Abstract
UV damage endonuclease (Uve1p) from Schizosaccharomyces pombe was initially described as a DNA repair enzyme specific for the repair of UV light-induced photoproducts and proposed as the initial step in an alternative excision repair pathway. Here we present biochemical and genetic evidence demonstrating that Uve1p is also a mismatch repair endonuclease which recognizes and cleaves DNA 5′ to the mispaired base in a strand-specific manner. The biochemical properties of the Uve1p-mediated mismatch endonuclease activity are similar to those of the Uve1p-mediated UV photoproduct endonuclease. Mutants lacking Uve1p display a spontaneous mutator phenotype, further confirming the notion that Uve1p plays a role in mismatch repair. These results suggest that Uve1p has a surprisingly broad substrate specificity and may function as a general type of DNA repair protein with the capacity to initiate mismatch repair in certain organisms.
There are multiple processes by which DNA single-base mismatches are produced in cells. The most common of these is the misincorporation of nucleotides by DNA polymerase during replication. Mismatches can also arise following deamination of cytosine to uracil, forming U/G mispairs, or upon recombination between homeologous sequences (30). To correct this type of DNA anomaly, cells have developed several mechanisms for mismatch repair (MMR) that are essential for maintaining the integrity of the genome. In addition to mediating the repair of single base mismatches, MMR functions in maintaining the stability of simple DNA repeat tracts during replication including insertions caused by slippage loops in the primer strand and deletions caused by failure to repair loops in the template strand. In Escherichia coli, several different MMR pathways have been identified, and these have served as models in other organisms. The E. coli MutHLS (long-patch MMR) system repairs all single-base mispairs except C/C mismatches (28). Repair is initiated by binding of MutS and MutL to the mismatch, followed by a MutH-mediated incision of the nonmethylated DNA strand at hemimethylated GATC sites. The nicked strand is then degraded past the site of mismatch, and DNA polymerase fills in the resulting gap (28, 33, 37). The very short patch MMR system of E. coli recognizes G/T mismatches at sites where cytosine is methylated by the Dcm methylase and restores them to G/C pairs (25, 26, 49). This pathway requires the Vsr endonuclease in addition to MutS and MutL (18). A third type of mismatch correction in E. coli is mediated by the MutY protein, which initiates conversion of A/G mispairs to C/G pairs by an N-glycosylase and an associated apurinic/apyrimidinic lyase activity (32).
Biochemical and genetic studies have demonstrated that eukaryotes possess nick-directed MMR capabilities which appear to be similar in a number of respects to the E. coli long-patch MMR system (11, 29). Most of our current information concerning eukaryotic MMR has come from studies of Saccharomyces cerevisiae and humans. S. cerevisiae possesses numerous genes encoding proteins with similarities to E. coli MutS and MutL, but only a subset of these (MSH2, MSH6, PMS1, and MLH1) are thought to function as bona fide base MMR proteins (7). Other S. cerevisiae homologs, such as MSH3, MSH4, and MSH5, are thought to function in loop repair and/or recombination. In addition, non-MutS/MutL homologs such as RTH1 (RAD27) and EXO1 appear to function in loop repair (7). In humans, at least six different genes have been identified that encode proteins related to the bacterial MutS and MutL proteins. The products of inherited mutations in four of these genes (MSH2, MLH1, PMS1, and PMS2) are associated with hereditary nonpolyposis colon cancer and confer microsatellite sequence instability in cells containing such mutations (5, 13, 22, 31). Thus, in both S. cerevisiae and humans, there are a variety of proteins which likely function in several different pathways for the repair of single-base mismatches and loops for the maintenance of genomic stability.
The fission yeast Schizosaccharomyces pombe has also served as a useful model system for eukaryotic DNA repair systems. In contrast to S. cerevisiae and humans, much less is known about MMR in S. pombe. The mutL homolog pms1 has been recently identified (14, 36). Disruption of the S. pombe pms1 gene confers a spontaneous mutator phenotype, reduction of spore viability, and a increase in postmeiotic segregation, indicating that it plays a role in MMR (36). Two other genes, swi4 and swi8, are homologs of S. cerevisiae MSH3 and MSH2, respectively, and it has been proposed that they may mediate roles in loop repair and, in the case of swi8, correction of single-base mismatches (7). S. pombe exonuclease 1 (encoded by the exo1 gene) is a meiotically induced 5′-to-3′ double-stranded DNA exonuclease, is a homolog of the S. cerevisiae EXO1 gene product, and has been proposed to play a role in mutation avoidance and MMR (38–40). Genetic analysis of meiotic recombination events has indicated the existence of at least two pathways responsible for MMR in S. pombe: a major, long-patch MMR system (mediated by msh1 and pms1) which recognizes all mismatch combinations except C/C; and a minor, short-patch MMR system which recognizes all combinations, including C/C mismatches (34, 35). Further support for these observations was provided by the discovery of two distinct mismatch-binding activities in S. pombe crude cell extracts (15). Recently, the S. pombe nucleotide excision repair (NER) genes rhp14, swi10, and rad16 (homologs of the S. cerevisiae RAD14, RAD10, and RAD1 genes, respectively) have been identified as components of the short-patch MMR system and function independently of msh2 pms1 (16). Taken together, the available genetic and limited biochemical data suggest that S. pombe possesses multiple pathways for conducting MMR.
We (4, 10, 17) and others (45, 48) have described an alternative excision repair pathway which exists in S. pombe and was proposed to be highly specific for cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs), the major toxic and mutagenic UV light-induced DNA photoproducts. UV damage endonuclease (Uve1p; also called UVDE), the enzyme initiating this alternative excision repair pathway, incises duplex DNA immediately 5′ to the sites of damage (4, 20). The S. pombe gene uve1, encoding this protein, as well as two similar genes from Neurospora crassa and Bacillus subtilis have been identified, indicating that this repair pathway exists in both prokaryotes and eukaryotes (8, 41, 42). Recently, we reported the overexpression, purification, and initial enzymological characterization of GΔ228-Uve1p, a fully active, truncated form of Uve1p (20). Because CPDs and 6-4PPs differ significantly with respect to the structural distortions that they induce in duplex DNA, it seemed reasonable to expect that Uve1p might recognize other types of DNA damage. A recent report on the thermodynamic and base-pairing properties of DNA dodecamer duplexes containing CPDs and 6-4PPs at sites of adjacent thymines (TT) indicate that proper Watson-Crick base pairing with the opposite AA is disrupted for both of these photoproducts (19). These observations raised the possibility that Uve1p recognizes single base mismatches resulting in disruption of normal Watson-Crick base pairing in duplex DNA.
In the present study, we report a novel mismatch endonuclease activity in S. pombe mediated by Uve1p. We present biochemical and genetic evidence indicating that Uve1p, an enzyme previously considered to be involved exclusively in the repair of UV-induced DNA photoproducts, is an initiating enzyme in an S. pombe MMR pathway. These results suggest that Uve1p (i) has a broad substrate specificity range, (ii) may be the initiating repair enzyme for a general excision repair pathway, and (iii) is likely to be a component of MMR pathways that exist in both prokaryotes and eukaryotes.
MATERIALS AND METHODS
Strains and vectors.
S. cerevisiae DY150 (Clontech) was used for protein expression. The S. cerevisiae expression vector pYEX4T-1 was obtained from Clontech. S. pombe strains used in this study were 972 (h−S) (24), PRS301 (h−S pms1::ura4+) (36), and Sp30 (h−S ade6-210 leu-32 ura4-D18) (9). Sp362 (h−S ade6-210 leu1-32 ura4-D18 uve1::ura4+) was constructed by transforming Sp30 with a linearized, genomic uve1+ fragment derived from pgUV2 (8) in which nucleotides 215 (EcoRI) to 1045 (ClaI) of uve1+ were replaced with the ura4+ gene. Extracts of Sp362 contained no detectable Uve1p activity against CPD-30mer (data not shown). Cultures were grown in pombe minimal medium (24) with glutamate (3.75 g/liter) replacing ammonium chloride as the nitrogen source (12) supplemented with 150 mg each of adenine, leucine, and uracil per liter (PMALUg). Solid medium was prepared by addition of agar (20 g/liter). l-Canavanine sulfate was sterilized prior to addition to the medium.
Purification of Uve1p and other mismatch endonucleases.
GFL-Uve1p and GΔ228-Uve1p (full-length and truncated Uve1p fused to glutathione S-transferase [GST]) were cloned and expressed in the pYEX4T-1 S. cerevisiae expression system to generate N-terminal GST-Uve1p fusion proteins as previously described (20). GΔ228-Uve1p was purified by glutathione-Sepharose affinity chromatography as previously described (20). GFL-Uve1p was found to rapidly lose activity following this purification step and was subsequently used in experiments as a crude extract preparation which showed greater stability. S. cerevisiae cells transformed with vector alone (expressing the GST tag only) were subjected to a parallel purification procedure, with the resulting GST used as a control for possible contaminating activities copurified from the expression system. Thrombin cleavage of GΔ228-Uve1p to generate Δ228-Uve1p followed by purification was carried out as previously described (20). Purified mismatch repair endonuclease, E. coli endonuclease V (44), was a gift from Yoke Wah Kow (Atlanta, Ga.).
Uve1p substrate preparations.
The CPD-30mer Uve1p substrate (20) containing a centrally embedded, cis-syn TT CPD was a gift from John-Stephen Taylor (St. Louis, Mo.). All other oligonucleotide substrates (Table 1) for mismatch endonuclease experiments were synthesized by Operon, Inc. (Alameda, Calif.), or IDT, Inc. (Coralville, Iowa). All oligonucleotides were gel purified and subjected to DNA sequence analysis for sequence confirmation. Oligonucleotides were 5′ end labeled with polynucleotide kinase (PNK) by using 50 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham) as previously described (4). 3′-end-labeled oligonucleotides were prepared by incubating 10 pmol of the indicated oligonucleotide with 10 U of terminal deoxynucleotidyltransferase (TdT; Promega) and 50 μCi of [α-32P]ddATP (3,000 Ci/mmol; Amersham) as previously described (4).
TABLE 1.
Base mismatch- and CPD-containing oligonucleotides used in this study
| Oligonucleotide | Sequence | Strand designation(s) |
|---|---|---|
| XY-31mera | 5′ GTACCCGGGGATCCTCCXAGTCGACCTGCA 3′ | GX, AX, TX, CX (X = G, A, T, or C) |
| 3′ CATGGGCCCCTAGGAGGYTCAGCTGGACGT 5′ | GY, AY, TY, CY (Y = G, A, T, or C) | |
| CX/AY-41mer | 5′ CGTTAGCATGCCTGCACGAACTAAGCAATTCGTAATGCATT 3′ | CX |
| 3′ GCAATCGTACGGACGTGCTTAATTCGTTAAGCATTACGTAA 5′ | AY | |
| CPD-30merb | 5′ CATGCCTGCACGAAT^TAAGCAATTCGTAAT 3′ | 30 D |
| 3′ GTACGGACGTGCTTA ATTCGTTAAGCATTA 5′ | 30 C |
Series of 16 different duplex oligonucleotides containing all possible base pair/mispair combinations between G, A, T, and C. In text, * denotes labeled strand (e.g., *CX/AY-31mer corresponds to C/A mismatch with the C-containing X strand as the labeled strand).
Contains a CPD designated T^T.
Uve1p activity assays.
Reactions with GΔ228-Uve1p were carried out by incubating approximately 100 fmol of labeled oligonucleotide substrate with 100 to 150 ng of purified GΔ228-Uve1p in 20 mM HEPES (pH 6.5)–10 mM MgCl2–1 mM MnCl2–150 mM NaCl for 20 min at 37°C (10 to 20 μl, final volume). Reactions with crude preparations of GFL-Uve1p were carried out with 20 to 30 μg of cell extract incubated with the appropriate substrate in 20 mM HEPES (pH 7.5)–100 mM NaCl–10 mM MgCl2–1 mM MnCl2 at 37°C for 20 min. The reaction products were processed by extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitation, resuspension, and analysis on 20% denaturing (7 M urea) polyacrylamide (DNA sequencing) gels as previously described (20). The DNA species corresponding to the uncleaved substrate and Uve1p-mediated DNA strand scission products were analyzed and quantified by PhosphorImager (Molecular Dynamics model 445SI) analysis and autoradiography.
Kinetics experiments were carried out with 331 nM GΔ228-Uve1p and 5 to 50 nM *CX/AY-31mer under otherwise standard reaction conditions (described above) for 0 to 5 min at 37°C. The apparent Km and Kcat values were determined from Lineweaver-Burk plots of averaged data from three individual experiments.
5′-terminal analysis.
GΔ228-Uve1p was incubated with 3′-end-labeled *CX/AY-31mer under standard reaction conditions at 37°C for 20 min. The ethanol-precipitated reaction products were incubated with 10 U of calf intestinal phosphatase (CIP; Promega) at 37°C for 30 min or with 10 U of T4 PNK (New England Biolabs) and 50 pmol of ATP as previously described (4). The reaction products were analyzed on 20% denaturing polyacrylamide gels as described above for Uve1p activity assays. Differences in electrophoretic mobilities between kinase-treated and untreated DNA strand scission products indicated the presence or absence of a preexisting 5′-phosphoryl group (4).
3′-terminal analysis.
To determine the chemical nature of the 3′ terminus of GSTΔ228-Uve1p-mediated DNA strand scission products, 5′-end-labeled *CX/AY-31mer was incubated with GΔ228-Uve1p as described above. The ethanol-precipitated, resuspended reaction products were then treated with 10 U of TdT and ddATP as previously described (4). Samples were processed and analyzed on polyacrylamide gels as described above for 5′-terminal analysis.
Establishment of optimal pH for mismatch endonuclease activity.
To determine the pH optimum for Uve1p-mediated mismatch cleavage, 100 fmol of 3′-end-labeled *CX/AY-31mer was incubated with approximately 100 ng of GΔ228-Uve1p plus 10 mM MgCl2 and 1 mM MnCl2 in 20 mM reaction buffers varying in pH sodium citrate (pH 3.0 to 6.0), HEPES KOH (pH 6.5 to 8.0), and sodium carbonate (pH 9.0 to 10.6). The reaction products were analyzed on a 20% denaturing polyacrylamide gel, and the optimal pH was calculated as previously described for Uve1p cleavage of CPD-30mer (20).
Substrate competition assay.
End-labeled *CX/AY-31mer was generated by annealing 3′-end-labeled CX with unlabeled strand AY. Unlabeled nonspecific (nonmismatch) competitor GX/CY-31mer was made by annealing strand GX to strand CY, resulting in a duplex oligonucleotide with a G/C base pair instead of a C/A mispair. Unlabeled CX/AY-31mer, a mismatch-containing specific competitor, was generated as described above. CPD-30mer, a well-characterized substrate for Uve1p, was used as an additional unlabeled, putative specific competitor. 3′-end-labeled *CX/AY-31mer (0.1 pmol) was incubated with 100 ng of purified GΔ228-Uve1p and increasing amounts (0.1 to 2.0 pmol) of either putative specific (CX/AY-31mer or CPD-30mer) or nonspecific (GX/CY-31mer) competitor. The competition reactions were processed and analyzed on 20% denaturing gels as described above. The DNA species corresponding to the uncleaved *CX/AY-31mer and the DNA strand scission products were quantified by PhosphorImager (Molecular Dynamics model 445SI) analysis.
Colony formation assays for canavanine resistance.
To determine sensitivity to l-canavanine, 10 ml of PMALUg was inoculated with 100 μl of the indicated saturated culture and grown to mid-log phase at 25°C; 200 cells were plated onto PMALUg plates with various concentrations of l-canavanine sulfate (0, 0.075, 0.22, 0.75, 2.2, 7.5, 22, and 75 μg/ml), and incubated at 30°C. Colonies were counted after 4 days, and viability was normalized against the 0-g/ml plate for each strain. Colony formation assays (mutation frequency) were conducted for each strain by plating 107 cells from saturated cultures onto 24 PMALUg plates supplemented with l-canavanine sulfate (75 μg/ml). Colonies were counted after 8 days of incubation at 30°C. Mean mutation frequencies were calculated by the method of the median (23).
RESULTS
GΔ228-Uve1p recognizes 12 possible base mismatch combinations.
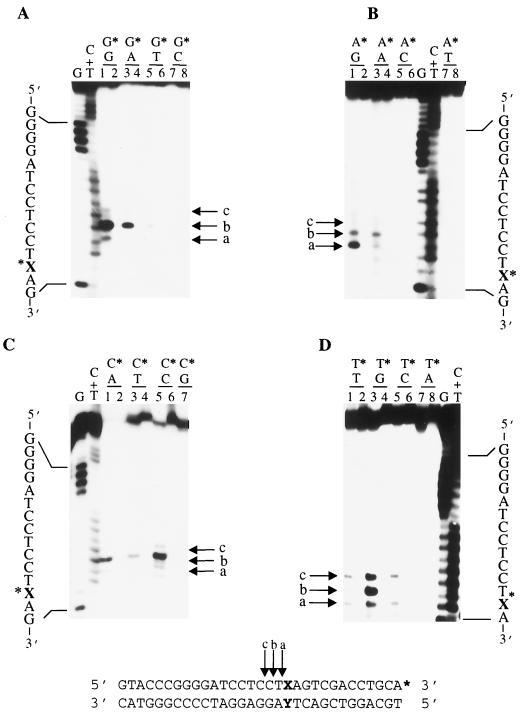
Previously, we reported that an overexpressed, GST-tagged version of Uve1p (GΔ228-Uve1p) containing an N-terminal 228-amino-acid deletion is active on UV photoproducts (CPDs and 6-4PPs) and stable in a purified form (20). In contrast, overexpressed GST-tagged, full length Uve1p (GFL-Uve1p) was found to be active and stable in crude preparations but rapidly lost activity following purification. Because of substantial structural differences between CPDs and 6-4PPs, it is not obvious what features of damaged DNA Uve1p recognizes. One possibility is that Watson-Crick base pairing is disrupted for the 3′ pyrimidines in both CPDs and 6-4PPs, suggesting that Uve1p might target its activity to mispaired bases in duplex DNA (19). We therefore investigated the ability of purified GΔ228-Uve1p to cleave duplex oligonucleotides containing all possible combinations of single-base mispairs embedded within the same flanking sequence context. For these studies, we used a collection of mismatch-containing oligonucleotides (series XY-31mer) which were designed so as to generate all possible mismatch combinations (Table 1). Strands GX, AX, TX, and CX were 3′ end labeled and then annealed to strand GY, AY, TY, or CY prior to incubation with purified GΔ228-Uve1p. Reaction products were analyzed on DNA sequencing-type gels (Materials and Methods). The ability of GΔ228-Uve1p to cleave all 12 possible mispair combinations is shown in Fig. 1. No DNA strand cleavage was observed for duplex substrates containing normal Watson-Crick G/C or A/T base pairs.
FIG. 1.
GΔ228-Uve1p recognizes 12 different base mismatch combinations. The 3′-end-labeled oligonucleotide series X/Y-31mer (sequence given at bottom; asterisk indicates labeled strand and labeled terminus) was used to assess Uve1p cleavage activity on 16 different base pair and base mispair combinations (Table 1). Base mispairs are indicated above numbered lanes, with asterisks denoting bases on the labeled strand for G series (A), A series (B), C series (C), and T series (D) treated with purified GΔ228-Uve1p (odd-numbered lanes) or mock reactions (even-numbered lanes). Reaction products were analyzed on DNA sequencing-type gels. Arrows indicate Uve1p cleavage sites immediately (arrow a) and one (arrow b) and two (arrow c) nucleotides 5′ to the mismatch site. G and C+T base-specific chemical cleavage DNA sequencing ladders were run in adjacent lanes as nucleotide position markers.
The sites of GΔ228-Uve1p-mediated mismatch-specific DNA cleavage were identified in each case by comparing the electrophoretic mobilities of the DNA strand scission products to those of a DNA sequencing ladder obtained by base-specific chemical cleavage. Arrows a, b, and c indicate the DNA strand scission products corresponding to cleavage by GΔ228-Uve1p immediately (position 0), one (position −1), or two (position −2) nucleotides 5′ to the site of the mismatch, respectively (Fig. 1). These sites of GΔ228-Uve1p-mediated endonucleolytic cleavage were confirmed in similar experiments using 5′-end-labeled GX, AX, TX, and CX strands in the mismatch substrates (not shown). In addition, GFL-Uve1p (in crude cell extracts) recognized and cleaved *CX-AY-31mer in the same manner as GΔ228-Uve1p (Fig. 2B). The preferred sites of cleavage and the efficiency with which each mismatch is recognized by GΔ228-Uve1p is variable and depends on the type of base mispair presented to the enzyme. Within the sequence context examined, GΔ228-Uve1p exhibited strong cleavage at the *C/C (asterisk denotes labeled strand base) site and at *C/A and *G/G sites (30 to 40% cleavage of substrate relative to that observed for CPD-30mer), moderate cleavage at *G/A, *A/G, and *T/G sites (10 to 25% cleavage relative to CPD-30mer), and weak cleavage at *G/T, *A/A, *A/C, *C/T, *T/T, and *T/C sites (5 to 10% cleavage relative to CPD-30mer). These differences in the extent of cleavage were reproducible and observed in three separate experiments. These results suggest that the GΔ228-Uve1p mismatch endonuclease activity may have a preference for certain base mismatch combinations (e.g., *C/A) over others (e.g., *T/C). However, these experiments do not rule out the possibility that the extent of cleavage observed is also influenced by the sequence flanking the mismatch.
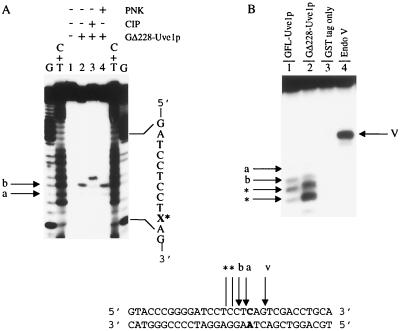
FIG. 2.
Nature of Uve1p-generated DNA strand scission products and activity of full-length Uve1p. (A) Analysis of 5′ termini of Uve1p-generated DNA cleavage products with *CX/AY-31mer. 3′-end-labeled oligonucleotide with C/A mismatch (sequence on bottom) was reacted with GΔ228-Uve1p and then further treated with PNK or CIP as indicated. Lane 1 represents buffer treatment only. X* indicates base mismatch site. Arrows a and b indicate sites of Uve1p cleavage. (B) Full-length Uve1p possesses mismatch endonuclease activity. 5′-end-labeled duplex *CX/AY-31mer was incubated with crude extracts of cells expressing either GFL-Uve1p (lane 1) or GΔ228-Uve1p (lane 2) and cells expressing the GST tag alone (lane 3) or with E. coli endonuclease (Endo) V, a known mismatch endonuclease (lane 4). Arrows indicate cleavage sites immediately (arrow a) and one nucleotide (arrow b) 5′ to the mismatch site. Arrow V indicates E. coli endonuclease V cleavage 3′ to the mismatch site and was used as a position reference. Bands below arrows (indicated by asterisks) correspond to shortened products due to a weak 3′-to-5′ exonuclease activity present in the Uve1p preparations (see text for details). Reaction products on DNA sequencing-type gels were analyzed as described for Fig. 1.
Kinetic studies were carried out with GΔ228-Uve1p and *CX/AY-31mer (Materials and Methods). The Km and Kcat values were determined to be 390 nM and 2.2 min−1, respectively. In comparison to the kinetic parameters of GΔ228-Uve1p for CPD-30mer (20), the Km for *CX/AY-31mer is 8-fold higher and the Kcat is 10-fold higher, indicating that although GΔ228-Uve1p binding to the mismatch-containing substrate is weaker than binding to a CPD-containing substrate, the catalytic efficiency of the enzyme is higher for the *CX/AY-31mer substrate.
Nature of the Uve1p mismatch endonuclease-generated DNA cleavage products.
Uve1p has been shown to incise DNA containing CPDs and 6-4PPs directly 5′ to the photoproduct site, generating products containing 3′-hydroxyl and 5′-phosphoryl groups (4). We wished to determine whether similar 3′ and 5′ termini were produced following Uve1p-mediated cleavage of base mismatch-containing substrates. To study this, the DNA strand scission products generated by GΔ228-Uve1p cleavage of 3′-end-labeled *CX/AY-31mer (CX strand labeled) (Table 1) were further treated with CIP, which removes 5′-terminal phosphoryl groups from substrate DNA. The major sites of Uve1p-mediated DNA cleavage relative to the base mispair site were found to be at positions 0 and −1 (Fig. 2A, lane 2). CIP treatment of these DNA cleavage products resulted in species that had retarded electrophoretic mobilities compared to non-CIP-treated DNA cleavage products, indicating a decrease in charge corresponding to removal of 5′-terminal phosphoryl groups (Fig. 2A, lanes 2 and 3). In addition, GΔ228-Uve1p mismatch endonuclease-generated DNA cleavage products were resistant to phosphorylation by PNK, an expected result if the 5′ termini already contain phosphoryl groups (Fig. 2A, lane 4). Electrophoretic mobility shift analysis utilizing 5′-end-labeled *CX/AY-31mer, TdT, and [α-32P]ddATP) resulted in addition of a single ddAMP to the 3′ end of GΔ228-Uve1p-generated DNA cleavage products and indicates the presence of a 3′-hydroxyl terminus (data not shown). We conclude from these results that the 3′ and 5′ termini of the products of GΔ228-Uve1p-mediated cleavage of substrates containing single-base mismatches are identical to those generated following cleavage of substrates containing CPDs or 6-4PPs.
To verify that the Uve1p mismatch endonuclease activity observed was not the result of trace endonucleolytic contamination from the S. cerevisiae expression system and to determine whether full-length Uve1p was also capable of mismatch endonuclease activity, extracts from cells overexpressing GFL-Uve1p, GΔ228-Uve1p, and GST tag alone were tested for their abilities to cleave 5′-end-labeled *CX/AY-31mer. Both GFL-Uve1p and GΔ228-Uve1p cleaved the base mismatch-containing substrate at positions 0, −1, and −2 (Fig. 2B). We also observed a weak 3′-to-5′ exonucleolytic activity associated with both crude GFL-Uve1p preparations and purified GΔ228-Uve1p which shortened the Uve1p-mediated cleavage products by one to three nucleotides (Fig. 2B, lanes 1 and 2). These shorter products are not due to additional cleavages by Uve1p mismatch endonuclease activity, as they are not observed in identical experiments with 3′-end-labeled substrates (not shown). Purified Δ228-Uve1p obtained following thrombin cleavage of the GST tag also possessed mismatch endonuclease activity (data not shown). In contrast, no cleavage of mismatch-containing substrates was observed when extracts from cells transfected with vector expressing only the GST tag were tested. We conclude from these results that GFL-Uve1p and its more stable, truncated version, GΔ228-Uve1p, both possess mismatch endonuclease activities.
GΔ228-Uve1p mismatch endonuclease and GΔ228-Uve1p UV photoproduct endonuclease exhibit similar properties and compete for the same substrates.
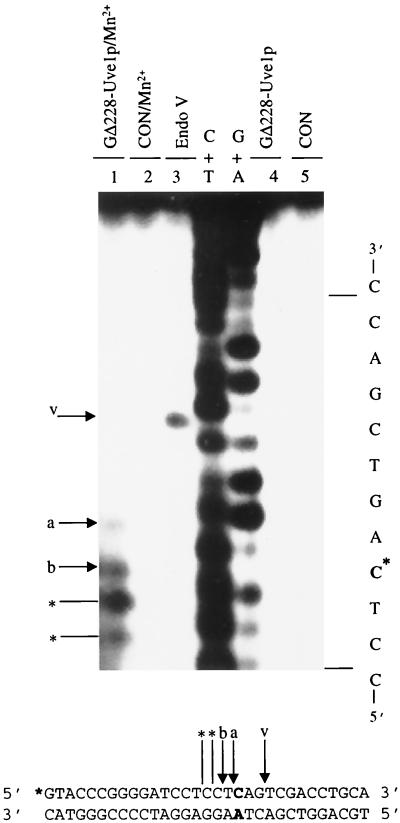
We have previously reported that GΔ228-Uve1p requires divalent cations for activity and exhibits optimal activity against UV photoproducts in the presence of 10 mM MgCl2 and 1 mM MnCl2 (19). Omission of divalent cations from the reaction buffer resulted in abolishing GΔ228-Uve1p mismatch endonuclease activity on 5′-end-labeled *CX/AY-31mer (Fig. 3). The pH optimum for GΔ228-Uve1p mismatch endonuclease activity on this same substrate was found to be 6.5 (not shown), which corresponds to the pH where optimal activity is observed against UV photoproducts (20).
FIG. 3.
GΔ228-Uve1p requires divalent cations for mismatch recognition. 5′-end-labeled duplex *CX/AY-31mer was incubated with GΔ228-Uve1p (lanes 1 and 4) or buffer only (control [CON]; lanes 2 and 5) in the presence (lanes 1 and 2) or absence (lanes 4 and 5) of Mn2+. C* indicates base mismatch site. Arrows a and b indicate Uve1p cleavage positions. E. coli endonuclease (Endo) V-reacted oligonucleotide (arrow v, lane 3) and C+T and G+A sequencing ladders included as nucleotide position markers are marked. Bands below arrow b (indicated by asterisks) correspond to shortened products due to 3′-to-5′ exonuclease activity (described in the legend to Fig. 2).
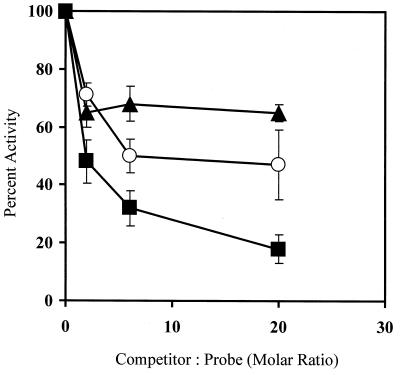
To further confirm that the mismatch endonuclease activity was mediated by GΔ228-Uve1p, a substrate competition experiment was performed with CPD-30mer, a known Uve1p substrate which contains a centrally located UV photoproduct (CPD). Addition of increasing amounts of unlabeled CPD-30mer resulted in a significant, concentration-dependent decrease in GΔ228-Uve1p-mediated mismatch endonuclease activity against 3′-end-labeled *CX/AY-31mer (C/A mispair) (Fig. 4). In contrast, increasing amounts of the undamaged GX/CY-31mer (G/C base pair) had only a modest inhibitory effect and did not increase with increasing amounts of added oligonucleotide, indicating nonspecific binding to Uve1p within this concentration range. Increasing amounts of added, unlabeled CX/AY-31mer showed a moderate inhibition but was not as effective as CPD-30mer. The effective competition by CPD-30mer for mismatch endonuclease activity suggests that both base mismatch and UV photoproduct endonuclease activities are associated with GΔ228-Uve1p.
FIG. 4.
GΔ228-Uve1p mismatch endonuclease and GΔ228-Uve1p UV photoproduct endonuclease compete for the same substrates. GΔ228-Uve1p was incubated with 3′-end-labeled duplex *CX/AY-31mer (Table 1) in the presence of increasing amounts of unlabeled duplex CPD-30mer (squares), duplex GX/CY-31mer (triangles), or duplex CX/AY-31mer (circles). The Uve1p-mediated DNA cleavage products were analyzed on DNA sequencing gels, and the extent of strand scission was quantified by PhosphorImager analysis (Materials and Methods). Uve1p activity is expressed as percentage of the cleavage observed relative to that observed in the absence of any competitor (defined as 100% activity). The error bars indicate the mean ± standard deviation from three separate experiments.
Uve1p incises only one strand of a duplex containing a base mismatch.
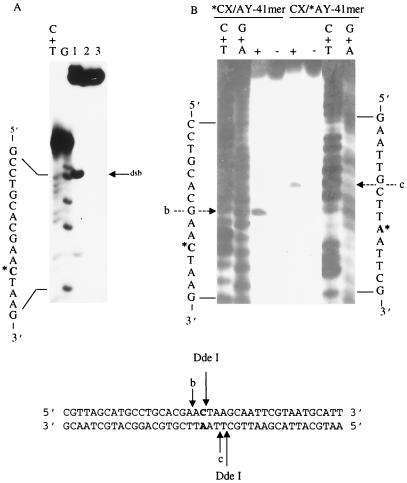
Since Uve1p recognizes all possible base mismatch combinations, it was of interest to determine whether the enzyme could incise both strands on the same molecule, resulting in a DNA double-strand break. To investigate this, an oligonucleotide (*CX/AY-41mer) was designed such that the base mispair was placed in the center of the oligonucleotide. GΔ228-Uve1p was incubated with 3′-end-labeled *CX/AY-41mer under standard conditions, and the DNA strand scission products were analyzed on both nondenaturing and denaturing gels (Fig. 5). In the event that GΔ228-Uve1p created a DNA double-strand break by incising 5′ to the base mismatch site on the two complementary strands, the resulting products would be similar in electrophoretic mobility to those created by the restriction enzyme DdeI (which cleaves adjacent to the mismatch) when analyzed on a nondenaturing polyacrylamide gel. In contrast, if GΔ228-Uve1p incises on either (but not both) complementary strand, then the resulting product would be a full-length duplex containing a single-strand nick which would comigrate with uncut duplex *CX/AY-41mer on a nondenaturing gel. Nondenaturing gel analysis of GΔ228-Uve1p-treated *CX/AY-41mer generated a product with an electrophoretic mobility identical to that of the untreated duplex with no products detected, corresponding to those created by a double-strand break (Fig. 5A). Denaturing gel analysis revealed a GΔ228-Uve1p-generated DNA strand scission product resulting from a single-strand break of the labeled strand of either *CX/AY-41mer or CX/*AY-41mer. Together with the nondenaturing gel analysis, these results indicate that within the GΔ228-Uve1p substrate population, nicks occur on one or the other strand but not both strands (Fig. 5B). We conclude from these results that GΔ228-Uve1p is capable of nicking only one of the two strands containing a base mismatch and does not make double-strand breaks in duplex DNA.
FIG. 5.
Uve1p incises only one strand of a duplex containing a base mismatch. (A) 3′-end-labeled *CX/AY-41mer was incubated with restriction enzyme DdeI (lane 1), GΔ228-Uve1p (lane 2), or buffer (lane 3). The reaction products were analyzed on a nondenaturing gel as described in the text for the presence of DNA double-strand break products (arrow dsb). Arrows b and c indicate the primary cleavage site for Uve1p on this substrate. (B) 3′-end-labeled *CX/AY-41mer or CX/*AY-41mer was incubated with GΔ228-Uve1p (+ lanes) or buffer (− lanes) and analyzed on denaturing, DNA sequencing-type gels as described in the text. Arrows b and c indicate positions of major Uve1p cleavage events relative to the mismatched base (asterisk) position. G+A and C+T base-specific sequencing ladders are included in outside lanes as nucleotide position markers.
uve1 null mutants exhibit a mutator phenotype.
We have examined the spontaneous mutation rate of uve1::ura4+ disruption mutants as assayed by the ability to form colonies resistant to the toxic arginine analog l-canavanine. Uptake of l-canavanine in S. pombe is mediated by an arginine permease encoded by the can1+ gene (12). Mutations in can1+ eliminate the uptake of l-canavanine, and mutant cells are able to form colonies on medium supplemented with l-canavanine, whereas wild-type cells cannot. We have compared the rates of spontaneous mutagenesis at the can1+ locus in uve1::ura4+ disruption mutants (Sp362) to those of both a negative control (wild-type strain 972) and a positive control, pms1::ura4+ (Materials and Methods). The pms1 gene product is a homolog of E. coli MutL, and loss of pms1 causes a strong mitotic mutator phenotype and increased postmeiotic segregation (36).
To determine the relative sensitivity of each yeast strain to l-canavanine, 200 cells from mid-log-phase cultures were plated onto PMALUg plates supplemented with increasing concentrations of l-canavanine. The strains were equally sensitive to l-canavanine (not shown). All strains were viable in the presence of lower concentrations of l-canavanine up to and including 2.2 μg/ml, while concentrations higher than this were toxic to all strains. However, the colonies which grew in the presence of 2.2 μg of l-canavanine per ml were smaller in diameter than the colonies which grew in the presence of lower concentrations.
The mean mutation rate of each of these strains was examined by fluctuation analysis. Single colonies from PMALUg medium were used to inoculate liquid cultures which were grown to saturation; 107 cells were plated onto PMALUg plates containing 75 μg of l-canavanine sulfate per ml, and the canavanine-resistant colonies were counted after incubation at 30°C for 8 days. We determined the number of colonies on 12 separate plates for each of three experiments; the data are summarized in Table 2. Both uve::ura4+ and pms::ura4+ strains showed an elevated number of resistant colonies compared to the wild type. The mutation rates (Table 2) for wild-type, uve1::ura4+, and pms::ura4+ strains were calculated by the method of the median (23). The uve1::ura4+ mutants have a mutation rate approximately 6.5-fold higher than that of the wild type and 2-fold lower than that of the pms::ura4+ strain, indicating that loss of Uve1p confers a mutator phenotype upon cells.
TABLE 2.
Spontaneous mutation rates of uve1 and pms1 null mutants
| Genotype | Distribution of canavanine-resistant colo-nies/plate
|
Median no. of colonies/ 107 cells | Calculated mutation frequency (mean ± SE) | |||
|---|---|---|---|---|---|---|
| 0–2 | 3–34 | 35–86 | >86 | |||
| Wild type | 18 | 16 | 2 | 0 | 2.5 | 1.5 × 10−7 ± 2.5 × 10−8 |
| uve1::ura4+ | 4 | 14 | 8 | 10 | 34.5 | 9.7 × 10−7 ± 4.2 × 10−8 |
| pms1::ura4+ | 0 | 8 | 10 | 18 | 86.5 | 2.0 × 10−6 ± 5.0 × 10−8 |
DISCUSSION
It has been recognized for a number of years that the repair of the major UV photoproducts, CPDs and 6-4PPs, in S. pombe is mediated by more than one excision repair pathway. The evidence for this was initially based on the finding that S. pombe NER mutants were still proficient in the removal of both CPDs and 6-4PPs from genomic DNA (27). The discovery of the Uve1p-mediated excision repair pathway provided a biochemical basis for these observations (4, 41). Subsequent investigations showed that mutants lacking NER and Uve1p were highly sensitive to UV light and were deficient for removal of CPDs and 6-4PPs from DNA (48). The discovery that Uve1p could recognize both CPDs and 6-4PPs, DNA lesions which induce quite different structural distortions in duplex DNA, raised the issue of the structural basis for DNA damage recognition by Uve1p. One possible explanation came from a recent thermodynamic and base-pairing study that compared structures of matched and mismatched DNA dodecamer duplexes containing cis-syn CPD and 6-4PP of TT (19). The covalent interaction between adjacent bases in the dimer pair prevents the 3′ pyrimidine from pairing correctly with the complementary base. Such disruption of Watson-Crick base pairing provided a possible explanation for the structural distortion recognized by Uve1p. Hence we tested the ability of Uve1p to incise DNA containing all 12 possible base mismatch combinations and found that each was a substrate for this enzyme.
The finding that Uve1p recognizes all potential DNA base mispair combinations indicates that, in addition to its UV photoproduct cleavage activity, it is a diverse mismatch endonuclease with broad substrate specificity. In this regard, Uve1p is similar to E. coli endonuclease V (43), an S. cerevisiae and human “all-type” mismatch endonuclease (6, 46), and calf thymus topoisomerase I (47), which also recognize all potential base mismatch combinations. These enzymes incise DNA at each of the 12 base mispairs with variable efficiencies and either to the 5′ (human all-type mismatch endonuclease) or 3′ (E. coli endonuclease V) sides of a mismatch. Uve1p shows a preference for *C/C and *C/A mispairs, a property similar to that of the human all-type mismatch endonuclease (46). In contrast, the strong preference of Uve1p for *G/G mispairs is a property which distinguishes Uve1p from all other mismatch endonucleases identified to date. The biochemical properties of Uve1p-mediated mismatch cleavage and the spontaneous mutator phenotype displayed by uve1 null mutants suggest that Uve1p is involved in MMR in vivo. Uve1p-generated incision 5′ to the base mismatch site could be followed by a 5′-to-3′ exonuclease activity such as that mediated by S. pombe exonuclease I (40) or the FEN-1 homolog Rad2p (1), followed by resynthesis and ligation.
S. pombe possesses at least two distinct MMR systems; the relationship of Uve1p to either of these is not known at present, nor is it known whether Uve1p functions in a distinct MMR system. The proposed major MMR pathway does not recognize C/C mismatches and has relatively long (approximately 100-nucleotide) repair tracts (34). Uve1p is thought to participate in a relatively short patch repair process which utilizes Rad2p (a FEN-1 homolog) DNA polymerase δ, DNA ligase, and accessory factors (1, 2). Based on these properties, it is unlikely that Uve1p is involved in a long-tract MMR system. The second, presumably less frequently utilized pathway recognizes all potential base mismatch combinations and has a repair tract length of about 10 nucleotides (34). Recently, this second pathway, which recognizes C/C mismatches, was shown to be mediated by the NER proteins Rhp14p, Swi10p, and Rad16p, which function in damage recognition and incision (16). Based on these observations, our results suggest that Uve1p may mediate a third, distinct MMR system in S. pombe.
To further address the role of Uve1p in MMR, we tested whether a mutant lacking Uve1p activity had an increased spontaneous mutation frequency, a predictable phenotype associated with MMR deficiencies (22). A uve1::ura4+ disruption strain was found to possess a mutator phenotype in the l-canavanine resistance assay, although at levels slightly lower than that observed for a pms1::ura4+ disruption strain. In the mutation fluctuation analysis, a wider range of mutant colonies was observed for uve1::ura4+ than for pms1::ura4+, suggesting that the pathways leading to mutation due to elimination of uve1 and pms1 are likely to be mechanistically different.
What is the structural basis for lesion recognition by Uve1p? Previous studies with Uve1p have focused exclusively on its role in the repair of UV light-induced DNA damage, resulting in the notion that this enzyme functions in the repair of UV photoproducts exclusively. In view of the findings reported here, it might be desirable to consider renaming this enzyme to reflect its overall properties. The results of this study clearly indicate a much broader involvement of Uve1p in S. pombe DNA repair and suggest that many other types of DNA lesions may be recognized by this versatile repair protein. For example, we have recently found that Uve1p recognizes and incises DNA substrates containing uracil, dihydrouracil, cis-platinum-induced adducts, as well as small base bulges (3, 21). The molecular basis for substrate recognition by Uve1p is not obvious but could in part be due to disruption of normal Watson-Crick base pairing and the corresponding changes expected in the electronic characteristics of the major and minor grooves of B-DNA. Structural studies of Uve1p associated with its substrates would provide important information regarding how this enzyme recognizes and accesses DNA damage.
ACKNOWLEDGMENTS
We thank Angela Avery, Yoke Wah Kow, and Gerald Shadel for helpful discussions.
This work was supported by NIH grants CA73041 (P.W.D.), CA72647 (G.A.F.), and ES07940 (G.A.F. and S.D.) and by Medical Research Council of Canada grant MT-14352 (S.D.) S.D. is a Cancer Care Ontario Scientist. J.L.A.F. is a Queen’s University R. S. McLaughlin Fellow.
REFERENCES
- 1.Alleva J L, Doetsch P W. Characterization of Schizosaccharomyces pombe Rad2 protein, a FEN-1 homolog. Nucleic Acids Res. 1998;26:3645–3650. doi: 10.1093/nar/26.16.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alleva, J. L., and P. W. Doetsch. Unpublished data.
- 3.Avery, A. M., B. Kaur, J. S. Taylor, J. A. Mello, J. M. Essigmann, and P. W. Doetsch. Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 4.Bowman K K, Sidik K, Smith C A, Taylor J S, Doetsch P W, Freyer G A. A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res. 1994;22:3026–3032. doi: 10.1093/nar/22.15.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 6.Chang D Y, Lu A L. Base mismatch-specific endonuclease activity in extracts from Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4761–4766. doi: 10.1093/nar/19.17.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crouse G F. Mismatch repair systems in Saccharomyces cerevisiae. In: Nickoloff J A, Hoekstra M F, editors. DNA repair in prokaryotes and lower eukaryotes. 1. DNA damage and repair. Totowa, N.J: Humana Press, Inc.; 1998. pp. 411–448. [Google Scholar]
- 8.Davey S, Nass M L, Ferrer J V, Sidik K, Eisenberger A, Mitchell D L, Freyer G A. The fission yeast UVDR DNA repair pathway is inducible. Nucleic Acids Res. 1997;25:1002–1008. doi: 10.1093/nar/25.5.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey S, Han C S, Ramer S A, Klassen J C, Jacobsen A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G A. Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom’s syndrome disease gene. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doetsch P W. What’s old is new: an alternative DNA excision repair pathway. Trends Biochem Sci. 1994;20:384–386. doi: 10.1016/s0968-0004(00)89084-2. [DOI] [PubMed] [Google Scholar]
- 11.Fang W H, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 12.Fantes P, Creanor J. Canavanine resistance and the mechanism of arginine uptake in the fission yeast Schizosaccharomyces pombe. J Gen Microbiol. 1984;130:3265–3273. doi: 10.1099/00221287-130-12-3265. [DOI] [PubMed] [Google Scholar]
- 13.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 14.Fleck O, Michael H, Heim L. The swi4+ gene of Schizosaccharomyces pombe encodes a homologue of mismatch repair enzymes. Nucleic Acids Res. 1992;20:2271–2278. doi: 10.1093/nar/20.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleck O, Schar P, Kohli J. Identification of two mismatch-binding activities in protein extracts of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5289–5295. doi: 10.1093/nar/22.24.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleck O, Lehmann E, Schar P, Kohli J. Involvement of nucleotide excision repair in msh2 pms1-independent mismatch repair. Nat Genet. 1999;21:314–317. doi: 10.1038/6838. [DOI] [PubMed] [Google Scholar]
- 17.Freyer G A, Davey S, Ferrer J V, Martin A M, Beach D, Doetsch P W. An alternative eukaryotic DNA excision repair pathway. Mol Cell Biol. 1995;15:4572–4577. doi: 10.1128/mcb.15.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennecke F, Kolmar H, Brundl K, Fritz H J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 19.Jing Y, Kao J F, Taylor J S. Thermodynamic and base-pairing studies of matched and mismatched DNA dodecamer duplexes containing cis-syn, (6-4) and Dewar photoproducts of TT. Nucleic Acids Res. 1998;26:3845–3853. doi: 10.1093/nar/26.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur B, Avery A M, Doetsch P W. Expression, purification, and characterization of ultraviolet endonuclease from Schizosaccharomyces pombe. Biochemistry. 1998;37:11599–11604. doi: 10.1021/bi981008c. [DOI] [PubMed] [Google Scholar]
- 21.Kaur, B., and P. W. Doetsch. Unpublished data.
- 22.Kolodner R D. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 23.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1943;49:264–284. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 24.Leupold U. Genetical methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:169–177. [Google Scholar]
- 25.Lieb M, Allen E, Read D. Very short patch mismatch repair in phage lambda: repair sites and length of repair tracts. Genetics. 1986;114:1041–1060. doi: 10.1093/genetics/114.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieb M. Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J Bacteriol. 1987;169:5241–5246. doi: 10.1128/jb.169.11.5241-5246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCready S, Carr A M, Lehmann A R. Repair of cyclobutane pyrimidine dimers and 6-4 photoproducts in the fission yeast Schizosaccharomyces pombe. Mol Microbiol. 1993;10:885–890. doi: 10.1111/j.1365-2958.1993.tb00959.x. [DOI] [PubMed] [Google Scholar]
- 28.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 29.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 30.Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 31.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 32.Porello S L, Leyes A E, David S S. Single turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 33.Radman M, Dohet C, Jones M, Doutriaux M P, Laengle-Rouault F, Maenhaut-Michel G, Wagner R. Processing of mispaired and unpaired bases in heteroduplex DNA in E. coli. Biochimie. 1985;67:745–752. doi: 10.1016/s0300-9084(85)80162-0. [DOI] [PubMed] [Google Scholar]
- 34.Schar P, Kohli J. Marker effects of G to C transversions on intragenic recombination and mismatch repair in Schizosaccharomyces pombe. Genetics. 1993;133:825–835. doi: 10.1093/genetics/133.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schar P, Munz P, Kohli J. Meiotic mismatch repair quantified on the basis of segregation patterns in Schizosaccharomyces pombe. Genetics. 1993;133:815–824. doi: 10.1093/genetics/133.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schar P, Baur M, Schneider C, Kohli J. Mismatch repair in Schizosaccharomyces pombe requires the mutL homologous gene pms1: molecular cloning and functional analysis. Genetics. 1997;146:1275–1286. doi: 10.1093/genetics/146.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su S S, Lahue R S, Au K G, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 38.Szankasi P, Smith G R. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 39.Szankasi P, Smith G R. A single-stranded DNA exonuclease from Schizosaccharomyces pombe. Biochemistry. 1992;31:6769–6773. doi: 10.1021/bi00144a017. [DOI] [PubMed] [Google Scholar]
- 40.Szankasi P, Smith G R. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 41.Takao M, Yonemasu R, Yamamoto K, Yasui A. Characterization of a UV endonuclease gene from the fission yeast Schizosaccharomyces pombe and its bacterial homolog. Nucleic Acids Res. 1996;24:1267–1271. doi: 10.1093/nar/24.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yajima H, Takao M, Yasuhira S, Zhao J H, Ishii C, Inoue H, Yasui A. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J. 1995;14:2393–2399. doi: 10.1002/j.1460-2075.1995.tb07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao M, Kow Y W. Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′-endonuclease from Escherichia coli. J Biol Chem. 1994;269:31390–31396. [PubMed] [Google Scholar]
- 44.Yao M, Kow Y W. Further characterization of Escherichia coli endonuclease V. J Biol Chem. 1997;272:30774–30779. doi: 10.1074/jbc.272.49.30774. [DOI] [PubMed] [Google Scholar]
- 45.Yasui A, McCready S J. Alternative repair pathways for UV-induced DNA damage. Bioessays. 1998;20:291–297. doi: 10.1002/(SICI)1521-1878(199804)20:4<291::AID-BIES5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 46.Yeh Y C, Chang D Y, Masin J, Lu A L. Two nicking enzyme systems specific for mismatch-containing DNA in nuclear extracts from human cells. J Biol Chem. 1991;266:6480–6484. [PubMed] [Google Scholar]
- 47.Yeh Y C, Liu H F, Ellis C A, Lu A L. Mammalian topoisomerase I has base mismatch nicking activity. J Biol Chem. 1994;269:15498–15504. [PubMed] [Google Scholar]
- 48.Yonemasu R, McCready S J, Murray J M, Osman F, Takao M, Yamamoto K, Lehman A R, Yasui A. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:1553–1558. doi: 10.1093/nar/25.8.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zell R, Fritz H J. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987;6:1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]