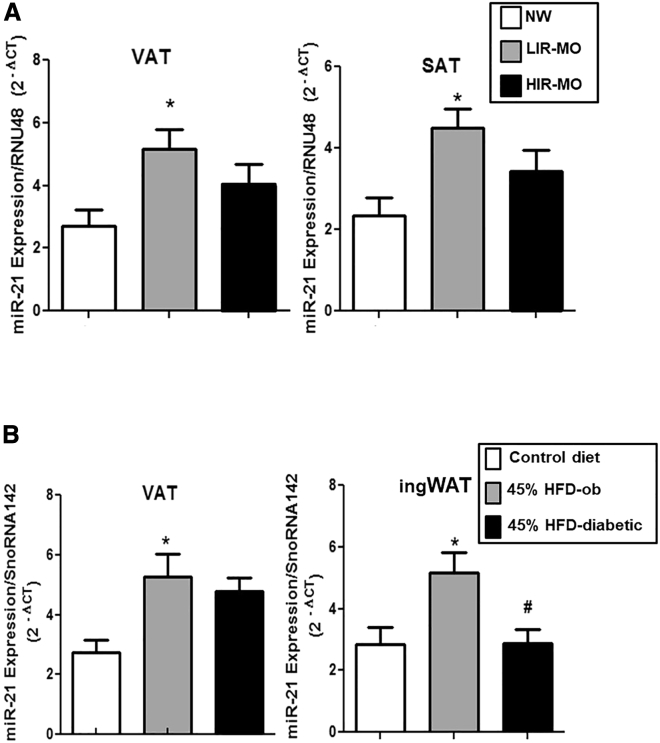
miR-21 expression levels were analyzed in both visceral AT (VAT) and subcutaneous AT (SAT) from obese humans and mice with and without T2D or IR. VAT and SAT miR-21 expression levels were significantly higher in the morbidly obese (MO) group with a low degree of IR (LIR-MO) compared to healthy normoglycemic NW individuals. Although a moderate downward trend was seen when compared with LIR-MO, no significant differences were detected in VAT or SAT from MO subjects with a high degree of IR (HIR-MO) in comparison with healthy NW or LIR-MO (Figure 1A). The C57BL/6J strain of mice (susceptible to developing obesity and metabolic disturbances after HFD feeding for several weeks) was used to mimic human obesity and diabetes.27 The study was carried out in three groups of mice: mice fed with control diet (control), non-diabetic obese mice (45% HFD for 8 weeks; 45% HFD-ob), and obese diabetic mice (45% HFD for 14 weeks; 45% HFD-diabetic) (Figure S1A). At 60 days, the body weight gain of mice fed 45% HFD groups began to be significantly greater than that of control mice (Figure S1B). At the end of the study, the HFD-diabetic group showed significant glucose and insulin intolerance compared to the control group (Figure S1C). Similar to human results, miR-21 expression levels in VAT and inguinal (ing)WAT from 45% HFD-ob mice were significantly higher compared to control mice. Notably, miR-21 levels in ingWAT from 45% HFD-diabetic mice were significantly lower compared to 45% HFD-ob mice (Figure 1B).
Figure 1.
miR-21 expression levels in VAT and SAT from humans and mice
miR-21 expression was measured in (A) VAT and SAT from non-diabetic normoweight (NW) subjects (n = 7), normoglycemic morbidly obese (MO) subjects with a low degree of insulin resistance (IR; LIR-MO; n = 9), and MO subjects with a high degree of IR (HIR-MO; n = 9) and in (B) VAT and inguinal white adipose tissue (AT; ingWAT) from C57BL/6J mice fed a low-fat diet (10% kcal fat; control diet; n = 9), non-diabetic obese mice fed a high-fat diet (HFD; 45% kcal fat) during 8 weeks (45% HFD-ob; n = 8), and diabetic obese mice fed a HFD (45% kcal fat) during 14 weeks (45% HFD-diabetic; n = 8) by quantitative real-time PCR using RNU48 (human) and SnoRNA142 (mice) as reference genes to calculate 2−Δct according to the manufacturer’s indications. Data are expressed as the mean ± SEM. ∗p ˂ 0.05 versus NW or control; #p ˂ 0.05 versus 45% HFD-ob according to one-way ANOVA with Bonferroni’s correction.
In silico analysis of miR-21 potential targets
The results described above (Figure 1) suggest that miR-21 could be involved in AT functionality regulation. In this line, Database for Annotation, Visualization and Integrated Discovery (DAVID) and Protein Analysis through Evolutionary Relationships (PANTHER) Classification System studies show that miR-21 is involved in browning, angiogenesis, apoptosis, and adipogenesis processes (Tables S1 and S2), which are well known to be related to AT functionality and then to obesity and associated metabolic alterations.28,29
The effect of miR-21 mimic on the expression levels of genes involved in angiogenesis, apoptosis, thermogenic, and browning processes in 3T3-L1-differentiated adipocytes
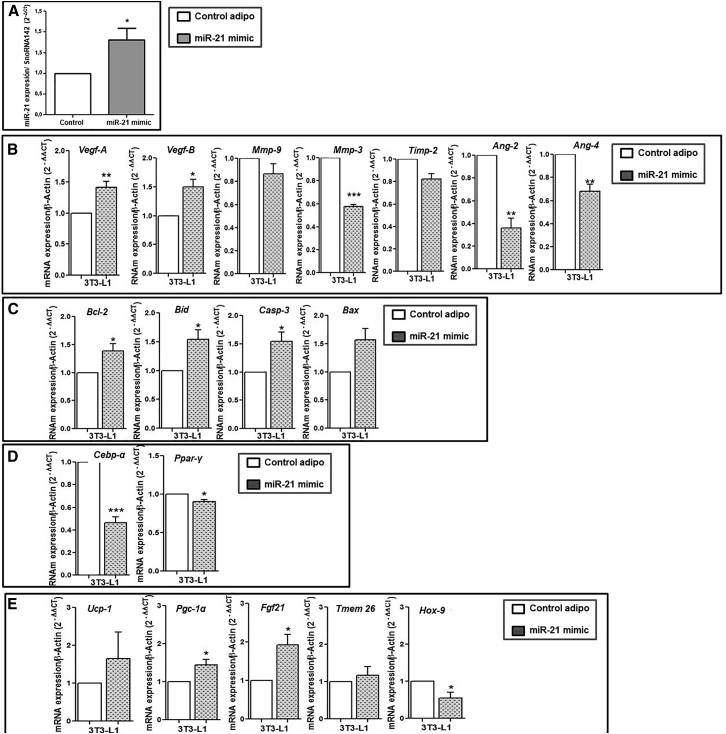
To better understand the relationship of miR-21 with the processes that control AT functionality, we transfected 3T3-L1-differentiated adipocytes with the miR-21 mimic to check its effects on the in silico-predicted miR-21 target genes. In vitro treatment of 3T3-L1 cells with the miR-21 mimic was validated by the significant increase of miR-21 levels in treated cells compared to controls (Figure 2A). miR-21 mimic led to a significant increase in angiogenic genes, mainly Vegf-A and Vegf-B, and decreased Ang-2, Ang-4, and the anti-angiogenic Timp-3 genes compared to control cells (Figure 2B). Moreover, the miR-21 mimic led to a significant increase of both the anti-apoptotic Bcl-2 and the pro-apoptotic Casp-3 and Bid, compared to controls (Figure 2C). However, both Ppar-γ and Cebp-α gene expressions were significantly decreased with miR-21 mimic treatment compared to control (Figure 2D), whereas genes involved in BAT differentiation, thermogenesis, and browning, mainly Ucp1, Fgf21, Pgc-1α, and Tmem26, showed an increase with mimic treatment compared to controls. Moreover, the expression of Hoxc-9 decreased with mimic treatment (Figure 2E).
Figure 2.
In vitro effect of the miR-21 mimic on the mRNA expression of angiogenic, apoptotic, adipogenic, browning, and thermogenesis markers in 3T3-L1 adipocytes
Differentiated 3T3-L1 cells were treated with miR-21 mimic 5 nM or control mimic (n = 6 per group) for 48 h. (A) miR-21 levels measured by quantitative real-time PCR using SnoRNA142 as reference miRNA (2−ΔΔct). Data are expressed as the mean ± SEM. ∗p < 0.05 miR-21 mimic versus control according to the Student’s t test. The expression levels of miR-21 were also measured in cell cultures in the presence of control mimic and transfect, and no differences were observed compared to those obtained in control adipo (3T3-L1 differentiated into adipocytes) (data not shown). mRNA levels of angiogenic (B), apoptotic (C), adipogenic (D), and browning and thermogenesis (E) genes were measured by quantitative real-time PCR using β-actin as a reference gene (2−ΔΔct). ∗p ˂ 0.05 and ∗∗p ˂ 0.01 versus control mimic. Data are expressed as the mean ± SEM. Student’s t test was used for statistical analysis.
In vivo sustained treatment with the miR-21 mimic decelerated weight gain without affecting glucose and insulin tolerance in HFD obese mice
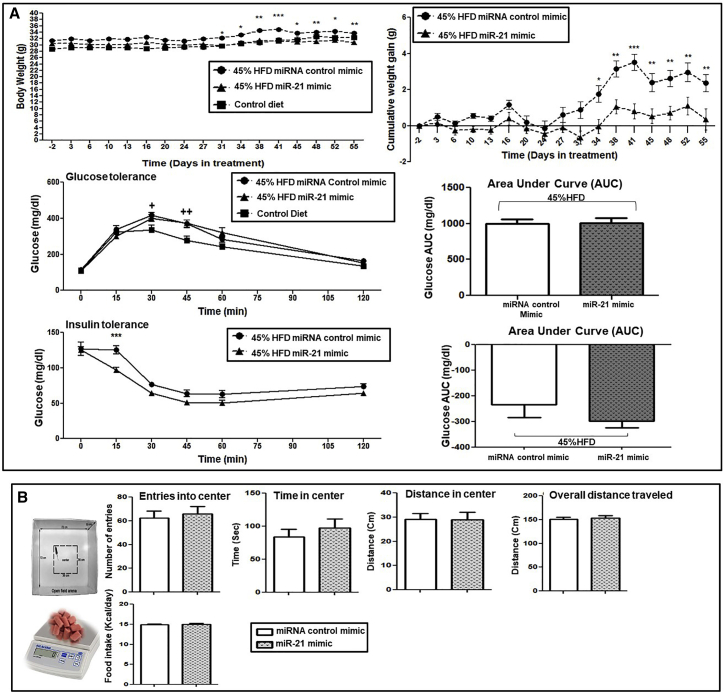
One of the most relevant findings of our in vitro study on 3T3-L1 adipocytes is that the transfection with the miR-21 mimic induces the expression of genes related to the thermogenic program and WAT browning. WAT browning and thermogenesis activation have been proposed as a therapeutic option for manage obesity.30 Subsequently, we aimed at analyzing these processes in an animal model of obesity treated with the miR-21 mimic. For this purpose, we treated HFD-induced obese mice with the miR-21 mimic (Figure 3). To be more precise, the C57BL/6J strain of mice (susceptible to developing obesity and metabolic disturbances after HFD feeding for several weeks, mimicking human obesity and diabetes27) was used as detailed below. We treated C57BL/6J obese mice (45% HFD) with 0.5 μg of miR-21 mimic or its corresponding miRNA control mimic for an additional period of 8 weeks under HFD (Figure 3). HFD-induced weight and glucose changes previous to miR-21 administration are depicted in Figure S2. The administration of the miR-21 mimic led to a significant deceleration of 45% HFD-induced weight gain compared to the miRNA control mimic (Figure 4A). As illustrated in the body weight and cumulative weight gain graphs, the group treated with the miR-21 mimic did not significantly gain weight upon miR-21 administration, whereas mice treated with the mimic control did. However, no significant amelioration was observed regarding insulin and glucose tolerance with the treatment with the in vivo miR-21 mimic (Figure 4A). No changes were observed with vehicle (jetPEI) treatment compared to the control (data not shown).
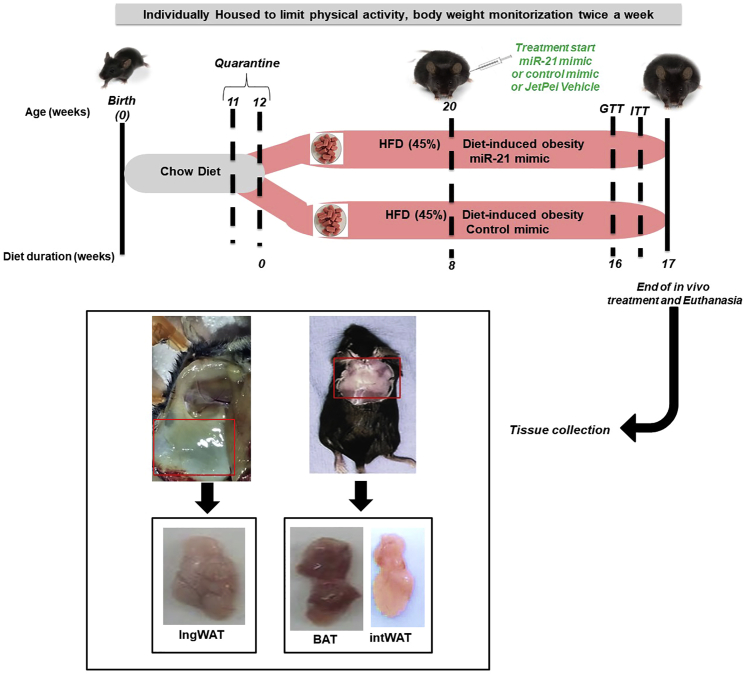
Figure 3.
Study design and workflow for in vivo miR-21 mimic treatment of HFD-induced obese mice
C57BL/6J mice (11 weeks old at arrival) were fed with chow diet for 1 week (quarantine period) and then switched to a HFD (45% kcal from saturated fat) from day 0 (start of the experiment) for 17 weeks. Twice a week, body weight was monitored in awake mice. During these 17 weeks of 45% HFD diet, after the first 8 weeks, mice were injected subcutaneously with miR-21 mimic (0.5 μg, n = 8) or control miRNA mimic (0.5 μg, n = 9) three times a week for further 8 weeks. Additionally, a subgroup of 45% HFD mice was injected with jetPEI (0.086 μL, n = 4) with the same timing as miR-21 mimic or control mimic (not shown in the graph). At week 16, the glucose tolerance test (GTT) was measured, and 2 days later, the insulin tolerance test (ITT) was measured. Mice were sacrificed by cervical dislocation at the end of the experiment at week 17. Then, blood samples, interscapular brown AT (BAT), interscapular WAT (intWAT), and ingWAT were collected and stored at −80°C until analysis. Mice fed a 45% HFD naive (n = 8) and treated with jetPEI (n = 4) were also included and weighed (data not shown). The control diet was taken as a reference for body weight gain and glucose tolerance in non-obese mice during the time course of the whole study.
Figure 4.
The effects of in vivo miR-21 mimic treatment on weight gain, GTT and ITT, food intake, and locomotor activity in HFD-induced obese mice
After 8 weeks of implementing 45% HFD, the mice were injected subcutaneously with 0.5 μg of miR-21 mimic or control mimic three times a week for another 8 weeks. (A) Body weight was monitored twice a week for the 16 weeks. Cumulative weight gain was calculated using the weight before the start of the treatment for each mouse as the basal point (0 g). AUC was calculated using the basal glucose levels before tolerance tests for each mouse as the basal point (0). GTT and ITT were carried out during the last week of treatment (week 16) by injecting 2 g/kg of D-glucose or 0.5 U/kg of insulin, respectively, after 10−12 h of fasting. Blood samples were collected from the tail vein at 0 (basal), 15, 30, 45, 60, and 120 min, and glucose was measured with a glucometer. (B) Open-field test (OFT) was performed to measure the locomotor activity of obese mice treated with miR-21 mimic or control mimic. Mice were moved to the experimental room and kept there for 30 min before starting the test. Locomotor activity was monitored in the open-field arena by video tracking for 10 min, and the time and distance walked in the center, entries into the center, and the overall distance traveled were measured. 5 weeks after miR-21 or control mimic treatment, a food intake test was carried out by measuring the mouse body weight and food pellets for 5 days. Daily kcal consumption was calculated based on the HFD. Data are expressed as the mean ± SEM. ∗p ˂ 0.05 versus control mimic. +p < 0.05 versus control diet. 2+p < 0.01 versus control diet. &p ˂ 0.05 versus the basal point (0). Repeated ANOVA measurements and Student’s t test were used for statistical analysis.
In vivo effect of sustained treatment with the miR-21 mimic on weight loss was independent of locomotor activity or food-intake changes in HFD obese mice
To understand whether miR-21-induced weight gain deceleration was due to a change in locomotor activity or food intake, we performed two tests: the open-field test (OFT) and the intake test. The OFT showed that there were no differences in the number of entrances to the central zone, in the time spent in the central zone, in the distance traveled in the central zone, or the total distance traveled, indicating that no significant differences were recorded in locomotor activity between miR-21 mimic-treated mice and control miRNA mimic-treated mice (Figure 4B). In addition, no significant difference in daily calorie intake was observed between miR-21 mimic- and control mimic-treated mice (Figure 4B).
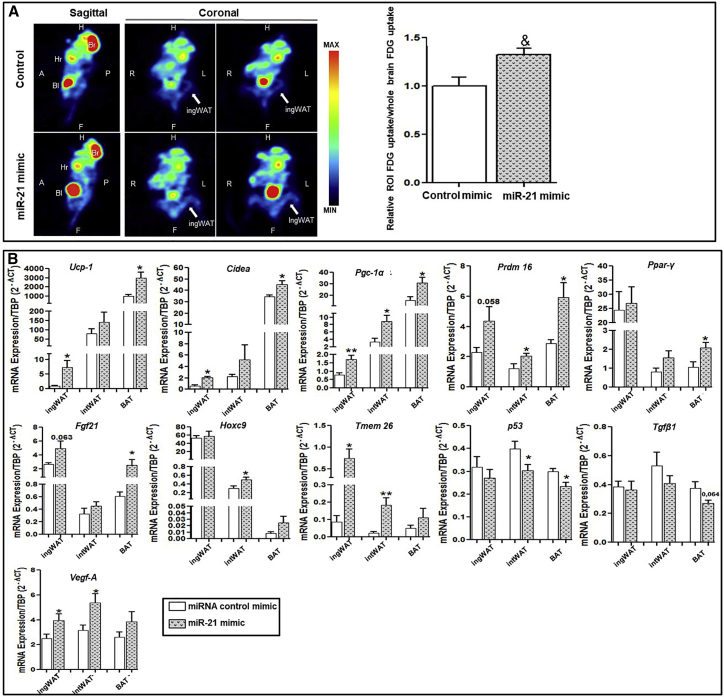
miR-21 mimic in vivo treatment-induced thermogenesis and 18F-fluorodeoxyglucose ([18F]-FDG) uptake in ingWAT
45% HFD mice, both under treatment with miR-21 mimic and control mimic, underwent18 [18F]-FDG positron emission tomography-computed tomography (PET-CT). [18F]-FDG PET-CT unveiled a significant increase in [18F]-FDG uptake in the ingWAT of miR-21 mimic-treated mice compared to control miRNA mimic-treated mice (Figure 5A). As it is well described that subcutaneous WAT adipocytes of rodents are prone to the conversion into thermogenic beige adipocytes and higher expression of uncoupling protein 1 (UCP1) and other brown fat cell markers in response to various stimuli, we further examined the expression levels of thermogenic and browning-specific genes in WAT and BAT upon miR-21 treatment. Thermogenic markers (Pgc-1α, Ucp1, Cidea, Fgf21, and Prdm16) and beige adipocyte-specific markers (Tmem26 and Hoxc9) were analyzed in ingWAT, interscapular WAT (intWAT), and BAT. Consistent with the increased [18F]-FDG intake observed in miR-21 mimic-treated mice, Ucp1, Cidea, Pgc-1α, Prdm16, and Fgf21 gene expression showed an increase with the miR-21 mimic compared to that of the control mimic in both BAT and ingWAT (Figure 5B). Moreover, intWAT displayed specifically increased levels of Pgc-1α and Prdm16 in miR-21 mimic-treated mice compared to levels in control mimic mice. The Ppar-γ gene expression showed a significant increase in BAT from miR-21 mimic-treated mice compared to control mice, whereas no significant changes were observed in intWAT. Notably, the expression of beige adipocyte markers Hoxc9 and Tmem26 was significantly higher in ingWAT and intWAT from miR-21 mimic-treated mice compared to control mice.
Figure 5.
Positron emission tomography (PET) analysis of thermogenic activity and browning induction and AT mRNA expression analysis
(A) Quantitative analysis of ingWAT 18F-fluorodeoxyglucose ([18F]-FDG) uptake imaging from control mimic or miR-21-treated mice was performed by PET (n = 6 per group). [18F]-FDG sagittal (left) and coronal (middle and right) PET images of control mimic (top) and miR21 mimic-treated (bottom) mice are shown. The average [18F]-FDG uptake was normalized to the whole brain average [18F]-FDG uptake. PET data were analyzed using PMOD v.3.3 software. Data are expressed as mean ± SEM. &p = 0.005 according to the paired Student’s t test. White arrows indicate ingWAT. P, posterior; A, anterior; L, left; R, right; H, head; F, foot; Br, brain; Hr, heart; Bl, bladder. (B) mRNA levels of browning, thermoregulatory, and angiogenic markers, as well as Tgf-β1 and p53 from ingWAT, intWAT, and BAT from mice treated with miR-21 mimic or control mimic (n = 7 per group), were measured by quantitative real-time PCR using TATA sequence binding protein (TBP) as a reference gene (2−Δct). ∗p ˂ 0.05 versus control mimic. Data are expressed as the mean ± SEM according to the Student’s t test.
Besides having a relevant role in the modulation of angiogenesis and being validated as a gene target for miR-21, VEGF-A is well known to play an important role in the regulation of energy homeostasis and obesity and IR prevention by inducing thermogenesis in BAT and browning activation in WAT.31,32 Accordingly, our data demonstrated that the miR-21 in vivo treatment induced an increase in Vegf-A mRNA expression levels in both ingWAT and intWAT tissues compared to that in the control treatment (Figure 5B). In addition, the gene expression of Tgf-β1 and p53 (reported to inhibit the differentiation and thermogenesis in BAT, to regulate the formation of beige cells in WAT, and to be associated with obesogenic pathways, T2D and IR33,34) was significantly downregulated with miR-21 in vivo treatment compared to that in control mimic mice (Figure 5B).
In order to ascertain whether in vivo miR-21 mimic treatment increased miR-21 expression levels in the tissues of interest, the expression levels of miR-21 were analyzed in VAT, ingWAT, BAT, and plasma from miR-21 mimic and control groups. Figure S3 shows a significant increase in miR-21 levels with in vivo miR-21 mimic treatment, significantly so in VAT, ingWAT, and BAT, compared to controls, confirming that miR-21 treatment led to an increase in the levels of this miRNA in the analyzed tissues.
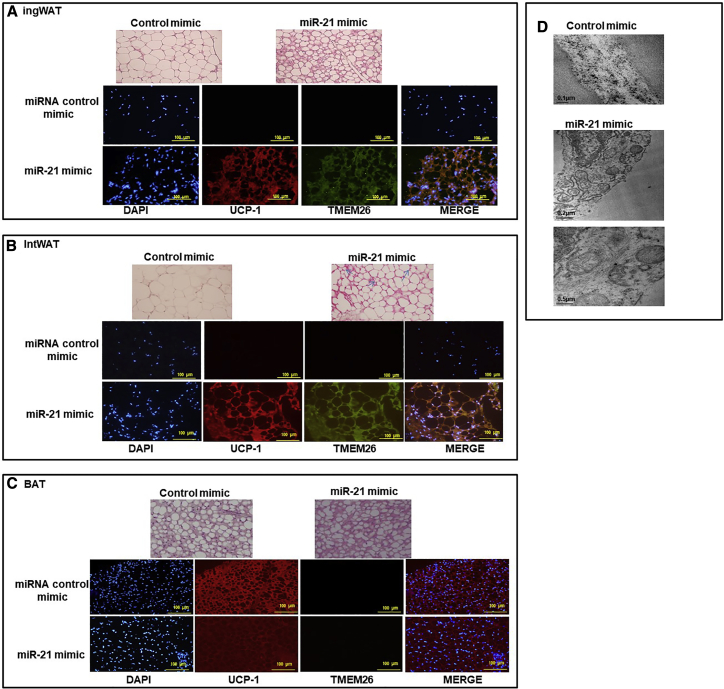
miR-21 mimic in vivo treatment induced the appearance of brown-like adipocytes with numerous small lipid droplets and mitochondria in WAT
Hematoxylin-eosin staining revealed that the intWAT and ingWAT of miR-21 mimic in vivo-treated mice accumulated a large number of multilocular adipocytes, whereas the intWAT and ingWAT from control miRNA mimic-treated mice had mostly large adipocytes with unilocular lipid inclusions (Figures 6A and 6B). Moreover, immunofluorescence staining showed that intWAT and ingWAT from miR-21 mimic in vivo-treated mice displayed a higher UCP1 and TMEM26 protein expression compared to control mimic. A co-expression of both TMEM26 and UCP1 in both ingWAT and intWAT from miR-21 mimic-treated mice and control mice was detected (merged image; Figure 6B). In BAT from miR-21 mimic-treated mice, multilocular adipocytes were present. In addition, immunostaining showed that UCP1 displayed a clear expression signal in both mice groups, whereas no signal corresponding to TMEM26 was detected (Figure 6C). Moreover, transmission electron microscopy (TEM) was used to compare the ultrastructure of the multilocular adipocytes and their mitochondria from miR-21 mimic ingWAT with that from control miRNA mimic ingWAT. As shown in Figure 6D, the presence of numerous mitochondria are larger in ingWAT from miR-21 mimic-treated mice compared to control mice.
Figure 6.
Histological analysis of WAT and BAT, confocal imaging of browning and thermogenic proteins, and transmission electron microscopy (TEM) of white adipocytes
After in vivo treatment with miR-21 mimic or control mimic, ingWAT (A), intWAT (B), and BAT (C) samples were stained by hematoxylin and eosin (×20). ingWAT (A), intWAT (B), and BAT (C) were immunostained with goat anti-UCP1 (red), rabbit anti-TMEM26 (green), and nuclei stained with DAPI (blue). Images were visualized by confocal microscopy (×20). (D) TEM images of cell organelles from ingWAT of mice treated with miR-21 mimic or control mimic. The micrographs are presented with scale bars of 0.5 μm and 0.2 μm.
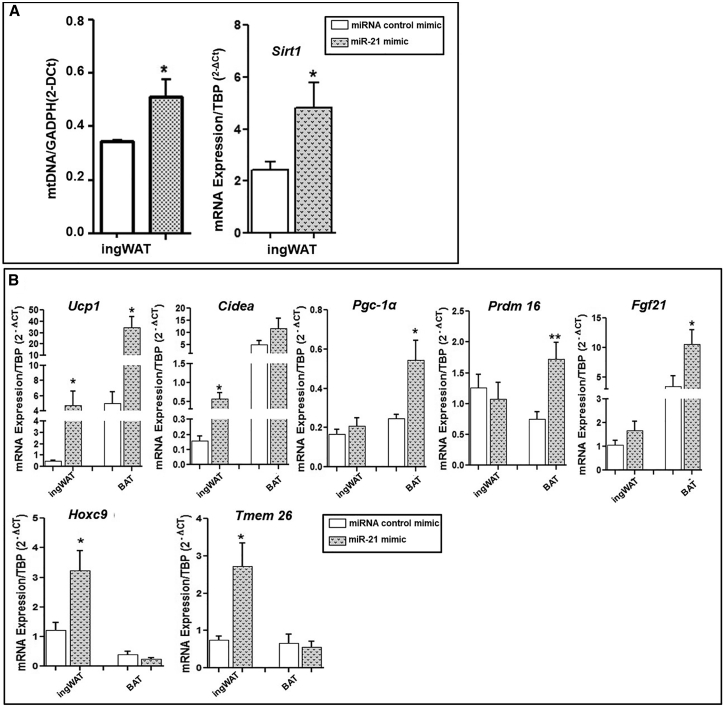
miR-21 mimic in vivo treatment increased mitochondrial DNA (mtDNA) copy number and sirtuin 1 (Sirt1) mRNA expression levels in ingWAT
We further confirmed the increase in the number of mitochondria in ingWAT from miR-21 mimic-treated mice compared to controls by quantifying the copy number of the mtDNA, which reflects the abundance of mitochondria within a cell (Figure 7A). We also analyzed Sirt1 mRNA expression, as it is well known that Sirt1 is a protein that plays a pivotal role in promoting mitochondrial biogenesis and metabolic control by means of PGC-1α deacetylation.35 In agreement with our results of mtDNA copy number and microscopy analysis, in vivo miR-21 mimic treatment induced a significant increase in Sirt1 expression levels in ingWAT compared to the control mimic (Figure 7A).
Figure 7.
In vivo effect of miR-21 mimic on mtDNA and Sirtuin 1 (Sirt1) expression levels and in vitro effect of miR-21 mimic on the mRNA expression of browning and thermogenesis markers of AT explants
(A) After in vivo treatment with miR-21 mimic or control mimic, Cox1 as a mitochondrial gene and Gapdh were measured by quantitative real-time PCR, and the relative mtDNA content was calculated the formula: mtDNA = 2−Δct, where ΔCt = Ct Cox1 − Ct Gapdh. Sirt1 mRNA expression was measured from the ingWAT of mice treated with miR-21 mimic or control mimic by quantitative real-time PCR using TBP as a reference gene (2−Δct). Data are expressed as the mean ± SEM. ∗p ˂ 0.05 versus control mimic according to the Student’s t test. (B) Fresh BAT and ingWAT explants from mice fed a control diet (10% kcal fat) were incubated for 48 h at 37°C with 5 nM miR-21 mimic or control mimic (n = 7). The mRNA expression of browning and thermoregulatory markers was measured by quantitative real-time PCR using TBP as a reference gene (2−Δct). Data are expressed as the mean ± SEM. ∗p ˂ 0.05 and ∗∗p ˂ 0.01 versus control mimic according to the Student’s t test.
Ex vivo treatment with miR-21 mimic induced thermogenic and browning gene expression in ingWAT and BAT mice explants
The effects of miR-21 mimic were analyzed in vitro in AT explants from mice fed with control diet (Figure 7B). ingWAT explants in vitro treated with miR-21 mimic displayed a significant increase in Ucp1, Cidea, Hoxc9, and Tmem26 gene expression compared to explants treated with control mimic. A significant increase in Ucp1 and Prdm16 gene expression in BAT explants upon miR-21 mimic treatment compared to control mimic was also detected, confirming the thermogenesis activation and browning induction related to the in vivo miR-21 treatment in mice.
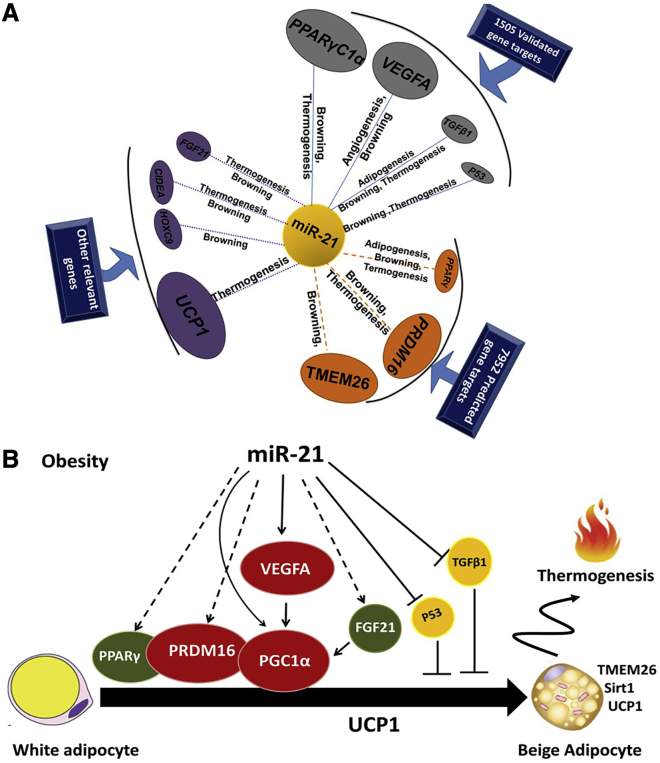
The effect of miR-21 on browning in WAT could be associated with the signaling pathways mediated by Vegf-A, Pgc-1α, Prdm16, Ppar-γ, Fgf21, Tgf-β1, and p53 genes, which lead to Ucp1, Cidea, and Tmem26 gene transcription induction
As described above, we have experimentally shown that Pgc-1α, Vegf-A, Tgf-β1, and p53 are potential target genes for miR-21 (Figure 8A). Up to date, Ppar-γ, Prdm16, and Tmem26 genes were predicted, but not validated, miR-21 target genes, according to our in silico analyses (Tables S1 and S2). PANTHER and GeneCodis3 (Gene Annotations Co-occurrence Discovery) showed that these target genes are mostly involved in thermogenesis and browning processes (Figure 8A). We anticipate that Ucp1, Hoxc9, Cidea, and Fgf21 genes could be potential key regulators for thermogenesis and browning (Figure 8A). In addition, inhibition of Tgf-β1 and p53 could also be another pathway regulated by miR-21 through which miR-21 regulates WAT function (Figure 8B).
Figure 8.
Schematic potential target genes and model pathways that might be involved in the miR-21 effects in AT
(A) Schema summarizing the interactions among miR-21, biologic processes, and the target genes. The validated and predicted target genes of miR-21 were obtained using miRTarBase 4.0, TarBase v.8, and miRWalk 2.0. The PANTHER Classification System and GeneCodis3 (Gene Annotations Co-occurrence Discovery) were applied to annotate the biological processes of the predicted targets. The interactions among miR-21, biologic processes, and the target genes were visualized with Cytoscape v.3.2.1 software. (B) Scheme showing the possible signaling pathways by which miR-21 could act in the activation of thermogenesis and browning of WAT of mice.
TargetScan tool analysis, which allows prediction of the binding sites of miRNA to its mRNA target,36 showed that miR-21 has effective binding sites only for Ucp1 (context ++ score percentile: 97%), with a probability of conserved targeting (PCT) value <0.1 , denoting the probability of segmentation conserved for a single target site (Table S3), indicating elevated evolutionary conservation.
Discussion
We here describe evidence showing that miR-21 is related to obesity independently of the glycemic state and is involved in AT functionality by regulating a number of genes and biological processes including thermogenesis, browning, angiogenesis, VEGF signaling, apoptosis, and adipogenesis, suggesting a pivotal role of miR-21 in obesity and its related metabolic alterations, such as T2D and IR, by modulating AT physiology. While investigating the effects of the previously reported increase of mir-21 levels in WAT in obesity and T2D,22,23 we came across that the mimetization of miR-21 may be a potentially effective therapy for obesity. Specifically, the administration of the miR-21 mimic in obese mice delayed the HFD-induced weight gain independently of food intake or physical activity. Concordantly, these effects were accompanied by the activation of WAT browning and the BAT thermogenic program corroborated by changes in metabolic activity, protein expression, and cellular ultrastructure. These data place the miR-21 mimic as a potential therapeutic candidate for obesity that inhibits weight gain.
We found that miR-21 is increased in obese subjects and mice without T2D (but not in diabetic obese) compared to NWs. Consistent with our findings, previous studies reported that miR-21 is highly expressed in human SAT and correlated with body mass index (BMI).22 In addition, an increase in miR-21 has been observed in the VAT of HFD-fed C57BL/6J mice compared to that of low-fat diet (LFD)-fed mice.37 Overall, these data support the potential role of miR-21 in the regulation of WAT functionality and expansion and suggest a differential regulation of this miRNA depending on the glycemic status in obese patients and mice. In line with this, Ling et al.38 showed that miR-21 was downregulated in insulin-resistant adipocytes and that treatment with the miR-21 precursor reverses high glucose and high insulin-induced IR in 3T3-L1 adipocytes, and the authors from this study suggest that miR-21 may be a therapeutic target for metabolic diseases such as obesity and T2D. Moreover, the in silico study highlighted that miR-21 has target genes that control a variety of processes well known to control AT functionality and expansion. Specifically, it is well described that deregulation of WAT apoptosis, adipogenesis, and angiogenesis is depicted to underlie AT dysfunction in obesity, which in turn increases the risk of T2D and IR.39 Here, we demonstrate that miR-21 mimic treatment regulates WAT angiogenesis through increasing the expression levels of the pro-angiogenic factors Vegf-A and Vegf-B. Vegf-A is a key regulator of angiogenesis and a validated target gene of miR-21, and its overexpression in AT has been reported to protect against diet-induced obesity and IR.40 Vegf-B has also been associated with an enhanced insulin delivery and function in AT, resulting in an improvement of metabolic health in obesity.41 In agreement with the pro-angiogenic role of miR-21, it has been previously reported that miR-21 triggers other angiogenic mediators, such as Timp-3, Ang2, and Ang4, in several cell types, which are in turn involved in WAT angiogenesis regulation and are associated with metabolism.42,43
We also found that the miR-21 mimic upregulates Bcl2, Bid, and Casp-3 genes in adipocytes, which have been well described to play a pivotal role in controlling the impairment of insulin signaling in human AT through the regulation of inflammatory processes and the development of obesity-associated IR.28,44 Surprisingly, in adipocytes, the miR-21 mimic appears to significantly reduce the expression levels of both Cebp-α and Ppar-γ, which are the master transcriptional regulators of adipogenesis,45 whereas increasing the expression of Pgc-1α and Fgf21, genes well known to play a relevant role in BAT thermogenesis and WAT browning.46,47 In line with these findings, a previous study showed that miR-21 controls adipogenic differentiation of human AT-derived stem cells. Specifically, this miRNA is increased at the early phase of adipogenesis but drops at later stages of differentiation. Notably, when the expression of this miRNA is experimentally maintained during adipogenesis, adipocyte differentiation is inhibited.24 Altogether, these results suggest that miR-21 mimic treatment not only regulates the key mediators of WAT functionality and expansion but also promotes the shift toward the thermogenic adipocyte cell lineage while impairing differentiation toward the white adipocyte cell lineage.
Sustained exogenous administration of the miR-21 mimic in obese mice abolished HFD-induced weight gain. It is well known that obesity and body weight are associated with the energy balance between food intake and energy expenditure.48 Within this context, the fact that the in vivo miR-21 mimic treatment did not affect the locomotor activity or calorie intake compared to controls suggests a direct involvement of miR-21 in the mechanisms regulating fat accumulation related to the activation of thermogenesis and browning, two processes that have arisen as potential targets for the management of obesity and its related diseases.49 Several studies have shown that the activity of brown and beige adipocytes is associated with resistance to obesity in several mouse models.50, 51, 52 In addition, it is well known that increased activity of brown and beige fat not only affects the weight gain of mice but also improves systemic metabolism, including the improvement in glucose tolerance and insulin sensitivity.50,52, 53, 54 Many miRNAs have been reported to be related to the regulation of gene mediators of beige and brown adipocyte differentiation.55, 56, 57 However, the involvement of miR-21 in the regulation of thermogenesis and browning gene expression has not been described. Here, we show that the miR-21 mimic plays a pivotal role in obesity and weight gain control through browning and thermogenesis activation involving the induction of Tmem26, Pgc-1α, Ucp1, Prdm16, Fgf21, Cidea, Ppar-γ, and Vegf-A gene expression and the mitochondrial biogenesis process.
Our in silico prediction study identified validated miR-21 target genes, including Vegf-A, Ppar-gc-1a, Tgf-β1, and p53. These genes are primarily involved in the regulation of browning thermogenesis, adipogenesis, and angiogenesis,31,33,34 as well as predicted (non-validated) miR-21 target genes, including Ppar-γ, Prdm16, and Tmem26, which are primarily known to be key regulators of browning and thermogenesis in AT19,52,58 (Figure 8A). We also provided experimental evidence through our in vivo, ex vivo, and in vitro studies of gene targets of miR-21 in WAT, namely, Ucp1, Hoxc9, Cidea, and Fgf21. Interestingly, the TargetScan tool identified miR-21 binding sites within Ucp1 mRNA. Further experiments should be carried out to ascertain whether Hoxc9, Cidea, and Fgf21 are direct target genes of miR-21.
Our comprehensive study allowed us to build a potential regulatory model, suggesting that the miR-21 mimic regulates browning and thermogenesis through several ways: (1) VEGF-A signaling pathway, (2) p53 signaling pathway, and (3) TGF-β1 pathway (Figure 8B). In line with our suggestions, the Vegf-A pathway has been described to be regulated by miR-2159,60 and has also been described to induce the expression of Pgc-1α and Ucp1 in both mouse BAT and WAT,31 and then Vegf-A regulates energy homeostasis and has a protective effect against obesity and IR, not only by angiogenesis regulation but also through the induction of BAT thermogenesis and WAT browning.31,32 As illustrated in Figure 8B, the activation of Ucp1 by miR-21 would involve Fgf21, Prdm16, and Ppar-γ pathways. In fact, Fgf21 has been well described to induce thermogenesis in BAT and browning in WAT by activating Pgc-1α.46,47 Prdm 16 and Pgc-1α have also been shown to induce the expression of Ucp1 and other thermogenic components and to regulate energy homeostasis and obesity.61, 62, 63 Moreover, miR-21 acts on browning and thermogenic processes through the inhibition of p53 and Tgf-β1, two validated miR-21 target genes that were downregulated in both the in vivo and in vitro miR-21 mimic treatment. These two target genes have been well described to inhibit differentiation and thermogenesis in BAT and to regulate the formation of beige cells in WAT and are related to obesity, T2D, and IR.33,34
Altogether, our data suggest the benefits of increasing miR-21 levels by using a synthetic mimic in AT as a potential therapeutic strategy against weight gain. In line with the positive effects exerted by the increase of miR-21, other authors demonstrated that the miR-21 precursor reverses high glucose and high insulin-induced IR in 3T3-L1 adipocytes and proposed miR-21 as a new therapeutic target for metabolic diseases such as T2D and obesity.38 By contrast, Seeger et al.64 showed that long-term inhibition of miR-21 by LNA-21 led to reduction of obesity in transgenic db/db mice through phosphatase and tensin (PTEN) and TGF-β receptor inhibition in AT. In addition, the effect of miR-21 inhibition seems not to be so clear, as Kim et al.24 reported that miR-21 inhibition by 2′-O-methyl-antisense miRNA increased TGF-β receptor levels as contrary to Seeger et al.64 Regardless of these controversial previous findings, the present study found that the use of the miR-21 mimic delayed weight gain in our diet-induced obesity mouse model and also showed that the inhibition of weight gain, which is independent of physical activity or food intake, could be explained, at least in part, by promoting BAT activity and WAT browning. These findings warrant further research on the use of the miR-21 mimic as a potential therapeutic option for obesity,
In conclusion, this study adds novel findings that highlight the role of the miR-21 mimic as a promising potential therapeutic strategy for limiting weight gain by modulating WAT and BAT physiology.
Materials and methods
Participants for the human study on miR-21 expression
MO subjects (BMI > 40 kg/m2), recruited at the Endocrinology Service of the Virgen de la Victoria University Hospital (Málaga, Spain) who underwent bariatric surgery, were included in the study and classified according to the American Diabetes Association (ADA) criteria65 and the homeostasis model assessment of the IR (HOMA-IR) index66 in (1) normoglycemic LIR-MO subjects (n = 9; fasting plasma glucose < 100 mg/dL and HOMA-IR < 3.5) and (2) prediabetic HIR-MO subjects (n = 9; fasting plasma glucose > 110 mg/dL and HOMA-IR > 7).
In addition, normoglycemic NW control subjects with a low degree of IR (BMI = 18.5−24 kg/m2; fasting plasma glucose < 110 mg/dL and HOMA-IR < 3.5) who underwent hiatal hernia or cholelithiasis surgery, age-matched to the LIR-MO and HIR-MO groups, and did not report changes in their body weight at least 3 months prior to the recruitment were included in the study (NW group; n = 7).
The exclusion criteria were the following: (1) T2D with medical treatment, (2) major cardiovascular disease in the 6 months prior to inclusion in the study, (3) evidence of acute or chronic inflammatory disease, (4) infectious diseases, and (5) refusal of the patient to participate in the study. Written, informed consent was received from participants prior to participation. The experimental protocol was reviewed and approved by the Ethics and Research Committee of the Virgen de la Victoria Clinical University Hospital (Málaga, Spain) and carried out in accordance with the principles of the Declaration of Helsinki.
The biochemical and anthropometric characteristics of the study subjects are shown in Table S4.
AT biopsies for miRNA analyses were obtained from subcutaneous and visceral (omental) areas during surgery and were frozen and stored at −80°C until analysis.
Generation of diet-induced obese and diabetic mice and study design for miR-21 expression studies
The C57BL/6J strain of mice used in the present study is susceptible to developing obesity and metabolic disturbances after being fed a HFD for several weeks.27 As we previously described, this mouse strain on a HFD for 10 weeks led to the development of a phenotype mimicking that of human obesity and pre-diabetes.27 Briefly, C57BL/6J mice (11 weeks old at arrival) were purchased from Charles River, France, and were allowed to acclimatize in the animal facility for 1 week prior to the experiments. Mice were singly housed under a 12-h light/dark cycle (8:00 p.m. lights off) in a room with controlled temperature (21°C ± 2°C) and humidity (50% ± 10%) and free access to pelleted chow (Standard Rodent Diet A04, SAFE; Panlab, Barcelona, Spain) and water. The study was carried out in three groups of mice: control, 45% HFD-ob, and 45% HFD-diabetic (Figure S1A). Specifically, to obtain 45% HFD-ob mice, a group of eight mice were fed a control diet containing 10% of kilocalorie (kcal; control diet) (D12450; Research Diets) for 6 weeks and then were fed, for further 8 weeks, a HFD (D12451; Research Diets, New Brunswick, NJ, USA) containing 45% of kcal from saturated fat (HFD; 45% HFD). To obtain diabetic obese mice, a group of eight mice were fed a HFD containing 45% of kcal from saturated fat for 14 weeks (45% HFD-diabetic). A control group of mice were fed a control diet containing 10% of kcal from fat (control) for 14 weeks (Figure S1A). Twice a week, body weight was monitored. As previously described,27 after 14 weeks of feeding, glucose and insulin tolerance was assessed by intraperitoneal glucose tolerance test (GTT) (and insulin tolerance test [ITT]) (Figure S1C). Then, mice were euthanized by cervical dislocation. Tissues were immediately collected for further histological and biochemical analyses. The European Union recommendations (2010/63/EU) on animal experimentation were followed. All procedures were approved by the Ethics Committee of the University of Malaga (authorization no. 2012-0061A).
The number of animals necessary for this study was determined by the G∗Power program, calculating sample size using a probability of a false-positive finding α = 0.05 and a power 1-β = 0.95 to detect 3% significant difference in weight of the animals and according to a previous study.27
Effect of miR-21 on differentiated 3T3-L1 adipocytes
The 3T3-L1 cell line (ATCC-CL-173) was cultured in DMEM/F12/10% fetal calf serum (FCS) at 37°C, 95% humidity, and 5% CO2 (n = 6). Differentiation of 3T3-L1 into adipocytes was induced by incubation in DMEM/F12/10% FCS supplemented with 0.5 mM isobutylmethylxanthine, 1 μM dexamethasone, and 1.67 μM insulin for 48 h and then in DMEM/F12/10% FCS supplemented with 1.67 μM insulin for 5 days. 7 days after adipogenic induction, the adipocytes were incubated with 5 nM miR-21 mimic (Syn-mmu-miR-21-5p miScript miRNA Mimic, MSY0000530; QIAGEN) or control mimic (AllStars Negative [Neg.] Control siRNA [small interfering RNA], 5 nmol, 1027289; QIAGEN) for 48 h. DharmaFECT Transfection Reagent was used to transfect cells.
Study design for in vivo treatment of 45% HFD-fed mice with miR-21 mimic
To obtain obese mice, a group of mice were fed a HFD containing 45% of kcal from saturated fat for 8 weeks (45% HFD). Body weight was monitored twice a week. After 8 weeks of feeding, glucose tolerance was assessed by intraperitoneal GTT (Figure S3). Cumulative weight gain was calculated by subtracting the weight obtained each time from posterior weight and by using the initial weight before starting diet-induced obesity as a basal point (0 g) for each mouse (Figure S3). Glucose area under the curve (AUC) was calculated using basal glucose after 10−12 h of fasting as the basal point (0) for each mouse (Figure S3). In this step of the in vivo study, the 45% HFD mice group was compared in parallel with a control group of age-matched mice, which were fed a control diet containing 10% of kcal from fat (LFD) in order to ensure that the 45% HFD mice group had significant differences in weight and glucose tolerance compared to 10% of the LFD group (control diet) after 8 weeks of diet, which indicates that miR-21 in vivo treatment can be started. The control diet group was then discarded, as the study was focused mainly on obese mice. The obese mice were separated into three subgroups: (1) a group treated three times a week for 8 weeks with subcutaneous injection of 0.5 μg miR-21 mimic (5′-UAGCUUAUCAGACUGAUGUUGA-3′; CONmiR mimic in vivo, M-00303-0100; Riboxx) dissolved in jetPEI vehicle (201-50G; Polyplus Transfection; n = 8); (2) a group treated three times a week for 8 weeks with mimic control (AllStars Neg. Control siRNA; QIAGEN) dissolved in jetPEI vehicle (n = 9); and (3) a group treated three times a week for 8 weeks with jetPEI vehicle (n = 4). Cumulative weight gain was calculated using the last weight before treatment as a basal point and assigned as 0 g (Figure 4A). In vivo jetPEI (Polyplus Transfection) is a powerful polymer-based reagent that forms stable and positively charged transfection complexes with nucleic acids (including miRNAs) facilitating its interaction with the cell membrane and endocytosis in its target tissues. In vivo miRNA mimic (CONmiR; Riboxx) is constituted by a mature miRNA sequence (guide strand) and its complementary sequence (passenger strand) supplied with RNAi-cap technology, which allows the loading of guide strand to the RISC complex and enhances the stability of mimic miRNA (up to 72 h in serum). Mimic has no homology with any human gene sequence and any known miRNAs or RNA, and its sequence is not provided by the manufacturer. All animals were sacrificed by cervical dislocation. Blood and AT biopsies, BAT, intWAT, ingWAT, and VAT were obtained during dissection and stored at −80°C. The number of mice in each experiment is stated in the figure legends (Figure 3). The number of animals necessary for this study was determined by the G∗Power program, calculating sample size using a probability of a false-positive finding α = 0.05 and a power 1-β = 0.95 to detect 3% significant difference in weight of the animals and according to a previous study.27
In vitro treatment of explants with miR-21 mimic
For the ex vivo study, fresh inguinal SAT and BAT explants (50 mg, n = 7) from mice fed a control diet (LFD, 10% kcal from fat) were dissected and cut into small pieces (5−10 mg). All explants were preincubated with PBS supplemented with 5% BSA for 30 min and then in 199 media supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin for 1 h at 37°C. Then, 5 nM miR-21 mimic (Syn-mmu-miR-21-5p miScript miRNA Mimic, MSY0000530; QIAGEN) or control mimic (AllStars Neg. Control siRNA, 5 nmol, 1027289; Sigma-Aldrich) was added, and explants were incubated at 37°C for 48 h. DharmaFECT Transfection Reagent was used to transfect AT explants.
GTT and ITT
Both the GTT and ITT were assessed after 8 weeks and 17 weeks of diet (control diet and diet-induced obesity) in the generation of diet-induced obese mice (Figure S1). In the in vivo study, GTT was assessed in the mice groups’ treatment at week 8 (before the start of treatment) and at the end of the treatment study (Figures 4A and S2).
Mice were injected intraperitoneally with 2 g/kg of D-glucose (Sigma-Aldrich, St. Louis, MO, USA) after 10−12 h of fasting before starting the treatment with miR-21 mimic and at the end of treatment. Blood glucose was measured at 0 (basal), 15, 30, 45, 60, and 120 min from the tail vein using a glucometer (Accu-Chek; Roche Diagnostics, Barcelona, Spain).
ITT was performed by injecting 0.5 U/kg of insulin intraperitoneally (Humulin, France) after 10−12 h of fasting. Blood drops were collected from the tail vein, and glucose was measured with a glucometer at 0 (basal), 15, 30, 45, 60, and 120 min. The glucose AUC was calculated using the basal point (0) as the basal glucose level for each mouse.
Food intake and OFTs
5 weeks after treatment, a food intake test was performed by weighing mice and food pellets for 5 days. Daily kcal consumption was calculated based on the HFD.
OFT was performed to measure the locomotor activity of obese mice treated with miR-21 mimic and control mimic. Mice were moved to the experimental room and kept there for 30 min before starting the test. Then, each mouse was placed in the middle of the open-field arena for 10 min, and variables such as the time and distance walked in the center, entries into the center, and overall distance traveled were measured using video-tracking SMART software.
PET
[18F]-FDG PET imaging was performed at the Unidad de Imagen Molecular, Centro de Investigaciones Médico-Sanitarias (CIMES), Spain.67 Animals (n = 6; 34.33 ± 3.2 g) fasted overnight were anesthetized by inhalation of a mixture of isoflurane/oxygen (5% for induction and 2% for maintenance) and placed prone on a PET scanner bed 1 h after intraperitoneal radiotracer administration (10.36 ± 2.22 MBq) to perform a static acquisition of 30 min. Images were subsequently reconstructed using an iterative 3D row action maximum likelihood algorithm (3D RAMLA). Corrections for dead time decay and random coincidences were applied. Images were reconstructed on a 128 × 128 × 120 matrix, where the voxel size equals 1 × 1 × 1 mm. PET images were normalized using the whole brain average uptake with PMOD software (3.3 PMOD Technologies, Zurich, Switzerland). The inguinal AT region of interest (ROI) was manually drawn.
Animals were kept at 25°C to maintain BAT activity as low as possible and to ensure that differences in thermogenic activity were only due to treatment. The blood glucose concentration (88.83 ± 6.88 mg/dL) was determined before tracer injection using a glucose-level meter with test strips (Accu-Chek Aviva Nano; Roche, Mannheim, Germany).
Histochemistry and immunofluorescence
AT samples were fixed in 4% formaldehyde for 48 h and then placed and processed in a paraffin Spin Tissue Processor (TP) STP 120 to allow the infiltration of paraffin into the tissue. Infiltrated samples were then embedded in paraffin using the paraffin-embedding center (EG1150H; Leica, Nussloch, Germany). The tissue was cut into sections up to 5 μm thick. After sample deparaffinization, nuclear staining was carried out with Harris Hematoxylin (H&H), followed by cytoplasmic staining with a mixture of eosin and 0.2% glacial acetic acid. The slides were dehydrated through a series of increasing grades of ethanol solutions. All sections were photographed using an Olympus BX61 microscope (Olympus, Tokyo, Japan).
Immunofluorescence was performed by incubating the samples in PBS plus 0.3% Triton X-100, 10% donkey serum (DS), and 10% sheep serum (SS) for 1 h. Then, the sections were incubated in a mixture of primary antibodies including rabbit anti-TMEM26 (1:50, NBP2-27334; Novus Biologicals Europe, Abingdon, UK) and goat anti-UCP1 (1:75, SAB2501082; Merck KGaA, Darmstadt, Germany) overnight at 4°C. The slides were incubated at room temperature for 2 h with the fluorescent-labeled secondary anti-rabbit immunoglobulin G (IgG)-fluorescein isothiocyanate (FITC) antibodies (1:50, F7512; Merck KGaA, Darmstadt, Germany) suspended in PBS + 5% DS and then incubated at room temperature for 2 h in anti-goat IgG heavy chains (H) and light chain (L) of the antibody (H+L) tetramethyl rhodamine isothiocyanate (TRITC; 1:50, ab6522; Abcam, Oxford, UK) suspended in PBS + 5% SS. Finally, FluoroShield 4′,6-diamidino-2-phenylindole (DAPI) medium was added. All sections were photographed using an Olympus BX61 microscope. Fluorescence photomicrographs were captured with a digital camera (DP70; Olympus, Tokyo, Japan) and software DP Controller (1.2.1.108; Olympus, Tokyo, Japan).
TEM
Small pieces of tissue were fixed with 2.5% glutaraldehyde solution for 75 h. Sections were washed in phosphate buffer (pH 7.4) and placed in an Automatic Sample Processor (electron microscope [EM] TP; Leica, Nussloch, Germany) for 25 h and 55 min. The processor performs washes in phosphate buffer (PB), followed by application of 2% osmium in PB, washed in distilled water, and acetone in increasing gradation from 25% to 100%. The processing continues with the inclusion in Epon resin in different percentages mixed with acetone, up to 100% pure resin. The re-cutting and semithin cuts were carried out using a glass blade in a standard range of 300 nm. To determine the areas for ultrathin cuts, the sections were placed on standard glass slides and stained with toluidine blue (89640; Merck KGaA, Darmstadt, Germany). Finally, the sections were placed on 300 mesh copper grids. The study of cell organelles was performed under a transmission microscope with a voltage of 80 kV (Libra-120; Zeiss, Oberkochen, Germany), and the images were obtained using iTEM software (Olympus, Tokyo, Japan).
miRNA extraction and quantitative real-time PCR
miRNA was isolated from AT samples and 3T3-L1 cells using the miRvana miRNA isolation kit (AM1561; Ambion, Spain) according to the manufacturer’s protocol. For plasma samples, automated extraction was carried out using Maxwell 16 Promega and the Maxwell 16 miRNA Tissue kit (AS 1470; Promega, Spain), according to the manufacturer’s recommendations. The miRNA concentration was measured using a Nanodrop ND-2000 (Thermo Fisher Scientific, USA) or Quantifluor RNA kit (E3310; Promega, Spain) and Quantus Fluorometer (Promega, Spain). 5 ng of miRNA was converted to cDNA by the specific TaqMan MicroRNA Reverse Transcription (RT) kit (Thermo Fisher Scientific, USA) and was carried out using specific primers for each miRNA (looped RT primer TaqMan small RNA assays), namely, hsa-miR-21 (000397), control miRNA assay snoRNA-142 (001231), and control miRNA assay RNU48 (001006). qRT-PCR reactions were carried out using specific TaqMan probes and an Agilent Mx3005P qPCR system. During amplification, the cycle threshold (Ct) value was determined, and specific signals were normalized with stable expression (BestKeeper) of the reference gene snoRNA-142 (mouse) or RNU48 (human) using the formula 2−Δct (ΔCT = Ct of target gene−Ct of reference gene) and 2−ΔΔCT (ΔΔCT = (Ct of target gene−Ct of reference gene)mimic miR21 − (Ct of target gene−Ct of reference gene)control adipo).
mRNA extraction and quantitative real-time PCR
Total RNA was extracted from ATs with the Qiazol RNeasy Lipid Tissue mini kit (QIAGEN, Valencia, CA, USA) or with RNA-Stat 60 Reagent (Ams Biotechnology, Abingdon, UK) for 3T3-L1 cells, according to the manufacturer’s recommendations. Then, 1, 2, or 4 μg of total RNA was converted to cDNA using RT (Transcriptor Reverse Transcriptase 20 U/μL, 03531287001; Roche). Next, qRT-PCR amplification was performed using specific TaqMan Gene Expression Assays, 10 ng of cDNA, and Brilliant III Ultra-Fast qPCR Master Mix. Specific signals were normalized to the constitutively expressed TATA sequence binding protein (TBP; Mm 00446973; Thermo Fisher Scientific) for AT using the formula 2−Δct or to β-actin (mouse ACTB, 452341E; Thermo Fisher Scientific) for 3T3-L1 cells using the formula 2−ΔΔct. The TaqMan probes used in this study are Vegf-A (Mm 00437306-m1), Vegf-B (Mm 00442102_m1), Ucp1 (Mm 01244861-m1), Tmem26 (Mm 01173641-m1), Pgc-1α (Mm 01208835), Prdm16 (Mm 00712556-m1), Cidea (Mm 00432554-m1), Ppar-γ (Mm 00440940-m1), P53 (Mm01731290-g1), Tgf-β1 (Mm01178820-m1), Fgf21 (Mm 00840165-g1), Hoxc9 (Mm 00433972-m1), Sirt1 (Mm01168521-m1), Mmp-9 (Mm 00442991_m1), Timp-2 (Mm 00441825_m1), Timp-3 (Mm 00441826-m1), Cebp-α (Mm 00514283 s1), Bcl-2 (Mm 00477631-m1), Bid (Mm 00432073_m1), and Casp-3 (Mm 01195085-m1).
DNA extraction and qPCR
Total DNA was extracted from ingSAT using the DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s recommendations. The DNA concentration was measured using a Nanodrop ND-2000 (Thermo Fisher Scientific, USA). Relative mtDNA content of ingSAT was analyzed by qPCR. Multiplex qPCR amplification was performed using specific probes, Brilliant III Ultra-Fast qPCR Master Mix, 160 ng of DNA, and reference dye. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as a nuclear gene (nuclear DNA [nDNA], Endogenous Control, VIC/MGB [minor groove binder] probe, primer limited; Thermo Fisher Scientific) and Cox1 as a mitochondrial gene (mtDNA, Mm04225243_g1). The relative mtDNA content was calculated using the following formula: mtDNA = 2−Δct, where ΔCt = Ct Cox1 − Ct Gapdh.
Prediction of putative miRNA target genes
The miRTarBase version (v.)4.0 (https://mirtarbase.cuhk.edu.cn/∼miRTarBase/miRTarBase_2019/php/index.php), TarBase v.8 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8), and miRWalk v.2.0 (https://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html) databases were used for prediction of validated and non-validated target genes of miR-21. The PANTHER Classification System (http://pantherdb.org/) and the GeneCodis3 (https://genecodis.genyo.es/) were applied to annotate the biological processes of the predicted targets.
Interactions among miRNAs, biological processes, and miR-21 target genes
The interactions among miR-21, biologic processes, and the target genes were visualized with Cytoscape software v.3.2.1 (https://cytoscape.org/).
Statistical analyses
Statistical analyses and graphics were carried out using GraphPad Prism 5.00.288 and IBM SPSS Statistics 22. All data are expressed as the mean ± SEM. Comparisons among multiple groups were performed by one-way ANOVA with Bonferroni’s post hoc test, and comparisons between two groups were performed by Student’s t test. GTT, ITT, and body weight were analyzed by repeated two-way ANOVA measurements. P ˂ 0.05 was considered statistically significant. In the PET study, significant differences between groups were determined using a 2-tailed 2-sample unequal variance Student’s t test in Microsoft Excel. A p < 0.05 was considered to indicate statistical significance.
Acknowledgments
The authors wish to thank all of the subjects for their collaboration. CIBER Fisiopatología de la Obesidad y Nutrición (CIBERobn) is part of the “Instituto de Salud del Carlos III” (ISCIII) project. This work was supported, in part, by grants from ISCIII and co-funded by Fondo Europeo de Desarrollo Regional (FEDER)-Unión Europea (UE) (PI18/00785) and Consejería de Salud, Junta de Andalucía (PI-0092-2017), Spain, and co-funded by FEDER. R.E.B. and F.J.B.-S. are under a contract from the “Nicolas Monarde” (C-0030-2016, RC-0005-2016) program from the Servicio Andaluz de Salud, Regional Ministry of Health of the Andalusian Government, Andalusia, Spain. S.L. was a recipient of postdoctoral grant Plan Andaluz de Investigación Desarrollo e Innovación (DOC-01138) from Consejería de Economía, Empresas y Universidades. A.-M.G. was a recipient of a postdoctoral grant Plan Propio de Investigación, Transferencia y Divulgación Científica, from Málaga University. M.C.-P. was recipient of a postdoctoral grant Juan de la Cierva Formación (FJCI-2017-32194) from the Ministerio de Ciencia, Innovación y Universidades (Spain), and postdoctoral research grant Plan Andaluz de Investigación, Desarrollo e Innovación (PAIDI 2020) (DOC_00448) from the Consejeria de Economía, Industria, Conocimiento y Universidades (Junta de Andalucía), Spain, co-funded by FEDER.
Author contributions
R.E.B. and F.J.T. designed the research. R.E.B. performed experiments, analyzed the data, and wrote the paper. S.L. and A.-M.G. performed in vivo, in vitro, ex vivo, and molecular biology experiments and bioinformatics analysis. W.O.O. carried out in vitro culture of preadipocytes. R.M.G.-P. and S.L. carried out histologic and microscopic analysis. M.F.-C. carried out PET analysis. S.Y.R.-Z. and F.J.B.-S. carried out animal models of obesity generation. C.L.G. performed mtDNA analyses. M.C.-P. and H.Z. contributed to manuscript writing and editing. M.C.-P., A.H., and N.H. contributed to statistical, data analysis, and interpretation. G.Oand J.S. contributed to human subjects’ recruitment and clinical analysis.
Declaration of interests
The authors have no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.06.019.
Contributor Information
Francisco J. Tinahones, Email: fjtinahones@uma.es.
Rajaa El Bekay, Email: relbekay@gmail.com.
Supplemental information
References
- 1.Hruby A., Hu F.B. The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics. 2015;33:673–689. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colman E., Golden J., Roberts M., Egan A., Weaver J., Rosebraugh C. The FDA’s assessment of two drugs for chronic weight management. N. Engl. J. Med. 2012;367:1577–1579. doi: 10.1056/NEJMp1211277. [DOI] [PubMed] [Google Scholar]
- 3.Gautron L., Elmquist J.K., Williams K.W. Neural control of energy balance: translating circuits to therapies. Cell. 2015;161:133–145. doi: 10.1016/j.cell.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deiuliis J.A. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016;40:88–101. doi: 10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie H., Sun L., Lodish H.F. Targeting microRNAs in obesity. Expert Opin. Ther. Targets. 2009;13:1227–1238. doi: 10.1517/14728220903190707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong H., Ma M., Liang T., Guo L. Role of MicroRNAs in Obesity-Induced Metabolic Disorder and Immune Response. J. Immunol. Res. 2018;2018:2835761. doi: 10.1155/2018/2835761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mollaei H., Safaralizadeh R., Rostami Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019;234:12369–12384. doi: 10.1002/jcp.28058. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z. The guideline of the design and validation of MiRNA mimics. Methods Mol. Biol. 2011;676:211–223. doi: 10.1007/978-1-60761-863-8_15. [DOI] [PubMed] [Google Scholar]
- 10.Beg M.S., Brenner A.J., Sachdev J., Borad M., Kang Y.-K., Stoudemire J., Smith S., Bader A.G., Kim S., Hong D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez A., Ezquerro S., Méndez-Giménez L., Becerril S., Frühbeck G. Revisiting the adipocyte: a model for integration of cytokine signaling in the regulation of energy metabolism. Am. J. Physiol. Endocrinol. Metab. 2015;309:E691–E714. doi: 10.1152/ajpendo.00297.2015. [DOI] [PubMed] [Google Scholar]
- 13.Chouchani E.T., Kajimura S. Metabolic adaptation and maladaptation in adipose tissue. Nat. Metab. 2019;1:189–200. doi: 10.1038/s42255-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajer G.R., van Haeften T.W., Visseren F.L.J. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur. Heart J. 2008;29:2959–2971. doi: 10.1093/eurheartj/ehn387. [DOI] [PubMed] [Google Scholar]
- 15.Friesen M., Cowan C.A. Adipocyte Metabolism and Insulin Signaling Perturbations: Insights from Genetics. Trends Endocrinol. Metab. 2019;30:396–406. doi: 10.1016/j.tem.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Harms M., Seale P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 17.Villarroya F., Vidal-Puig A. Beyond the sympathetic tone: the new brown fat activators. Cell Metab. 2013;17:638–643. doi: 10.1016/j.cmet.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Reitman M.L. How Does Fat Transition from White to Beige? Cell Metab. 2017;26:14–16. doi: 10.1016/j.cmet.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J., Cohen P., Cinti S., Spiegelman B.M. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karbiener M., Scheideler M. MicroRNA functions in brite/brown fat - Novel perspectives towards anti-obesity strategies. Comput. Struct. Biotechnol. J. 2014;11:101–105. doi: 10.1016/j.csbj.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekar D., Hairul Islam V.I., Thirugnanasambantham K., Saravanan S. Relevance of miR-21 in HIV and non-HIV-related lymphomas. Tumour Biol. 2014;35:8387–8393. doi: 10.1007/s13277-014-2068-9. [DOI] [PubMed] [Google Scholar]
- 22.Keller P., Gburcik V., Petrovic N., Gallagher I.J., Nedergaard J., Cannon B., Timmons J.A. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 2011;11:7. doi: 10.1186/1472-6823-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guglielmi V., D’Adamo M., Menghini R., Cardellini M., Gentileschi P., Federici M., Sbraccia P. MicroRNA 21 is up-regulated in adipose tissue of obese diabetic subjects. Nutr. Healthy Aging. 2017;4:141–145. doi: 10.3233/NHA-160020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y.J., Hwang S.J., Bae Y.C., Jung J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- 25.Lee E.H., Park H.J., Jeong J.H., Kim Y.J., Cha D.W., Kwon D.K., Lee S.H., Cho J.Y. The role of asporin in mineralization of human dental pulp stem cells. J. Cell. Physiol. 2011;226:1676–1682. doi: 10.1002/jcp.22498. [DOI] [PubMed] [Google Scholar]
- 26.Richart A., Loyer X., Néri T., Howangyin K., Guérin C.L., Ngkelo A., Bakker W., Zlatanova I., Rouanet M., Vilar J. MicroRNA-21 coordinates human multipotent cardiovascular progenitors therapeutic potential. Stem Cells. 2014;32:2908–2922. doi: 10.1002/stem.1789. [DOI] [PubMed] [Google Scholar]
- 27.Romero-Zerbo S.Y., Ruz-Maldonado I., Espinosa-Jiménez V., Rafacho A., Gómez-Conde A.I., Sánchez-Salido L., Cobo-Vuilleumier N., Gauthier B.R., Tinahones F.J., Persaud S.J., Bermúdez-Silva F.J. The cannabinoid ligand LH-21 reduces anxiety and improves glucose handling in diet-induced obese pre-diabetic mice. Sci. Rep. 2017;7:3946. doi: 10.1038/s41598-017-03292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tinahones F.J., Coín Aragüez L., Murri M., Oliva Olivera W., Mayas Torres M.D., Barbarroja N., Gomez Huelgas R., Malagón M.M., El Bekay R. Caspase induction and BCL2 inhibition in human adipose tissue: a potential relationship with insulin signaling alteration. Diabetes Care. 2013;36:513–521. doi: 10.2337/dc12-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tinahones F.J., Coín-Aragüez L., Mayas M.D., Garcia-Fuentes E., Hurtado-Del-Pozo C., Vendrell J., Cardona F., Calvo R.M., Obregon M.J., El Bekay R. Obesity-associated insulin resistance is correlated to adipose tissue vascular endothelial growth factors and metalloproteinase levels. BMC Physiol. 2012;12:4. doi: 10.1186/1472-6793-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito M. Human brown adipose tissue: regulation and anti-obesity potential. Endocr. J. 2014;61:409–416. doi: 10.1507/endocrj.ej13-0527. [DOI] [PubMed] [Google Scholar]
- 31.Elias I., Franckhauser S., Bosch F. New insights into adipose tissue VEGF-A actions in the control of obesity and insulin resistance. Adipocyte. 2013;2:109–112. doi: 10.4161/adip.22880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X., Ji Y., Zhang L., Zhang Y., Zhang S., An Y., Liu P., Zheng Y. Resistance to obesity by repression of VEGF gene expression through induction of brown-like adipocyte differentiation. Endocrinology. 2012;153:3123–3132. doi: 10.1210/en.2012-1151. [DOI] [PubMed] [Google Scholar]
- 33.Hallenborg P., Fjære E., Liaset B., Petersen R.K., Murano I., Sonne S.B., Falkerslev M., Winther S., Jensen B.A.H., Ma T. p53 regulates expression of uncoupling protein 1 through binding and repression of PPARγ coactivator-1α. Am. J. Physiol. Endocrinol. Metab. 2016;310:E116–E128. doi: 10.1152/ajpendo.00119.2015. [DOI] [PubMed] [Google Scholar]
- 34.Yadav H., Quijano C., Kamaraju A.K., Gavrilova O., Malek R., Chen W., Zerfas P., Zhigang D., Wright E.C., Stuelten C. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang B.L. Sirt1 and the Mitochondria. Mol. Cells. 2016;39:87–95. doi: 10.14348/molcells.2016.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao M. Construction of an miRNA-Regulated Pathway Network Reveals Candidate Biomarkers for Postmenopausal Osteoporosis. Comput. Math. Methods Med. 2017;2017:9426280. doi: 10.1155/2017/9426280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chartoumpekis D.V., Zaravinos A., Ziros P.G., Iskrenova R.P., Psyrogiannis A.I., Kyriazopoulou V.E., Habeos I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE. 2012;7:e34872. doi: 10.1371/journal.pone.0034872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ling H.Y., Hu B., Hu X.B., Zhong J., Feng S.D., Qin L., Liu G., Wen G.B., Liao D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes. 2012;120:553–559. doi: 10.1055/s-0032-1311644. [DOI] [PubMed] [Google Scholar]
- 39.Blüher M. Adipose tissue dysfunction in obesity. Exp. Clin. Endocrinol. Diabetes. 2009;117:241–250. doi: 10.1055/s-0029-1192044. [DOI] [PubMed] [Google Scholar]
- 40.Elias I., Franckhauser S., Ferré T., Vilà L., Tafuro S., Muñoz S., Roca C., Ramos D., Pujol A., Riu E. Adipose tissue overexpression of vascular endothelial growth factor protects against diet-induced obesity and insulin resistance. Diabetes. 2012;61:1801–1813. doi: 10.2337/db11-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karaman S., Hollmén M., Yoon S.Y., Alkan H.F., Alitalo K., Wolfrum C., Detmar M. Transgenic overexpression of VEGF-C induces weight gain and insulin resistance in mice. Sci. Rep. 2016;6:31566. doi: 10.1038/srep31566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J., Ni S., Cao Y., Zhang T., Wu T., Yin X., Lang Y., Lu H. The Angiogenic Effect of microRNA-21 Targeting TIMP3 through the Regulation of MMP2 and MMP9. PLoS ONE. 2016;11:e0149537. doi: 10.1371/journal.pone.0149537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An Y.A., Sun K., Joffin N., Zhang F., Deng Y., Donzé O., Kusminski C.M., Scherer P.E. Angiopoietin-2 in white adipose tissue improves metabolic homeostasis through enhanced angiogenesis. eLife. 2017;6:e24071. doi: 10.7554/eLife.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alkhouri N., Gornicka A., Berk M.P., Thapaliya S., Dixon L.J., Kashyap S., Schauer P.R., Feldstein A.E. Adipocyte apoptosis, a link between obesity, insulin resistance, and hepatic steatosis. J. Biol. Chem. 2010;285:3428–3438. doi: 10.1074/jbc.M109.074252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White U.A., Stephens J.M. Transcriptional factors that promote formation of white adipose tissue. Mol. Cell. Endocrinol. 2010;318:10–14. doi: 10.1016/j.mce.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coskun T., Bina H.A., Schneider M.A., Dunbar J.D., Hu C.C., Chen Y., Moller D.E., Kharitonenkov A. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149:6018–6027. doi: 10.1210/en.2008-0816. [DOI] [PubMed] [Google Scholar]
- 47.Fisher F.M., Kleiner S., Douris N., Fox E.C., Mepani R.J., Verdeguer F., Wu J., Kharitonenkov A., Flier J.S., Maratos-Flier E., Spiegelman B.M. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slawik M., Vidal-Puig A.J. Adipose tissue expandability and the metabolic syndrome. Genes Nutr. 2007;2:41–45. doi: 10.1007/s12263-007-0014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peirce V., Carobbio S., Vidal-Puig A. The different shades of fat. Nature. 2014;510:76–83. doi: 10.1038/nature13477. [DOI] [PubMed] [Google Scholar]
- 50.Cederberg A., Grønning L.M., Ahrén B., Taskén K., Carlsson P., Enerbäck S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 51.Kopecky J., Clarke G., Enerbäck S., Spiegelman B., Kozak L.P. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J. Clin. Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H.M., Erdjument-Bromage H. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bordicchia M., Liu D., Amri E.Z., Ailhaud G., Dessì-Fulgheri P., Zhang C., Takahashi N., Sarzani R., Collins S. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J. Clin. Invest. 2012;122:1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boström P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C., Rasbach K.A., Boström E.A., Choi J.H., Long J.Z. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mori M., Nakagami H., Rodriguez-Araujo G., Nimura K., Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012;10:e1001314. doi: 10.1371/journal.pbio.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L., Xie H., Mori M.A., Alexander R., Yuan B., Hattangadi S.M., Liu Q., Kahn C.R., Lodish H.F. Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trajkovski M., Ahmed K., Esau C.C., Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 2012;14:1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- 58.Petrovic N., Walden T.B., Shabalina I.G., Timmons J.A., Cannon B., Nedergaard J. Chronic peroxisome proliferator-activated receptor γ (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei Z., Li B., Yang Z., Fang H., Zhang G.M., Feng Z.H., Huang B. Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS ONE. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu L.Z., Li C., Chen Q., Jing Y., Carpenter R., Jiang Y., Kung H.F., Lai L., Jiang B.H. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS ONE. 2011;6:e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 62.Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D., Langin D. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 63.Villanueva C.J., Vergnes L., Wang J., Drew B.G., Hong C., Tu Y., Hu Y., Peng X., Xu F., Saez E. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seeger T., Fischer A., Muhly-Reinholz M., Zeiher A.M., Dimmeler S. Long-term inhibition of miR-21 leads to reduction of obesity in db/db mice. Obesity (Silver Spring) 2014;22:2352–2360. doi: 10.1002/oby.20852. [DOI] [PubMed] [Google Scholar]
- 65.American Diabetes Association 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care. 2019;42(Suppl 1):S13–S28. doi: 10.2337/dc19-S002. [DOI] [PubMed] [Google Scholar]
- 66.Wallace T.M., Levy J.C., Matthews D.R. Use and abuse of HOMA modeling. Diabetes Care. 2004;27:1487–1495. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 67.Prieto E., Collantes M., Delgado M., Juri C., García-García L., Molinet F., Fernández-Valle M.E., Pozo M.A., Gago B., Martí-Climent J.M. Statistical parametric maps of 18F-FDG PET and 3-D autoradiography in the rat brain: a cross-validation study. Eur. J. Nucl. Med. Mol. Imaging. 2011;38:2228–2237. doi: 10.1007/s00259-011-1905-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.