To the Editor,
Since August 2020, a series of cases of reinfection by a phylogenetically distinct variant of SARS‐CoV‐2 have been reported, raising pertinent questions on the heterogeneity of the natural immune response to SARS‐CoV‐2 infection that may not uniformly confer protective immunity to all individuals. 1 , 2 , 3 , 4 , 5 Specifically, reinfection seems more likely to occur in individuals whose immune system has been weakened by underlying comorbidities or therapies. 6 , 7 , 8 Here, we report a case of a 52‐year‐old male patient suffering from transitional cell carcinoma of the renal pelvis and ureter who was infected at two separate times with two genetically distant SARS‐CoV‐2 strains, with the reappearance of the first strain four months after the first infection. The patient's past medical history and treatments are summarized in Figure S1. On June 23, 2020 (Day 0), he had a cough and fever and was diagnosed with COVID‐19 by SARS‐CoV‐2 reverse transcriptase‐polymerase chain reaction (RT‐PCR) assay of a nasopharyngeal swab specimen (cycle threshold, C t, values for SARS‐CoV‐2 E, RdRp, and N genes ranged from 25 to 26) (Figure 1A). Chest X‐ray did not reveal any abnormality, and his clinical conditions improved with resolution of cough and fever within 2 weeks. On Days 35 and 36, two consecutive nasopharyngeal swabs resulted negative for SARS‐CoV‐2 infection. In the next few months, the patient did not show any respiratory symptoms. However, the deterioration of his cancer condition leading to urinary tract infection and sepsis required further hospitalization. On Day 110, the patient had a fever caused by an ongoing Escherichia coli‐induced sepsis. RT‐PCR assay of a nasopharyngeal swab resulted positive again, causing concern for a recurrence of COVID‐19 (C t values of 34 and 36 for E and N genes, and over 40 for the RdRp gene). An abdominal computed tomography scan performed on Day 113 showed thrombosis of the inferior vena cava, of the right iliac vein, and of both femoral veins. On Day 115, the patient died from septic shock and respiratory failure.
Figure 1.
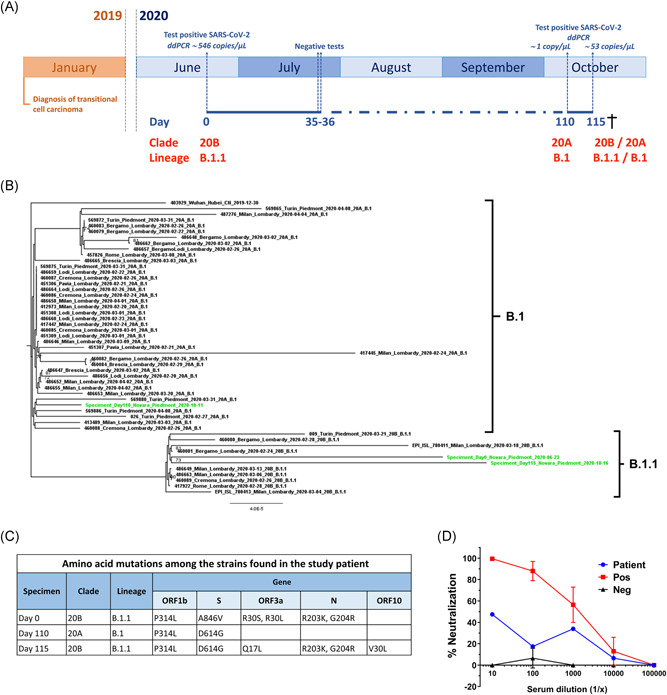
(A) Timeline of clinical presentations and SARS‐CoV‐2 testing, including viral loads (copies/μl) and the strains found in the study patient. Timing of relevant clinical events, such as the outcome of diagnostic tests, is shown. (B) Phylogenomic analyses of described SARS‐CoV‐2 strains in the study patient. The tree was constructed by the maximum likelihood method. Clade information as inferred by Nextstrain and Pangolin nomenclatures is shown. (C) Viral genome classification and amino acid mutations were identified according to Nextclade and Pangolin among the three specimens harvested on Days 0, 110, and 115. (D) Serum neutralizing assay against rVSV‐SARS‐CoV‐2‐SΔ21 with a sample harvested at Day 110. Data are representative of two independent experiments performed in duplicate. Error bars represent the standard deviation. Patient (blue dot), normal human serum (Neg) (black triangle), positive serum = COVID‐19 convalescent serum (Pos) (red square)
Quantitative SARS‐CoV‐2 viral loads by droplet digital PCR detected 546, 1, and 53 copies/μl on Days 0, 110, and 115 nasopharyngeal swabs, respectively (Figure 1A). Whole‐genome sequencing and phylogenetic analysis of RNA from the first two specimens showed that the viral genome found at Day 0 could be grouped in the Nextstrain clade 20B and Pangolin lineage B.1.1, while the strain isolated on Day 110 belonged to the Nextstrain clade 20A and Pangolin lineage B.1 (Figure 1B). However, when we sequenced the RNA from the third sample harvested on Day 115, we detected again the Nextstrain clade 20B, suggesting that the first infection strain had never been cleared completely. With regard to amino acid changes, by analyzing minority variants in the Day 115 specimen, the mutations R203K and G204R, which distinguish B.1 and B.1.1 lineages, were the predominant ones until 65% coverage, but below this cut‐off, we were also able to detect significant levels of the wild‐type virus (Figure 1C). Furthermore, the D614G variant was always present in specimens isolated on Days 110 and 115, whereas it was absent, even as a minority variant, in the specimen harvested on Day 0. No evidence of recombination events was observed. Phylogenetic analysis was congruent with both persistent infection with B.1.1 strains (specimens from Days 0 and 115) and reinfection with B.1 strain on Day 110.
The patient's blood was only available on Day 110 and resulted negative for the presence of SARS‐CoV‐2 genome by droplet digital PCR analysis. Rapid immunochromatographic test on blood resulted positive for immunoglobulin G (IgG) anti‐SARS‐CoV‐2 N protein. Very low levels of IgG anti‐SARS‐CoV‐2 spike protein were found in this sample (1200 AU/ml with the low threshold < 2.544 AU/ml). In addition, anti‐receptor binding domain antibodies were determined by a different enzyme‐linked immunosorbent assay, which confirmed the presence of very low reactivity. Consistently, the neutralizing activity performed using the replication‐competent chimeric VSV expressing the SARS‐CoV‐2 spike protein (rVSV‐SARS‐CoV‐2‐SΔ21) was very low when compared to convalescent positive control. The inhibitory concentration (IC50) of the positive control was 0.0007 while the patient's serum was a thousand times less potent (IC50 = 0.1) and barely reached 50% of neutralization at the lowest serum dilution of 1:10 (Figures 1D and S2). 9 The fact that VSV harbors the D614 form of the spike protein—the same found in the strain isolated on Day 0—and that the G614 form is reportedly unable to interfere with the neutralizing titer 10 rules out any detection bias of our approach, indicating that the patient did fail to mount an appropriate neutralizing humoral response.
Overall, this case highlights the concerning risk of reinfection in cancer patients who fail to mount an efficient neutralizing humoral response along with the underlying existence of persistent asymptomatic/undetectable infection.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Methodology: Gloria Griffante. Resources: Paolo Ravanini, Luigi Castello, Carlo Cattrini, and Alessia Mennitto. Investigation: Gloria Griffante, Alessia Lai, Annalisa Bergna, and Paolo Ravanini. Data curation: Cinzia Borgogna and Marco De Andrea. Formal analysis: Cinzia Borgogna, Gloria Griffante, Alessia Lai, and Annalisa Bergna. Visualization: Cinzia Borgogna and Marco De Andrea. Conceptualization: Marisa Gariglio and Alessandra Gennari. Writing—original draft: Cinzia Borgogna and Marisa Gariglio. Writing—review and editing: Marco De Andrea, Alessia Lai, Massimo Galli, Gianguglielmo Zehender, Luigi Castello, Carlo Cattrini, Alessia Mennitto, and Alessandra Gennari. Supervision: Marisa Gariglio, Massimo Galli, and Gianguglielmo Zehender.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
The authors thank Marcello Arsura for critically reviewing the manuscript. This study was funded by the AGING Project – Department of Excellence – DIMET, Università del Piemonte Orientale.
REFERENCES
- 1. Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS‐CoV‐2: a case study. Lancet Infect Dis. 2020;21:52‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prado‐Vivar B, Becerra‐Wong M, Guadalupe JJ, et al. A case of SARS‐CoV‐2 reinfection in Ecuador. Lancet Infect Dis. 2020;23:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gupta V, Bhoyar RC, Jain A, et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS‐CoV‐2. Clin Infect Dis. 2020. ciaa1451 10.1093/cid/ciaa1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. To KK, Hung IF, Ip JD, et al. COVID‐19 re‐infection by a phylogenetically distinct SARS‐coronavirus‐2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020. ciaa1275 10.1093/cid/ciaa1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Elslande J, Vermeersch P, Vandervoort K, et al. Symptomatic SARS‐CoV‐2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020. ciaa1330 10.1093/cid/ciaa1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS‐CoV‐2 in an immunocompromised host. N Engl J Med. 2020;383:2291‐2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS‐CoV‐2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183:1901‐1912.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Babiker A, Marvil C, Waggoner JJ, Collins MH, Piantadosi A. The importance and challenges of identifying SARS‐CoV‐2 reinfections. J Clin Microbiol. 2020;59(4):e02769‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Case JB, Rothlauf PW, Chen RE, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication‐competent VSV‐SARS‐CoV‐2 and a clinical isolate of SARS‐CoV‐2. Cell Host Microbe. 2020;28:475‐485.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zou J, Xie X, Fontes‐Garfias CR, et al. The effect of SARS‐CoV‐2 D614G mutation on BNT162b2 vaccine‐elicited neutralization. npj Vaccines. 2021;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.


