Abstract
In previous studies, we detected a major, unidentified Egr response element (ERE) binding complex in brain extracts. We now report that this complex contains a truncated isoform of Egr3 generated by use of an alternate translation start site at methionine 106. Furthermore, the ERE binding complex previously thought to contain full-length Egr3 includes several isoforms generated by initiation at other internal methionines. Full-length and truncated (missing residues 1 to 105) Egr3 isoforms differ in the ability to stimulate transcription directed by a tandem repeat of two EREs but not by a single ERE. Taken together, our results indicate that alternative translation start sites are used to generate Egr3 isoforms with distinct transcriptional properties.
Recent studies indicate that the Egr family of transcription regulatory factors plays an important role in mediating changes in gene expression triggered by cell surface receptor activation. Members of this family are induced as part of the immediate-early gene response in a wide variety of cell types, allowing them to regulate subsequent waves of gene expression (11). The four identified members of this family share a highly conserved DNA binding domain that binds to a common DNA consensus sequence, referred to as the Egr response element (ERE) (4, 35).
The high levels of expression of several Egr family members in adult brain (40) combined with compelling evidence that the immediate-early gene response plays a critical role in synaptic plasticity (20, 24) have heightened interest in this transcription factor family in the nervous system. Both Egr1 and Egr3 mRNAs are robustly induced in hippocampal dentate granule cells by afferent synaptic stimulation that elicits long-term potentiation of this pathway, raising the possibility that they mediate changes in gene expression critical for long-lasting changes in synaptic efficacy (6, 39, 41). In addition, Egr family members are also induced in brain neurons by natural stimuli that induce plasticity (21, 22, 26, 27, 29, 38).
In recent studies aimed at defining the trans factors expressed in brain that bind to the ERE, we found a previously unidentified ERE binding complex (28). In gel shift assays performed on hippocampal extracts, we detected three major ERE binding complexes. The bands displaying the slowest and intermediate mobilities were identified as complexes containing Egr1 and Egr3, respectively. However, the third major complex eluded our initial attempts to establish its identity. As the DNA sequence binding specificity of this novel complex matched that displayed by other Egr family members, we suspected that it might represent a fifth Egr family member or a novel isoform of one of the known members.
To determine whether this putative Egr family member was induced by neuronal stimulation, we have in previous studies (28) examined its response to electrically induced seizure activity. In contrast to the Egr1 complex, which reaches maximal levels at 1 h and returns to basal levels by 4 h, this unidentified complex displayed a delayed response to stimulation similar to that found for the Egr3 complex. Both of these delayed complexes were unchanged at 1 h, reached peak levels at 4 to 6 h, and returned to basal levels by 18 h. The parallel time course of these delayed ERE binding complexes prompted us to consider the possibility that the novel complex represents a second isoform of Egr3. This view was strengthened by the observation that incubation of recombinant Egr3 with the ERE probe yielded two complexes that comigrated with the delayed bands detected in vivo. Since an N-terminal Egr3 antibody that disrupted the upper band generated by recombinant Egr3 and its endogenous counterpart did not affect the lower bands detected in vivo or in vitro, we hypothesized that they contain a shorter isoform of Egr3 lacking an N-terminal segment. Herein, we test this model and provide evidence that several truncated Egr3 isoforms are generated in vivo by utilization of multiple translation initiation sites. Furthermore, we demonstrate that Egr3 isoforms that contain or lack the N-terminal segment differ in the ability to activate transcription.
MATERIALS AND METHODS
Animal treatments.
Male Sprague-Dawley rats (200 to 250 g; Harlan, Indianapolis, Ind.) were used to assess the expression of Egr3 under control conditions as well as following stimulation with either maximal electroconvulsive seizure (MECS), administered as described previously (28), or cocaine (30 mg per kg of body weight, intraperitoneally). Wild-type and Egr3 knockout mice were injected with pentylenetetrazol (50 mg kg−1, intraperitoneally) to induce seizure activity. Egr3 knockout mice (37) and wild-type littermates were generously provided by Warren Tourtellotte and Jeff Milbrandt. Egr3 knockout mice were generated by gene targeting that resulted in the deletion of the DNA binding domain and a 3.4-kb portion of the 3′ untranslated region of the Egr3 gene.
Antibody generation.
To generate antibodies directed against an internal epitope of Egr3, a PCR-amplified Egr3 internal fragment (Egr3-INT) spanning amino acids 101 to 189 was cloned into the pGEX-2T (Pharmacia; Piscataway, N.J.) glutathione S-transferase expression vector. Fusion protein purification, immunization, test bleeds, and production bleeds were performed as described previously (28).
Electrophoretic mobility shift assay.
Brain extracts were prepared by Dounce homogenization as previously described (28) except that we included pepstatin (1 mg ml−1) in the harvest buffer. In the processing of rat brains, tissue was homogenized immediately following dissection; mouse brain tissue was processed in a similar fashion except that whole brains were frozen after removal from the skull and then stored at −80°C until homogenization just prior to analysis by gel shift assay.
Two methods were used to prepare extracts from hEK-293 cells, with similar results. In one method, Dounce homogenization was used to obtain a high-salt extract of total cellular protein from hEK-293 cells. The other method (19) entails isolation of nuclei prior to the high-salt extraction step. Briefly, cells were rinsed and then incubated in phosphate-buffered saline (PBS)–0.5 mM EDTA to remove the cells from the dish. Cells were collected, pelleted, and resuspended in PBS. The cells were then incubated for 15 min at 4°C in 400 μl of a low-salt buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 μg of leupeptin ml−1, 1 mM phenylmethylsulfonyl fluoride, 3 mM dithiothreitol). Following incubation, 25 μl of 10% Nonidet P-40 was added to the extract, which was then centrifuged at 14,000 × g for 10 min to pellet the nuclei. The cytoplasmic proteins present in the supernatant were collected, aliquoted, and stored at −80°C. Next, the nuclei were incubated in a high-salt buffer (50 mM HEPES [pH 7.4], 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 10% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride, 3 mM dithiothreitol) for 30 min at 4°C to solubilize the DNA binding proteins. Extracts were centrifuged at 14,000 × g for 10 min, and the supernatant containing nuclear proteins was collected, aliquoted, and stored at −80°C. Protein concentrations were determined by using a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.).
Gel shift assays were conducted as described previously (28). Briefly, double-stranded oligonucleotides containing the canonical ERE, 5′-CTA GGA GCG GGG GCG CTC ATG-3′ (bold letters indicate the ERE), were end labeled and purified. In the standard binding reaction, the probe (0.1 to 1 ng of DNA corresponding to ∼40,000 cpm) is incubated at room temperature for 15 min with 10 to 20 μg of protein in the presence of calf thymus DNA (10 μg ml−1). As indicated, this reaction was modified by addition of 0.5 to 1 μl of antibody specific to Egr3-NT or Egr3-INT (αEgr3-NT or αEgr3-INT). The mutant ERE oligonucleotide, 5′-CTA GGA GCG GGT GCG CTC ATG-3′, which contains a single nucleotide substitution (underlined), was used to eliminate binding of the nonspecific band in mouse forebrain and hEK-293 cell extracts.
Immunoblot analysis.
Samples were solubilized by addition of 0.5 volume of 3× sodium dodecyl sulfate (SDS) sample buffer (75 mM Tris-HCl [pH 6.8], 2.5% SDS, 6% 2-mercaptoethanol, 12% glycerol) followed by sonication if needed. Then 20 to 40 μg of extract protein was separated by SDS-polyacrylamide gel electrophoresis, transferred, and processed for immunoblotting as described previously (28). Blots were probed with αEgr3-INT (1:500 dilution).
Plasmid constructs and mutagenesis.
For in vitro transcription/translation experiments, Egr3 cDNA constructs were cloned into pBluescript (Stratagene, La Jolla, Calif.). Recombinant Egr3 protein was synthesized using the Promega (Madison, Wis.) TnT kit. The cytomegalovirus (CMV)-driven eukaryotic expression vector pCB6, containing the full-length Egr3 insert (31), and the 1× ERE and 2× ERE luciferase reporter constructs (7) were provided by the laboratory of Jeff Milbrandt (Washington University, St. Louis, Mo.).
To generate mutants that resulted in substitution of leucine or alanine for Met, primers containing the desired mutations were designed to overlap a suitable, adjacent restriction site if available. This mutated primer and a vector-specific primer were used in a PCR to generate the desired mutation in Egr3. If a convenient restriction site was not present in the vicinity of the mutation, we used splicing by the overlapping extension method (12) to generate point mutants. Mutated PCR products were ligated into the vector of interest after both were digested with the appropriate restriction enzymes, and the vector was additionally treated with calf intestinal phosphatase (New England Biolabs, Beverly, Mass.). All ligation reactions were carried out with a Rapid DNA ligation kit (Boehringer Mannheim, Indianapolis, Ind.). All mutant Egr3 inserts were sequenced in their entirety to verify that the desired mutation had been achieved and that no inadvertent mutations were introduced during this process.
Cell culture, transfection, and reporter assays.
We used hEK-293 cells for the transfection and reporter experiments, as these cells lacked detectable levels of Egr family expression in routine gel shift or immunoblot assays. hEK-293 cells were maintained in 10-cm-diameter dishes at 37°C and 5% CO2 in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, glutamine (2 mM), and a penicillin-streptomycin mixture (50 U ml−1). At 24 h prior to transient transfection via calcium phosphate precipitation (3), cells were passed into either 10-cm (mutagenesis studies)- or 38-mm (reporter studies)-diameter six-well dishes. For mutagenesis studies, cells were transfected with 5 μg of Egr3 expression plasmid, rinsed 10 to 16 h later, and harvested 24 h after rinsing. For reporter assays, we transfected cells with much lower levels of Egr3 expression plasmid (50 ng) to ensure that the values obtained were in the linear range of the luciferase assay. In addition, cells were transfected with 100 ng of reporter plasmid and 20 ng of β-galactosidase (β-Gal) plasmid (to monitor transfection efficiency) and left undisturbed for 10 to 16 h after initiation of the transfection process. To control for background reporter activity derived from endogenous hEK-293 cell proteins, we conducted parallel transfection experiments with the luciferase reporter, the β-Gal construct, and an empty CMV vector. We consistently found that there is little detectable background reporter activity in the hEK-293 cells under these conditions. Cells were washed twice with warm medium and then fed with fresh medium; 24 h later, cells were rinsed twice with warm PBS and harvested in 1× reporter lysis buffer (Promega) and placed into 1.5-ml tubes on ice. Extracts were vortexed for 10 s and centrifuged for 5 min at 14,000 × g. Supernatants were collected, aliquoted, and used for both the luciferase and luminescent β-Gal assays, conducted according to the protocols of the manufacturers (Promega and Clontech, Palo Alto, Calif.). For each, both luciferase and β-Gal assays were performed in triplicate, and average values were used in further analysis. To help control for variability in transfection efficiency, β-Gal activity was used to normalize the luciferase values obtained.
RESULTS
αEgr3-INT recognizes both delayed ERE binding complexes.
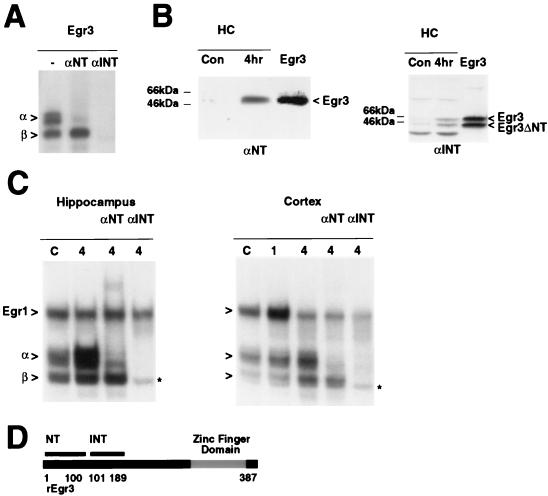
To test the possibility that the faster-migrating, delayed ERE DNA binding complex detected in vivo contains a truncated isoform of Egr3, we generated an antibody directed against an internal portion of the Egr3 protein that spanned amino acid residues 101 to 189 (αEgr3-INT [Fig. 1D]). To evaluate these antibodies, we first performed gel shift assays on recombinant Egr3 synthesized in vitro. As described previously, incubation of recombinant Egr3 with the ERE probe generates two bands, referred to as α and β (Fig. 1A), and the αEgr3-NT antibody disrupts complex α but not complex β. In contrast, αEgr3-INT inhibits both complexes. Thus, the faster-migrating β complex likely contains a shorter form of Egr3 missing the N-terminal domain recognized by αEgr3-NT. Immunoblot analysis of recombinant Egr3 and endogenous Egr3 in hippocampal extracts provided direct confirmation of this interpretation (Fig. 1B). αEgr3-NT detects a single protein band in hippocampal extracts that is induced following a seizure stimulus and comigrates with recombinant Egr3. In contrast, αEgr3-INT detects two recombinant Egr3 protein bands, an upper band or longer isoform also recognized by αEgr3-NT and a lower band corresponding to a shorter form that reacts only with αEgr3-INT. As expected, both were induced following a seizure stimulus (Fig. 1B), as found for the corresponding α and β gel shift complexes (28).
FIG. 1.
Identification of a truncated Egr3 isoform. (A) αEgr3-INT detects two Egr3 isoforms in a gel shift analysis of recombinant Egr3 incubated either without antibody (−) or with αEgr3-NT (αNT) or αEgr3-INT (αINT). α and β refer to the slower- and faster-migrating Egr3-containing complexes, respectively. (B) Immunoblots of recombinant Egr3 run adjacent to control (Con) and 4-h post-MECS rat hippocampal extracts (HC) probed with αEgr3-NT (1:1,000 dilution) and labeled αNT (left) and of similar extracts probed with αEgr3-INT (1:500 dilution) and labeled αINT (right). The band labeled Egr3 is recognized by αEgr3-NT and αEgr3-INT. In contrast, the band labeled Egr3ΔNT is detected only by αEgr3-INT, indicating that it is truncated at the N terminus. (C) αEgr3-INT abolishes both delayed ERE binding complexes in vivo in ERE gel shift assays using 32P-labeled ERE oligonucleotide probe on control and MECS-treated hippocampal (left) or cortical (right) extracts. α and β refer to Egr3-containing complexes. The slowest-migrating band in the lane labeled αNT (left) is a supershifted Egr3α complex. The slowest-migrating band in all other lanes is Egr1. The asterisk refers to a nonspecific band that does not display sequence-specific ERE binding activity. (D) Schematic representation of rat Egr3 (rEgr3) and the segments used to generate the αEgr3-NT (amino acids 1 to 100) and αEgr3-INT (amino acids 101 to 189) antibodies. The zinc finger DNA binding domain is highlighted in gray.
To determine directly whether the β complex detected in vivo contains the shorter Egr3 isoform, we added αEgr3-INT to the gel shift reaction mixture. As found for recombinant Egr3, αEgr3-INT disrupted binding of both α and β complexes whereas αEgr3-NT selectively blocked the binding activity of complex α (Fig. 1C). Taken together, these results provide compelling evidence that the β ERE binding complex observed in vivo contains a truncated isoform of Egr3 that is missing the N-terminal domain recognized by αEgr3-NT. As expected, the αEgr3-INT antibody does not affect the residual nonspecific band that migrates in proximity with the β complex. In previous studies, we found that this band does not contain a specific ERE binding complex, since it is displaced by a mutant ERE oligonucleotide not recognized by Egr family members (28).
The truncated Egr3 isoform is widely expressed in the brain.
To assess whether the shorter Egr3 isoform is also expressed in other brain regions or is restricted to the hippocampus, we conducted similar studies of extracts harvested from the cerebral cortex. In gel shift assays of cortical extracts from control and MECS-treated rats, we observed that three distinct ERE binding complexes (Fig. 1C) were increased following stimulation. This pattern of ERE DNA binding activity closely mimics that observed in the hippocampus (28). At 1 h following MECS, the uppermost complex is induced, returning to control levels by 4 h, when elevations in the α and β complexes are apparent. As found previously in studies using hippocampal extracts, the slowest-migrating ERE binding complex in the cortex contains Egr1, as its binding activity is selectively abolished by an Egr1 antibody (data not shown), and the α complex contains Egr3, since it is selectively eliminated by αEgr3-NT. In addition, both α and β complexes are blocked by αEgr3-INT.
To determine if this pattern of ERE binding activities is also induced by other stimuli, we performed gel shifts with the ERE probe on striatal extracts following administration of cocaine, as this treatment has been shown to trigger rapid increases in both Egr1 and Egr3 mRNAs in this region (1, 25, 41). In these gel shift assays, we detected the same progression of ERE binding complexes and antibody sensitivities (data not shown) found in the hippocampus and cortex following MECS stimulation. Taken together, the data from the hippocampus, cortex, and striatum indicate that neuronal stimulation elicits the delayed induction of two distinct Egr3-containing complexes; one corresponds to an Egr3 isoform that contains the N terminus, while the other isoform does not.
Egr3 knockout mice lack the α and β complexes.
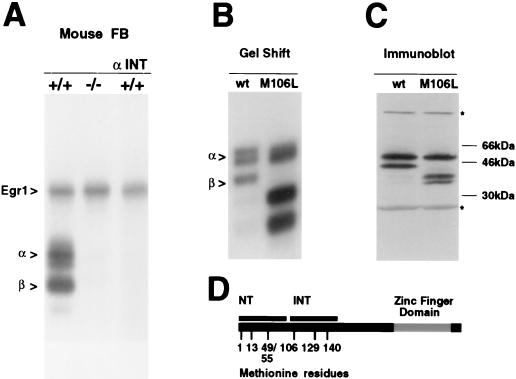
The recent generation of Egr3 knockout mice (37) enabled us to perform a rigorous test of our interpretation of these antibody studies. If our view is correct, then the α and β complexes should be absent in extracts prepared from these mice. The pattern of ERE binding complexes detected in forebrain extracts prepared from wild-type mice was identical to that observed in the rat brain (Fig. 2A). However, as predicted, extracts prepared from Egr3 knockout mice lacked the α and β complexes, which were also abolished by the αEgr3-INT antibody in extracts prepared from wild-type mouse brain (Fig. 2A). Of note, the Egr1 complex was unchanged in both wild-type and Egr3 knockout mice. Thus, these studies support the conclusion that the α and β complexes found in both rat and mouse brain correspond to Egr3 isoforms that include and lack the N terminus, respectively.
FIG. 2.
Egr3 knockout mice lack α and β complexes. (A) Pattern of ERE binding complexes detected in 4-h postseizure forebrain (FB) extracts from wild-type (+/+) and Egr3 knockout (−/−) mice. In the right lane, labeled αINT, wild-type extracts were incubated with αEgr3-INT. α and β refer to specific Egr3-containing complexes in panels A and B. We eliminated binding of the nonspecific band in mouse brain extracts by inclusion of 100-fold excess of unlabeled mutant ERE oligonucleotide. (B) Mutation of Met 106 abolishes expression of truncated Egr3 isoform, determined by a gel shift assay of recombinant Egr3 synthesized in vitro from wild-type (wt) and mutated (M106L) Egr3 cDNA templates. (C) Immunoblot of recombinant extracts containing wild-type or mutated (M106L) Egr3 probed with αEgr3-INT (1:500 dilution). The asterisks refer to cross-reacting proteins that are present in the rabbit reticulocyte preparations. (D) Schematic representation of rat Egr3 protein depicting Met residues 1, 13, 49, and 55 within the NT epitope and Met residues 106, 129, and 140 within the INT epitope. The zinc finger DNA binding domain is shown in gray.
Mutation of Met 106 blocks formation of the truncated Egr3 isoform in vitro.
Since both Egr3 isoforms are expressed in vitro from the same cDNA template, we suspected that the shorter isoform is generated via a posttranscriptional mechanism rather than alternative splicing. Either proteolytic cleavage of full-length Egr3 or utilization of an alternative translation start site could yield a truncated Egr3 isoform with an N terminus located just prior to or within the internal segment targeted by αEgr3-INT (amino acids 101 to 189). However, we favored the latter alternative since we reliably obtain the shorter isoform in vitro despite the presence of protease inhibitors. Furthermore, prolonging the incubation does not lead to production of higher levels of the shorter isoform at the expense of the longer isoform, as might be expected if generation of the shorter isoform depended on proteolytic cleavage of the longer one.
To test the possibility that the shorter isoform is generated by utilization of an alternative translation start site, we checked whether mutating candidate Met residues eliminated production of this isoform in vitro. We focused on the Met residues located at positions 106, 129, and 140 since they are conserved in rats and mice and could serve as alternative translation initiation sites that generate Egr3 isoforms recognized by αEgr3-INT but not αEgr3-NT. We first tested Met 106 by generating an Egr3 point mutant construct, Egr3M106L, containing a substitution of Leu for Met at residue 106. Recombinant Egr3 was synthesized in vitro from both wild-type and Egr3M106L mutant constructs and used in immunoblot and gel shift experiments. Immunoblots probed with αEgr3-INT demonstrated that the M106L mutation led to a selective loss of the shorter isoform (Fig. 2C). Also, gel shift assays revealed that this mutation abolished the Egr3-containing complex β (Fig. 2B). These results indicate that the shorter Egr3 isoform is made by initiating translation at M106. Unexpectedly, we found that the αEgr3-INT antibody detected two lower-molecular-weight protein bands in immunoblots of Egr3M106L recombinant extracts. Furthermore, two additional ERE binding complexes were observed in the gel shift assay. Presumably, elimination of the preferred start site at M106 favors utilization of the downstream Met residues at positions 129 and 140. Examination of additional mutants described below supports this inference.
Use of additional translation start sites.
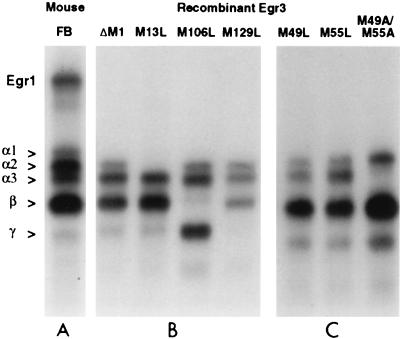
In the course of conducting these studies, we optimized conditions for separating the Egr3 DNA binding complexes and noticed that a complex previously thought to contain only full-length Egr3 could be reliably resolved into three distinct complexes, α1, α2, and α3 (Fig. 3). In addition to the initial Met, there are three Met residues at positions 13, 49, and 55 located within the N-terminal segment targeted by αEgr3-NT (Fig. 2D). Since our analysis of the β complex indicates that it contains a shorter Egr3 isoform derived from initiation at Met 106, we wondered whether the α complex might include additional complexes formed by truncated versions of Egr3 that start at Met residues near the N terminus. To address this possibility, we made a series of mutations targeting the potential internal translation start sites and examined their effects on the pattern of ERE binding complexes generated in vitro.
FIG. 3.
Mutation of specific Met codons abolishes expression of Egr3 isoforms in vitro. (A) Pattern of ERE binding complexes detected in wild-type mouse forebrain (FB), included to highlight the close similarity between Egr3 complexes expressed in vivo and in vitro. (B and C) Gel shift assays of recombinant Egr3 synthesized in vitro from the mutated versions of the Egr3 cDNA indicated above the lanes. All Egr3 cDNAs shown in panels B and C are in the Egr3ΔM1 vector backbone and thus do not express Egr3α1.
In initial studies, we demonstrated that deletion of Met 1 eliminates the α1 complex (Fig. 3), which represents the ERE binding complex containing a full-length Egr3. Using this construct, Egr3ΔM1, as a template, we generated a series of constructs with point mutations at the next five Met residues (at positions 13, 49, 55, 106, and 129). The M13L mutation abolished formation of the α2 complex. Since we had already shown that formation of the β complex was dependent on M106, we predicted that either M49 or M55 would be critical for generating the α3 complex. Unexpectedly, analysis of the M49L and M55L mutants (Fig. 3) revealed that each individual mutation did not completely abolish the α3 complex. However, the M49L mutation partially blocked the α3 band whereas the M55L mutant had little or no effect. In contrast, the M49A/M55A double mutant resulted in a complete abolition of the α3 complex (Fig. 3). These findings suggest that both M49 and M55 are utilized and that we are unable to resolve their corresponding complexes. Accordingly, we refer to the α3 complex as being derived from M49/55.
As shown in Fig. 2B, the Egr3M106L mutation eliminates the β complex as well as enhancing synthesis of faster-migrating ERE binding complexes thought to reflect initiation at M129 and M140. Close inspection of the complexes detected in vivo as well as in vitro revealed the consistent presence of a minor band that migrates ahead of the β complex and comigrates with the major complex generated by the M106L mutant. This band, labeled γ, is induced by neuronal stimulation (Fig. 1C) disrupted by αEgr3-INT (Fig. 1C and 2A) and absent in Egr3 knockout mice (Fig. 2A), indicating that it is a bona fide, albeit minor, ERE binding complex present in vivo. It is abolished by the M129L mutation, indicating that it contains a truncated form of Egr3 initiated at this Met. Furthermore, these findings support our contention that the M106L mutation enhances initiation at distal Met residues. Although this mutational analysis provides strong evidence that the multiple Egr3 complexes detected in vivo reflect translation initiation at internal Met residues, we wanted to examine whether these in vitro findings accurately describe the situation in intact cells.
Egr3 isoform expression abolished by start site mutations in intact cells.
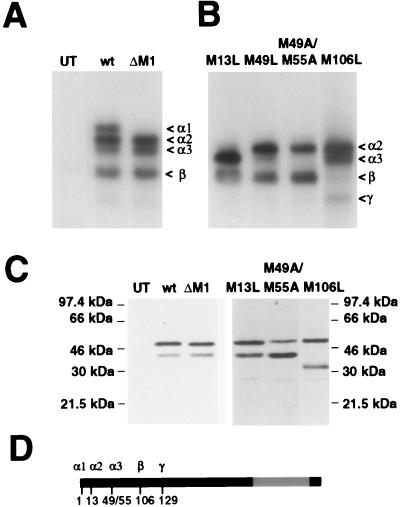
In preliminary gel shift experiments, we found that transient transfection of hEK-293 cells with the full-length Egr3 construct (CMVEgr3wt) yielded a pattern of Egr3 complexes (Fig. 4A) similar to that found in the brain. To assess the effect of mutating putative start sites in intact cells, we prepared a series of point mutants (M13L, M49L, M49A/M55A, and M106L) in the Egr3ΔM1 backbone used for the in vitro experiments. Cells transfected with the CMVEgr3ΔM1 construct lacked the full-length Egr3 α1 complex (Fig. 4A). The M13L and M106L mutations abolished the α2 and β complexes, respectively (Fig. 4B). Also, the M49L mutation partially suppressed the α3 complex, while the M49A/M55A double mutant completely abolished this band (Fig. 4B).
FIG. 4.
Mutagenesis studies of Egr3 in intact cells. (A) ERE binding complexes detected in hEK-293 cell extracts that were untransfected (UT) or transfected with CMVEgr3wt (wt [wild-type]) or ΔM1 cDNA. (B) Autoradiograph of a similar gel shift experiment performed on hEK-293 cell extracts transfected with CMVEgr3M13L, M49L, M49A/M55A, and M106L cDNAs. The mutated constructs in panel B are in the CMVEgr3ΔM1 backbone and thus do not express Egr3α1. The γ complex appears when M106 is mutated to Leu. (C) Immunoblot analysis using αEgr3-INT (1:500 dilution) performed on the nuclear extracts used for gel shift assays. αEgr3-INT detects two immunoreactive bands in CMVEgr3wt-transfected cells that correspond to the α (upper) and β (lower) gel shift complexes. The M106L mutation abolishes expression of the lower band and leads to the generation of a faster-migrating band that corresponds to that complex. (D) Schematic representation of the translation start sites of the Egr3 isoforms detected in vivo, Egr3α1 (M1), α2 (M13), α3 (M49/55), β (M106), and γ (M129). The zinc finger DNA binding domain is indicated in gray.
In previous immunoblot studies of recombinant Egr3 with αEgr3-INT, we found that the M106L mutation led to a selective loss of the truncated Egr3 isoform that corresponds to the β complex in gel shifts. To test whether the CMVEgr3M106L mutant had a similar effect in intact cells, we performed immunoblotting with αEgr3-INT on the transfected hEK-293 extracts used for the gel shift studies. In contrast to CMVEgr3wt, which drives expression of two αEgr3-INT immunoreactive bands, CMVEgr3M106L-transfected cells exhibited a selective loss of the shorter Egr3 isoform (Fig. 4C). Also, in M106L mutant extracts, αEgr3-INT detects a faster-migrating band that corresponds to the γ complex. Of note, immunoblots performed on extracts of the CMVEgr3ΔM1, CMVEgr3M13L, and CMVEgr3M49A/M55A mutants revealed that neither of these mutations completely abolished immunoreactivity of the upper band recognized by αEgr3-INT. Although we detect slight alterations in the mobility of the upper band in immunoblots of these mutant extracts, the multiple Egr3 isoforms present could not be consistently resolved into discrete bands (Fig. 4C). In summary, studies using intact cells corroborate our in vitro observations that the Egr3 isoforms (Fig. 4D) identified in vivo are generated through initiation at multiple translation start sites, i.e., M1, M13, M49/M55, M106, and M129.
Egr3 isoform transcriptional activity.
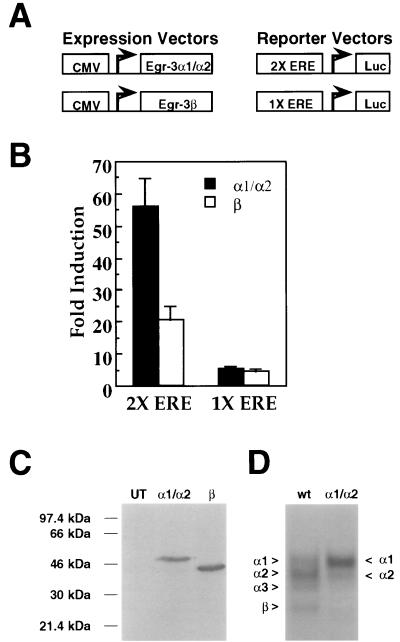
Since the domains of Egr3 mediating transcriptional activation have not been defined, we wanted to test whether the truncated Egr3β isoform is transcriptionally active and, if so, how it compares to full-length Egr3 (Egr3α1). As an initial attempt to express full-length Egr3 in the absence of truncated isoforms, we altered the sequence surrounding the initial start codon such that it contains the two most important Kozak consensus residues (A at −3 and G at +4 [17]). Although this increased the proportion of the α1 complex, substantial amounts of the α3 and β complexes were generated as well. Accordingly, we prepared the CMVEgr3M49A/M106A mutant construct in the CMVEgr3wt backbone that retains Met residues at positions 1, 13, 55, and 129. To express Egr3β (Fig. 5C), we generated CMVEgr3Δ1-105/M129L, which contains the preferred Kozak residues surrounding the Met 106 start codon and a mutated M129. To test whether these constructs could successfully drive expression of the desired Egr3 isoforms, we transfected hEK-293 cells with equal amounts of the cDNAs encoding Egr3α1 and Egr3β and prepared immunoblots of nuclear extracts from these cells. As shown in Fig. 5C, Egr3α1 and Egr3β are expressed at comparable levels when equivalent amounts of their respective cDNAs are transfected into cells. Since an immunoblot does not allow us to determine the relative expression of each Egr3α isoform, we performed parallel gel shift experiments on these extracts to monitor the individual expression of Egr3 isoform DNA binding activity. The CMVEgr3M49A/M106A construct achieved the desired result, yielding predominant expression of the α1 complex and only residual expression of Egr3α2 (Fig. 5D). Thus, we refer to this form as Egr3α1/α2.
FIG. 5.
Transcriptional activity of Egr3 isoforms. (A) Schematic representation of the expression and reporter vectors used in the reporter assays. The 1X ERE and 2X ERE reporter constructs contained either one or two copies of an ERE (GCG GGG GCG) upstream of the firefly luciferase gene. (B) Bar graph showing fold induction in luciferase activity induced by transfection with either CMVEgr3M49A/M106A (encoding Egr3α1/α2) or CMVEgr3Δ1-105/M129L (encoding Egr3β) over background reporter activity (taken from extracts of hEK-293 cell transfected with an empty CMV vector) for the 2X (left) and 1X (right) EREs. Error bars represent standard errors of the means. Statistical analysis of these data demonstrate that the Egr3 constructs tested differ significantly in their effects on the 2X ERE but not the 1X ERE (P < 0.0005 and P > 0.5, respectively; Student’s t test). (C) Immunoblot of untransfected hEK-293 cells run adjacent to hEK-293 cells transfected with 5 μg of CMVEgr3M49A/M106A (encoding Egr3α1/α2) or CMVEgr3Δ1-105/M129L (encoding Egr3β) probed with αEgr3-INT (1:500 dilution). (D) Representative gel shift experiment using a 32P-labeled ERE oligonucleotide probe on extracts of hEK-293 cells untransfected (UT) or transfected with 5 μg of CMVEgr3wt (wt [wild type]) or CMVEgr3M49A/M106A (α1/α2).
We next examined the ability of Egr3α1/α2 and Egr3β to drive expression of luciferase reporter constructs (Fig. 5A) containing either one (1X ERE) or two (2X ERE) EREs in the proximal promoter region. When we compared these isoforms on the 2X ERE promoter, we found that Egr3α1/α2 was more effective than Egr3β at driving reporter activity (Fig. 5B). In contrast, when we tested Egr3 isoform transcriptional output on a promoter containing a single copy of the ERE, Egr3α1/α2 expression and Egr3β expression had comparable effects (Fig. 5B). Thus, these data suggest that the transcriptional activity of the Egr3α1/α2 and Egr3β isoforms display different properties depending on the promoter context.
DISCUSSION
In this study, we conducted experiments aimed at identifying a major ERE binding complex expressed in the brain, defining how it is generated, and understanding its role in regulating ERE-mediated transcription. In the first phase of the study, we confirmed our hypothesis that the unidentified ERE complex contained a truncated form of Egr3 missing the N terminus. This conclusion is based on its absence from extracts prepared from Egr3 knockout mice as well as studies with an antibody directed to an internal segment of Egr3. In the second phase of the study, we obtained evidence that the shorter isoform is generated by utilization of an internal translation start site located at M106 and that additional isoforms of Egr3 are generated by utilization of other Met residues as well. In the last phase of this investigation, we compared the functional activities of Egr3 isoforms that contain or lack the N-terminal segment and found that they display distinct transactivation properties.
Although we initially identified multiple Egr3 isoforms in hippocampal extracts, gel shift studies on extracts prepared from other brain regions indicate that they express a similar array of truncated isoforms. In addition, heterologous expression of Egr3 in hEK-293 cells generated a pattern of Egr3 binding complexes that was indistinguishable from the pattern displayed in the brain. In the periphery, Egr3 is induced by T-cell activation (23). In preliminary studies, we have detected a similar pattern of Egr3 expression in a murine T-cell hybridoma, 2B4.11. Accordingly, coexpression of full-length and truncated Egr3 isoforms appears to be a general feature of this transcript.
Previous in situ hybridization studies have demonstrated induction of all four Egr family members in the hippocampus following electroconvulsant stimulation (2, 5, 8, 41). Thus, it is surprising that all of the major ERE binding complexes observed in gel shift studies contain only Egr1 and Egr3. The absence of prominent Egr2 and Egr4 binding complexes in these assays could in principle reflect low levels of protein expression or inhibitory factors that block binding activity. In any case, these findings suggest that Egr1 and Egr3 are the dominant family members mediating ERE-driven transcription in the hippocampus.
The demonstration that mutations targeting individual Met residues block expression of Egr3 isoforms provides compelling evidence that they are generated by utilization of internal translation start sites. Although viruses commonly utilize this mechanism to express variant proteins, it has been seldom implicated with regard to cellular proteins (32). A priori initiation at internal start sites can be attributed either to internal entry of the ribosomal complex, instead of its attachment to the 5′ cap, or to leaky ribosomal scanning, i.e., ignoring potential start sites presumably because the flanking sequence deviates from the rules governing faithful initiation. As shown in Table 1, the leaky scanning mechanism may apply to Egr3. However, it is unclear if this fully accounts for the ability of the translation initiation complex to skip the first Met. Substituting nucleotides at key sites surrounding the initial start codon to comply with these rules enhanced usage of the Egr3 initial Met but did not prevent generation of truncated products.
TABLE 1.
Nucleotide sequences surrounding Egr3 initiation sitesa
| Determination | Sequence | No. of matches | ||||
|---|---|---|---|---|---|---|
| −3 | +4 | |||||
| Consensus | GCC | GCC | ACC | ATG | G | |
| G | A | |||||
| rEgr3 | ||||||
| Met 1b | GGG | AGT | GCT | ATG | A | 4/10 |
| Met 13 | CCG | GTG | ACC | ATG | A | 6/10 |
| Met 49 | TAC | ACT | CAG | ATG | G | 3/10 |
| Met 55 | GAG | AAG | GTG | ATG | G | 3/10 |
| Met 106 | ATT | AGC | CTC | ATG | A | 3/10 |
| Met 129 | ACG | GCT | AGC | ATG | G | 6/10 |
The preferred 13-nucleotide Kozak consensus is shown in bold above the rat Egr3 (rEgr3) sequence. Residues −3 and +4 of the Kozak sequence are highlighted, as these are considered to be the two most important residues. While A at −3 and G at +4 are preferred, either purine present at these sites serves as a proinitiation signal. Residues shown underlined and in bold indicate identity to the consensus sequence. The number of perfect matches excluding the ATG start codon is indicated on the right.
For the initial methionine, we used the human Egr3 sequence, as the reported rat Egr3 sequence does not include any 5′ untranslated region.
Although alternative splicing is widely recognized as a mechanism used to generate multiple protein isoforms from a single transcription factor gene, only a few instances in which alternative translation start sites are utilized have been reported. Interestingly, two members of the C/EBP family, C/EBPβ (9) and C/EBPα (30), employ this strategy to generate truncated versions with markedly different transcriptional properties. As the splicing mechanism is not available to these intronless genes, the alternative translation start site strategy appears to be used instead. This teleological rationale presumably applies to Egr3 as well, since it contains only two exons, as found for all Egr family genes (8).
Our comparison of the functional activities of Egr3 isoforms that retain or lack the N-terminal segment indicates that deletion of this segment has little effect on Egr3’s ability to stimulate transcription of genes containing a single copy of the ERE in their promoter. In contrast, deletion of the N-terminal segment impairs its ability to stimulate transcription when two copies of the ERE are present. Accordingly, the N-terminal domain appears to play an important role in boosting expression of genes containing multiple EREs. Based on these findings, it is tempting to speculate that altering the ratio of Egr3α and Egr3β isoforms may provide a mechanism for modulating Egr target genes with multiple EREs such as platelet-derived growth factor A chain (15, 33), prohormone convertase 2 (13), and synapsin I (36) without affecting those containing a single ERE, e.g., platelet-derived growth factor B chain (14), transforming growth factor (10, 16), luteinizing hormone beta chain (18), and FasL (23). In this scenario, shifts in the functional activities of full-length and truncated Egr3 isoforms could be achieved directly by changes in their protein levels. Indeed, preliminary findings (26a) suggest that there is some developmental regulation of the pattern of Egr3α isoform expression. Egr3α1 is present at relatively high levels in embryonic day 17 rat cortex, and its levels decrease to near adult levels by postnatal day 8. In contrast, Egr3α2 DNA binding activity is low in embryonic day 17 cortex but its levels are significantly increased in postnatal day 8 cortex. Alternatively, as the N-terminal truncation does not impinge on the domain mediating suppression of Egr3 by NAB proteins (31, 34) located just N terminal to the DNA binding domain, it is conceivable that changes in the relative sensitivities of full-length and truncated Egr isoforms to NAB proteins could be used to alter their activities.
Although the Egr family DNA binding domain has been extensively characterized, little is known about how Egr family members act to stimulate the transcriptional apparatus. Our initial analysis of naturally occurring truncations of Egr3 has revealed interesting differences in the activation properties of these isoforms and underscores the need to conduct more extensive studies aimed at defining the activation domains contained within these proteins and how they interact with the transcriptional apparatus. Progress in this direction will be helpful in elucidating the functional significance of the Egr3 isoforms expressed in vivo as well as in understanding how the Egr family orchestrates changes in gene expression in response to cellular stimulation.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grants from the National Institute of Drug Abuse (DA00266 and DA00358 [J.M.B.] and DA05753 [K.J.O.]) and an NARSAD independent investigator award (J.M.B.).
We thank E. Eipper, D. Ginty, S. S. Wang, and P. Worley for use of equipment, helpful discussions, and advice; D. Ahn, Y. S. Kwon, and W. Z. Tang for expert technical assistance; Jeff Milbrandt and Warren Tourtellotte for generously providing reagents and samples; and D. Ginty for critical reading of the manuscript.
REFERENCES
- 1.Bhat R V, Cole A J, Baraban J M. Chronic cocaine treatment suppresses basal expression of zif268 in rat forebrain: in situ hybridization studies. J Pharmacol Exp Ther. 1992;263:343–349. [PubMed] [Google Scholar]
- 2.Bhat R V, Worley P F, Cole A J, Baraban J M. Activation of the zinc finger encoding gene krox-20 in adult rat brain: comparison with zif268. Brain Res Mol Brain Res. 1992;13:263–266. doi: 10.1016/0169-328x(92)90034-9. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci USA. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole A J, Abu-Shakra S, Saffen D W, Baraban J M, Worley P F. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55:1920–1927. doi: 10.1111/j.1471-4159.1990.tb05777.x. [DOI] [PubMed] [Google Scholar]
- 6.Cole A J, Saffen D W, Baraban J M, Worley P F. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 7.Crosby S D, Puetz J J, Simburger K S, Fahrner T J, Milbrandt J. The early response gene NGFI-C encodes a zinc finger transcriptional activator and is a member of the GCGGGGGCG (GSG) element-binding protein family. Mol Cell Biol. 1991;11:3835–3841. doi: 10.1128/mcb.11.8.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosby S D, Veile R A, Donis-Keller H, Baraban J M, Bhat R V, Simburger K S, Milbrandt J. Neural-specific expression, genomic structure, and chromosomal localization of the gene encoding the zinc-finger transcription factor NGFI-C. Proc Natl Acad Sci USA. 1992;89:4739–4743. doi: 10.1073/pnas.89.10.4739. . (Erratum, 89:6663.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 10.Dey B R, Sukhatme V P, Roberts A B, Sporn M B, Rauscher III F J, Kim S J. Repression of the transforming growth factor-beta 1 gene by the Wilms’ tumor suppressor WT1 gene product. Mol Endocrinol. 1994;8:595–602. doi: 10.1210/mend.8.5.8058069. [DOI] [PubMed] [Google Scholar]
- 11.Gashler A, Sukhatme V P. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 12.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 13.Jansen E, Ayoubi T A, Meulemans S M, Van De Ven W J. Regulation of human prohormone convertase 2 promoter activity by the transcription factor EGR-1. Biochem J. 1997;328:69–74. doi: 10.1042/bj3280069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khachigian L M, Lindner V, Williams A J, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 15.Khachigian L M, Williams A J, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 16.Kim S J, Park K, Rudkin B B, Dey B R, Sporn M B, Roberts A B. Nerve growth factor induces transcription of transforming growth factor-beta 1 through a specific promoter element in PC12 cells. J Biol Chem. 1994;269:3739–3744. [PubMed] [Google Scholar]
- 17.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S L, Sadovsky Y, Swirnoff A H, Polish J A, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 19.Lin K I, Lee S H, Narayanan R, Baraban J M, Hardwick J M, Ratan R R. Thiol agents and Bcl-2 identify an alphavirus-induced apoptotic pathway that requires activation of the transcription factor NF-kappa B. J Cell Biol. 1995;131:1149–1161. doi: 10.1083/jcb.131.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linden D J. A protein synthesis-dependent late phase of cerebellar long-term depression. Neuron. 1996;17:483–490. doi: 10.1016/s0896-6273(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 21.Mack K J, Mack P A. Induction of transcription factors in somatosensory cortex after tactile stimulation. Brain Res Mol Brain Res. 1992;12:141–147. doi: 10.1016/0169-328x(92)90077-o. [DOI] [PubMed] [Google Scholar]
- 22.Mello C V, Clayton D F. Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J Neurosci. 1994;14:6652–6666. doi: 10.1523/JNEUROSCI.14-11-06652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittelstadt P R, Ashwell J D. Cyclosporin A-sensitive transcription factor Egr-3 regulates Fas ligand expression. Mol Cell Biol. 1998;18:3744–3751. doi: 10.1128/mcb.18.7.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montarolo P G, Goelet P, Castellucci V F, Morgan J, Kandel E R, Schacher S. A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 25.Moratalla R, Robertson H A, Graybiel A M. Dynamic regulation of NGFI-A (zif268, egr1) gene expression in the striatum. J Neurosci. 1992;12:2609–2622. doi: 10.1523/JNEUROSCI.12-07-02609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris M E, Viswanathan N, Kuhlman S, Davis F C, Weitz C J. A screen for genes induced in the suprachiasmatic nucleus by light. Science. 1998;279:1544–1547. doi: 10.1126/science.279.5356.1544. [DOI] [PubMed] [Google Scholar]
- 26a.O’Donovan, K. J. Unpublished data.
- 27.O’Donovan K J, Tourtellotte W G, Milbrandt J, Baraban J M. The Egr family of transcription regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 28.O’Donovan K J, Wilkens E P, Baraban J M. Sequential expression of Egr-1 and Egr-3 in hippocampal granule cells following electroconvulsive stimulation. J Neurochem. 1998;70:1241–1248. doi: 10.1046/j.1471-4159.1998.70031241.x. [DOI] [PubMed] [Google Scholar]
- 29.Okuno H, Miyashita Y. Expression of the transcription factor Zif268 in the temporal cortex of monkeys during visual paired associate learning. Eur J Neurosci. 1996;8:2118–2128. doi: 10.1111/j.1460-9568.1996.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 30.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo M W, Sevetson B R, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 33.Silverman E S, Khachigian L M, Lindner V, Williams A J, Collins T. Inducible PDGF A-chain transcription in smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am J Physiol. 1997;273:H1415–H1426. doi: 10.1152/ajpheart.1997.273.3.H1415. [DOI] [PubMed] [Google Scholar]
- 34.Svaren J, Sevetson B R, Apel E D, Zimonjic D B, Popescu N C, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swirnoff A H, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiel G, Schoch S, Petersohn D. Regulation of synapsin I gene expression by the zinc finger transcription factor zif268/egr-1. J Biol Chem. 1994;269:15294–15301. [PubMed] [Google Scholar]
- 37.Tourtellotte W G, Milbrandt J. Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat Genet. 1998;20:87–91. doi: 10.1038/1757. [DOI] [PubMed] [Google Scholar]
- 38.Wallace C S, Withers G S, Weiler I J, George J M, Clayton D F, Greenough W T. Correspondence between sites of NGFI-A induction and sites of morphological plasticity following exposure to environmental complexity. Brain Res Mol Brain Res. 1995;32:211–220. doi: 10.1016/0169-328x(95)00076-5. [DOI] [PubMed] [Google Scholar]
- 39.Wisden W, Errington M L, Williams S, Dunnett S B, Waters C, Hitchcock D, Evan G, Bliss T V, Hunt S P. Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron. 1990;4:603–614. doi: 10.1016/0896-6273(90)90118-y. [DOI] [PubMed] [Google Scholar]
- 40.Worley P F, Christy B A, Nakabeppu Y, Bhat R V, Cole A J, Baraban J M. Constitutive expression of zif268 in neocortex is regulated by synaptic activity. Proc Natl Acad Sci USA. 1991;88:5106–5110. doi: 10.1073/pnas.88.12.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagata K, Kaufmann W E, Lanahan A, Papapavlou M, Barnes C A, Andreasson K I, Worley P F. Egr3/Pilot, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalizes with Egr1/zif268. Learn Mem. 1994;1:140–152. [PubMed] [Google Scholar]