Abstract
Severe acute respiratory syndrome coronavirus (SARS‐CoV‐2) has affected all inhabited continents, and India is currently experiencing a devastating second wave of coronavirus disease‐2019 (COVID‐19). Here, we examined the duration of clearance of SARS‐CoV‐2 in respiratory samples from 207 infected cases by real‐time reverse‐transcription polymerase chain reaction (RT‐PCR). A substantial proportion of COVID‐19 positive cases with cycle threshold (Ct) values more than or equal to 31 (45.7%) were subsequently tested negative for SARS‐CoV‐2 RNA within 7 days of initial detection of the viral load. A total of 60% of all the patients with COVID‐19, irrespective of their Ct values, cleared SARS‐CoV‐2 RNA within 14 days of the initial detection. Longitudinal assessment of RT‐PCR test results in individuals requiring 15–30 days to clear SARS‐CoV‐2 RNA showed a significant reduction of the viral load in samples with high or intermediate viral loads (Ct values ≤ 25 and between 26 and 30, respectively) but the follow‐up group with low viral RNA (Ct values ≥ 31) exhibited a stable viral load. Together, these results suggest that COVID‐19 positive cases with Ct values more than or equal to 31 require reduced duration to clear SARS‐CoV‐2, and thus, a shorter isolation period for this group might be considered to facilitate adequate space in the COVID Care Centres and reduce the burden on healthcare infrastructure.
Keywords: COVID 19, cycle threshold value, isolation, RT‐PCR, SARS‐CoV‐2
1. INTRODUCTION
Coronavirus disease‐2019 (COVID 19) caused by severe acute respiratory syndrome coronavirus (SARS‐CoV‐2) is a serious global health threat that the world is facing since December 2019. India reported the first confirmed case of COVID‐19 on January 30, 2020, and the cases rose exponentially in the next 8 months, the highest peak was recorded in the middle of September and then the caseload started declining for the next few months. However, India is witnessing the second wave of COVID‐19 pandemic since the first half of April 2021. The raging wave has greatly impacted the healthcare system with a deficiency in beds, oxygen, and medical supplies. The respiratory illness is transmitted in the community through respiratory droplets and contacts of infected people. 1 , 2 SARS‐CoV‐2 infection could be asymptomatic or symptomatic and the viral load is not associated with disease severity. 3 The most common symptoms reported in patients are fever, fatigue, sore throat, dry cough, and breathlessness whereas headache, dizziness, abdominal pain, diarrhea, nausea, and vomiting are the less common symptoms. 4
Real‐time reverse‐transcription polymerase chain reaction (RT‐PCR) test has been routinely used as the diagnostic test to detect SARS‐CoV‐2 nucleic acids in respiratory specimen swabs from throat, nasopharynx, and nose of individuals suspected of COVID‐19 during the acute phase of infection. Different viral target genes have been used for the detection of SARS‐CoV‐2, including spike (S), nucleocapsid (N), RNA‐dependent RNA polymerase (RdRp), open reading frame (ORF) 1 (O), and envelope (E). A fully automated assay, such as the Cobas platform uses a two‐target RT‐PCR for detecting SARS‐CoV‐2: O‐gene, a nonstructural region (ORF‐1) that is specific for SARS‐CoV‐2 (target 1) and E‐gene, a structural envelope region that is common to all Sarbecovirus subgenus (target 2). 5 , 6 Cycle threshold (Ct) values in RT‐PCR have been used to measure the amplification needed for the target viral gene to cross the threshold and are inversely related to the viral load. 7 Detection of viral load is important to prevent the potential transmission of infection. A period of 14 days' isolation is generally recommended throughout the world to prevent the spread of SARS‐CoV‐2. Increased age was positively associated with prolonged RT‐PCR positivity. 8 Although SARS‐CoV‐2 RNA shedding can be detected for an extended period, the duration of a live virus is relatively short‐lived. 9 In this study, we compared the duration of SARS‐CoV‐2 viral RNA in individuals with different Ct values and evaluated the dynamics of viral load in cases that required 15–30 days to clear SARS‐CoV‐2 viral RNA.
2. MATERIALS AND METHODS
A total of 2233 nasopharyngeal swab specimens from outpatients and inpatients were collected in HiViral Transport Medium (Himedia) from COVID Care Centres in Mumbai. These samples were transferred to the laboratory under cold chain conditions within 12 h.
The Cobas SARS‐CoV‐2 real‐time RT‐PCR assay was performed on the Cobas 6800 platform as per the manufacturer's instructions (Roche Molecular Diagnostics). Briefly, after loading the samples, nucleic acid extraction and subsequent real‐time RT‐PCR on the ORF1/a nonstructural region (O‐gene) that is specific for SARS‐CoV‐2 (target 1), a structural envelope gene (E‐gene) that is common to all Sarbecovirus subgenus (target 2) and an internal control RNA (noninfectious RNA in bacteriophage) are automatically performed by the platform. If detected, a Ct or cycle threshold value is obtained for each gene.
Statistical analyses were performed using GraphPad Prism version 5.0, the Mann–Whitney test was used as the test of significance, and Spearman's rank correlation was used for correlation analysis.
3. RESULTS
A total of 1500 (67%) samples collected from the COVID Care Centres were tested positive for SARS‐CoV‐2. Of these positive cases, 207 individuals were followed until the viral load was reduced to an undetectable level by RT‐PCR. Based on the Ct values, it was observed that a decreased Ct value or higher viral load was linked to extended SARS‐CoV‐2 RNA shedding. As shown in Figure 1, a proportion of 79.2% cases with Ct values less than or equal to 25 compared with 39.5% cases with Ct values between 26%–30% and 33.6% cases with Ct values more than or equal to 31 required 15–30 days to clear SARS‐CoV‐2 viral load. We also found that 17.1% and 28.6% of individuals having low viral load (Ct values ≥ 31) were able to clear SARS‐CoV‐2 RNA within 3 days and 7 days, respectively (Figure 1). The Ct values of SARS‐CoV‐2 were positively correlated with the duration of the viral clearance (p < 0.0001 and r = 0.77). Other factors, such as the symptomatic COVID‐19, underlying medical condition (UMC), and age were not correlated with the duration of virus clearance (data not shown).
Figure 1.
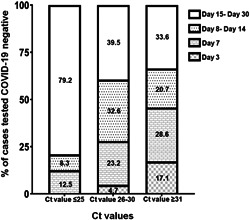
Distribution of COVID‐19 cases that tested negative when followed up over a period of 30 days at 3 days, 7 days, between 8 and 14 days, and between 15 and 30 days. Stacked graph bar chart depicts the COVID‐19 cases that tested negative for SARS‐CoV‐2 RNA in individuals with Ct values of ≤25, 26–30, and ≥31, respectively at the mentioned follow‐up days. COVID‐19, coronavirus disease 2019; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Furthermore, samples from 61 cases that required 15–30 days to clear SARS‐CoV‐2 were analyzed. The median age of this group was 50 years (22–77 years) with almost an equal ratio of symptomatic and asymptomatic cases, and 44 out of 61 (72%) individuals did not have any underlying medical conditions (Table 1). Notably, previous RT‐PCR tests between 8 and 14 days for individuals having high and intermediate viral load with initial Ct values of less than or equal to 25 and Ct values between 26 and 30, respectively showed a significant reduction in the SARS‐CoV‐2 (Figure 2A,B). However, cases having low viral load with Ct values more than 31 showed a consistent level of SARS‐CoV‐2 RNA between 8 and 14 days before becoming undetectable to RT‐PCR (Figure 2C).
Table 1.
Description of the study group with prolonged SARS‐CoV‐2 viral shedding
| Ct values | Clinical findings | Underlying medical condition (UMC) | Age (in years) | Total follow‐up cases | |||||
|---|---|---|---|---|---|---|---|---|---|
| Symptomatic | Asymptomatic | With UMC | Without UMC | >60 | 45–60 | 30–45 | 18–30 | ||
| Ct ≤ 25 | 8 | 11 | 3 | 16 | 2 | 9 | 4 | 4 | 19 |
| Ct 26–30 | 6 | 7 | 4 | 9 | 0 | 7 | 3 | 3 | 13 |
| Ct ≥ 31 | 14 | 15 | 10 | 19 | 13 | 9 | 7 | 0 | 29 |
Figure 2.
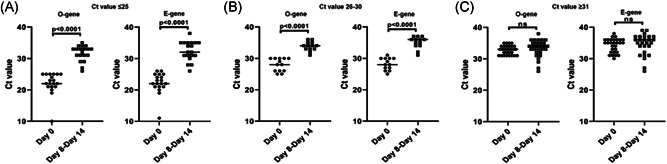
Cycle threshold (Ct) values for the O and E genes at two‐time intervals of Day 0 (initial day of testing) and Day 8–14 in follow‐up patients that required 15–30 days to clear SARS‐CoV‐2 viral load. O‐gene stands for ORF1, a nonstructural region that is specific for SARS‐CoV‐2 (target 1) and E‐gene stands for a structural envelope gene that is common to all Sarbecovirus subgenus (target 2). Scatter plot showing Ct values for O and E genes with (A) Ct values of ≤25 (n = 19), (B) Ct values between 26 and 30 (n = 13), and (C) Ct values of ≥31 (n = 29). RT‐PCR tests between 8 and 14 days revealed a significantly lowered SARS‐CoV‐2 viral load in individuals with initial Ct values of ≤25 and Ct values between 26 and 30 (p < 0.0001), but there was no significant change in the viral load of individuals with initial Ct values of ≥31 (nonsignificant). Each dot represents an individual and the horizontal lines indicate mean values. The Mann–Whitney test was used as the test of significance. RT‐PCR, reverse‐transcription polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
4. DISCUSSION
Detection of viral nucleic acid by RT‐PCR is considered as a gold standard viral diagnostic assay. The use of automated RT‐PCR assays for mass screening of individuals for SARS‐CoV‐2 has the advantage of minimal hands‐on time and accuracy of results over the conventional RT‐PCR. An automated assay, such as the Cobas 6800 test has the additional benefit of having a lower limit of detection. 6 In this study, we have used automated SARS‐CoV‐2 assay on the Cobas 6800 platform to examine the dynamics of SARS‐CoV‐2 viral load and duration of RNA shedding
We observed that individuals with an increased Ct value or lower viral load could clear SARS‐CoV‐2 viral RNA in a short duration, for instance, 66.4% of cases with Ct value more than or equal to 31 could clear the viral load within 14 days of initial detection. In contrast, only a small fraction of individuals (20.8%) with a Ct value less than or equal to 25 were able to clear the viral load during the same period. A total of 40% of all the patients with COVID‐19, irrespective of their Ct values, were positive for SARS‐CoV‐2 RNA even after 14 days of the initial detection. Nonetheless, detection of viral RNA by RT‐PCR fails to determine the presence of a replicative virus or viral infectivity. Several studies have shown that higher SARS‐CoV‐2 Ct values correspond to nonreplicative or noninfectious viral RNA, as determined by viral culture. 10 , 11 , 12 , 13 Longitudinal assessment of RT‐PCR test results in individuals requiring 15–30 days to clear SARS‐CoV‐2 RNA showed that groups with initial high viral load (Ct values ≤25) and intermediate viral load (Ct values 26–30) exhibited a significant reduction of viral load between 8 and 14 days. However, the case of the individuals having initial lower viral load with Ct values more than or equal to 31 showed consistent viral load at 8–14 days followed by the absence of SARS‐CoV‐2 RNA between days 15–‐30. Overall, a sizeable proportion of COVID‐19 positive individuals with Ct values more than or equal to 31 (45.7%) were able to clear SARS‐CoV‐2 RNA within 7 days and subsequent re‐testing of the individuals (33.6%) requiring a long time to clear the viral RNA (15–30 days) showed no significant changes in the viral load before becoming undetectable until Days 15–30. Additionally, we found that cases with Ct values more than or equal to31 and requiring 15–30 days to clear SARS‐CoV‐2 have a considerable proportion (76%) of individuals above 45 years of age and around 66% of them were without any underlying medical conditions, suggesting prolonged RNA shedding in these individuals.
A small fraction of individuals with initial Ct values of more than or equal to 31 that showed prolonged SARS‐CoV‐2 RNA shedding (15–30 days), albeit without any significant change in their viral load between 8 and 14 days, maybe considered for subsequent home quarantine after 7 days, if there is no severity in the disease symptoms. Positive/negative SARS‐CoV‐2 diagnosis with Ct values could serve as a test‐based strategy and potential guide for patient management. In conclusion, our findings suggest categorization of SARS‐CoV‐2 positive cases based on their Ct values and subsequent considerations for home quarantine for individuals with less viral load. This approach would support dealing with the COVID‐19 pandemic and its successive waves that have overburdened the healthcare system and COVID Care Centres.
CONFLICT OF INTERESTS
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Dimpu Gogoi conceived the study. Clara Aranha and Dimpu Gogoi collated the data and performed the data analysis. Testing and reporting of all the patient samples were carried out under the supervision of Vainav Patel and Vikrant Bhor. Dimpu Gogoi and Clara Aranha drafted the manuscript. All authors reviewed the manuscript and approved it for submission.
ACKNOWLEDGMENTS
We thank ICMR‐NIRRH COVID‐19 team members for their support in testing and reporting SARS‐CoV‐2 infection. We also thank the Director, ICMR‐NIRRH for her continued support.
Aranha C, Patel V, Bhor V, Gogoi D. Cycle threshold values in RT‐PCR to determine dynamics of SARS‐CoV‐2 viral load: An approach to reduce the isolation period for COVID‐19 patients. J Med Virol. 2021;93:6794‐6797. 10.1002/jmv.27206
REFERENCES
- 1. Liu J, Liao X, Qian S, et al. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis. 2020;26(6):1320‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biguenet A, Bouiller K, Marty‐Quinternet S, Brunel AS, Chirouze C, Lepiller Q. SARS‐CoV‐2 respiratory viral loads and association with clinical and biological features. J Med Virol. 2021;93(3):1761‐1765. [DOI] [PubMed] [Google Scholar]
- 4. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poljak M, Korva M, Knap Gašper N, et al. Clinical evaluation of the cobas SARS‐CoV‐2 test and a diagnostic platform switch during 48 hours in the midst of the COVID‐19 pandemic. J Clin Microbiol. 2020;58(6):e00599‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pujadas E, Ibeh N, Hernandez MM, et al. Comparison of SARS‐CoV‐2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS‐CoV‐2 test and a laboratory‐developed real‐time RT‐PCR test. J Med Virol. 2020;92(9):1695‐1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tom MR, Mina MJ. To interpret the SARS‐CoV‐2 test, consider the cycle threshold value. Clin Infect Dis. 2020;71(16):2252‐2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhattacharya B, Kumar R, Meena VP, et al. SARS‐CoV‐2 RT‐PCR profile in 298 Indian COVID‐19 patients: a retrospective observational study. Pathog Dis. 2021;79(1):ftaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta‐analysis. Lancet Microbe. 2021;2(1):e13‐e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS‐CoV‐2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663‐2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laferl H, Kelani H, Seitz T, et al. An approach to lifting self‐isolation for health care workers with prolonged shedding of SARS‐CoV‐2 RNA. Infection. 2021;49(1):95‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. [DOI] [PubMed] [Google Scholar]


