Summary
A neuronal circuit of ∼150 neurons modulates rhythmic activity-rest behavior of Drosophila melanogaster. While it is known that coherent ∼24-hr rhythms in locomotion are brought about when 7 distinct neuronal clusters function as a network due to chemical communication amongst them, there are no reports of communication via electrical synapses made up of gap junctions. Here, we report that gap junction proteins, Innexins play crucial roles in determining the intrinsic period of activity-rest rhythms in flies. We show the presence of Innexin2 in the ventral lateral neurons, wherein RNAi-based knockdown of its expression slows down the speed of activity-rest rhythm along with alterations in the oscillation of a core-clock protein PERIOD and the output molecule pigment dispersing factor. Specifically disrupting the channel-forming ability of Innexin2 causes period lengthening, suggesting that Innexin2 may function as hemichannels or gap junctions in the clock circuit.
Subject areas: Biological sciences, Physiology, Neuroscience, Behavioral neuroscience
Graphical abstract
Highlights
-
•
Gap junction protein Innexin2 modulates adult Drosophila activity rhythms
-
•
Innexin2 is expressed and functions in the ventral lateral neurons
-
•
Innexin2 levels influence PERIOD protein oscillations
-
•
Innexin2 affects amplitude of cycling of PDF in s-LNv neuronal terminals
Biological sciences; Physiology; Neuroscience; Behavioral neuroscience
Introduction
Drosophila melanogaster is widely used as a model organism in circadian biology due to its robust, easily quantifiable behaviors, and relatively few neurons regulating them (Allada and Chung, 2010). The adult D. melanogaster circadian circuit is composed of about 150 neurons distributed bilaterally in the brain and divided into the lateral and dorsal neuronal clusters (reviewed in Sheeba, 2008). Each cell has a ticking molecular clock composed of a self-sustained transcriptional translational feedback loop made up of several genes, among which the core clock genes are Clock, cycle, period, and timeless. The period of molecular oscillations in mRNA and protein within the pacemaker neurons mirror the period of rhythmic locomotor activity-rest behavior (Hardin, 2005). Several previous studies have shown that these individual clusters function as a network, although it is not entirely clear how these neuronal clusters with distinct intrinsic periodicities (Yoshii et al., 2009), together bring about one coherent period of the behavioral activity rhythm. Early studies of Drosophila clock neuronal network have shown that under constant darkness and temperature (DD 25°C), the small ventral lateral neurons (s-LNv) and core clock in these cells are necessary for the persistence of activity-rest rhythms (Helfrich-Förster, 1998; Renn et al., 1999). s-LNv release neuropeptide Pigment dispersing factor (PDF) in the dorsal part of the brain via their projections in a time-of-day dependent manner (Park et al., 2000). PDF receptor (PdfR) is widely distributed in most clock neurons in the circuit (Hyun et al., 2005; Mertens et al., 2005), most of them being responsive to PDF (Shafer et al., 2008), thus establishing its role as an important synchronizing factor in the circadian circuit. Apart from PDF, several other neuropeptides and neurotransmitters have been implicated to play diverse roles in the chemical communication among circadian neurons (Beckwith and Ceriani, 2015).
Across organisms and behaviors, most studies have focused on the role played by chemical synapses among neurons in a circuit, even though electrical and chemical synapses have been known to co-exist in neural networks of most organisms (Pereda, 2014; Nagy et al., 2018). Electrical synapses are made up of gap junctions which are tightly coupled clusters of proteins forming intercellular membrane spanning channels connecting the cytoplasm of adjacent cells. These channels facilitate electrical coupling of adjacent cells through diffusion of ions, metabolites and cyclic nucleotides (Faber and Pereda, 2018). Connexins form gap junctions in chordates; Innexins were identified as the structural proteins of gap junctions in invertebrates and Pannexins which are structurally similar to Innexins are found in some invertebrates and chordates and mostly function as hemichannels (Beyer and Berthoud, 2018). Drosophila melanogaster has eight members of the Innexin family named Innexin1-8 (Bauer et al., 2005). Functions of several of the Innexin classes have been characterized extensively during development and recently in behaviors exhibited by adult flies (Güiza et al., 2018).
Studies which suggest a role for gap junctions in modulating circadian behavior have been mostly undertaken in mammals. The mammalian circadian clock cells which are part of the suprachiasmatic nucleus (SCN) express a number of different Connexins (Welsh and Reppert, 1996; Colwell, 2000; Rash et al., 2007). Gap junction coupling in the intact SCN tissue has been demonstrated with tracer molecules and by electrical stimulation and recording membrane potential in neighboring cells (Jiang et al., 1997; Colwell, 2000; Shinohara et al., 2000). Gap junctions are also involved in synchronous firing of coupled cells in the SCN neuronal network (Wang et al., 2014). Specifically, knockout of Connexin36 (Cx36) in mice, blocks intercellular electrical coupling between SCN neurons and results in lower amplitude of circadian locomotor activity rhythms and a decrease in overall activity levels under constant conditions (Long et al., 2005). However, a later study shows that the absence of Cx36 does not affect the synchronous PER protein oscillations in SCN neuronal network although the Cx36 knockout mice have lengthened period of wheel-running activity as compared to control mice (Diemer et al., 2017). While electrical coupling among SCN neurons is important for synchronous firing, as well as for output behavior, the underlying mechanisms are poorly understood. To the best of our knowledge, in invertebrates, there has been only one study in the cockroach species, Leucophaea maderae, where, use of gap junction blockers results in desynchronized firing of accessory medulla neurons, the anatomical site of their circadian pacemakers (Schneider and Stengl, 2006). However, in the D. melanogaster circadian pacemaker circuit, there have been no reports of a systematic investigation of the roles played by gap junctions in modulating circadian rhythms.
We performed a screen to identify if gap junction genes have functions in the Drosophila circadian circuit. We found that the levels of two gap junction genes innexin1 (ogre) and Innexin2 determine an integral clock property, its free-running period. We demonstrate that Innexin2 is expressed in the circadian pacemaker neurons and knockdown of its levels causes the rhythm in locomotor activity to slow down. We also observe that oscillation of the core molecular clock protein PERIOD is delayed in most circadian neurons upon Innexin2 knockdown along with a change in the levels of neuropeptide PDF in the s-LNv dorsal projections. Thus, we provide evidence of a functional role for gap junction proteins in the circadian pacemaker circuit of D. melanogaster and suggest possible mechanisms for the same.
Results
RNAi knockdown screen of Innexins in clock neurons
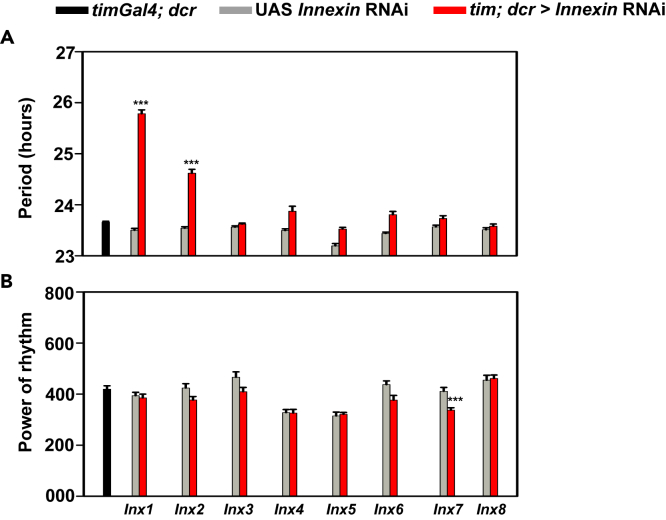
To examine the role of Innexins in the fly circadian network, we performed a RNAi knockdown screen where we downregulated the expression of each of the eight classes of Innexin genes using a broad Gal4 driver (timA3Gal4) (Kaneko and Hall, 2000) that targets all the ∼150 known circadian neurons and examined rhythm properties under constant darkness (DD 25°C) (Figure 1; Table 1). We observed a significant lengthening of free-running period both in case of knockdown of innexin1 (inx1) and Innexin2 (Inx2), as compared to their parental controls (Figures 1A, 2A, left, and 2B; Table 2), while no change in period was detected upon knockdown of the other 6 Innexin classes suggesting that Innexin1 and Innexin2 play important roles in determining the period of free-running activity-rest rhythm. We also quantified the power of the rhythm, which is indicative of the robustness of the underlying clock (Klarsfeld et al., 2003). We found a significant decrease in the power of the rhythm in case of knockdown of Innexin7 in one trial (Figure 1B). However, this result was not consistent across multiple replicate experiments, and hence it is not discussed here. Importantly, we found no difference in the power of the rhythm in case of innexin1 or Innexin2 knockdown despite significant lengthening of period (Figures 1B and 2A, right), suggestive of the fact that the robustness of the clock was not affected. Here we detail our findings on the role of Innexin2 in the circadian clock network.
Figure 1.
RNA interference screen of Innexins in the clock neurons under DD 25°C
(A) Mean free-running period of flies with individual Innexin (Innexin 1-8) genes knocked down are being plotted along with their common Gal4 control (timGal4; dcr) and respective UAS controls (UAS Innexin RNAi).
(B) Power of rhythm in case of individual knockdown of all the eight classes of Innexins along with their relevant parental controls are being plotted (n > 20 flies for each genotype). See Table 1 for more details. Error bars are SEM. Free-running period and power values are determined using Chi-square periodogram for a period of 8 days. All statistical comparisons were performed using one-way ANOVA with genotype as a fixed factor, followed by post-hoc analysis using Tukey’s Honest Significant Difference (HSD) test.
Table 1.
Effect of knockdown of Innexin1-8 in circadian pacemakers using timGal4 driver on free-running rhythm properties
| Genotype | Period ± SEM | POR ± SEM | % Rhythmicity |
|---|---|---|---|
| w; timGal4; dcr | 23.65 ± 0.01 | 419.2 ± 13.19 | 100 |
| tim; dcr > Inx1 RNAi | 25.78 ± 0.07 | 385.3 ± 14.55 | 100 |
| tim; dcr > Inx2 RNAi | 24.61 ± 0.07 | 377 ± 13.5 | 100 |
| tim; dcr > Inx3 RNAi | 23.62 ± 0.01 | 409.4 ± 16.76 | 100 |
| tim; dcr > Inx4 RNAi | 23.87 ± 0.09 | 325.8 ± 14.28 | 100 |
| tim; dcr > Inx5 RNAi | 23.52 ± 0.03 | 321.2 ± 6.9 | 95.23 |
| tim; dcr > Inx6 RNAi | 23.8 ± 0.06 | 376.5 ± 18.17 | 95.23 |
| tim; dcr > Inx7 RNAi | 23.73 ± 0.05 | 337.2 ± 91.18 | 100 |
| tim; dcr > Inx8 RNAi | 23.58 ± 0.04 | 461.3 ± 13.44 | 100 |
| UAS Inx1 RNAi | 23.5 ± 0.03 | 393.8 ± 13.24 | 100 |
| UAS Inx2 RNAi | 23.54 ± 0.02 | 424.2 ± 16.55 | 92.59 |
| UAS Inx3 RNAi | 23.56 ± 0.02 | 466.1 ± 20.9 | 100 |
| UAS Inx4 RNAi | 23.49 ± 0.03 | 327.5 ± 11.34 | 95.83 |
| UAS Inx5 RNAi | 23.19 ± 0.04 | 314.6 ± 14.84 | 96 |
| UAS Inx6 RNAi | 23.43 ± 0.02 | 437.2 ± 14.4 | 100 |
| UAS Inx7 RNAi | 23.56 ± 0.03 | 410.9 ± 51.2 | 96.15 |
| UAS Inx8 RNAi | 23.5 ± 0.03 | 454.6 ± 19.17 | 100 |
Table representing the average period (±SEM), power of the rhythm (POR) (±SEM) and % rhythmicity values of all the experimental (tim; dcr > Inx1-8 RNAi) lines used for the screen and their respective parental controls (w; timGal4; dcr) and UAS control (UAS Inx 1-8 RNAi).
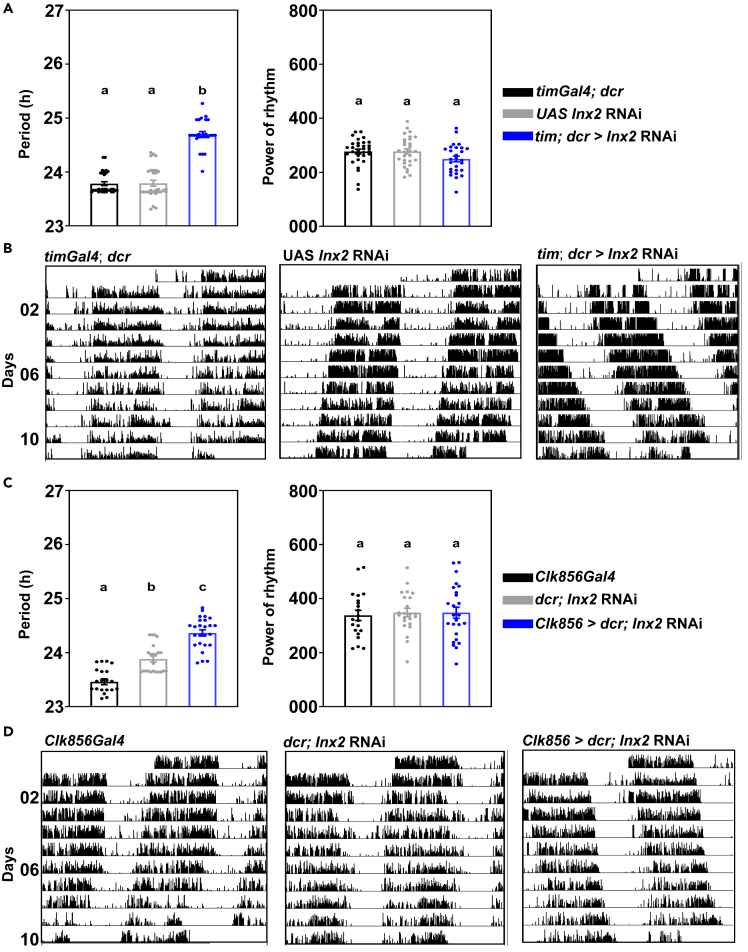
Figure 2.
Innexin2 knockdown in all clock neurons lengthens free-running period
(A) Free-running period (left) of experimental flies (tim; dcr > Inx2 RNAi) (n = 27) is significantly longer than both Gal4 control (n = 31) and UAS control (n = 29) flies, (one-way ANOVA, post-hoc Tukey’s test, p < 0.001) while the power of the chi-square periodogram (right) of the experimental flies is not significantly different from the parental controls.
(B) Representative double-plotted actograms of individual flies of each indicated genotype under constant darkness. On each day, activity is depicted as black vertical bars across the time of day which is double plotted such that 2 consecutive days are shown on the x axis.
(C) Free-running period (left) of experimental flies (Clk856 > dcr; Inx2 RNAi) (n = 27) is significantly longer than its Gal4 control (n = 21) and UAS control (n = 23) genotypes (one-way ANOVA, post-hoc Tukey’s test, p < 0.001), whereas the power of rhythm (right) is not different from controls.
(D) Representative double-plotted actograms of individual flies of each indicated genotype under constant darkness. On each day, activity is depicted as black vertical bars across the time of day which is double plotted such that 2 consecutive days are shown on the x axis. Error bars are SEM, period and power values are determined using Chi-square periodogram for a period of 8 days. All statistical comparisons were performed using one-way ANOVA with genotype as a fixed factor, followed by post-hoc analysis using Tukey’s Honest Significant Difference (HSD) test. Data plotted is from one out of 3 independent experiments. See Table 2; Figure S1 for more details.
Table 2.
Table depicting the genotype, no. of replicates, sample size, period and power of rhythm values when Innexin2 is downregulated in different clock neuronal subsets
| Genotype | N | n | Period ± SEM | Power of rhythm ± SEM |
|---|---|---|---|---|
| tim; dcr (Gal4 cont.) | 3 | 30 | 23.6 ± 0.01 | 419.2 ± 13.1 |
| 31 | 23.7 ± 0.03 | 276.8 ± 8.82 | ||
| 29 | 23.7 ± 0.06 | 268 ± 7.1 | ||
| UAS Inx2 RNAi | 3 | 25 | 23.5 ± 0.02 | 424.2 ± 16.5 |
| 29 | 23.7 ± 0.05 | 277.3 ± 10 | ||
| 29 | 23.7 ± 0.04 | 292.7 ± 12.2 | ||
| tim dcr > Inx2 RNAi | 3 | 18 | 24.6 ± 0.07 | 377 ± 13.5 |
| 27 | 24.7 ± 0.04 | 249.3 ± 10.52 | ||
| 18 | 24.65 ± 0.07 | 271.7 ± 13.7 | ||
| Clk856Gal4 (Gal4 cont.) | 3 | 21 | 23.4 ± 0.05 | 337.8 ± 18.8 |
| 29 | 23.6 ± 0.06 | 303.9 ± 11.7 | ||
| 22 | 23.7 ± 0.06 | 244.1 ± 9.19 | ||
| dcr: Inx2 RNAi (UAS cont.) | 3 | 23 | 23.8 ± 0.05 | 348.1 ± 15.7 |
| 29 | 23.3 ± 0.05 | 263.3 ± 11.7 | ||
| 22 | 24 ± 0.05 | 273.5 ± 13.1 | ||
| Clk856 > dcr; Inx2 RNAi | 3 | 27 | 24.3 ± 0.05 | 347.9 ± 20.4 |
| 26 | 24.5 ± 0.05 | 364.2 ± 14.5 | ||
| 27 | 24.3 ± 0.04 | 272.5 ± 11.36 | ||
| tim; tubGal80ts (Gal4 cont.; DD 29°C) | 3 | 18 | 24.2 ± 0.05 | 359.4 ± 24.9 |
| 21 | 24 ± 0.05 | 430.4 ± 23.5 | ||
| 23 | 24 ± 0.04 | 265.2 ± 13.9 | ||
| dcr; Inx2 RNAi (UAS cont.; DD 29°C) | 3 | 15 | 23.3 ± 0.09 | 302.7 ± 37.3 |
| 14 | 23.3 ± 0.03 | 551 ± 21.4 | ||
| 18 | 23.5 ± 0.04 | 384 ± 19.2 | ||
| tim; tubGal80ts> dcr; Inx2 RNAi (DD 29°C) | 3 | 19 | 25.1 ± 0.09 | 533.9 ± 14.9 |
| 27 | 24.9 ± 0.04 | 526.8 ± 15.1 | ||
| 27 | 24.7 ± 0.05 | 260.9 ± 9.7 | ||
| tim; tubGal80ts (Gal4 cont.; DD 19°C) | 2 | 22 | 23.7 ± 0.08 | 269.8 ± 13.6 |
| 26 | 23.6 ± 0.06 | 248.8 ± 8 | ||
| dcr; Inx2 RNAi (UAS cont.; DD 19°C) | 2 | 22 | 24 ± 0.09 | 279.4 ± 11.5 |
| 21 | 23.7 ± 0.06 | 232.7 ± 9.38 | ||
| tim; tubGal80ts> dcr; Inx2 RNAi (DD 19°C) | 2 | 23 | 23.5 ± 0.05 | 231 ± 10.5 |
| 26 | 23.7 ± 0.05 | 239 ± 12.5 | ||
| dvpdfGal4 (Gal4 cont.) | 26 | 24.1 ± 0.04 | 307.5 ± 17.2 | |
| dcr; Inx2 RNAi (UAS cont.) | 23 | 23.8 ± 0.03 | 363.9 ± 12.9 | |
| dvpdf > dcr;Inx2 RNAi | 16 | 25.7 ± 0.16 | 234.4 ± 11 | |
| pdfGal4 (Gal4 cont.) | 2 | 30 | 24.1 ± 0.04 | 418.4 ± 11.35 |
| 23 | 24.2 ± 0.06 | 244.5 ± 10.98 | ||
| dcr: Inx2 RNAi (UAS cont.) | 2 | 26 | 23.65 ± 0.04 | 371.6 ± 17.7 |
| 22 | 24 ± 0.05 | 273.5 ± 13.1 | ||
| pdf > dcr; Inx2 RNAi | 2 | 29 | 25 ± 0.03 | 414.6 ± 13.3 |
| 25 | 25.4 ± 0.05 | 287.4 ± 9.92 | ||
| LNdGal4 (Gal4 cont.) | 2 | 13 | 24 ± 0.06 | 524 ± 34.9 |
| 24 | 24 ± 0.03 | 413.4 ± 14.9 | ||
| dcr; Inx2 RNAi (UAS cont.) | 2 | 28 | 23.8 ± 0.04 | 416.8 ± 17.8 |
| 26 | 23.7 ± 0.04 | 288.9 ± 13.1 | ||
| LNd > dcr; Inx2 RNAi | 2 | 18 | 23.8 ± 0.05 | 450.2 ± 26.1 |
| 22 | 23.8 ± 0.04 | 345.5 ± 14.9 | ||
| Clk4.1MGal4 (Gal4 cont.) | 16 | 23.5 ± 0.05 | 359.2 ± 15.7 | |
| Clk4.5FGal4 (Gal4 cont.) | 17 | 23.7 ± 0.06 | 294.6 ± 13.2 | |
| dcr; Inx2 RNAi (UAS cont.) | 16 | 23.8 ± 0.04 | 385.9 ± 18.2 | |
| Clk4.1M > dcr; Inx2 RNAi | 24 | 23.7 ± 0.04 | 358.8 ± 9.97 | |
| Clk4.5F > dcr; Inx2 RNAi | 14 | 23.8 ± 0.04 | 372.9 ± 19.7 | |
| tim; pdfGal80 (Gal4 cont.) | 2 | 23 | 23.9 ± 0.06 | 334.3 ± 17.5 |
| 30 | 24.3 ± 0.06 | 328.2 ± 16.5 | ||
| dcr; Inx2 RNAi (UAS cont.) | 2 | 29 | 23.3 ± 0.05 | 263.3 ± 11.7 |
| 27 | 23.8 ± 0.08 | 317.4 ± 15.9 | ||
| tim; pdfGal80 > dcr; Inx2 RNAi | 2 | 25 | 24 ± 0.04 | 310.2 ± 12.4 |
| 29 | 24.25 ± 0.07 | 275.2 ± 14.25 | ||
| timGal4 (Gal4 cont.) | 28 | 24 ± 0.05 | 362.9 ± 13.3 | |
| pdfGal4 (Gal4 cont.) | 29 | 24.3 ± 0.03 | 393.7 ± 8.79 | |
| UAS RFP-Inx2 (UAS cont.) | 27 | 23.9 ± 0.05 | 385.5 ± 14.2 | |
| tim > RFP-Inx2 | 30 | 24.5 ± 0.05 | 429.1 ± 11.7 | |
| pdf > RFP-Inx2 | 31 | 24.6 ± 0.05 | 427.8 ± 13.6 |
Table representing the genotype, no. of replicates, no. of flies, average period (±SEM), average power of the periodogram (±SEM) values in case of Innexin2 knockdown in different subsets of clock neurons.
timA3Gal4 drives expression in many cells in the brain, most of the circadian pacemaker neurons, neurons in the optic lobe and some glial cells (Kaneko and Hall, 2000). Hence, we also used another driver, Clk856Gal4 which has a narrower expression pattern as compared to timGal4, but targets most clock neurons, except maybe few DNs (Gummadova et al., 2009). Innexin2 knockdown using Clk856Gal4 also resulted in the lengthening of free-running period of experimental flies as compared to their respective parental controls (Figures 2C, left and 2D; Table 2). Consistent with previous results, no significant difference in the power of the rhythm was observed in case of Innexin2 knockdown using the Clk856Gal4 (Figure 2C, right). We also observed period lengthening with a second Innexin2 RNAi construct (BL 80409) which is present on a different chromosome suggesting that the period lengthening phenotype seen is an effect of Innexin2 knockdown and not because of positional effects of transgene insertion (Figures S1A and S1B).
Innexin2 functions in the adult brain to modulate the free-running period of locomotor activity rhythm
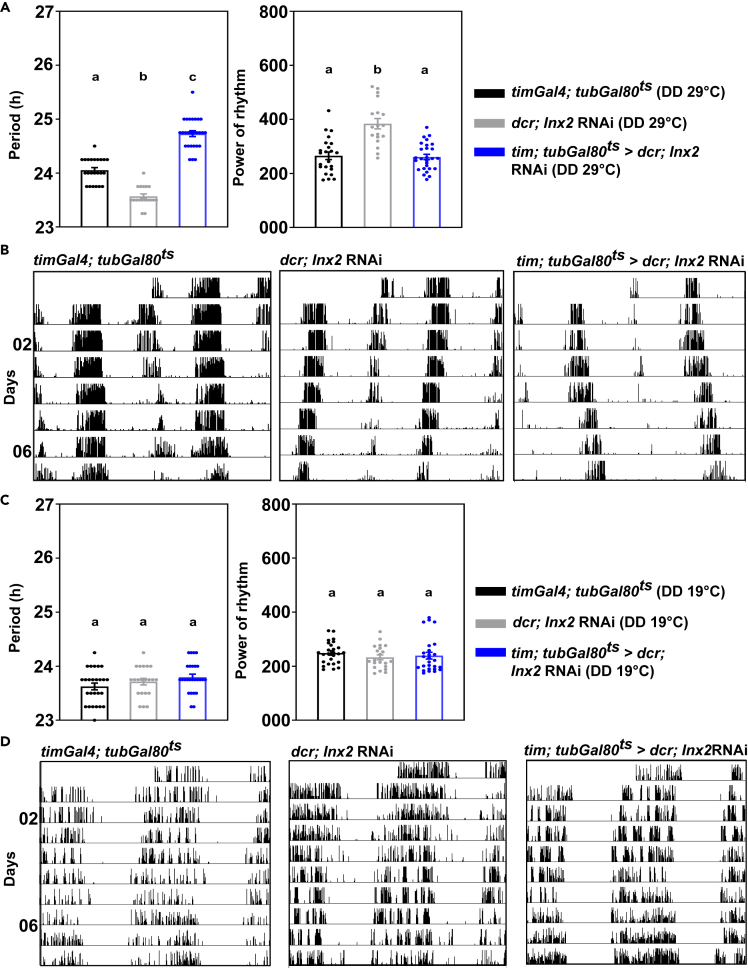
Since several previous studies have shown that Innexin2 plays crucial roles during several developmental processes in Drosophila (Bauer et al., 2002, 2004; Holcroft et al., 2013) we asked if the period lengthening seen in our experiments is due to defects in development of the circadian pacemaker circuit or due to roles played by Innexin2 in the mature, adult circuit. To distinguish between the two possibilities, we temporally restricted Innexin2 knockdown to the adult stages using the TARGET system (McGuire et al., 2004). TARGET system allows for temporal expression of gene of interest under UAS control using a temperature-sensitive Gal80ts, which is a repressor of Gal4. At a permissive temperature (19°C), Gal80ts is active and represses Gal4, thus UAS Innexin2 RNAi will not be expressed when flies are kept at this temperature. At restrictive temperatures (29°C), Gal80ts will be inactive; hence Gal4 will drive the expression of UAS Innexin2 RNAi in the cells of interest. Significant lengthening of period of activity rhythm was observed in experimental flies as compared with controls even when Innexin2 knockdown in clock neurons was restricted to the adult stages (Figures 3A, left and 3B; Table 2), suggesting that Innexin2 has roles in the adult circadian circuit to determine the period of free-running rhythms. Under such adult-specific Innexin2 knockdown, the power of the rhythm of experimental flies was found to be significantly lower only compared with its UAS control (Figure 3A, right). As a complementary experiment, we also restricted the knockdown of Innexin2 to only the developmental stages using the TARGET system. In this case, we do not observe a significant lengthening of period in the experimental flies as compared with its controls (Figures 3C, left and 3D; Table 2), with no change in the power of the rhythm (Figure 3C, right) thus strengthening the conclusion that Innexin2 functions in the adult Drosophila circadian circuit to determine free-running period. The efficiency of the system in repressing Gal4 was verified by expressing UAS-eGFP under the same driver, timGal4; tubGal80ts and assessing GFP expression by immunohistochemistry in the larval (L3) stage at 19°C and adult stages at 29°C. In the larval stages, we detect a faint trace of GFP in 1 out of 3–4 s-LNvs/hemisphere in a small fraction of brain samples (3/9) hemispheres, suggesting that tubGal80ts may have been unable to completely repress the expression of the Innexin2 RNAi construct during development (Figure S2).
Figure 3.
Innexin2 functions in the circadian circuit in adult stages to modulate free-running period
(A) Free-running period (left) of activity rhythm when Innexin2 knockdown in all the clock neurons is restricted to adult stages (tim; tubGal80ts > dcr; Inx2 RNAi), (n = 27) is significantly longer compared with its Gal4 control (n = 23) and UAS control (n = 18) flies (one-way ANOVA, post-hoc Tukey’s test at p < 0.001). Power of rhythm (right) of the experimental flies was found to be significantly lower only compared with the UAS control (one-way ANOVA, post-hoc Tukey’s test, p < 0.001). Data plotted is from one of 3 independent experiments.
(B) Representative double-plotted actograms of individual flies of each indicated genotype under constant darkness. On each day, activity is depicted as black vertical bars across the time of day which is double plotted such that 2 consecutive days are shown on the x axis.
(C) Free-running period (left) of activity rhythm in case of development-specific knockdown of Innexin2 in all clock neurons (tim; tubGal80ts > dcr; Inx2 RNAi) (n = 26) was not significantly different from its GAL4 control (n = 26) and UAS control (n = 21) genotypes, (one-way ANOVA, post-hoc Tukey’s test, p > 0.05) Power of rhythm (right) of the experimental flies was not different from both the parental control genotypes. Data representative from 2 independent experiments.
(D) Representative double-plotted actograms of individual flies of each indicated genotype under constant darkness. On each day, activity is depicted as black vertical bars across the time of day which is double plotted such that 2 consecutive days are shown on the x axis. Error bars are SEM, period and power values are determined using Chi-square periodogram for a period of 8 days in DD 29°C (for panel A and B) and DD 19°C (for panel C and D), respectively. All statistical comparisons were performed using one-way ANOVA with genotype as a fixed factor, followed by post-hoc analysis using Tukey’s Honest Significant Difference (HSD) test. See Table 2; Figure S2 for more details.
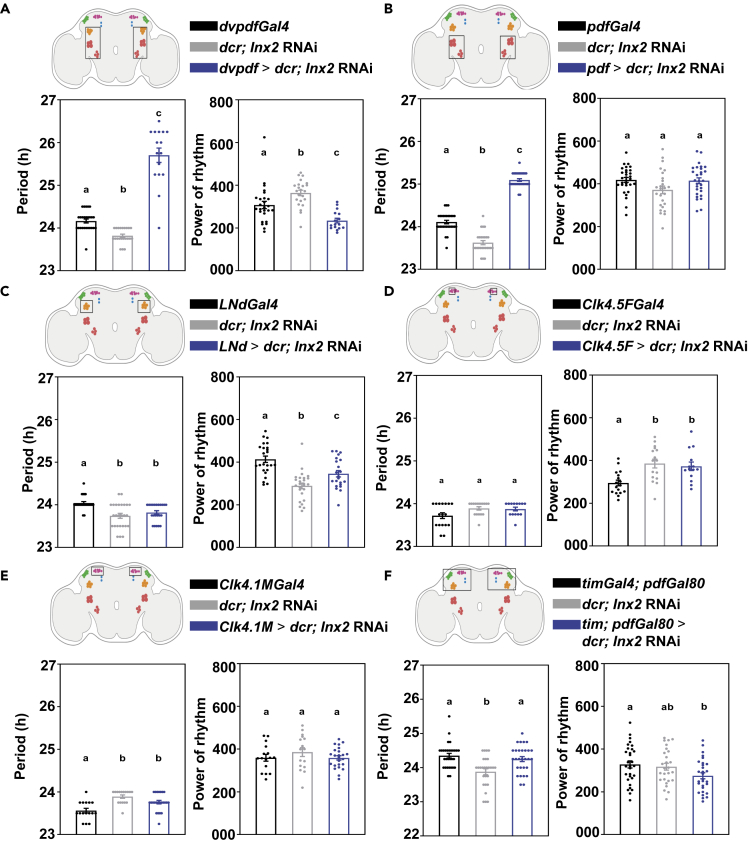
Innexin2 in ventral lateral neurons is important in determining the period of free-running rhythms
Since the knockdown of Innexin2 in all clock neurons lengthens period, we proceeded to downregulate its expression in smaller, distinct subsets of clock neurons to determine the functional role of Innexin2 in each of these subsets. First, we used dvpdfGal4 which targets the small and large ventral lateral neurons (s-LNv and l-LNv, respectively), as well as 4 dorsal lateral neurons (LNd). Knockdown of Innexin2 with dvpdfGal4 results in experimental flies having significantly longer period than both its parental genotypes (Figure 4A, left; Table 2). In this case, we also observed a significant decrease in the power of rhythm of the experimental flies (Figure 4A, right). Next, we used pdfGal4 which is only expressed in the ventral lateral neurons to knock down the levels of Innexin2. We found that the experimental flies have significantly longer period than their parental controls (Figure 4B; Table 2), suggesting that Innexin2 functions in the ventral lateral neurons in the circadian circuit. The period lengthening obtained in this case was similar to what was observed with Innexin2 knockdown using timGal4. To determine the contribution of Innexin2 in LNds to period lengthening, we downregulated the expression of Innexin2 using LNdGal4 (Bulthuis et al., 2019), which is strongly expressed in all 6 LNd cells but not in any ventral lateral neurons except for faint expression in some l-LNvs (Figure S3A). Knockdown of Innexin2 using LNdGal4 did not significantly change the free-running period or power of rhythm as compared with both parental controls (Figure 4C; Table 2), suggesting that, under free-running conditions, Innexin2 functions in the ventral neuronal subsets among the lateral neurons. We also downregulated Innexin2 expression in the dorsal neuronal subsets (DN1ps) using the Clk4.1M and Clk 4.5FGal4s but found no significant difference in the period or power of rhythms from their respective parental controls (Figures 4D and 4E; Table 2). Finally, to eliminate the contribution of other clock neurons apart from the ventral lateral neurons, we downregulated Innexin2 expression in all clock neurons except the LNvs using tim; pdfGal80. tim; pdfGal80 > Inx2 RNAi flies do not show a significantly lengthened period compared with both the controls (Figure 4F, left; Table 2) and there was no change in the power of the rhythm (Figure 4F, right), suggesting that the function of Innexin2 in the ventral lateral subset is critical for the maintenance of speed of activity-rest rhythm. The efficiency of the pdfGal80 construct in suppressing Gal4 expression in the ventral lateral neurons was verified via immunohistochemistry using a GFP marker (Figure S3B). Innexin2 is widely expressed in glial cells in Drosophila both during developmental stages and in the adult brain (Holcroft et al., 2013; Chaturvedi et al., 2014; Farca Luna et al., 2017). To examine if Innexin2 in glial cells contribute to modulate the free-running period, we downregulated its expression using two glial-specific drivers, repoGal4 (pan-glial driver) and alrmGal4 (expressed in astrocyte-like glia) (Ou et al., 2016). Fewer flies were obtained in case of Innexin2 knockdown using repoGal4, probably because of developmental lethality, as reported by previous studies (Holcroft et al., 2013; Chaturvedi et al., 2014). Knockdown of Innexin2 using the above glial specific drivers did not affect the free-running period of experimental flies as compared with control flies (Figure S4). No significant difference was observed in the power of rhythms of experimental flies as compared to parental controls (Figure S4), suggesting that Innexin2 in glial cells is probably not involved in modulating the free-running period. Innexin2, being a gap junction protein can function in cells either in the form of intercellular channels or hemichannels (Holcroft et al., 2013). Alternatively, many gap junction proteins, including Innexin2 have been shown to have channel-independent functions like cell adhesion and signaling roles in gene regulation (Dbouk et al., 2009; Elias and Kriegstein, 2008; Richard and Hoch, 2015). To distinguish between these two roles of Innexin2 in the clock neurons, we used a previously characterized mutant, where a reporter gene is fused with Inx2 (UAS RFP-Inx2) such that it interferes with the folding of the N-terminal domain which is essential for channel formation, thus affecting only the channel-based functions of Innexin2 (Nakagawa et al., 2010; Spéder and Brand, 2014). Expression of RFP-Inx2 with timGal4 lengthens the period of free-running rhythms in experimental flies as compared with controls without a significant change in the power of the rhythm (Figure S5; Table 2). Similarly, expression of RFP-Inx2 under pdfGal4 driver also lengthens the free-running period without any change in the power of the rhythm (Figure S5; Table 2). Taken together, these results suggest that Innexin2 could function as gap junctions or hemichannels in the ventral lateral neurons and a disruption of its channel forming domain lengthens the free-running period.
Figure 4.
Innexin2 knockdown in ventral lateral neurons lengthens free-running period
Depiction of adult Drosophila brains with the box indicating the circadian neuronal subsets targeted.
(A) Free-running period (left) of experimental flies (dvpdf > dcr; Inx2 RNAi) (n = 16) is significantly longer than both its Gal4 control (n = 26) and UAS control (n = 23) genotypes (one-way ANOVA with post-hoc Tukey’s test, p < 0.001) Power of rhythm (right) of the experimentals is significantly lower than both the parental genotypes (one way ANOVA with post-hoc Tukey’s test, p < 0.001).
(B) Free-running period (left) of experimental flies (pdf > dcr; Inx2 RNAi) (n = 29) is significantly longer than both its Gal4 control (n = 30) and UAS control (n = 26) genotypes, (one-way ANOVA with post-hoc Tukey’s test, p < 0.001). Power of rhythm (right) of the experimental flies is not significantly different from its parental genotypes.
(C) Free-running period (left) of experimental flies (LNd > dcr; Inx2 RNAi) (n = 22) is only significantly different from its Gal4 control (n = 24) and not from its UAS control (n = 26) genotype (one-way ANOVA, post-hoc Tukey's test, p < 0.01). Power of rhythm (right) of the experimentals is significantly different from its parental genotypes (one-way ANOVA, post-hoc Tukey's test, p < 0.05).
(D) Free-running period (left) of experimental flies (Clk 4.5F > dcr; Inx2 RNAi) (n = 14) is not significantly different from its Gal4 control (n = 17) and UAS control (n = 16) genotypes. Power of rhythm (right) of the experimental flies is significantly different only from its Gal4 control (one-way ANOVA with post-hoc Tukey’s test, p < 0.01).
(E) Free-running period (left) of experimental flies (Clk 4.1M > dcr; Inx2 RNAi) (n = 24) is significantly different only from its Gal4 control (n = 16) and not from its UAS control (n = 16) genotype (one-way ANOVA, post-hoc Tukey's test, p < 0.01). Power of rhythm of experimental flies is not different from its Gal4 and UAS control genotypes.
(F) Free-running period (left) of experimental flies (tim; pdfGal80 > dcr; Inx2 RNAi) (n = 29) is significantly different only from its UAS control (n = 27) and not from its Gal4 control (n = 30) genotype (one-way ANOVA, post-hoc Tukey's test, p < 0.01). Power of rhythm (right) of experimental flies is only significantly different from its Gal4 control genotype (one-way ANOVA, post-hoc Tukey's test, p < 0.05). Error bars are SEM, period values are determined using Chi-square periodogram for a period of 8 days. All statistical comparisons were performed using one-way ANOVA with genotype as a fixed factor, followed by post-hoc analysis using Tukey’s Honest Significant Difference (HSD) test. See Table 2; Figure S3, for more details. Adult brain depictions are created with BioRender.
Distribution of Innexin2 in the circadian pacemaker circuit
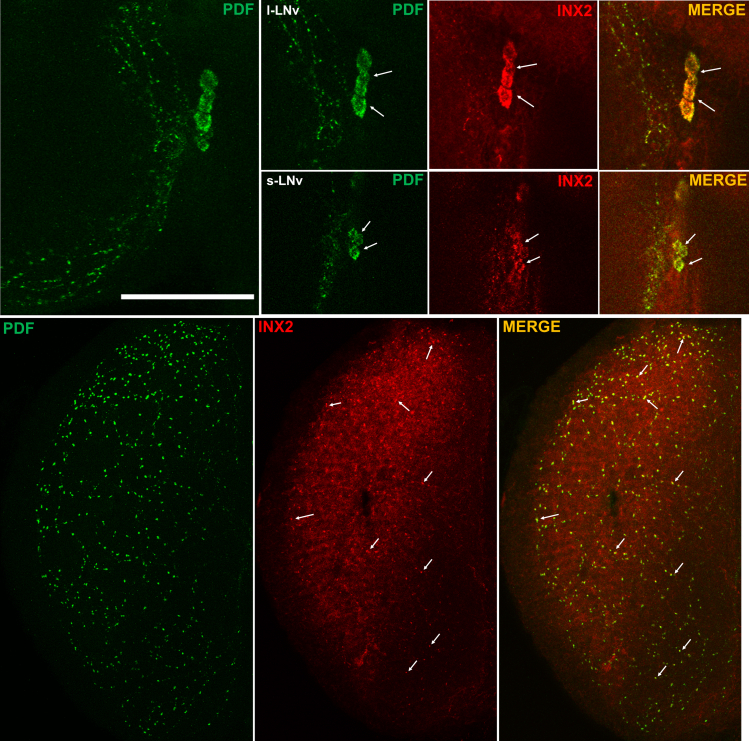
To investigate the distribution of Innexin2 protein in the adult Drosophila circadian circuit, we performed immunohistochemistry using a previously characterized anti-Innexin2 antibody (Bohrmann and Zimmermann, 2008). To examine the co-localization of Innexin2 with the clock neurons, we dissected the brains of wild-type flies and stained them with antibodies against Period (PER), Innexin2, and PDF. In accordance with our behavioral results, we find that among clock neurons, Innexin2 is expressed only in the ventral lateral neuronal subset i.e. the small and large ventral neurons (Figures 5, top panel and S6). Additionally, we also consistently observe Innexin2 in optic lobes co-localized with PDF marking the varicosities of l-LNv projections. Multiple such points of co-localization are marked in the image (Figure 5, lower panel, arrowheads). Thus, taken together, our results from behavioral and immunohistochemistry experiments suggest that Innexin2 protein expressed in the ventral lateral neuronal subsets of the circadian pacemaker circuit is critical in determining the period of free-running rhythms.
Figure 5.
Innexin2 is expressed in the small and large ventral lateral neurons in the circadian pacemaker circuit
Representative images of Drosophila adult brains showing the distribution of INX2 protein among clock cells. Flies were dissected at ZT12 and stained with anti-INX2 antibody and co-stained with anti-PDF for identification and co-localization with ventral lateral neurons. INX2 was found to be predominantly localized to the small and large ventral lateral neurons (s-LNv and l-LNv) among the clock neurons (arrows, top panel). INX2 was also found to be present in the varicosities of l-LNv projections in the optic lobes (arrowheads, bottom panel). Brightness and contrast of representative images were adjusted to facilitate better visualization. Arrows are used to indicate s-LNvs and l-LNvs. Scale-bar represents 55 μm, n = 12 brain samples were imaged. See also Figure S6.
The efficiency of the RNAi construct used for knockdown was examined in s-LNv and l-LNv using immunohistochemistry. While we find that Innexin2 levels were significantly reduced in the experimental flies in s-LNv, we could only observe a trend in case of l-LNvs (Figure S7). This could be because of our inability to detect a significant decrease in the levels with this given antibody concentration as Innexin2 levels are much higher in l-LNvs. Perhaps, a lower antibody concentration can be used for detecting differences in this case.
Knockdown of Innexin2 delays the phase of PERIOD protein oscillation in the circadian clock network and leads to higher levels of PDF accumulation in the s-LNv dorsal terminals
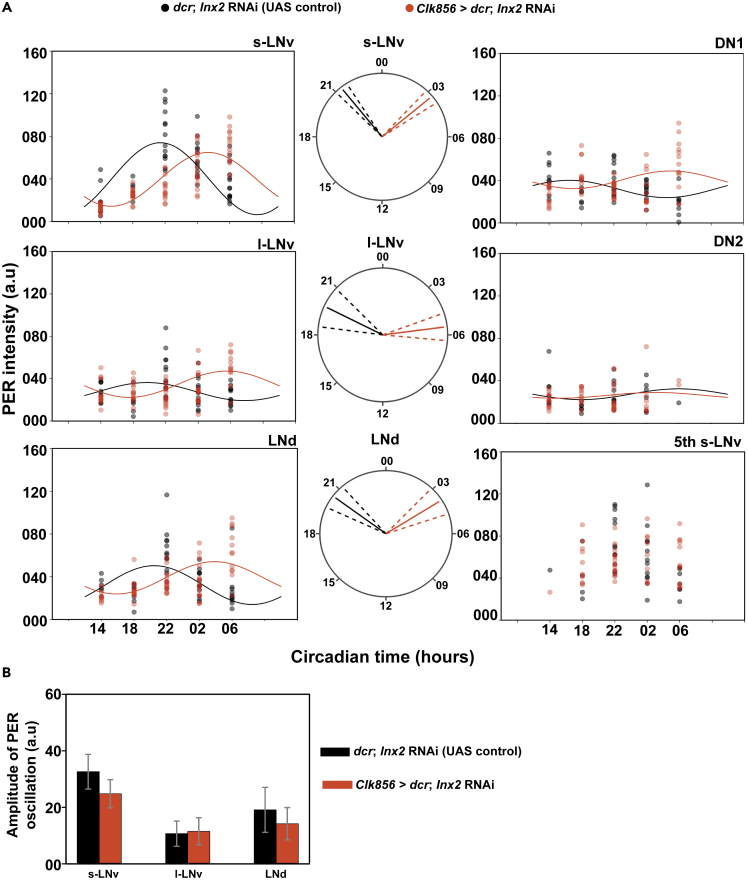
In order to examine the mechanism by which Innexin2 influences the period of free-running rhythms in Drosophila, we tracked the phase of oscillation of the core molecular clock protein, PERIOD (PER) in 6 circadian pacemaker cell clusters and also the levels of the clock output neuropeptide PDF in the s-LNv dorsal projections over a 24-hr cycle on the third day after introducing both control (dcr; Inx2 RNAi) and experimental (Clk856 > dcr; Inx2 RNAi) flies into constant darkness (DD). Although Innexin2 is present only in the ventral lateral neurons, upon Innexin2 downregulation, the oscillation of PERIOD protein is phase-delayed in other clock neuronal clusters in the circuit. Using a COSINOR-based curve-fitting method (Lee Gierke and Cornelissen, 2016), we found a significant 24-hr rhythm in PER oscillation in s-LNv in case of both control and experimental flies (Figure 6A; Table 3). The phase of PER oscillation in case of experimental flies was significantly delayed compared to control flies (Figures 6A and S8), suggesting that Innexin2 knockdown results in a delay in the core molecular clock protein oscillation in the s-LNv. The amplitude of oscillation upon Innexin2 downregulation was not found to be different from the controls (Figure 6B). In case of l-LNv, we could detect a significant 24-hr period for both control and experimental flies (Figure 6A and Table 3). The phase of oscillation was also significantly delayed in experimental flies as compared to controls (Figure 6A). The amplitude of PER oscillation in the l-LNv of experimental flies was not found to be different from the controls (Figure 6B). However, in control flies, consistent with previous reports, the amplitude of oscillation in l-LNv was found to be significantly lower than that of the s-LNv (Figure S9). In case of fifth s-LNv, we were unable to detect cells at timepoints of lower intensities; hence a COSINOR-based analysis was not performed for this cell type. Nevertheless, a scatterplot of control and experimental intensities are represented (Figure 6A). In case of LNd, both the control and experimental flies show a significant 24-hr rhythm in PER oscillation (Figure 6A; Table 3). The phase of oscillation of PER in experimental flies was found to be significantly delayed compared to controls (Figure 6A). The amplitude of oscillation in LNd was not found to be different between the control and experimental flies (Figure 6B). In case of DN1, although we could detect a significant 24-hr period in control flies, the amplitude of the oscillation was highly dampened (Figures 6A and S9; Table 3). In experimental flies however, DN1 do not show significant rhythmicity in PER oscillation and had overall low amplitude (Figure 6A; Table 3). PER levels in DN2 does not show a significant 24-hr rhythmicity and is highly dampened in both control and experimental flies (Figure 6A; Table 3).
Figure 6.
Knockdown of Innexin2 delays the oscillation of PER in clock neuronal subsets
(A) Scatterplots of PER staining intensities in each of the bilaterally located six distinct neuronal clusters of the circadian pacemaker network of both the control (dcr; Inx2 RNAi) and experimental (Clk856 > dcr; Inx2 RNAi) flies plotted at different time-points over a 24-hr cycle on third day of DD. Each dot represents the mean PER intensity value averaged over both the hemispheres of one brain. The black and red lines are the best fit COSINE curve from the parameters that were extracted using COSINOR analysis. Polar-plots are depicting the acrophase of PER oscillation in control (dcr; Inx2 RNAi) (black lines) and experimental flies (Clk856 > dcr; Inx2 RNAi) (red lines) in 3 distinct neuronal clusters of the circadian pacemaker network where both control and experimental genotypes show a significant 24-hr rhythm in COSINOR analysis. The acrophase values obtained after COSINOR curve fitting are shown as solid lines and the error (95% CI values) is depicted as dashed lines around the mean for all the cell types. Non-overlapping error values indicate that phase values of experimental flies are significantly different from controls as seen in the case of s-LNv, l-LNv, and LNd.
(B) Amplitude values obtained from COSINOR curve fits are plotted for control and experimental flies for those cell groups which show significant 24-hr rhythms (s-LNv, l-LNv and LNd). Error bars are 95% CI values calculated from the standard error obtained from COSINOR analysis. Overlapping error bars indicate that amplitude values of experimental flies are not significantly different from controls. COSINOR analysis was implemented using the CATCosinor function from the CATkit package written for R (Lee Gierke and Cornelissen, 2016). See Table 3; Figures S8 and S9 for more details. n > 7 brain samples both in case of control and experimental flies in most cell types (s-LNv, l-LNv, LNd, DN1) and all time points except for DN2.
Table 3.
Parameters obtained on fitting a COSINOR curve of 24-hr periodicity on the PER intensity data obtained on DD day3 for each of the circadian neuronal subsets and PDF intensity in s-LNv dorsal projections over a 24-hr cycle
| Cell type | p value | PR | Amplitude ± SE | Phase ± SE |
|---|---|---|---|---|
| s-LNv control | <0.05 | 53.7 | 33.82 ± 4.28 | −321.1 ± 6.15 |
| s-LNv experimental | <0.05 | 62.8 | 25.3 ± 2.59 | −50.12 ± 7.13 |
| l-LNv control | <0.05 | 11.8 | 8.52 ± 3.19 | −295.46 ± 18.38 |
| l-LNv experimental | <0.05 | 28.7 | 12.41 ± 2.64 | −82.94 ± 12.05 |
| LNd control | <0.05 | 23.8 | 18.16 ± 4.77 | −306.69 ± 11.78 |
| LNd experimental | <0.05 | 32.9 | 15.11 ± 2.94 | −57.67 ± 13.89 |
| DN1 control | <0.05 | 13.88 | 8.19 ± 3.01 | −247.28 ± 20 |
| DN1 experimental | >0.05 | 9.88 | 8.3 ± 3.48 | −75.34 ± 25.3 |
| DN2 control | >0.05 | 3.58 | 5.07 ± 4.99 | −89.82 ± 36.5 |
| DN2 experimental | >0.05 | 1.94 | 2.72 ± 3.38 | −47.2 ± 67.5 |
| PDF in DP control | <0.05 | 35.36 | 1.17 ± 0.2 | −52.71 ± 11.39 |
| PDF in DP experimental | <0.05 | 58.73 | 2.34 ± 0.25 | −65.41 ± 6.63 |
Table representing the parameters obtained after fitting a COSINE curve on the PER and PDF intensity data obtained over a 24-hr period for all the circadian neuronal subsets on third day of constant darkness. The parameters represented are p-values and percent rhythm (PR) to test for significant 24-hr periodicity, and the amplitude and phase values along with their respective standard errors (SE).
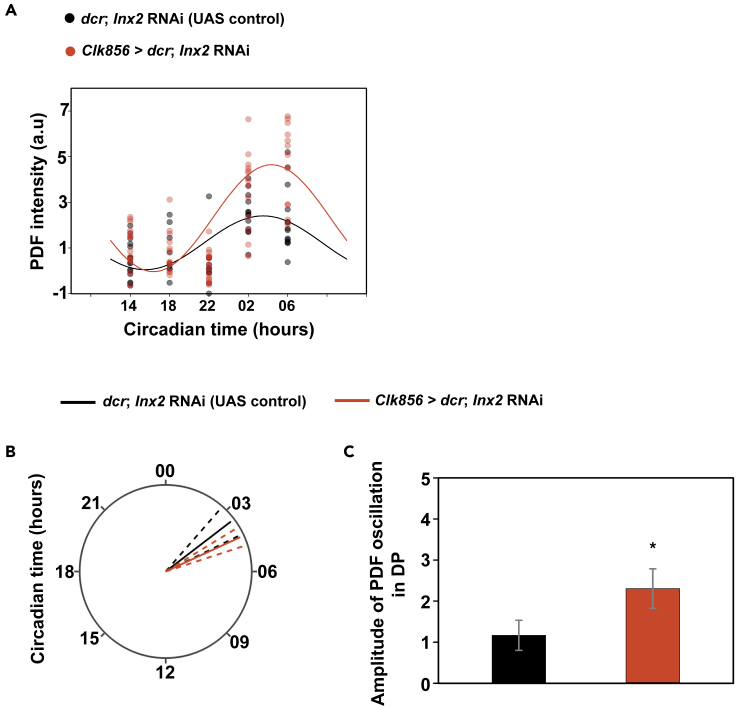
Since PDF is an important neuropeptide in the circadian pacemaker circuit for synchronization of clock neurons and acts as an output signal, we examined whether Innexin2 knockdown has an effect on the PDF levels or oscillations in the s-LNv dorsal terminal. We found that, both control and experimental flies show a robust 24-hr oscillation in PDF levels in the dorsal projections (Figure 7A; Table 3). In contrast to PER oscillation, there was no significant difference in the phase of PDF oscillation in experimental flies as compared to the control flies (Figures 7B and S10; Table 3). However, we observed that PDF levels and amplitude was significantly higher in experimental flies as compared with the control flies (Figures 7C and S10; Table 3), suggesting that Innexin2 levels could affect neuropeptide release from the s-LNv.
Figure 7.
Innexin2 knockdown affects the amplitude of PDF oscillation in the s-LNv dorsal projection
(A) Scatterplots of PDF intensity in the s-LNv dorsal projection of both the control (dcr; Inx2 RNAi) and experimental (Clk856 > dcr; Inx2 RNAi) flies plotted at different time-points over a 24-hr cycle on the third day of DD. Each dot represents the mean PDF intensity value averaged over both the hemispheres of one brain. The black and red lines are the best-fit COSINE curve from the parameters that were extracted from the COSINOR analysis.
(B) Polar-plots depicting the acrophase of PDF oscillation in control (dcr; Inx2 RNAi) and experimental flies (Clk856 > dcr; Inx2 RNAi). The phase values obtained after COSINOR curve fitting are shown as solid lines and the error values (95% CI) are depicted as dashed lines around the mean. Overlapping error bands indicate that the experimental phases are not significantly different from controls.
(C) Amplitude values obtained from COSINOR curve fits are plotted for control and experimental flies. Error bars are 95% CI values calculated from the standard error obtained from COSINOR analysis. Non-overlapping error bars indicate that experimental lines are significantly different from controls. COSINOR analysis was implemented using the CATCosinor function from the CATkit package written for R (Lee Gierke and Cornelissen, 2016) See Table 3; Figure S10 for more details. n > 9 brain samples both in case of control and experimental flies for all timepoints.
Discussion
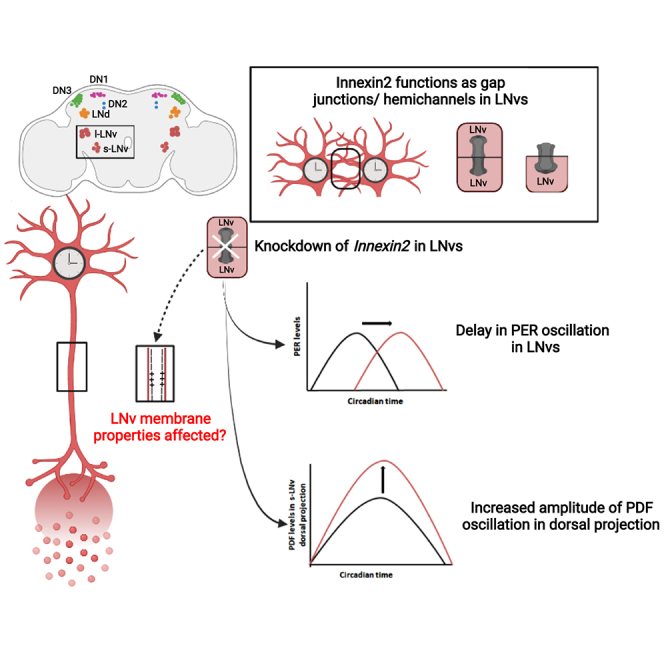
The neuronal and molecular mechanisms underlying circadian rhythms have been extensively studied in Drosophila melanogaster for many years now. Yet, we do not have a complete understanding of how free-running behavioral rhythms with a near 24-hr periodicity are generated by the network. Although membrane properties have been shown to be important for circadian behavior in Drosophila and there are some reports from mammalian studies on the role played by gap junctions in SCN neural network, there have been no systematic studies investigating the importance of electrical synapses in the circadian pacemaker circuit in Drosophila. We report here that gap junction genes play important roles in the Drosophila circadian pacemaker circuit to influence the period of free-running rhythms. Our RNAi screen revealed that Innexin2 levels in the LNv, a critical subset of the circadian network can affect the speed of the clock and alter the phase of oscillation of the core-clock protein PER and the amplitude of oscillation of the output neuropeptide PDF in the s-LNv dorsal projection.
Innexin2 functions in the mature, adult circadian circuit to influence free-running periodicity
Most of the studies which focus on the role of Innexins in Drosophila have described it extensively during development (Güiza et al., 2018), however in the recent past, several reports have implicated their roles in mature, adult nervous systems to modulate complex behaviors like learning, memory, and sleep (Liu et al., 2016; Troup et al., 2018; Wu et al., 2011). We find that Innexin2 plays predominant roles in the adult circadian circuit to influence the period of free-running rhythms. We also show that loss of Innexin2 during development in clock neurons does not affect the free-running period as adults. While previous studies have shown critical roles for Innexin2 in various aspects of nervous system development (Holcroft et al., 2013), our studies do not detect any defects in pace-making function of clock neurons due to the absence of Innexin2 in most clock neurons during the developmental stages. Interestingly, Innexin2 knockdown with dvpdfGal4 (Figure 4A), gives a greater period lengthening compared with other drivers. Since the only difference between dvpdf and pdfGal4 driver expression in the adult brain is that the former also targets the LNd neuronal subset (Bahn and Lee, 2009), we used LNdGal4 (Bulthuis et al., 2019) to target all 6 LNds, which did not result in significant period lengthening as compared with controls. Furthermore, we do not observe any Innexin2 expression in the LNd neurons. Therefore we speculate that the difference in period lengthening seen here is due to difference in the strength of RNAi mediated knockdown by the drivers.
Innexin2 phase-shifts the molecular clocks in circadian neurons
We found that, among clock neurons, Innexin2 is present and functions solely in the small and large ventral lateral neuronal subsets. Several previous studies have shown the importance of s-LNv and PDF in the circadian network under constant darkness to generate free-running rhythms of near 24-hr periodicity (Renn et al., 1999; Grima et al., 2004; Stoleru et al., 2004; Park et al., 2000; Yoshii et al., 2009; Sheeba et al., 2008a, 2008b; Yao and Shafer, 2014; Dissel et al., 2014; Schlichting et al., 2019; Delventhal et al., 2019). Since, Innexin2 is present in the s-LNv and influences the free-running period, we investigated the underlying mechanism. As a first step toward the same, we examined the phase of oscillation of molecular clock protein PERIOD on the third day of DD in both control and experimental flies in which Innexin2 was knocked down in all clock neurons. We found that the phase of the molecular clock is delayed in most clock neurons even though Innexin2 was only found to be present in the LNv. This is similar to what has been observed in case of Cx36 mutants in SCN where lack of Cx36 affects period of behavioral rhythms but does not affect the amplitude or synchrony among molecular clocks in the SCN slices (Long et al., 2005; Diemer et al., 2017). Phase of PER oscillations in case of s-LNv, l-LNv, and LNd cell types in experimental flies were found to be significantly delayed as compared with control flies. Although the amplitude of PER oscillations was not different between the experimental and control flies in each of these cell types, amplitude of oscillations in l-LNv in control flies were significantly lower than the s-LNv. It has been observed in several previous studies that amplitude of PER oscillation in the l-LNv dampens under constant darkness (Yang and Sehgal, 2001; Shafer et al., 2002; Peng et al., 2003; Roberts et al., 2015) similar to our experiment. In case of LNd, we find that the molecular clock in Innexin2 knockdown flies is delayed as compared with controls. Although the amplitude of oscillations in LNds in control flies is not significantly different from s-LNvs, we do observe lower amplitude values. This could be because LNds are a heterogeneous cell group which are differentially coupled to the s-LNvs (Yao and Shafer, 2014) and the low amplitude values could be the result of averaging PER intensity across these different cell types. In case of DN1, we observe highly dampened rhythms in control flies, similar to previous studies which have reported less robust rhythms, with patterns of dampening amplitude and loss of coherent rhythmicity in DN1 over 6 days in DD (Roberts et al., 2015; Yoshii et al., 2009). Lack of rhythmicity in DN1 in case of experimental flies could be explained by the fact that these cells receive conflicting signals from s-LNv and LNd with different periodicities (Zhang et al., 2010). In case of DN2, neither the control nor the experimental flies show significant 24-hr rhythmicity but have highly dampened rhythms. Although PDFR is expressed in DN2, molecular clock in DN2 was found to be independent of the control of s-LNv and do not seem to have profound effects on rhythmic activity-rest behavior in DD (Stoleru et al., 2005). Thus, overall our results indicate that Innexin2 influences the free-running period via the molecular clock and reduced Innexin2 levels result in delay in clock protein oscillations across the circuit. Since Innexin2 is present and functions only in the LNvs, we hypothesize that it affects the phase of molecular clocks in the LNv, which is then transmitted to the entire network possibly via PDF.
Mechanism of action of Innexin2 in circadian neurons
The mechanism of action of gap junctions have been well-studied in case of Connexins and not so much in case of Innexins, although the basic structure and function of the two types of proteins are similar (Beyer and Berthoud, 2018). Gap junctions in nervous systems are known to facilitate synchronous firing of neurons which can also affect the release of neuropeptide and neurotransmitters. Other than being involved in electrical coupling, gap junctions also facilitate cell to cell passage of secondary messengers and small molecules with less than 1 kDa size or they can act as hemichannels and facilitate transport between cells and extracellular matrix (Nielsen et al., 2012).
How does a gap junction protein localized to the membrane of LNv affect the phase of molecular clock protein oscillations in the network and the free-running period? We propose two possible hypotheses for the mechanism by which Innexin2 in LNv membrane could affect the molecular clock oscillations in circadian neurons. The first possibility is that Innexin2 could be involved in electrical coupling and synchronous neuronal firing among the LNv and its absence results in desynchronized firing which could affect the release of PDF from the LNv. This hypothesis is supported by previous studies in mammals which show that SCN neurons of gap junction mutants fire action potentials in a desynchronized manner (Long et al., 2005). Similar desynchronization of clock neuronal firing was also reported in invertebrates when gap junction blockers were applied to the bath while recording from accessory medulla region (Schneider and Stengl, 2006). While the significance of synchronized firing of circadian neurons for rhythmic behavior is unclear, examples from other instances have suggested that a pre-synaptic group of neurons are most efficient in driving post-synaptic sites when their firing is synchronized (Long et al., 2005). In our experiments, we find that the amplitude of PDF oscillation and level of PDF in the s-LNv dorsal projections is significantly higher when Innexin2 is downregulated. Several previous studies have shown that PDF acts to lengthen the period of the circadian network and that overexpression or ectopic expression of PDF in the dorsal protocerebrum lengthens the period of activity rhythms, leads to desynchronization of activity-rest behavior and molecular clocks in the circadian neurons (Helfrich-Förster et al., 2000; Wülbeck et al., 2008; Shafer and Yao, 2014).
An alternate possibility could be that Innexin2 is necessary to maintain the appropriate membrane potential of the LNv at specific times of the day or to regulate the number, frequency, or pattern of action potential firing in these cells. Disruption of the temporal pattern of firing in case of Innexin2 knockdown could translate into a delay in the molecular clocks in LNvs which in turn could be transmitted to other neurons in the circuit via PDF. Previous studies have shown that membrane excitability states of the LNv can affect both the core molecular clock and properties of activity-rest rhythms (Nitabach et al., 2002, 2006; Mizrak et al., 2012). Indeed, there is some evidence to suggest that gap junctions affect the frequency of firing of action potentials in the l-LNv membrane. A study by Cao and Nitabach (Cao and Nitabach, 2008) using gap junction blocker Carbenoxolone in the bath showed that blocking electrical synapses in the l-LNvs reduces the frequency of firing of action potentials in these cells. Presently, it is not clear how these changes in firing frequency may alter the core molecular clock in circadian pacemakers; however studies performed on neurons from the dorsal root ganglion suggest that expression levels of many genes are highly affected by firing frequency (Fields et al., 1997; Lee et al., 2017).
These two hypotheses cannot be considered as mutually exclusive and the actual mechanism of gap junctional modulation could be a combination of the two. While current experiments do not allow us to test these hypotheses, further experiments need to be done to address the following questions. Do Innexin2 mutant flies exhibit time-of-day dependent oscillations in membrane potential in the LNv? Does knockdown of Innexin2 affect the absolute membrane potential value of LNv as compared with controls or does it affect the synchronous firing of those neurons? These questions can be addressed by electrophysiological recording of the LNv although recording from the s-LNv can be technically challenging. An alternate approach would be to measure the spontaneous calcium activity rhythms in the LNvs, as well as all the other cells of the circuit in a time-of-day-dependent manner which can reveal changes in the membrane and biochemical states in circadian pacemaker neurons (Liang et al., 2016).
Apart from their roles in forming gap junctions, Innexin2 can also function as an intercellular signaling molecule in the circadian neurons. Do Innexin2 form gap junctions/hemichannels in the LNv? From our experiments with the Innexin2 mutant which selectively abolishes channel-based functions (Spéder and Brand, 2014), we can infer that Innexin2 probably works as gap junctions or hemichannels in these cells, although their other cellular roles cannot be completely excluded. Gap junction hemichannels can be classified as homomeric or heteromeric, i.e. composed of the same or different classes of gap junction proteins, respectively. Intercellular channels are called homotypic if the hemichannel is made up of a single protein and heterotypic if each of the two hemichannels are made of different type of proteins (Faber and Pereda, 2018). Previous studies have reported that Innexin2 can form functional heteromeric gap junctions with Innexin1, Innexin3, or Innexin4 and form homotypic gap junctions with itself to facilitate passage of ions and/or secondary messengers or small molecules (Bauer et al., 2003; Bohrmann and Zimmermann, 2008; Holcroft et al., 2013) or it could function as hemichannels (Holcroft et al., 2013). Since, we do not observe any period lengthening in case of Innexin3 or Innexin4 knockdown, the only possibilities are Innexin2 forming functional heteromeric gap junction with Innexin1 or homotypic gap junctions with itself. Alternatively, Innexin2 can also function as hemichannels and facilitate coupling between the cell and extracellular matrix. However, this needs further validation. The recently developed technique by Wu et al. (Wu et al., 2019) to demonstrate functional electrical coupling among cells – Pairing Actuators and Receivers to optically ISolate gap junctions could prove to be useful. Application of this method to determine functional electrical coupling among the clock neurons would be important to answer these questions.
Taken together, our findings highlight a hitherto unknown role for Innexins in the adult circadian pacemaker circuit of D. melanogaster in determining free-running period of activity rhythms and reveals that circadian timekeeping is brought about by a combination of electrical and chemical synapses in the underlying neuronal network.
Limitations of the study
-
•
This study relies on RNAi-mediated knockdown of Innexin2. Hence, it is yet unknown how the complete loss of Innexin2 affects clock function. Since homozygous mutants of Innexin2 are lethal, tissue-specific knockout of Innexin2 using the CRISPR-Cas9 system would be helpful.
-
•
Knockdown of Innexin2 affects the levels and amplitude of PDF cycling in the s-LNv dorsal terminals, it is also associated with changes in phase of downstream neurons. While we speculate that altered PDF cycling in s-LNv results in the changes seen downstream, whether it is causal is yet unknown.
-
•
Our results suggest a role for Innexin2, a membrane protein in modulating the phase of a core circadian clock protein; however, at this stage we provide no evidence for changes in membrane properties nor mechanistic information regarding how clock speed is affected.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-PER (Rabbit) | Obtained from Jeffrey Hall, Brandeis University | N/A |
| Anti-PDF (C7, Mouse) | Developmental Studies Hybridoma Bank (DSHB) | RRID: AB_760350 AB_2315084 |
| Anti-Innexin2 (cytoplasmic loop; Guinea pig) | Obtained from Michael Hoch and Reinhard Bauer, University of Bonn | Bohrmann and Zimmermann (2008) |
| Anti-GFP (chicken) | Life Technologies | Cat. No. A10262; RRID: AB_2534023 |
| Experimental models: Organisms/strains | ||
| UAS Innexin1 RNAi | Bloomington Stock Centre | RRID:BDSC_44048 |
| UAS Innexin2 RNAi | Bloomington Stock Centre | RRID:BDSC_42645 |
| UAS Innexin3 RNAi | Bloomington Stock Centre | RRID:BDSC_60112 |
| UAS Innexin4 RNAi | Bloomington Stock Centre | RRID:BDSC_27674 |
| UAS Innexin5 RNAi | Bloomington Stock Centre | RRID:BDSC_28042 |
| UAS Innexin6 RNAi | Bloomington Stock Centre | RRID:BDSC_44663 |
| UAS Innexin7 RNAi | Bloomington Stock Centre | RRID:BDSC_26297 |
| UAS Innexin8 RNAi | Bloomington Stock Centre | RRID:BDSC_57706 |
| UAS eGFP | Bloomington Stock Centre | RRID:BDSC_6874 |
| UAS tubGal80ts | Bloomington Stock Centre | RRID:BDSC_7017 |
| UAS Innexin2 RNAi | Bloomington Stock Centre | RRID:BDSC_80409 |
| Clk4.1MGal4 | Bloomington Stock Centre | RRID:BDSC_36316 |
| Clk4.5FGal4 | Bloomington Stock Centre | RRID:BDSC_37526 |
| UAS dicer-2 (II chromosome) | Bloomington Stock Centre | RRID:BDSC_24650 |
| UAS dicer-2 (III chromosome) | Bloomington Stock Centre | RRID:BDSC_24651 |
| repoGal4 | Bloomington Stock Centre | RRID:BDSC_7415 |
| alrmGal4 | Bloomington Stock Centre | RRID:BDSC_67032 |
| tim(A3)Gal4 | Obtained from Todd Holmes, UC Irvine | (Kaneko and Hall, 2000) |
| pdfGal4 | Obtained from Todd Holmes, UC Irvine | (Renn et al., 1999) |
| Clk856Gal4 | Obtained from Orie Shafer, ASRC, CUNY | (Gummadova et al., 2009) |
| pdfGal80 | Obtained from Helfrich-Forster, University of Wurzburg | Stoleru et al. (2004) |
| dvpdfGal4 | Obtained from Michael Rosbash, Brandeis University | (Bahn and Lee, 2009) |
| LNdGal4 | Obtained from Daniel Cavanaugh, Loyola University | (Bulthuis et al., 2019) |
| UAS RFP-Inx2 | Obtained from Andrea Brand, University of Cambridge | (Spéder and Brand, 2014) |
| Software and algorithms | ||
| Graphpad Prism 9.0 | Graphpad software | SCR_002798 https://www.graphpad.com/scientific-software/prism/ |
| ClockLab | Actimetrics | https://actimetrics.com/products/clocklab/ |
| RhythmicAlly | Abhilash and Sheeba, 2019 | https://github.com/abhilashlakshman/RhythmicAlly |
| DAM Filescan | Trikinetics | (Pfeiffenberger et al., 2010) |
| Fiji-ImageJ | Schindelin et al., 2012 | https://imagej.net/software/fiji/ |
| Cosinor function from Chronomics Analysis Toolkit (CAT) | (Lee Gierke and Cornelissen, 2016) | https://rdrr.io/cran/CATkit/man/CatCosinor.html |
| ActogramJ | Schmid et al., 2011 | https://bene51.github.io/ActogramJ/ |
| Other | ||
| Drosophila Activity Monitor (DAM) | Trikinetics | (Pfeiffenberger et al., 2010) |
| DAM Drosophila Environmental monitors | Trikinetics | (Pfeiffenberger et al., 2010) |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sheeba Vasu (sheeba@jncasr.ac.in).
Materials availability
• No new reagents are generated in this study.
• Further information and requests for resources and reagents should be directed to the lead contact, Sheeba Vasu (sheeba@jncasr.ac.in).
Experimental model and subject details
Fly lines and husbandry
All genotypes were reared on standard cornmeal medium under LD (12 hr Light: 12 hr Dark) cycles and 25°C, unless specified otherwise. The transgenic lines used in this study are UAS Innexin1 RNAi, UAS Innexin2 RNAi, UAS Innexin2 RNAi (BL 80409), UAS Innexin3 RNAi, UAS Innexin4 RNAi, UAS Innexin5 RNAi, UAS Innexin6 RNAi, UAS Innexin7 RNAi, UAS Innexin8 RNAi, UAS eGFP, UAS tubGal80ts, Clk4.1MGal4, Clk4.5FGal4, pdfGal4, tim(A3)Gal4, Clk856Gal4, pdfGal80, dvpdfGal4, LNdGal4, repoGal4, alrmGal4, UAS RFP-Inx2. See key resources table for more details on the transgenic lines and their source.
For experiments involving temporal knockdown during the adult stages, the flies were reared at a permissive temperature of 19°C from embryonic stages till 3 days after eclosion to allow for repression of Gal4 by tubGal80ts and facilitate proper development including the final pruning of synaptic connections in the nervous system. The flies were then transferred to LD 12:12 at 29°C and then assayed under constant darkness and restrictive temperature of 29°C. For experiments involving temporal knockdown of Innexin2 during developmental stages, the flies were reared at restrictive temperature of 29°C from embryonic stages till 3 days after eclosion to allow for Gal4 expression. The flies were then transferred to LD 12:12 at 19°C and then assayed under constant darkness and permissive temperature of 19°C.
Method details
Locomotor activity rhythm assay
Individual virgin male flies (4–6 days old) were housed in glass tubes (length 65 mm, diameter 7 mm) with corn food on one end and cotton plug on the other end. Locomotor activity was recorded using the Drosophila Activity Monitors (DAM, Trikinetics, Waltham, United States of America). Experiments were conducted in incubators manufactured by Sanyo (Japan) or Percival (USA) with controlled light and temperature conditions.
Immunohistochemistry
Brains from adult male Drosophila (4–6 days old) were dissected in ice-cold Phosphate buffered saline (PBS) and fixed immediately after dissection in 4% Paraformaldehyde (PFA) for 30 min. The fixed brains were then treated with blocking solution (10% horse serum) for 1-hr at room temperature and additional 6-hr at 4°C (additional incubation is only given in case of staining with anti-PER antibody to reduce staining of non-specific background elements), followed by incubation with primary antibodies at 4°C for 24–48 hr. The primary antibodies used were anti-PER (rabbit, 1:20,000, kind gift from Jeffrey Hall, Brandeis University), anti-PDF (mouse, 1:5000, C7, DSHB), anti-GFP (chicken, 1:2000, Invitrogen), anti-INNEXIN2 (guinea pig, 1:50, kind gift from Michael Hoch, University of Bonn). After incubation, the brains were given 6–7 washes with 0.5% PBS + Triton X-(PBT) after which they were incubated with Alexa-fluor conjugated secondary antibodies for 24-hr at 4°C. The following secondary antibodies were used, goat anti-chicken 488 (1:3000, Invitrogen), goat anti-rabbit 488 (1:3000, Invitrogen), goat anti-mouse 546 (1:3000, Invitrogen), goat anti-mouse 647 (1:3000, Invitrogen), goat anti-guinea pig 546 (1:3000, Invitrogen). The brain samples were further washed 6–7 times with 0.5% PBT, cleaned and mounted on a clean, glass slide in mounting media (7:3 glycerol: PBS). Exact same procedure was followed for experiments where immunostaining of larval brains (L3 stage) was required.
Quantification and statistical analysis
Activity data analysis
Raw data obtained from the DAM system were scanned and binned into activity counts of 15 min interval using DAM Filescan. Data was analyzed using the CLOCKLAB software (Actimetrics, Wilmette, IL) or RhythmicAlly (Abhilash and Sheeba, 2019). Values of period and power of rhythm were calculated for a period of 7–8 days using the Chi-square periodogram with a cut-off of p = 0.05. The period and power values of all the flies for a particular experimental genotype were compared against the parental controls using one-way ANOVA with genotype as the fixed factor followed by post-hoc analysis using Tukey’s Honest Significant Difference (HSD) test. The details on the statistical comparisons and the no. of flies used in a given experiment are indicated in the respective figure legends section. Representative actograms were generated using ActogramJ plugin of ImageJ (Schmid et al., 2011).
Image acquisition and analysis
The slides prepared for immunohistochemistry were imaged using confocal microscopy in a Zeiss LSM880 microscope with 20×, 40× (oil-immersion) or 63× (oil-immersion) objectives. Image analysis was performed using Fiji software (Schindelin et al., 2012). In the samples, clock neurons were classified based on their anatomical locations and expression of PER/PDF. PER intensity in these neurons were measured by selecting the slice of the z stack which shows maximum intensity, drawing a Region of Interest (ROI) around the cells and measuring their intensities. 3–6 separate background values were also measured around each cell and the final intensity was taken as the difference between the cell intensity and the background. For quantification of PDF in the dorsal projections, a rectangular box was drawn as ROI starting from the point where the PDF projection turns into the dorsal brain and intensity was measured. 3–6 background values were also measured around the projection. The intensity values obtained from both the hemispheres for each cell type for each brain was averaged and used for statistical analysis. We used a COSINOR based curve-fitting method (Cornelissen, 2014) to estimate different aspects of rhythmicity like presence of a 24-hr periodicity, phase and amplitude values of the oscillation. COSINOR analysis was implemented using the CATCosinor function from the CATkit package written for R (Lee Gierke and Cornelissen, 2016).
Acknowledgments
We are grateful to Michael Hoch and Reinhard Bauer for providing us anti-INX2 (cytoplasmic loop, guinea pig) antibody and to Jeffrey Hall for sharing the anti-PER (rabbit) antibody, and Orie Shafer, Todd Holmes, Michael Rosbash, Daniel Cavanaugh, Andrea Brand, and Charlotte Helfrich-Forster for kindly sharing fly lines; Jaimin Bhatt, Ankit Sharma, and Sushma Rao for help with dissections for the immunohistochemistry experiment; Ms Keerthana for help with imaging and Rajanna and Muniraju for technical assistance. We thank Abhilash Lakshman for useful discussions regarding statistical analysis of immunohistochemistry experiment data; and for help in plotting polar plots using R. We thank 2 anonymous reviewers and Abhilash Lakshman for comments and suggestions on a previous version of the manuscript. We also thank Gaiti Hasan for useful discussions and inputs at various stages of this project. This work was supported by JNCASR Intramural funds, a core research grant from DST-SERB (CRG/2019/006802) to VS and a fellowship from DST-INSPIRE to AR.
Author contributions
A.R., conceptualization, methodology, investigation, formal analysis, and writing-original draft; V.S., conceptualization, methodology, supervision, writing-review and editing, and funding acquisition.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103011.
Supplemental information
Data and code availability
-
•
All data will be made available upon email request to the lead contact, Sheeba Vasu (sheeba@jncasr.ac.in).
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyse the data reported in this paper will be available upon email request to the lead contact, Sheeba Vasu (sheeba@jncasr.ac.in).
References
- Abhilash L., Sheeba V. RhythmicAlly: your R and shiny–based open-source ally for the analysis of biological rhythms. J. Biol. Rhythms. 2019;34:551–561. doi: 10.1177/0748730419862474. [DOI] [PubMed] [Google Scholar]
- Allada R., Chung B.Y. Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J.H., Lee G., Jae H Park Comparative analysis of Pdf-mediated circadian behaviors between Drosophila melanogaster and D. virilis. Genetics. 2009;181:965–975. doi: 10.1534/genetics.108.099069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R., Lehmann C., Fuss B., Eckardt F., Hoch M. The Drosophila gap junction channel gene innexin 2 controls foregut development in response to Wingless signalling. J. Cell Sci. 2002;115:1859–1867. doi: 10.1242/jcs.115.9.1859. [DOI] [PubMed] [Google Scholar]
- Bauer R., Lehmann C., Martini J., Eckardt F., Hoch M. Gap junction channel protein innexin 2 is essential for epithelial morphogenesis in the Drosophila embryo. Mol. Biol. Cell. 2004;15:2992–3004. doi: 10.1091/mbc.E04-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R., Löer B., Ostrowski K., Martini J., Weimbs A., Lechner H., Hoch M. Intercellular communication: the Drosophila innexin multiprotein family of gap junction proteins. Chem. Biol. 2005;12:515–526. doi: 10.1016/j.chembiol.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Bauer R., Martini J., Lehmann C., Hoch M. Cellular distribution of innexin 1 and 2 gap junctional channel proteins in epithelia of the Drosophila embryo. Cell Commun. Adhes. 2003;10:221–225. doi: 10.1080/cac.10.4-6.221.225. [DOI] [PubMed] [Google Scholar]
- Beckwith E.J., Ceriani M.F. Communication between circadian clusters: the key to a plastic network. FEBS Lett. 2015;589:3336–3342. doi: 10.1016/j.febslet.2015.08.017. [DOI] [PubMed] [Google Scholar]
- Beyer E.C., Berthoud V.M. Gap junction gene and protein families: connexins, innexins, and pannexins. Biochim. Biophys. Acta Biomembr. 2018;1860:5–8. doi: 10.1016/j.bbamem.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrmann J., Zimmermann J. Gap junctions in the ovary of Drosophila melanogaster: localization of innexins 1, 2, 3 and 4 and evidence for intercellular communication via innexin-2 containing channels. BMC Dev. Biol. 2008;8:1–12. doi: 10.1186/1471-213X-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulthuis N., Spontak K.R., Kleeman B., Cavanaugh D.J. Neuronal activity in non-LNv clock cells is required to produce free-running rest: activity rhythms in Drosophila. J. Biol. Rhythms. 2019;34:249–271. doi: 10.1177/0748730419841468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Nitabach M.N. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R., Reddig K., Li H.S. Long-distance mechanism of neurotransmitter recycling mediated by glial network facilitates visual function in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2812–2817. doi: 10.1073/pnas.1323714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell C.S. Rhythmic coupling among cells in the suprachiasmatic nucleus. J. Neurobiol. 2000;43:379–388. doi: 10.1002/1097-4695(20000615)43:4<379::aid-neu6>3.0.co;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen G. Cosinor-based rhythmometry. Theor. Biol. Med. Model. 2014;11:1–24. doi: 10.1186/1742-4682-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk H.A., Mroue R.M., El-Sabban M.E., Talhouk R.S. Connexins: a myriad of functions extending beyond assembly of gap junction channels. Cell Commun. Signal. 2009;7:1–17. doi: 10.1186/1478-811X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delventhal R., O’Connor R.M., Pantalia M.M., Ulgherait M., Kim H.X., Basturk M.K., Canman J.C., Shirasu-Hiza M. Dissection of central clock function in drosophila through cell-specific CRISPR-mediated clock gene disruption. Elife. 2019;8:1–24. doi: 10.7554/eLife.48308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer T., Landgraf D., Noguchi T., Pan H., Moreno J.L., Welsh D.K. Cellular circadian oscillators in the suprachiasmatic nucleus remain coupled in the absence of connexin-36. Neuroscience. 2017;357:1–11. doi: 10.1016/j.neuroscience.2017.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S., Hansen C.N., Özkaya Ö., Hemsley M., Kyriacou C.P., Rosato E. The logic of circadian organization in Drosophila. Curr. Biol. 2014;24:2257–2266. doi: 10.1016/j.cub.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias L.A., Kriegstein A.R. Gap junctions: multifaceted regulators of embryonic cortical development. Trends Neurosci. 2008;31:243–250. doi: 10.1016/j.tins.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber D.S., Pereda A.E. Two forms of electrical transmission between neurons. Front. Mol. Neurosci. 2018;11:1–11. doi: 10.3389/fnmol.2018.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farca Luna A.J., Perier M., Seugnet L. Amyloid precursor protein in drosophila glia regulates sleep and genes involved in glutamate recycling. J. Neurosci. 2017;37:4289–4300. doi: 10.1523/JNEUROSCI.2826-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields R.D., Eshete F., Stevens B., Itoh K. Action potential-dependent regulation of gene expression: temporal specificity in Ca2+, cAMP-responsive element binding proteins, and mitogen-activated protein kinase signaling. J. Neurosci. 1997;17:7252–7266. doi: 10.1523/JNEUROSCI.17-19-07252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Chélot E., Xia R., Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Güiza J., Barría I., Sáez J.C., Vega J.L. Innexins: expression, regulation, and functions. Front. Physiol. 2018;9:1–9. doi: 10.3389/fphys.2018.01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummadova J.O., Coutts G.A., Robert N., Glossop J. Analysis of the Drosophila clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. J. Biol. Rhythms. 2009;24:353–367. doi: 10.1177/0748730409343890. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. The circadian timekeeping system of Drosophila. Curr. Biol. 2005;15:714–722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J. Comp. Physiol. A Sens. Neural Behav. Physiol. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C., Täuber M., Park J.H., Mühlig-Versen M., Schneuwly S., Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J. Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcroft C.E., Jackson W.D., Lin W.H., Bassiri K., Baines R.A., Phelan P. Innexins ogre and inx2 are required in glial cells for normal postembryonic development of the Drosophila central nervous system. J. Cell Sci. 2013;126:3823–3834. doi: 10.1242/jcs.117994. [DOI] [PubMed] [Google Scholar]
- Hyun S., Lee Y., Hong S.T., Bang S., Paik D., Kang J., Shin J., Lee J., Jeon K., Hwang S. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Jiang Z.G., Yang Y.Q., Allen C.N. Tracer and electrical coupling of rat suprachiasmatic nucleus neurons. Neuroscience. 1997;77:1059–1066. doi: 10.1016/s0306-4522(96)00539-8. [DOI] [PubMed] [Google Scholar]
- Kaneko M., Hall J.C. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A., Leloup J.C., Rouyer F. Circadian rhythms of locomotor activity in Drosophila. Behav. Process. 2003;64:161–175. doi: 10.1016/s0376-6357(03)00133-5. [DOI] [PubMed] [Google Scholar]
- Lee Gierke C., Cornelissen G. Chronomics analysis toolkit (CATkit) Biol. Rhythm Res. 2016;47:163–181. [Google Scholar]
- Lee P.R., Cohen J.E., Iacobas D.A., Iacobas S., Fields R.D. Gene networks activated by specific patterns of action potentials in dorsal root ganglia neurons. Sci. Rep. 2017;7:1–14. doi: 10.1038/srep43765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Holy T.E., Taghert P.H. Synchronous drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science (80-. ) 2016;351:976–981. doi: 10.1126/science.aad3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yang X., Tian J., Gao Z., Wang M., Li Y., Guo A. Gap junction networks in mushroom bodies participate in visual learning and memory in Drosophila. Elife. 2016;5:1–18. doi: 10.7554/eLife.13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M.A., Jutras M.J., Connors B.W., Burwell R.D. Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat. Neurosci. 2005;8:61–66. doi: 10.1038/nn1361. [DOI] [PubMed] [Google Scholar]
- McGuire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Science's STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Mertens I., Vandingenen A., Johnson E.C., Shafer O.T., Li W., Trigg J.S., De Loof A., Schoofs L., Taghert P.H. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Mizrak D., Ruben M., Myers G.N., Rhrissorrakrai K., Gunsalus K.C., Blau J. Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Curr. Biol. 2012;22:1871–1880. doi: 10.1016/j.cub.2012.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy J.I., Pereda A.E., Rash J.E. Electrical synapses in mammalian CNS: past eras, present focus and future directions☆. Biochim. Biophys. Acta Biomembr. 2018;1860:102–123. doi: 10.1016/j.bbamem.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Maeda S., Tsukihara T. Structural and functional studies of gap junction channels. Curr. Opin. Struct. Biol. 2010;20:423–430. doi: 10.1016/j.sbi.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Nielsen M.S., Axelsen L.N., Sorgen P.L., Verma V., Delmar M., Holstein-Rathlou N.H. Gap junctions. Compr. Physiol. 2012;2:1981–2035. doi: 10.1002/cphy.c110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach M.N., Blau J., Holmes T.C. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach M.N., Wu Y., Sheeba V., Lemon W.C., Strumbos J., Zelensky P.K., White B.H., Holmes T.C. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J. Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Gao Z., Song L., Ho M.S. Analysis of glial distribution in Drosophila adult brains. Neurosci. Bull. 2016;32:162–170. doi: 10.1007/s12264-016-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Helfrich-Förster C., Lee G., Liu L., Rosbash M., Hall J.C. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Stoleru D., Levine J.D., Hall J.C., Rosbash M. Drosophila free-running rhythms require intercellular communication. Plos Biol. 2003;1:32–40. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereda A.E. Electrical synapses and their functional interactions with chemical synapses. Nat. Rev. Neurosci. 2014;15:250–263. doi: 10.1038/nrn3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C., Lear B., Keegan K., Allada R. Processing circadian data collected from the Drosophila Activity Monitoring (DAM) system. Drosophila Neurobiology. Cold Spring Harbor. 2010 doi: 10.1101/pdb.prot5519. [DOI] [PubMed] [Google Scholar]
- Rash J.E., Olson C.O., Pouliot W.A., Davidson K.G.V., Yasumura T., Furman C.S., Royer S., Kamasawa N., Nagy J.I., Dudek F.E. Connexin36 vs. connexin32,“miniature” neuronal gap junctions, and limited electrotonic coupling in rodent suprachiasmatic nucleus. Neuroscience. 2007;149:350–371. doi: 10.1016/j.neuroscience.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S.C.P., Park J.H., Rosbash M., Hall J.C., Taghert P.H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Richard M., Hoch M. Drosophila eye size is determined by innexin 2-dependent decapentaplegic signalling. Dev. Biol. 2015;408:26–40. doi: 10.1016/j.ydbio.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Roberts L., Leise T.L., Noguchi T., Galschiodt A.M., Houl J.H., Welsh D.K., Holmes T.C. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr. Biol. 2015;25:858–867. doi: 10.1016/j.cub.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting M., Díaz M.M., Xin J., Rosbash M. Neuron-specific knockouts indicate the importance of network communication to drosophila rhythmicity. Elife. 2019;8:1–20. doi: 10.7554/eLife.48301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B., Helfrich-Förster C., Yoshii T. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J. Biol. Rhythms. 2011;26:464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- Schneider N.L., Stengl M. Gap junctions between accessory medulla neurons appear to synchronize circadian clock cells of the cockroach Leucophaea maderae. J. Neurophysiol. 2006;95:1996–2002. doi: 10.1152/jn.00835.2005. [DOI] [PubMed] [Google Scholar]
- Shafer O.T., Kim D.J., Dunbar-Yaffe R., Nikolaev V.O., Lohse M.J., Taghert P.H. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O.T., Rosbash M., Truman J.W. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O.T., Yao Z. Pigment-dispersing factor signaling and circadian rhythms in insect locomotor activity. Curr. Opin. Insect Sci. 2014;1:73–80. doi: 10.1016/j.cois.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V. The Drosophila melanogaster circadian pacemaker circuit. J. Genet. 2008;87:485–493. doi: 10.1007/s12041-008-0071-x. [DOI] [PubMed] [Google Scholar]
- Sheeba V., Kaneko M., Sharma V.K., Holmes T.C. The Drosophila circadian pacemaker circuit: Pas de Deux or Tarantella? Crit. Rev. Biochem. Mol. Biol. 2008;43:37–61. doi: 10.1080/10409230701829128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V., Sharma V.K., Gu H., Chou Y.T., O’Dowd D.K., Holmes T.C. Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J. Neurosci. 2008;28:217–227. doi: 10.1523/JNEUROSCI.4087-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K., Hiruma H., Funabashi T., Kimura F. GABAergic modulation of gap junction communication in slice cultures of the rat suprachiasmatic nucleus. Neuroscience. 2000;96:591–596. doi: 10.1016/s0306-4522(99)00556-4. [DOI] [PubMed] [Google Scholar]
- Spéder P., Brand A.H. Gap junction proteins in the blood-brain barrier control nutrient-dependent reactivation of Drosophila neural stem cells. Dev. Cell. 2014;30:309–321. doi: 10.1016/j.devcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Nawathean P., Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Troup M., Yap M.H.W., Rohrscheib C., Grabowska M.J., Ertekin D., Randeniya R., Kottler B., Larkin O., Munro K., Shaw P.J., van Swinderen B. Acute control of the sleep switch in drosophila reveals a role for gap junctions in regulating behavioral responsiveness. Elife. 2018;7:1–22. doi: 10.7554/eLife.37105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.H., Chen N., Wang J.H. The coupling features of electrical synapses modulate neuronal synchrony in hypothalamic superachiasmatic nucleus. Brain Res. 2014;1550:9–17. doi: 10.1016/j.brainres.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Welsh D.K., Reppert S.M. Gap junctions couple astrocytes but not neurons in dissociated cultures of rat suprachiasmatic nucleus. Brain Res. 1996;706:30–36. doi: 10.1016/0006-8993(95)01172-2. [DOI] [PubMed] [Google Scholar]
- Wu C.L., Shih M.F.M., Lai J.S.Y., Yang H.T., Turner G.C., Chen L., Chiang A.S. Heterotypic gap junctions between two neurons in the drosophila brain are critical for memory. Curr. Biol. 2011;21:848–854. doi: 10.1016/j.cub.2011.02.041. [DOI] [PubMed] [Google Scholar]
- Wu L., Dong A., Dong L., Wang S.Q., Li Y. PARIS, an optogenetic method for functionally mapping gap junctions. Elife. 2019;8:1–22. doi: 10.7554/eLife.43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wülbeck C., Grieshaber E., Helfrich-Förster C. Pigment-dispersing factor (PDF) has different effects on Drosophila’s circadian clocks in the accessory medulla and in the dorsal brain. J. Biol. Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]
- Yang Z., Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yao Z., Shafer O.T. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Wülbeck C., Sehadova H., Veleri S., Bichler D., Stanewsky R., Helfrich-Förster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J. Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chung B.Y., Lear B.C., Kilman V.L., Liu Y., Mahesh G., Meissner R.A., Hardin P.E., Allada R. DN1p circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr. Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data will be made available upon email request to the lead contact, Sheeba Vasu (sheeba@jncasr.ac.in).
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyse the data reported in this paper will be available upon email request to the lead contact, Sheeba Vasu (sheeba@jncasr.ac.in).