Abstract
Background:
Bordetella pertussis, a highly contagious respiratory. Notably, the resurgence of pertussis has recently been associated with the lacking production of vaccine virulence factors. This study aimed to screen pertactin (Prn) and filamentous hemagglutinin (Fha) production in Iran with 50 years’ whole cell vaccine (WCV) immunization program.
Methods:
Overall, 130 B. pertussis isolates collected from Pertussis Reference Laboratory of Iran during 2005–2018. Real-time PCR was performed by targeting IS481, ptxP, IS1001 and IS1002 for species confirmation of B. pertussis. Western-blot was used to evaluate the expression of virulence factors (pertactin and filamentous hemagglutinin).
Results:
All tested B. pertussis isolates expressed Prn and all except two isolates expressed Fha. We have sequenced genomes of these strains and identified differences compared with genome reference B. pertussis Tohama I.
Conclusion:
Many countries reporting Prn and Fha-deficiency due to acellular vaccine (ACV) pressure. Our results demonstrate in a country with WCV history, Fha-deficient isolates may rise independently. However, Prn-deficient isolates are more under the ACV pressure in B. pertussis isolates. Continues surveillance will provide a better understanding of the effect of WCV on the evolution of the pathogen deficiency.
Keywords: Whooping cough, Bordetella pertussis, Iran, Whole-cell vaccine, Filamentous hemagglutinin, Pertactin
Introduction
Bordetella pertussis is the causative agent of whooping cough, a highly contagious respiratory disease most severe in infants and young children (1). Introduction of whole-cell vaccine (WCV) during the 1950s significantly reduced the morbidity and mortality of pertussis globally. However, due to the side effect of WCV, acellular vaccine (ACV) with three [pertussis toxin (Ptx), pertactin (Prn) and filamentous hemagglutinin (Fha)] to five component (additional Fimbriae 2 and 3) was replaced in many countries (2, 3). Despite the long history of pertussis immunization and dramatic decline in the mortality and morbidity of whooping cough in newborns, epidemics still occur in populations with high vaccination rate (4). A rise in the incidence of pertussis has been reported with epidemics in Europe, Australia and the US in the last decade (5–7). The re-emergence of disease in the world might be due to increased awareness of the disease, waning vaccine-induced immunity, pathogen adaptation to vaccination, antigenic divergence and changes in gene expression, by either up or down-regulation of antigenic factors (8–11). Notably, the resurgence of pertussis has recently been associated with the emergence and spread of isolates carrying pertussis toxin promoter allele 3 (ptxP3) and lacking the production of major vaccine antigens especially pertactin in countries like Australia, Japan, the USA and European countries administrating ACV (12–15). Prn defined is under ACV vaccine pressure (16). The introduction of vaccination applied a new selective pressure to the circulating B. pertussis populations. Isolates collected during the pre-vaccine era were mainly harboring ptxP1, ptxA2 and prn1 alleles. After introduction of WCV, prn2 and ptxP3 were reported sporadically but spread fast after replacing the ACV and now predominant circulating isolates harboring ptxP3, ptxA1, prn2 alleles (17).
In Iran, the whole-cell pertussis vaccine (WCV) has been administered since 1950s (18–20). Immunization program for pertussis in Iran recommends three primary doses of whole-cell diphtheria–tetanus–pertussis (DTwP) vaccine at 2, 4, 6 months and a booster in 18 months and 6-y-old children. Despite high pertussis vaccination coverage (96% since 2000), we had reduction in cases of infection but again raised and Iranian population has experienced pertussis resurgence since 2007 and reached its peak in 2012 and 2013 (21, 22). In previous study allelic variations of Iranian B. pertussis under WCV pressure were described such as pertussis toxin (ptxA), we found four allele (ptxA1, ptxA2, and ptxA4, ptxA5), four ptxP alleles (ptxP1, ptxP2, ptxP3 and ptxP4) the promoter of the pertussis toxin operon, three pertactin alleles (prn1, prn2, prn9), two filamentous hemagglutinin alleles, a major adhesin involved in colonization (fhaB1, fhaB2) and finally three fim3 alleles (fim3-1, fim3-2, fim3-3) and two fim2 alleles (fim2-1, fim2-2) encoding the fimbrial proteins Fim2 and Fim3 (23).
Iranian predominant circulating isolates harboring ptxP3, ptxA1, prn2, fim2-1, fim3-2 alleles. Allele profiles of recently collected B. pertussis isolates in Iran are similar to countries with ACV vaccination with reports of Prn negative isolates (23). In this study, we investigated Fha and Prn expression of the 130 B. pertussis clinical isolates available in Pertussis Reference Laboratory of Pasteur Institute of Iran during 2005–2018.
Materials and Methods
Bacterial Isolates and molecular identification
In the current study, 130 B. pertussis clinical samples were obtained from Pertussis Reference Laboratory of Pasteur Institute of Iran from Jan 2005 to Oct 2018. Isolates belonged to 18 different provinces in Iran from patients aged 20 d to 50 years old (supplementary File 1).
All B. pertussis isolates were sub-cultured on Bordet-Gangue agar supplemented with 15% defibrinated sheep blood, incubated at 36°C for 72 hours. B. pertussis isolates were confirmed by a combination of colony morphology, Gram stain and conventional biochemical tests and use of specific Bordetella antiserums (Difco B. Pertussis Antiserum, Rabbit serum for slide agglutination).
DNA extraction was performed using High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany). Real-time PCR was performed by targeting IS481, ptxP, IS1001 and IS1002 for species confirmation of B. pertussis. Primers and probes are available in Table 1.
Table 1:
Specific primers sequences targeting IS481, ptxP, IS1001 and IS1002
| Target region | Primer and probe sequence (5′-3′) |
|---|---|
| ptxP | PT1a: 5′-GCATGCGTGCAGATTCGTC -3′ PT2a: 5′-CTCTGCGTTTTGATGGTGCCTAT -3′ PT-FAM:6FAM-AATCCAACACGGCATGAACGCTCCTTC--BHQ2 |
| IS481 | IS481-F 5′ ATCAAGCACCGCTTTACCC 3′ IS481-R 5′ TTGGGAGTTCTGGTAGGTGTG 3′ IS481-FAM : 6FAM-AATGGCAAGGCCGAACGCTTCA BHQ1 |
| IS1001 | IS1001-F 5′ CCAGAGCCGTTTGAGTTCGT 3′ IS1001-R 5′ AATTGCTGCAAGCCAACCA 3′ IS1001-CY5 : CY5-ACATAGACCGTCAGCAG-BHQ-3 3′ |
| IS1002 | IS1002-F 5 ′CTAGGTCGAGCCCTTCTTGTTAAC 3′ IS1002-R 5′ GCGGGCAAGCCACTTGTA 3′ IS1002-FAM : 6FAM-CTACGTCCAGTTCTGTTGCATCACCC-BHQ-1 3′ |
Amplification was carried out in a total volume of 30 μl containing 1X master mix (Roche Applied Science), 7.5 μM of each primers and probe and 5μl of extracted DNA (24).
Confirmation of prn (regions I and II), and fha genes in all isolates were performed according to standardized recommendations for B. pertussis PCR (25–27). The primers were described in Table 2.
Table 2:
PCR forward and reverse primers used in fha and prn amplification
| Gene Name | Sequence 5′→ 3 |
|---|---|
| fha | F: GGTTCAGAGCGTCAACAGC R: CTCACCAGCTTCGCAACG |
| prn-1 | F: GCCAATGTCACGGTCCAA R: GCAAGGTGATCGACAGGG |
| prn-2 | F: AGCTGGGCGGTTCAAGGT R: CGGATTCAGGCGCAACTC |
Fim serotyping
We assessed fimbriae 2 and 3 (Fim) expression with monoclonal antisera slide agglutination assays (anti-B. pertussis fimbriae monoclonal antibody, NIBSC code: 06/120, UK) (28).
Western Immunoblotting
Bacteria sub-cultured on Bordet Gengou agar with 15% sheep blood at 36 °C for 72 hours. Pure colony was suspended in Phosphate-buffered saline (PBS) and inactivated in 55 °C for 30 minutes. B. pertussis strain Tohama I (Gene Bank accession number BX470248) was used as a positive control and Klebsiella (ATCC 13883) as negative control. Forty microliter of the cell suspension was added to 20 μ of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiled for 10 minutes. Bacterial proteins were separated by 10% SDS polyacrylamide gradient gel (Bio-Rad California, USA) for 2 h at 100 V.
Proteins were transferred to polyvinylidene fluoride (PVDF) membranes at 300 V for 1 hour. Membranes were blocked with skim milk powder in PBS and incubated overnight. The 220-kDa Fha protein and 69-kDa Prn were detected using B. pertussis anti-FHA serum (sheep) (NIBSC code: 97/564) and B. pertussis anti - 69kD serum (sheep) (NIBSC code: 97/558) at a 1:1,000 dilution for 1 h at room temperature, after 3 washes with washing buffer (PBS and Tween 20) membrane were incubated with a horseradish peroxidase (HRP)-conjugated rabbit anti-sheep immunoglobulin (Avecina Research Institute, Tehran, Iran) (29). After a final wash, membranes were developed with Metal Enhanced DAB Substrate (3, 3′-Diaminobenzidine, Sigma-Aldrich, Germany) to visualize the antigen-antibody complex.
Genome sequence and structure variation
For whole-genome sequencing, genomic DNA was extracted and purified from pure culture using phenol-chloroform method (30). DNA libraries were constructed using Nextera XT kit (Illumina, San Diego, USA) according to manufacturer’s protocol and sequenced on the Illumina NextSeq instrument using 2x150 bp paired-end protocol. The contigs were aligned to the reference B. pertussis strain Tohama I (Gene Bank accession number BX470248) using progressive Mauve (ver. 2.3.1) and the de novo assembly of the raw reads were performed using SPAdes ver. 3.13.0 (31, 32). To extract Single nucleotide polymorphism (SNPs) a combination of Burrows-Wheeler Alignment (BWA) tools (ver. 0.7.5), Samtools (ver. 0.1.19) and progressive Mauve (33, 34) were used. Briefly, the filtered SNPs from mapping were compared to the SNPs exported by progressive Mauve and final SNPs were selected for further analysis. The contigs were compared to the reference B. pertussis strain Tohama I using progressive Mauve to identify deletions.
Results
Description of the isolates
Overall, 130 B. pertussis isolates were selected based on year and state of isolation. Isolates were collected from 18 provinces mostly from Tehran, Mazandaran and East Azerbaijan. The majority of isolates (62%, 80 from 130) were collected from patients under 6 months -old with incomplete vaccination. The results of the Real-time PCR showed the presence of ptxP, IS481, IS1002 in pertussis isolates and no IS1001 were detected in 130 clinical isolates.
Filamentous hemagglutinin and pertactin genes were detected in all samples and prn2 allele and fhaB1 allele were described as the predominant allele type with frequency up to 90% in the clinical isolates.
Antigen expression
Serotyping showed both fimbrial protein type 2 and 3 expressions among 80% of 130 clinical isolates. The western blot analysis showed that all tested clinical B. pertussis isolates expressed Prn. However, all except two isolates (IR34, IR178) expressed Fha (Fig. 1). IR34 as Fha deficient isolate was collected in 2012 from 5 yr old patient in Hamedan provinces (Table 3).
Fig. 1:
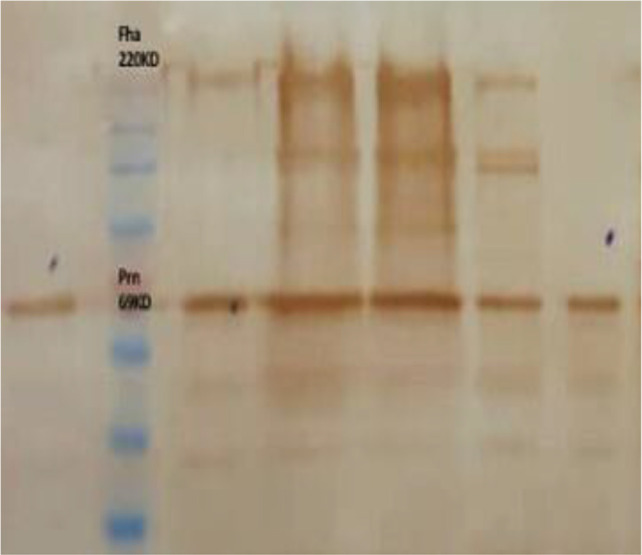
Fha-deficient detection and reduced Fha express by western blotting
Table 3:
Characterization of Fha-deficient isolates
| Strain name | Western result of Fha | Year | State | Age of child | Vaccination status | Fim Serotyping Fim2/fim3 | Allelic profiles | Detected mutations |
|---|---|---|---|---|---|---|---|---|
| Tohama | + | 1950 | - | - | - | +/− | ptxP1, ptxA2, prn1, fim2-1, fim3-2 | - |
| Vaccine 134 | + | - | - | - | - | +/+ | ptxP1, ptxA2, prn1, fim2-1, fim3-2 | - |
| Vaccine 509 | + | - | - | - | - | +/+ | ptxP2, ptxA4, prn7, fim2-2, fim3-1 | - |
| IR34 | Fha− | 2012 | Hamedan | 5 year | + | +/+ | ptxP1, ptxA2, prn1, fim2-1, fim3-2 | fhaB: Large deletion (330bp), position 6423 |
| IR178 | Fha− | 2018 | Tehran | 1 month | - | −/+ | ptxP3, ptxA1, prn2, fim2-1, fim3-2 | fhaB: deletion G, position 1087 |
Sequence analysis of Fha-deficient isolates
Whole-genome sequencing (WGS) was performed for two isolates (IR34 and IR178) that did not express Fha to find out the mechanisms of disruption. Sequence reads based on Illumina sequencing, were mapped against reference genome B. pertussis Tohama I. Single nucleotide polymorphisms (SNP) (Table 4), gene loss and insertion sequence disruption were investigated in these two isolates.
Table 4:
Number of sequence variation for both Fha-negative isolates based on whole genome sequencing
| WGS details | IR34 (FHA−) | IR178 (FHA−) |
|---|---|---|
| SNP | 2 | 66 |
| Synonymous | 1 | 26 |
| Non synonymous | 1 | 29 |
| Intragenic | - | 11 |
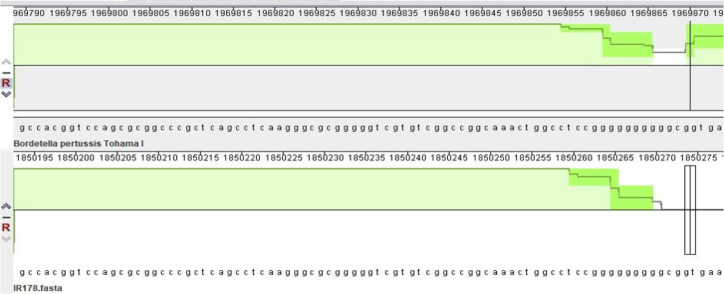
The gene loss analysis of Fha-deficient IR34 shows a 334 bp deletion at position 6423 in fhaB gene (supplementary file -2). IR34 is most closely related to reference genome B. pertussis Tohama I in PFGE dendrogram (23) with the same allelic profiles and differed from Tohama I in genome sequence by only 2 SNP in the genome (supplementary file -3). Clinical isolate IR178, had 66 SNPs and checked manually for short insertion and deletion and we detected one base deletion (G) at position 1087 of the fhaB gene (BP1879), a homopolymeric tract that might affect produces a truncated FhaB protein (Fig. 2) (35). No insertion sequence disruption related to expression of Fha was detected in IR34 and IR178.
Fig. 2:
Nucleotide deletion identified in IR178
Discussion
In the present study, we used western blot to investigate the effect of the WCV on expression of two major virulence factors, pertactin and filamentous hemagglutinin. B. pertussis encodes a wide range of virulence factors including pertactin, pertussis toxin, filamentous hemagglutinin and fimbriae 2 and 3 which play important roles in pathogenicity (36–38) and are included in acellular pertussis vaccine.
Several different alleles of the virulence factors have been identified in the world including, 11 alleles of ptxA, 21 alleles of ptxP, 17 alleles of prn, two alleles of fim2 and six alleles of fim3. It was observed isolates circulating in high vaccine coverage regions were different from the vaccine strains used for the production of WCV or ACV. Pertactin and Fha, are two important virulence factors and major adhesions of B. pertussis (39). Pertactin, originally known as the 69-kDa protein, is a surface-associated protein exported to the outer membrane and promotes attachment to tracheal epithelial cells and participates in invasion (40). Filamentous hemagglutinin is understood to function as both a surface-associated and secreted protein (36, 41). Fha plays critical roles in adherence to respiratory epithelium and contributes to persistence of infection in the respiratory tract (42, 43).
Unlike WCV, the ACV contains only a small number of bacterial proteins involved in pathogenicity such as Ptx, Prn, Fha, Fim2 and Fim3 (44, 45). While antigenic divergence was frequently observed in WCV, the ACV led to the increased circulation of clinical isolates not producing PRN. The emergence and expansion Prn-deficiency is the result of ACV immunity pressure proven in many studies (13, 15, 44, 46).
The lack of PRN did not impact the virulence of B. pertussis in the murine model of respiratory infection or in humans by comparing clinical symptoms in infants (47). In recent years, the spread of Prn deficiency has primarily occurred in the predominant genetic profile defined by prn2, ptxP3, ptxA1(14). Previous virulence factor analyses of clinical isolates collected in Iran, have indicated that the predominant B. pertussis genotype harbored, prn2, ptxP3, ptxA1 (23), which is similar to countries with ACV program (48–50). While the allele profile of recent isolates in Iran is the same as countries with ACV immunization programs, our study showed no prn-deficient isolate in Iran with WCV history that may confirm the ACV pressure on Prn deficiency.
Our results are consistent with the finding of other countries such as Russia and Poland, showing all isolates express Prn (51). However, in Poland, isolates lacking Prn production was reported and concluded, the Prn negative isolates, might be a result of an overflow of the bacteria from other countries (46). While most of neighbor countries around Iran still use WCV. Furthermore, there is no information about the allelic profile of clinical B. pertussis isolates collected from neighbor countries.
In terms of Fha expression, we found two clinical B. pertussis isolates (IR34, IR178) among 130 isolates that did not express Fha. Globally, Fha negative isolates had changed within the homopolymeric G tract (HPTs) in fhaB (8). In B. Pertussis one of the most common mechanisms of phase variation is occur in HPTs (35, 52). We have sequenced genomes of these strains and identified differences and gene loss compared with reference genome B. pertussis Tohama I. This study reports the first fha-deficient isolate in Iran. Based on data from sequencing, analysis identified herein IR178 isolate include deleted G at position 1087 occurred in HPTs, this mutation has been reported previously (41). In Weigand study, they were identified one isolate with severely reduced Fha production, exhibited the same mutations in homopolymeric tract within fhaB (41). In Australia, Zheng found Fha-negative isolate by Western immunoblotting with the same HPTs mutation in ptxP3 prn negative strain (53).
The adaptation of B. pertussis to its host proceeded mostly through gene loss (54). IR34 has large genome deletion at position 6423 in fhaB. These two isolates originated from different lineages. IR34 with similar PFGE pattern and allelic profile to the Tohama I (prn1, ptxP1, ptxA2). The genotyping of IR34 showed a genetic distinction between the isolates from predominant profile in the world that resulted from ACV. On the other hand, IR178 which had predominated in PFGE pattern (unpublished data) with prn2, ptxP3, ptxA1 profile, similar to predominant profiles in countries with ACV and distinction with vaccine seed strain B. p134 that use in Iran.
Conclusion
Many countries reporting Prn and Fha-deficiency in clinical B. pertussis isolates due to ACV-vaccine pressure. ACV-induced immunity is focused on just a few proteins, creating stronger selection pressure for strains with different allelic profile from vaccine seeds and isolates not expressing ACV-antigen proteins due to their better fitness in population. In a country with WCV history, no Prn-deficient isolates were found confirming the ACV pressure effect on the Prn deficiency in B. pertussis isolates. However, Fha deficiency may be under the pressure of either ACV or WCV continues surveillance of B. pertussis will provide a better understanding of the effect of WCV, on the evolution of the pathogen deficiency and emphasis the importance of continued surveillance of other major pertussis virulence factors and optimize strategies to reduce the incidence of pertussis.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This study was supported by institute Pasteur of Iran (Grant No: B_9112). We would also like to show our gratitude to all the staff in Pertussis Reference Laboratory of institute Pasteur of Iran.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Sáfadi MAP. (2015). Pertussis in young infants: a severe vaccine-preventable disease. Autops Case Rep, 5(2):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Advani A, Hallander HO, Dalby T, et al. (2013). Pulsed-field gel electrophoresis analysis of Bordetella pertussis isolates circulating in Europe from 1998 to 2009. J Clin Microbiol, 51(2):422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausiello CM, Cassone A. (2014). Acellular pertussis vaccines and pertussis resurgence: revise or replace? : mBio, 5(3):e01339–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherry JD. (2012). Epidemic pertussis in 2012—the resurgence of a vaccine-preventable disease. N Engl J Med, 367(9):785–7. [DOI] [PubMed] [Google Scholar]
- 5.Safarchi A, Octavia S, Wu SZ, et al. (2016). Genomic dissection of Australian Bordetella pertussis isolates from the 2008–2012 epidemic. J Infect, 72(4):468–77. [DOI] [PubMed] [Google Scholar]
- 6.Sealey KL, Harris SR, Fry NK, et al. (2015). Genomic analysis of isolates from the United Kingdom 2012 pertussis outbreak reveals that vaccine antigen genes are unusually fast evolving. J Infect Dis, 212(2):294–301. [DOI] [PubMed] [Google Scholar]
- 7.Winter K, Glaser C, Watt J, Harriman K. (2014). Pertussis epidemic--California, 2014. MMWR Morb Mortal Wkly Rep, 63(48):1129–32. [PMC free article] [PubMed] [Google Scholar]
- 8.Bart MJ, Harris SR, Advani A, et al. (2014). Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio, 5(2):e01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiappini E, Stival A, Galli L, De Martino M. (2013). Pertussis re-emergence in the post-vaccination era. BMC Infect Dis, 13:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Q, Mertsola J. (2008). Factors contributing to pertussis resurgence. Future Microbiol, 3(3):329–39. [DOI] [PubMed] [Google Scholar]
- 11.Syed MA. (2017). Choosing from Whole Cell and Acellular Pertussis Vaccines-Dilemma for the Developing Countries. Iran J Public Health, 46(2):272–3. [PMC free article] [PubMed] [Google Scholar]
- 12.Lam C, Octavia S, Bahrame Z, et al. (2012). Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis. Infect Genet Evol, 12(2):492–5. [DOI] [PubMed] [Google Scholar]
- 13.Lam C, Octavia S, Ricafort L, et al. (2014). Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis, 20(4):626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawloski L, Queenan A, Cassiday P, et al. (2014). Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol, 21(2):119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeddeman A, Van Gent M, Heuvelman C, et al. (2014). Investigations into the emergence of pertactin-deficient Bordetella pertussis isolates in six European countries, 1996 to 2012. Euro Surveill, 19(33):20881. [DOI] [PubMed] [Google Scholar]
- 16.Belcher T, Preston A. (2015). Bordetella pertussis evolution in the (functional) genomics era. Pathog Dis, 73(8): ftv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchez V, Hegerle N, Strati F, et al. (2015). New data on vaccine antigen deficient Bordetella pertussis isolates. Vaccines (Basel), 3(3):751–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikbin VS, Ahmadi NJ, Hosseinpour M, et al. (2015). Virulence factors variation among Bordetella pertussis isolates in Iran. Int J Mol Cell Med, 4(2):138–42. [PMC free article] [PubMed] [Google Scholar]
- 19.Zarei S, Jeddi-Tehrani M, Zeraati H, et al. (2009). Short term reactogenicity of a triple Diphtheria-Tetanus-Whole cell pertussis vaccine in Iranian infants. Iran J Public Health, 38(1):100–11. [Google Scholar]
- 20.Gouya MM. (2009). Expanded programme on immunization in Iran: last 3 decades achievements from 1979 to 2008. Iran J Public Health, 38(Suppl 1):81. [Google Scholar]
- 21.Moradi-Lakeh M, Esteghamati A. (2013). National Immunization Program in Iran: whys and why nots. Hum Vaccin Immunother, 9(1):112–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khazaei S, Ayubi E, Mansori K, Khazaei S. (2016). Pertussis incidence by time, province and age group in Iran, 2006–2011. Iran J Public Health, 45(11):1525–7. [PMC free article] [PubMed] [Google Scholar]
- 23.Heravi FS, Nikbin VS, Lotfi MN, et al. (2018). Strain variation and antigenic divergence among Bordetella pertussis circulating strains isolated from patients in Iran. Eur J Clin Microbiol Infect Dis, 37(10):1893–1900. [DOI] [PubMed] [Google Scholar]
- 24.Lotfi MN, Nikbin VS, Nasiri O, et al. (2017). Molecular detection of Bordetella holmesii in two infants with pertussis-like syndrome: the first report from Iran. Iran J Microbiol, 9(4):219–23. [PMC free article] [PubMed] [Google Scholar]
- 25.Mooi F, Hallander H, Von König CW, et al. (2000). Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis, 19(3):174–81. [DOI] [PubMed] [Google Scholar]
- 26.van Loo IH, Heuvelman KJ, King AJ, Mooi FR. (2002). Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J Clin Microbiol, 40(6):1994–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottero D, Gaillard ME, Fingermann M, et al. (2007). Pulsed-field gel electrophoresis, pertactin, pertussis toxin S1 subunit polymorphisms, and surfaceome analysis of vaccine and clinical Bordetella pertussis strains. Clin Vaccine Immunol, 14(11):1490–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guiso N, von Konig CW, Becker C, Hallander H. (2001). Fimbrial typing of Bordetella pertussis isolates: agglutination with polyclonal and monoclonal antisera. J Clin Microbiol, 39(4):1684–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghasemi A, Salari MH, Zarnani AH, et al. (2013). Immune reactivity of Brucella melitensis–vaccinated rabbit serum with recombinant Omp31 and DnaK proteins. Iran J Microbiol, 5(1):19–23. [PMC free article] [PubMed] [Google Scholar]
- 30.Octavia S, Lan R. (2006). Frequent recombination and low level of clonality within Salmonella enterica subspecies I. Microbiology (Reading), 152(Pt 4):1099–1108. [DOI] [PubMed] [Google Scholar]
- 31.Darling AE, Mau B, Perna NT. (2010). ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One, 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol, 19(5):455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gogol EB, Cummings CA, Burns RC, Relman DA. (2007). Phase variation and microevolution at homopolymeric tracts in Bordetella pertussis. BMC Genomics, 8:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheller EV, Cotter PA. (2015). Bordetella filamentous hemagglutinin and fimbriae: critical adhesins with unrealized vaccine potential. Pathog Dis, 73(8):ftv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carbonetti NH. (2016). Bordetella pertussis: new concepts in pathogenesis and treatment. Curr Opin Infect Dis, 29(3):287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AM, Guzmán CA, Walker MJ. (2001). The virulence factors of Bordetella pertussis: a matter of control. FEMS Microbiol Rev, 25(3):309–33. [DOI] [PubMed] [Google Scholar]
- 39.Preston A. (2016). The role of B. pertussis vaccine antigen gene variants in pertussis resurgence and possible consequences for vaccine development. Hum Vaccin Immunother, 12(5):1274–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leininger E, Roberts M, Kenimer JG, et al. (1991). Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence of mammalian cells. Proc Natl Acad Sci U S A, 88(2):345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigand MR, Pawloski LC, Peng Y, et al. (2018). Screening and genomic characterization of filamentous hemagglutinin-deficient Bordetella pertussis. Infect Immun, 86(4):e00869–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hegerle N, Guiso N. (2013). Epidemiology of whooping cough & typing of Bordetella pertussis. Future Microbiol, 8(11):1391–403. [DOI] [PubMed] [Google Scholar]
- 43.De Gouw D, Diavatopoulos DA, Bootsma HJ, et al. (2011). Pertussis: a matter of immune modulation. FEMS Microbiol Rev, 35(3):441–74. [DOI] [PubMed] [Google Scholar]
- 44.Hegerle N, Paris A-S, Brun D, et al. (2012). Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect, 18(9):E340–6. [DOI] [PubMed] [Google Scholar]
- 45.Guiso N. (2009). Bordetella pertussis and pertussis vaccines. Clin Infect Dis, 49(10):1565–9. [DOI] [PubMed] [Google Scholar]
- 46.Polak M, Zasada AA, Mosiej E, et al. (2019). Pertactin-deficient Bordetella pertussis isolates in Poland—a country with whole-cell pertussis primary vaccination. Microbes Infect, 21(3–4):170–5. [DOI] [PubMed] [Google Scholar]
- 47.Bodilis H, Guiso N. (2013). Virulence of pertactin-negative Bordetella pertussis isolates from infants, France. Emerg Infect Dis, 19(3):471–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hallander H, Advani A, Riffelmann M, et al. (2007). Bordetella pertussis strains circulating in Europe in 1999 to 2004 as determined by pulsed-field gel electrophoresis. J Clin Microbiol, 45(10):3257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shuel M, Jamieson FB, Tang P, et al. (2013). Genetic analysis of Bordetella pertussis in Ontario, Canada reveals one predominant clone. Int J Infect Dis, 17(6):e413–7. [DOI] [PubMed] [Google Scholar]
- 50.Octavia S, Sintchenko V, Gilbert GL, et al. (2012). Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010. J Infect Dis, 205(8):1220–4. [DOI] [PubMed] [Google Scholar]
- 51.Kurova N, Njamkepo E, Brun D, Tseneva G, Guiso N. (2010). Monitoring of Bordetella isolates circulating in Saint Petersburg, Russia between 2001 and 2009. Res Microbiol, 161(10):810–5. [DOI] [PubMed] [Google Scholar]
- 52.Orsi RH, Bowen BM, Wiedmann M. (2010). Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics, 11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Z, Octavia S, Luu LDW, et al. (2019). Pertactin-Negative and Filamentous Hemagglutinin-Negative Bordetella pertussis, Australia, 2013–2017. Emerg Infect Dis, 25(6):1196–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J, Zhang Y, Buboltz AM, et al. (2012). Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics, 13:545. [DOI] [PMC free article] [PubMed] [Google Scholar]