Abstract
Background
Mirror-image pain (MIP), which develops from the healthy body region contralateral to the actual injured site, is a mysterious pain phenomenon accompanying many chronic pain conditions, such as complex regional pain syndrome (CRPS). However, the pathogenesis of MIP still remains largely unknown. The purpose of this study is to perform an expression profiling to identify genes related to MIP in an animal model of CRPS-I.
Methods
We established a rat chronic post-ischemic pain (CPIP) model to mimic human CRPS-I. RNA-sequencing (RNA-Seq), bioinformatics, qPCR, immunostaining, and animal behavioral assays were used to screen potential genes in the contralateral dorsal root ganglia (DRG) that may be involved in MIP.
Results
The CPIP model rats developed robust and persistent MIP in contralateral hind paws. Bilateral DRG neurons did not exhibit obvious neuronal damage. RNA-Seq of contralateral DRG from CPIP model rats identified a total 527 differentially expressed genes (DEGs) vs sham rats. The expression changes of several representative DEGs were further verified by qPCR. Bioinformatics analysis indicated that the immune system process, innate immune response, and cell adhesion were among the mostly enriched biological processes, which are important processes involved in pain sensitization, neuroinflammation, and chronic pain. We further identified DEGs potentially involved in pain mechanisms or enriched in small- to medium-sized sensory neurons or TRPV1-lineage nociceptors. By comparing with published datasets summarizing genes enriched in pain mechanisms, we sorted out a core set of genes which might contribute to nociception and the pain mechanism in MIP.
Conclusion
We provided by far the first study to profile gene expression changes and pathway analysis of contralateral DRG for the studying of MIP mechanisms. This work may provide novel insights into understanding the mysterious mechanisms underlying MIP.
Keywords: RNA-Seq, pain, CRPS-I, dorsal root ganglia, inflammation, cytokine
Introduction
Mirror-image pain (MIP) is a mysterious pain phenomenon which is accompanied with many clinical pain conditions.1 MIP develops from the healthy body region which is contralateral to the actual injured site.1–3 MIP is typically characterized by increased mechanical hypersensitivity on the uninjured mirror-image body side.4 It can be triggered in response to light touch/pressure stimuli, which are normally perceived as innocuous.5 Patients with chronic pain, such as complex regional pain syndrome (CRPS) and atypical facial pain, etc., can develop MIP to the unaffected body side.6,7
Complex regional pain syndrome type-I (CRPS-I) is a chronic pain condition that usually impacts the extremities of the patients.8 It is usually triggered by an initial injury, including tissue ischemia, fractures, or surgery in the extremity and develops into a chronic pain condition that impacts the life quality of the patients.9–11 One of the clinical features of CRPS-I is the MIP developed in the unaffected extremities.7,12,13 CRPS-I patients developed hypersensitivity to capsaicin, thermal, and mechanical stimuli in the unaffected extremities.12 Furthermore, we and others also observed thermal and mechanical hypersensitivity in the unaffected hind limb of a rat model of CRPS-I, suggesting the occurrence of MIP in animal models.14–18
At present, the pathogenesis of MIP still remained largely unknown. However, certain possible mechanisms have been proposed to contribute to MIP.19 Spinal glial activation has been observed in the contralateral spinal cord dorsal horn of MIP model animals.15,17,20 Pharmacological blocking spinal glia metabolism and proinflammatory cytokines (including TNF-α, IL-1β, and IL-6) significantly alleviates MIP, suggesting spinal glial and proinflammatory cytokines are involved in MIP.2 In addition to the central mechanism, peripheral mechanisms have also been proposed for MIP.4,21 It is reported that TNF-α produced from ipsilateral injured DRG can diffuse to the contralateral side via cerebrospinal fluid and activates satellite cells to produce NGF, which promotes nociceptor excitability on the contralateral side and induces MIP.4 Although these studies provide evidence suggesting central or peripheral glia and proinflammatory cytokines are important for MIP pathogenesis, it still remains unclear how contralateral DRG is affected in chronic pain conditions and exerts pain signals that may contribute to MIP.
In order to study the molecular mechanisms underlying MIP, we performed a genome-wide expression profiling of DRG innervating contralateral hind limbs of a rat model of CRPS-I by means of RNA-Seq. We obtained gene expression profiles and identified differentially expressed genes (DEGs) in CPIP model rats vs sham rats. We studied the major functions or signaling pathways that these DEGs may be involved in. We further screened the genes which might contribute to nociception and pain processing during MIP. Thus, this study provides novel insights into understanding the peripheral mechanisms contributing to MIP.
Materials and Methods
Animals
Male adult Sprague-Dawley rats weighing 300–320 g were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Sciences. Rats were randomly assigned to each group and housed in the Laboratory Animal Center of Zhejiang Chinese Medical University accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) under standard environmental conditions (12 hour light–dark cycles and 24±2°C). Rats had free access to food and water. The animals were given 1 week to adapt to the new environment before formal experiment. All experimental procedures were carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) and approved by the Animal Ethics Committee of Zhejiang Chinese Medical University (Permission Number: ZSLL-2017-183).
CPIP Rat Model Establishment
The CPIP model was established by prolonged hind paw ischemia and reperfusion as reported before.16,22 Anesthesia was induced by intraperitoneal injection of 50 mg/kg of sodium pentobarbital and was maintained at a dose of 20 mg/kg/h. Glycerin was applied to the ankle joints of rats. Afterwards, a Nitrile 70 Durometer O-ring (Gaohui Rubber & Plastic Co., Ltd., Shanghai, China) with an inner diameter of 7/32 inch (5.5 mm) was tightly passed around the right hind limb just proximal to the ankle joint. The O-ring was cut off 3 hours later for reperfusion. Sham rats received the same anesthesia procedure but a cut O-ring, which could not block blood flow, was applied to the same part as the CPIP model rats.
Determination of Mechanical Allodynia
Before baseline test, rats were habituated to the testing environment every day for 3 consecutive days. Rats were individually placed on an elevated mesh floor, covered with transparent Plexiglas chambers. Rats were habituated for 30 minutes before the test. The mechanical allodynia was measured using a series of von Frey filaments (ranging from 0.6–26 g, UGO Basile, Italy) applied perpendicularly to the mid-plantar surface of the hind paws, with sufficient force to bend the filament slightly for 5 seconds according to methods we previously used.23 Nocifensive responses were identified as an abrupt withdrawal of the paw and licking and vigorously shaking in response to the stimulation. The threshold was determined by the “Up–Down” testing paradigm, and the 50% paw withdrawal threshold (PWT) was calculated by the nonparametric Dixon test.24,25 Behavior tests were all conducted by an experimenter blinded to experimental conditions.
Measurement of Hind Paw Edema
Edema was observed as an increase in hind paw diameter and measured by a digital caliper, and was calculated as the difference between the basal value and the test value as described in our previous study.15 Each rat was measured three times and the mean value was calculated.
Tissue Collection and RNA Extraction
At day 7, rats were deeply anesthetized with sodium pentobarbital at a dose of 50 mg/kg and were perfused transcardially with 200 mL 0.9% saline (4°C). After the perfusion, the left (contralateral) side L4-6 DRG segments were collected and were immediately preserved in the RNA later solution (Thermo Fisher, USA). Total RNA was extracted using the Trizol reagent (Thermo Fisher, USA) following the manufacturer’s instructions. DNase I (Thermo Fisher, USA) was used to degrade contaminating DNA. The purity and concentration of the samples was assessed by a Nanodrop Spectrophotometer (NanoDrop Products, USA) and the RNA integrity was assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies, USA).
Immunofluorescence Staining
At day 7, rats were deeply anesthetized with sodium pentobarbital at a dose of 50 mg/kg and were perfused transcardially with 200 mL 4% formaldehyde after 200 mL 0.9% saline (4°C). The bilateral L4-6 DRGs was harvested and post-fixed in the same fixative for 4 hours (4°C) before transferring to 15% and 30% sucrose for 48 hours for dehydration. A few days later, the DRG were serially cut into 8-μm thick transverse sections on a frozen microtome (Cryostar NX50, Thermo Scientific, USA) and mounted on adhesive slides as eight sets of every 10th serial section. All the slides were blocked with 5% normal donkey serum in PBS for 1 hour at 37°C and then incubated overnight at 4°C with corresponding primary antibodies. The primary antibodies used were rabbit anti-ATF3 (#HPA001562, Sigma, USA) and mouse anti-NeuN (#ab104224, Abcam, UK). On the 2nd day, the sections were rinsed with PBS and incubated for 1 hour with a mixture of corresponding secondary antibodies. A Nikon A1R laser scanning confocal microscope (Nikon, Japan) was used to capture fluorescence images. Uniform microscope settings were maintained throughout all image capture sessions in order for quantitative fluorescence intensity analysis. All stained sections were examined and analyzed in a blinded manner. Five images were randomly selected per rat tissue and averaged and then compared according to methods described in our previous studies.26
RNA-Seq Library Establishment and RNA-Seq
We isolated the total mRNAs of four rats of each group and used them to construct sequencing libraries. mRNAs were purified from total RNA using oligo (dT)-attached magnetic beads using two rounds of purification. Following purification, mRNA was fragmented into small pieces using fragmentation reagent under elevated temperature. Subsequently, first-strand cDNA was generated by random hexamer-primed reverse transcription, followed by a second-strand cDNA synthesis. The synthesized cDNA was subjected to end-repair and then was 3ʹ adenylated. Adaptors were ligated to the ends of these 3ʹ adenylated cDNA fragments. This process is to amplify the cDNA fragments with adaptors from previous step. PCR products are purified with the SPRI beads and dissolved in EB solution. The double stranded PCR products were heat denatured and circularized by the splint oligonucleotide sequence. The single-strand circle DNA (ssCir DNA) was formatted as the final library. The library was validated by the Agilent 2100 Bioanalyzer System (Agilent Technologies, USA). The DNA nanoballs were loaded into the patterned nanoarray and single-end 50 base reads were generated in the way of being sequenced by synthesis. Finally, the fragments were enriched by PCR amplification to construct a library ready for sequencing using the BGI SEQ-500 platform (BGI Group, Shenzhen, China). The RNA-Seq dataset has been deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus repository with accession number GSE174299.
Bioinformatics Analysis
Primary sequencing data produced by RNA-Seq (raw reads) were subjected to quality control (QC). The information of total reads and mapping ratio reads is shown in Table 1. Raw reads were filtered into clean reads using internal software SOAP nuke (version 1.5.2), as follows: Remove reads in which unknown bases (N) are more than 10%; Remove reads with adaptors; Remove low quality reads (we define the low quality read as the percentage of base which quality is < 15 and > 50% in a read). QC of alignment was performed to determine if re-sequencing was needed. If the alignment result passed QC, downstream analysis including gene expression, differentially expressed genes, cluster analysis, Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and Gene Set Enrichment Analysis (GSEA) analysis were then performed.
Table 1.
Sequences of the Primers Used for qPCR Validation of RNA-Seq Dataset
| Gene Name | Gene ID | Primer Sequence (5′ −3′) | Amplicon Size (bp) |
|---|---|---|---|
| Gpr183 | 679975 | F: 5ʹ-GCAGAACCCACTGACCGAGAAATC-3’ | 83 |
| R: 5ʹ-ACAAGACGAACACGCCGATGATG-3’ | |||
| Tlr8 | 684440 | F: 5ʹ-CGCCTTGACCGTCTGTGGAATG-3’ | 142 |
| R: 5ʹ-GGCTCTGAGGCAAGTTGAGGAATG-3’ | |||
| Tbx18 | 315870 | F: 5ʹ-GCAAGGCAACACAAGTTCTTCAGC-3’ | 98 |
| R: 5ʹ-GAGAGGAACAGGAGGAGCCAGAC-3’ | |||
| Egfr | 24329 | F: 5ʹ-CACTACGCCGCCTGCTTCAAG-3’ | 145 |
| R: 5ʹ-ACTGTGCCAAATGCTCCTGAACC-3’ | |||
| Cxcl13 | 498335 | F: 5ʹ-CGACTTTGAAAGGTTGCTTGTA-3’ | 219 |
| R: 5ʹ-ACACTGGATGAATAGGAAACGT-3’ | |||
| Slfn1 | 688900 | F: 5ʹ-GCTGAGAGTGACCACAGGCATTG-3’ | 113 |
| R: 5ʹ-AGCCCAAGACAGACTCCCACAG-3’ | |||
| Gli2 | 304729 | F: 5ʹ-ACCTCCATCACCGTGCCTACC-3’ | 144 |
| R: 5ʹ-AGTCTTGACCTTGCTCCGCTTATG-3’ | |||
| Stk32b | 305431 | F: 5ʹ-AGAAGTGCGTGGAGAGGGATGAG-3’ | 96 |
| R: 5ʹ-AGTACCACAGGTTCACCAGGAAGG-3’ | |||
| Asgr2 | 29403 | F: 5ʹ-CACGGAGGTAGCAGGAATGACAAC-3’ | 136 |
| R: 5ʹ-AGGTCAGGGTTCGCAGATCCAG-3’ | |||
| Smoc2 | 292401 | F: 5ʹ-AGGAAGCCAAGCAACCCAAGAATG-3’ | 117 |
| R: 5ʹ-CTACCAGCACACACCAGCAGTATC-3’ | |||
| Fcgr1a | 295279 | F: 5ʹ-CCTGGCTTACGGCTTTACTTCTCC-3’ | 140 |
| R: 5ʹ-CCGTCCTCCGTGGCTACCTC-3’ | |||
| Osr2 | 315039 | F: 5ʹ-CGGCAAAGTGTTCAGGCGAAAC-3’ | 139 |
| R: 5ʹ-TGGAAGGCGTGGAGAGGTGTC-3’ | |||
| Sned1 | 316638 | F: 5ʹ-CTGCGTCCGTGCCTCAATGG-3’ | 89 |
| R: 5ʹ-GTGAAGCCAGCGAGACAGGAAC-3’ | |||
| Siglec1 | 311426 | F: 5ʹ-ACTCCAGACCTGACCACCTTCTTG-3’ | 119 |
| R: 5ʹ-GCCAATGTGAGACCTCCGTGTG-3’ | |||
| Myl1 | 56781 | F: 5ʹ-AGCAGACTGAGAGGAGCAGGAAG-3 | 103 |
| R: 5ʹTTGGTGTTGATGAGGCTGGTGTTC-3 | |||
| Myom2 | 306616 | F: 5ʹ-AAGACAACCACCAACGCTCACTG-3’ | 107 |
| R: 5ʹ-GCCACGATTCTTCTCAGCAAATGC-3’ | |||
| Capg | 297339 | F: 5ʹ-GGAGAGGAGCCAGCCGAGATG-3’ | 97 |
| R: 5ʹ-CGTTGGTCTGGTCCGCTGTAATG-3’ | |||
| Retsat | 246298 | F: 5ʹ-GAGCGTGCGGTGTCAGTGTG-3’ | 128 |
| R: 5ʹ-TGGCAGGTAGCGGACAGACTC-3’ |
Cluster Analysis and Screening of Differentially Expressed Genes
Methods were adopted as before.27 Briefly, distances of expressed genes were calculated using the Euclidean method.28 The sum of the squared deviations algorithm was used to calculate distance. The cluster analysis and heat map visualization of gene expression patterns was performed using the “pheatmap” package in the R software of Bioconductor. Differentially expressed mRNAs with statistical significance were identified through Volcano Plot filtering. The threshold required for the results to be considered significant was as follows: q-value≤0.001 and fold change≥1.5 or ≤0.667.
Functional Enrichment Analysis of DEGs
Functional enrichment analysis was performed through the functional annotation package “cluster Profiler” in R studio software (RStudio, Boston, MA, USA). GO and KEGG enrichment analysis were also conducted. For each enriched function term, the Q-value of enriched functions and the Q-value by multiple testing corrections were calculated by “cluster Profiler” package in R studio software. The GO functional and KEGG pathway enrichment analysis were performed for DEGs using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tools (http://www.geneontology.org and http://www.genome.jp/kegg).
GSEA Analysis
Gene set enrichment analysis (GSEA) was performed according to methods previously described.29 We chose the top 10 most significantly upregulated pathways from GSEA analysis. The gene sets were downloaded from the Molecular Signatures Database v7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). A FDR≤0.25 and NES>1.0 were adopted as the criterion for judging significance.29
Real-Time Quantitative PCR (qPCR) Analysis
According to the manufacturer’s instruction, the extracted total RNA from the DRGs was reverse transcribed into cDNA using random hexamers primers (TaKaRa Bio Inc., China). The sequences of all primers used are shown in Table 1. qPCR was performed using SYBR Green I Master (Roche, Switzerland) in 20 µL reaction system according to the manufacturer’s protocol. qPCR was performed with the LightCycler 480 real-time PCR system (Roche, Switzerland). Each reaction was performed in triplicate and normalized to β-actin gene expression. The CT value of each well was determined using the LightCycler480 System software and the average of the triplicates was calculated. The relative quantification was determined by the ΔΔCT method.30,31
Source of Microarray Data
Pain genes (PG): The pain gene list was retrieved from a previous study using the same list to predict potential genes related to mediating masseter inflammation-induced pain.32 This pain gene list contains 685 genes, which mainly includes central and peripheral inflammatory responses and pain mood and affect, human genes related to transmission of pain signals (see Suppl Table 1 for the full list of genes).
Sensory neuronal genes (SN): The sensory neuronal gene list was retrieved from a previous study by magnetic cell sorting (MACS) to separate neuronal cells from non-neuronal cells from mouse DRG.33 According to this study, MACS successfully segregated neuronal cells from non-neuronal cells from mouse DRGs. This procedure can enrich the neuronal population up to 95% compared with 10% before sorting. Transcriptome analysis further confirmed that a lot of genes were enriched in the neuronal population collected through MACS. We selected genes that showed an increase in expression by >40% according to the criterion described by Chung et al32 (see Suppl Table 2 for the full list of genes).
TRPV1-lineage neuronal genes (VL): The TRPV1-lineage neuronal genes list was retrieved from a study which identified a group of genes enriched in TRPV1 expressing nociceptive sensory neurons.34 By using mouse lines selectively labeling or ablating TRPV1 lineage neurons, Goswami et al34 identified a group of genes enriched in VL sensory neurons. We screened genes that are enriched >3-fold in VL neurons compared to non-VL neurons according to a study by Chung et al32 (see Suppl Table 3 for the full list of genes).
Protein–Protein Interaction (PPI) Network Analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) is used to provide information regarding predicted and experimental interactions of proteins and the prediction method of this database is from neighborhood, gene fusion, co-occurrence, co-expression experiments, databases, and text mining. By setting the Combination score >0.7 as the reliability threshold value, the web based STRING database (http://string-db.org/) was used to produce PPI predictions after uploading the union gene list to the search bar.35 Based on the interplayed relationships, a PPI network was established and then visualized using the Cytoscape software.36 The connectivity degree of each protein, namely the number of proteins it connected, was calculated to evaluate its importance in this network.
Statistical Analysis
Data in graphs are expressed as means±SEM. One- or two-way ANOVA followed by Tukey’s post-hoc test was used for comparisons among groups ≥3. Student’s t-test was used for comparisons between two groups. The data from different groups were verified for normality using Kolmogorov–Smirnov test. If not normally distributed, a non-parametric test (Mann Whitney U-test) was used. The comparison is considered significantly different if the p-value is less than 0.05.
Results
The Establishment of a Rat Model of CRPS-I and the Evaluation of the Animals’ Nocifensive Behavior
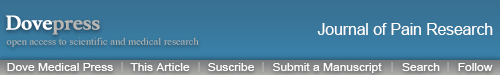
We first established the rat CPIP model to mimic human CRPS-I following methods described before.16,22 An O-ring with 7/32 inch internal diameter was tightly passed around the right hind limb just proximal to the ankle joint and applied for 3 hours. During the ligation, the right hind limb displayed obvious cyanosis (an indication of tissue hypoxia) and edema (Figure 1A). The edema persisted for 3 days and gradually returned back to normal (Figure 1B and C). The PWT (paw withdrawal threshold) of ipsilateral hind paws of CPIP model group rats was significantly reduced on Day 1 compared with the sham group and lasted over 7 days (Figure 1D and F). Moreover, the contralateral hind paw also showed a significant reduction in PWT on Day 1 compared with sham rats and persisted over 7 days (Figure 1E and G), which is a clear sign of MIP. The contralateral mechanical allodynia developed in all CPIP model rats we had observed. These results were consistent with previous studies,15–17 demonstrating the successful establishment of the CPIP model and the presence of MIP in model rats.
Figure 1.
The CPIP model rats displayed remarkable and prolonged mechanical allodynia in both ipsilateral and contralateral hind paws. (A) Representative pictures of the rat hind paw taken before/during CPIP model establishment and 20 minutes, 1 day, and 7 days after model establishment. The arrow indicates the position of the O-ring. (B and C) Edema evaluation of the ipsilateral (B) and contralateral (C) hind paw. (D and E) 50% paw withdraw threshold (PWT) of ipsilateral (D) and contralateral (E) hind paw. (F and G) Summary of the normalized area under the curve (AUC) as in panels (D and E), respectively. n=10 rats/group. **p<0.01 vs sham group. Student’s t-test or two-way ANOVA followed by Tukey post-hoc test was used for statistical analysis.
Abbreviation: CPIP, chronic post-ischemic pain.
Pathological Evaluation of Ipsilateral and Contralateral DRG Neurons Innervating the Hind Paws
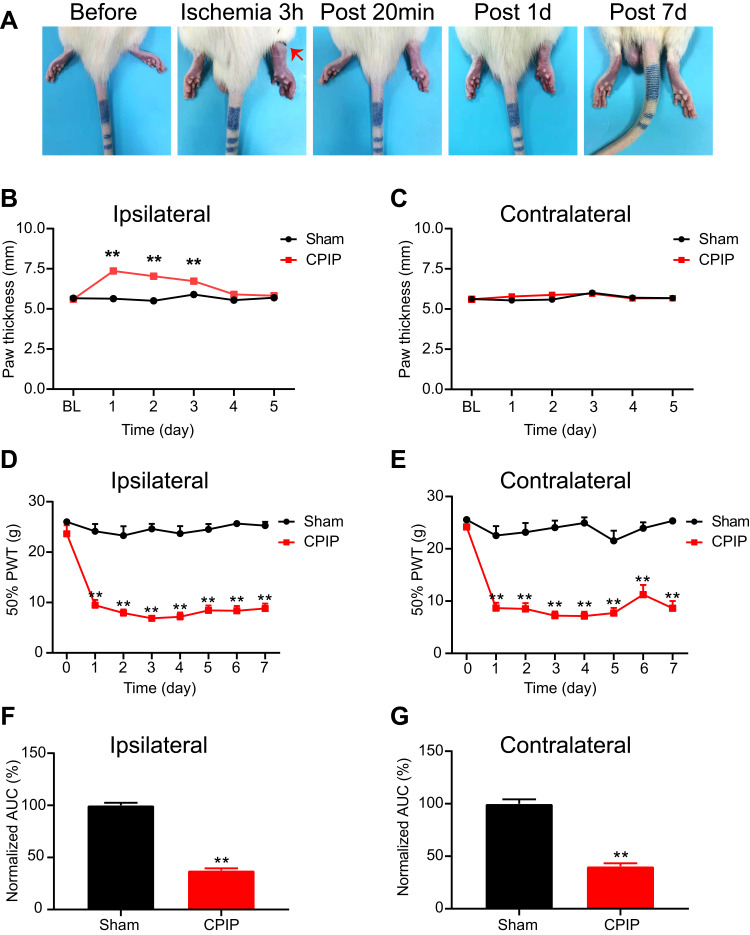
We sacrificed the rats on Day 7 and harvested bilateral L4–6 DRG. We first performed immunostainings to examine whether any neuronal damage occurred in the DRG. We used ATF3, a widely used marker for neuronal damage, for the examination. As shown in Figure 2A–C, we did not identify any ATF3 positively stained neurons in ipsilateral or contralateral L4–6 DRG of CPIP model rats compared with sham rats. For positive control, in one of our recent studies, we stained the DRG with ATF3 from paclitaxel-treated rats, a rat model for chemotherapy-induced peripheral neuropathy and well known for its peripheral neuropathy. We found the number of ATF3 positively expressed neurons was significantly increased in DRG of paclitaxel-treated rats vs control rats.27 Therefore, these results indicated no neuronal damage occurred in bilateral DRG neurons of the CPIP model rats.
Figure 2.
The CPIP model rats did not show any neuronal damage in bilateral DRG neurons innervating the hind limbs. (A) Representative immunostaining pictures showing ATF3 antibody (green) staining of ipsilateral and contralateral DRG from the sham group and CPIP model group. NeuN was used to label neurons (red). (B) Summary of the number of ATF3 positive neurons per observation field in ipsilateral DRG. (C) Summary of the number of ATF3 positive neurons per observation field in contralateral DRG. Scale bar indicates 100 μm. n=6 rats/group.
Abbreviations: CPIP, chronic post-ischemic pain; DRG, dorsal root ganglia.
Examination of Gene Expression Profile Changes in Contralateral DRG from CPIP Model Rats via RNA-Seq
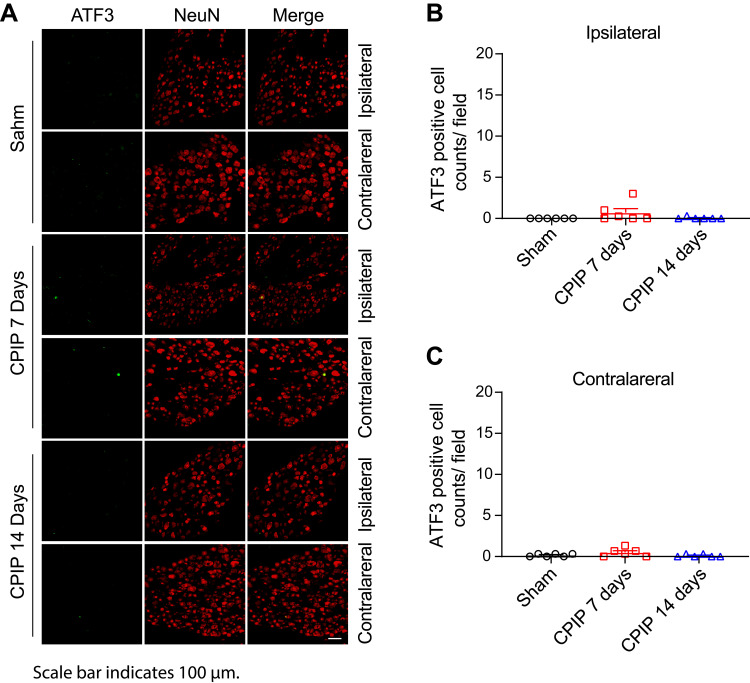
To gain further insights into the mechanisms underlying MIP of CPIP model rats, we harvested contralateral L4-6 DRG at Day 7 from CPIP model and sham rats and analyzed the gene expression profiles via RNA-Seq. We managed to obtain high quality RNA for RNA-Seq analysis (Suppl Figure 1). RNA-Seq generated approximately 23.9 million raw reads from each sample. The ratio of clean reads reached above 98.0% (Table 2). Over 90% of bases had a quality score ≥Q30 and over 94% of the clean reads were mapped to the rat genome. In all, a total of 23, 267 genes were successfully mapped and identified from RNA-Seq (Suppl Table 4). We then filtered out the differentially expressed genes (DEGs) with the criteria of fold change ≥1.5 or ≤0.667 and q-value≤0.001 (Suppl Table 5). We displayed the DEGs of the CPIP group vs sham group in the volcano plot as shown in Figure 3A. In all, 527 DEGs (including 245 up- and 282 down-regulated) were identified from the CPIP group vs sham group (Figure 3A). We further summarized and plotted the identified DEGs in a heat map as shown in Figure 3B. Cluster analysis indicated a high level of consistency within CPIP or sham group samples, but a clear separation between these two groups (Figure 3B). These results indicated that we have obtained a high quality RNA-Seq dataset that can be further used for bioinformatics analysis.
Table 2.
The Information of Total Reads and Mapping Ratio for Sham and CPIP Groups in RNA-Seq
| Sample | Total Raw Reads (M) | Total Clean Reads (M) | Clean Reads Q30 (%) | Clean Reads Ratio (%) | Total Mapping (%) |
|---|---|---|---|---|---|
| CPIP1 | 23.92 | 23.74 | 93.65 | 99.22 | 94.65 |
| CPIP2 | 23.92 | 23.74 | 93.86 | 99.21 | 94.83 |
| CPIP3 | 23.92 | 23.73 | 93.96 | 99.2 | 94.81 |
| CPIP4 | 23.92 | 23.74 | 93.15 | 99.24 | 94.47 |
| Sham1 | 23.92 | 23.66 | 93.21 | 98.88 | 94.08 |
| Sham2 | 23.92 | 23.73 | 94.01 | 99.19 | 94.75 |
| Sham3 | 23.92 | 23.73 | 93.59 | 99.2 | 94.66 |
| Sham4 | 23.92 | 23.75 | 93.58 | 99.27 | 94.71 |
Figure 3.
Gene expression profiling by RNA-Seq in contralateral DRG of CPIP model rats. (A) Volcano plot illustrating gene expression profiles in contralateral DRG of CPIP model group vs sham group. Red and blue dots show up- and down-regulated DEGs, respectively. Gray dots indicate non-DEGs. (B) Heat map showing the hierarchical clustering of DEGs from CPIP and sham groups and within groups.
Abbreviations: RNA-Seq, RNA-sequencing; DRG, dorsal root ganglia; CPIP, chronic post-ischemic pain; DEGs, differentially expressed genes.
Analysis of DEGs from Contralateral DRG of CPIP Model Rats
Among the DEGs we identified, a certain number of genes were reported to participate in pain mechanisms, which include: Cxcl14 (C-X-C motif chemokine 14 precursor, fold change=3.2), Cxcl13 (C-X-C motif chemokine 13 precursor, fold change=2.6), Tlr5 (Toll-like receptor 5, fold change=2.6), Ngf (Nerve growth factor, fold change=2.3), and Tlr8 (Toll-like receptor 8, fold change=1.8). We further analyzed the RNA-Seq dataset and identified 167 genes (63 up-regulated and 104 down-regulated gene) showing expression changes >10-fold, including Tsr2 (TSR2 Ribosome Maturation Factor, fold change=109,177.8), Aph1b (Gamma-Secretase Subunit APH-1B, fold change=436.9), etc. In total, 124 genes had expression changes between 5- and 10-fold, including 40 up-regulated and 84 down-regulated genes. The top 20 up- or 20 down-regulated DEGs are summarized in Tables 3 and 4. These data suggest that the DEGs are probably correlated with MIP pathogenesis.
Table 3.
The Detailed Information of the Top 20 Up-Regulated DEGs
| Upregulated Gene | Gene ID | Location | log2 Fold-Change (CPIP/Sham) | Q Value | Official Gene Name (NCBI) |
|---|---|---|---|---|---|
| Tsr2 | 317418 | 4:20141406–20146082 | 16.73631777 | 3.05E-48 | TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| LOC100912618 | 100912618 | 4:5867350–5869085 | 10.69826964 | 1.77E-12 | ubiquitin-conjugating enzyme E2 variant 1-like |
| LOC100910581 | 100910581 | 4:21987993–21989200 | 9.114037693 | 6.00E-51 | Protein phosphatase 1 regulatory subunit 35-like |
| Aph1b | 300802 | 4:72508008–72598921 | 8.771010005 | 5.97E-42 | aph-1 homolog B, gamma secretase subunit |
| LOC103691556 | 103691556 | 4:144191722–144211986 | 7.608251098 | 8.58E-60 | Mothers against decapentaplegic homolog 9 |
| LOC102550456 | 102550456 | 4:21748602–22021862 | 7.14204003 | 4.65E-16 | TSC22 domain family protein 4-like |
| LOC108349606 | 108349606 | 4:166282044–166282845 | 6.69115347 | 3.43E-12 | 60S ribosomal protein L7a-like |
| LOC100911233 | 100911233 | 4:33876697–33909836 | 6.417167166 | 2.65E-10 | Cell division cycle and apoptosis regulator protein 1-like |
| LOC100912399 | 100912399 | 4:43938500–44000199 | 6.383284264 | 4.34E-10 | Mitogen-activated protein kinase kinase kinase 1-like |
| Potef | 684969 | 4:24780521–24783147 | 6.285874308 | 1.68E-09 | POTE ankyrin domain family, member F |
| Hmgn5 | 100910616 | 4:80134557–80142902 | 5.972626604 | 7.99E-08 | High mobility group nucleosome binding domain 5 |
| LOC108348106 | 108348106 | 4:90957132–90962541 | 5.585249332 | 3.78E-06 | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 |
| LOC100909795 | 100909795 | 4:78550661–78577178 | 5.500305695 | 7.81E-06 | Colorectal mutant cancer protein-like |
| LOC100911564 | 100911564 | 4:16314107–16317575 | 5.395697586 | 1.82E-05 | Glycine N-methyltransferase-like |
| LOC100361265 | 100361265 | 4:71676810–71681062 | 5.30501238 | 1.26E-09 | rCG64283-like |
| RGD1584023 | 690146 | 4:94827988–94840706 | 5.00539585 | 2.68E-04 | Similar to zinc finger protein 11B |
| RGD1560795 | 297402 | 4:116933277–116936466 | 4.968015878 | 3.36E-04 | similar to Sepiapterin reductase (SPR) |
| LOC102552708 | 102552708 | 4:134996–135398 | 4.877490295 | 5.64E-04 | DNA-directed RNA polymerase II subunit GRINL1A-like |
| Zp2 | 81828 | 4:189899906–189911650 | 4.462124948 | 2.03E-05 | zona pellucida glycoprotein 2 |
| Slc25a36l1 | 364991 | 4:37019755–37022003 | 3.968951847 | 6.98E-10 | Solute carrier family 25 (pyrimidine nucleotide carrier), member 36-like 1 |
Table 4.
The Detailed Information of the Top 20 Down-Regulated DEGs
| Downregulated Gene | Gene ID | Location | log2 Fold-Change (CPIP/Sham) | Q Value | Official Gene Name (NCBI) |
|---|---|---|---|---|---|
| LOC100909761 | 100909761 | 4:78661643–78681196 | −8.814802024 | 5.59E-86 | Myotilin-like |
| Myot | 291605 | 4:35573978–35593541 | −8.321037008 | 2.02E-32 | Myotilin |
| LOC100910177 | 100910177 | 4:52919250–52919654 | −8.043339285 | 1.43E-27 | Dolichol-phosphate mannosyl transferase subunit 3-like |
| Mb | 59108 | 4:118101633–118108864 | −7.245234184 | 0 | Myoglobin |
| Myh2 | 691644 | 4:53711895–53738164 | −7.020133868 | 0 | Myosin heavy chain 2 |
| LOC108348161 | 108348161 | 4:52767635–52778693 | −6.986899856 | 1.22E-14 | Phytanoyl-CoA hydroxylase-interacting protein |
| LOC103694902 | 103694902 | 1:2148–29792 | −6.605321706 | 1.95E-196 | ubiquitin-conjugating enzyme E2 B |
| LOC103690173 | 103690173 | 1:3738–18580 | −6.475307726 | 1.07E-10 | Zinc finger protein 280B-like |
| LOC100359951 | 100359951 | 4:1012044–1012567 | −6.336444056 | 8.02E-10 | Ribosomal protein S20-like |
| LOC100909700 | 100909700 | 4:81609177–81627763 | −6.201682182 | 4.93E-09 | CD177 antigen-like |
| Txlnb | 308622 | 4:12915781–12961680 | −6.187131229 | 3.36E-34 | Taxilin beta |
| Cacng1 | 29658 | 4:95921479–95934170 | −6.167231672 | 4.38E-17 | Calcium voltage-gated channel auxiliary subunit gamma 1 |
| Myl1 | 56781 | 4:73937820–73958480 | −6.018352729 | 0 | Myosin, light chain 1 |
| Nmrk2 | 100912521 | 4:11382966–11386170 | −5.997306671 | 5.91E-08 | Nicotinamide riboside kinase 2 |
| LOC100361543 | 100361543 | 4:37261428–37262470 | −5.95800371 | 9.12E-08 | RhoA activator C11orf59-like |
| Myl2 | 363925 | 4:39947717–39966734 | −5.951502981 | 9.79E-08 | Myosin light chain 2 |
| Igfn1 | 304823 | 4:52791184–52823037 | −5.912190057 | 7.21E-85 | Immunoglobulin-like and fibronectin type III domain containing 1 |
| Tnnc2 | 296369 | 4:161243606–161246543 | −5.911770486 | 0 | Troponin C2, fast skeletal type |
| Ucp3 | 25708 | 4:165482912–165495895 | −5.87994972 | 3.84E-14 | Uncoupling protein 3 |
| LOC100911166 | 100911166 | 4:112519789–112524281 | −5.703837223 | 1.25E-06 | RNA pseudouridylate synthase domain-containing protein 2-like |
RNA-Seq Data Validation by Means of qPCR
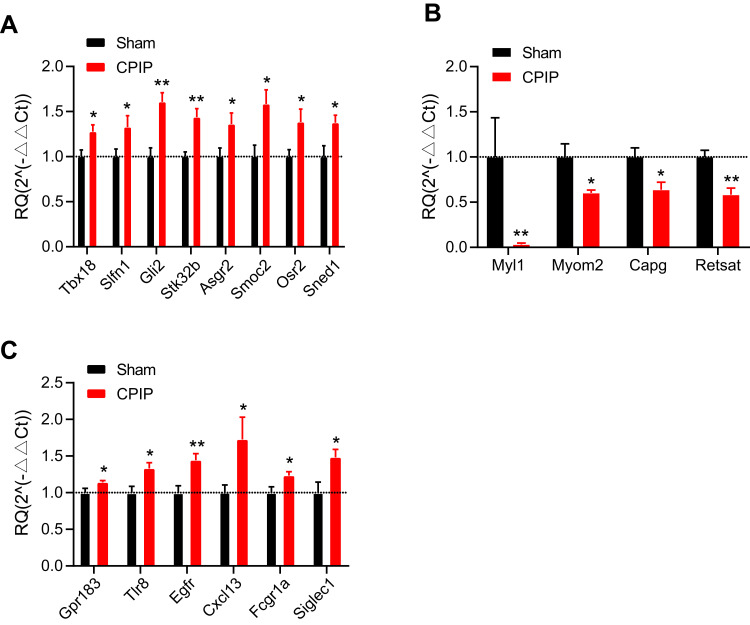
We then set to validate the RNA-Seq dataset by means of qPCR. We randomly selected eight up-regulated genes (Tbx18, Slfn1, Gli2, Stk32b, Asgr2, Smoc2, Osr2, and Sned1) and four down-regulated genes (Myl1, Myom2, Capg, and Retsat) from the DEGs list and verified their expression by qPCR. The results of qPCR showed that the expression of Tbx18, Slfn1, Gli2, Stk32b, Asgr2, Smoc2, Osr2, and Sned1 were all significantly up-regulated, whereas Myl1, Myom2, Capg, and Retsat were all significantly down-regulated in contralateral DRG of CPIP model rats vs sham rats (Figure 4A and B), which is consistent with the RNA-Seq dataset. We then proceeded to evaluate some representative genes which were known to contribute to pain or neuroinflammation. These genes include Gpr183, Tlr8, Egfr, Cxcl13, Fcgr1a, and Siglec1. qPCR testing showed that the expression of these genes were all significantly increased in CPIP model rats vs sham rats, which is consistent with the RNA-Seq result (Figure 4C). Therefore, these results indicated that the RNA-Seq dataset we obtained was reliable.
Figure 4.
Validating the RNA-Seq results by means of qPCR. (A and B) qPCR validation of the expression of some randomly selected up-regulated (A) and down-regulated (B) DEGs from contralateral DRG. (C) qPCR validation of the expression of some representative genes involved in pain and neuroinflammation. n=7 rats/group. *p<0.05, **p<0.01 vs sham group. Student’s t-test was used for comparisons.
Abbreviations: RNA-Seq, RNA-sequencing; DEGs, differentially expressed genes; DRG, dorsal root ganglia.
Function and Pathway Analysis of DEGs from Contralateral DRGs of CPIP Model Rats
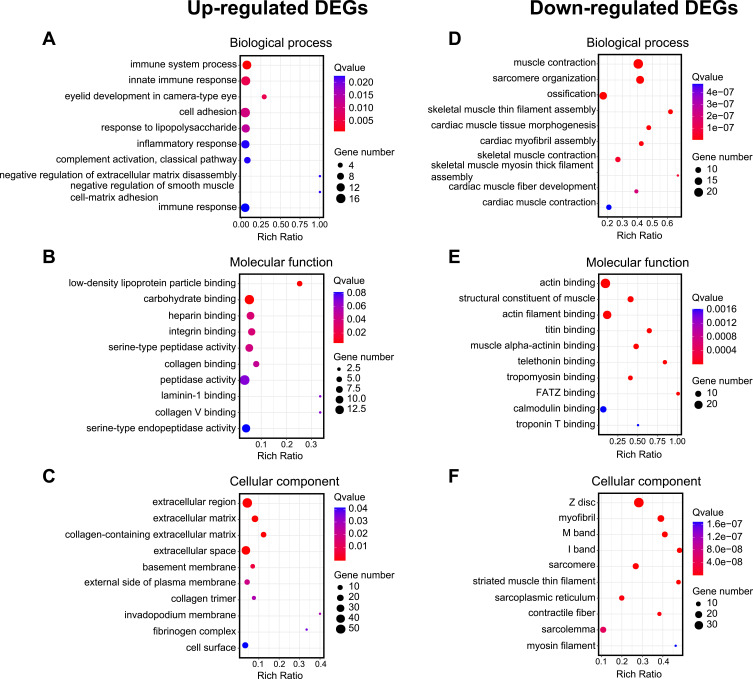
In order to gain more insights into the molecular mechanisms underlying MIP of CRPS-I, gene ontology (GO) enrichment analysis was performed to analyze the DEGs from contralateral DRGs of CPIP model rats vs sham rats. GO enrichment analysis indicated that the mostly enriched biological process among up-regulated DEGs included immune system process, innate immune response, and cell adhesion, etc. (Figure 5A). The mostly enriched molecular function among up-regulated DEGs included low-density lipoprotein particle binding, carbohydrate binding, and heparin binding, etc. (Figure 5B). The mostly enriched cellular function among up-regulated DEGs included the extracellular region and extracellular matrix, etc. (Figure 5C). On the other hand, the mostly enriched biological process among down-regulated DEGs included muscle contraction, sarcomere organization, and ossification, etc. (Figure 5D). The mostly enriched molecular function of down-regulated DEGs included actin binding, a structural constituent of muscle and actin filament binding, etc. (Figure 5E). The mostly enriched cellular function of down-regulated DEGs included Z disc, myofibril, and M band, etc. (Figure 5F). The DEGs allocated to each category of GO analysis are summarized in Suppl Table 6. These results indicated that immune response occurred in contralateral DRG and may possibly be related with MIP pathogenesis.
Figure 5.
GO enrichment analysis of identified DEGs from contralateral DRG. (A–C) Bubble plots showing the top 10 mostly enriched biological processes, molecular functions and cellular components of up-regulated DEGs. (D–F) Bubble plots showing the top 10 mostly enriched biological processes, molecular functions, and cellular components of down-regulated DEGs. Larger bubbles indicate higher number of genes and vice versa. The color of each bubble reflects the significance as indicated (q-value).
Abbreviations: GO, gene ontology; DEGs, differentially expressed genes; DRG, dorsal root ganglia.
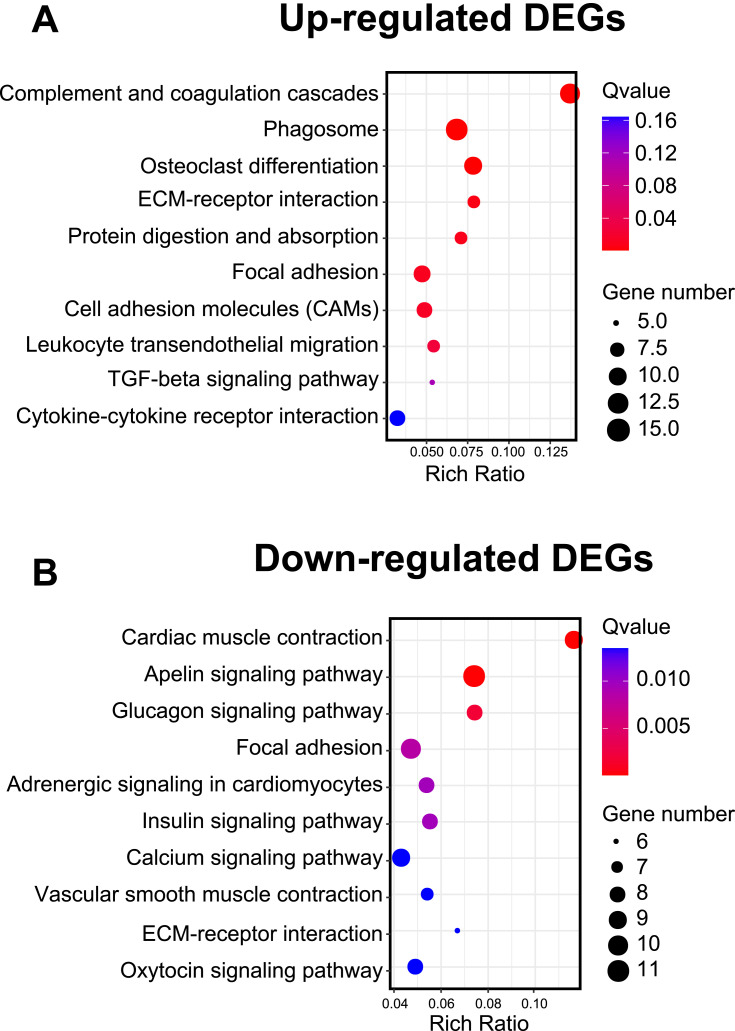
We then performed KEGG analysis to analyze the DEGs from contralateral DRG of the CPIP model rats. KEGG analysis indicated that the up-regulated DEGs were involved in complement and coagulation cascades, phagosome and osteoclast differentiation, etc. (Figure 6A). In contrast, the down-regulated DEGs were involved in cardiac muscle contraction, apelin signaling pathway and glucagon signaling pathway, etc. (Figure 6B, Suppl Table 7). We further performed gene set enrichment analysis (GSEA) of the up-regulated DEGs. Suppl Figure 2 lists the most significantly enriched pathways of these DEGs (NES>1.0, FDR≤0.25), including complement and coagulation cascades and Toll-like receptor signaling pathway, etc.
Figure 6.
KEGG pathway analysis of identified DEGs from contralateral DRG. (A) Bubble plots showing the top 10 pathways of up-regulated DEGs identified by KEGG. (B) Bubble plots showing the top 10 pathways of down-regulated DEGs identified by KEGG. Larger bubbles indicate higher number of genes and vice versa. The color of each bubble reflects significance as indicated (q-value).
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; DRG, dorsal root ganglia.
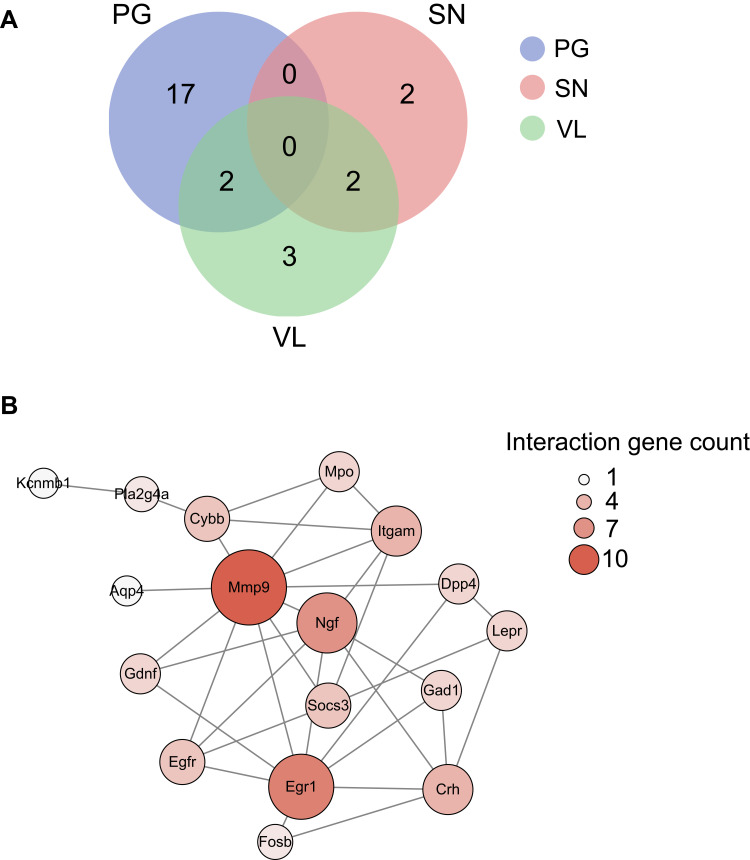
Next, we aim to focus our analysis more on the genes related with pain processing. We were able to retrieve three datasets that summarized genes enriched in small-diameter primary sensory DRG neurons (SN), TRPV1-lineage nociceptors (VL), and pain-related genes (PG), respectively.32 We then compared the DEGs we identified from RNA-Seq with these three datasets. We identify that 19 DEGs were related with PG and Stoml3, Fosb, and Gdnf are among the genes most up-regulated (Figure 7A, Suppl Table 8). We identified four DEGs were related with SN and Eno3 showed the most down-regulation (Figure 7A, Suppl Table 8). We identified seven genes were related with VL and Fosb showed the most up-regulation (Figure 7A, Suppl Table 8). PPI analysis was conducted on these 26 genes and the major hub genes identified from the network included Mmp9, Egr1, Ngf, etc. (Figure 7B). These data suggest that these specific DEGs related with pain, small-diameter primary sensory DRG neurons, and TRPV1-lineage nociceptors may probably contribute to MIP mechanisms.
Figure 7.
Analysis of DEGs potentially involved in pain mechanisms. (A) Overlapped DEGs with genes involved in pain processing (PG, in blue), genes enriched in TRPV1-lineage DRG neurons (VL, in green) and genes enriched in small-diameter DRG neurons (SN, pink) illustrated via Venn diagram. (B) The construction of PPI network of DEGs which showed overlapping with PG, VL, and SN gene pools. Deeper color and larger circle indicate more protein–protein interactions and vice versa.
Abbreviations: DEGs, differentially expressed genes; DRG, dorsal root ganglia; PPI, protein–protein interaction.
Discussion
In this study, we established the rat CPIP model to replicate CRPS-I. We found the model rats developed obvious MIP in contralateral hind paws. We then performed expression profiling of the DRG innervating the contralateral hind limb of the model rats by RNA-Seq. We identified the DEGs and validated the expression by means of qPCR. We then studied the major functions or signaling pathways that these DEGs were involved in. Lastly, by comparing with published datasets that summarized genes enriched in pain mechanisms, we further sorted out the genes which might contribute to nociception and pain processing. Thus, we provided by far the first study to profile gene expression changes and performed pathway analysis of contralateral DRG for investigating MIP mechanisms.
CRPS-I is a chronic and devastating pain condition that usually affects the extremities of the patients. One feature of CRPS-I patients is the symptom of MIP.7,12 CRPS-I patients may develop increased sensitivity to chemical, thermal, and mechanical stimuli in bilateral extremities.12 In order to facilitate mechanistic studies of CRPS-I, a rat chronic post-ischemic pain (CPIP) model was developed by applying prolonged hind paw ischemia and reperfusion to mimic CRPS-I.16 The rat CPIP models exhibit several key features that resemble CRPS-I, including early hind paw edema, vascular disturbance and blood flow dysfunction in the hind paw, persistent bilateral mechanical allodynia and heat hyperalgesia, increased sensitivity to capsaicin treatment, etc.15,16,22,37 At present, the CPIP model has become a commonly used animal model for mechanistic studies of CRPS-I. We and others found that the CPIP model rats develop robust MIP symptoms in the unaffected hind paw, including mechanical and thermal hypersensitivities.15–17 Therefore, in the present study, we established the rat CPIP model and made good use of it for a mechanistic study of MIP.
We first performed a pathological analysis to examine whether there was any neuronal damage which occurred in DRG innervating bilateral hind paws of CPIP model rats. Immunostaining showed that the expression of ATF3, a well-established neuronal damage marker, is not altered in ipsilateral or contralateral DRG of CPIP model rats, suggesting bilateral DRG neurons remain intact in CPIP model rats. In an earlier study, Coderre et al found no evidence of degeneration in bilateral tibial nerves of CPIP model rats.16 These findings are consistent with the clinical definition that CRPS-I patients have no known or identifiable peripheral nerve injury. Thus, this result excludes the possibility that neuronal damage may contribute to MIP in CPIP model rats.
Peripheral mechanisms have been proposed to contribute to MIP. For example, in a spinal nerve ligation (SNL) animal model for neuropathic pain, TNF-α can be produced from ipsilateral injured DRG and diffuses to the contralateral side via cerebrospinal fluid. TNF-α then activates satellite cells to produce NGF, which in turn promotes nociceptor excitability on the contralateral side and induces MIP.4 Although these findings suggest that peripheral satellite glial cells and pro-inflammatory cytokines are important for MIP, it still remains unclear how contralateral DRG is affected during MIP. Therefore, we performed a genome-wide expression profiling of the DRG that innervate the contralateral hind limbs of the rat CPIP model by means of RNA-Seq.
According to GO enrichment analyses of DEGs in contralateral DRG, we found that the mostly enriched biological processes of up-regulated genes were mainly involved in the immune system process, innate immune response, cell adhesion, response to lipopolysaccharide, inflammatory response, etc. This information indicates that immune and inflammatory responses, which can cause neuroinflammation and pain, take place in contralateral DRG. This finding is similar with our recent study which identified lipopolysaccharide and inflammatory response as the predominant biological processes in ipsilateral DRG of CPIP model rats.14 It is known that that neuroinflammation, including inflammatory cytokine/chemokine release, glial cell activation, and inflammatory cell infiltration in the sensory nerve system, make important contributions to the initiation and maintaining phase of chronic pain.38–41 Therefore, this result suggests that neuroinflammation in contralateral DRG may constitute an important mechanism for MIP of CPIP model rats.
In addition, KEGG pathway analysis indicated that one of the mostly enriched pathways that these DEGs participate in is “phagosome”. This finding is similar with our recent studies showing that phagosome is a major signaling pathway which occurred in ipsilateral DRG and spinal cord dorsal horn (SCDH) of CPIP model rats.14,17 Similar findings were also reported in SCDH of an animal model of neuropathic pain.42 Itgam is a well-established marker gene for monocytes and macrophages and is involved in the phagosome process. In this study, we found that Itgam gene expression is significantly up-regulated in contralateral DRG of CPIP model rats by RNA-Seq. This implies that macrophage or monocyte infiltration may likely take place in contralateral DRG of CPIP model rats. Macrophages or monocytes can infiltrate into peripheral tissues and sensory ganglia, where they release an array of inflammatory cytokines and reactive oxygen species (ROS) products.43–46 These products activate or sensitize peripheral sensory neurons to produce or enhance pain through neuro-immune crosstalk.39 Therefore, we speculate that macrophage or monocyte infiltration may occur in contralateral DRG, which could contribute to MIP in CPIP model rats. Further studies using specific markers for macrophages or monocytes will be needed to confirm their infiltrations into contralateral DRG.
In our study, the RNA-Seq identified that Ngf gene is significantly up-regulated in contralateral DRG of CPIP model rats. NGF is a neurotrophic factor and neuropeptide implicated in a wide range of development and maintenance functions. NGF can be released upon tissue inflammation and is implicated in chronic pain.47 NGF sensitizes nociceptors and promotes their excitability by up-regulating TRPV1 and Nav1.8 channel expression and activity via acting on the TrkA receptor.48,49 In a previous study, Cheng et al4 found that TNF-α produced from ipsilateral injured DRG of a rat spinal nerve ligation model can diffuse to the contralateral side via cerebrospinal fluid and activates satellite cells to produce NGF, which promotes nociceptor excitability on the contralateral side and induces MIP. Their study suggested that NGF expression is up-regulated in contralateral DRG and contributes to the MIP mechanism. In the present study, we found that the Ngf gene was significantly up-regulated in the contralateral DRG of CPIP model rats, a result consistent with Cheng et al’s findings. Thus, our result suggests that NGF may be potentially involved in mediating MIP of CPIP model rats.
We continued to explore other potential genes involved in MIP. By means of RNA-Seq and qPCR validation, we found that chemokine Cxcl13 gene expression showed significant up-regulation in contralateral DRG. CXCL13 is known as a B-cell-attracting chemokine that acts upon its receptor CXCR5 to exert biological functions. Recent studies identified that the CXCL13/CXCR5 axis is present in peripheral sensory ganglia and spinal cord and contributes to chronic pain.50–52 Genetic knockout or knockdown of CXCL13/CXCR5 significantly reduced chronic pain as well as glial cell over-activation in SCDH in animal models of neuropathic pain.52 Intrathecal CXCL13 injection can provoke thermal and mechanical hypersensitivities in naïve animals.52 Moreover, it is found that CXCl13 is increased in DRG neurons of CFA-induced inflammatory model mice and acts upon CXCR5 in DRG neurons to activate the p38-MAP kinase, which enhances Nav1.8 channel expression and contributes to chronic pain.51 Therefore, our findings suggest CXCL13 released from contralateral DRG may be another possible contributor to MIP of CPIP model rats.
Single-cell RNA-sequencing is a powerful technique that has greatly facilitated the identification of molecular profiles of heterogeneous cells, especially on the transcription level of single cells. Recently, two groups published single-cell RNA-sequencing results of DRG neurons.53,54 These studies provide novel insights into understanding the biological function of somatosensory neurons, particularly for the nociceptors, and suggest that neuron types are defined by their transcriptomic, morphological, and functional characteristics. We hope that single-cell RNA-sequencing may be applied in the MIP study in the future, which could provide detailed transcriptome profiles of single nociceptors in conditions of MIP.
In conclusion, to our knowledge, this is the first study that performs expression profiling of gene expression changes in contralateral DRG of an animal model of CRPS-I by RNA-Seq to explore MIP mechanisms. This study identifies a number of genes and signaling pathways in contralateral DRG that may possibly contribute to MIP. Thus, this study may provide novel insights into understanding the mysterious mechanisms underlying MIP.
Acknowledgment
We thank the Academy of Chinese Medicine Sciences, Zhejiang Chinese Medical University for the technical support to our work.
Funding Statement
This project was supported by the National Natural Science Foundation of China (81873365&81603676), Zhejiang Provincial Natural Science Funds for Distinguished Young Scholars (LR17H270001), Zhejiang Provincial Natural Science Funds (LQ21H270004), Research Projects from Zhejiang Chinese Medical University (Q2019J01, 2021JKZDZC07, KC201943), Natural Science Foundation of Hebei Province (H20206360), Projects from Health and Family Planning Commission of Hebei Province (20201069) and GuangDong Natural Science Foundation (2018A030313092).
Abbreviations
CRPS-I, Complex regional pain syndrome type-I; MIP, Mirror-image pain; CPIP, Chronic post-ischemic pain; RNA-Seq, RNA-sequencing; qPCR, Real-time quantitative PCR; DRG, Dorsal root ganglia; DEGs, Differentially expressed genes; PWT, Paw withdrawal threshold; QC, Quality control; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene set enrichment analysis; PG, Pain genes; SN, Sensory neuronal genes; VL, TRPV1-lineage neuronal genes; PPI, Protein–protein interaction; STRING, Search Tool for the Retrieval of Interacting Genes; BP, Biological processes; MF, Molecular function; CC, Cellular component; ROS, reactive oxygen species.
Data Sharing Statement
The key data are contained in the figures, tables, and additional files. The datasets used and/or analyzed during this study can be further obtained from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
All experimental procedures were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978) and approved by the Animal Ethics Committee of Zhejiang Chinese Medical University (Permission Number: ZSLL-2017-183).
Author Contributions
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest for this work.
References
- 1.Huang D, Yu B. The mirror-image pain: an unclered phenomenon and its possible mechanism. Neurosci Biobehav Rev. 2010;34:528–532. doi: 10.1016/j.neubiorev.2009.10.011 [DOI] [PubMed] [Google Scholar]
- 2.Milligan ED, Twining C, Chacur M, et al. Spinal glia and proinflammatory cytokines mediate mirror-image neuropathic pain in rats. J Neurosci. 2003;23:1026–1040. doi: 10.1523/JNEUROSCI.23-03-01026.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kambiz S, Brakkee EM, Duraku LS, Hovius SE, Ruigrok TJ, Walbeehm ET. Mirror-image pain after nerve reconstruction in rats is related to enhanced density of epidermal peptidergic nerve fibers. Exp Neurol. 2015;267:87–94. doi: 10.1016/j.expneurol.2015.02.033 [DOI] [PubMed] [Google Scholar]
- 4.Cheng CF, Cheng JK, Chen CY, et al. Mirror-image pain is mediated by nerve growth factor produced from tumor necrosis factor alpha-activated satellite glia after peripheral nerve injury. Pain. 2014;155:906–920. doi: 10.1016/j.pain.2014.01.010 [DOI] [PubMed] [Google Scholar]
- 5.Rotpenpian N, Tapechum S, Vattarakorn A, et al. Evolution of mirror-image pain in temporomandibular joint osteoarthritis mouse model. J Appl Oral Sci. 2021;29:e20200575. doi: 10.1590/1678-7757-2020-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hampf G, Bowsher D, Wells C, Miles J. Sensory and autonomic measurements in idiopathic trigeminal neuralgia before and after radiofrequency thermocoagulation: differentiation from some other causes of facial pain. Pain. 1990;40:241–248. doi: 10.1016/0304-3959(90)91121-X [DOI] [PubMed] [Google Scholar]
- 7.Maleki J, LeBel AA, Bennett GJ, Schwartzman RJ. Patterns of spread in complex regional pain syndrome, type I (reflex sympathetic dystrophy). Pain. 2000;88:259–266. doi: 10.1016/S0304-3959(00)00332-8 [DOI] [PubMed] [Google Scholar]
- 8.Ott S, Maihofner C. Signs and symptoms in 1043 patients with complex regional pain syndrome. J Pain. 2018;19:599–611. doi: 10.1016/j.jpain.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 9.Birklein F, Ajit SK, Goebel A, Perez R, Sommer C. Complex regional pain syndrome - phenotypic characteristics and potential biomarkers. Nat Rev Neurol. 2018;14:272–284. doi: 10.1038/nrneurol.2018.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coderre TJ, Bennett GJ. A hypothesis for the cause of complex regional pain syndrome-type I (reflex sympathetic dystrophy): pain due to deep-tissue microvascular pathology. Pain Med. 2010;11:1224–1238. doi: 10.1111/j.1526-4637.2010.00911.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urits I, Shen AH, Jones MR, Viswanath O, Kaye AD. Complex regional pain syndrome, current concepts and treatment options. Curr Pain Headache Rep. 2018;22:10. doi: 10.1007/s11916-018-0667-7 [DOI] [PubMed] [Google Scholar]
- 12.Terkelsen AJ, Gierthmuhlen J, Finnerup NB, Hojlund AP, Jensen TS. Bilateral hypersensitivity to capsaicin, thermal, and mechanical stimuli in unilateral complex regional pain syndrome. Anesthesiology. 2014;120:1225–1236. doi: 10.1097/ALN.0000000000000220 [DOI] [PubMed] [Google Scholar]
- 13.Veldman PH, Reynen HM, Arntz IE, Goris RJ. Signs and symptoms of reflex sympathetic dystrophy: prospective study of 829 patients. Lancet. 1993;342:1012–1016. doi: 10.1016/0140-6736(93)92877-V [DOI] [PubMed] [Google Scholar]
- 14.Yin C, Hu Q, Liu B, et al. Transcriptome profiling of dorsal root ganglia in a rat model of complex regional pain syndrome type-I reveals potential mechanisms involved in pain. J Pain Res. 2019;12:1201–1216. doi: 10.2147/JPR.S188758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Q, Wang Q, Wang C, et al. TRPV1 channel contributes to the behavioral hypersensitivity in a rat model of complex regional pain syndrome type 1. Front Pharmacol. 2019;10:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coderre TJ, Xanthos DN, Francis L, Bennett GJ. Chronic post-ischemia pain (CPIP): a novel animal model of complex regional pain syndrome-type I (CRPS-I; reflex sympathetic dystrophy) produced by prolonged hindpaw ischemia and reperfusion in the rat. Pain. 2004;112:94–105. doi: 10.1016/j.pain.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 17.Chen R, Yin C, Hu Q, et al. Expression profiling of spinal cord dorsal horn in a rat model of complex regional pain syndrome type-I uncovers potential mechanisms mediating pain and neuroinflammation responses. J Neuroinflammation. 2020;17:162. doi: 10.1186/s12974-020-01834-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Zheng X, Liu B, et al. Electroacupuncture alleviates mechanical allodynia of a rat model of CRPS-I and modulates gene expression profiles in dorsal root ganglia. Front Neurol. 2020;11:580997. doi: 10.3389/fneur.2020.580997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jancalek R. Signaling mechanisms in mirror image pain pathogenesis. Ann Neurosci. 2011;18:123–127. doi: 10.5214/ans.0972.7531.11183010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi HS, Roh DH, Yoon SY, et al. Microglial interleukin-1beta in the ipsilateral dorsal horn inhibits the development of mirror-image contralateral mechanical allodynia through astrocyte activation in a rat model of inflammatory pain. Pain. 2015;156:1046–1059. doi: 10.1097/j.pain.0000000000000148 [DOI] [PubMed] [Google Scholar]
- 21.Su YS, Mei HR, Wang CH, Sun WH. Peripheral 5-HT3 mediates mirror-image pain by a cross-talk with acid-sensing ion channel 3. Neuropharmacology. 2018;130:92–104. doi: 10.1016/j.neuropharm.2017.11.044 [DOI] [PubMed] [Google Scholar]
- 22.Hu Q, Zheng X, Chen R, et al. Chronic post-ischemia pain model for complex regional pain syndrome type-I in rats. J Vis Exp. 2020;155:e60562. doi: 10.3791/60562 [DOI] [PubMed] [Google Scholar]
- 23.Chai W, Tai Y, Shao X, et al. Electroacupuncture alleviates pain responses and inflammation in a rat model of acute gout arthritis. Evid Based Complement Alternat Med. 2018;2018:2598975. doi: 10.1155/2018/2598975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9 [DOI] [PubMed] [Google Scholar]
- 25.Dixon WJ. Efficient analysis of experimental observations. Annu Rev Pharmacol Toxicol. 1980;20:441–462. doi: 10.1146/annurev.pa.20.040180.002301 [DOI] [PubMed] [Google Scholar]
- 26.Hu Q, Zheng X, Li X, et al. Electroacupuncture alleviates mechanical allodynia in a rat model of complex regional pain syndrome type-I via suppressing spinal CXCL12/CXCR4 signaling. J Pain. 2020;21:1060–1074. doi: 10.1016/j.jpain.2020.01.007 [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Yin C, Liu B, et al. Transcriptome profiling of long noncoding RNAs and mRNAs in spinal cord of a rat model of paclitaxel-induced peripheral neuropathy identifies potential mechanisms mediating neuroinflammation and pain. J Neuroinflammation. 2021;18:48. doi: 10.1186/s12974-021-02098-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Ding H, Zhou X, Lin H, Chou KC. iRNA(m6A)-PseDNC: identifying N(6)-methyladenosine sites using pseudo dinucleotide composition. Anal Biochem. 2018;561–562:59–65. doi: 10.1016/j.ab.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin C, Liu B, Wang P, et al. Eucalyptol alleviates inflammation and pain responses in a mouse model of gout arthritis. Br J Pharmacol. 2020;177:2042–2057. doi: 10.1111/bph.14967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Chung MK, Park J, Asgar J, Ro JY. Transcriptome analysis of trigeminal ganglia following masseter muscle inflammation in rats. Mol Pain. 2016;12:1744806916668526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakur M, Crow M, Richards N, et al. Defining the nociceptor transcriptome. Front Mol Neurosci. 2014;7:87. doi: 10.3389/fnmol.2014.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goswami SC, Mishra SK, Maric D, et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J Pain. 2014;15:1338–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15. doi: 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Tao X, Huang P, et al. N-methyl-d-aspartate receptor subunit 2B on keratinocyte mediates peripheral and central sensitization in chronic post-ischemic pain in male rats. Brain Behav Immun. 2020;87:579–590. doi: 10.1016/j.bbi.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donnelly CR, Andriessen AS, Chen G, et al. Central nervous system targets: glial cell mechanisms in chronic pain. Neurotherapeutics. 2020;17:846–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda M, Huh Y, Ji RR. Roles of inflammation, neurogenic inflammation, and neuroinflammation in pain. J Anesth. 2019;33:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen R, Yin C, Fang J, Liu B. The NLRP3 inflammasome: an emerging therapeutic target for chronic pain. J Neuroinflammation. 2021;18:84. doi: 10.1186/s12974-021-02131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji RR, Chamessian A, Zhang YQ. Pain regulation by non-neuronal cells and inflammation. Science. 2016;354:572–577. doi: 10.1126/science.aaf8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du H, Shi J, Wang M, An S, Guo X, Wang Z. Analyses of gene expression profiles in the rat dorsal horn of the spinal cord using RNA sequencing in chronic constriction injury rats. J Neuroinflammation. 2018;15:280. doi: 10.1186/s12974-018-1316-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin C, Liu B, Li Y, et al. IL-33/ST2 induces neutrophil-dependent reactive oxygen species production and mediates gout pain. Theranostics. 2020;10:12189–12203. doi: 10.7150/thno.48028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trevisan G, Benemei S, Materazzi S, et al. TRPA1 mediates trigeminal neuropathic pain in mice downstream of monocytes/macrophages and oxidative stress. Brain. 2016;139:1361–1377. doi: 10.1093/brain/aww038 [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Xie W, Zhang J, et al. Key role of CCR2-expressing macrophages in a mouse model of low back pain and radiculopathy. Brain Behav Immun. 2021;91:556–567. doi: 10.1016/j.bbi.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Liu H, Hamel KA, et al. Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat Commun. 2020;11:264. doi: 10.1038/s41467-019-13839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denk F, Bennett DL, McMahon SB. Nerve growth factor and pain mechanisms. Annu Rev Neurosci. 2017;40:307–325. doi: 10.1146/annurev-neuro-072116-031121 [DOI] [PubMed] [Google Scholar]
- 48.Dib-Hajj SD, Black JA, Cummins TR, Kenney AM, Kocsis JD, Waxman SG. Rescue of alpha-SNS sodium channel expression in small dorsal root ganglion neurons after axotomy by nerve growth factor in vivo. J Neurophysiol. 1998;79:2668–2676. doi: 10.1152/jn.1998.79.5.2668 [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Huang J, McNaughton PA. NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. EMBO J. 2005;24:4211–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang BC, Liu T, Gao YJ. Chemokines in chronic pain: cellular and molecular mechanisms and therapeutic potential. Pharmacol Ther. 2020;212:107581. [DOI] [PubMed] [Google Scholar]
- 51.Wu XB, Cao DL, Zhang X, et al. CXCL13/CXCR5 enhances sodium channel Nav1.8 current density via p38 MAP kinase in primary sensory neurons following inflammatory pain. Sci Rep. 2016;6:34836. doi: 10.1038/srep34836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang BC, Cao DL, Zhang X, et al. CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J Clin Invest. 2016;126:745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li CL, Li KC, Wu D, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26:83–102. doi: 10.1038/cr.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, Ginty DD. Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron. 2019;103:598–616 e7. doi: 10.1016/j.neuron.2019.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]