Abstract
Favipiravir is a broad‐spectrum inhibitor of viral RNA‐dependent RNA polymerase (RdRp) currently being used to manage COVID‐19. Accumulation of mutations in severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RdRp may facilitate antigenic drift, generating favipiravir resistance. Focussing on the chain‐termination mechanism utilized by favipiravir, we used high‐throughput interface‐based protein design to generate > 100 000 designs of the favipiravir‐binding site of RdRp and identify mutational hotspots. We identified several single‐point mutants and designs having a sequence identity of 97%–98% with wild‐type RdRp, suggesting that SARS‐CoV‐2 can develop favipiravir resistance with few mutations. Out of 134 mutations documented in the CoV‐GLUE database, 63 specific mutations were already predicted as resistant in our calculations, thus attaining ˜ 47% correlation with the sequencing data. These findings improve our understanding of the potential signatures of adaptation in SARS‐CoV‐2 against favipiravir.
Keywords: drug resistance, favipiravir, fitness, nsp12, protein design, RNA‐dependent RNA polymerase, SARS‐CoV‐2
Abbreviations
COVID‐19, coronavirus disease
EUA, emergency use authorization
nsp12, nonstructural protein 12
RdRp, RNA‐dependent RNA polymerase
RMSD, root mean square fluctuation
SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
SNP, single nucleotide polymorphism
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RNA‐dependent RNA polymerase (RdRp or nonstructural protein 12 (nsp12)‐nsp7‐nsp8 complex) has emerged as a critical drug target for treating coronavirus disease (COVID‐19). It is an ideal target because it is the central enzyme involved in the virus replication and transcription, and its enzymatic counterpart is absent in host cells. The recent detailed characterization of structural and biochemical properties of SARS‐CoV‐2 RdRp has allowed the design and development of specific RdRp inhibitors [1, 2]. The major drug approved for treating hospitalized patients with COVID‐19 is the RdRp inhibitor remdesivir, which received emergency use authorization (EUA) in several countries [3]. RdRps are highly conserved enzymes across different coronaviruses, with ˜ 96% sequence identity between SARS‐CoV and SARS‐CoV‐2 [4]. All coronavirus RdRps have conserved three‐dimensional (3D) right‐handed structure, being organized into palm, thumb and finger subdomains, as well as show conserved active site motifs. These features can be used to develop broad‐spectrum inhibitors for these and other RNA viruses.
Favipiravir (6‐fluoro‐3‐oxo‐3,4‐dihydropyrazine‐2‐carboxamide) is a broad‐spectrum drug originally developed to act against the influenza virus. In addition, it is effective against a wide variety of RNA viruses, including flaviviruses, hantaviruses, arenaviruses, phleboviruses, noroviruses, enteroviruses, foot‐and‐mouth disease virus, Western equine encephalitis virus, Ebola virus, rabies and respiratory syncytial virus [5, 6]. For COVID‐19, oral therapy with favipiravir has been recommended in several countries, such as Russia, India, Japan, Saudi Arabia, Thailand and Kenya, and has been proven useful in managing mild‐to‐moderate cases of COVID‐19 [7, 8, 9]. Furthermore, more than 40 clinical trials of favipiravir are in progress in various countries, including Japan, Italy, UK, USA, Canada, France, Germany, India, Thailand, Russia, Egypt and Saudi Arabia, to treat patients with COVID‐19 (https://clinicaltrials.gov). Favipiravir treatment could rapidly clear viral load and induce faster clinical improvement. In India currently, it is a major antiviral presently used for the clinical management and treatment of COVID‐19 [10, 11]. Favipiravir’s mode of action is suggested to have a mix of both chain terminator and mutator events. In the first case, favipiravir (prodrug) acts as a nucleoside analog that is metabolically activated via ribosylation and phosphorylation and is converted into favipiravir ribofuranosyl‐5B‐triphosphate (favipiravir‐RTP). It is then incorporated into the nascent viral RNA by viral RdRp resulting in chain termination or accumulation of deleterious mutations [12, 13, 14]. Favipiravir binds to the active site of the viral RdRp, being mistaken as a purine nucleotide, transforming the binding pocket into a catalytically nonproductive conformation. It stacks onto the 3′ nucleotide of the primer strand and forms a noncanonical base pair with the template RNA strand using its amide group (Fig. 1A–C). Therefore, favipiravir functions as a chain terminator, inhibiting viral RdRp by the termination of chain elongation at the site of incorporation. In the second mode of action (mutator), favipiravir exerts its antiviral effect through lethal mutagenesis, where its insertion into the viral RNA induces C‐to‐U and G‐to‐A conversion in the SARS‐CoV‐2 genome [15]. Recently, a cryo‐EM structure of the SARS‐CoV‐2 RdRp in complex with favipiravir‐RTP and template primer double‐stranded RNA has been determined (PDB ID: 7AAP), and this allowed us to execute this study with a focus on the chain‐termination mechanism of favipiravir [16].
Fig. 1.
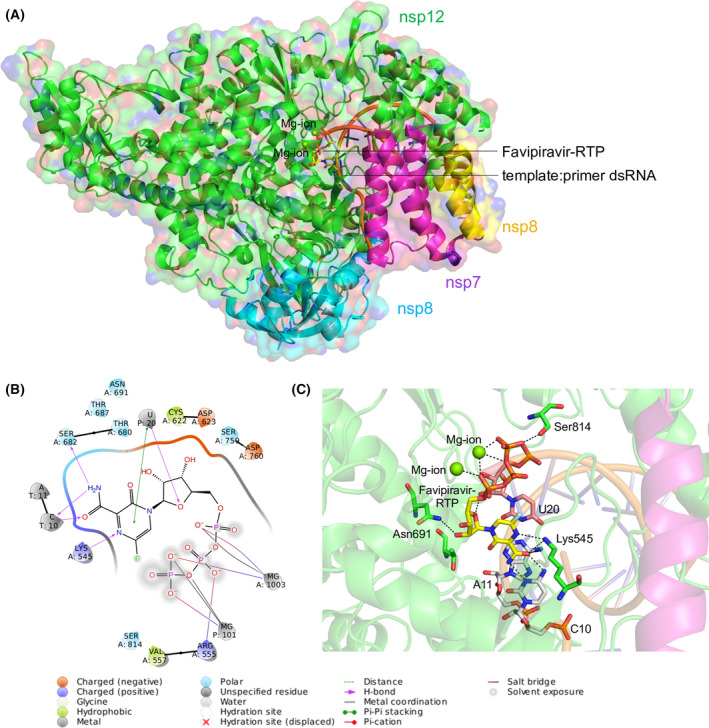
Structure of the SARS‐CoV‐2 RdRp complex. (A) The cryo‐EM structure of SARS‐CoV‐2 RdRp in complex with the template:primer dsRNA and favipiravir‐RTP. The nsp7, nsp8 and nsp12 domains are labelled with respective colours, and catalytic Mg2+ ion is shown. The template:primer dsRNA and favipiravir‐RTP are shown as a cartoon and stick model respectively. (B) Ligand interaction diagram of favipiravir‐RTP bound to the template:primer dsRNA. (C) Intermolecular interactions and interacting residues of nsp12 and RNA bases of favipiravir‐RTP (yellow) and Mg2+ (green sphere) coordinating the favipiravir‐RTP. Hydrogen bonds are shown as black dashes.
Since its emergence in December 2019, SARS‐CoV‐2 has undergone more than 50 000 mutations and recombination events, as compared to the Wuhan reference genome (NC_045512.2; http://cov‐glue.cvr.gla.ac.uk/#/home). Until December 2020, at least 198 sites in the viral genome have already undergone recurrent, independent mutations [17]. These 198 sites were associated with changes in 290 amino acids, with 232 composed of nonsynonymous and 58 of synonymous mutations. Although the rate is lower than that of influenza and human immunodeficiency virus (HIV), SARS‐CoV‐2 is presently accumulating at least two mutations per month in its genome [18]. The continuously emerging mutations are a cause of concern, as they could hinder the development of long‐lasting and effective therapeutics. These concerns are supported by the observations of the natural favipiravir resistance emergence in Chikungunya virus [19] and enterovirus 71 [20], and development of the favipiravir resistance by the pandemic H1N1 influenza A virus in laboratory studies [21]. Chances of the emergence of favipiravir‐resistant SARS‐CoV‐2 strains increase due to its intensive use as the COVID‐19 pandemic progresses. We recently identified potential hotspot residues that could contribute to remdesivir and molnupiravir resistance in SARS‐CoV‐2, as well as identified potential signatures of adaptation in SARS‐CoV‐2 main protease against boceprevir and telaprevir [22, 23]. We believe the knowledge of such mutants is crucial in the design of new and more effective drugs that can guide clinicians to properly manage and administer the treatment regimen. This study used a high‐throughput interface design strategy to predict potential mutation hotspots and resistance sites in the favipiravir‐binding site in the RdRp of SARS‐CoV‐2. The work can help develop exhaustive strategies for robust antiviral design and discovery.
Methods
Identification of favipiravir‐interacting residues in the nsp12
The cryo‐EM structure of SARS‐CoV‐2 RdRp (nsp7‐nsp8‐nsp12) in complex with the template:primer dsRNA and favipiravir‐RTP (PDB code: 7AAP) was used to determine the interactions between nsp12 and favipiravir. This analysis identified 72 nsp12 residues interacting with favipiravir. The complex structure was subsequently processed using the Molecular Operating Environment (MOE 2018.01, v2021.03).
MOE‐based resistance scan design methodology
To identify potential single‐point mutations of nsp12 that could develop resistance against favipiravir, MOE’s resistance scan methodology was used (MOE 2018.01, v2021.03) [24]. The prepared complex structure was used as an input, and 41 nsp12 residues that interacted with favipiravir were designed with naturally sampled SNPs, whereas the remaining residues that interacted with RNA were not selected for the design. During the design, the template:primer dsRNA was retained in the complex structure. Due to the flexibility of the designed site, the rotamer explorer option of the ensemble protein design protocol was enabled. During this stage, an root mean square fluctuation (RMSD) cut‐off of 0.5 Å was used, and the energy window, conformation limit and fix residues, farther than, were set to default values of 10 kcal·mol−1, 25 K and 4.5 Å respectively. Subsequently, a total of 350 designs were generated for in‐depth analysis. The affinity and stability of the designed complexes were examined. The relative binding affinity of the mutated proteins with respect to the wild‐type protein (dAffinity) was calculated, where dAffinity represents the Boltzmann average of the relative affinities of the ensemble. This method offered an indirect computational fitness test model, sampling only nondeleterious mutations that were more likely to arise naturally.
Rosetta‐based structural refinement and ligand‐based interface design
The complex structure prepared using MOE was next used for Rosetta ligand‐based interface design [25, 26, 27, 28]. First, parameters for favipiravir compatible with the Rosetta force field were generated by adding OpenEye's AM1‐BCC charges and following Rosetta’s parameter generation protocol. The parameter file contains definitions of ligand topology, atom types, partial charges, rotatable bonds etc., compatible with Rosetta design experiments. Second, the complex was energy minimized and refined using Rosetta relax [26]. During this step, several structures were generated with a goal to obtain the lowest energy structure; however, as the energy differences were not significantly distinct among the generated structures, the lowest energy complex structure of the favipiravir‐bound complex was eventually selected for the targeted interface design experiment. The Rosetta macromolecular modelling suite was used to redesign nsp12 with backbone flexibility [25]. In this step, 41 residues of nsp12 that constituted the favipiravir‐interacting site were designed with the naturally sampled SNPs, whereas the remaining residues that interact with RNA and all other residues of nsp7, nsp8 and nsp12 were allowed to only repack without design. In the design experiments, a modified RosettaScript was used to sample the favipiravir‐interacting residues of nsp12, which considered minor movements of the backbone of nsp12 to prevent steric clashes with ligands after introducing mutations [27]. The Rosetta all‐atom force field with Monte‐Carlo‐simulated annealing was used to sample the designs. A total of 100 000 designs of favipiravir‐bound complexes were generated, and the Rosetta total score, RMSD from the initial structure, Rosetta interface delta (denoting the binding affinities between the designed nsp12 and favipiravir) and the per cent sequence identities from the initial nsp12 sequence were analysed to understand the mutational landscapes and effect of mutations on the physicochemical characteristics of nsp12 and the overall complex.
Validation of the design protocol
The accuracy and predicting ability of our MOE‐based and Rosetta‐based interface design protocol were confirmed on a well‐studied remdesivir‐resistant nsp12 mutant, named V557L of SARS‐CoV [29]. As described in our recent study [23], first, we used the MOE resistance scan methodology, where Val557 was designed with A, D, E, G, I, L, M and F, and the affinities between designed nsp12 and remdesivir were calculated. Second, we used the Rosetta interface design protocol, where Val557 was designed with naturally sampled SNP residues, although the remaining nsp12 residues were only repacked without design. In this run, 5000 designs were generated and analysed for the designed mutations with their respective total scores. Finally, the resistance mutations from our MOE‐generated designs were compared with the known SARS‐CoV‐2 nsp12 sequences available in the CoV‐GLUE database [30]. The frequency of the mutations retrieved from CoV‐GLUE was compared with the designs in the favipiravir‐binding site of nsp12 to determine the accuracy of our design methodology.
Mutational landscapes of the top‐ranked affinity‐attenuating and affinity‐enhancing designs
WebLogo was used to obtain and plot the type and frequency of designed amino acid residues in the 41 favipiravir‐interacting residues of nsp12 from the top‐ranked affinity‐enhanced and affinity‐attenuated designs [31].
Calculation of intermolecular interactions between favipiravir‐bound nsp12 designs
The intermolecular interactions between top‐ranked affinity‐attenuating and affinity‐enhancing favipiravir‐nsp12 designs were identified using the Arpeggio web server [32]. The total number of interactions obtained was cumulative of van der Waals interactions, hydrogen bonds, proximal contacts, polar contacts, hydrophobic contacts, aromatic contacts and carbonyl interactions. The interaction figures were created using Schrödinger Maestro (Schrödinger Release 2016–4: Maestro, Schrödinger, New York, NY, USA).
Normal mode analysis of native nsp12 and certain affinity‐attenuating designs
The normal mode analysis of the nsp12 subunit of RdRp was carried out by (a) computing the atomic displacement of each C⍺ atom for the first six lowest‐frequency nontrivial modes and (b) obtaining the correlation matrix of nsp12 using the WEBnm@ server [33, 34]. Next, an analysis of the protein dynamics was carried out by (c) computing the normalized deformation energies and (4) squared atomic fluctuations for certain affinity‐attenuating designs by considering the native nsp12 as control.
Results
Analysis of nsp12‐favipiravir‐interacting residues and selection of nsp12 residues for designing
The cryo‐EM structure of SARS‐CoV‐2 RdRp in complex with favipiravir‐RTP revealed the interacting residues between nsp12 and favipiravir (Fig. 1A) [35]. A total of 72 residues of nsp12 were found to be interacting with favipiravir, of which those that interacted with the template:primer dsRNA were identified as essential for the catalytic activity of RdRp. Overall, 41 residues of nsp12 were identified and subjected to design; these were primarily located within a 6 Å distance of favipiravir. Certain critical residues, such as Asn691, Lys545 and Ser814, formed hydrogen bonds with favipiravir. Furthermore, favipiravir formed hydrogen bonds with C10, metal coordination of Mg2+ and a π‐π stacking with U20 of the template:primer dsRNA (Fig. 1B,C).
Identification of resistance mutations from MOE‐based design experiments
Next, we designed 41 shortlisted residues of nsp12 using resistance mutation scan methodology employed in Molecular Operating Environment (MOE), where each residue was mutated with only a single nucleotide polymorphism (SNP) of the wild‐type sequence. As a result, the mutations were confined to SNPs to resemble the variations that may occur naturally during the evolution of the virus. Table 1 shows a list of SNPs sampled and designed for 41 favipiravir‐interacting residues of nsp12. A total of 350 single‐point mutants potentially associated with the resistance development were generated, and the affinity between the designed proteins and favipiravir was calculated. The relative binding affinity of the mutated proteins in comparison with the wild‐type protein (dAffinity) was calculated. Here, a large positive increase in the dAffinity value indicates that the mutation had a lower affinity to favipiravir and hence could become easily resistant to the ligand (favipiravir).
Table 1.
Favipiravir‐interacting nsp12 residues that are designed with corresponding SNPs of the wild‐type sequence. A limited number of amino acids for each position were sampled because they are more likely to happen naturally over the evolution of the protein than others
| S. No | Nsp12‐favipiravir‐interacting and designed wild‐type residues | Sampled SNPs in designs | S. No | Nsp12‐favipiravir‐interacting and designed wild‐type residues | Sampled SNPs in designs |
|---|---|---|---|---|---|
| 1 | His439 | RNDQLPY | 22 | Arg624 | NCQGHILKMPSTW |
| 2 | Asp452 | ANEGHYV | 23 | Ala625 | DEGPSTV |
| 3 | Tyr456 | NDCHFS | 24 | Met626 | RILKTV |
| 4 | Met542 | RILKTV | 25 | Arg631 | NCQGHILKMPSTW |
| 5 | Tyr546 | NDCHFS | 26 | Val662 | ADEGILMF |
| 6 | Ala547 | DEGPSTV | 27 | Lys676 | RNQEIMT |
| 7 | Ser549 | ACLFPTWY | 28 | Gly679 | ARDCESWV |
| 8 | Ala550 | DEGPSTV | 29 | Thr680 | ARNIKMPS |
| 9 | Lys551 | RNQEIMT | 30 | Ser681 | ACLFPTWY |
| 10 | Arg553 | NCQGHILKMPSTW | 31 | Ala690 | DEGPSTV |
| 11 | Ala554 | DEGPSTV | 32 | Ser692 | ACLFPTWY |
| 12 | Arg555 | NCQGHILKMPSTW | 33 | Phe694 | CILSYV |
| 13 | Thr556 | ARNIKMPS | 34 | Asn695 | DHIKSTY |
| 14 | Gly616 | ARDCESWV | 35 | Ala762 | DEGPSTV |
| 15 | Trp617 | RCGLS | 36 | Val763 | ADEGILMF |
| 16 | Asp618 | ANEGHYV | 37 | Lys798 | RNQEIMT |
| 17 | Tyr619 | NDCHFS | 38 | Trp800 | RCGLS |
| 18 | Pro620 | ARQHLST | 39 | His810 | RNDQLPY |
| 19 | Lys621 | RNQEIMT | 40 | His816 | RNDQLPY |
| 20 | Cys622 | RGFSWY | 41 | Asp833 | ANEGHYV |
| 21 | Asp623 | ANEGHYV |
350 nsp12 designs revealed that the dAffinity ranged from –0.54 to 0.52 kcal·mol−1 (Fig. S1), out of which 152 designs showed positive dAffinity values, indicating their reduced affinity towards favipiravir. The dAffinity values of the 41 favipiravir‐interacting residues with their corresponding single‐point resistant mutations are shown in Figs 2 and 3. Furthermore, a stringent dAffinity cut‐off revealed that 13 mutants had dAffinities higher than 0.2 kcal·mol−1 (Fig. 4). This dAffinity cut‐off was considered for a clear interpretation of affinity‐attenuating mutants and distinguishing them from other mutants. These mutants with very high dAffinity values can be significant for the development of the resistance against favipiravir. Notably, most of these 13 mutants originated from mutations in four nsp12 residues, namely His439, Cys622, Asp623 and Thr680 (Fig. 4). Furthermore, certain residues, such as Asp452, Tyr456, Met542, Tyr546, Ala554, Trp617, Tyr619, Cys622, Arg624, Gly679 and Trp800, were susceptible to developing resistance during evolution if an immune and drug response was mounted on SARS‐CoV‐2, as indicated by their affinity profiles (Figs 2, 3, 4). Using this method, we carried out a representative computational fitness test to sample only those mutations that were not lethal to the virus and were more likely to develop naturally to assist RdRp in maintaining structural and functional integrity.
Fig. 2.

Relative binding affinities of designs from the favipiravir‐interacting nsp12 residues. Radar plots (1–32) showing the MOE‐derived relative binding affinities (dAffinities) for favipiravir‐interacting nsp12 residues and their single‐point mutant designs.
Fig. 3.
Relative binding affinities of designs from favipiravir‐interacting nsp12 residues. The radar plots for (33–41) interacting residues.
Fig. 4.
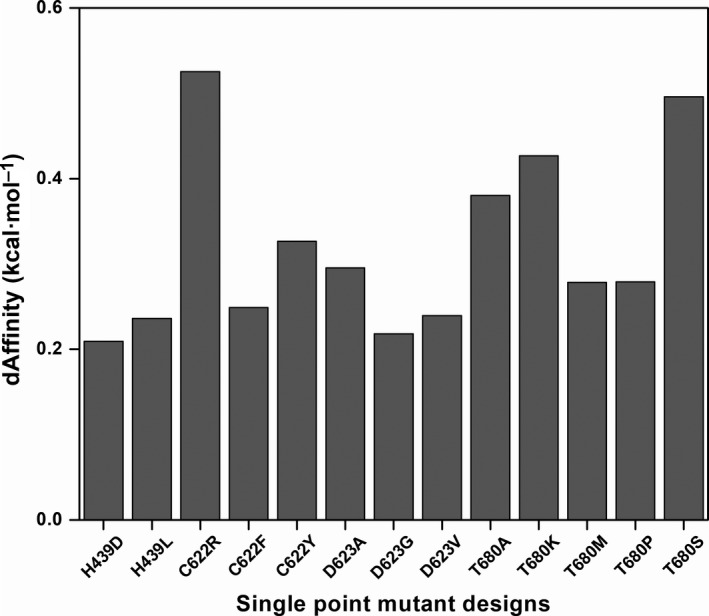
Relative binding affinities of the potential designs for favipiravir resistance. Bar plot showing the dAffinities for the most plausible favipiravir‐interacting nsp12 residues and their single‐point mutant designs. These designs exhibited dAffinities higher than 0.2 kcal·mol−1, suggesting potential resistance to favipiravir. Here, a large positive increase in the dAffinity value indicates that the mutation decreased the affinity of nsp12 towards favipiravir and hence could develop resistance for the ligand. This dAffinity cut‐off was considered for a clear interpretation of affinity‐attenuating mutants and distinguishing them from other mutants.
Rosetta‐based design of favipiravir‐binding nsp12 region and associated physicochemical features
After we identified single‐point mutants that has the potential to induce favipiravir resistance, Rosetta‐based ligand interface design of the favipiravir‐binding site of nsp12 in the nsp7‐nsp8‐nsp12‐dsRNA complex was performed [25]. This was conducted to obtain the plausible hotspot residues of nsp12 at once and better understand the mutational landscape profile during the emergence of drug resistance. Although 72 residues of nsp12 were found to interact with favipiravir, 41 were selected to design with backbone flexibility, as they did not interact with the template:primer dsRNA and were not involved in the catalytic activity. In this step, each of the 41 residues was mutated based on the corresponding SNPs to resemble the mutations that are more likely to evolve naturally during the evolution of the protein (Table 1). During the design experiment, nsp12 residues other than the interfacial residues were only repacked. A total of 100 000 designs were generated, and their physicochemical parameters were analysed. The 100 000 designs enclosing the favipiravir‐binding site of nsp12 were categorized into affinity‐enhancing and affinity‐attenuating designs based on their binding affinities and control values.
First, 100 000 designs were examined for Rosetta total scores vs. their root mean square deviations (RMSDs). The Rosetta total score is the weighted sum of several energy terms, including physical forces like van der Waals’ interactions, electrostatics and other statistical terms. Next, RMSD is computed between the designed structures with respect to the wild‐type nsp7‐nsp8‐nsp12‐template:primer dsRNA complex structure with favipiravir‐RTP. It was observed that nearly all designs retained RMSDs below 1.5 Å, and more than half of them retained RMSDs below 1 Å, suggesting they did not considerably change from the initial complex structure during designing 41 interface residues of nsp12 and introducing mutations (Fig. 5A). At this stage, we performed a control run, where 41 residues of nsp12 were only repacked without designing. This distinguished the affinity‐attenuating and affinity‐enhancing designs, indicating that the affinity‐attenuating designs showed a minor increase in RMSDs in their overall structures and comparatively unfavourable energetics due to the introduction of missense mutations.
Fig. 5.
Structural and physicochemical parameters from the ligand‐based Rosetta interface design of the favipiravir–RdRp interacting complex. (A) Rosetta total score vs. RMSD of 100 000 designs of the favipiravir‐bound RdRp complex. (B) Rosetta total score vs. interface delta (binding affinity) of the designs between favipiravir and the RdRp complex. (C) Interface delta vs. RMSD of the designs. (D) Interface delta vs. per cent sequence identity of favipiravir‐interacting residues of the RdRp complex shows the distribution of designs. Blue to red colour indicates designs with low to high binding affinities between favipiravir and RdRp complex. In panels (A–C), the dashed boxes represent the control values, where favipiravir‐interacting residues were not designed. Further, the top‐scored affinity‐enhancing and affinity‐attenuating designs are highlighted in green and red boxes respectively.
Second, the designs were analysed for binding affinities between the designed nsp12 with favipiravir (represented as interface delta) vs. the Rosetta total scores. The affinity‐attenuating designs had considerably lower binding affinities and lower total scores than the affinity‐enhancing and control designs (Fig. 5B). This suggested that unfavourable energetics and lower total scores influenced the binding affinity in the affinity‐attenuating designs.
Third, we computed the RMSDs vs. interface delta for the 100 000 designs. We found that several affinity‐attenuating designs with lower binding affinities displayed higher RMSDs of over 1.2 Å, indicating that the corresponding mutations destabilized the interactions (Fig. 5C). However, several affinity‐attenuating designs with lower binding affinities retained RMSDs as compared to the affinity‐enhancing and control designs as well, indicating that residue‐specific side‐chain movements could have contributed to the reduced binding affinities in those cases.
Finally, the interface delta of the designs was evaluated against the percentage sequence identities of the generated designs. Interestingly, although nsp12 has 41 residues interacting with favipiravir, only a few of them were potentially hotspot residues and prone to mutations (Fig. 5D). On average, the designs retained a sequence identity of 97%–98% compared to the wild‐type structure. The affinity‐attenuating designs with lower binding affinities were less conserved than their counterparts (Fig. 5D), demonstrating that nsp12 developed resistance against favipiravir with only selected mutations at a few hotspot residues whenever SARS‐CoV‐2 encounters evolutionary and immune/drug response.
Mutational landscape and sequence variations in favipiravir‐nsp12 binding designs
The top 50 designs from each category (affinity‐enhancing and affinity‐attenuating designs) were further analysed and compared to determine the key differences in the mutations and sequence diversities (Fig. 6). Detailed analysis of the mutational landscape profile revealed that in both categories, residues His439, Asp452, Tyr456, Met542, Tyr546, Ala547, Ser549, Ala550, Lys551, Arg553, Ala554, Arg555, Thr556, Lys621, Cys622, Ala625, Arg631, Val662, Lys676, Gly679, Thr680, Ser681, Ala690, Ser692, Phe694, Asn695, Val763, Trp800, His810, His816 and Asp833 were identically sampled (Fig. 6). However, certain wild‐type residues, such as Trp617, Asp618, Tyr619, Asp623 and Ala762, exhibited the most diverse sequence variations between the two categories. Furthermore, few other residues, such as Gly616, Pro620, Arg624, Met626 and Lys798, showed mutations with a relatively lower number of sequence variations between the affinity‐attenuating and affinity‐enhancing designs (Fig. 6). Therefore, a total of 10 residue sites exhibited sequence variations between the affinity‐attenuating and affinity‐enhancing designs. Notably, these residues that showed sequence variations and were highly susceptible to mutations belonged to the palm domain of RdRp. In addition, in the affinity‐attenuating designs, residues at positions 617, 618, 619, 623 and 762 were sampled to a high number of extremely diverse amino acids (Fig. 6).
Fig. 6.
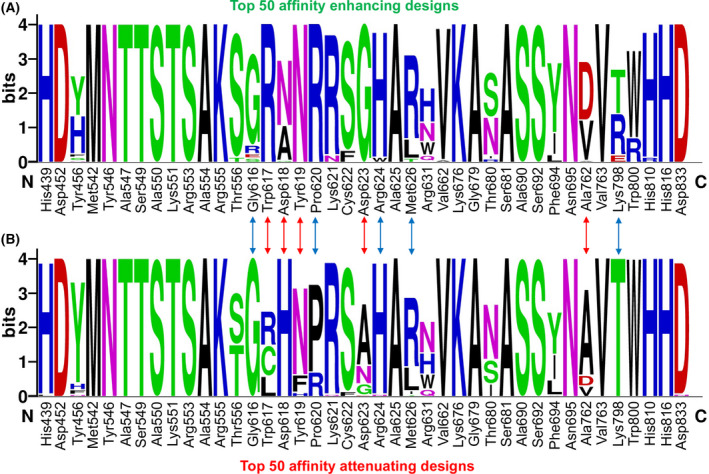
Sequence logos showing the type and frequency of designed favipiravir‐nsp12 interacting residues. Sequence logos of the 50 top‐scored (A) affinity‐enhancing vs. (B) affinity‐attenuating designs are displayed. The X‐axis represents the native nsp12 residue index, and the Y‐axis shows the sequence conservation at that position. The height of symbols indicates the relative frequency of a particular amino acid at that position. The red arrow denotes residues that experienced diverse sequence variations with a relatively higher number of sampled amino acids, whereas the blue arrow denotes residues that exhibited diverse mutations but with fewer amino acids sampled.
A comparison of MOE‐derived single‐point resistance‐developing mutants with the Rosetta‐generated mutational landscapes demonstrated that both methods predicted certain commonly occurring mutants, such as D623A and D623G in affinity‐attenuating designs (Figs 2, 3, 4, and 6). Moreover, Rosetta‐based designs revealed a few more mutation types and hotspot residues. This sequence‐specific conservation and diversity of the favipiravir‐bound nsp12 designs indicated that, in the future, these hotspot residues could undergo selective mutations to establish a resistance to favipiravir and similar drugs.
Validation of design protocols
To confirm the predictive ability and accuracy of the MOE‐based and Rosetta‐based design methodologies, a well‐defined remdesivir‐resistant SARS‐CoV mutant (V557L) was considered. Chikungunya virus and H1N1 influenza A virus have been shown to develop resistance against favipiravir. However, since their 3D structures are not available, the findings were validated with the SARS‐CoV V557L mutant, as Val557 is conserved in the nsp12 of SARS‐CoV‐2. To evaluate whether our MOE and Rosetta design methodologies are able to rank L557 among the low‐affinity designs, we scanned residues in the 557 position with other amino acids and realized that V557L was ranked as the low‐affinity mutant in the design calculations (Fig. S2). These observations validated the capability of our design methodologies to score and rank‐order the affinity‐attenuating designs that can develop favipiravir resistance in SARS‐CoV‐2. However, because V557L is a remdesivir‐resistant mutation of SARS‐CoV, the SARS‐CoV‐2 may not mutate Val557 residue to acquire favipiravir resistance. Instead, it could use any other hotspot residues to develop resistance. Besides, since remdesivir acts as a chain terminator, this control experiment offered only this aspect of the drug’s mode of action and subsequent computational scores. Finally, mutations of nsp12 in the favipiravir‐binding site of the RdRp were obtained from the CoV‐GLUE database, and their frequency of occurrence was determined. We observed that out of 134 mutations obtained from the CoV‐GLUE database (in the favipiravir‐binding site of nsp12, which are also part of our designed residues), 63 mutations were already predicted as resistant, as shown in our MOE‐based design calculations, thereby attaining ˜ 47% match with the sequencing data (Fig. 7). Notably, many high‐frequency mutations such as A554S, A554T, A554V, W617C, Y619C and H810Y were already found to be favipiravir‐resistant in our MOE‐generated design calculations (Fig. 7). However, it should be noted that the frequencies of the mutations recorded in the CoV‐GLUE database may not correlate directly with the calculated dAffinities. It is possible that as the pandemic advances and more sequencing data become available, other sampled mutations from our designs may evolve and correlate with the new sequences. In conclusion, the control experiments validated the design methodology to score and rank‐order the affinity‐attenuating designs.
Fig. 7.
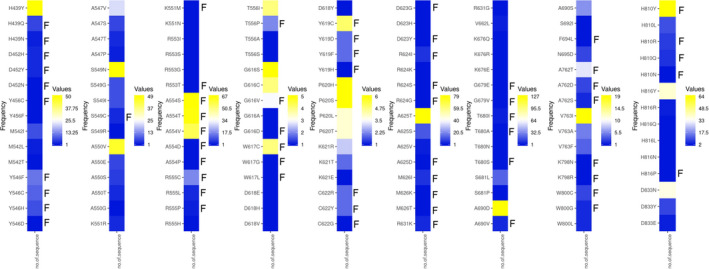
Heat maps showing the mutations and their frequencies in the favipiravir‐binding site of nsp12. Mutations in the favipiravir‐binding site of SARS‐CoV‐2 nsp12 obtained from the CoV‐GLUE database are presented. The frequencies of the mutations in the reported COVID‐19 cases ranging from lower to higher numbers are shown from blue to yellow. The mutants that developed resistance towards favipiravir are marked as ‘F’ adjacent to the mutants. Out of 134 mutations obtained from the CoV‐GLUE database (in the favipiravir‐binding site of nsp12), 63 were predicted as resistant in our design computations, thus attaining ˜ 47% correlation with the sequencing data
Interactions between favipiravir and nsp12 in the top‐ranked affinity‐attenuating and affinity‐enhancing designs
We examined the intermolecular interactions between favipiravir and nsp12 in the top‐scored affinity‐attenuating and affinity‐enhancing designs. The top‐scored affinity‐enhancing design had 237 interactions, while the affinity‐attenuating design had 223 interactions (Table 2). The van der Waals and proximal interactions played a significant role in decreasing the affinity between favipiravir and nsp12 in the affinity‐attenuating designs (Fig. 8A,B). A ligand interaction diagram from the affinity‐attenuating and affinity‐enhancing designs showed that the lack of a hydrogen bond between Asn691‐favipiravir significantly contributed to the lower binding affinity in the affinity‐attenuating designs (Fig. 8C,D).
Table 2.
Detailed intermolecular interactions formed between favipiravir and nsp12 binding residues in the top‐scoring affinity‐enhancing and affinity‐attenuating design
| Types of interactions | Favipiravir‐nsp12 | |
|---|---|---|
| Affinity‐enhancing design | Affinity‐attenuating design | |
| Van der Waals interactions | 3 | 1 |
| Proximal interactions | 217 | 196 |
| Polar contacts | 8 | 8 |
| Hydrogen bonds | 5 | 6 |
| Ionic interactions | 2 | 10 |
| Metal complex interactions | 2 | 2 |
| Total number of interactions | 237 | 223 |
Fig. 8.
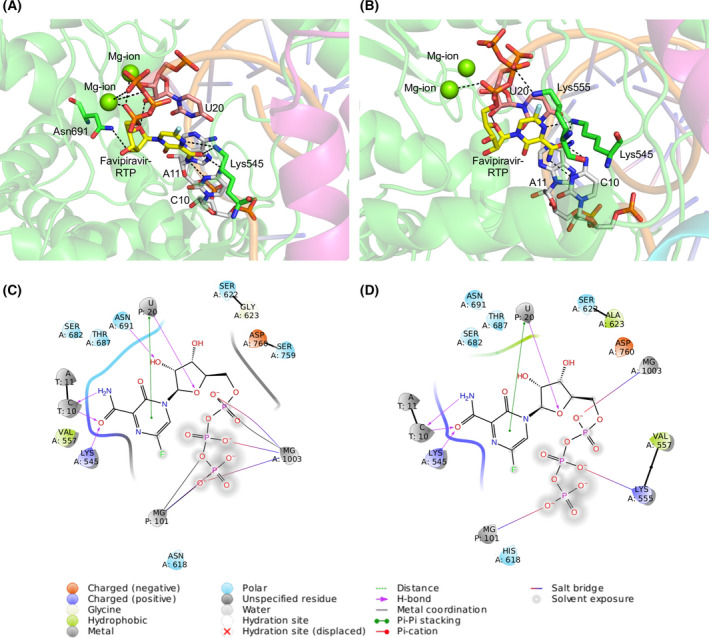
Intermolecular interactions between favipiravir and the RdRp complex for the top‐scoring designs. Intermolecular interactions between favipiravir and the RdRp complex for the top‐scored (A) affinity‐enhancing designs and (B) affinity‐attenuating designs. Interacting residues of corresponding nsp12 (green) and RNA bases with favipiravir (yellow) and Mg2+ (green sphere) coordinating the favipiravir‐RTP are shown. Hydrogen bonds are shown as black dashes. (C) 2D ligand interaction diagrams of RdRp‐favipiravir complex for the top‐scored affinity‐enhancing and (D) affinity‐attenuating design. The intermolecular interactions are labelled below.
Dynamics and local flexibility of wild‐type nsp12 and certain affinity‐attenuating designs
To understand the contribution of nsp12 regions/sites to the large amplitude movements of RdRp of SARS‐CoV‐2, we carried out the normal mode analysis. First, the atomic displacements for the six lowest‐frequency nontrivial modes were computed, which referred to the normalized square of displacement of each C⍺ atom so that the sum of all residues was 100 (Fig. 9A). The higher values corresponded to the highly displaced regions, and peak clusters on the plots indicated significantly displaced regions. Notably, residues 120–400 and 450–800 were less displaced, being primarily engaged in local flexibility in nsp12 (Fig. 9A). Second, the residue correlation matrix highlighting the correlated movement of C⍺ atoms in nsp12 revealed that residues 450–780 of nsp12 showed a highly correlated coupled motion (Fig. 9B). Third, the normalized deformation energies per C⍺ atom averaged over the normal modes were obtained for the native nsp12 and certain affinity‐attenuating designs. This quantifies the energy contribution for each C⍺ atom to the intrinsic motion of nsp12, which revealed that the designs did not experience much change in the contribution per energy as a result of mutations (Fig. 9C). Finally, the squared atomic fluctuations for each C⍺ atom were computed, normalized so that the sum of all C⍺ in a sequence is 100. This profile for the native nsp12 and certain affinity‐attenuating designs revealed that nsp12 exhibits interdomain motions, and residues 120–400 and 450–800 were largely stable and unaffected due to mutations (Fig. 9D). Overall, this analysis suggests that the mutations possibly affect the favipiravir‐binding sites and proximal sites rather than the overall dynamics of the whole protein.
Fig. 9.
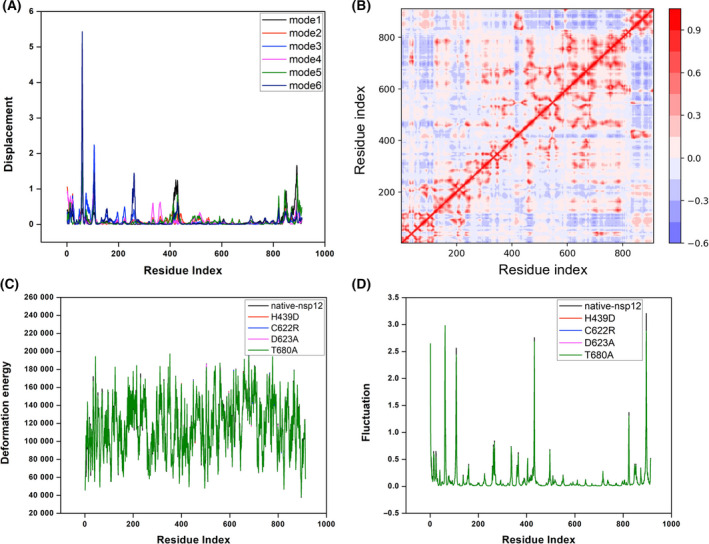
Normal mode analysis of native nsp12 and certain affinity‐attenuating designs. (A) The atomic displacement of each C⍺ atom for modes 1–6. (B) The residue correlation matrix showing the correlated movement of the C⍺ atoms in nsp12. In the plot, each cell indicates the coupling of two residues ranging from −1 (anticorrelated, blue) to 0 (uncorrelated) to 1 (correlated, red), thus representing correlated motions. (C) The normalized deformation energies per C⍺ averaged over the normal modes for the native nsp12, and certain affinity‐attenuating designs are shown and labelled. (D) The squared atomic fluctuations for each C⍺ atom are normalized and shown so that the sum of all C⍺ in a sequence is 100. The profiles for the native nsp12 and certain affinity‐attenuating designs are labelled to understand the effect of mutations.
Discussion
A combination of different strategies encompassing multiple vaccines and antiviral therapeutics is required for the effective and complete management of COVID‐19. Recently, several vaccine candidates have received EUA for COVID‐19 prevention in several countries. However, because RNA viruses mutate and recombine rapidly, preparing long‐lasting vaccines and continually effective therapeutics is challenging, as evident from the dramatic shifts in vaccine use worldwide. The majority of vaccines in the pipeline focus on the S‐protein. Recent data from several countries, such as the UK, South Africa and Australia, demonstrate accumulating mutations in the S‐protein, allowing the virus to escape immune recognition, thus rendering the vaccines less effective or even ineffective. This could jeopardize the success of approved COVID‐19 vaccines and the chances of accomplishing herd immunity. These findings suggest that combining different strategies, including multiple vaccines and antivirals, will be required to effectively manage COVID‐19.
SARS‐CoV‐2 jumped to humans in late 2019 via a possible transmission through bats, civets or pangolin. Therefore, SARS‐CoV‐2 may not be completely adapted to its human host due to its recent association with this host. However, since its emergence, the SARS‐CoV‐2 has undergone a considerable number of mutations and recombination events. Although the majority of mutations detected were neutral, these are gradually accumulating, leading to increased genomic diversity. Although the current rate of sequence variations among SARS‐CoV‐2 isolates is modest, this RNA virus has ample opportunity to generate multiple variants because of the many rounds of viral replication resulting in viral mutations over the course of the pandemic (https://www.gisaid.org/). All this increases the probability of the emergence of more infectious strains in the future [36]. According to the population genetics theory, the neutral and beneficial mutations that affect viral fitness can reach higher frequencies. The development and spread of a mutation are governed by several factors, including population growth, founder effects, range expansion and random genetic drift, as well as by a potential positive selection pressure to confer enhanced transmissibility or drug and immune resistance. Therefore, SARS‐CoV‐2 may evolve into phenotypically distinct lineages as it establishes itself as an endemic human pathogen. The nonsynonymous D614G mutation in the viral S‐protein was the first mutation detected in early March 2020 that rapidly dominated by July 2020 [37]. Although the D614G mutant isolate is not linked to increased mortality or clinical severity, it is more transmissible [38, 39] and enhances the viral replication rate [40, 41]. This was followed by the discovery of several mutations in the S‐protein RBD in mink and humans associated with mink farms [42]. By December 2020, a new lineage, B.1.1.7 (also called 501Y.V1), emerged in the UK and is spreading rapidly with a higher transmission rate [43]. This B.1.1.7 variant includes several mutations, of which eight mutations are in the S‐protein [44]. Another lineage, B.1.351 (also called 501Y.V2), which is rapidly spreading in South Africa, has nine mutations in the S‐protein. A third strain, P.1 (also called 501Y.V3), emerged in Brazil in December 2020. This P.1 variant contains a unique constellation of ten mutations, including three mutations in the receptor‐binding domain of the spike protein: K417T, E484K and N501Y. In March 2021, a highly transmissible variant, named B.1.617, which has around 15 mutations, emerged in India. Reports claim that this variant is responsible for the much severe second wave of infection in India. Because most of the current vaccines target the viral S‐protein antigen, mutations in the S‐protein can mediate virus escape from host antibodies and possibly weaken the efficacy of the vaccine. Therefore, multiple pharmacological options should be available for the effective management of COVID‐19.
Favipiravir and remdesivir are the key drugs currently used to treat patients with COVID‐19 in several countries [45]. They are being evaluated in clinical trials globally for treating SARS‐CoV‐2 infection. Favipiravir is considerably cheaper than remdesivir and is, therefore, more affordable for developing and poorer countries. Like other drug therapies, a major challenge with the widespread use of remdesivir and favipiravir could be the potential development of resistance [22]. These drugs are nucleoside analogs and target the RdRp of SARS‐CoV‐2 to stop viral replication and transcription. With the accumulation of mutations, an antigenic drift in response to the host immune pressure could facilitate the emergence of resistance against the antivirals [46]. Although no natural favipiravir‐resistant mutants have been found in influenza viruses [14, 47, 48, 49, 50, 51], an in vitro study has reported that the K229R mutation in the influenza virus RdRp induces favipiravir resistance [21]. Although the K229R mutation reduces the polymerase activity, the fitness cost of this mutation is compensated by a P653L mutation in RdRp [21]. Therefore, P653L mutation can restore RdRp activity while maintaining favipiravir resistance. A combination of K229R and P653L mutations resulted in a virus strain that was 30‐times less susceptible to favipiravir than the wild‐type virus while maintaining the replication kinetics. Notably, significant favipiravir resistance was reported from experimental evolution studies in the enterovirus 71 [20] and Chikungunya virus [19]. Overall, these data indicate a potentially universal mechanism for favipiravir resistance functioning under evolutionary and survival pressure. The present study will help better understand the structural dynamics of susceptible hotspots and resistance mutations and their possible effects on the efficacy of favipiravir. The validation will require close monitoring of SARS‐CoV‐2 evolution using multiple high‐throughput genome analyses, surveillance and SNP evaluation during and in the coming days of the pandemic.
Limitations of the study
This study had a few limitations. First, computationally predicted resistance mutations of nsp12 against favipiravir should be validated using biochemical experiments to determine the gain/loss of the binding affinity between nsp12 designs and favipiravir to get a complete understanding of the evolution of favipiravir‐resistant mutations. Second, as the Rosetta‐based interface design protocol aims to design mutants with improved binding affinity to favipiravir, a more appropriate design methodology and scoring function is required to scan for resistance mutations (designs that reduce binding affinity to a ligand). Long‐timescale molecular dynamics (MD) simulations may be performed for examining the mutational effects on the conformational dynamics and stability of the protein [52]. Despite these limitations, to validate that our design methodology correctly predicted the affinity‐attenuating designs interpreting the resistance mutations, we conducted adequate control experiments on a SARS‐CoV remdesivir‐resistant mutant, which is a chain‐termination drug. Further, using the publicly available SARS‐CoV‐2 nsp12 sequences, we ensured that the potential single‐point mutations characterizing the favipiravir‐resistant mutants are identified from our design experiments. Finally, the MOE‐based resistance design approach was employed to validate that the potential single‐point mutants signifying the resistant mutations are identified and predicted quantitatively.
Conclusions
Pandemics have been affecting humanity for centuries. One positive aspect of the COVID‐19 pandemic has been the unprecedented availability of scientific and technological advances that allowed the rapid development of therapeutic strategies. The SARS‐CoV‐2 has acquired only modest genetic variations to date with no specific mutation contributing to drug resistance. However, it is impossible to rule out the future development of resistance against the currently used drug regimens. Considering the critical importance of defining possible signatures for adaptation in SARS‐CoV‐2 against the current drugs, we conclude that our work will provide grounds for a better understanding of the favipiravir resistance and COVID‐19 management and may offer insights into the structural strategies for the development of more effective therapeutics.
Author contributions
AKP carried out all the design experiments, data generation and analysis. JD, PS and VNU helped in the analysis. AKP and TT conceived the study, participated in its design and coordination and drafted the manuscript. All authors read and approved the final manuscript.
Supporting information
Fig. S1. Relative binding affinities of all the 350 designs from the favipiravir‐interacting nsp12 residues.
Fig. S2. MOE‐ and Rosetta‐based binding affinity scores for the V557L remdesivir‐resistant mutant of SARS‐CoV.
Acknowledgements
The authors are grateful to Dr. Kam Y.J. Zhang (Laboratory for Structural Bioinformatics, RIKEN, Yokohama) for his continuous support and valuable suggestions for improving the manuscript. The authors acknowledge RIKEN ACCC for the Hokusai supercomputing resources.
Edited by Urs Greber
Data accessibility
Research data pertaining to this article are located at figshare.com: https://doi.org/10.6084/m9.figshare.14554239.v2
The data that support the findings of this study are available from the corresponding author [timir.tripathi@gmail.com] upon reasonable request.
References
- 1. Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L et al. (2020) Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science 368, 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D and Cramer P (2020) Structure of replicating SARS‐CoV‐2 polymerase. Nature 584, 154–156. [DOI] [PubMed] [Google Scholar]
- 3. Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M et al. (2020) Structural basis for inhibition of the RNA‐dependent RNA polymerase from SARS‐CoV‐2 by remdesivir. Science 368, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF and Barnard DL (2013) Favipiravir (T‐705), a novel viral RNA polymerase inhibitor. Antiviral Res 100, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shiraki K and Daikoku T (2020) Favipiravir, an anti‐influenza drug against life‐threatening RNA virus infections. Pharmacol Ther 209, 107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen PJ, Chao CM and Lai CC (2020) Clinical efficacy and safety of favipiravir in the treatment of COVID‐19 patients. J Infect 82, 186–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sreekanth Reddy O and Lai WF (2020) Tackling COVID‐19 using remdesivir and favipiravir as therapeutic options. ChemBioChem 22, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Misra N (2021) A short review on important drugs under clinical trial against Covid‐19. Mini Rev Med Chem 21, 1666–1678. [DOI] [PubMed] [Google Scholar]
- 10. Joshi S, Parkar J, Ansari A, Vora A, Talwar D, Tiwaskar M, Patil S and Barkate H (2021) Role of favipiravir in the treatment of COVID‐19. Int J Infect Dis 102, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Udwadia ZF, Singh P, Barkate H, Patil S, Rangwala S, Pendse A, Kadam J, Wu W, Caracta CF and Tandon M (2021) Efficacy and safety of favipiravir, an oral RNA‐dependent RNA polymerase inhibitor, in mild‐to‐moderate COVID‐19: A randomized, comparative, open‐label, multicenter, phase 3 clinical trial. Int J Infect Dis 103, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sangawa H, Komeno T, Nishikawa H, Yoshida A, Takahashi K, Nomura N and Furuta Y (2013) Mechanism of action of T‐705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother 57, 5202–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jin Z, Smith LK, Rajwanshi VK, Kim B and Deval J (2013) The ambiguous base‐pairing and high substrate efficiency of T‐705 (Favipiravir) Ribofuranosyl 5'‐triphosphate towards influenza A virus polymerase. PLoS ONE 8, e68347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baranovich T, Wong SS, Armstrong J, Marjuki H, Webby RJ, Webster RG and Govorkova EA (2013) T‐705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol 87, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shannon A, Selisko B, Le NTT, Huchting J, Touret F, Piorkowski G, Fattorini V, Ferron F, Decroly E, Meier C et al. (2020) Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS‐CoV‐2 lethal mutagenesis. Nat Commun 11, 4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naydenova K, Muir KW, Wu LF, Zhang Z, Coscia F, Peet MJ, Castro‐Hartmann P, Qian P, Sader K, Dent K et al. (2021) Structure of the SARS‐CoV‐2 RNA‐dependent RNA polymerase in the presence of favipiravir‐RTP. Proc Natl Acad Sci USA 118, e2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, Owen CJ, Pang J, Tan CCS, Boshier FAT et al. (2020) Emergence of genomic diversity and recurrent mutations in SARS‐CoV‐2. Infect Genet Evol 83, 104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callaway E (2020) The coronavirus is mutating ‐ does it matter? Nature 585, 174–177. [DOI] [PubMed] [Google Scholar]
- 19. Delang L, Segura Guerrero N, Tas A, Quérat G, Pastorino B, Froeyen M, Dallmeier K, Jochmans D, Herdewijn P, Bello F et al. (2014) Mutations in the chikungunya virus non‐structural proteins cause resistance to favipiravir (T‐705), a broad‐spectrum antiviral. J Antimicrob Chemother 69, 2770–2784. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y, Li G, Yuan S, Gao Q, Lan K, Altmeyer R and Zou G (2016) In vitro assessment of combinations of enterovirus inhibitors against enterovirus 71. Antimicrob Agents Chemother 60, 5357–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldhill DH, Te Velthuis AJW, Fletcher RA, Langat P, Zambon M, Lackenby A and Barclay WS (2018) The mechanism of resistance to favipiravir in influenza. Proc Natl Acad Sci USA 115, 11613–11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Padhi AK, Shukla R, Saudagar P and Tripathi T (2021) High‐throughput rational design of the remdesivir binding site in the RdRp of SARS‐CoV‐2: implications for potential resistance. iScience 24, 101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Padhi AK and Tripathi T (2021) Targeted design of drug binding sites in the main protease of SARS‐CoV‐2 reveals potential signatures of adaptation. Biochem Biophys Res Commun 555, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vilar S, Cozza G and Moro S (2008) Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem 8, 1555–1572. [DOI] [PubMed] [Google Scholar]
- 25. Moretti R, Bender BJ, Allison B and Meiler J (2016) Rosetta and the design of ligand binding sites. Methods Mol Biol 1414, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alford RF, Leaver‐Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K et al. (2017) The rosetta all‐atom energy function for macromolecular modeling and design. J Chem Theory Comput 13, 3031–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fleishman SJ, Leaver‐Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G et al. (2011) RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaufmann KW and Meiler J (2012) Using RosettaLigand for small molecule docking into comparative models. PLoS ONE 7, e50769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, Stevens LJ et al. (2020) An orally bioavailable broad‐spectrum antiviral inhibits SARS‐CoV‐2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12, eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singer J, Gifford R, Cotten M and Robertson D (2020) CoV‐GLUE: a web application for tracking SARS‐CoV‐2 genomic variation. Preprints.
- 31. Crooks GE, Hon G, Chandonia JM and Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jubb HC, Higueruelo AP, Ochoa‐Montaño B, Pitt WR, Ascher DB and Blundell TL (2017) Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J Mol Biol 429, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hollup SM, Salensminde G and Reuter N (2005) WEBnm@: a web application for normal mode analyses of proteins. BMC Bioinformat 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tiwari SP, Fuglebakk E, Hollup SM, Skjærven L, Cragnolini T, Grindhaug SH, Tekle KM and Reuter N (2014) WEBnm@ v2.0: web server and services for comparing protein flexibility. BMC Bioinformat 15, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naydenova K, Muir KW, Wu L‐F, Zhang Z, Coscia F, Peet MJ, Castro‐Hartmann P, Qian P, Sader K, Dent K et al. (2021) Structure of the SARS‐CoV‐2 RNA‐dependent RNA polymerase in the presence of favipiravir‐RTP. Proc Natl Acad Sci U S A 118, e2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Padhi AK and Tripathi T (2020) Can SARS‐CoV‐2 accumulate mutations in the S‐protein to increase pathogenicity? ACS Pharmacol Transl Sci 3, 1023–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B et al. (2020) Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell 182, 812–827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yurkovetskiy L, Wang X, Pascal KE, Tomkins‐Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C et al. (2020) Structural and functional analysis of the D614G SARS‐CoV‐2 spike protein variant. Cell 183, 739–751.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF et al. (2020) Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell 184, 64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes‐Garfias CR et al. (2020) Spike mutation D614G alters SARS‐CoV‐2 fitness. Nature 592, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, Leist SR, Schäfer A, Nakajima N, Takahashi K et al. (2020) SARS‐CoV‐2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M et al. (2020) Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science 371, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Darbrera G, O'Toole Á et al. (2021). Transmission of SARS‐CoV‐2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. [Google Scholar]
- 44. Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E et al. (2020) Preliminary genomic characterisation of an emergent SARS‐CoV‐2 lineage in the UK defined by a novel set of spike mutations. December 18, 2020. https://virological.org/t/preliminary‐genomic‐characterisation‐of‐an‐emergent‐sars‐cov‐2‐lineage‐inthe‐uk‐defined‐by‐a‐novel‐set‐of‐spike‐mutations/563
- 45. Mishra SK and Tripathi T (2020) One year update on the COVID‐19 pandemic: Where are we now? Acta Trop 214, 105778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pauly MD, Procario MC and Lauring AS (2017) A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. Elife 6, e26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y et al. (2002) In vitro and in vivo activities of anti‐influenza virus compound T‐705. Antimicrob Agents Chemother 46, 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daikoku T, Yoshida Y, Okuda T and Shiraki K (2014) Characterization of susceptibility variants of influenza virus grown in the presence of T‐705. J Pharmacol Sci 126, 281–284. [DOI] [PubMed] [Google Scholar]
- 49. Bank C, Renzette N, Liu P, Matuszewski S, Shim H, Foll M, Bolon DNA, Zeldovich KB, Kowalik TF, Finberg RW et al. (2016) An experimental evaluation of drug‐induced mutational meltdown as an antiviral treatment strategy. Evolution 70, 2470–2484. [DOI] [PubMed] [Google Scholar]
- 50. Marathe BM, Wong SS, Vogel P, Garcia‐Alcalde F, Webster RG, Webby RJ, Najera I and Govorkova EA (2016) Combinations of oseltamivir and T‐705 extend the treatment window for highly pathogenic influenza A(H5N1) virus infection in mice. Sci Rep 6, 26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takashita E, Ejima M, Ogawa R, Fujisaki S, Neumann G, Furuta Y, Kawaoka Y, Tashiro M and Odagiri T (2016) Antiviral susceptibility of influenza viruses isolated from patients pre‐ and post‐administration of favipiravir. Antiviral Res 132, 170–177. [DOI] [PubMed] [Google Scholar]
- 52. Padhi AK, Rath SL and Tripathi T (2021) Accelerating COVID‐19 Research Using Molecular Dynamics Simulation. J Phys Chem B 125, 9078–9091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Relative binding affinities of all the 350 designs from the favipiravir‐interacting nsp12 residues.
Fig. S2. MOE‐ and Rosetta‐based binding affinity scores for the V557L remdesivir‐resistant mutant of SARS‐CoV.
Data Availability Statement
Research data pertaining to this article are located at figshare.com: https://doi.org/10.6084/m9.figshare.14554239.v2
The data that support the findings of this study are available from the corresponding author [timir.tripathi@gmail.com] upon reasonable request.


