Abstract
Problem
The utility of the polymerase chain reaction (PCR) cycle threshold (C t) values in the management of patients with coronavirus disease 2019 (COVID‐19) remains controversial.
Methods
We assessed the correlation of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) C t values in nasopharyngeal swab samples with the oxygen requirements at the time of sample collection. Specimens were tested with the Simplexa PCR platform, which targets the SARS‐CoV‐2 ORF1ab and S genes.
Results
We identified 23 COVID‐19 patients with 49 C t values available. While C t values from ORF1ab and S genes were highly correlated for a given specimen, there was no correlation between C t values for any of these target genes and the oxygen requirements of the patient at the time of sample collection. We found no differences in the initial nor the nadir C t values between survivors and non‐survivors or mild/moderate versus severe/critical illness at the maximum point of illness.
Conclusion
SARS‐CoV‐2 C t values have limited value in the management of COVID‐19.
Keywords: COVID‐19, hematopoietic cell transplantation, oxygen, SARS‐CoV‐2/Ct value
1. INTRODUCTION
The cycle threshold (C t) in reverse transcription polymerase chain reaction (RT‐PCR) refers to the number of cycles needed to amplify viral RNA to reach a detectable level. C t values are inversely correlated with the viral load and the ability to grow severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in vitro;1, 2, 3 the probability of isolating SARS‐CoV‐2 in cell culture is 77, 24, and less than 3% for clinical specimens with C t values between 10 and 20, 20–30, and 30–40, respectively.1, 2, 4 The C t value‐based estimates of viral load have been used to predict disease progression, infer transmissibility and differentiate active viral replication from prolonged virus shedding.5 At our transplant center, for example, we have used the C t value in a case‐by‐case basis to clear transplant candidates to undergo urgently needed conditioning/lymphodepleting therapies under the assumption that curative transplantation benefits outweigh the potential risks of COVID‐19 in those with a high C t value (e.g., >35), symptom resolution and are greater than 3–4 weeks from diagnosis. We have also used low C t values in patients with a compatible clinical syndrome (e.g., worsening respiratory status) to guide clinical decisions such as administering a second course of remdesivir. However, definitive data to support the predictive value of C t values in these situations are lacking.5 C t values generated by the currently available qualitative PCR assays do not reliably correspond to specific RNA concentrations and are not consistent across platforms. In addition, C t values from viral RNA can vary depending on the method of specimen collection, specimen source, transport, and the time from infection to collection to analysis.6 Current guidance from the Centers for Disease Control and Prevention recommends against the use of C t values in the clinical setting.
2. METHODS
To further explore the clinical utility of C t values, we assessed the correlation of SARS‐CoV‐2 C t values in nasopharyngeal swab samples with the oxygen requirements at the time of sample collection for PCR testing, in a cohort of hematological patients with COVID‐19 managed at a large academic center. We identified 23 patients with COVID‐19 with a total of 49 C t values available (Supplementary Table 1). Specimens were tested with The DiaSorin Molecular Simplexa COVID‐19 Direct real‐time RT‐PCR assay, which has a C t cutoff less than 40 and targets the SARS‐CoV‐2 ORF1ab (encoding 16 nonstructural proteins) and S (encoding the structural spike glycoprotein) genes. Data were summarized using descriptive statistics. Mann–Whitney or Kruskal–Wallis tests were used where appropriate. All tests were two‐sided and p < .05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism Software, Inc, version 7.03.
3. RESULTS
Patients underwent a median of two PCR tests (range, 1–7) with C t value available. While C t values from ORF1ab and S genes were highly correlated for a given specimen (Figure 1A), there was no apparent correlation between C t values for any of these SARS‐CoV‐2 target genes and the oxygen requirements of the patient at the time of sample collection (Figure 1B,C). When plotting C t values by the fraction of inspired oxygen (FiO2) strata (0.21 vs. 0.24‐0.28 vs. >0.4; Figure 1D,G) or simply by room air versus any oxygen supplementation (Figure 1E,H) there was no difference in the SARS‐CoV‐2 C t values by oxygen requirements at the time of PCR testing. The trajectory of the C t values over time is shown in Figure 1F,I.
Figure 1.
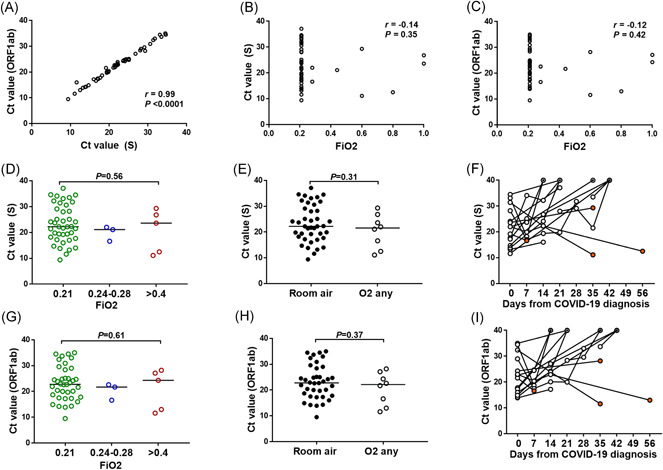
Lack of correlation between SARS‐CoV‐2 PCR cycle threshold (C t) values and oxygen requirements at the time of PCR testing. (A–C) Spearman correlation between S and ORF1ab gene targets on the same clinical specimen (A), and between each SARS‐CoV‐2 gene target and the recorded fraction of inspired oxygen (FiO2) (B–C) at the time of sample collection for PCR testing. (D–I) Distribution of SARS‐CoV‐2 C t values across requirements of FiO2 strata at the time of PCR testing for S (D–E) and ORF1ab (G–H) genes. Bars corresponds to the median C t value. p value was calculated using Mann–Whitney or Kruskal–Wallis tests where appropriate for comparison between groups. Trajectory of C t values over time for patients with 2 or more PCR swabs available (F, I). All negative values were given a value of 40 indicated with circled dot. Three patients with negative PCR documented on routine testing obtained more than 6 weeks from previous positive test were considered outliers and not included in the figure. Orange circles correspond to the last C t value available if patient is deceased. PCR, polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
We next compared the initial SARS‐CoV‐2 C t value (i.e., on the PCR obtained at the time of COVID‐19 diagnosis) by survival, disease severity and need for admission (Figure 2A,B) and observed no significant difference in the initial C t values for the SARS‐CoV‐2 gene target tested between survivors and non survivors, or mild/moderate versus severe/critical disease at the maximum point of illness. Patients who required hospital admission had lower initial C t values than those who did not (Figure 2A,B).
Figure 2.
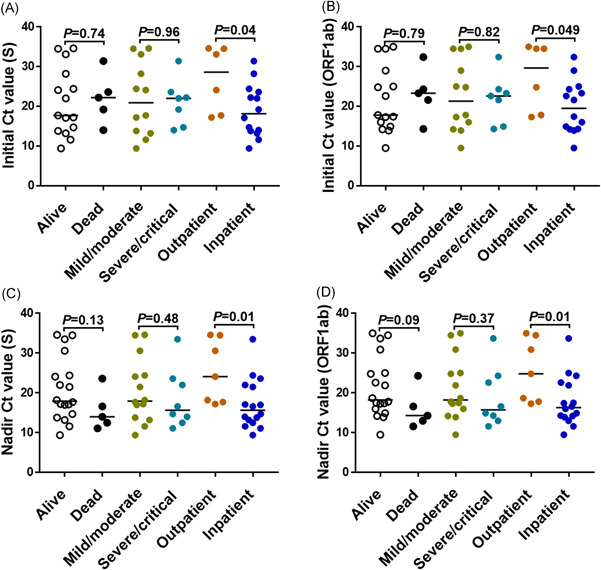
Lower SARS‐CoV‐2 PCR C t value among COVID‐19 patients requiring hospital admission. Initial (A–B) and nadir (C–D) SARS‐CoV‐2 C t values by clinical outcome are shown for the gene targets specified in the y‐ axis. COVID‐19 severity was defined as follows: mild (the clinical symptoms are mild and no pneumonia manifestations can be found in imaging); moderate (pneumonia manifestations on imaging); severe (tachypnea with respiratory rate >30 per minute, hypoxia requiring FiO2 > 0.4 to attain SpO2 > 94%, a ratio of arterial partial pressure of oxygen to fraction of inspired oxygen [PaO2/FiO2]<300 mmHg, and/or lung infiltrates >50%); and critical (respiratory failure requiring mechanical ventilation, septic shock and/or multiorgan failure). Data for maximum COVID‐19 severity missing in one patient admitted at an outside hospital. p value was calculated using Mann–Whitney for comparison between groups. COVID‐19, coronavirus disease 2019; C t, cycle threshold; PCR, polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
We also performed analysis using the lowest C t value available (as a surrogate marker of the peak viral load) for a given patient and found no differences in the nadir C t values between survivors and nonsurvivors or mild/moderate versus severe/critical illness at the maximum point of illness (Figure 2C,D). The group of patients who required admission to the hospital due to COVID‐19, however, exhibited lower C t values than those managed in the outpatient setting (Figure 2C,D).
4. DISCUSSION
Admission SARS‐CoV‐2 viral load among hospitalized patients with COVID‐19 independently correlates with the risk of intubation and in‐hospital mortality.7, 8 Those with C t values less than 25 (i.e., high viral load) had a three‐fold increased risk of intubation and five to six‐fold increased risk of mortality.7, 8 C t value has been also used to assess response to antiviral therapy.9 However, several studies have shown that nasopharyngeal SARS‐CoV‐2 C t values are not associated with COVID‐19 severity and do not support a predictive role for the C t value in the clinical setting.10, 11, 12, 13, 14 For example, in a report that included a total of 414 throat swabs collected from 94 patients, there was no obvious difference in viral loads across disease severity.10 Another study showed similar initial and peak viral loads between patients with mild and those with severe COVID‐19.11 In addition, SARS‐CoV‐2 C t values from asymptomatic patients are similar to those in symptomatic patients,12, 13, 14 which might account for the high rates of asymptomatic transmission.15
Here we further explored the clinical utility of C t values and observed no correlation between C t values from the nasopharyngeal PCR samples and disease severity as measured by the oxygen requirements. Although C t values were significantly lower among patients requiring hospital admission, we observed no differences in the initial and nadir C t values when segregating the cohort by COVID‐19 severity or mortality outcomes.
Our study has limitations including small sample size, and that observations obtained with the ORF1ab and S target genes using the Simplexa PCR platform cannot be extrapolated to other target genes and/or platforms. Likewise, our cohort consisted of hematological patients and validation of our findings in other patient populations requires further study. Despite such limitations, to our knowledge, this is the first study assessing the correlation between C t values and FiO2 requirements at the time of PCR testing. We conclude that SARS‐CoV‐2 C t values have limited value in the management of hospitalized patients with COVID‐19.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
All authors contributed to study design, data analysis, and writing of manuscript.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
The authors are indebted to all the patients who participated in the study and to Orlando Bracho for assistance with data collection.
Camargo JF, Lin RY, Komanduri KV. Lack of correlation between the SARS‐CoV‐2 cycle threshold (C t) value and clinical outcomes in patients with COVID‐19. J Med Virol. 2021;93:6059‐6062. 10.1002/jmv.27171
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1.Gniazdowski V, Morris CP, Wohl S, et al. Repeat COVID‐19 molecular testing: correlation of SARS‐CoV‐2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2020. 10.1093/cid/ciaa1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young BE, Ong SWX, Ng LFP, et al. Viral dynamics and immune correlates of COVID‐19 disease severity. Clin Infect Dis. 2020. 10.1093/cid/ciaa1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663‐2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS‐CoV‐2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Infectious Diseases Society of America . IDSA and AMP joint statement on the use of SARS‐CoV‐2 PCR cycle threshold (Ct) values for clinical decision‐making. 2021; https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-amp-statement.pdf
- 6.Rhoads D, Peaper DR, She RC, et al. College of American Pathologists (CAP) Microbiology Committee perspective: caution must be used in interpreting the cycle threshold (Ct) value. Clin Infect Dis. 2020;72:685. 10.1093/cid/ciaa1199 [DOI] [PubMed] [Google Scholar]
- 7.Magleby R, Westblade LF, Trzebucki A, et al. Impact of SARS‐CoV‐2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2020. 10.1093/cid/ciaa851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westblade LF, Brar G, Pinheiro LC, et al. SARS‐CoV‐2 viral load predicts mortality in patients with and without cancer who are hospitalized with COVID‐19. Cancer Cell. 2020;38(5):661‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joo EJ, Ko JH, Kim SE, et al. Clinical and virologic effectiveness of remdesivir treatment for severe coronavirus disease 2019 (COVID‐19) in Korea: a Nationwide Multicenter Retrospective Cohort Study. J Korean Med Sci. 2021;36(11):e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26(5):672‐675. [DOI] [PubMed] [Google Scholar]
- 11.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Kim T, Lee E, et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS‐CoV‐2 infection in a Community Treatment Center in the Republic of Korea. JAMA Intern Med. 2020;180(11):1447‐1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ra SH, Lim JS, Kim GU, Kim MJ, Jung J, Kim SH. Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS‐CoV‐2 infection. Thorax. 2021;76(1):61‐63. [DOI] [PubMed] [Google Scholar]
- 14.Louie JK, Stoltey JE, Scott HM, et al. Comparison of symptomatic and asymptomatic infections due to severe acute respiratory coronavirus virus 2 (SARS‐CoV‐2) in San Francisco long‐term care facilities. Infect Control Hosp Epidemiol. 2020:1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oran DP, Topol EJ. The proportion of SARS‐CoV‐2 infections that are asymptomatic: a systematic review. Ann Intern Med. 2021;174(5):655‐662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


