Abstract
Tat protein strongly activates transcription from the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) by enhancing the elongation efficiency of RNA polymerase II complexes. Tat-mediated transcriptional activation requires cellular cofactors and specific cis-acting elements within the HIV-1 promoter, among them a functional TATA box. Here, we have investigated the mechanism by which one of these cofactors, termed CA150, regulates HIV-1 transcription in vivo. We present a series of functional assays that demonstrate that the regulation of the HIV-1 LTR by CA150 has the same functional requirements as the activation by Tat. We found that CA150 affects elongation of transcription complexes assembled on the HIV-1 promoter in a TATA-box-dependent manner. We discuss the data in terms of the involvement of CA150 in the regulation of Tat-activated HIV-1 gene expression. In addition, we also provide evidence suggesting a role for CA150 in the regulation of cellular transcriptional processes.
Regulation of RNA polymerase II (RNAP II) transcription initiation is accomplished by the involvement of at least three different types of factors. General transcription factors (GTFs) assemble near the start site of transcription and direct the basal level of promoter expression. For example, the TATA-binding protein (TBP) initiates assembly of transcription complexes by binding to the TATA box sequence in TATA-containing, class II gene promoters. A second class of factors are either activators or repressors of transcription which normally bind to specific DNA sequences and regulate the rate of RNAP II transcription initiation. Finally, there are adapter proteins (also called cofactors or mediators), which provide a link between DNA-bound transcription factors and the GTFs and can positively or negatively regulate transcription (20, 62). The joint action of GTFs, activators and/or repressors, and adapters allows efficient initiation of transcription and subsequent elongation by RNAP II complexes. Transcription elongation is also a target for gene regulation, and many factors that stimulate elongation and read-through have been identified (54). The establishment of an elongation-competent transcription complex is a process that probably involves phosphorylation of the C-terminal heptapeptide repeat domain (CTD) of the largest subunit of RNAP II (12), which consists of multiple repeats of the sequence YSPTSPS.
Genetic and biochemical approaches with yeast, and more recently with mammals, have shown that RNAP II, GTFs, and adapters can be found in preassembled complexes generically termed RNAP II holoenzyme (38). The holoenzyme composition varies depending on the protocol for purification, and there are indications that multiple holoenzyme complexes may exist. Importantly, holoenzyme is responsive to transcription activators (6, 26, 32), and recruitment of holoenzyme to a promoter is sufficient for gene activation (1, 15, 17, 25).
The human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) contains the elements of a prototypic class II eukaryotic regulatory unit. It contains enhancer and promoter elements where binding sites for many transcription factors have been identified (24). The core HIV-1 promoter consists of a canonical TATA box and two elements further downstream, which appear to be necessary for transcriptional activity (63). Other relevant elements include two and three tandem DNA-binding sites for the inducible NF-κB and the ubiquitous Sp1 transcription factors, respectively. Despite the presence of binding sites for multiple transcription activators, transcription directed by the HIV-1 promoter is very weak in vivo, and robust transcription requires the expression of the viral transactivator Tat, which increases the level of transcription more than 100-fold. Several studies have analyzed the contribution of upstream elements to basal and activated HIV-1 transcription. In particular, basal transcription is dependent more on NF-κB sites than on Sp1 elements, whereas the opposite effect is seen for Tat-activated transcription. TATA box sequences are required for Tat activation but have a smaller contribution to basal activity from the HIV-1 promoter (4, 29, 44).
Tat activates transcription by binding to an RNA structure called the trans-activation response (TAR) element, a regulatory region located at the 5′ ends of all viral mRNAs (10). In the absence of Tat, the HIV-1 LTR produces mainly short, nonpolyadenylated transcripts, whereas in the presence of Tat, there is a substantial increase in the level of longer, polyadenylated transcripts. Tat affects the elongation efficiency of RNAP II complexes formed on the HIV-1 promoter, and the mechanism of this regulation has been a matter of intense investigation. Tat associates with a CTD kinase (21, 22), and the CTD of RNAP II is required for Tat trans activation (9, 43, 61). These findings suggest that phosphorylation of the RNAP II CTD can be increased by Tat. This is particularly relevant because increasing CTD phosphorylation by a CTD kinase has been shown to stimulate the elongation efficiency of RNAP II (27). The identification of positive transcription elongation factor b (P-TEFb), as the protein complex that binds to Tat and assembles onto the HIV-1 TAR element, has been a key step towards our understanding of the mechanism of transcription activation of the HIV-1 LTR by Tat (10). P-TEFb is comprised minimally of a CTD kinase called CDK9, which is part of the Tat-associated kinase (34, 60, 66), and its cyclin component, termed cyclin T1, which binds directly to Tat (58). Immunodepletion of CDK9 inhibited basal and Tat-activated transcription in vitro (64, 66), and its overexpression abrogated Tat trans activation in cells (19). Immunodepletion of cyclin T1 decreased basal and Tat activation in vitro (58). Importantly, transfection of a human cyclin T1-expressing plasmid into mouse cells, which are unable to respond to Tat via the HIV-1 TAR element, rescued Tat transcriptional activation (58). Based on these findings, it has been proposed that Tat recruits P-TEFb to the HIV-1 TAR element, thus allowing the phosphorylation of the CTD and/or other components of the transcription apparatus by CDK9 and increasing the elongation competence of RNAP II complexes.
In addition to P-TEFb, other factors have been implicated in Tat-mediated transcriptional activation. CA150 (coactivator of 150 kDa [50]) and Tat-SF1 (Tat stimulatory factor 1 [65]) are nuclear proteins which were purified by using in vitro transcription systems and Tat affinity chromatography. Depletion of these proteins from HeLa nuclear extracts specifically decreased Tat trans activation, and transient-transfection experiments with either protein carried out in HeLa cells resulted in alterations in the Tat-activated response. The fact that CA150 and Tat-SF1 were isolated by Tat affinity chromatography, while no direct binding between these factors and Tat has been reported, suggests that CA150 and Tat-SF1 affect Tat trans activation indirectly. Very little is known about the contribution of CA150 and Tat-SF1 to Tat-mediated transcription activation of the HIV-1 promoter. In this study, we delineated the role of CA150 in the regulation of RNAP II transcription by using HIV-1 and other TATA-box-containing promoters. We have found a selective role of CA150 in transcription from certain promoters and have shown that CA150 regulates HIV-1 LTR transcription in a TATA-box-dependent fashion. In addition, we provide evidence demonstrating that CA150 regulates transcription from the HIV-1 LTR, at least in part, by affecting the elongation efficiency of RNAP II complexes. The relevance of these findings to the regulation of HIV-1 gene expression is discussed.
MATERIALS AND METHODS
Plasmids.
pEFBOST7CA150 expressed the full-length CA150 protein under the control of the polypeptide chain elongation factor 1α promoter (37). PCR was used to obtain two fragments representing the 5′ and 3′ portions of the CA150 gene by using the CA150 cDNA plasmid (50) as a template. Pfu polymerase (Stratagene, La Jolla, Calif.) was used in this and subsequent PCRs. The following oligonucleotides were used: 5′-GGGAGATCTTGATGGCCCAACAGCAGGCCTTGAGG-3′ (forward) with 5′-GCTTTAACAGGCTCATCTTC-3′ (reverse) and 5′-GGGGAGCCCAAAGAAGAGGAGATGACT-3′ (forward) with 5′-GGGAGATCTGATGCCCCTATGGAAGAGTATTTA-3′ (reverse). Each PCR fragment contained an overlapping BclI restriction site and BglII (underlined) ends. BclI- and BglII-digested PCR fragments were ligated together into BamHI-digested pEFBOST7HRH1 plasmid (42) to create pEFBOST7CA150. The expressed CA150 protein contains the 11-amino-acid T7 epitope tag at its amino terminus.
The reporter constructs HIV-CAT (promoter sequences from bp −640 to +82), CMV-IE-CAT (promoter sequences from bp −467 to +71), and RSV-CAT (promoter sequences from bp −290 to +34) are the same plasmid containing the HIV-1, the cytomegalovirus (CMV) major immediate-early (IE), and the Rous sarcoma virus (RSV) promoter sequences, respectively, linked to the chloramphenicol acetyltransferase (CAT) gene, and they have been previously described (3, 11). SV40 (simian virus 40 early)-CAT is the pSV2CAT plasmid (promoter sequences from bp −271 to +69), and it was kindly provided by Cristina Hernández-Munain (Duke University). pcTat, expressing the 86-amino-acid Tat protein under the control of the CMV promoter, has been previously described (33). HSV-TK-LUC contains the herpes simplex virus (HSV) thymidine kinase (TK) promoter (sequences from bp −107 to +57), and it was kindly provided by Antonio A. Postigo (Washington University, St. Louis, Mo.). The minimal HIV-1 promoter plasmids (NF/SP [wild type] and NF-R) and the HIV-CAT mutated vectors where the TATA box has been changed (SV40-TATA and m-TATA) or deleted (d-TATA) have been described elsewhere (4) and were kindly provided by Kuan-Teh Jeang (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). The 5′ deletion mutants of the α4 integrin promoter have been described previously (45, 47), and they were kindly provided by A. A. Postigo. To express human TBP, we used pCGNTBP, which adds a hemagglutinin (HA) epitope tag to the amino terminus of the expressed protein, and it was kindly provided by William P. Tansey (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.) (51).
Cell culture and transfections.
We used 293T cells, a line of transformed human epithelial kidney cells into which the gene for the SV40 T antigen has been introduced (13). Transient-transfection experiments involving this cell line required a careful titration and balance of the different promoters tested. Transfections were carried out with at least two different preparations of each plasmid DNA purified by using kits from Qiagen Inc. (Santa Clarita, Calif.). 293T cells were grown in Dulbecco’s modified Eagle medium (Gibco/BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (Hyclone, Logan, Utah) and penicillin-streptomycin to 100 U and 100 μg per ml, respectively (Gibco/BRL). Transfections were performed in 35-mm-diameter plates (Becton Dickinson Labware, Franklin Lakes, N.J.). Each well was seeded with 5 × 105 cells approximately 20 h prior to transfection. Cells were grown to approximately 50% confluence and were transfected with the amounts indicated in the figure legends by using calcium phosphate. The reporter vector HSV-TK-LUC (10 ng per well) was used as an internal control for transfection, and yeast tRNA carrier (Sigma, St. Louis, Mo.) was used to keep constant the total amount of nucleic acids. Cells in each well were carefully rinsed with phosphate-buffered saline (PBS) and replenished with fresh medium 16 h after transfection, and they were collected approximately 40 h after transfection. Cells were rinsed with PBS and then lysed by three cycles of freezing in dry ice and ethanol and thawing at 37°C in a water bath. After centrifugation, the supernatant was used immediately or stored at −80°C for future use. The cell extracts were normalized on the basis of protein concentration by using the Bradford method with bovine gamma globulin as a standard (Bio-Rad Laboratories, Hercules, Calif.). CAT assays were done by the diffusion method of Neumann and coworkers (41). Typically, 100 μg of cell protein was used in each CAT assay. Luciferase activity was measured at room temperature with a fixed amount of protein of individual cell extracts, luciferase assay reagents from Promega Corp. (Madison, Wis.), and a semiautomatic luminometer (LUMAT LB 9507; EG&G Berthold/Wallac Inc., Gaithersburg, Md.). Values from one representative experiment are shown throughout the paper. Similar results were obtained in at least three independent transfection experiments in which relative CAT activities varied less than 20% between experiments.
Antibodies and Western blotting.
Antibodies against CA150 have been described previously (50). Antibodies against the T7 and HA tags were purchased from Novagen (Madison, Wis.) and Berkeley Antibody Co. (Richmond, Calif.), respectively, and used accordingly to their specifications. TBP-specific serum was obtained from Upstate Biotechnology (Lake Placid, N.Y.).
Analysis of protein expression was carried out with whole-cell lysates from 293T cells. Briefly, cells were washed twice with PBS, and the cell pellets were incubated with cold radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 7.5]) containing phenylmethylsulfonyl fluoride at 50 μg/ml during 30 min with gentle hand shaking in a water-ice bucket. After spinning at 10,000 × g for 10 min at 4°C, the lysate was removed to a clean tube and used for protein analysis. For Western blot analysis, proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to an Immobilon-P (Millipore Corp., Bedford, Mass.) membrane, and then incubated with the specific antiserum. After washing, the membrane was incubated with a peroxidase-conjugated secondary antibody (Amersham, Arlington Heights, Ill.), and bound antibodies were detected by enhanced chemiluminescence (Amersham).
RNA purification and RT-PCR.
Total cellular RNA was isolated from transfected 293T cells by the method of Chomczynski and Sacchi (8). After isolation, RNA samples were treated with RQ1 DNase I (Promega Corp.) according to the manufacturer’s specifications and then phenol-chloroform extracted and precipitated. Reverse transcription (RT) reaction mixtures contained 4 μg of total RNA, 100 mM KCl, and 18 μM specific primers. The primers used were 5′-AAGCTTTATTGAGGCTTAAGCAGT-3′ and 5′-GAAAACGGGGGCGAAGAA-3′. These primers were used to synthesize cDNAs of 82 and 542 nucleotides, respectively. RT annealing reaction mixtures were placed in boiling water for 1 min, then placed at 50°C for 20 min, and then placed on ice. RT extension reaction mixtures contained 250 mM Tris-HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2, 10 mM dithiothreitol, 500 μM each deoxynucleoside triphosphate, and 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco) and were incubated at 37°C for 1 h. One microliter of the appropriate dilution of each RT reaction mixture was amplified in a 50-μl PCR mixture containing 200 μM (each) dATP, dGTP, and dTTP; 50 μM dCTP; 0.5 μl of [α-32P]dCTP (3,000 Ci/mmol; ICN Biomedicals, Irvine, Calif.); 1 μM each primer; and 2.5 U of cloned Pfu DNA polymerase (Stratagene) with the manufacturer’s reaction buffer. The primers used were 5′-GGGTCTCTCTGGTTAGAC-3′ (forward) and the same oligonucleotide used in the RT reactions to measure transcripts of 82 nucleotides (reverse). Amplification reaction conditions consisted of an initial denaturation step at 94°C for 2 min followed by 20 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The reaction was finished by a final 10-min extension at 72°C. PCR products were resolved on nondenaturing 8% acrylamide-bisacrylamide (30:1)–Tris-borate-EDTA gels. Electrophoresis was at 7 V/cm for approximately 5 h, followed by drying and exposure to Amersham Hyperfilm-MP. Analysis was performed with a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager.
RESULTS
Overexpression of CA150 represses HIV-1 basal and Tat-activated transcription in vivo.
Several laboratories have studied the role of Tat cofactors in the regulation of Tat-mediated transcription activation of the HIV-1 promoter. In those studies, an effect on Tat trans activation was observed when Tat-SF1 or P-TEFb components were depleted from nuclear extracts or overexpressed in cells. In some of these studies, an alteration of the basal activity of the promoter (i.e., that in the absence of Tat) has been reported (58, 64, 65), thus suggesting that these factors may also be important for maintaining the basal level of HIV-1 transcription and/or are able to interact with basal factors. We previously reported the inhibition of Tat-activated transcription upon depletion of CA150 from nuclear extracts and by overexpression of a deletion mutant CA150 protein in cells (50). We sought to further analyze the contribution of CA150 to HIV-1 gene regulation.
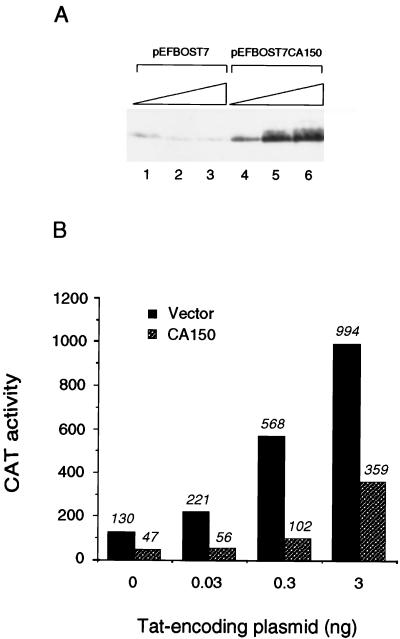
In an effort to overexpress CA150 protein, we performed transient-transfection experiments with several human cell lines as well as a number of different expression vectors (data not shown). A full-length CA150 protein was overexpressed in 293T cells by using the promoter of the human polypeptide chain elongation factor 1α gene contained in the pEFBOS vector (37) (Fig. 1A). This expression vector directed the synthesis of CA150 efficiently in 293T cells; therefore, it was chosen for further studies. pEFBOS vector was modified (42) to include the T7 epitope tag upstream of the cloned gene, which allows the detection of the expressed protein by using specific antibodies against the tag. To investigate the role of CA150 in HIV-1 transcription, we sought to analyze the effect of the overexpression of a full-length CA150 protein on the activity of the HIV-1 promoter in the absence and presence of the viral Tat activator in 293T cells. Cotransfection of an HIV LTR CAT reporter plasmid with a Tat-expressing construct activated transcription of the HIV-1 promoter in a dose-dependent manner. We observed a marked decrease in the activities of both basal and Tat-activated transcription of the HIV-1 promoter when CA150 protein was overexpressed in these cells (Fig. 1B).
FIG. 1.
Overexpression of CA150 inhibits basal and Tat-activated transcription from the HIV-1 LTR. (A) Analysis of CA150 protein expression. Cells were transfected with 1, 2, and 3 μg of vector alone (pEFBOST7) (lanes 1 to 3) and the same amounts of the CA150-expressing construct (pEFBOST7-CA150) (lanes 4 to 6). Whole-cell lysates were prepared, and proteins were resolved by SDS-PAGE and transferred to a membrane. Antibodies against the protein were used to localize CA150. (B) Effect of CA150 overexpression on basal and Tat-activated transcription from the HIV-1 promoter. The levels of CAT activity in extracts from cells that were cotransfected with 0.1 μg of HIV-1 LTR reporter vector and the indicated amounts of Tat in the presence of 3 μg of vector alone or the CA150-expressing construct were measured. Transfection and activity assays were done as described in Materials and Methods.
Several possibilities can explain the repression of the activity of the HIV-1 promoter observed upon overexpression of CA150. CA150 could be a general transcription repressor, or it could affect the activity of the HIV-1 promoter indirectly. For example, the repression could be due to the sequestering of an RNAP II GTF (so-called squelching), or the effect could be more specific to the HIV-1 promoter (e.g., disruption of the normal stoichiometry of a specific complex formed at the promoter).
Inhibition of transcription by overexpressed CA150 depends on specific promoter sequences.
To study the effect of the overexpression of CA150 on transcription carried out by RNAP II, we performed similar transient-transfection experiments with several viral TATA-box-containing promoters. Of five promoters tested (HIV-1, CMV IE, early SV40, RSV, and HSV TK), only the HIV-1 promoter was specifically repressed by CA150 (Table 1). These data indicate that CA150 is not a general repressor of transcription and suggest that the repression mediated by the overexpression of CA150 is not due to the squelching of a GTF or disruption of a complex required for general transcription. The fact that several of the constructs used in these experiments differ only in the promoter elements (HIV-1, CMV IE, or RSV [see Materials and Methods]) also suggests that the observed repression by CA150 depends on specific promoter sequences.
TABLE 1.
Effect of CA150 on basal transcription from different viral promotersa
| CA150 | Reporter plasmid | CAT activityb | LUC activityc | Corrected CAT activityd | Fold inhibition |
|---|---|---|---|---|---|
| Absente | HIV-CAT | 675.2 | 5,995 | 675.2 | |
| CMV-IE-CAT | 323.2 | 4,375 | 323.2 | ||
| SV40-CAT | 802.7 | 5,053 | 802.7 | ||
| RSV-CAT | 463.2 | 3,185 | 463.2 | ||
| Present | HIV-CAT | 151.3 | 6,385 | 142.7 | 4.7 |
| CMV-IE-CAT | 238.0 | 3,590 | 285.6 | 1.1 | |
| SV40-CAT | 533.7 | 5,704 | 485.1 | 1.6 | |
| RSV-CAT | 746.4 | 5,309 | 466.5 | 1.0 |
Cells were cotransfected with 0.1 μg of the reporter plasmids HIV-CAT, SV40-CAT, and RSV-CAT, or with 0.01 μg of CMV-IE-CAT, in the presence of 3 μg of the CA150-expressing construct. Transfection and activity assays were done as described in Materials and Methods. All results shown are the averages of data from duplicate specimens in a representative experiment. Transfections and activity assays were performed at least three times for each plasmid with similar results.
CAT activity is the value of the slope of the linear function obtained by plotting counts per minute of acetylated chloramphenicol versus time (41).
Luciferase (LUC) activity was obtained by measuring light emission in the same samples with a luminometer. Values are per microgram of cell extract. The HSV TK promoter was driving luciferase expression. Cells were cotransfected with 0.01 μg of this plasmid in the presence of the indicated constructs.
Corrected CAT activities are normalized to luciferase activity.
Empty vector was used to balance the promoter concentration.
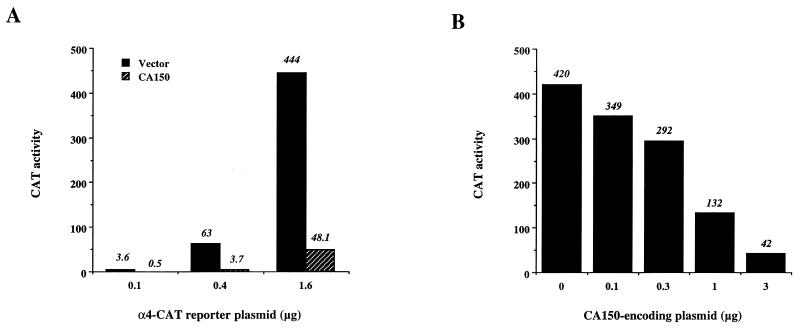
We also found that the cellular α4 integrin promoter was greatly repressed by CA150 (Fig. 2A). The protein product of the α4 integrin gene is a member of the integrin family that mediates attachment of lymphoid and myeloid cells to extracellular matrixes and is also involved in cell-cell interactions. In addition to the immune system, α4 integrin is also expressed in a developmentally regulated pattern in skeletal muscle, where a role in its normal development has been proposed (48). Overexpression of CA150 repressed transcription from the TATA-box-containing α4 integrin promoter in a dose-dependent manner (Fig. 2B). These results indicate that overexpression of CA150 can interfere with transcription carried out by RNAP II from certain TATA-box-containing promoters.
FIG. 2.
Overexpression of CA150 inhibits the activity of the α4 integrin promoter. (A) The indicated amounts of the α4 integrin reporter vector were cotransfected into 293T cells in the presence of 3 μg of vector alone or the CA150-expressing construct. (B) The same cotransfection experiment was carried out with a fixed amount of α4 integrin reporter vector (1.6 μg) and the indicated concentrations of the CA150-expressing plasmid. Empty vector was used to keep the total amount of DNA constant. Transfection and activity assays were done as described in Materials and Methods.
TATA box sequences are critical in the regulation of the HIV-1 promoter by CA150.
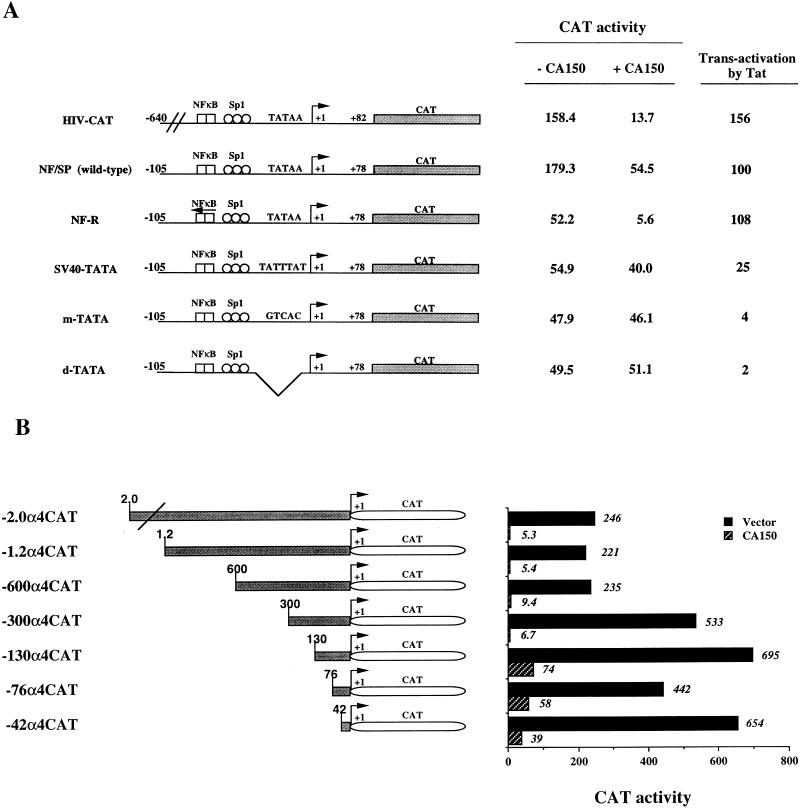
To study which regions of the HIV-1 promoter are important for repression by overexpressed CA150, we performed transient-transfection experiments with a series of 5′ promoter deletion constructs. We used a set of HIV-1 constructs that were originally described by Berkhout and Jeang (4). NF/SP (wild type) contains a minimal HIV-1 promoter with Sp1 and NF-κB sites (up to bp −105). The basal activity of this promoter was similar to the activity of the HIV-1 promoter containing complete enhancer sequences (up to bp −640) (Fig. 3A). We carried out cotransfection experiments with either the HIV-1 construct containing complete enhancer sequences (HIV-CAT) or the deletion mutant construct (NF/SP) and the CA150-expressing vector. We observed an inhibition of promoter activity with both HIV-1 constructs, which indicates that most of the HIV-1 LTR is dispensable for CA150-dependent inhibition (Fig. 3A). We noticed a reproducible lower level of repression with the NF/SP (wild-type) construct, which could indicate an effect of upstream sequences in CA150 inhibition (see below). Further deletion of the NF-κB and Sp1 sites produced a minimal promoter with an almost undetectable level of basal activity in 293T cells (data not shown).
FIG. 3.
Repression mediated by overexpression of CA150 requires specific promoter sequences. (A) Effect of CA150 overexpression on the activity of HIV-1 LTR constructs: CA150 overexpression inhibits HIV-1 promoter activity in a TATA-box-dependent manner. A schematic representation of the HIV-1 promoter constructs is shown. Numbers on the constructs indicate the amount of viral sequences present. NF-R has both NF-κB sites in a reverse orientation (arrow on top). The indicated HIV-1 LTR constructs (0.1 μg) were cotransfected into 293T cells in the presence of 3 μg of vector alone (−CA150) or CA150-expressing vector (+CA150). Numbers indicate CAT activity from transfected-cell extracts (see Materials and Methods). The transcription activity from HIV-CAT, NF/SP (wild-type), and NF-R constructs was repressed by overexpressing CA150. Changing the TATA box sequence to TATTTAT (SV40-TATA) or to the random sequence GTCAC (m-TATA) or using a construct where the TATA box was deleted (d-TATA) abrogated the repression mediated by overexpression of CA150. The abilities of these constructs to respond to Tat are shown in the last column. 293T cells were cotransfected with 0.1 μg of the indicated HIV-1 reporter constructs, 25 ng of pcTat, and 10 ng of HSV-TK-LUC expression plasmid. The level of Tat activation of NF/SP (wild-type) plasmid was set at 100. (B) Effect of CA150 overexpression on the activity of the α4 integrin promoter. A schematic representation of the 5′ α4 integrin reporter constructs is shown. Numbers in the construct name indicate the amount of 5′ flanking region present. The 5′ α4 integrin reporter constructs were cotransfected into 293T cells in the presence of 3 μg of vector alone or the CA150-expressing construct. The approximately threefold reduction in transcription activity of the constructs containing sequences upstream to bp +600 has been described previously (45). Transfection and activity assays were done as described in Materials and Methods.
To determine whether the altered activity of the HIV-1 promoter upon CA150 overexpression was mediated by the TATA box, we performed cotransfection experiments with the rest of the constructs shown in Fig. 3A. The NF-R construct has both NF-κB elements in a reverse orientation, and its activity was approximately one-third of that of HIV-CAT or NF/SP (wild type), in agreement with the results of Berkhout and Jeang (4) (Fig. 3A). As shown with HIV-1 constructs with enhancer sequences, transient cotransfection of this construct with CA150 showed a marked decrease of its activity (Fig. 3A). We next tested HIV-1 CAT reporter constructs with the TATA element replaced by TATTTAT (SV40-TATA), replaced by the random sequence GTCAC (m-TATA), or deleted (d-TATA). Previous work with these TATA mutant constructs showed that the transcription start site was shifted by only a few bases (4). The basal activities of these mutant HIV-1 promoters were one-third of the activity of the wild-type construct and similar to the activity of the NF-R vector (Fig. 3A). Strikingly, the transcription activities of these three mutant constructs were not repressed by CA150 in cotransfection experiments (Fig. 3A). These results indicated that the HIV-1 TATA box is important in the transcription repression of the HIV-1 promoter mediated by overexpression of CA150. The quantitative difference observed with the NF/SP construct is consistent with a necessary, although possibly not sufficient, role for the HIV-1 TATA box element in the repression mediated by overexpression of CA150.
We also sought to analyze transcription activity of the mutant HIV-1 constructs in the presence of Tat. As shown in Fig. 3A, those constructs with a wild-type TATA box (HIV-CAT, NF/SP [wild type], and NF-R) were strongly activated by Tat. Replacement of the TATA element with the SV40 TATA box (SV40-TATA) produced a construct that was fourfold less activated by Tat. Changing the TATA box to the random sequence GTCAC (m-TATA) or deleting the TATA box (d-TATA) produced constructs that lost the ability to respond to Tat (Fig. 3A). Our results confirm previous studies using these constructs (4) and strengthen the importance of the HIV-1 TATA box in Tat-mediated transcriptional activation. Interestingly, HIV-1 constructs which are fully responsive to Tat were also repressed by overexpressed CA150, and HIV-1 constructs which are nonresponsive to Tat were also unaffected by overexpressed CA150 (Fig. 3A). Therefore, it appears that CA150-mediated repression and Tat trans activation have the same promoter structure requirement, which is critically dependent on the TATA box. This also may indicate that Tat and CA150 are capable of modifying the same RNAP II transcription complexes assembled on the HIV-1 TATA box.
We have shown above that transcription from the α4 integrin promoter was also repressed by overexpression of CA150 (Fig. 2). In order to dissect the promoter elements necessary for repression by CA150 overexpression, we used a series of 5′ deletions of the α4 integrin promoter (Fig. 3B). Previous experiments utilizing these constructs demonstrated the presence of two sites for the zinc finger/homeodomain ZEB protein at bp −361 and −399 that diminished the activity of the α4 integrin promoter in a variety of cell lines tested (45). We also found an approximately threefold reduction in promoter activity when the constructs containing those sites were used in 293T cells (Fig. 3B, −600, −1.2, and −2.0 α4CAT constructs). Transient-cotransfection experiments using the α4 5′ deletion constructs and CA150 expression vector indicated that CA150 repression was mediated by the core promoter sequences, consisting of only a canonical TATA box sequence (−42α4CAT in Fig. 3B). This finding ruled out the requirement for upstream promoter elements in the mechanism of CA150 repression and indicated that sequences downstream of position −42 are required for the inhibition by CA150 on the α4 integrin promoter.
Overexpression of TBP can alleviate CA150-mediated transcription repression.
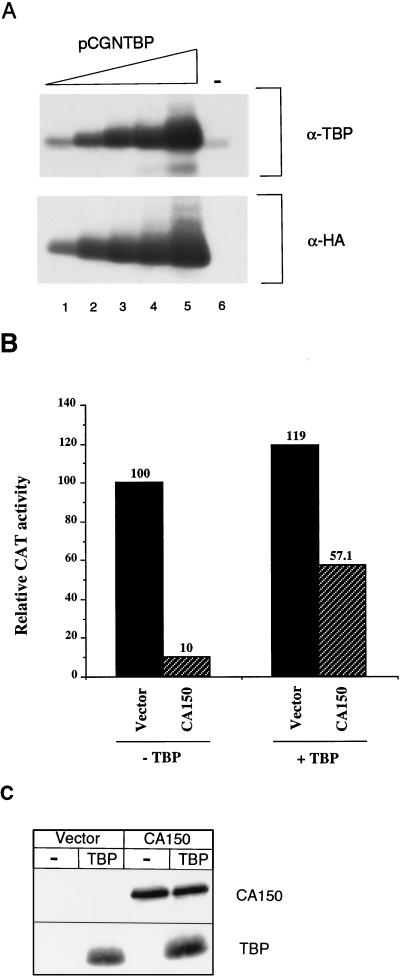
Overexpression of CA150 did not cause a general repression of all RNAP II transcription but instead was restricted to certain types of promoters, including the HIV LTR and α4 integrin promoter (Table 1 and Fig. 2). These results suggest that the mechanism by which the overexpression of CA150 represses transcription is not due to generalized sequestration of TBP, thereby inhibiting efficient transcription initiation. The fact that changes in TATA box sequences disrupted the repression observed in HIV-1 transcription (Fig. 3A), however, was suggestive of a role of TBP in the mechanism of CA150 action. For example, CA150 could be affecting specific TBP-containing complexes. Thus, we sought to investigate the effect of the overexpression of TBP on the repression of the HIV-1 promoter mediated by CA150. We used the mammalian expression vector pCGNTBP (51), which adds a 15-amino-acid HA epitope tag to the amino terminus of the expressed TBP (Fig. 4A). Cotransfection of pCGNTBP with the HIV-CAT reporter vector did not significantly change (1.2-fold increase) the basal activity of the HIV-1 promoter under these conditions (Fig. 4B). Overexpression of TBP relieved repression by CA150 (Fig. 4B) (10-fold versus 2-fold repression in the absence or presence of overexpressed TBP, respectively). This was not due to a reduction in expression of the CA150 protein, as CA150 expression was not decreased as analyzed by Western blotting with T7 tag-specific antibodies (Fig. 4C). HA-specific antibodies were used to analyze the overexpressed TBP (Fig. 4C).
FIG. 4.
TBP can alleviate CA150-mediated repression of the HIV-1 promoter. (A) Analysis of TBP expression. Fifty micrograms of extracts from 293T cells transfected with 1, 10, 50, 100, and 500 ng of pCGNTBP (lanes 1 to 5) or mock transfected (lane 6) was subjected to SDS-PAGE and Western blot analysis. Antibodies against TBP (α-TBP) and the HA epitope tag (α-HA) were used to visualize the proteins. (B) HIV-CAT reporter plasmid (0.1 μg) was cotransfected into 293T cells with 0.1 μg of pCGN (empty vector) (−TBP) or pCGNTBP (+TBP) in the presence of 3 μg of vector alone (black bars) or CA150-expressing construct (hatched bars). Transfection and activity assays were done as described in Materials and Methods. (C) Analysis of CA150 and TBP protein expression in the transfected cells. Twenty micrograms of transfected-cell extracts was used in SDS-PAGE and Western blot analysis to analyze protein expression. Antibodies against the T7 and HA epitope tags were used to visualize CA150 and TBP, respectively.
Overexpression of CA150 decreases transcription elongation from the HIV-1 promoter.
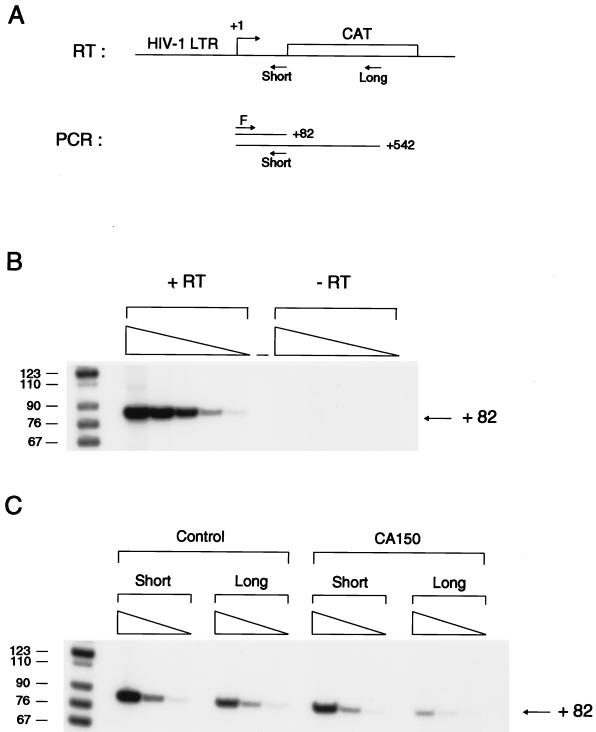
Our data above show a TATA box dependence for CA150-mediated HIV-1 LTR repression (Fig. 3A). As mentioned, the TATA box element is also essential for the specific stimulation of transcription elongation of the HIV-1 LTR by Tat. Based on these findings, together with data suggesting a role for CA150 in Tat trans activation (50), we hypothesized a role for CA150 in transcription elongation from the HIV-1 LTR. In order to test this possibility, the effect of CA150 on transcription directed from the HIV-1 LTR was further analyzed by quantifying the transcripts synthesized in transfected 293T cells. A quantitative RT-PCR protocol was designed to measure transcripts of different lengths derived from the HIV-1 promoter (Fig. 5A). Two specific oligonucleotides were used such that each one would serve to synthesize cDNA fragments of 82 and 542 nucleotides, respectively, in the RT reaction. Hence, this assay allowed us to measure the relative amounts of transcription complexes that reached nucleotides +82 and +542 of the nascent transcript.
FIG. 5.
Overexpression of CA150 reduces transcriptional elongation from the HIV-1 LTR. RNA from transfected 293T cells was isolated and subjected to RT-PCR analysis. (A) Representation of the RT-PCR assay used. A schematic map of the relevant regions in the HIV-CAT reporter construct is shown. The start site of transcription (+1) is indicated with an arrow. The relative positions of the specific primers used in the RT reaction mixture are shown. These RT specific primers measured transcription complexes that reached nucleotides +82 (Short) and +542 (Long) relative to the +1 start site of transcription. For PCR, the RT reaction mixtures were subsequently amplified with the same set of primers (F and Short) to compare directly the radioactive signals (see Materials and Methods). (B) RT-PCR assay. PCR products were linear over the range of concentrations used. RNA from cells transfected with the control plasmid was isolated and reverse transcribed in the presence (+RT) or absence (−RT) of reverse transcriptase enzyme. One microliter of the RT reaction mixture and subsequent fivefold dilutions were subjected to amplification by PCR and visualized as described in Materials and Methods. Molecular size markers (in base pairs) are indicated on the left. The position of the labeled PCR product is also indicated. (C) Inhibition of HIV-1 transcriptional elongation by overexpressed CA150. RNA from cells transfected with the control plasmid or CA150 expression construct (CA150) was isolated and reverse transcribed by using the short and long RT specific primers. RT reaction mixtures were subsequently amplified by the PCR approach described for panel A. Molecular size markers (in base pairs) are indicated on the left. The position of the labeled PCR product is also indicated. Quantification was performed to yield values for the radioactive signals from PCR products (see Materials and Methods and Results).
RNA from transfected 293T cells was isolated and reverse transcribed with these specific primers. RT reaction mixtures were subsequently amplified by PCR with the same set of primers to compare directly the radioactive signals (Fig. 5A) (see Materials and Methods). The RT-PCR assay was linear over the range of concentrations used (Fig. 5B). Figure 5C shows the results from a representative experiment. The level of +82 transcripts was affected less than twofold, versus a fivefold decrease in the level of +542 transcripts. The same results were obtained in RT-PCR analysis with samples from a second independent transfection experiment (data not shown). Based on these results, we conclude that the overexpression of CA150 repressed transcription from the HIV-1 LTR, at least in part, by decreasing the level of elongation-competent transcription complexes.
DISCUSSION
It has recently been demonstrated that activation of the HIV-1 promoter by Tat requires the assembly of a functional Tat–P-TEFb complex on the TAR element (34, 58, 60, 66). The interaction between Tat and cyclin T1, one of the components of P-TEFb, has recently been investigated by two groups (5, 16). Less information is available on the roles that other Tat cofactors play in this process (28, 50, 65). CA150 was purified by using an in vitro transcription system and Tat affinity chromatography (50). In vitro and in vivo experiments suggested that CA150 is necessary, although not sufficient, for Tat-mediated transcriptional activation of the HIV-1 promoter (50). Here, we have used a series of functional assays to further evaluate the role of CA150 in HIV-1 transcription.
First, we demonstrated that overexpression of CA150 protein reduces the activity of HIV-1 basal and Tat-activated transcription of the HIV-1 LTR. This inhibition is specific, since the activity of other promoters remained unchanged (Table 1), arguing against depletion of a limiting GTF (i.e., squelching). Next, we showed that the inhibition mediated by CA150 overexpression depends on a specific TATA box sequence in the HIV-1 promoter (Fig. 3A). It is clear from the work of several laboratories that the assembly of Tat-responsive, elongation-competent RNAP II transcription complexes depends critically on the TATA box element within the HIV-1 LTR. The HIV-1 LTR also specifies a TATA-box-independent transcription complex, which cannot be activated by Tat (our data and references 4, 29, and 44). Therefore, the HIV-1 promoter can specify two different types of transcription complexes, which respond differently to Tat. In our experiments, changes in the HIV-1 TATA box sequence had a minor effect on basal transcription (about one-third of the wild-type activity) and caused a severe abrogation of Tat trans activation. Our data also show that HIV-1 constructs able to be strongly activated by Tat (HIV-CAT, NF/SP [wild type], and NF-R) were also repressed by CA150 but that altered HIV-1 TATA box constructs (SV40-TATA, m-TATA, and d-TATA) were neither activated by Tat nor repressed by CA150 (Fig. 3A). These data indicate that CA150 overexpression affects the specific RNAP II complexes whose assembly critically depends on the HIV-1 TATA box and which are also responsive to Tat.
How does overexpression of CA150 inhibit HIV-1 transcription? In the absence of overexpressed CA150, TBP nucleates the assembly of the components of an active preinitiation complex (PIC), probably by recruiting an RNAP II holoenzyme (46). The formation of this PIC is critically dependent on a functional (wild-type) HIV-1 TATA box element. These transcription complexes are also Tat responsive; therefore, they can elongate efficiently. Transcription complexes formed on mutated HIV-1 TATA box elements may be devoid of critical components and therefore be affected by neither overexpressed CA150 nor Tat. Overexpression of CA150 could alter the activity of the Tat-responsive, TATA-dependent transcription complex, maybe by sequestering one of its components which is necessary for efficient transcriptional elongation. This hypothesis is consistent with previously observed interactions between CA150 and components of RNAP II holoenzyme complexes (50). More recently, we have detected an interaction between CA150 and proteins known to promote RNAP II transcriptional elongation, which supports this model further (19a). According to this, overexpression of CA150 results in the formation of TATA-box-dependent transcription complexes that are unable to elongate efficiently. Indeed, our data show that overexpressed CA150 can decrease the amount of transcripts derived from the HIV-1 LTR and that this defect is at the elongation level (Fig. 5). At present, it is not known whether CA150 affects the elongating RNAP II complex directly or during the formation of an active, elongation-competent PIC (e.g., by disrupting or sequestering the RNAP II holoenzyme or critical components of this complex). Identification and purification of CA150-binding factors within the transcription complex will help us to understand how this protein exerts its regulation and the mechanism underlying repression.
We have also shown that TBP overexpression may counteract CA150 inhibition of the HIV-1 LTR (Fig. 4B). As mentioned before, the specific repression on transcription from certain types of promoters by overexpressed CA150 (Table 1 and Fig. 2) suggested that repression was not due to a direct effect on a limiting GTF. These data, together with experiments showing no physical interaction between CA150 and TBP in vitro (unpublished data), make it unlikely that this GTF is the direct target of the repression. The mechanism by which TBP relieves the inhibition mediated by overexpressed CA150 is unknown. Synergy between Gal4-TBP and Tat has been reported recently (31, 59). Those data may indicate that TBP increases the formation of Tat-responsive transcription complexes whose assembly is dependent on a functional (wild-type) HIV-1 TATA box element. In our experiments, overexpressed TBP may counteract CA150 repression by increasing the formation of these transcription complexes. To date, it is not clear if recruitment of TBP activates the basal level of the HIV-1 promoter (31, 59). Clarification of this will be important to determine whether the Tat-responsive RNAP II complexes are the target of CA150.
Our results agree with the notion that HIV-1 TATA box sequences affect the elongation of transcripts initiated from the HIV-1 LTR. Sequences within promoter regions have been shown to affect late events in transcription in other systems. For example, the efficiency with which RNAP terminates transcription at a given termination site can be modulated by sequences linked to prokaryotic promoters (52). In addition, promoter sequences required for efficient elongation and premature termination have been defined in the human c-myc gene (36).
The transcription repression mediated by CA150 on the HIV-1 promoter has similarities to effects seen upon overexpression of the adenovirus E1A gene product. Overexpression of E1A protein leads to an inhibition of basal and Tat-activated transcription of the HIV-1 LTR in vivo (53, 56, 57) and in vitro (49). E1A did not affect the RSV promoter in vitro (49). Strikingly, E1A repression of the HIV-1 promoter was dependent on a functional TATA box sequence (53). Mutational analyses have identified the amino terminus and the CR1 region of E1A protein to be important for its repressive effect (49, 53, 56, 57). These mutations did not disrupt the binding of E1A to TBP (which lies within the CR3 region in E1A); thus, it is unlikely that TBP mediates the repression. The same studies established a good correlation between repression and binding of E1A to CBP-p300. CBP and p300 are nuclear proteins that participate in a variety of transcription pathways and cell growth control (14, 18). They interact with transcriptional activators as well as repressors; therefore, it has been suggested that promoters regulated by molecules requiring CBP-p300 would be repressed by E1A. CBP-p300 associates with RNAP II holoenzyme and interacts with RNAP II (7, 39, 40). E1A may disrupt interactions between transcription factors and RNAP II that are mediated by CBP-p300. Interestingly, CBP-p300 has been shown recently to interact with Tat and regulate the activity of the HIV-1 promoter (2, 23, 35). It will be of interest to test whether overexpression of CBP-p300 can abrogate CA150-mediated repression of the HIV-1 promoter.
Another protein capable of repressing transcription from many viral and cellular promoters whose initiation is dependent on the presence of a TATA box is p53 (30). A proline-rich motif in p53 has been shown to be essential for repression (55). Similarly, the amino and CR1 regions of E1A protein, which are involved in its repressive effects, likewise contain many prolines. These data may explain the specific inhibition of Tat-activated, but not basal, transcription from the HIV-1 promoter upon overexpression of a truncated CA150 protein in HeLa cells (50). The truncated CA150 protein contained a deletion in the amino-terminal part which included, among other motifs, the polyproline-rich region, which may be important for repression of HIV-1 basal transcription activity. In fact, we have found that deletion of the polyproline-rich region of CA150 reduced CA150-mediated repression by 50% in 293T cells (unpublished results). We cannot rule out, however, an effect of the different cell lines used in these studies. Mutational studies are in progress to elucidate the roles of different regions and motifs of CA150 in the transcriptional repression by overexpression of this protein. Together, these data implicate proline-rich regions in transcription repression. Further investigation into the mechanism by which these otherwise unrelated proteins inhibit transcription will shed light on their specific roles in regulating gene expression.
Finally, the α4 integrin promoter is also repressed by CA150 overexpression (Fig. 2 and 3B). This finding suggests that CA150 regulates transcription of cellular genes. Nucleotide sequence analysis of the HIV-1 and α4 integrin minimal promoters reveals similarities only at the TATA box motif (TATAA and TATA for the HIV-1 and α4 integrin promoters, respectively). The other promoters used in this study contained TATA box sequences that were more divergent from the consensus (TATATAA, TATTTAT, TATTTAA, and TATTAA for the CMV, early SV40, RSV, and HSV-TK-LUC promoters, respectively). The overexpression of CA150 may affect a specific subset of factors whose assembly requires certain types of TATA box sequences. Whether the HIV-1 and α4 integrin promoters are transcriptionally regulated through similar mechanisms will be the focus of further investigation.
ACKNOWLEDGMENTS
We thank C. Hernández-Munain and members of the M. A. Garcia-Blanco laboratory for their help during the course of this work. We also thank P. Bohjanen, R. Carstens, A. Goldstrohm, C. Hernández-Munain, and Y. Liu for critical review of the manuscript. We also thank K.-T. Jeang for HIV-1 promoter constructs, A. Postigo for α4 integrin promoter constructs, and W. Tansey for pCGNTBP.
This research was supported by a grant from the NIH to M.A.G.-B. C.S. is supported by the NIAID Research Training Program in AIDS to the Division of Infectious Diseases at Duke University Medical Center. We also acknowledge the Keck Foundation for support to the Levine Science Research Center.
REFERENCES
- 1.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 2.Benkirane M, Chun R F, Xiao H, Ogryzko V V, Howard B H, Nakatani Y, Jeang K T. Activation of integrated provirus requires histone acetyltransferase. J Biol Chem. 1998;273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 3.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, Jeang K T. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 7.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Chun R F, Jeang K T. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus 1 and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 11.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 12.Dahmus M. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 13.Dubridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 15.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 16.Garber M E, Ping W, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudreau L, Adam M, Ptashne M. Activation of transcription in vitro by recruitment of the yeast RNA polymerase II holoenzyme. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 18.Giles R H, Peters D J M, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 19.Gold M O, Yang X, Herrmann C H, Rice A P. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Goldstrohm, A., C. Suñé, and M. A. Garcia-Blanco. Unpublished data.
- 20.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrmann C H, Rice A P. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 23.Hottiger M O, Nabel G J. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 25.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 26.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 27.Lee J M, Greenleaf A L. Modulation of RNA polymerase II elongation efficiency by c-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 28.Li X-Y, Green M R. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Welsh T M, Peterlin B M. The human immunodeficiency virus type 1 long terminal repeat specifices two different transcription complexes, only one of which is regulated by Tat. J Virol. 1993;67:1752–1760. doi: 10.1128/jvi.67.4.1752-1760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mack D H, Vartikar J, Pipas J M, Laimins L A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 31.Majello B, Napolitano G, De Luca P, Lania L. Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J Biol Chem. 1998;273:16509–16516. doi: 10.1074/jbc.273.26.16509. [DOI] [PubMed] [Google Scholar]
- 32.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 33.Malim M H, Hauber J, Fenrick R, Cullen B R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 34.Mancebo H, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzio G, Tyagi M, Gutierrez M I, Giacca M. HIV-1 Tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. J Biol Chem. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meulia T, Krumm A, Spencer C, Groudine M. Sequences in the human c-myc P2 promoter affect the elongation and premature termination of transcripts initiated from the upstream P1 promoter. Mol Cell Biol. 1992;12:4590–4600. doi: 10.1128/mcb.12.10.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 40.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann J R, Morency C A, Russian K O. A novel rapid assay for chloramphenicol acetyl transferase gene expression. BioTechniques. 1987;5:444–447. [Google Scholar]
- 42.Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto H, Sheline C T, Corden J L, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen H S, Rosen C A. Contribution of the TATA motif to Tat-mediated transcriptional activation of human immunodeficiency virus gene expression. J Virol. 1992;66:5594–5597. doi: 10.1128/jvi.66.9.5594-5597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Postigo A A, Sheppard A M, Mucenski M L, Dean D C. c-Myb and Ets proteins synergize to overcome transcriptional repression by ZEB. EMBO J. 1997;16:3924–3934. doi: 10.1093/emboj/16.13.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranish J A, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosen G D, Barks J L, Iademarco M F, Fisher R J, Dean D C. An intricate arrangement of binding sites for the Ets family of transcription factors regulates activity of the integrin gene promoter. J Biol Chem. 1994;269:15652–15660. [PubMed] [Google Scholar]
- 48.Rosen G D, Sanes J R, LaChance R, Cunningham J M, Roman J, Dean D C. Roles for the integrin VLA-4 and its counter receptor VCAM-1 in myogenesis. Cell. 1992;69:1107–1119. doi: 10.1016/0092-8674(92)90633-n. [DOI] [PubMed] [Google Scholar]
- 49.Song C-Z, Loewenstein P M, Green M. Repression in vitro, by human adenovirus E1A protein domains, of basal or Tat-activated transcription of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:2907–2911. doi: 10.1128/jvi.69.5.2907-2911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suñé C, Hayashi R, Liu Y, Lane W S, Young R A, Garcia-Blanco M A. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tansey W P, Ruppert S, Tjian R, Herr W. Multiple regions of TBP participate in the response to transcriptional activators in vivo. Genes Dev. 1994;8:2756–2769. doi: 10.1101/gad.8.22.2756. [DOI] [PubMed] [Google Scholar]
- 52.Telesnitsky A P W, Chamberlin M J. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J Mol Biol. 1996;205:315–330. doi: 10.1016/0022-2836(89)90343-4. [DOI] [PubMed] [Google Scholar]
- 53.Tsang S X, Morris G F, Lu M, Morris C B. TATA-dependent repression of human immunodeficiency virus type-1 transcription by the adenovirus E1A 243R oncoprotein. Oncogene. 1996;12:819–826. [PubMed] [Google Scholar]
- 54.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 55.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventura A M, Arens M Q, Srinivasan A, Chinnadurai G. Silencing of human immunodeficiency virus long terminal repeat expression by an adenovirus E1a mutant. Proc Natl Acad Sci USA. 1990;87:1310–1314. doi: 10.1073/pnas.87.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei P, Garber M E, Fang S, Fischer W H, Jones K A. A novel CDK9 associated c-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 59.Xiao H, Lis J T, Jeang K T. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol Cell Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Gold M O, Tang D N, Lewis D E, Aguilar-Cordova E, Rice A P, Herrmann C H. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 63.Zenzie-Gregory B, Sheridan P, Jones K A, Smale S T. HIV-1 core promoter lacks a simple initiator element but contains a bipartite activator at the transcription start site. J Biol Chem. 1993;268:15823–15832. [PubMed] [Google Scholar]
- 64.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Q, Sharp P A. Tat-SF1: cofactor for stimulation of transcription elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M, Price D. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]