Abstract
A high‐throughput, fully automated antigen detection test for SARS‐CoV‐2 is a viable alternative to reverse‐transcription polymerase chain reaction (RT‐qPCR) for mass screening during outbreaks. In this study, we compared RT‐qPCR for viral load and the VITROS® SARS‐CoV‐2 Antigen Test with reference to the results of the LUMIPULSE® SARS‐CoV‐2 Ag Test. Of 128 nasopharyngeal swab specimens taken from patients suspected of being infected with SARS‐CoV‐2, 49 were positive and 79 were negative according to RT‐qPCR. Consistent dose‐dependent detection with VITROS® assay was successfully achieved when using nasopharyngeal swab specimens with Ct values of 32.0 or lesser, whereas the CLEIA‐based LUMIPULSE® assay was able to detect lower viral loads compared with the VITROS® assay. Our results show that the performance of the VITROS® assay was satisfactory for the diagnosis of contagious COVID‐19 patients in the clinical setting.
Highlights The performance of the VITROS® SARS‐CoV‐2 Antigen Test was sufficient for the diagnosis of contagious COVID‐19. This test showed high sensitivity and specificity in the detection of SARS‐CoV‐2 in samples with a Ct value of 32 or less.
Keywords: CLEIA, COVID‐19, nasopharyngeal swab, RT‐qPCR, SARS‐CoV‐2
1. INTRODUCTION
Nucleic acid amplification (NAA) by reverse‐transcription polymerase chain reaction (RT‐qPCR) has been used to diagnose COVID‐19 and has the advantage of being able to monitor replication status based on viral load, but the testing procedure is complicated and relatively time‐consuming. In addition, a positive result based on NAA does not always allow for a definitive conclusion as to whether the patient is contagious. 1 Recently, the emergence of SARS‐CoV‐2 variants such as P.1 and B.1.351, which contain a mutation repertoire within the Spike gene, has raised the possibility of emerging reinfections. 2 , 3 , 4 Furthermore, a previous report demonstrated that RT‐qPCR can detect noninfectious SARS‐CoV‐2 RNA in nasopharyngeal (Np) swab specimens in recovered adult patients up to 3 months after onset. 5 To overcome these problems, high‐throughput diagnostic technology designed to detect active SARS‐CoV‐2 infection has become essential.
Recently, the VITROS® SARS‐CoV‐2 Antigen Test (Ortho Clinical Diagnostics) was developed as a new diagnostic technology based on a two‐step chemiluminescent enzyme immunoassay (CLEIA) and can be used to perform up to 130 tests per hour for the detection of active SARS‐CoV‐2 infection. This is a fully automated antigen detection test designed to detect active viral infection and is the first high‐throughput COVID‐19 antigen test to receive Emergency Use Authorization (EUA) from the US Food and Drug Administration (FDA). Recently, Favresse et al. 6 demonstrated that the VITROS® SARS‐CoV‐2 Antigen Test completely aligned with RT‐qPCR for Ct values up to 33 and found that the test had high specificity (100%). However, evaluation in the clinical setting remains limited, so further studies are needed to confirm the real‐world effectiveness of this technology.
In this study, we compared RT‐qPCR in terms of viral load with two rapid antigen tests, namely, the VITROS® SARS‐CoV‐2 Antigen Test and the CLEIA‐based LUMIPULSE® SARS‐CoV‐2 Ag Test (Fujirebio), to investigate their performance in the analysis of clinical Np swab specimens. 7
2. MATERIALS AND METHODS
Np swab specimens were collected from patients suspected of being infected with SARS‐CoV‐2 at Saitama Medical University Hospital, Saitama, Japan, between December 22, 2020, and January 7, 2021. Each specimen was well suspended in 1000 μl of phosphate‐buffered saline (PBS) and used for the following analysis of COVID‐19. Asymptomatic carriers were defined as confirmed COVID‐19 patients with no history of clinical signs or symptoms on admission. The day of onset was defined as the first day of symptoms caused by COVID‐19 in symptomatic patients or the day of the first positive RT‐qPCR result using Np swab specimens in asymptomatic carriers.
The definitive diagnosis of COVID‐19 was confirmed by RT‐qPCR according to the nationally recommended protocol, using RNA extracted from 140 μl of the abovementioned suspension. 8 Briefly, RNA extraction was performed with the QIAamp Viral RNA Mini kit (Qiagen) and RNA was finally eluted with 60 μl of Buffer AVL according to the manufacturer's instructions. Conventional RT‐qPCR for specific amplification of the N2 gene of SARS‐CoV‐2 was performed using TaqMan‐based real‐time PCR with the following sets of primers and probe (2.4 μM of forward primer, 5′‐AAATTTTGGGGACCAGGAAC‐3′; 3.2 μM of reverse primer, 5′‐TGGCAGCTGTGTAGGTCAAC‐3′; 0.4 μM of probe; 5′‐FAM‐ATGTCGCGCATTGGCATGGA‐BHQ‐3′). RT‐qPCR amplification was performed using the QuantiTect Probe RT‐PCR Kit (Qiagen). Briefly, 5 μl of the extracted RNA was added to the amplification mixture, and distilled water was added to a final volume of 25 μl. Amplification was performed under the following conditions: Reverse transcription at 50°C for 30 min; initial denaturation at 95°C for 15 min; and 40 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 60 s. Positive RNA controls were prepared in 10‐fold serial dilutions ranging from 5.0 × 105 to 5.0 × 100 copies/reaction using Vitro synthesized SARS‐CoV‐2 RNA, which was kindly provided by the National Institute of Infectious Diseases, Japan. A calibration assay was carried out in parallel to create a calibration curve by RT‐qPCR. 8
The VITROS® SARS‐CoV‐2 Antigen Test was also performed using the above Np swab specimens suspended in PBS. Briefly, 200 μl of the PBS suspension was added to 50 μl of the included VITROS® SARS‐CoC‐2 antigen sample treatment solution. Then, the assay was automatically performed with the VITROS® 3600 automated immunoassay analyzer (Ortho Clinical Diagnostics). The analytical results were reported as signal/cutoff (S/C) values, where 1.0 or greater was defined as a positive test result and less than 1.0 as negative.
The LUMIPULSE® SASR‐CoV‐2 Ag Test was performed with the LUMIPULSE® G1200 (Fujirebio) according to the manufacturer's instructions. Briefly, each Np swab specimen was centrifuged at 20 000g for 5 min, and the supernatant was used for subsequent analysis. The amount of SARS‐CoV‐2 antigen was determined based on the quantitative intensity of the reaction signal, where an antigen level of 1.34 pg/ml or above was defined as a positive test result and below 1.34 pg/ml as negative. 7
3. RESULTS
A total of 128 Np swab specimens were collected from patients suspected of being infected with SARS‐CoV‐2. Of the 128 Np swab specimens, 49 (38.3%) were found to be positive by RT‐qPCR, and the median Ct was 30.0 (interquartile range [IQR]: 24.0–35.0). The median age of patients diagnosed with COVID‐19 was 58 years (IQR: 47–74) and 23 of these patients (46.9%) were women.
In the VITROS® SARS‐CoV‐2 Antigen Test, all RT‐qPCR‐negative samples (n = 79) gave negative test results (100% specificity), whereas antigens were detected in 37 of 49 RT‐qPCR‐positive samples (75.5% sensitivity). The median Ct value of 12 RT‐qPCR positive and VITROS® SARS‐CoV‐2 Antigen Test negative results was 36.0 (IQR: 34.7–37.3). Therefore, the overall concordance with RT‐qPCR was 90.6% (116/128). In the LUMIPULSE® SASR‐CoV‐2 Ag Test, the analysis revealed antigen reactivity in 1 of 79 RT‐qPCR‐negative samples (98.7% specificity), whereas antigens were detected in 43 of 49 RT‐qPCR‐positive samples (87.8% sensitivity). The median Ct value of six RT‐qPCR positive and LUMIPULSE® SASR‐CoV‐2 Ag Test negative results was 37.1 (IQR: 36.8–37.5). Therefore, the overall concordance with RT‐qPCR was also 94.5% (121/128).
Figure 1A,B shows the correlations between the Ct value obtained by RT‐qPCR and the amounts of SARS‐CoV‐2 antigens determined by the VITROS® SARS‐CoV‐2 Antigen Test and LUMIPULSE® SASR‐CoV‐2 Ag Test, respectively. Consistent dose‐dependent detection was successfully achieved when Np swab specimens with Ct values of 32.0 or lesser were used. All six samples that were positive according to the LUMIPULSE® SASR‐CoV‐2 Ag Test and negative according to the VITROS® SARS‐CoV‐2 Antigen Test had a Ct value of over 32.0. Table 1 summarizes the antigen detection rates for each quantified viral load from Np swab specimens according to the two tests. The detection rate was 85.5% (34/40) for 50.0 copies/reaction or over, 96.8% (30/31) for 250.0 copies/reaction or over, and 100% (29/29) for 500.0 copies/reaction or over in the VITROS® SARS‐CoV‐2 Antigen Test. The detection rate was 97.5% (39/40) for 50.0 copies/reaction or over, 100% (31/31) for 250.0 copies/reaction or over, and 100% (29/29) for 500.0 copies/reaction or over in the LUMIPULSE® SASR‐CoV‐2 Ag Test.
Figure 1.
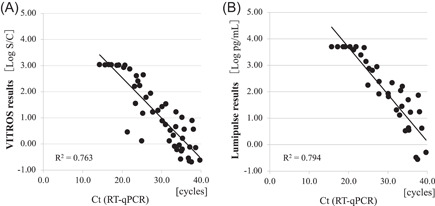
Correlations between Ct values obtained with reverse‐transcription polymerase chain reaction (RT‐qPCR) and antigen levels obtained by each automated antigen‐detection test. The antigen level in (A) the VITROS® SARS‐CoV‐2 Antigen Test and (B) the LUMIPULSE® SASR‐CoV‐2 Ag Test were plotted relative to RT‐qPCR Ct value. The diagonal line shows the cut‐off value
Table 1.
Positivity rate of SARS‐CoV‐2 antigen by viral loads in nasopharyngeal swab specimens
| Viral load (n) (copies/reaction) | The VITROS® SARS‐CoV‐2 Antigen Test | The LUMIPULSE® SASR‐CoV‐2 Ag Test | p Value | |||
|---|---|---|---|---|---|---|
| Positive samples | Detection rate (%) | Positive samples | Detection rate (%) | |||
| Total | n = 49 | n = 37 | 75.5 | n = 43 | 87.8 | 0.12 |
| ≥50 | n = 40 | n = 34 | 85.0 | n = 39 | 97.5 | 0.05 |
| ≥250 | n = 31 | n = 30 | 96.8 | n = 31 | 100 | 0.32 |
| ≥500 | n = 29 | n = 29 | 100 | n = 29 | 100 | NS |
Abbreviation: NS, not significant.
To assess the sensitivity of these assays according to the number of days after onset, 49 COVID‐19 patients were divided into two groups: 9 days or under after onset and over 10 days after onset. The positivity rate of each period wwas analyzed and the results are shown in Table 2.
Table 2.
Positivity rates of SARS‐CoV‐2 antigen by the number of days after onset
| Days after onset | p Value | ||
|---|---|---|---|
| ≤9 days (n = 35) | ≥10 days (n = 14) | ||
| VITROS® SARS‐CoV‐2 Antigen Test | 85.7% (30/35) | 50.0% (7/14) | 0.02 |
| LUMIPULSE® SASR‐CoV‐2 Ag Test | 97.1% (34/35) | 64.3% (9/14) | 0.01 |
4. DISCUSSION
The VITROS® SARS‐CoV‐2 Antigen Test can detect SASR‐CoV‐2 within 48 min and is capable of processing up to 130 Np swab specimens per hour. In this study, we demonstrated that the sensitivity of the VITROS® SARS‐CoV‐2 Antigen Test was 100% in samples with Ct values below 32.0. It should be noted that, unlike the LUMIPULSE® SASR‐CoV‐2 Ag Test, the VITROS® SARS‐CoV‐2 Antigen Test exhibits high specificity without the need for additional pretreatment procedures such as high‐speed centrifugation. The VITROS® SARS‐CoV‐2 Antigen Test realizes high‐throughput and rapid testing, does not require skilled technicians or multistep procedures, and can be performed using equipment already installed in many laboratories. Thus, this test is a viable alternative to RT‐qPCR and is suitable for mass screening during outbreaks.
Previous reports demonstrated that a “positive” NAA result including RT‐qPCR reflects only the detection of viral RNA fragments and does not always indicate the presence of viable virus particles. 9 This is a clinically important point. In a larger cohort, Singanayagam et al. 10 reported that 8% of samples with Ct values above 35.0 were positive for virus culture. In addition, previous reports have also demonstrated that high Ct levels were associated with noninfectious SARS‐CoV‐2. 9 , 11 In our study, the VITROS® SARS‐CoV‐2 Antigen Test exhibited 100% sensitivity in Np swab specimens with a viral load above 500 copies/reaction or Ct values below 32.0, as reported previously. 6 However, the CLEIA‐based LUMIPULSE®◻ SASR‐CoV‐2 Ag Test was able to detect lower viral loads compared with the VITROS® SARS‐CoV‐2 Antigen Test, but there was no statistically significant difference between the two tests (Table 2). These results indicate that the VITROS® SARS‐CoV‐2 Antigen Test may be effective in detecting viable virus particles and thus may be useful for selecting contagious COVID‐19 patients for infection control. However, it is still necessary to pay attention to false‐negative results when using the VITROS® SARS‐CoV‐2 Antigen Test because different RT‐qPCR assays may yield different Ct values with the same RNA load.
In this study, the diagnostic agreement between RT‐qPCR and both tests was significantly higher in samples collected in the early phase after symptom onset (9 days or under) than in the late phase (over 10 days). Previous studies have demonstrated that no live virus is isolated from culture 9 days after symptom onset despite the prolonged RNA shedding detected with Np swab specimens. 12 , 13 Of the five discordant samples that were positive for RT‐qPCR but negative for the VITROS® SARS‐CoV‐2 Antigen Test in the present study, all samples had Ct values above 33.0 and appeared to be in a noncontagious recovery period with low viral loads.
In clinical practice, detecting the shedding of infectious live viruses is not only related to the diagnosis of COVID‐19 but also to infection control in hospitals, including the decision on when to discharge patients. To summarize, our results show that the performance of the VITROS® SARS‐CoV‐2 Antigen Test was satisfactory for the diagnosis of contagious COVID‐19 patients in the clinical setting. The test exhibited high sensitivity and specificity in the detection of SARS‐CoV‐2 in samples with a Ct value of ≤32 without the need for additional pretreatment procedures.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ETHICAL APPROVAL
The study design and protocol were reviewed and approved by the Institutional Review Board of Saitama Medical University Hospital (Approval No. 20153.01).
AUTHOR CONTRIBUTIONS
Nanako Matsuzaki, Masaru Matsuoka, Shinichi Takeuchi, Kazuo Imai, Norihito Tarumoto, Shigefumi Maesaki, and Takuya Maeda conceived and designed the study. Nanako Matsuzaki, Yuta Orihara, Masahiro Kodana, and Rieko Kawamura performed molecular analysis. Yutaro Kitagawa and Masaru Matsuoka performed automatic immunoassay. All authors contributed to the interpretation of data and to drafting sections of the manuscript. All the authors approved the final manuscript.
Matsuzaki N, Orihara Y, Kodana M, et al. Evaluation of a chemiluminescent enzyme immunoassay‐based high‐throughput SARS‐CoV‐2 antigen assay for the diagnosis of COVID‐19: the VITROS® SARS‐CoV‐2 Antigen Test. J Med Virol. 2021;93:6778‐6781. 10.1002/jmv.27153
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Lanser L, Bellmann‐Weiler R, Öttl KW, et al. Evaluating the clinical utility and sensitivity of SARS‐CoV‐2 antigen testing in relation to RT‐PCR Ct values. Infection. 2020;13:1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome‐related coronavirus 2 (SARS‐CoV‐2) lineage with multiple spike mutations in South Africa. medRxiv. 2020. 10.1101/2020.12.21.20248640 [DOI] [Google Scholar]
- 3. Rani PR, Imran M, Lakshmi JV. Symptomatic reinfection of SARS‐CoV‐2 with spike protein variant N440K associated with immune escape. J Med Virol. 2021;93(7):4163‐4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramírez JD, Muñoz M, Ballesteros N, et al. Phylogenomic evidence of reinfection and persistence of SARS‐CoV‐2: first report from Colombia. Vaccines. 2021;9(3):282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention . Duration of isolation and precautions for adults with COVID‐19. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html. Accessed June 29, 2021
- 6. Favresse J, Gillot C, Oliveira M, et al. Head‐to‐head comparison of rapid and automated antigen detection tests for the diagnosis of SARS‐CoV‐2 infection. J Clin Med. 2021;10(2):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hirotsu Y, Maejima M, Shibusawa M, et al. Comparison of automated SARS‐CoV‐2 antigen test for COVID‐19 infection with quantitative RT‐PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shirato K, Nao N, Katano H, et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV‐2019) in Japan. Jpn J Infect Dis. 2020;73:304‐307. [DOI] [PubMed] [Google Scholar]
- 9. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. [DOI] [PubMed] [Google Scholar]
- 10. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT‐PCR cycle threshold values in cases of COVID‐19, England, January to May 2020. Euro Surveill. 2020;25:2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS‐CoV‐2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta‐analysis. Lancet Microbe. 2021;2(1):e13‐e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spinner CD, Gottlieb RL, Criner GJ, et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID‐19: a randomized clinical trial. JAMA. 2020;324(11):1048‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


