Abstract
The main protease (Mpro) is the key enzyme of nCOVID-19 and plays a decisive role that makes it an attractive drug target. Multiple analysis of crystal structures reveals the presence of W1, W2, and W3 water locations in the active site pocket of Mpro; W1 and W2 are unstable and are weakly bonded with protein in comparison to W3 of Mpro-native. So, we adopt the water displacement method to occupy W1 or W2 sites by triggering HCQ or its analogs to inactivate the enzyme. Virtual screening is employed to find out best analogs of HCQ, molecular docking is used for water displacement from catalytic region of Mpro, and finally, MD simulations are conducted for validation of these findings. The docking study reveals that W1 and W2 are occupied by respective atoms of ZINC28706440 whereas W2 by HCQ and indacaterol. Finally, MD results demonstrate (i) HCQ occupies W1 and W2 positions, but its analogs (indacaterol and ZINC28706440) are inadequate to retain either W1 or W2 (ii) His41 and Asp187 are stabilized by W3 in Mpro-native and His41, Cys145 and HCQ by W7 in ZINC28706440, and W4, W5, and W6 make water mediated bridge between indacaterol with His41. The structural, dynamical, and thermodynamic (WFP and J value) profiling parameters suggest that W3, W4, and W7 are prominent in their corresponding positions in comparison with W5 and W6. The final results conclude that ZINC28706440 may act as a best analog of HCQ with acceptable physico-chemical and toxicological scores and may further be synthesized for experimental validation.
Keywords: Hydroxychloroquine (HCQ), Structural analogs of HCQ, Computational binding study, MD Simulation, Water displacement method, Main protease, nCOVID-19
Abbreviations: SARS, severe acute respiratory syndrome; CoV, coronavirus; nCOVID-19, novel coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; HCQ, hydroxychloroquine; Mpro, main protease; 3CLpro, 3C-like protease; HTS, high-throughput screening; WFP, water finding probability
Graphical abstract
1. Introduction
The World Health Organization (WHO) declared on March 11th, 2020, that the outbreak of “Novel CoronaVirus Disease 2019” (nCOVID-19) had become a pandemic and was one of the highly pathogenic that infected humans [1]. Since the onset of the pandemic, there were more than 200 million people confirmed nCOVID-19 cases globally, leading to over more than 4 million deaths were reported to WHO [2]. A single or combined test that provides virological and serological output would be ideal to identify individuals who had been infected. The novel coronavirus had a rapid spread to most of the countries till date and caused deadly deaths with a 2% mortality rate. Risk factors for mortality include advanced age, obesity, diabetes, and hypertension. However, there are no clinically approved or specific therapeutic drugs that are available for nCOVID-19 [3]. Intensive research on SARS-CoV-2 is urgently required for the treatment of nCOVID-19. The test, trace, and treat (T3) programs will become mandatory and may act as effective drugs for safe therapies [4].
Vaccines might not be enough to quell the pandemic, although many repurposed drug candidates are being evaluated, many are redundant, and lack a strong rationale for clinical development [5]. Earlier in the course of the disease, due to lack of specific treatment for nCOVID-19, pre-clinical investigation explored the usage of hydroxychloroquine (HCQ), an anti-malarial drug, against nCOVID-19. Many clinical reports and research articles have explored the effectiveness of HCQ for the treatment of nCOVID-19 [[6], [7], [8], [9]]. However, HCQ is a well-known to have some complications and side-effects in some cases. The cause for a failure of HCQ treatment should be investigated by testing the isolated SARS-CoV-2 strains of the non-respondents and analyzing their genome that may be associated with the metabolism of HCQ. Medicine like remdesivir, HCQ, lopinavir, and interferon regimens had a small or no effect on hospitalized patients with nCOVID-19, as recorded by overall mortality, initiation of ventilation, and duration of hospital stay [10]. The nCOVID-19 Treatment Guidelines Panel recommended against the use of chloroquine or HCQ with or without azithromycin for the treatment of nCOVID-19 in hospitalized and nonhospitalized patients. Furthermore, the Panel also advised against the use of high-dose chloroquine (600 mg twice daily for 10 days) for the treatment of nCOVID-19. However, despite demonstrating antiviral activity in some in vitro systems, HCQ with or without azithromycin did not reduce upper or lower respiratory tract viral loads or demonstrate clinical efficacy in a rhesus macaque model [11]. The trial enrollment ended early on 5th June 2020, after an independent data-monitoring committee recommended reviewing the unblinded data, and the investigators and trial-steering committee also concluded that the data showed no beneficial effect of hydroxychloroquine [12,13].
According to the phylogenetic and genomic information, the coronaviruses are genotypically categorized into four genera: α-CoV, β- CoV, γ- CoV, and δ-CoV. Furthermore, its β‐type is subdivided into four viral lineages of A, B, C, and D. However, CoV has a substantial single‐stranded RNA virus genome (27–32 kb) [14] and its virion contains two major components: genomic RNA and a protein capsid. The RNA genome of n-Cov-19 has 29891 nucleotides in length that encode 9860 amino acids. Moreover, its genome contains the following items: two flanking untranslated regions (UTRs), a single open reading frame (7096-aa), a non-structural polyprotein (7096-aa), four structural proteins – Spike (S) (1273-aa), Envelope (E) (75-aa), Membrane (M) (222-aa), Nucleocapsid (N) (419-aa), and five accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) [15].
The main protease (Mpro) or 3C-like protease (3CLpro) is another key enzyme in the life cycle of nCOVID-19, playing a pivotal role in mediating viral replication, and transcription that makes it an attractive drug target for this virus [16]. Mpro resembles the structure of cysteine proteases with two antiparallel β-barrels domains [17]. The active site of this enzyme comprises a catalytic dyad His41 and Cys145, and lacks the third catalytic residue, which is replaced by a stable water molecule [18] (Fig. S1). This Cys-His catalytic dyad of this enzyme is located in a cleft between the domains I and II. The N-terminal residues (res. ID. from 1 to 7) of Mpro are considered to play an eminent role in the proteolytic activity, whereas the –C terminal (domain III) is reported to be required for dimerization [19]. Crystal or NMR structures for Mpro protein with HCQ or its analogs are not yet available because the determination of such complex structures may be difficult. In the face of global economic recession, the developments of infrastructure for such experimental trails are really challenging for most of the researchers due to high cost. Thus, the approach adopted in our investigation is to search structural analogs of HCQ for possible inhibition of SARS-CoV-2. The functional differences between HCQ and its analogs may translate into performance variances that could potentially influence the decision process in either HCQ or its analogs and can ultimately become a viable therapeutic agent for nCOVID-19.
As we know, water molecules play a vital role in protein structure, enzyme catalysis, protein architecture, conformational stability, and protein plasticity, stabilization of salt bridges, ligand binding, and selectivity for specific interactions [[20], [21], [22], [23], [24], [25], [26], [27]]. However, drug repurposing has been a strategy adopted by several research groups to provide effective treatment in a short period. Therefore, few computational studies have been reported including a massive virtual screening, molecular docking, followed by molecular dynamics (MD) simulation for obtaining new anti-COVID molecules. The static structural data do not explain the functional and dynamical nature of water molecules in the Mpro protein, changes of geometrical and electronic variations at the ligand binding site after binding HCQ. In this context, it is important to trace the stepwise movement of the biochemical mechanism of Mpro from native to ligand bound state, its dynamics, and conformational changes using MD simulation. During the MD simulations, the movements of ligands in respect of active site residue could explain interesting dynamical phenomena that are generally inaccessible in crystal structure of Mpro.
The estimation of binding free energy for the ligand may play a decisive role to achieve experimentally determined value for target proteins, but accurate binding energy calculation of ligand remains a formidable challenge. In structure-based drug design, the free energy perturbation and thermodynamic integration methods are not commonly used to compute binding energy calculation of ligand because these methods are implemented numerically, require sufficient statistical sampling, are time-consuming, and are computationally expensive [28]. Hence, linear interaction energy (LIE) and molecular mechanics/Poisson−Boltzmann surface area (MM/PBSA) are the alternative and popular computational methods that are broadly used in structure-based drug discovery to calculate the binding energy of ligand in a fast and rational way [29]. In this study, we can therefore evaluate and compare the performance of LIE-D and MM/PBSA methods with respect to their ability and efficacy. In the current work, we investigated whether the LIE-D can achieve similar accuracy compared to MM/PBSA in calculating binding affinities of HCQ and its analogs? These approaches are efficient, less time consuming, and are also computationally inexpensive than other methods mentioned above.
Numerous computational methods like molecular dynamics simulations with inhomogeneous fluid solvation theory [30] allow us to identify the specific geometrical and electronic locations of invariant water molecules in the active site of the protein and also compute their thermodynamic properties [31]. Multiple analyses of crystal structures of the native form of Mpro protein provide suggestions into which water molecules can be targeted for displacement. The medicinal chemistry methods explain that tightly bound and well-ordered water molecules may gain entropy and are difficult to displace them by ligands, whereas weakly bound water molecules are mostly favorable for replacement by ligands that may expect to increase binding affinities [32]. To the best of our knowledge this is the first computational study that explores the results of molecular docking on the basis of crystal water displacement in the active site pocket of Mpro and these data are validated by MD analysis by focusing on conformational dynamics of HCQ and its analogs. Moreover, the integrated findings of single trajectory simulations are the starting structures which were obtained from molecular docking study from specific binding poses of HCQ or its analogs. No MD simulation study has yet been reported on structural transition of Mpro from its native to HCQ or its analogs bound state to capture the conformational transitions. Our computational techniques act as a complementary approach to synthesis of new compounds or repurposing of drugs. The vision of present study is to evaluate the inhibitors that are structurally similar to HCQ, and it will be compared to Mpro protein in the complex. In this aspect, our computational study provides a testable hypothesis for the binding mechanism of HCQ or its structurally similar analogs that may be synthesized experimentally for further validation and also guide us to solve the crystal structure of the Mpro with HCQ bound complex.
2. Materials and methods
2.1. Structures collection and identification of conserved water molecules at catalytic region of Mpro
In the present study, the eleven (PDB ID: 6YB7, 6Y84, 7ALH, 7ALI, 7BRO, 7JR3, 7KPH, 7C2Y, 7CWB, 7JUN, and 7JVZ) ligand-free crystal structures of the main protease were obtained from the Protein Data Bank [33]. The PDB ID: 6YB7 (resolution 1.25 Å) was taken as a template for further molecular docking and MD simulation study because it adopted an open conformation to accommodate ligands in the active site (Table S1). Three conserved water molecules (W1, W2, and W3) were identified at the catalytic region of these ligand-free crystal structures using the 3dSS program [34]; besides, the conserved positions of these water molecules were also verified by the Swiss PDB Viewer program [35]. Two water molecules (oxygen atoms) or two atoms (oxygen atoms between two water molecules or oxygen atom of water molecule and specific atom of ligand) whose center-to-center distance was within 1.80 Å, [36], in between reference (PDB ID: 6YB7), and movable structures were assigned as conserved [37,38]. The W1 is H-bonded with ND1 of His41 in 6YB7, 6Y84, 7ALH, 7ALI, 7BRO, 7JR3, and 7KPH crystal structures, W2 is also H-bonded only with W1, and W3 makes H-bonded with His41 and Asp187 in 7C2Y, 7CWB, 7JUN, and 7JVZ; therefore, they were assigned as conserved water positions at the catalytic region of native Mpro protein. The solvent-accessible surface area (SASA) of W1, W2, and W3 water molecules were measured using the in-house TCL script with a probe radius of 1.4 Å. The normalized B factor (BNorm) for all conserved water molecules were calculated using the formula, BNorm = (Bi –)/σ(B), where Bi is the B factor of each atom of protein, is the mean B factor of the protein molecule, and σ(B) is the standard deviation of the B factors [39].
2.2. Virtual screening and QSAR property analysis
The SMILES [CCN(CCO)CCCC(C)NC1=C2C CC(Cl) CC2=NC C1] of HCQ was obtained from the Drug-Bank database (v5.1.5) [40] and it was used for high throughput screening to investigate structural analogs of HCQ by Swiss Similarity program [41]. Four chemical libraries (Ligands from PDB, ChEMBL, Zinc-drug like, and Zinc-lead like) were chosen for the high throughput screening (HTS) to achieve the best structural analogs of HCQ. After HTS, the fifty structural analogs of HCQ are observed with reasonably good similarity scores like HCQ and finally, the best 14 HCQ analogs (mentioned in the supplementary section of Fig. S2) were obtained on the basis of similarity scores for further molecular docking study with 6YB7 crystal.
The Osiris property explorer [42] and Swiss-ADME [43] programs have been used to compare the pharmacokinetics and drug-likeness scores between the HCQ molecule and its analogs (14 compounds). Each molecule was filtered out on the basis of six molecular properties (cLogP, solubility, molecular weight, TPSA, drug-likeness, and drug-score) from the Osiris program and four characters (molar refractivity, Lipinski, bioavailability score, and synthetic accessibility) from Swiss-ADME program.
2.3. Molecular docking study
Displacing the W1 (PDB ID: W639) and W2 (PDB ID: W864) positions by HCQ or its analogs from the 6YB7 crystal structure is the main aim of the present docking study. The W1 and W2 locations are weakly bound and have less interaction energy with protein, hence they may displace well, in contrast the W3 which is tightly bound to protein with strong interaction energy and it seems to be unfavorable for water displacement. The “A” chain was obtained (excluding water molecules) from 6YB7 crystal structure and it was considered as a receptor. The HCQ molecule and its 14 structural analogs were also taken as a ligand for further molecular docking study using the AutoDock Vina program (v 1.1.2) [44]. The required PDBQT file of the receptor protein and 15 ligand molecules were assigned Kollman united atom charges [45] and Gasteiger partial atomic charges, respectively [46]. To obtain all the possible binding orientations, rigid and flexible dockings were performed not only for HCQ but also for its 14 analogs, since they should have a similar binding mode. Therefore, all the rotatable bonds of each ligand were kept to be flexible during docking, and residues inside the binding pocket were made rigid initially, and then flexible docking was performed by keeping the side-chains of His41 of the receptor protein made flexible. Grid point spacing was set at 1 Å and grid points 30 were taken in each direction of grid box and the concern box was centered at ND1 of His41 (in 6YB7). Best five binding modes of each ligand were identified after cluster analysis according to their binding affinity. The molecular docking results produced that the drug HCQ, indacaterol, and ZINC28706440 show a desirable result and these three docked structures were further consider for molecular dynamics simulation study.
2.4. Molecular dynamics simulations and trajectory analysis
The initial docked structure of HCQ, indacaterol, ZINC28706440, and native conformation of 6YB7 were obtained for further MD simulation studies to identify the dynamic properties of water molecules in the ligand-binding pocket of Mpro protein and also to decipher its structural changes from native to different ligand-bound conformations. Using the AutoPSF module of Visual Molecular Dynamics (VMD v.1.9.3.) program [47], missing hydrogen atoms were added to each complex structure. Furthermore, the ligand HCQ, indacaterol, and ZINC28706440 were parameterized using the SwissParam program [48] to obtain topology and parameter files for the CHARMM force field [49]. All these structures were then solvated in a cubic box of around 10,000 TIP3P water molecules extending at least 10 Å from the protein surface. Sodium and chloride ions were employed to neutralize the overall charge of the system; the resulting system consisted of around 33,000 atoms. MD simulation was performed for four solvated structures (HCQ, indacaterol, and ZINC28706440, and Mpro-native) using the Nanoscale Molecular Dynamics (NAMD v.2.11) program [50] by assigning the CHARMM-36 all-atom force field (with map correction) [51] for protein and the CHARMM General Force Field (CGenFF) for all ligands [52]. Three step minimizations were adopted to stabilize the system using the steepest-descent method. Initially, energy minimizations were performed on the solvent molecules for 5000 steps with the fixed solute. Two additional minimization processes were applied; restrained (CA atoms of protein) and unrestrained minimizations were carried out for 2000 and 1000 cycles, respectively. Subsequently, water molecules and ions were equilibrated for 2 ns by fixing solute to ensure the removal of any remaining steric clashes. Stepwise heating was carried out from 0 to 310 K for 50 ps in three steps at constant volume and with a gradually reduced restraint. Then each system was equilibrated for 500 ps under NPT restraints followed by NVT conditions reverting to the NPT ensemble for a period of 1 ns [53]. We ensured that each system was brought to equilibrium before continuing our simulations by verifying that each system reached a point where the energy fluctuations and RMSD were stable and PCA data are not scattered over a larger area [54].
To mimic physiological conditions, the temperature was kept at 310 K using Langevin dynamics with a damping coefficient of 5 ps−1. The pressure was maintained at 1 atm using the Langevin piston Nose–Hoover method, with a piston period of 100 fs and a decay time of 50 fs. During the simulations, a Nosé-Hoover Langevin piston barostat [55] and Langevin thermostat [56] were used to enforce constant pressure and temperature. The SHAKE algorithm [57] was used to keep bonds involving H atoms at their equilibrium length, allowing a 2 fs time step. The van der Waals interactions were truncated at 12.0 Å with switching from 10.0 Å. Electrostatic interactions were modeled accordingly with a dielectric constant of 1.0 throughout the equilibration and production runs. The standard Particle–Mesh Ewald method was used with periodic boundary conditions to compute the long-range electrostatic interaction of the system by specifying an appropriate Particle–Mesh Ewald grid size. Finally, production runs of all-atom MD for each structure were performed in the NPT ensemble for 50 ns. The atomic coordinates were recorded at every 2 ps for further data analysis (25,000 frames). Each MD trajectory, especially 12,500 recorded snapshots (frame Id. 12,500 to 25,000) were considered for further data analysis using the VMD program. Average root mean square deviations (RMSD) and root mean square fluctuations (RMSF) of CA atoms of MD structures (12,500 frames) were calculated (for the reference structure) using an in house TCL script.
2.5. Identification of water sites in MD structures and calculation of their thermodynamic and structural dynamic properties
To investigate the identification of water sites (W) between each ligand and His41 residue in the catalytic region of native MD structure, several amino acids were identified, especially relevant, in determining the binding with those water molecules (Table 1 ). Water molecules (W3 to W7) were investigated from corresponding HCQ, indacaterol, and ZINC28706440 bound MD structures by the clustering algorithm of WATCLUST [58] program in VMD. The clustering parameters of watnumbermin, dist, dr, WFRr, and pop were assigned as 15% of the total frame, 50.0, 0.16, 0.20, 0.60, and 0.90 Å, respectively. The visual analysis of the WATCLUST results provide us identification of each water site (W) that has maximum WFP values and is occupied with the H-bonding distance, either, between each ligand and their adjacent atoms of the protein or only respective atoms of protein. A water molecule is defined as being inside that corresponding water site if its oxygen atom distance to the water center is less than 0.6 Å, a value approximately corresponding to a volume of 1 Å3 for the water site [59]. However, among the five water sites (W3 to W7) in MD structures, W3, W4, and W7 are found in both the X-ray and MD structures, whereas W1 and W2 were observed only in X-ray. Therefore, the remaining W5 and W6 were assigned a new numbering scheme to distinguish them from water molecules in X-ray structures.
Table 1.
Thermodynamic and structural parameters of water molecules at the catalytic site (ligand binding) in different conformations of MD simulated structures of Mpro protein.
| MD-Structures | Conserved Water Sites | Reference Residue ID |
EWP | ½ EW−bulk | ΔE | WFP | J |
|---|---|---|---|---|---|---|---|
| Native | W3 | H41, D187, H164 | −18.75 | −3.50 | −15.25 | 24.00 | −1.90 |
| HCQ | W3 | H41, D187, H164 | −20.73 | 0.15 | −20.88 | 26.00 | −1.95 |
| Indacaterol | W4 | H41 | −8.27 | −4.29 | −2.82 | 6.16 | −1.09 |
| W5 | W4, W6 | 1.60 | −6.14 | 7.74 | 1.89 | −0.38 | |
| W6 | S46, LIG | −1.92 | −5.45 | 2.37 | 3.58 | −0.76 | |
| ZINC28706440 | W7 | H41, C145 | −10.49 | −3.61 | −6.88 | 11.00 | −1.43 |
EWP, Conserved Water with protein; EW−bulk; Conserved Water with bulk water; LIG; indacaterol.
*The calculation is defined previously in Methods in Section 2.5. The energies (EWP, EW−bulk, ΔE, WFP, and J interaction value) are in kcal/mol.
To calculate the average potential energy, combine, with the interaction of water molecules inside the water site (W) with protein and the remaining solvent, electrostatic and van der Waals interactions were computed. The interaction energy of W3 to W7 water sites in the HCQ, indacaterol, and ZINC28706440 bound structures were calculated by sampling of 25,000 snapshots. For each snapshot, we computed the average interaction energy with both the protein and all other (bulk) water molecules with the VMD program using the namd-energy plugin [60]. We calculated the energy ΔE shown in the following equation
| ΔE = EWP- ½ (EW-bulk) |
where ΔE is the energy difference, EWP is the mean interaction energy between a water molecule in the hydration site and protein, and EW−bulk is the mean total interaction energy of a water molecule in bulk.
The water site is a region where water molecules are more likely to be found than in bulk. Therefore, the probability of finding water molecules or WFP is assigned as a function in volume V normalized with respect to the bulk solvent density for each MD structures. Using the inverse Boltzmann relationship, we obtain the following equation and characterize each water site from each conformation and also quantify this magnitude in energetic terms, by defining J as
| J = −RT ln (WFP) |
To compare this parameter for different water sites, the values were reported in Table 1 and were calculated as an arbitrarily chosen volume of 1 Å3 [61].
2.6. Analysis of ligand binding pocket
The Pocket Volume Measurer program (POVME: version 3.0) [62] was employed to identify and characterize the volume, shape, and size of ligand binding pocket in four MD structures. Using POVME, we quantitatively compared the volume of ligand-binding pockets as a function of time, which is inaccessible experimentally. The program uses a single inclusion sphere by assigning the ND1 atom at the active site residue (His41) as a center of the inclusion sphere, which encompasses the entire binding cleft of ligand. The POVME algorithm calculates the volume of pocket by subtracting the space occupied by protein atoms in each frame from the inclusion sphere volume. This procedure was used for all MD structures after superimposing each one and aligning their trajectories to their first frame. Around 500 frames from each trajectory were used to measure the average volume of each pocket. The size of the inclusion sphere was set to 11 Å, and the remaining values of van der Waals radius, grid spacing, and contiguous cutoff parameters were set to 4.50, 0.50, and 1.09 Å, respectively [62].
2.7. Binding energy calculation using MM/PBSA and LIE-D method
The binding energy of ligand HCQ, indacaterol, and ZINC28706440 were evaluated by the results from protein−ligand MD trajectories using the molecular mechanics combined with the Poisson−Boltzmann and surface area (MM/PBSA) method in the program Calculation of Free Energy (CaFE) [63]. The single-trajectory approach is more widespread owing to its simplicity, precision, efficiency, and accuracy compared to the multi-trajectory setup. The MD trajectories of the protein-ligand complex as obtained in the one-average strategy were used to evaluate each free energy term on ΔGBind within the following equation:
| ΔGBind = ΔGcomplex- (ΔGprotein+ ΔGligand). |
ΔGcomplex represents the free energy of the complex, and ΔGprotein and ΔGligand indicate the free energies of the unbound protein and ligand, respectively. Three different energy terms are calculated by the MM/PBSA method which includes energy difference between the complex and separate receptor and ligand molecule in the gas phase from NAMD. In MM/PBSA, polar solvation free energy was calculated using the APBS program [64] by solving the Poisson−Boltzmann equation by assigning the PB radii as bondi, PB boundary condition as mdh (multiple Debye-Huckel), and PB chgm (charges are mapped to the grids) as spl2 (cubic B-spline). The nonpolar term (ΔGnonpolar) is often approximated by a solvent accessible surface area (SASA) and were estimated by assigning SA radii as bondi using a simple empirical relation: (ΔGnonpolar) = γA + b, where A is the solvent-accessible surface area that was estimated using the VMD program, gamma (γ) and b are empirical constants, and in this work, we used 0.0054 and 0.92 kcal/mol for γ and b, respectively. The change in conformational entropy (−TΔS) is generally computed by normal-mode analysis on a set of conformational snapshots taken from MD simulations. Due to the high computational demand and large standard error compared to other contributing terms, the entropic term was neglected in the MM/PBSA models when only the relative binding free energies of similar ligands are needed and presented in this work [65].
By default, the dielectric constant of solute (εsolute) was set to 1 to examine the sensitivity of calculated binding free energies to the solute dielectric constant. The 2,500 frames were used to estimate the contribution of the binding free energy. In the current work, we evaluated the LIE-D method [66] to achieve the binding energy of each ligand to compare the result from the MM/PBSA model. The binding free energies of ligand HCQ, indacaterol, and ZINC28706440 were estimated from protein–ligand MD structures using the modified linear interaction energy method (LIE-D) in the CaFE plugin of VMD. LIE-D is widely used to calculate non-covalent binding free energies of ligands to proteins. To estimate the binding free energy of ΔG (kcal/mol), we used the following scoring function:
| ΔGbindLIE-D = β (Vele-complex - Vele-ligand) + α (Vvdw-complex – Vvdw-ligand) + (−0.95 [β (Vele-complex - Vele-ligand) - α (Vvdw-complex – Vvdw-ligand)]- 2.06) |
The coefficients α and β are adjustable empirical parameters for the van der Waals and electrostatic contribution of the non-bonded interaction energy to the free energy of binding, respectively. The balance (difference) between electrostatic (polar) and van der Waals (non-polar) contributions to the binding free energy was defined as D = β (V ele-complex - V ele-ligand) - α (V vdw-complex – V vdw-ligand) [66]. The final 5 ns trajectories from each MD simulation were considered for calculating the binding energy of each ligand using LIE-D. The empirical α and β coefficients were initially set to 0.18 and 0.44, respectively.
3. Results and discussion
3.1. Analysis of ligand-binding pocket in Mpro-crystal structures
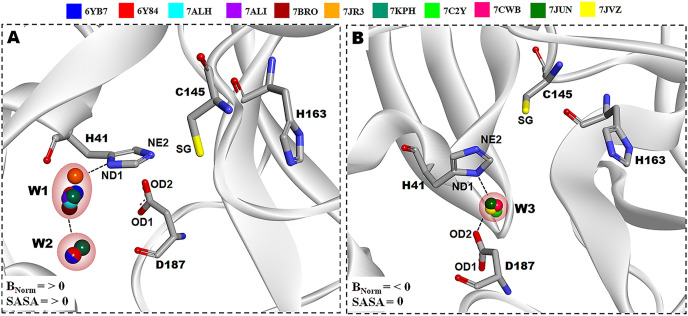
The eleven crystal structures of the Mpro-native are obtained from the Protein Data Bank either in ligand-bound or ligand-free forms. These structures have been independently solved by several research groups using PHENIX or REFMAC or BUSTER or nCNS refinement programs at 6.1–7.5 pH. Only those structures determined in C 1 2 1 space groups were considered in the present computational study to eliminate crystal lattice bias (except PDB ID: 7C2Y in P 21 21 21) because insufficiencies of structural data in different space groups may not conclude a comparative analogous study. In this context, the present study revealed the existence of W1, W2, and W3 water molecules at the catalytic region of the Mpro which may have plausible roles in the structure, function, and stabilization of Mpro-native. Furthermore, the superimposed complex structures between 6YB7, 6Y84, 7ALH, 7ALI, 7BRO, 7JR3, and 7KPH have clearly displayed the presence of W1 and W2 water molecules those are occupied a bit far (<3.50 Å) from Asp187, in contrast, to the superpositions of 7C2Y, 7CWB, 7JUN, and 7JVZ structures showing the position of W3 site that makes H-bond with Asp187. Interestingly, W1 forms H-bonds with ND1 of His41 and oxygen of W2, W2 associates only W1 site, and W3 makes H-bonds with ND1 of His41 and OD2 of Asp187 (presented in Fig. 1 ). The geometrical positions of W639, W615, W573, W563, W499, W484, and W629 are found to be conserved in the W1 site with BNorm value from 0.42 to 5.07 Å2 and average SASA value ∼8.97 Å2, moreover, W864, W800, and W691 are located at W2 position with BNorm ranging from 2.24 to 3.89 Å2 and average of SASA value ∼23.87 Å2, and similarly, W346, W441, W409, and W429 are occupied in W3 position with BNorm values from −0.78 to −1.54 Å2 and average of SASA value 0 0.0 Å2 . Thus, the BNorm of W1 and W2 positions are >0.0 Å2 and SASA value > 2.5 Å2 (except for few structures, PDB ID: 6Y84, 7JR3, and 7KPH), on the contrary, the corresponding value for BNorm in W3 site is < 0.0 Å2 and SASA value ∼ 0.0 Å2 (detail mentioned in Table S2). Especially, the average Pearson's correlation coefficient rp (R2) between BNorm and SASA values in W1 and W2 are −0.61 (0.37) and −0.78 (0.61), respectively, that represents an opposite correlations. Moreover, water molecules with an accessible surface area <2.5 Å2 were considered to be buried water molecules [67]. Hence, the observed atomic displacement parameters (a.d.p) usually referred as BNorm and SASA values of water molecules revealed that the W3 site is considerably stable, invariant, and deeply buried in crystal structures whereas the locations of W1 and W2 are dynamic and highly exposed. According to suggestion from Carugo et al. [68], if BNorm is greater than or ∼ 1.20 Å2 for any hydration site, then that will be fully occupied by water molecule. This information indicates the W3 location in PDB ID: 7CWB and 7JUN can also be occupied fully by a water molecule.
Fig. 1.
Superposition of 11 native conformations of X-ray structures reveal the positions of conserved water molecules inside the catalytic pocket in Mpro protein. (A) Water molecule W1 and W2 is conserved with respect of D187 in PDB ID: 6YB7, 6Y84, 7ALH, 7ALI, 7BRO, 7JR3, and 7KPH. (B) W3 is conserved and also makes H-bond with respect of D187 in PDB ID: 7C2Y, 7CWB, 7JUN, and 7JVZ.
The crystallographic structural parameters (c.s.p.) such as Matthews coefficient, solvent content, and calculated mean B-factors of protein are found maximum (2.83, 56.48, and 33.56 Å2) in 7C2Y and minimum (1.91, 35.50, and 16.87 Å2) in 6YB7. Moreover, the observed ratio of the number of protein atoms with respect to water molecules (NPROT/NHOH) in 11 Mpro-native (at room temperature with 1.25–2.50 Å resolution) are ranges from 5.34 to 51.45 that are suggesting 7JVZ ensemble is actually random compared to remaining structures [69] (mentioned in Table S2). Remarkably, the average Pearson's correlation coefficient rp (R2) between BNorm and Matthews coefficient in W1, W2, and W3 are −0.18 (0.03), −0.83 (0.69), and 0.62 (0.39) whereas the rp (R2) between BNorm and NPROT/NHOH in W1, W2, and W3 are −0.79 (0.63), −0.99 (0.98), and 0.13 (0.02). In the present study, an attempt has been made to establish a relationship between a.d.p. of three water molecules and c.s.p of 11 crystal structures that indicates 6YB7 is the stable structure but its W1 and W2 site are considerably unstable compared to W3.
3.2. Binding mode prediction of HCQ, its analogous and analysis of their pharmacological properties
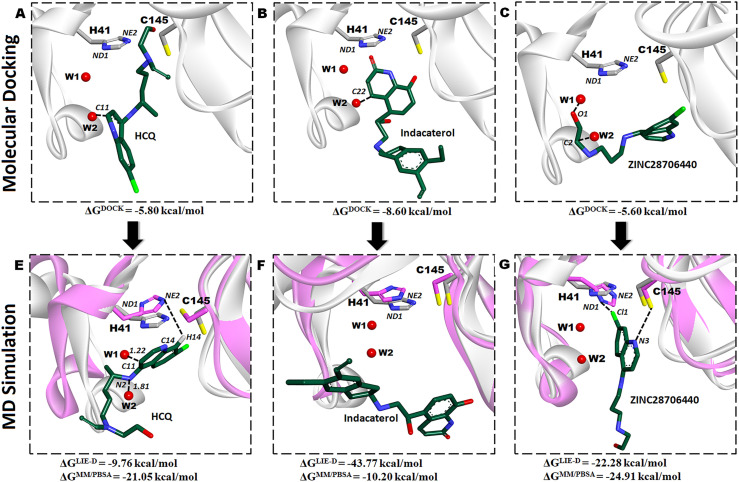
Our computational drug repurposing workflow against Mpro-native was started with a docking of 4 FDA-approved drugs and 11 new ligands (analogous HCQ) using AutoDock Vina program (v1.1.2). In this approach, we elucidate how the ligands are occupying the positions of W1 or W2 in the binding site? In water-based drug discovery technique, the docking results can be considered a “good fit” if ligands grip the location of water sites and occupy those positions accordingly [31]. To do so, we exhaustively searched the proper and accurate orientation of each docked ligand, so that corresponding atoms of the respective ligand can occupy the positions of water sites in docked structure. To probe the validity of this procedure, we obtained the docked structures accordingly. The HCQ, indacaterol, and ZINC28706440 were only considered for further study because they were displaced of those water molecules in the binding pocket of Mpro whereas remaining 12 HCQ analogs were not feasible. Analysis of molecular docking results showed HCQ, indacaterol, and ZINC28706440 formed short contacts with ND1 of His41 and their binding energies were −5.80, −8.60, and −5.60 kcal/mol respectively (Table S3). Furthermore, HCQ, indacaterol, and ZINC28706440 have three; five, and four H-bonding interactions with the residues of Mpro respectively, and they also get short contacts with Cys145 (except ZINC28706440). The indacaterol has high affinity for Mpro protein rather than remaining 14 ligands, but HCQ and ZINC28706440 have similar binding affinities in docking study.
To assess the statistical significance of this docking result, the physico-chemical and toxicity comparison was performed for 15 ligands. In the present study, OSIRIS Property Explorer was used to predict the drug-likeness score of all compounds and comparing them with the drug HCQ. The program calculated the risks of side effects, such as mutagenic, irritant, tumorigenic, and reproductive effects, as well as drug-relevant properties. Moreover, the overall drug-score was estimated from summation of cLogP, solubility, TPSA, drug-likeness, drug-score, and molar refractivity parameters. Interestingly, the potential drug-likeness score of HCQ, chloroquine, methylergometrin, indacaterol, and ZINC77520192 were significantly higher (ranging from 6.54 to 8.03) than remaining ligands. However, except HCQ, other three compounds, produced, insignificant docking results. However, the values of physico-chemical parameters referred to as drug-score, cLogP, TPSA, drug-likeness, and synthetic-accessibility are 0.48, 3.08, 48.39, 6.54, and 2.82 in HCQ, 0.66, 2.81, 81.59, 2.12, and 3.68 in indacaterol, and 0.50, 1.60, 57.18, 1.74, and 2.01 in ZINC28706440, respectively (Table S4). Mostly, the drug-score of HCQ, indacaterol, and ZINC28706440 were significantly higher (ranging from 0.48 to 0.66) and their predicted in silico toxicity-risk properties were also higher than other ligands (Table S5).
3.3. Analysis of MD trajectories
3.3.1. Ligand- Mpro interaction profiling
The findings of MD simulations have been explored in terms of dynamics and provide new insights to discriminate the structural and functional properties between native and ligand bound Mpro conformations. We observed that HCQ interacts with His41, Cys145, His164, Val186, Arg188, and Gln189, indacaterol with His41, Ser46, Glu47, Asn142, and Gln189, and ZINC28706440 with His41, His163, His164, Glu166, Arg188, and Gln189. Ligand-residue interactions profiling were measured on final 2,500 MD-structures and were presented in Table S6. During simulation, conformational changes upon binding HCQ, indacaterol, and ZINC28706440 to Mpro protein suggest that HCQ and ZINC28706440 have similar binding pose and identical interactions with His41, His164, Arg188, and Gln189 but indacaterol has different interactional patterns in respect of His41 (Fig. S3).
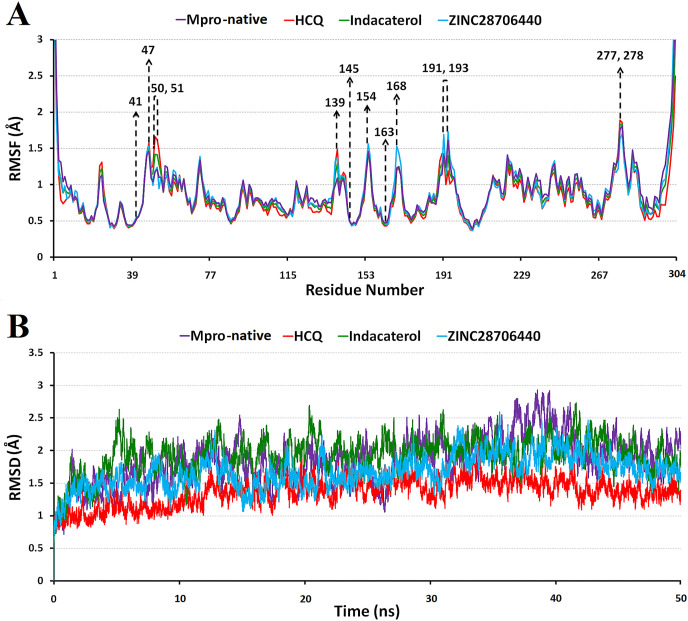
The present investigation focuses on the residues of ligand-binding pocket (His41, Cys145, and His163) along with non-catalytic sites and seems significant. The flexibility (RMSF) of each residue in the Mpro-native, HCQ, indacaterol, and ZINC28706440-bound MD structures are described accordingly, where the N- and –C terminal regions of this protein exhibit more flexibility (upper panel of Fig. 2 ). In MD structure, atomic fluctuation (CA atom only) of Mpro-native is 0.39–3.00 Å, HCQ is 0.37–2.50 Å, indacaterol is 0.38–3.00 Å, and ZINC28706440 is 0.37–3.00 Å. The RMSF values for active site residues of His41, Cys145, and His163 are 0.54, 0.50, and 0.48 Å in Mpro-native, 0.53, 0.49, and 0.43 Å in HCQ, 0.54, 0.50, and 0.46 Å in indacaterol, and 0.56, 0.49, and 0.45 Å in ZINC28706440, Whereas, residues like Glu47, Leu50, Asn51, Ser139, Tyr154, Pro168, Ala191, Ala193, Asn277, and Gly278 have reasonably high values ranging from 1.73 to 1.89 Å. Therefore, the atomic deviations of these residues in Mpro-native and indacaterol structures seem more flexible than remaining structures. Moreover, the comparative trajectory analysis of RMSD (CA atom) with respect to time (ns) among the MD structures of Mpro-native, HCQ, indacaterol, and ZINC28706440-bound forms indicate that the RMSD value is within 0.54–2.92 Å in Mpro-native, 0.55 to 2.17 Å in HCQ, 0.51 to 2.73 Å in indacaterol, and 0.52 to 2.59 Å in ZINC28706440-bound structures (lower panel of Fig. 2). A comparative analysis of these four MD structures show the average RMSD of Mpro-native (1.84 Å) and indacaterol (1.93 Å) have large fluctuations and display more dynamic nature than HCQ (1.37 Å) and ZINC28706440 (1.68 Å) -bound structure.
Fig. 2.
Top: Comparison of RMSF (root-mean-square fluctuation) (Å) of CA atoms in MD structure of Mpro-native (violet), HCQ (red), indacaterol (green), and ZINC28706440 (blue) bound form. The final 25,000 frames for each conformation were used. Bottom: Comparison of RMSD (root-mean-square deviation) (Å) of CA atoms in MD structure of Mpro-native (violet), HCQ (red), indacaterol (green), and ZINC28706440 (blue) bound form.
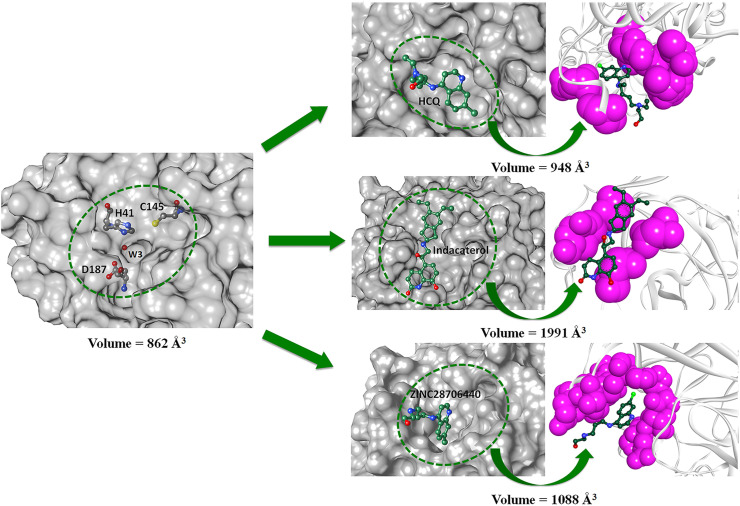
The average volumes of ligand-binding pockets, especially the free space are ∼820 Å3 in Mpro-native crystal structures. As a rough approximation, we calculated the size of the pocket required to bind a ligand by measuring the volume of a “minimal sphere.” During the conformational transition from Mpro-native to HCQ or indacaterol or ZINC28706440, the volume of ligand-binding pocket increases from 862 to 948 Å3 in HCQ, 1991 Å3 in indacaterol, and 1088 Å3 in ZINC28706440 (Fig. 3 ). Therefore, MD data suggest that the volume of the ligand-binding pocket expands significantly in ligand-bound conformation relative to Mpro-native, even though, the volumes are enlarged similarly in HCQ or ZINC28706440 than indacaterol.
Fig. 3.
Approximate volume (Å3) of the binding pocket for ligand HCQ, indacaterol, and ZINC28706440 during structural transitions from native to ligand-bound form in the MD simulation. Each molecular surface of the panel marks by a green circle represents ligand-binding groove of the Mpro protein. The changes in volume of each MD structure in (A) native Mpro, (B) HCQ, (C) indacaterol, and (D) ZINC28706440 are marked below the structures.
3.3.2. Structural, dynamical, and thermodynamic characterization of water molecules
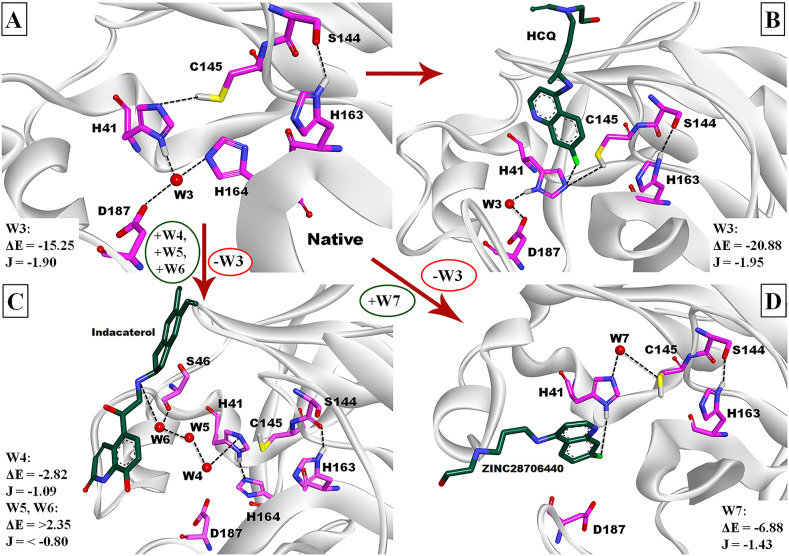
The present MD simulation studies of ligand-binding pocket demonstrate and confirm the positions of five (W3 to W7) water molecules in the protein. The thermodynamic and structural properties of these water molecules were characterized and mentioned in Methods in Section 2.5. These water sites establish that the H-bonds with ligands or residues or conserved water molecules (described in Table 1) and by assigning them as references (explained in the method section), we were capable to find these water sites with significant water occupation probability. Due to the inherent mobility of water molecules in MD structures, it is difficult to determine and visualize their positions. Therefore, to identify the presence of water molecules in the ligand-binding pocket in X-ray crystallographic structure this is the actual source for obtaining structural information. We therefore compare the water positions in MD conformations with their respective crystal structures in Mpro-native. We noticed that the positions of water site W3 (W604 in 6Y84), W4 (W861 in 6Y84), and W7 (W816 in 6Y84) were available in both MD and X-ray structures whereas W5 and W6 water locations were only produced during MD simulation. If the crystal structures of Mpro do not show any explicit water molecules in the ligand-binding site, this does not necessarily mean that they are really absent in structures. Perhaps, these water sites are either in rapid exchange with bulk solvent or relatively mobile within the binding site. Therefore, they were inaccessible in crystallographic studies, but may still play a major role in the protein function.
As described in Table 1, water site W3, W4, and W7 have high WFP values and ranging from 26.00 to 6.16 because they have, (i) strong EWP (−20.73 to −8.27 kcal/mol), and (ii) weak EW−bulk interaction energy (0.15 to −4.29 kcal/mol), therefore, their ΔE (−20.88 to −2.82 kcal/mol) are also high. In contrast, the W5 and W6 sites contain less WFP values (lower than 4.00) with high EW−bulk (−6.14 and −5.45 kcal/mol), and weak EWP interaction energy (1.60 and −1.92 kcal/mol), as a result, ΔE are also weak (7.74 and 2.37 kcal/mol). The corresponding WFP values expressed in the terms of interaction energy (J) are distinct and are also important. W3, W4, and W7 are also highly prominent and stable in their corresponding positions because they have the lowest J energy value ranging from −1.95 to −1.09 kcal/mol in comparison with W5 and W6 whose corresponding values are −0.38 and −0.76 kcal/mol. Remarkably, the average Pearson's correlation coefficient rp (R2) of W3 with W4, W3 with W7, and W4 with W7 are significantly high [0.97 (0.94) to 0.99 (0.99)] in respect of EWP, EW−bulk, and WFP and explains these corresponding water sites could be present predominately in MD structures.
The ligand-binding site is stabilized by either W3 or W7 in Mpro-native, HCQ, and ZINC28706440-bound conformation, whereas W4, W5, and W6 water sites are observed in indacaterol. To identify the interactions of water molecules with ligand or catalytic residues in each MD-structure, we observed the region of His163---Ser144-Cys145---His41---W3---Asp187 in Mpro-native, His163---Ser144-Cys145/HCQ---His41---W3---Asp187 in HCQ, His163---Ser144-Cys145---W4---His41---His164/Asp187 and His41---W4---W5---W6---indacaterol in indacaterol, and His163---Ser144-Cys145---W7---His41---ZINC28706440 in ZINC28706440-bound conformation (Fig. 4 ). To explain the possible role of each water molecule from each MD structure, we assumed that W3 water can act as a catalytic partner of Mpro by stabilizing the thiolate–imidazolium (Cys145-SH---NE-His41) and imidazole-chlorine (HCQ-Cl---NE-His41) interactions in the Mpro-native and HCQ structure respectively. Meanwhile, W4 makes H-bond with the imidazole group of His41, W5 also makes a water mediated bridge between W4 and W6, and W6 interacts with indacaterol (N1 atom). Moreover, W7 is observed to occupy Cys-His dyad in ZINC28706440 to inactivate the thiolate–imidazolium ion-pair.
Fig. 4.
MD simulations of native, HCQ, indacaterol, and ZINC28706440 inside the active-site pocket of Mpro protein. Catalytic residues His41, Cys145, His163, and ligands are highlighted by the stick models, while water molecules represent in the pink ball. (A) The interactions of His163---Ser144-Cys145---His41---W3---Asp187 are shown inside the active-site pocket in Mpro-native, (B) His163---Ser144-Cys145/HCQ---His41---W3---Asp187 are shown in HCQ-bound MD structure, (C) His163---Ser144-Cys145---W4---His41---His164/Asp187 and His41---W4---W5---W6---indacaterol in indacaterol-bound MD structure, and (D) His163---Ser144-Cys145---W7---His41---ZINC28706440 in ZINC28706440-bound conformation.
3.3.3. Occupying the positions of W1 or W2 by ligand in X-ray crystal structure
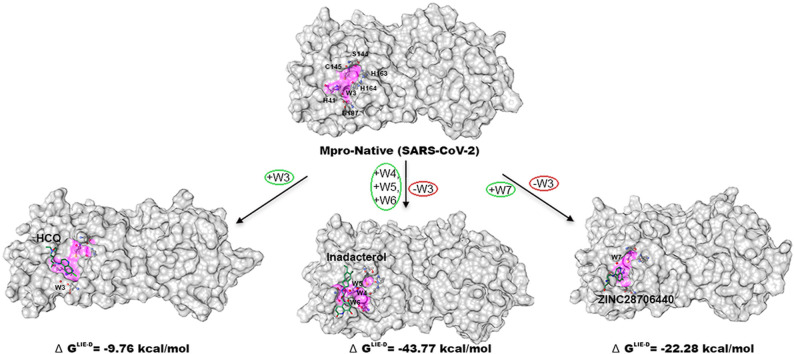
In the present study, we investigated experimentally known data in order to produce the crystallographically predicted conserved water molecules in Mpro-native on a basis of statistical and thermodynamic analysis. Thus, the interaction energy of W1 and W2 sites in 6YB7 and W3 in 7CWB crystal structure are computed accordingly. The corresponding values expressed in the terms of interaction energy ΔE for W1, W2, and W3 are −4.79, −3.90, and-17.19 kcal/mol while their interaction energy with protein (ΔEWP) are −2.93, −1.98, and −17.43 kcal/mol respectively. The displacement of W1 or W2 water site from the catalytic region could inactivate the Mpro enzyme and these water sites are less stable and dynamic due to BNorm > 0 Å2, are highly exposed with solvent-accessible surface areas because of SASA value > 0 Å2, and their electrostatic (ΔE) and bulk water (EW−bulk) energy are higher than W3 position. Therefore, taking up the positions of W1 or W2 by the proposed ligand in Mpro using molecular docking study will be a challenging task. This structural result can help us for more detailed physico-chemical characterizations of the conserved water molecules that play an important role in molecular docking study. For investigating water displacement approach regarding the ligand-water interactions; we superimposed the HCQ, indacaterol, and ZINC28706440 on the 6YB7 crystal structure. Findings from this docking study reveals the C11 of HCQ and C22 of indacaterol occupy the position of W2, while C2 and O1 of ZINC28706440 grip the locations of W1 and W2, respectively. Molecular docking study may produce insignificant information regarding the rearrangements of W1, W2, and W3 water molecules after binding to the ligand, thus, MD-simulation is the best alternative option to justify the adjustment of this water in terms of dynamics at the active site of pocket. Finally, the MD-simulation suggests that the C11 and N2 atoms in HCQ obtain the positions of W1 and W2, respectively, and binding energy of HCQ comparatively augments the docked conformation (−5.80 to −9.76 kcal/mol). Consistently, indacaterol grip the W2 location during docking but it cannot occupy either W1 or W2 position during MD-simulation and gets more stable by losing energy (−8.60 to −43.77 kcal/mol). Similarly, ZINC28706440 captures the locations of W1 and W2 in the docking study but it moves away from these water locations in MD-simulation and obtains stability by releasing the energy from −5.60 to −22.28 kcal/mol (Fig. 5 and Table S7).
Fig. 5.
Top: Superimposed complex structures of HCQ, indacaterol, and ZINC28706440 (docked conformation) with Mpro-native (PDB ID: 6YB7). The C11 of HCQ and C22 of indacaterol occupy the W2 location, but C2 and O1 of ZINC28706440 take the W1 and W2 sites, respectively. Bottom: Superposition of the HCQ, indacaterol, and ZINC28706440 bound-MD conformations on 6YB7 crystal structure show the C11 and N2 atom of HCQ grip the W1 and W2 locations, but indacaterol, and ZINC28706440 are remotely occupied without displacing W1 and W2 site.
3.3.4. Comparative analysis of LIE-D and MM/PBSA energy of ligand for Mpro
The LIE-D and MM/PBSA binding free energies and their decompositions for the three ligands were compared and mentioned in Table 2 . As a model in MM/PBSA, the atomic radii plays a significant role in the performance of solvation energy calculations. In the present work, we used bondi radii because the chlorine atom (in HCQ and ZINC28706440) is parameterized accordingly and assigned the value 1.76 Å [70].
Table 2.
The binding free energy (in kcal/mol) of the ligand HCQ, Indacaterol, and ZINC28706440 are calculated by the LIE-D and MM/PBSA method. The parameters α and β are used in LIE-D, whereas SD is calculated from MM/PBSA.
| LIE-D |
MM/PBSA |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α | β | ΔEvdw | ΔEele | ΔGLIE−D | ΔEvdw | ΔEele | ΔEPB | ΔESA | ΔGpol | ΔGnp | ΔGMM/PBSA | SD | Lig-RMSD | ||
| HCQ | 50ns | 0.18 | 0.44 | −30.54 | −10.05 | −9.92 | −30.54 | −9.82 | 24.05 | −4.52 | 14.22 | −35.06 | −20.84 | 3.31 | 1.84 |
| 49ns | 0.18 | 0.44 | −30.82 | −12.05 | −10.85 | −30.82 | −11.87 | 25.53 | −4.57 | 13.65 | −35.40 | −21.75 | 2.44 | 1.73 | |
| 48ns | 0.18 | 0.44 | −30.16 | −12.60 | −10.97 | −30.16 | −12.05 | 23.99 | −4.49 | 11.94 | −34.66 | −22.72 | 3.54 | 1.65 | |
| 47ns | 0.18 | 0.44 | −25.83 | −9.04 | −8.62 | −25.83 | −8.72 | 19.28 | −4.21 | 10.55 | −30.05 | −19.49 | 3.84 | 1.85 | |
| 46ns | 0.18 | 0.44 | −27.17 | −8.04 | −8.43 | −27.17 | −8.24 | 19.13 | −4.18 | 10.88 | −31.35 | −20.46 | 4.47 | 1.65 | |
| Indacaterol | 50ns | 0.18 | 0.44 | −11.55 | −156.31 | −70.86 | −11.55 | −182.79 | 183.73 | −3.56 | 0.93 | −15.12 | −14.18 | 3.44 | 2.18 |
| 49ns | 0.18 | 0.44 | −8.55 | −122.39 | −55.39 | −8.55 | −145.90 | 148.30 | −2.81 | 2.39 | −11.36 | −8.96 | 5.77 | 2.10 | |
| 48ns | 0.18 | 0.44 | −9.09 | −68.87 | −31.94 | −9.08 | −94.45 | 100.45 | −2.51 | 5.99 | −11.60 | −5.61 | 3.68 | 1.97 | |
| 47ns | 0.18 | 0.44 | −13.00 | −79.66 | −37.39 | −13.00 | −107.87 | 112.73 | −3.18 | 4.86 | −16.18 | −11.32 | 3.86 | 1.85 | |
| 46ns | 0.18 | 0.44 | −13.40 | −47.39 | −23.26 | −13.40 | −88.49 | 94.03 | −3.08 | 5.53 | −16.48 | −10.95 | 2.24 | 1.72 | |
| ZINC28706440 | 50ns | 0.18 | 0.44 | −20.81 | −158.61 | −73.53 | −20.81 | −177.34 | 174.32 | −4.06 | −3.02 | −24.87 | −27.89 | 4.26 | 1.79 |
| 49ns | 0.18 | 0.44 | −19.26 | −154.63 | −71.50 | −19.26 | −175.83 | 174.37 | −4.09 | −1.46 | −23.35 | −24.82 | 5.05 | 1.82 | |
| 48ns | 0.18 | 0.44 | −22.31 | −149.70 | −69.88 | −22.31 | −172.53 | 171.10 | −4.12 | −1.42 | −26.43 | −27.86 | 4.08 | 1.39 | |
| 47ns | 0.18 | 0.44 | −22.71 | −138.38 | −64.97 | −22.71 | −170.96 | 173.19 | −4.10 | 2.22 | −26.81 | −24.58 | 5.46 | 1.69 | |
| 46ns | 0.18 | 0.44 | −26.33 | −79.24 | −39.60 | −26.33 | −104.53 | 115.51 | −4.04 | 10.97 | −30.37 | −19.40 | 2.82 | 1.87 | |
*The SD column shows the standard deviation of the various terms in the MM/PBSA method, Lig-RMSD indicates root-mean-square deviations of ligand−atom positions (Å) between initial pose and each MD structures.
We observed that the average lig-RMSD is 1.74 in HCQ, 1.96 in indacaterol, and 1.71 Å in ZINC28706440. The lig-RMSD is < 2 Å suggesting a stable conformation of ligand thus it reflects HCQ, indacaterol and ZINC28706440 that are relatively stable. For MM/PBSA, the mean value of electrostatic energy (ΔE ele +ΔG PB) is 12.26 in HCQ, 3.95 in indacaterol, and 1.46 kcal/mol in ZINC28706440 but for LIE-D, the value of ΔE ele is −10.36 in HCQ, −94.92 in indacaterol, and −136.11 (kcal/mol) in ZINC28706440. However, the electrostatic energies are significantly different for the three binding modes. The overall correlation value rp (R2) of ΔE ele +ΔG PB with ΔGMM/PBSA and ΔE ele with ΔGLIE−D are −0.43 (0.19) and 0.98 (0.95) in HCQ, 0.62 (0.39) and 1.00 in indacaterol, and 0.95 (0.90) and 1.00 in ZINC28706440. The contribution of electrostatic energy to ΔG in MM/PBSA method is poor in HCQ and strong in ZINC28706440 while the LIE-D predicts these energies contributions are strong and significant in the respective three ligands. Similarly, for the van der Waals and hydrophobic interaction energies (ΔE vdw+ ΔE SA) in MM/PBSA and ΔE vdw values in LIE-D are −33.30 and −28.90 in HCQ, −14.14 and −11.12 in indacaterol, and −26.37 and −22.28 kcal/mol in ZINC28706440. For the three binding modes, the van der Waals energies are very different, which means HCQ and ZINC28706440 have a good hydrophobic contact than indacaterol. The correlation value rp (R2) of ΔE vdw+ΔE SA with ΔGMM/PBSA and ΔE vdw with ΔGLIE−D are 0.80 (0.63) and 0.89 (0.80) in HCQ, 0.73 (0.53) and −0.31 (0.10) in indacaterol, −0.72 (0.52) and −0.92 (0.84) in ZINC28706440 that reveals this relationship of two energy terms with ΔG that are very high in HCQ and poor in ZINC28706440.
In this regard, it is concluded that both the electrostatic and the van der Waals interactions have significant role for binding of ligand, but the relative electrostatic interaction contributes more. The electrostatic energy and lig-RMSD of HCQ and ZINC28706440 are apparently similar and has been predicted by LIE-D. Our findings also support the results of different research groups [71] to indicate that the electrostatic interaction energy may be a good predictor for identifying best structural analogous of ligands and it is the predominant factor for determining the different binding orientations. Only when comparing the ΔG correlation results of HCQ versus indacaterol and HCQ versus ZINC28706440, we observed that the rp(R2) values are −0.74 (0.54) and 0.47 (0.22) in MM/PBSA and 0.41 (0.61) and 0.72 (0.52) in LIE-D. According to the correlation findings (rp and R2), the MM/PBSA and LIE-D methods suggest that the ΔG value of HCQ is correlated strongly with ZINC28706440 and insignificantly with indacaterol. Hence, HCQ has a strong correlation with ZINC28706440 than indacaterol and thus these statistical findings in respect of rp values also demonstrate that ZINC28706440 is the best structural analogous of HCQ.
4. Conclusion
The present study focuses on the ligand-binding pocket of the Mpro protein to characterize its stabilization and the role of water molecules during the enzymatic mechanism. The comprehensive analysis of multiple crystal structures of Mpro-native reveals the presence of three conserved water locations (W1, W2, and W3) that maintain the architecture of the active site pocket when ligands are absent in the enzyme. The crystallographic structural parameters of those concern sites and their interaction energy profiling conclude that the W1 and W2 are dynamic in nature, considerably unstable, weakly bonded with protein in low interaction energy, and are highly exposed. Whereas W3 position is apparently stable, invariant, tightly bonded with the protein by strong interaction energy, and deeply buried in crystal structures of the Mpro-native. Therefore, we have planned to displace W1 or W2 water sites by triggering HCQ or its analogs to inactivate the enzyme and thus virtual screening with molecular docking studies have been adopted to determine the analogs for HCQ. The findings demonstrated that indacaterol and ZINC28706440 are the best analogs of HCQ with reasonably acceptable physico-chemical and toxicological values. The docking study reveals that HCQ and indacaterol occupy the W2 position and ZINC28706440 grip the locations of W1 and W2 with binding energy -5.80, -8.60 and -5.60 kcal/mol. Finally, the MD-simulations results show that the HCQ obtains the positions of W1 and W2 whereas its analogs (indacaterol and ZINC28706440) could not retain them and leave the position of either W1 or W2. These ligands gain stability by releasing energy compared to HCQ from docking to MD simulation study. The MD simulation results again consistent with the existence of five water molecules in the ligand binding pocket of the enzyme, especially water site W3, which is observed in Mpro-native and HCQ, W4, W5, and W6 in indacaterol, and W7 in ZINC28706440-bound structure. The W3 and W7 stabilize the thiolate–imidazolium and imidazole-chlorine interaction in Mpro-native and HCQ respectively and W7 also stabilizes His41---Cys145 in ZINC28706440-bound conformation. Interestingly, W4, W5, and W6 in indacaterol make a water mediated H-bond between ligand and His41. During conformational transition from Mpro-native to indacaterol or ZINC28706440, W3 is departed from Mpro-native and W4, W5, and W6 enter the binding pocket in indacaterol and similarly W7 arrives in ZINC28706440-bound structure. The computational evidence encourages us to identify structural, dynamical, and thermodynamic discriminative features of ligand binding site for comparative analysis of Mpro-native with HCQ or its analogs. In this context, WFP, J value, and other thermodynamic profiling parameters suggest that W3, W4, and W7 are highly prominent in their corresponding positions in comparison with W5 and W6. Our results appeared to be significant and evident that we decided to explore our important findings with the medical community in the current pandemic situation. We, therefore, recommend that ZINC28706440 is the best structural analog for HCQ that should be further synthesized, and require proper experimental and pre-clinical investigation for the possible treatment of nCOVID-19.
Authors contributions
Saima Reyaz: Methodology, Software handling (Molecular Docking, Screening, and ADMET), Formal Analysis, Data Curation, Writing-Review & Editing. Alvea Tasneem: Methodology, Software handling (Molecular Docking, and MD Simulation), Formal Analysis, Data Curation and Validation, Writing-Review & Editing. Gyan Prakash Rai: Methodology, Software handling (Molecular Docking, Screening, and ADMET), Formal Analysis, Investigation, Writing Review and Editing. Hridoy R. Bairagya: Conceptualization, Supervision and full correspondence, writing original draft preparation.
Data availability statement
Supplementary data of this article is provided in the supplementary material.
Ethical approval
The manuscript does not contain experiments on animals and humans; hence ethical permission not required.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
HRB is thankful to C.S.I.R. (Council of Scientific and Industrial Research) for providing Senior Research Associateship (Pool Scientist scheme). HRB gratefully acknowledges the Department of Biophysics, A.I.I.M.S., New Delhi and S.R., A.T., and G.P.R. also thankfully acknowledges the Department of Computer Science, Jamia Millia Islamia, Central University-New Delhi, for providing computational resources. We also acknowledge the support of High-performance computing facility at I.I.T. Delhi for MD simulations study. We are grateful to Prof. David J. Wales for his comments on the original manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2021.108021.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization . 2020. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. 2020.https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 March 11. World Health Organization. [Google Scholar]
- 2.WHO COVID-19 dashboard. https://covid19.who.int/ Available online: accessed.
- 3.Martinez M.A. Lack of effectiveness of repurposed drugs for COVID-19 treatment. Front. Immunol. 2021;12:653. doi: 10.3389/fimmu.2021.653371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asselah T., Durantel D., Pasmant E., Lau G., Schinazi R.F. COVID-19: discovery, diagnostics and drug development. J. Hepatol. 2020;74:168–184. doi: 10.1016/j.jhep.2020.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartsch S.M., O'Shea K.J., Ferguson M.C., Bottazzi M.E., Cox S.N., Strych U., McKinnell J.A., Wedlock P.T., Siegmund S.S., Hotez P.J., Lee B.Y. medRxiv; 2020. How Efficacious Must a COVID-19 Coronavirus Vaccine Be to Prevent or Stop an Epidemic by Itself. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou D., Dai S.M., Tong Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020;75:1667–1670. doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautret P., Lagier J.C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derwand R., Scholz M. Does zinc supplementation enhance the clinical efficacy of chloroquine/hydroxychloroquine to win today's battle against COVID-19? Med. Hypotheses. 2020;142:109815. doi: 10.1016/j.mehy.2020.109815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO Solidarity Trial Consortium Repurposed antiviral drugs for COVID-19—interim WHO SOLIDARITY trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg E.S., Dufort E.M., Udo T., Wilberschied L.A., Kumar J., Tesoriero J., Weinberg P., Kirkwood J., Muse A., DeHovitz J., Blog D.S., Hutton B., Holtgrave D.R., Zucker H.A. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. J. Am. Med. Assoc. 2020;323:2493. doi: 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.RECOVERY Collaborative Group Effect of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;383:2030–2040. doi: 10.1056/nejmoa2022926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horby P.W., Mafham M., Linsell L., Bell J.L., Staplin N., Emberson J.R., Wiselka M., Ustianowski A., Elmahi E., Prudon B., Whitehouse A., Felton T., Williams J., Faccenda J., Underwood J., Baillie J.K., Chappell L., Faust S.N., Jaki T., Jeffery K., Lim W.S., Montgomery A., Rowan K., Tarning J., Watson J.A., White N.J., Juszczak E., Haynes R., Landray M.J. 2020. Effect of Hydroxychloroquine in Hospitalized Patients with COVID-19: Preliminary Results from a Multi-centre, Randomized, Controlled Trial. MedRxiv. [DOI] [Google Scholar]
- 14.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect. Genet. Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui W., Yang K., Yang H. Recent progress in the drug development targeting SARS-CoV-2 main protease as treatment for COVID-19. Front. Mol. Biosci. 2020;7:616341. doi: 10.3389/fmolb.2020.616341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandi T.K., Bairagya H.R., Mukhopadhyay B.P., Sekar K., Sukul D., Bera A.K. Conserved water-mediated H-bonding dynamics of catalytic Asn 175 in plant thiol protease. J. Biosci. 2009;34:27–34. doi: 10.1007/s12038-009-0006-6. [DOI] [PubMed] [Google Scholar]
- 18.Tasneem A., Rai G.P., Reyaz S., Bairagya H.R. The possible molecular mechanism of SARS-CoV-2 main protease: new structural insights from computational methods. Sci. Med. J. 2021;2:108–126. doi: 10.28991/scimedj-2020-02-si-11. [DOI] [Google Scholar]
- 19.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.2210/pdb1p9u/pdb. [DOI] [PubMed] [Google Scholar]
- 20.Bairagya H.R., Tasneem A., Rai G.P., Reyaz S. Structural and dynamical impact of water molecules at substrate-or product-binding sites in human GMPR enzyme: a study by molecular dynamics simulations. J. Phys. Chem. B. 2020;125:1351–1362. doi: 10.1021/acs.jpcb.0c08818. [DOI] [PubMed] [Google Scholar]
- 21.Bairagya H.R., Mukhopadhyay B.P., Bera A.K. Conserved water mediated recognition and the dynamics of active site Cys 331 and Tyr 411 in hydrated structure of human IMPDH‐II. J. Mol. Recogn. 2011;24:35–44. doi: 10.1002/jmr.1021. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay B.P., Ghosh B., Bairagya H.R., Bera A.K., Nandi T.K., Das S.B. Modeling study of rusticyanin-cytochrome C4 complex: an insight to possible H-bond mediated recognition and electron—transfer process. J. Biomol. Struct. Dyn. 2007;25:157–164. doi: 10.1080/07391102.2007.10507164. [DOI] [PubMed] [Google Scholar]
- 23.Bairagya H.R., Mukhopadhyay B.P. An insight to the dynamics of conserved water-mediated salt bridge interaction and interdomain recognition in hIMPDH isoforms. J. Biomol. Struct. Dyn. 2013;31:788–808. doi: 10.1080/07391102.2012.712458. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee A., Bairagya H.R., Mukhopadhyay B.P., Nandi T.K., Mishra D.K. Conserved water mediated H-bonding dynamics of Ser117 and Thr119 residues in human transthyretin–thyroxin complexation: inhibitor modeling study through docking and molecular dynamics simulation. J. Mol. Graph. Model. 2013;44:70–80. doi: 10.1016/j.jmgm.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti B., Bairagya H.R., Mallik P., Mukhopadhyay B.P., Bera A.K. An insight to conserved water molecular dynamics of catalytic and structural Zn+ 2 ions in matrix metalloproteinase 13 of human. J. Biomol. Struct. Dyn. 2011;28:503–516. doi: 10.1080/07391102.2011.10508591. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee A., Bairagya H.R., Mukhopadhyay B.P., Nandi T.K., Bera A.K. 2010. Structural Insight to Mutated Y116S Transthyretin by Molecular Dynamics Simulation.https://nopr.niscair.res.in/handle/123456789/10118 [PubMed] [Google Scholar]
- 27.Mukhopadhyay B.P., Bairagya H.R. Protein folding: grand challenge of nature. J. Biomol. Struct. Dyn. 2011;28:637–638. doi: 10.1080/073911011010524971. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Morin P., Wang W., Kollman P.A. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 29.Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z., Hou T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119:9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 30.Lazaridis T. Inhomogeneous fluid approach to solvation thermodynamics. 1. Theory. J. Phys. Chem. 1998;B102:3531–3541. doi: 10.1021/jp9723574. [DOI] [Google Scholar]
- 31.Wahl J., Smieško M. Thermodynamic insight into the effects of water displacement and rearrangement upon ligand modifications using molecular dynamics simulations. ChemMedChem. 2018;13:1325–1335. doi: 10.1002/cmdc.201800093. [DOI] [PubMed] [Google Scholar]
- 32.Bissantz C., Kuhn B., Stahl M. A medicinal chemist's guide to molecular interactions. J. Med. Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumathi K., Ananthalakshmi P., Roshan M.M., Sekar K., 3dSS 3D structural superposition. Nucleic Acids Res. 2006;34:W128–W132. doi: 10.1093/nar/gkl036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guex N., Peitsch M.C. SWISS‐MODEL and the Swiss‐Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 36.Ogata K., Wodak S.J. Conserved water molecules in MHC class-I molecules and their putative structural and functional roles. Protein Eng. Des. Sel. 2002;15:697–705. doi: 10.1093/protein/15.8.697. [DOI] [PubMed] [Google Scholar]
- 37.Bairagya H.R., Mishra D.K., Mukhopadhyay B.P., Sekar K. Conserved water-mediated recognition and dynamics of NAD+ (carboxamide group) to hIMPDH enzyme: water mimic approach toward the design of isoform-selective inhibitor. J. Biomol. Struct. Dyn. 2014;32:1248–1262. doi: 10.1080/07391102.2013.812982. [DOI] [PubMed] [Google Scholar]
- 38.Bairagya H.R., Mukhopadhyay B.P., Bhattacharya S. Role of the conserved water molecules in the binding of inhibitor to IMPDH-II (human): a study on the water mimic inhibitor design. J. Mol. Struc.-Theochem. 2009;908:31–39. doi: 10.1016/j.theochem.2009.04.037. [DOI] [Google Scholar]
- 39.Smith D.K., Radivojac P., Obradovic Z., Dunker A.K., Zhu G. Improved amino acid flexibility parameters. Protein Sci. 2003;12:1060–1072. doi: 10.1110/ps.0236203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoete V., Daina A., Bovigny C., Michielin O. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016;56:1399–1404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
- 42.Sander T. 2001. OSIRIS Property Explorer, Organic Chemistry Portal.https://www.organic-chemistry.org/prog/peo/ [Google Scholar]
- 43.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris G.M., Huey R., Olson A.J. Using autodock for ligand‐receptor docking. Curr. Protoc. Bioinf. 2008;24:8–14. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- 47.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 49.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S.A., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- 50.Kalé L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999;151:283–312. doi: 10.1006/jcph.1999.6201. [DOI] [Google Scholar]
- 51.Huang J., MacKerell A.D., Jr. CHARMM36 all‐atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., Mackerell A.D., Jr. CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manoharan P., Ghoshal N. Fragment-based virtual screening approach and molecular dynamics simulation studies for identification of BACE1 inhibitor leads. J. Biomol. Struct. Dyn. 2018;36:1878–1892. doi: 10.1080/07391102.2017.1337590. [DOI] [PubMed] [Google Scholar]
- 54.Grossfield A., Feller S.E., Pitman M.C. Convergence of molecular dynamics simulations of membrane proteins. Proteins: Struct. Funct. Bioinf. 2007;67:31–40. doi: 10.1002/prot.21308. [DOI] [PubMed] [Google Scholar]
- 55.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 56.Pimthon J., Willumeit R., Lendlein A., Hofmann D. Membrane association and selectivity of the antimicrobial peptide NK‐2: a molecular dynamics simulation study. J. Pept. Sci. 2009;15:654–667. doi: 10.1002/psc.1165. [DOI] [PubMed] [Google Scholar]
- 57.Allen M.P., Tildesley D.J. 2017. Computer Simulation of Liquids. Oxford Scholarship Online. [DOI] [Google Scholar]
- 58.Lopez E.D., Arcon J.P., Gauto D.F., Petruk A.A., Modenutti C.P., Dumas V.G., Marti M.A., Turjanski A.G. WATCLUST: a tool for improving the design of drugs based on protein-water interactions. Bioinformatics. 2015;31:3697–3699. doi: 10.1093/bioinformatics/btv411. [DOI] [PubMed] [Google Scholar]
- 59.Gauto D.F., Di Lella S., Estrin D.A., Monaco H.L., Martí M.A. Structural basis for ligand recognition in a mushroom lectin: solvent structure as specificity predictor. Carbohydr. Res. 2011;346:939–948. doi: 10.1016/j.carres.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 60.Huggins D.J., Marsh M., Payne M.C. Thermodynamic properties of water molecules at a protein–protein interaction surface. J. Chem. Theor. Comput. 2011;7:3514–3522. doi: 10.1021/ct200465z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Lella S., Martí M.A., Álvarez R.M.S., Estrin D.A., Ricci J.C.D. Characterization of the galectin-1 carbohydrate recognition domain in terms of solvent occupancy. J. Phys. Chem. B. 2007;111:7360–7366. doi: 10.1021/jp068989k. [DOI] [PubMed] [Google Scholar]
- 62.Wagner J.R., Sørensen J., Hensley N., Wong C., Zhu C., Perison T., Amaro R.E. POVME 3.0: software for mapping binding pocket flexibility. J. Chem. Theor. Comput. 2017;13:4584–4592. doi: 10.1021/acs.jctc.7b00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu H., Hou T. CaFE: a tool for binding affinity prediction using end-point free energy methods. Bioinformatics. 2016;32:2216–2218. doi: 10.1093/bioinformatics/btw215. [DOI] [PubMed] [Google Scholar]
- 64.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z., Hou T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119:9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 66.Miranda W.E., Noskov S.Y., Valiente P.A. Improving the LIE method for binding free energy calculations of protein–ligand complexes. J. Chem. Inf. Model. 2015;55:1867–1877. doi: 10.1021/acs.jcim.5b00012. [DOI] [PubMed] [Google Scholar]
- 67.Kanaujia S.P., Sekar K. Structural and functional role of water molecules in bovine pancreatic phospholipase A2: a data-mining approach. Acta Crystallogr. D Biol. Crystallogr. 2009;65:74–84. doi: 10.1107/S0907444908039292. [DOI] [PubMed] [Google Scholar]
- 68.Carugo O. Correlation between occupancy and B factor of water molecules in protein crystal structures. Protein Eng. 1999;12:1021–1024. doi: 10.1093/protein/12.12.1021. [DOI] [PubMed] [Google Scholar]
- 69.Carugo O., Bordo D. How many water molecules can be detected by protein crystallography? Acta Crystallogr. D Biol. Crystallogr. 1999;55:479–483. doi: 10.1107/S0907444998012086. [DOI] [PubMed] [Google Scholar]
- 70.Bondi A.V. van der Waals volumes and radii. J. Phys. Chem. 1964;68:441–451. doi: 10.1021/j100785a001. [DOI] [Google Scholar]
- 71.Keretsu S., Bhujbal S.P., Cho S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-74468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary data of this article is provided in the supplementary material.