Abstract
This study aimed to determine the frequency of SARS‐CoV‐2 RNA in serum and its association with the clinical severity of COVID‐19. This retrospective cohort study performed at Toyama University Hospital included consecutive patients with confirmed COVID‐19. The prevalence of SARS‐CoV‐2 RNAemia and the strength of its association with clinical severity variables were examined. Fifty‐six patients were included in this study. RNAemia was detected in 19.6% (11/56) patients on admission, and subsequently in 1.0% (1/25), 50.0% (6/12), and 100.0% (4/4) moderate, severe, and critically ill patients, respectively. Patients with RNAemia required more frequent oxygen supplementation (90.0% vs. 13.3%), ICU admission (81.8% vs. 6.7%), and invasive mechanical ventilation (27.3% vs. 0.0%). Among patients with RNAemia, the median viral loads of nasopharyngeal (NP) swabs that were collected around the same time as the serum sample were significantly higher in critically ill (5.4 log10 copies/μl; interquartile range [IQR]: 4.2–6.3) than in moderate‐severe cases (2.6 log10 copies/μl; [IQR: 1.1–4.5]; p = 0.030) and were significantly higher in nonsurvivors (6.2 log10 copies/μl [IQR: 6.0–6.5]) than in survivors (3.9 log10 copies/μl [IQR: 1.6–4.6]; p = 0.045). This study demonstrated a relatively high proportion of SARS‐CoV‐2 RNAemia and an association between RNAemia and clinical severity. Moreover, among the patients with RNAemia, the viral loads of NP swabs were correlated with disease severity and mortality, suggesting the potential utility of combining serum testing with NP tests as a prognostic indicator for COVID‐19, with higher quality than each separate test.
Keywords: COVID‐19, mortality, nasopharyngeal viral load, RNAemia, SARS‐CoV‐2, severity
1. INTRODUCTION
Knowledge of the viral load dynamics of SARS‐CoV‐2 in nasopharyngeal (NP) swabs and other tissues plays an important role in understanding the pathogenesis, transmission, and management of patients with coronavirus disease (COVID‐19). 1 Previous studies have found that the viral load in NP swab specimens peaks around the time of symptom onset and generally becomes undetectable about 2 or 3 weeks after symptom onset 2 , 3 ; however, there is evidence of prolonged virus detection in elderly patients, 4 with previous studies showing that the viral load in the respiratory specimens of symptomatic patients was similar to that of asymptomatic patients, 5 thus implying that the viral load in respiratory specimens may not objectively reflect disease severity. 6
On the other hand, some studies have reported that a high viral load in NP swabs was an independent risk factor for intubation and/or death. 7 , 8 Thus, the real impact of initial SARS‐CoV‐2 viral load in NP swabs on clinical outcomes is not been clearly elucidated.
Previous studies have indicated that SARS‐CoV‐2 RNA may also be detected in plasma and serum, 9 , 10 and serum SARS‐CoV‐2 viral RNA (termed RNAemia by the authors in a recent study) was detected in 15% of COVID‐19 patients. 11 Recent studies demonstrated that RNAemia is associated with disease severity and unfavorable clinical outcomes. 12 , 13 This suggests that extra‐respiratory spread of SARS‐CoV‐2 contributes to systemic inflammatory responses, which are an important factor in the systemic pathogenesis of COVID‐19, including thrombosis and coagulopathy. 13 , 14
However, the clinical implication with SARS‐CoV‐2 RNAemia detection have not been completely clarified and information regarding the use of NP swabs and blood samples to improve patient management is insufficient.
In this study, we conducted a retrospective, monocentric cohort study of consecutive COVID‐19 patients to investigate the frequency of SARS‐CoV‐2 RNA in serum and its association with the clinical severity of COVID‐19.
2. MATERIALS AND METHODS
2.1. Study design
This cohort study enrolled consecutive patients who were admitted to and/or from whom viral load measurements and blood tests were conducted at Toyama University Hospital with confirmed COVID‐19 from April 13, 2020, to September 28, 2020, were tested for SARS‐CoV‐2 RNA in serum collected at the initial visit. NP samples that were collected approximately at the same time of serum collection around admission were included in the study as paired NP samples if residual tested serum was collected within seven days before or after NP collection. Given that viral culture was not performed to determine virus viability, the presence of viral RNA in serum is referred to as RNAemia instead of viremia.
2.2. Data collection
A retrospective chart review was performed for all individuals in the study to identify basic demographic data, medical history, clinical presentation, and laboratory test results on hospital admission. The collected outcome data included the need for oxygen supplementation, admission to an intensive care unit (ICU), invasive mechanical ventilation, and all‐cause mortality. Chest computed tomography was carried out in all patients around admission and severity was divided into five categories: asymptomatic, mild (symptomatic patients without pneumonia), moderate (pneumonia patients without required oxygen supplementation), severe (required oxygen supplementation), and critically ill (requiring invasive mechanical ventilation, shock, and/or multiple organ dysfunction). The sample size was based on a convenience set of all available clinical samples, without formal power calculations.
2.3. Quantitative reverse transcriptase‐polymerase transcription assay (RT‐qPCR)
Serum samples and NP swabs were collected from all patients and RNA was extracted. NP swab specimens were pretreated with 500 µl of sputazyme (Kyokuto Pharmaceutical). After centrifugation at 20 000 g for 30 min at 4°C, the supernatant was used for RNA extraction. A total of 60 µl RNA solution was obtained from 140 µl of the supernatant or 140 µl of the serum using the QIAamp ViralRNA Mini Kit (QIAGEN) or Nippongene Isospin RNA Virus (Nippongene), according to the manufacturer's instructions. The viral loads of SARS‐CoV‐2 were quantified on the basis of an N2 gene‐specific primer/probe set by RT‐qPCR, according to the protocol of the National Institute of Infectious Diseases of Japan. 15 Quantification quality was controlled using AcroMetrix COVID‐19 RNA Control (Thermo Fisher Scientific). The detection limit was approximately 0.4 copies/µl (two copies/5 µl). When undetectable, viral load data were hypothetically calculated as −0.5 log10 copies/µl.
2.4. Statistical analysis
Continuous and categorical variables are presented as median (interquartile range [IQR]) and n (%), respectively. We used the Mann–Whitney U test, χ 2 test, or Fisher's exact test to compare the differences between patients with and without RNAemia. In all analyses, we preliminarily confirmed the effect of multicollinearity of the covariates used in the statistical analysis. Univariate logistic regression analysis was used to investigate variables potentially associated with clinical severity. Multivariate logistic regression analyses were performed with all the independent variables (p ≤ 0.20) on univariate analysis as well as with the main variable of RNAemia and variables deemed either clinically relevant or supported in the medical literature. Data were analyzed using JMP Pro version 14.2.0 software (SAS Institute Inc.).
2.5. Ethics approval
This study was performed in accordance with the Declaration of Helsinki and was approved by the ethical review board of the University of Toyama (approval no.: R2019167). Written informed consent was obtained from all patients.
3. RESULTS
Fifty‐six patients were included, with a median age of 54.5 years. A total of 24 (42.9%) patients were men, and eight (14.3%) patients had Charlson comorbidity index ≥2 (Table 1). Hypertension was the most common comorbidity, followed by dyslipidemia, diabetes, and bronchial asthma. Hypertension was significantly more frequent in patients with detectable RNAemia than in those without.
Table 1.
Demographic and clinical characteristics of 56 patients with paired nasopharyngeal and serum samples tested for SARS‐CoV‐2
| Characteristics | Overall patients, n = 56 | With RNAemia, n = 11 | Without RNAemia, n = 45 | p |
|---|---|---|---|---|
| Age, median, y | 54.5 | 78 | 50 | 0.0013 |
| 0–19, n (%) | 4 (7.1) | 0 (0.0) | 4 (7.14) | 0.58 |
| 20–64, n (%) | 32 (57.1) | 4 (36.4) | 28 (62.2) | 0.18 |
| ≥65, n (%) | 20 (35.7) | 7 (63.6) | 13 (28.9) | 0.042 |
| Sex | ||||
| Male, n (%) | 24 (42.9) | 5 (45.5) | 19 (42.2) | 1 |
| Presence of symptoms, n (%) | ||||
| Asymptomatic | 6 (10.7) | 0 (0.0) | 6 (13.3) | 0.33 |
| Symptomatic | 50 (89.3) | 11 (100.0) | 39 (86.7) | 0.33 |
| Mild | 9 (18.0) | 0 (0.0) | 9 (23.1) | 0.18 |
| Pneumonia | 41 (73.2) | 11 (100.0) | 30 (66.7) | 0.026 |
| Moderate | 25 (50.0) | 1 (9.1) | 24 (61.5) | 0.0046 |
| Severe | 12 (24.0) | 6 (54.6) | 6 (15.4) | 0.014 |
| Critically ill | 4 (8.0) | 4 (36.4) | 0 (0.0) | 0.0014 |
| Comorbidities, n (%) | ||||
| Hypertension | 13 (23.2) | 6 (54.6) | 7 (15.6) | 0.013 |
| Dyslipidemia | 9 (16.1) | 2 (18.2) | 7 (15.6) | 1 |
| Diabetes | 5 (8.9) | 1 (9.1) | 4 (8.9) | 1 |
| Bronchial asthma | 4 (7.1) | 0 (0.0) | 4 (8.9) | 0.58 |
| Charlson comorbidity index ≥ 2 | 8 (14.3) | 2 (18.2) | 6 (13.3) | 0.65 |
| Laboratory findings, median (IQR) | ||||
| White blood cell count (×103/μl) | 5.1 (3.7–5.9) | 5.1 (2.9–5.7) | 5.1 (3.7–5.9) | 0.77 |
| Neutrophil count (/μl) | 2780.1 (1991.4–3915.3) | 3099.8 (1918.6–4936.4) | 2672.1 (1996.2–3783.2) | 0.48 |
| Lymphocyte count (/μl) | 1234.2 (725.2–1801.0) | 1283.3 (861.2–2030.5) | 699.4 (430.3–1360.8) | 0.010 |
| Hemoglobin (g/dl) | 14.0 (13.0–15.5) | 13.3 (11.7–14.3) | 14.1 (13.1–15.6) | 0.11 |
| Platelet count (×103/μl) | 202.5 (133.8–234.5) | 186 (142–219) | 207 (128.5–250.5) | 0.34 |
| Albumin (g/dl) | 4.1 (3.4–4.4) | 4.2 (3.9–4.4) | 3.1 (2.9–3.3) | <0.0001 |
| AST (U/L) | 30 (19.5–45.5) | 41 (31–61) | 28 (18.5–36) | 0.0023 |
| ALT (U/L) | 22 (13.3–42.5) | 20 (12–42) | 26 (16–60) | 0.23 |
| Lactate dehydrogenase (U/L) | 224 (182–320) | 425 (279–559) | 213.5 (177.5–276.3) | <0.0001 |
| Total bilirubin (mg/dl) | 0.5 (0.3–0.6) | 7.06 (2.76–16.7) | 0.44 (0.06–1.77) | 0.0019 |
| Urea nitrogen (mg/dl) | 13.4 (10.5–18.6) | 23.3 (13.4–27.6) | 12.7 (10–15.7) | 0.0063 |
| Creatinine (mg/dl) | 0.80 (0.57–0.92) | 1.01 (0.56–1.19) | 0.78 (0.57–0.89) | 0.076 |
| CRP (mg/dl) | 0.7 (0.083–3.1) | 7.1 (2.8–16.7) | 0.4 (0.06–1.8) | 0.0019 |
| d‐dimer (μg/ml) | 0.7 (0.5–1.3) | 1.5 (1.4–3.2) | 0.7 (0.5–0.9) | <0.0001 |
| Treatment, n (%) | ||||
| Antiviral therapy | 19 (33.9) | 9 (81.8) | 10 (22.2) | 0.0004 |
| Favipiravir | 17 (30.4) | 8 (72.7) | 9 (20.0) | 0.0016 |
| Remdesivir | 4 (7.1) | 4 (36.4) | 0 (0.0) | 0.0009 |
| Antibiotic therapy | 15 (26.8) | 9 (81.8) | 6 (13.3) | <0.0001 |
| Clinical outcomes, n (%) | ||||
| Required oxygen supplementation | 16 (28.6) | 10 (90.9) | 6 (13.3) | <0.0001 |
| ICU admission | 12 (21.4) | 9 (81.8) | 3 (6.7) | <0.0001 |
| Invasive mechanical ventilation | 3 (5.4) | 3 (27.3) | 0 (0.0) | 0.006 |
| In‐hospital mortality | 3 (5.4) | 2 (18.2) | 1 (2.2) | 0.095 |
The serum tested for the detection of RNA was collected at the initial visit in all patients except for two (collected 2 or 5 days after hospital admission). Paired NP swabs were collected within one day before and after the day of serum collection in 51 (91.1%) patients. SARS‐CoV‐2 RNAemia was detected in 11 (19.6%) patients, and patients with detectable RNAemia were significantly older than those without RNAemia (78 vs. 50 years; p = 0.0013). Lymphocyte count and serum albumin level were significantly lower in patients with RNAemia than in those without RNAemia, and serum levels of aspartate aminotransferase (AST), lactate dehydrogenase, total bilirubin urea nitrogen, CRP, and d‐dimer were significantly higher in the patients with than in those without RNAemia.
The numbers of asymptomatic, mild, moderate, severe, and critically ill cases were 6, 9, 25, 12, and 4, respectively. Figure 1 shows the proportion of patients with detectable RNAemia, stratified by severity. The proportions of patients with RNAemia in moderate, severe, and critically ill cases were 1.0% (1/25), 50.0% (6/12), and 100.0% (4/4), respectively, but there were no asymptomatic and mild cases.
Figure 1.
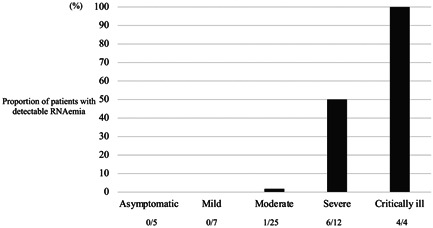
Proportion of patients with detectable RNAemia stratified by severity
In terms of clinical outcomes, patients with RNAemia required more frequent oxygen supplementation (90.0% vs. 13.3%; p < 0.0001) and ICU admission (81.8% vs. 6.7%; p < 0.0001), and also required invasive mechanical ventilation (27.3% vs. 0.0%; p < 0.0001). In addition, RNAemia was independently associated with an increased risk of requiring more frequent oxygen supplementation and ICU admission in univariate and multivariate conditional logistic regression analyses (Tables 1 and 2). RNAemia was also associated with required invasive mechanical ventilation in univariate analysis, but multivariate conditional logistic regression analysis could not be conducted due to complete separation because of strong relationships. In‐hospital mortality was more frequent in patients with SARS‐CoV‐2 RNAemia than in those without SARS‐CoV‐2 RNAemia; however, this difference was not significant (18.2% vs. 2.2%; p = 0.095), possibly because of the small sample size (Table 1).
Table 2.
Univariate and multivariate conditional logistic regression analyses of required oxygen supplementation and ICU admission
| Variables (required oxygen supplementation) | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |
| RNAemia | 65.0 (7.0–603.3) | <0.0001 | 232.3 (17.2–12468.5) | 0.0007 |
| Male | 0.73 (0.22–2.41) | 0.77 | 0.040 (0.00089–0.78) | 0.052 |
| Charlson comorbidity index (per 1‐point increment) | 1.85 (0.87–3.94) | 0.11 | 2.35 (0.67–10.57) | 0.20 |
| Hemoglobin (per 1‐g/dl increment) | 0.68 (0.47–0.98) | 0.026 | 1.19 (0.59–2.63) | 0.65 |
| Platelet count (per 1.0 × 103/μl increment) | 0.9918 (0.9850–0.9986) | 0.0067 | 0.9898 (0.9756–1.001) | 0.11 |
| Creatinine (per 1‐mg/dl increment) | 39.98 (2.274–702.6) | 0.0013 | 995.6 (1.286–4858026) | 0.063 |
| Variables (ICU admission) | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |
| RNAemia | 63.0 (9.2–433.4) | <0.0001 | 440.1 (22.3–91446.6) | 0.0022 |
| Male | 1.44 (0.39–5.33) | 0.74 | 0.81 (0.041–15.62) | 0.88 |
| Charlson comorbidity index (per 1‐point increment) | 2.19 (0.99–4.83) | 0.068 | 3.79 (0.91–30.01) | 0.11 |
| Platelet count (per 1.0 × 103/μl increment) | 0.9922 (0.9850–0.9995) | 0.013 | 0.9857 (0.9647–0.9998) | 0.092 |
| ALT (per 1‐U/L increment) | 1.014 (0.9908–1.037) | 0.053 | 1.025 (0.9541–1.082) | 0.37 |
| Creatinine (per 1‐mg/dl increment) | 48.13 (2.106–1099.6) | 0.0023 | 3.054 (0.001935–4174.8) | 0.75 |
There was no significant difference in viral loads in the NP swabs of patients with RNAemia (4.0 log10 copies/μl [IQR: 2.1–4.9]) and without RNAemia (4.2 log10 copies/μl [2.0–5.7]; p = 0.85) (Table 3). The overall median number of days of symptoms at the time of NP testing was six (IQR: 3–9), which was similar in moderately to critically ill patients with and without RNAemia.
Table 3.
Nasopharyngeal viral load in 56 patients with and without RNAemia
| Characteristics | Overall patients, n = 56 | With RNAemia, n = 11 | Without RNAemia, n = 45 | p |
|---|---|---|---|---|
| Viral load (log10 copies/μl) of nasopharyngeal swab collected around admission, median (IQR) | 4.2 (2.1–5.5) | 4.0 (2.1–4.9) | 4.2 (2.0–5.7) | 0.85 |
| Severity | ||||
| Moderate | 4.1 (2.0–5.3) | 0.7 | 4.2 (2.4–5.4) | 0.24 |
| Severe | 3.5 (2.1–4.9) | 3.2 (1.9–4.5) | 4.0 (2.1–5.5) | 0.47 |
| Critically ill | 5.4 (4.2–6.3) | 5.4 (4.2–6.3) | – | – |
| In‐hospital mortality | ||||
| Survivor | 4.2 (2.0–5.4) | 3.9 (1.6–4.6) | 4.3 (2.0–5.8) | 0.30 |
| Nonsurvivor | 6.0 (2.1–6.5) | 6.2 (6.0–6.5) | 2.1 | 0.22 |
In an in‐depth study of RNAemia patients, although the days after symptom onset at the time of NP testing were not significantly different, the median viral loads of NP swabs were significantly higher in critically ill (5.4 log10 copies/μl [IQR: 4.2–6.3]) than in moderate‐severe cases (2.6 log10 copies/μl [1.1–4.5]; p = 0.030) and in nonsurvivors (6.2 log10 copies/μl [IQR: 6.0–6.5]) than in survivors (3.9 log10 copies/μl [1.6–4.6]; p = 0.045).
4. DISCUSSION
In this cohort study, 56 consecutive patients were assessed for RNAemia detection in serum samples. These findings show that RNAemia was present in almost 20% of the samples tested. Previous studies have reported different rates of SARS‐CoV‐2 detection in serum (ranging from 10.4% to 74.1%), 1 , 8 , 9 , 16 , 17 , 18 whereas other studies did not find any patients or reported only 1% of RNAemia. 1 , 16 , 17 However, a direct comparison of these results is difficult because of the use of different testing protocols, including differences in specimen extraction volumes, sample types (plasma, serum, and whole blood), viral genomic targets (N, S, E genes, or ORF1ab region), variable timing of blood collection, and different patient populations. 9
Regarding the clinical importance of the presence of SARS‐CoV‐2 RNAemia, our results are consistent with previous reports, suggesting an association with ICU admission, invasive mechanical ventilation, and mortality. 1 , 19 , 20
However, the association between viral load in respiratory specimens and severity remains unclear and controversial in the literature. Multiple case series have reported that the viral load in asymptomatic patients is as high as that in asymptomatic patients, 5 , 18 while recent studies have found that NP viral load at admission is independently correlated with the risk of intubation and in‐hospital mortality. 8 This discrepancy may be attributed to the different patient populations, including the levels of severity.
In this study, there was no significant difference in the viral load in the NP swabs of patients with and without RNAemia. These results confirm those of Berastegui‐Cabrera et al. 1 and Prebensen et al., 21 who did not find any association between the viral load in NP samples and the presence of SARS‐CoV‐2 RNAemia. However, a recent study in ICU patients reported that the nasopharyngeal viral load was associated with RNAemia as well as 28‐day mortality. 12 Therefore, in some severe and critical cases, both NP viral load and RNAemia may be useful as prognostic markers of condition development and mortality.
To date, information regarding the combined use of NP swabs and blood samples to improve patient management is insufficient. To the best of our knowledge, no previous study investigates the prognostic potential of RNAemia combined with the viral load of NP swabs in hospitalized patients with COVID‐19. Among the patients with RNAemia in the present study, the higher viral load of NP swabs was correlated with severity and mortality. This result suggested that NP viral load may also be useful as an aid to provide a more accurate prediction of the severity and mortality of patients with RNAemia than RNAemia alone.
Although this study was not performed in a controlled setting, most serum and NP samples were collected on the same day as their admission, thus supporting a potential role for the early detection of individuals at risk of severe clinical illness. Thus, in addition to assessing the viral load of NP swabs, which is the technique most frequently used for COVID‐19 diagnosis, serum tests may be considered a complementary modality for the early identification of individuals who are likely to develop severe COVID‐19. 9
This study has several limitations inherent to its small sample size, retrospective design, and potential for confounding the detection of SARS‐CoV‐2 RNA from the serum, NP viral load, and clinical conditions that cannot be excluded. In addition, we did not conduct multivariate or age‐adjusted analyses owing to the strong multicollinearity between variables and the small sample size. Longitudinal analyses were also not performed because viral load dynamics could be modified by the treatment, including combinations of antivirals and antibiotics during hospitalization.
5. CONCLUSIONS
In conclusion, this study demonstrated a relatively high overall proportion of detectable SARS‐CoV‐2 RNAemia as well as a strong association between serum viral detection and severe clinical disease, including the need for oxygen supplementation and ICU admission, invasive mechanical ventilation, and the likelihood of in‐hospital mortality. In addition, among the patients with RNAemia, the viral loads of NP swabs were significantly higher in critically ill and nonsurvivor cases than in moderate‐severe and survivor cases. Further studies are required to investigate the prognostic potential of RNAemia and those combined with the viral load of NP swabs to facilitate early therapeutic and supportive interventions before severe clinical deterioration of COVID‐19.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Hitoshi Kawasuji, Yoshitomo Morinaga, and Yoshihiro Yamamoto planned the study design, contributed to data analysis, and wrote and revised the manuscript. Yoshitomo Morinaga, Hideki Tani, Yoshihiro Yoshida, Miyuki Kimura, and Hiroshi Yamada contributed to conduct and perform RT‐qPCR. Hitoshi Kawasuji, Yusuke Takegoshi, Makito Kaneda, Yushi Murai, Kou Kimoto, Akitoshi Ueno, Yuki Miyajima, Yasutaka Fukui, Ippei Sakamaki, and Yoshihiro Yamamoto contributed to obtaining informed consent, collecting specimens, and patient management. All the authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
This study was supported by the Research Program on Emerging and Re‐Emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED) under Grant JP20he0622035. The authors would like to thank Ai Kakumoto for her support in the COVID‐19 testing.
Kawasuji H, Morinaga Y, Tani H, et al. SARS‐CoV‐2 RNAemia with a higher nasopharyngeal viral load is strongly associated with disease severity and mortality in patients with COVID‐19. J Med Virol. 2021;94:147‐153. 10.1002/jmv.27282
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Berastegui‐Cabrera J, Salto‐Alejandre S, Valerio M, et al. SARS‐CoV‐2 RNAemia is associated with severe chronic underlying diseases but not with nasopharyngeal viral load. J Infect. 2020;82:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kawasuji H, Takegoshi Y, Kaneda M, et al. Transmissibility of COVID‐19 depends on the viral load around onset in adult and symptomatic patients. PLoS One. 2020;15(12):e0243597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larremore DB, Wilder B, Lester E, et al. Test sensitivity is secondary to frequency and turnaround time for COVID‐19 screening. Sci Adv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID‐19). Clin Infect Dis. 2020;71(15):799‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS‐CoV‐2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X, Zhao B, Qu Y, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71(8):1937‐1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de la Calle C, Lalueza A, Mancheño‐Losa M, et al. Impact of viral load at admission on the development of respiratory failure in hospitalized patients with SARS‐CoV‐2 infection. Eur J Clin Microbiol Infect Dis. 2021;40(6):1209‐1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pujadas E, Chaudhry F, McBride R, et al. SARS‐CoV‐2 viral load predicts COVID‐19 mortality. The Lancet Respiratory Medicine. 2020;8(9):e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hogan CA, Stevens BA, Sahoo MK, et al. High frequency of SARS‐CoV‐2 RNAemia and association with severe disease. Clin Infect Dis. 2020;51:114‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA. 2020;323(18):1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gutmann C, Takov K, Burnap SA, et al. SARS‐CoV‐2 RNAemia and proteomic trajectories inform prognostication in COVID‐19 patients admitted to intensive care. Nat Commun. 2021;12(1):3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang K, Wu L, Luo Y, Gong B. Quantitative assessment of SARS‐CoV‐2 RNAemia and outcome in patients with coronavirus disease 2019. J Med Virol. 2021;93(5):3165‐3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riel Dv, Embregts CWE, Sips GJ, et al. Temporal kinetics of RNAemia and associated systemic cytokines in hospitalized COVID‐19 patients. mSphere. 2021;6(3):e00311‐e00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shirato K, Nao N, Katano H, et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV‐2019) in Japan. Jpn J Infect Dis. 2020;73:304‐307. [DOI] [PubMed] [Google Scholar]
- 16. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walsh KA, Jordan K, Clyne B, et al. SARS‐CoV‐2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81(3):357‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagman K, Hedenstierna M, Gille‐Johnson P, et al. SARS‐CoV‐2 RNA in serum as predictor of severe outcome in COVID‐19: a retrospective cohort study. Clin Infect Dis. 2020. ciaa1285. 10.1093/cid/ciaa1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Veyer D, Kernéis S, Poulet G, et al. Highly sensitive quantification of plasma SARS‐CoV‐2 RNA shelds light on its potential clinical value. Clin Infect Dis. 2020. ciaa1196. 10.1093/cid/ciaa1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prebensen C, Hre PLM, Jonassen C, et al. SARS‐CoV‐2 RNA in plasma is associated with ICU admission and mortality in patients hospitalized with COVID‐19. Clin Infect Dis. 2020;65:351‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


