Abstract
This study aimed to systematically investigate if any of the available drugs in the electronic health record (EHR) can be repurposed as potential treatment for coronavirus disease 2019 (COVID‐19). Based on a retrospective cohort analysis of EHR data, drug‐wide association studies (DrugWAS) were performed on 9,748 patients with COVID‐19 at Vanderbilt University Medical Center (VUMC). For each drug study, multivariable logistic regression with overlap weighting using propensity score was applied to estimate the effect of drug exposure on COVID‐19 disease outcomes. Patient exposure to a drug between 3‐months prior to the pandemic and the COVID‐19 diagnosis was chosen as the exposure of interest. All‐cause of death was selected as the primary outcome. Hospitalization, admission to the intensive care unit, and need for mechanical ventilation were identified as secondary outcomes. Overall, 17 drugs were significantly associated with decreased COVID‐19 severity. Previous exposure to two types of 13‐valent pneumococcal conjugate vaccines, PCV13 (odds ratio (OR), 0.31, 95% confidence interval (CI), 0.12–0.81 and OR, 0.33, 95% CI, 0.15–0.73), diphtheria toxoid and tetanus toxoid vaccine (OR, 0.38, 95% CI, 0.15–0.93) were significantly associated with a decreased risk of death (primary outcome). Secondary analyses identified several other significant associations showing lower risk for COVID‐19 outcomes: acellular pertussis vaccine, 23‐valent pneumococcal polysaccharide vaccine (PPSV23), flaxseed extract, ethinyl estradiol, estradiol, turmeric extract, ubidecarenone, azelastine, pseudoephedrine, dextromethorphan, omega‐3 fatty acids, fluticasone, and ibuprofen. In conclusion, this cohort study leveraged EHR data to identify a list of drugs that could be repurposed to improve COVID‐19 outcomes. Further randomized clinical trials are needed to investigate the efficacy of the proposed drugs.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Drug repurposing methodologies have emerged as an attractive strategy to rapidly identify safe and effective treatments for coronavirus disease 2019 (COVID‐19). Despite recent advances, widely available treatments that can be used early in a patient’s illness to prevent hospitalization, progression to more severe outcomes, and long‐term complications have yet to be discovered.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ Can electronic health records be used to search for drug candidates that could be repurposed to treat COVID‐19?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ The study found 17 drug ingredients that are significantly associated with a decreased risk of death and other severe COVID‐19 outcomes. The study suggests that Streptococcus pneumoniae vaccines and diphtheria toxoid and tetanus toxoid vaccine, with or without acellular pertussis vaccine should not be delayed or discontinued due to the COVID‐19 pandemic as they may protect the general population from severe acute respiratory syndrome coronavirus 2 infection worldwide.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The list of drugs proposed by this study could provide additional insights into developing new candidates for COVID‐19 treatment.
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has triggered a pandemic infection leading to unprecedented excess mortality and adverse consequences to global economy. 1 , 2 According to the World Health Organization (WHO), SARS‐CoV‐2 has spread to over 222 countries and territories resulting in > 81 million infected individuals and > 1.8 million confirmed deaths as of December 2020. Although significant progress has been achieved to successfully develop and deploy safe and effective SARS‐CoV‐2 vaccines, 3 , 4 intensive scientific efforts are currently underway to discover treatments that improve COVID‐19 outcomes, particularly drug treatments that can be used early in a patient’s illness to prevent hospitalization or death. Recently, the antiviral drug remdesivir has been proven to reduce the recovery time of adult patients hospitalized with COVID‐19. 5 Another study indicated that use of dexamethasone reduces 28‐day mortality of patients hospitalized with COVID‐19 receiving mechanical ventilation or high‐flow oxygen. 6 Monoclonal antibodies have been shown to reduce the viral load and improve clinical outcomes in outpatients with mild or moderate COVID‐19. 7 Furthermore, several other drugs, including corticosteroids, antiviral therapies, immune‐modulators, and anticoagulants, are currently investigated as potential therapies for COVID‐19. 8 , 9 , 10 Despite recent advances, however, there is an urgent need for discovering safe and effective treatments that are able to prevent COVID‐19 progression and long‐term complications. 11
Because a de novo treatment usually requires many years to reach the market, involves significant costs, and has a low rate of success, drug repurposing methodologies have emerged as an attractive strategy to accelerate the discovery of novel COVID‐19 treatments. 12 , 13 Leveraging real‐world data from electronic health records (EHRs), we conducted a drug‐wide association study (DrugWAS) to systematically investigate all recorded drug exposures, including prescription drugs and dietary supplements, as potential COVID‐19 treatments. We hypothesized that drug exposures associated with a lower risk of death or severe COVID‐19 outcomes could identify candidates for further therapeutic study.
METHODS
Study design
DrugWAS is a high‐throughput method for independently investigating associations between drugs and disease outcomes. Whereas the method was designed to run each drug ingredient and COVID‐19 outcome association independently, some of the ingredients are highly correlated because they are available only in specific combinations. DrugWAS relies on a retrospective cohort analysis of data stored in the Vanderbilt University Medical Center (VUMC) Research Derivative, a daily updated database of identified EHR data restructured for research. Specific data elements extracted from the Research Derivative include demographics data, laboratory tests, drugs, clinical outcomes, comorbidities, and clinical notes. The study was approved by the institutional review board at VUMC.
Study population
The study included all patients who were tested at VUMC between March 9, 2020, and March 10, 2021, and were diagnosed with SARS‐CoV‐2 confirmed by polymerase chain reaction (PCR) assay (Figure 1 ). Being a large medical center in Middle Tennessee, and the sole provider of COVID‐19 testing early in the pandemic, VUMC tested a substantial number of individuals who never had a care visit at the medical center prior to their test. As the baseline clinical and drug exposure data for these patients was sparse, and the risk of treatment misclassification was high, patients without an encounter in the EHR within a 1‐year period prior to February 15, 2020, were excluded. The exclusion date corresponded to the date that the COVID‐19 pandemic was first detected in our geographical area. Although no age, gender, race, or ethnicity selection criteria were imposed, patients with missing demographic information (e.g., unknown race) were excluded from the study. At VUMC, SARS‐CoV‐2 PCR testing was initially limited to symptomatic individuals and a selected category of patients who were required PCR testing before their clinic visit as they needed to be physically present in a VUMC facility (e.g., pregnant women or patients scheduled for surgery). These patients, flagged as asymptomatic at the test time, were excluded from the main study because they may increase the false positive rate of COVID‐19 outcomes (e.g., admission to a hospital of an asymptomatic patient with COVID‐19 would be likely influenced by surgery rather than by the COVID‐19 diagnosis).
Figure 1.
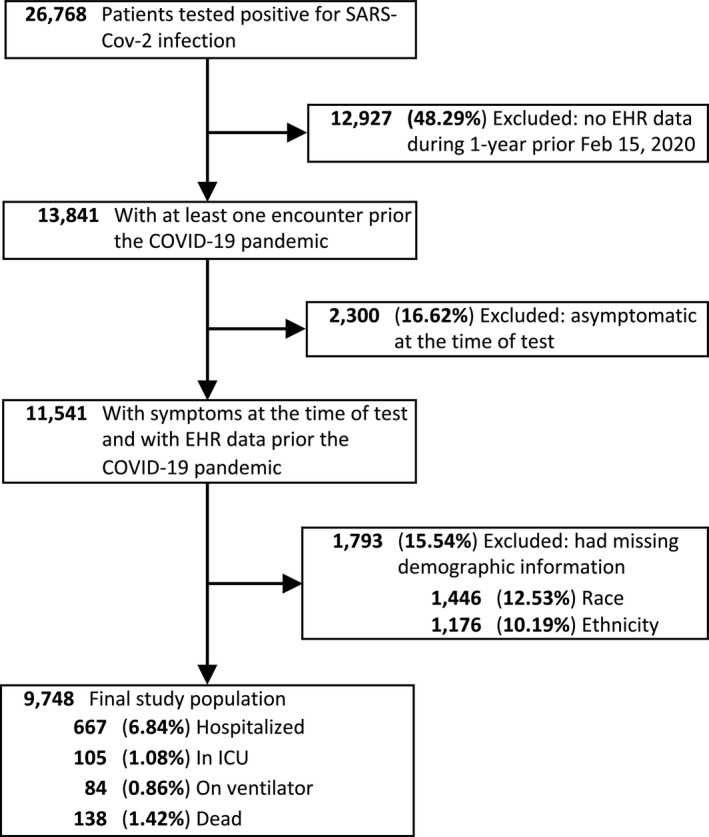
Selection of patients with COVID‐19 for the DrugWAS analysis. COVID‐19, coronavirus disease 2019; EHR, electronic health record; ICU, intensive care unit; SARS‐CoV‐2, severe acute respiratory syndrome‐coronavirus 2.
Exposure of interest
Patient exposure to a drug between November 15, 2019, (approximated as 3‐months prior to the pandemic arriving in our geographical area) and the first time a patient tested positive was selected as the exposure of interest for each drug‐outcome association study in DrugWAS (Figure S1 ). Longer drug exposure intervals starting 6‐months and 1‐year prior to the pandemic were selected for sensitivity analysis. An additional exploratory analysis investigated associations for a selected set of therapeutic categories (drug classes), including antidepressants, antihistamines, nonsteroidal anti‐inflammatory drugs, omega‐3 supplements, sigma‐1 receptor agonists, serotonin and norepinephrine reuptake inhibitors, selective serotonin reuptake inhibitors, and tricyclic antidepressants. Here, a positive drug class exposure was assigned to all patients who were exposed to at least one drug in the corresponding class. The drugs included in each category for this exploratory analysis are listed in Table S1 .
All generic and brand drugs recorded in the EHR for the final study population during the exposure interval were extracted from the drug table and normalized to drug ingredients using a previously developed drug normalization pipeline. 14 Natural language processing (NLP) was also used to extract drug information from free‐text notes. This process contributed to increasing the sample size of drug exposure cohorts and to identifying exposure of drugs that were not prescribed at VUMC (e.g., over‐the‐counter drugs or drugs prescribed by outside providers). For this, we used MedXN‐v1.0.3, 15 a high‐performance NLP drug extractor previously evaluated on the Vanderbilt EHR, 16 , 17 to parse > 1 million notes with dates between February 15, 2019, and PCR test time. There was no filter restriction by note type for NLP‐based drug extraction; thus, notes such as problem lists, clinical communications, and outpatient Rx order summaries were also included in this process. Diagnostic drug ingredients, excipients, and other nontherapeutic agents (e.g., placebo and inert ingredients) were excluded from the study. Drugs and supplements were assigned a drug class using the Lexicomp database.
Outcomes
Outcomes were extracted using EHR data from the time of the first positive test until March 10, 2021 (Figure S1 ). Based on the WHO guidelines on COVID‐19 severity scale, they were classified as: (1) never hospitalized, (2) hospitalized with mild conditions and without intensive treatment (hospitalized‐mild), (3) admitted to the intensive care unit (ICU), (4) on mechanical ventilation, and (5) dead. All‐cause of death was selected as the primary outcome. Two strategies were designed to combine hospitalized‐mild, admitted to the ICU, and on mechanical ventilation into multiple secondary outcomes: (1) cumulative severity, where a specific category is combined with more severe categories on the scale, and (2) exclusive severity, which includes only the patients from a specific category. For instance, the cumulative severity strategy for the ICU outcome also includes on ventilator and dead categories, whereas the exclusive severity for the same outcome includes only the patients in ICU (Figure S2 ). Of note, for the primary outcome, both strategies will generate the same severity group. All the severity groups corresponding to the primary and secondary outcomes were compared against non‐hospitalized, alive patients with COVID‐19.
Covariates
Age, sex, race, and ethnicity were selected to account for the differences in patient characteristics. Additionally, the weighted Elixhauser comorbidity score was chosen to account for the severity of medical conditions because multiple comorbidities have been shown to be associated with COVID‐19 outcomes. 18 The weighted Elixhauser comorbidity score represents a summary measure of patient comorbidities that has been commonly used for comorbidity adjustment in various epidemiological studies. 19 The covariate encoding this comorbidity score was computed by: (1) extracting the International Classification of Diseases, 9th/10th Revision, Clinical Modification (ICD‐9/10‐CM) billing codes from each patient record up to the PCR test time and determining their inclusion in any of the 31 Elixhauser comorbidity groups 20 ; (2) aggregating the severity scores derived from associations between the 31 comorbidity groups and the risk of in‐hospital death; and (3) categorizing the aggregated severity scores into 4 ordinal categories: < 0, 0, 1–4, and 5+. The impact of comorbidity adjustment has been further explored through sensitivity analysis where the categorical predictor encoding the weighted Elixhauser comorbidity score has been replaced with 14 individual comorbidity predictors representing chapters of the ICD‐10‐CM codes (Table S2 ).
Statistical analysis
Patient characteristics were reported as means and SDs for continuous variables and counts (percentages) for categorical variables.
For each drug studied, a propensity score method was used to adjust for differences between the patients exposed to the drug prior to being diagnosed with SARS‐CoV‐2 (exposed group) and those not exposed (unexposed group). The propensity score represents the probability of a patient being assigned to the exposed group conditional on the observed patient characteristics. In observational, nonrandomized studies, propensity score methods are used to balance the main patient characteristics across treatment groups, which is essential in reducing the bias in estimating treatment effects. 21 , 22 In DrugWAS, the propensity score adjustment played a critical role in reducing the likelihood of confounding (especially confounding by indication) because patients exposed to drugs are likely to have comorbidities as treatment indications that may also affect the study outcomes.
This study applied the overlap weighing with a propensity score method. The method has been shown to achieve high performance under different configurations, 23 and, recently, it has been successfully used in estimating the relationship between use of specific drugs and COVID‐19 outcomes. 24 , 25 Specifically, the propensity score for being exposed to a drug was estimated by a multivariable logistic regression model using age, sex, race, ethnicity, and weighted Elixhauser comorbidity score. Using the estimated propensity score, a weighted multivariable logistic regression (adjusted for drug exposure, age, sex, race, ethnicity, and weighted Elixhauser comorbidity score) was performed to estimate the effect of drug exposure on both primary and secondary outcomes, where each patient was weighted with the probability of the patient being assigned to the opposite exposure group. The estimates of adjusted odds ratio (OR) and 95% confidence intervals (CIs) were reported in the results. Associations corresponding to a specific outcome were performed for all drugs with at least 100 exposed patients of whom at least 5 had the outcome. All drugs with corresponding effect estimates indicating reduced severity risk (OR < 1) were reported as potential candidates for COVID‐19 treatment repurposing. No adjustment for multiple testing was performed. All statistical analyses were done in R, version 3.6.1.
RESULTS
Patients
The study included 9,748 patients infected with SARS‐CoV‐2. The mean age was 42 years and most patients were women (60.2%), White (84.2%), and non‐Hispanic or Latino (96.5%). From this cohort, 667 (6.84%) were hospitalized and 138 (1.42%) died (Figure 1 ). Among those who died, 13 did not have an inpatient visit at VUMC after they were diagnosed with SARS‐CoV‐2 infection. Although the weighted Elixhauser comorbidity score indicated a current state of health for most of the patients, 2,371 (24.3%) of them had severe comorbidity scores (Table 1 ). The hospitalized patients had a mean age of 60 years, and an increased percentage of men (50.4%), Blacks (23.2%), and severe comorbidity scores (57%).
Table 1.
Patient characteristics
| Characteristic | All patients | Hospitalized patients | ||
|---|---|---|---|---|
| N | % | N | % | |
| Total | 9,748 | 100 | 667 | 100 |
| Age, years a | 42 | 20 | 60 | 19 |
| Sex | ||||
| Men | 3,878 | 39.8 | 336 | 50.4 |
| Women | 5,870 | 60.2 | 331 | 49.6 |
| Race | ||||
| White | 8,212 | 84.2 | 495 | 74.2 |
| Black | 1,276 | 13.1 | 155 | 23.2 |
| Asian | 260 | 2.7 | 17 | 2.5 |
| Ethnicity | ||||
| Not Hispanic or Latino | 9,411 | 96.5 | 646 | 96.9 |
| Hispanic or Latino | 337 | 3.5 | 21 | 3.1 |
| Weighted Elixhauser comorbidity score | ||||
| < 0 | 1,414 | 14.5 | 71 | 10.6 |
| 0 | 4,600 | 47.2 | 147 | 22 |
| 1–4 | 1,363 | 14 | 69 | 10.3 |
| 5+ | 2,371 | 24.3 | 380 | 57 |
Reported as mean and SD.
Primary outcome
The analysis for the primary outcome consisted of 213 association studies between previous drug exposure and all‐cause of death (Tables S3 and S4 ). Propensity score overlap‐weighted logistic regression indicated that 58 drugs have lower death risk estimates (adjusted OR < 1). Among these, previous exposures to two types of 13‐valent pneumococcal conjugate vaccines, PCV13, (OR, 0.31, 95% CI, 0.12–0.81 and OR, 0.33, 95% CI, 0.15–0.73), diphtheria toxoid and tetanus toxoid vaccine (OR, 0.38, 95% CI, 0.15–0.93) are associated with a significantly decreased risk of death. After overlap weighting with propensity score for the first (and second) type of PCV13, the death rate was 2.2% (2%) in the exposed group compared with a death rate of 5.4% (4.9%) in the unexposed group. The death rates in the exposed vs. unexposed groups for diphtheria and tetanus toxoid vaccine were 0.9% vs. 2.2%, respectively. Figure 2 and Table S5 show additional details of the 58 drug studies for the primary outcome whereas Table S6 demonstrates that the overlap weighting method achieved a good balance in patient characteristics between the exposed and unexposed groups.
Figure 2.
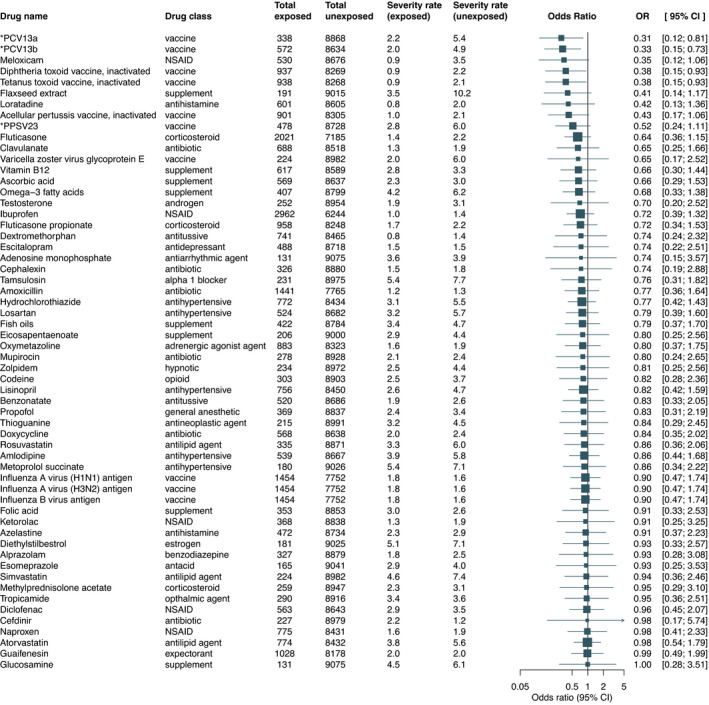
Association between drug exposure and all‐cause of death. Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) were estimated using propensity score overlap‐weighted logistic regression while death rates were computed using overlap weighting with propensity score. Abbreviations and acronyms: *PCV13a, 13‐valent pneumococcal conjugate vaccine with ingredients: Streptococcus pneumoniae serotype (1, 19A, 3, 5, 6A, 7F) capsular antigen diphtheria CRM197 protein conjugate vaccine; *PCV13b, 13‐valent pneumococcal conjugate vaccine with ingredients: Streptococcus pneumoniae serotype (14, 18C, 19F, 23F, 4, 6B, 9V) capsular antigen diphtheria CRM197 protein conjugate vaccine; *PPSV23, 23‐valent pneumococcal polysaccharide vaccine with ingredients: Streptococcus pneumoniae type (1, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 2, 20, 22F, 23F, 3, 33F, 4, 5, 6B, 7F, 8, 9N, 9V) capsular polysaccharide antigen; NSAID: nonsteroidal anti‐inflammatory drug. [Colour figure can be viewed at wileyonlinelibrary.com]
Secondary outcomes
The secondary outcome analyses led to the discovery of additional drugs as potential treatments for COVID‐19. Figure 3 summarizes the significant associations obtained across all COVID‐19 outcomes whereas Tables S7–S12 list the results of secondary analyses. In addition to the vaccines found in the primary analysis, previous exposure to acellular pertussis vaccine (OR, 0.41, 95% CI, 0.18–0.95) and 23‐valent pneumococcal polysaccharide vaccine, PPSV23 (OR, 0.46, 95% CI, 0.22–0.96) showed a protective effect for patients with COVID‐19 on ventilatory support (cumulative severity) or with less severe outcomes. Furthermore, use of flaxseed extract, ethinyl estradiol, estradiol, turmeric extract, ubidecarenone (also known as coenzyme Q10), azelastine, pseudoephedrine, dextromethorphan, omega‐3 fatty acids, fluticasone, and ibuprofen indicated a reduced risk for less severe COVID‐19 outcomes.
Figure 3.
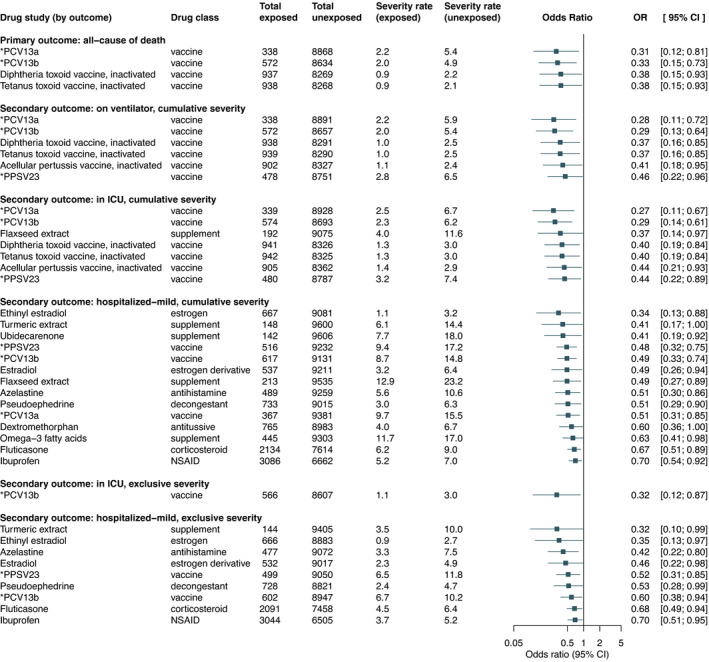
Significant associations grouped by outcome. Abbreviations and acronyms: *PCV13a, 13‐valent pneumococcal conjugate vaccine with ingredients: Streptococcus pneumoniae serotype (1, 19A, 3, 5, 6A, 7F) capsular antigen diphtheria CRM197 protein conjugate vaccine; *PCV13b, 13‐valent pneumococcal conjugate vaccine with ingredients: Streptococcus pneumoniae serotype (14, 18C, 19F, 23F, 4, 6B, 9V) capsular antigen diphtheria CRM197 protein conjugate vaccine; *PPSV23, 23‐valent pneumococcal polysaccharide vaccine with ingredients: Streptococcus pneumoniae type (1, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 2, 20, 22F, 23F, 3, 33F, 4, 5, 6B, 7F, 8, 9N, 9V) capsular polysaccharide antigen; CI, confidence interval; NSAID: nonsteroidal anti‐inflammatory drug; OR, odds ratio. [Colour figure can be viewed at wileyonlinelibrary.com]
Notably, some of the results are highly correlated because they correspond to drug ingredients that are available only in specific combinations. For example, some vaccine combinations contain diphtheria and tetanus vaccines (e.g., DT and Td vaccines) whereas other combinations contain diphtheria, tetanus, and acellular pertussis vaccines (e.g., DTaP and Tdap vaccines). Additional examples include PCV13, PPSV23, and Neisseria meningitidis vaccines whose specific ingredients are detailed in each table and figure where they are presented.
Risk trend analysis
A visual analysis showing the risk trends over the selected outcomes for all 58 drugs from the primary analysis is depicted in Figure 4 . Spline regression was used to visualize nonlinearity of risk trends across COVID‐19 outcomes ordered by increased severity (Figures S3 and S4 ). Drugs including PCV13, PPSV23, fluticasone, flaxseed extract, and ibuprofen show a constant low risk across the entire COVID‐19 severity scale, whereas drugs such as cephalexin and losartan indicate a decrease in effect estimates mainly for the primary outcome.
Figure 4.
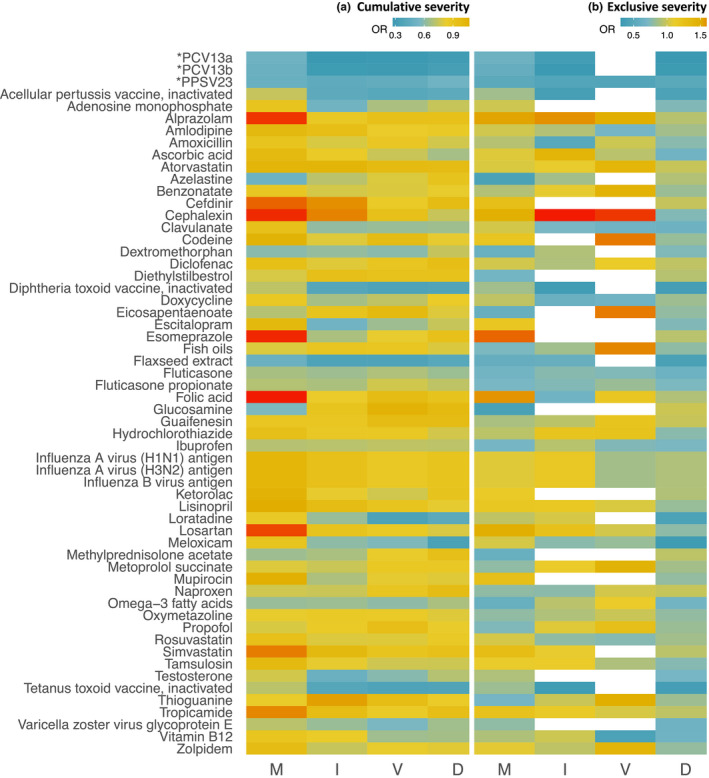
Heatmap of 58 drug associations across the COVID‐19 severity scale: hospitalized‐mild (M), ICU admission (I), on ventilation (V), and death (D). In (a) and (b) the secondary outcomes were extracted following the cumulative severity and exclusive severity strategies, respectively. In b, drug studies that did not meet the inclusion criteria are shown in white. Abbreviations: *PCV13a, 13‐valent pneumococcal conjugate vaccine with ingredients: Streptococcus pneumoniae serotype (1, 19A, 3, 5, 6A, 7F) capsular antigen diphtheria CRM197 protein conjugate vaccine; *PCV13b, 13‐valent pneumococcal conjugate vaccine with ingredients: Streptococcus pneumoniae serotype (14, 18C, 19F, 23F, 4, 6B, 9V) capsular antigen diphtheria CRM197 protein conjugate vaccine; *PPSV23, 23‐valent pneumococcal polysaccharide vaccine with ingredients: Streptococcus pneumoniae type (1, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 2, 20, 22F, 23F, 3, 33F, 4, 5, 6B, 7F, 8, 9N, 9V) capsular polysaccharide antigen; COVID‐19, coronavirus disease 2019; ICU, intensive care unit; OR, odds ratio.
Sensitivity analyses
Several sensitivity analyses were performed for the drugs found significantly associated with a COVID‐19 outcome. The analyses using longer drug exposure intervals starting 6‐months and 1‐year prior to the pandemic showed that most of the drugs remain significant (Tables S13 and S14 ). Similar trends are observed for the repeated analyses that include both symptomatic and asymptomatic patients (Table S15 ). Furthermore, the sensitivity analysis on comorbidity adjustment (where the weighted Elixhauser comorbidity score has been replaced by the 14 individual comorbidity predictors listed in Table S2 to estimate the propensity scores and adjust the weighted multivariable logistic regression models) shows that all the drugs remain significant (Table S16 ). In contrast, the sensitivity analysis where the exposure of interest was extracted using only the information in structured format (i.e., from the drug table) revealed that (1) most of the associations were not performed due to a small sample size of the corresponding drug exposure cohorts and (2) only two drugs, ethinyl estradiol and azelastine, remained significantly associated with a COVID‐19 outcome (Table S17 ). This analysis suggests that NLP methods can be used to better represent exposure cohorts in drug association studies because drug exposure information is highly represented in unstructured format (i.e., clinical text).
Inspired by our results at individual drug level and results from other recent studies, 13 , 26 , 27 , 28 we conducted an exploratory analysis to investigate associations of the drug classes listed in Table S1 . Here, similar to the sensitivity analysis on comorbidity adjustment, we replaced the weighted Elixhauser comorbidity score with the 14 individual comorbidity predictors listed in Table S2 . The main results of this analysis show that sigma‐1 receptor agonists are significantly associated with a reduced risk for all COVID‐19 outcomes. Furthermore, nonsteroidal anti‐inflammatory drug, antidepressants, selective serotonin reuptake inhibitors, and omega‐3 supplements show a protective effect for some of the outcomes (Table S18 ).
DISCUSSION
We presented a high‐throughput method to systematically investigate associations between drug exposures and COVID‐19 outcomes. Our study found 17 drug ingredients significantly associated with a decreased risk of death and other severe COVID‐19 outcomes. Moreover, 115 other drug ingredients indicated a protective effect for COVID‐19 outcomes. It is our hope that this proposed list of drug ingredients provides additional insights into developing efficient COVID‐19 treatments and would serve as a starting point for future prospective studies. Because their short‐ and long‐term adverse events have been already studied, the efficacy of these drug ingredients against COVID‐19 could be investigated rapidly in clinical trials. 29
For each drug study, overlap weighting with propensity score was implemented to adjust for confounding when comparing the drug exposed and unexposed groups. Another significant contribution of our study is the use of NLP to better capture drug exposure information from clinical notes. Notably, this process had a critical role in increasing the sample size of drug exposure cohorts and enabling the study of drugs, including dietary supplements, as potential COVID‐19 treatments.
Many of the significant drug ingredients found by our method have been previously studied as potential dug repurposing candidates for COVID‐19. A retrospective study using EHR data showed a decreased COVID‐19 fatality rate for women 50+ years old receiving estradiol therapy (OR, 0.33, 95% CI, 0.18–0.62). 30 Studies based on epidemiological data and in vitro analysis demonstrated potential benefits of azelastine to inhibit SARS‐CoV‐2 infection and reduce its viral activity. 26 , 31 In a network‐based bioinformatics analysis, pseudoephedrine was ranked as the best treatment candidate against COVID‐19. 32 In contrast to our results, dextromethorphan, a common over‐the‐counter cough suppressant, was found to have proviral activity on animal cells in laboratory settings; however, further controlled clinical studies are needed to validate the results in humans. 33 Of note, other studies suggest that sigma‐1 receptor agonists (which include drugs such as dextromethorphan and fluvoxamine) are essential inhibitors of cytokine production in COVID‐19. 10 , 28 This could also explain the main results of our exploratory analysis indicating that sigma‐1 receptor agonists are associated with a significantly decreased risk of all COVID‐19 outcomes. A pilot study in 100 patients hospitalized with COVID‐19 analyzed omega‐3 fatty acid levels in blood samples suggesting that patients with higher omega‐3 levels have a decreased risk of death. 34 Two network‐based bioinformatics approaches found fluticasone as a possible efficient treatment for COVID‐19, 13 , 35 while an in vitro study indicated that fluticasone does not suppress SARS‐CoV‐2 replication. 36 Concerns regarding the use of ibuprofen causing potential harm to patients with COVID‐19 had initially received significant attention. 37 More recent studies, however, found no significant evidence to suggest that ibuprofen is associated with severe COVID‐19 outcomes. 38 , 39 Moreover, an observational study using EHR data from 6 hospitals indicated that exposure to ibuprofen is associated with a lower risk of hospitalization due to COVID‐19 (OR, 0.73, 95% CI, 0.64–0.84). 40 Multiple clinical trials are currently underway to test the efficacy and safety of using estradiol (NCT04539626), omega‐3 fatty acids (NCT04647604, NCT04553705, and NCT04828538), and ibuprofen (NCT04334629, NCT04382768, and NCT04383899) against COVID‐19.
An important finding by our study is that recent exposure to various types of Streptococcus pneumoniae vaccines and diphtheria toxoid and tetanus toxoid vaccine, with or without acellular pertussis vaccine, is associated with a decreased risk of death and other severe COVID‐19 outcomes. This finding replicates the results of our preliminary analysis on 7,768 patients infected with SARS‐CoV‐2. 41 Recently, Streptococcus pneumoniae vaccines were also found to have a protective effect against COVID‐19 outcomes suggesting that pneumococci may interact with SARS‐CoV‐2 in the respiratory tract. 42 A double‐blind placebo‐randomized trial showed that the pneumococcal conjugate vaccine protects children against pneumococcal coinfections with seasonal coronaviruses. 43 Another suggestion was that pneumonia vaccination may prevent COVID‐19 exacerbation due to co‐infections or secondary bacterial infections. 44 , 45 Compared with Streptococcus pneumoniae vaccines, the results of diphtheria, tetanus, and acellular pertussis vaccines should be interpreted with caution because not all sensitivity analyses indicate their corresponding associations as significant. Nevertheless, in support of our results, an in silico study found that combinations of diphtheria, tetanus, and acellular pertussis vaccines induce potential cross‐reactive immunity to SARS‐CoV‐2. 46 The findings on vaccines may be in part explained by their ability to stimulate the immune system and provide immunologic protection against SARS‐CoV‐2. 47 They may also partly explain why specific categories of individuals for whom some of these vaccines are recommended (e.g., children and the youngest, pregnant women) are better protected against SARS‐CoV‐2. Overall, based on our results, we strongly advocate for these vaccines to be administered without delay when recommended because they may protect the general population from COVID‐19 worldwide.
To our knowledge, dietary supplements, such as turmeric extract, ubidecarenone, flaxseed extract, and omega‐3 fatty acids, have not been previously shown to be associated with a reduced COVID‐19 severity risk. However, due to their anti‐inflammatory properties, they have been proposed as alternative treatments to improve the clinical outcomes of patients with COVID‐19. 48 , 49 Yet, the dietary supplement results should be interpreted with caution because the corresponding ingredients are not evaluated by the US Food and Drug Administration for safety and effectiveness and are not intended to diagnose, treat, cure, or prevent any disease.
Limitations
Although overlap weighting with propensity score was applied to balance out the main patient characteristics between the drug exposed and unexposed groups, there may be unmeasured confounding factors that were not included in the propensity score model. Thus, whereas the bias may be reduced, confounding by indication is still possible due to unmeasured confounding factors. This is a major limitation, generally applicable to observational studies that lack randomization for drug exposure assignment.
Treatment misclassification was addressed by including only patients with at least one encounter in the EHR such that drug exposure information could be extracted for each patient who is SARS‐CoV‐2 positive during at least 3 months prior to diagnosis. However, EHR phenotyping pose many challenges, which could lead to inaccurately extracting the treatment status of the patient. 50 For example, drug exposure extraction from clinical text relies on accurate identification of text expressions that are negated (e.g., “he could not be on Coumadin because of history of GI bleed”) or hypothetical (e.g., “Zofran 4 mg PO once a day as needed for nausea”). 14 This is another reason for interpreting the results of over‐the‐counter drugs with caution because they are primarily extracted from clinical notes. Furthermore, assuming drug exposure information is accurately extracted for a patient, exposure of the drug at and after diagnosis time is not guaranteed. However, vaccine data in the EHR is not subject to this limitation.
Misclassification of COVID‐19 outcomes was addressed by excluding patients that were asymptomatic at test time. These patients could introduce bias in the study because, for instance, they may be admitted to the hospital for reasons other than COVID‐19. However, the sensitivity analysis, including both symptomatic and asymptomatic patients, showed that most of the associations remained significant. Additionally, despite including only patients with VUMC as their “medical home,” there could be a small number of patients among those classified as nonhospitalized who were in fact admitted to another hospital.
Finally, generalizability has yet to be proven on larger cohorts with a more heterogeneous study population. In this single‐site study, patients are predominantly White, non‐Hispanic or Latino, and relatively young (mean age of 42 years). Larger cohorts would also enable conducting subgroup analyses for a specific race, sex, age category, set of comorbidities, drug dose, drug route, or combination therapy.
CONCLUSIONS
Leveraging EHR data, the DrugWAS of COVID‐19 severity outcomes enable the discovery of drug ingredients that could be repurposed as potential treatments for COVID‐19. In addition to the prescription drugs available in structured format, extracting drug information from clinical notes using NLP facilitates the study of over‐the‐counter drugs on improving the recovery of patients with COVID‐19. The main results of this study suggest that it is essential for Streptococcus pneumoniae vaccines and diphtheria toxoid and tetanus toxoid vaccine, with or without acellular pertussis vaccine, to not be delayed or discontinued due to the COVID‐19 pandemic as they may provide protection against SARS‐CoV‐2. In the age of COVID‐19, we cannot dismiss the critical need to continue with excellent preventative medicine and public health measures established before this pandemic arrived. The efficacy of the identified drug ingredients needs to be further evaluated in prospective clinical trials using larger and more heterogenous study populations.
FUNDING
This work was supported by National Institutes of Health (NIH) grants P50GM115305, R01HG010863, R01AI150295, K23AI118804, R01GM124109, and UL1TR000445.
CONFLICT OF INTEREST
K.N.C. reported personal fees from Teva, personal fees from Optinose, personal fees from Novartis, personal fees from GlaxoSmithKline, personal fees from Blueprint Medicines, personal fees from Third Harmonic Bio, personal fees from Sanofi Pasteur, personal fees from Genentech, personal fees from Regeneron and personal fees from Ribon Therapeutics, outside the submitted work. J.F.P. reported personal fees from Color Genomics outside the submitted work. E.J.P. receives Royalties from UpToDate and consulting fees from Janssen, Vertex, Biocryst, and Regeneron outside of the submitted work. She is co‐director of IIID Pty Ltd that holds a patent for HLA‐B*57:01 testing for abacavir hypersensitivity, and has a patent pending for Detection of Human Leukocyte Antigen‐A*32:01 in connection with Diagnosing Drug Reaction with Eosinophilia and Systemic Symptoms without any financial remuneration and not directly related to the submitted work. All other authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
C.A.B., K.N.C., P.J.S., L.C., J.F.P., and E.J.P. wrote the manuscript. C.A.B., K.N.C., and E.J.P. designed the research. C.A.B. performed the research. C.A.B., K.N.C., L.C., and E.J.P. analyzed the data. K.N.C. and P.J.S. contributed to analytical tools.
ROLE OF THE FUNDER/SPONSOR
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Supporting information
Supplementary Material
References
- 1. Paules, C.I. , Marston, H.D. & Fauci, A.S. Coronavirus infections‐more than just the common cold. JAMA 323, 707–708 (2020). [DOI] [PubMed] [Google Scholar]
- 2. Nicola, M. et al. The socio‐economic implications of the coronavirus pandemic (COVID‐19): A review. Int. J. Surg. 78, 185–193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polack, F.P. et al. Safety and efficacy of the BNT162b2 mRNA COVID‐19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baden, L.R. et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beigel, J.H. et al. Remdesivir for the treatment of covid‐19 ‐ final report. N. Engl. J. Med. 383, 1813–1826 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.[No authors listed]. Dexamethasone in hospitalized patients with COVID‐19 ‐ preliminary report 10.1101/2020.06.22.20137273 [DOI]
- 7. Chen, P. et al. SARS‐CoV‐2 neutralizing antibody LY‐CoV555 in outpatients with COVID‐19. N. Engl. J. Med. 384, 229–237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanders, J.M. , Monogue, M.L. , Jodlowski, T.Z. & Cutrell, J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID‐19): A review. JAMA 323, 1824–1836 (2020). [DOI] [PubMed] [Google Scholar]
- 9. Wiersinga, W.J. , Rhodes, A. , Cheng, A.C. , Peacock, S.J. & Prescott, H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID‐19): A Review. JAMA 324, 782 (2020). [DOI] [PubMed] [Google Scholar]
- 10. Lenze, E.J. et al. Fluvoxamine vs placebo and clinical deterioration in outpatients with symptomatic COVID‐19: A randomized clinical trial. JAMA 324, 2292–2300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim, P.S. , Read, S.W. & Fauci, A.S. Therapy for early COVID‐19: A critical need. JAMA 324, 2149 (2020). [DOI] [PubMed] [Google Scholar]
- 12. Riva, L. et al. Discovery of SARS‐CoV‐2 antiviral drugs through large‐scale compound repurposing. Nature 586, 113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou, Y. , Hou, Y. , Shen, J. , Huang, Y. , Martin, W. & Cheng, F. Network‐based drug repurposing for novel coronavirus 2019‐nCoV/SARS‐CoV‐2. Cell Discov. 6, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bejan, C.A. , Wei, W.Q. & Denny, J.C. Assessing the role of a medication‐indication resource in the treatment relation extraction from clinical text. J. Am. Med. Inform. Assoc. 22, e162–e176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sohn, S. , Clark, C. , Halgrim, S.R. , Murphy, S.P. , Chute, C.G. & Liu, H. MedXN: an open source medication extraction and normalization tool for clinical text. J. Am. Med. Inform. Assoc. 21, 858–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weeks, H.L. et al. medExtractR: A targeted, customizable approach to medication extraction from electronic health records. J. Am. Med. Inform. Assoc. 27, 407–418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi, L. et al. Development of a system for postmarketing population pharmacokinetic and pharmacodynamic studies using real‐world data from electronic health records. Clin. Pharmacol. Therap. 107, 934–943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta, S. et al. Factors associated with death in critically Ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 180, 1436–1447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elixhauser, A. , Steiner, C. , Harris, D.R. & Coffey, R.N. Comorbidity measures for use with administrative data. Med. Care 36, 8–27 (1998). [DOI] [PubMed] [Google Scholar]
- 20. Quan, H. et al. Coding algorithms for defining comorbidities in ICD‐9‐CM and ICD‐10 administrative data. Med, Care 43, 1130–1139 (2005). [DOI] [PubMed] [Google Scholar]
- 21. D'Agostino, R.B. Jr & D'Agostino, R.B. Sr. Estimating treatment effects using observational data. JAMA 297, 314–316 (2007). [DOI] [PubMed] [Google Scholar]
- 22. Thomas, L.E. , Li, F. & Pencina, M.J. Overlap weighting a propensity score method that mimics attributes of a randomized clinical trial. JAMA 323, 2417–2418 (2020). [DOI] [PubMed] [Google Scholar]
- 23. Li, F. , Thomas, L.E. & Li, F. Addressing extreme propensity scores via the overlap weights. Am. J. Epidemiol. 188, 250–257 (2019). [DOI] [PubMed] [Google Scholar]
- 24. Mehta, N. et al. Association of use of angiotensin‐converting enzyme inhibitors and angiotensin II receptor blockers with testing positive for coronavirus disease 2019 (COVID‐19). JAMA Cardiol. 5, 1020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao, S.Y. , Petrache, I. , Fingerlin, T.E. & Maier, L.A. Association of inhaled and systemic corticosteroid use with Coronavirus Disease 2019 (COVID‐19) test positivity in patients with chronic pulmonary diseases. Respir Med 176, 106275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reznikov, L.R. et al. Identification of antiviral antihistamines for COVID‐19 repurposing. Biochem. Biophys. Res. Commun. 538, 173–179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoertel, N. et al. Association between antidepressant use and reduced risk of intubation or death in hospitalized patients with COVID‐19: results from an observational study. Mol Psychiatry 1–14 (2021). 10.1038/s41380-021-01021-4 [DOI] [PubMed] [Google Scholar]
- 28. Hoertel, N. et al. Association between FIASMAs and Reduced Risk of Intubation or Death in Individuals Hospitalized for Severe COVID‐19: an observational multicenter study. Clin. Pharmacol. Ther 10.1002/cpt.2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rubin, R. Study aims to identify drugs that could be repurposed for COVID‐19. JAMA 324, 2019 (2020). [DOI] [PubMed] [Google Scholar]
- 30. Seeland, U. et al. Evidence for treatment with estradiol for women with SARS‐CoV‐2 infection. BMC Med. 18, 369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konrat, R. et al. The anti‐histamine azelastine, identified by computational drug repurposing, inhibits SARS‐CoV‐2 infection in reconstituted human nasal tissue in vitro. bioRxiv 10.1101/2020.09.15.296228. [DOI] [PMC free article] [PubMed]
- 32. Li, X. et al. Network bioinformatics analysis provides insight into drug repurposing for COVID‐2019. Med. Drug Discov 10.1016/j.medidd.2021.100090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gordon, D.E. et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Asher, A. , Tintle, N.L. , Myers, M. , Lockshon, L. , Bacareza, H. & Harris, W.S. Blood omega‐3 fatty acids and death from COVID‐19: A pilot study. Prostaglandins Leukot. Essent. Fatty Acids 166, 102250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cava, C. , Bertoli, G. & Castiglioni, I. In silico discovery of candidate drugs against COVID‐19. Viruses 12, 404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsuyama, S. et al. The inhaled steroid ciclesonide blocks SARS‐CoV‐2 RNA replication by targeting the viral replication‐transcription complex in cultured cells. J Virol 95, e01648‐20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Day, M. Covid‐19: ibuprofen should not be used for managing symptoms, say doctors and scientists. BMJ 368, m1086 (2020). [DOI] [PubMed] [Google Scholar]
- 38. Rinott, E. , Kozer, E. , Shapira, Y. , Bar‐Haim, A. & Youngster, I. Ibuprofen use and clinical outcomes in COVID‐19 patients. Clin. Microbiol. Infec. 26, 1259.e5–1259.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kragholm, K. et al. Association between prescribed ibuprofen and severe COVID‐19 infection: A nationwide register‐based cohort study. Clin. Transl. Sci. 13, 1103–1107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castro, V.M. , Ross, R.A. , McBride, S. & Perlis, R.H. Identifying common pharmacotherapies associated with reduced COVID‐19 morbidity using electronic health records. medRxiv 10.1101/2020.04.11.20061994 [DOI]
- 41. Bejan, C.A. , Cahill, K.N. , Staso, P.J. , Choi, L. , Peterson, J.F. & Phillips, E.J. DrugWAS: Leveraging drug‐wide association studies to facilitate drug repurposing for COVID‐19. medRxiv 10.1101/2021.02.04.21251169 [DOI] [PMC free article] [PubMed]
- 42. Lewnard, J.A. et al. Prevention of COVID‐19 among older adults receiving pneumococcal conjugate vaccine suggests interactions between Streptococcus pneumoniae and SARS‐CoV‐2 in the respiratory tract. J. Infect. Dis 10.1093/infdis/jiab128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nunes, M.C. , Cutland, C.L. , Klugman, K.P. & Madhi, S.A. Pneumococcal conjugate vaccine protection against coronavirus‐associated pneumonia hospitalization in children living with and without HIV. mBio 12. 10.1128/mBio.02347-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Al‐Ani, A.H. et al. Review article: prevention, diagnosis and management of COVID‐19 in the IBD patient. Aliment. Pharmacol. Ther. 52, 54–72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sender, V. , Hentrich, K. & Henriques‐Normark, B. Virus‐induced changes of the respiratory tract environment promote secondary infections with streptococcus pneumoniae. Front. Cell. Infect. Microbiol. 11, 643326 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reche, P.A. Potential cross‐reactive immunity to SARS‐CoV‐2 from common human pathogens and vaccines. Front. Immunol. 11, 586984 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ietto, G. SARS ‐ CoV‐2: Reasons of epidemiology of severe ill disease cases and therapeutic approach using trivalent vaccine (tetanus, diphtheria and Bordetella pertussis). Med. Hypotheses 141, 109779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Faria Coelho‐Ravagnani, C. , Corgosinho, F.C. , Sanches, F.F.Z. , Prado, C.M.M. , Laviano, A. & Mota, J.F. Dietary recommendations during the COVID‐19 pandemic. Nutr. Rev. 1, 382–393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manoharan, Y. , Haridas, V. , Vasanthakumar, K.C. , Muthu, S. , Thavoorullah, F.F. & Shetty, P. Curcumin: a Wonder Drug as a Preventive Measure for COVID19 Management. Indian J. Clin. Bioche. 35, 373–375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeLozier, S. et al. Phenotyping coronavirus disease 2019 during a global health pandemic: lessons learned from the characterization of an early cohort. J. Biomed. Inform. 117, 103777 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


