Abstract
In T lymphocytes, the hematopoietic cytokine interleukin-2 (IL-2) uses phosphatidylinositol 3-kinase (PI 3-kinase)-induced signaling pathways to regulate E2F transcriptional activity, a critical cell cycle checkpoint. PI 3-kinase also regulates the activity of p70s6k, the 40S ribosomal protein S6 kinase, a response that is abrogated by the macrolide rapamycin. This immunosuppressive drug is known to prevent T-cell proliferation, but the precise point at which rapamycin regulates T-cell cycle progression has yet to be elucidated. Moreover, the effects of rapamycin on, and the role of p70s6k in, IL-2 and PI 3-kinase activation of E2Fs have not been characterized. Our present results show that IL-2- and PI 3-kinase-induced pathways for the regulation of E2F transcriptional activity include both rapamycin-resistant and rapamycin-sensitive components. Expression of a rapamycin-resistant mutant of p70s6k in T cells could restore rapamycin-suppressed E2F responses. Thus, the rapamycin-controlled processes involved in E2F regulation appear to be mediated by p70s6k. However, the rapamycin-resistant p70s6k could not rescue rapamycin inhibition of T-cell cycle entry, consistent with the involvement of additional, rapamycin-sensitive pathways in the control of T-cell cycle progression. The present results thus show that p70s6k is able to regulate E2F transcriptional activity and provide direct evidence for the first time for a link between IL-2 receptors, PI 3-kinase, and p70s6k that regulates a crucial G1 checkpoint in T lymphocytes.
The cytokine interleukin-2 (IL-2) controls T-cell cycle progression and differentiation. The essential and irreplaceable functions of IL-2 in the immune system and the potential use of this cytokine in immunotherapy has prompted clinical and pharmacological interest in IL-2 signal transduction (42). Triggering of the IL-2 receptor activates the Janus kinases (JAKs) 1 and 3 (1, 20, 29, 37, 47). Signaling cascades initiated by the action of these IL-2-induced tyrosine kinases include activation of Ras effector pathways (11, 18, 45), activation of the transcription factors STATs 3 and 5 (2, 17, 21, 24), and the regulation of phosphatidylinositol 3-kinase (PI 3-kinase) and the kinase Akt, also called protein kinase B (PKB) (34, 36).
A key event in the G1 phase of the cell cycle and an important checkpoint for mitogenesis is the transcriptional activation of E2Fs (15). Sites for E2F binding have been identified in many genes important for cell cycle regulation, such as the cyclin E gene (4). We have recently examined the signaling pathways used by IL-2 to regulate E2F transcriptional activity and have established that PI 3-kinase has a critical role in coupling the IL-2 receptor to E2F regulation. In quiescent G0/G1-arrested cells, the transcriptional activity of E2Fs is repressed by their binding of a pocket protein, of which three have been identified: pRb, p107, and p130 (15). The complexes between pocket proteins and E2Fs are controlled by protein phosphorylation: cyclin–cyclin-dependent kinase (cdk)-mediated phosphorylation of pocket proteins results in the release of E2F, resulting in loss of E2F repressor function, and allows E2F transcriptional activity. In T lymphocytes, PI 3-kinase signals control pRb and p130 hyperphosphorylation (5), which is regulated by cyclin D-cdk complexes. In turn, PI 3-kinase signals are required for IL-2 upregulation of cyclin D3. Moreover, PI 3-kinase signaling is necessary and sufficient for E2F transcriptional activity in T cells (5). The importance of PI 3-kinase for the regulation of pocket protein phosphorylation and hence for the regulation of E2Fs explains why activation of this lipid kinase is essential for the proliferative responses of lymphoid cells.
The proximal effectors of PI 3-kinase that regulate E2Fs have not yet been characterized. PI 3-kinase signaling pathways previously described in T cells include the MEK/ERK2 pathway (22). However, inhibition of the ERKs has no effect on IL-2 activation of E2Fs, excluding this pathway from any critical role in E2F responses to IL-2 (5). PI 3-kinase also couples the IL-2 receptor to signaling pathways inhibited by the drug rapamycin (34), a powerful and important immunosuppressant that blocks T-cell proliferation. The distal signaling pathways regulated by rapamycin that explain its antiproliferative actions in T cells have not been characterized. Rapamycin forms an inhibitory complex with the immunophilin FKBP12, and this complex regulates the activity of a protein termed mTOR (mammalian target for rapamycin) which has homology to protein and lipid kinases (6, 38). Direct targets for mTOR, including the initiation factor 4E binding protein 1 (4EBP1) (7), a repressor of eukaryotic initiation factor 4E (eIF4E), have been described (3). Phosphorylation of 4EBP1 releases the protein from eIF4E, allowing the initiation factor to form a productive mRNA cap binding complex. Rapamycin interactions with mTOR also regulate the activity of p70s6k, the kinase that phosphorylates the 40S ribosomal protein S6 (44). S6 is thought to be the only p70s6k substrate, and by controlling S6 phosphorylation, p70s6k regulates the translation of an essential family of mRNAs that contain an oligopyrimidine tract at their transcriptional start site (19). This mRNA subset includes transcripts encoding ribosomal proteins and protein synthesis elongation factors (27). The role of p70s6k in the suppressive actions of rapamycin on cell proliferation has been the subject of much debate. Recent analysis of embryonic stem cells containing a targeted deletion of p70s6k showed that loss of this enzyme did not cause a change in the rapamycin sensitivity of proliferative responses (23). However, a new S6 kinase gene, termed p70s6k2, which contains all the same regulatory motifs and rapamycin-regulated phosphorylation sites as p70s6k, has recently been identified. p70s6k2 catalytic activity, like that of p70s6k, is fully suppressed by rapamycin. Thus, the rapamycin sensitivity of p70s6k-null cells could be explained by the presence of a compensating, rapamycin-sensitive, functional analogue of p70s6k (41).
IL-2 regulation of p70s6k and E2Fs are both controlled by PI 3-kinase. Moreover, PI 3-kinase signals alone are sufficient to drive activation of p70s6k, just as they are sufficient to switch on E2F transcriptional activity. p70s6k is therefore a potential candidate to mediate PI 3-kinase responses for E2F regulation. In this context, it is well documented that rapamycin, which abrogates p70s6k activity, has an antiproliferative effect on T cells. Rapamycin blocks T-cell proliferation by delaying G1 transit times rather than by causing an absolute mitotic block (43). If rapamycin signaling pathways were important for IL-2 and PI 3-kinase activation of E2Fs, then this would be the molecular basis for the inhibitory effects of this drug on T-cell clonal expansion. Several studies have examined the effects of rapamycin on the phosphorylation of the pocket proteins (9, 14), but these have generally been performed with nonlymphoid cells and have yielded discrepant results. Rapamycin has also been described to prevent IL-2-induced loss of the cyclin-cdk inhibitor p27kip1 (32). However, the targeted degradation of p27kip1 is initiated by cyclin E-cdk2 phosphorylation (39). Hence, if rapamycin-treated T cells fail to correctly activate cyclin-cdk complexes, then p27kip1 levels would persist but as a consequence, not a cause, of failed cell cycle progression. To resolve this issue, an analysis of the effects of rapamycin on IL-2-induced pocket protein phosphorylation in T cells and an analysis of the effects of rapamycin on the transcriptional activation of E2Fs in T cells are required.
The objective of the present study was to use rapamycin and mutants of p70s6k to explore the role of p70s6k in IL-2 and PI 3-kinase regulation of E2F transcriptional activity. Our results show that IL-2- and PI 3-kinase-induced pathways for the regulation of E2F transcriptional activity are rapamycin sensitive. We show that rapamycin abrogates IL-2 activation of p70s6k. Overexpression of wild-type p70s6k, but not that of a kinase-dead mutant, increased E2F transcriptional activity. More importantly, expression of a p70s6k rapamycin-resistant mutant rescued the inhibition of E2F transcriptional activity by rapamycin. These results map cell cycle targets for rapamycin in T cells. They show also that p70s6k is able to regulate E2F transcriptional activity, and they provide direct evidence for the first time that p70s6k controls signals that regulate a G1 checkpoint in T lymphocytes.
MATERIALS AND METHODS
Reagents.
IL-2 was supplied by Chiron. Rapamycin was provided by G. Thomas. [14C]acetyl coenzyme A (at 50 mCi/mmol) and [γ-32P]ATP (5,000 Ci/mmol) were purchased from Amersham Corp. Antibodies for p130, E2F-1, and E2F-4 were purchased from Santa Cruz Biotechnology. Antibodies for pRb were obtained from Pharmingen.
Cell culture.
Kit225 cells (16), a human IL-2-dependent T-cell line, were maintained in RPMI medium supplemented with 20 ng of IL-2/ml in a 5% CO2 humidified incubator. In the absence of IL-2, these cells accumulate in the G1 phase of the cell cycle. The cells were deprived of IL-2 for 24 h prior to transfection by two washes in RPMI medium. For other experiments the cells were deprived of IL-2 for 72 h. Human peripheral blood-derived T lymphocytes were generated and maintained as described elsewhere (8).
Plasmids.
E2ACAT, originally described by Murthy et al. (31) and used subsequently by Mann and Jones (26), comprises bp −284 to +62 of the E2A promoter upstream of a chloramphenicol acetyltransferase (CAT) gene. Two E2F binding sites are present in E2ACAT (the first is between −29 and −21, and the second is between −82 and −66) and allow the E2A promoter to report and sensitively quantitate E2F transcriptional activity (5, 26, 33). E2CAT(E2F−) was a generous gift from J. R. Nevins (Howard Hughes Medical Institute, Duke University Medical Center, Durham, N.C.). It contains bp −85 to +40 from the E2A promoter, in which both E2F sites have been mutated, upstream of a CAT gene (25). The reporter plasmid GRRCAT contains five copies of the gamma interferon receptor response element upstream of the thymidine kinase (TK)-CAT gene (2).
The following expression plasmids have been described elsewhere: pEF, expressing rCD2p110 (35); a cytomegalovirus (CMV) plasmid expressing myc-tagged wild-type p70s6k (19); p70s6kD3E-E389, a CMV plasmid expressing myc-tagged rapamycin-resistant p70s6k (19); p70s6kQ100, expressing myc-tagged kinase-dead p70s6k (19); and pRbΔ (pEF), an Rb construct in which the p34cdc2 phosphorylation sites are mutated (13). Plasmid DNA was purified by cesium chloride density gradient centrifugation.
E2A DNA affinity precipitations.
For affinity precipitation of E2Fs from T-cell lysates, biotinylated double-stranded oligonucleotides which corresponded to the E2F binding sites in the E2A promoter (TAGTTTTCGCGCTTAAATTTGAGAAAGGGCGCGAAACTAGTC [E2F binding sites are underlined]) were synthesized. A mutant oligonucleotide, in which two nucleotides in each E2F binding site were mutated (shown in boldface), was used to check the specificity of binding. The sequence of the oligonucleotide was TAGTTTTCGATCTTAAATTTGAGAAAGGGTACGAAACTAGTC. Briefly, 107 cells were lysed in 1% Nonidet P-40 (NP-40) lysis buffer (50 mM Tris-HCl [pH 8.0], 1% NP-40, 150 mM NaCl, 0.1 mM EDTA, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of chymostatin/ml) for 30 min on ice. DNA binding proteins were isolated from extracts by incubation at 4°C for 2 h with 1 μg of double-stranded, 5′-biotinylated oligonucleotide coupled to 30 μl of a 50% suspension of streptavidin agarose (Sigma). Complexes were washed twice, after which protein was eluted from the beads with reducing sample buffer. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% polyacrylamide). E2A DNA binding complexes were analyzed by Western blotting and enhanced chemiluminescence (ECL; Amersham). For competing affinity-purified complexes, a double-stranded E2F oligonucleotide (not biotinylated) with the sequence ATTTAAGTTTCGCGCCCTTTCCAA (30) was used. Samples were preincubated with the oligonucleotide for 30 min prior to the addition of the biotinylated oligonucleotide.
p70s6k assays.
p70s6k assays were carried out by a method described by Reif et al. (34). Kit225 cells were transfected with myc epitope-tagged S6 kinase (S6k) constructs as described previously (34). Cells were then maintained under normal growth conditions for 16 h, counted, and treated with rapamycin for 20 min. Treatment was terminated by placing the samples on ice; they were then centrifuged and lysed in lysis buffer (120 mM NaCl, 50 mM Tris [pH 8.0], 20 mM NaF, 1 mM benzamidine, 1 mM EDTA, 6 mM EGTA, 7.5 mM inorganic pyrophosphate, 15 mM p-nitrophenyl phosphate, 1% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, and 0.1 mM Na3VO4). Postnuclear lysates were precleared with protein A cell suspension (Sigma) prior to incubation with 15 μg of 9E10 antibody precoupled to protein G-Sepharose beads. Immunoprecipitates were washed three times in lysis buffer and once in p70s6k assay buffer (50 mM morpholinepropanesulfonic acid [MOPS] [pH 7.2], 5 mM MgCl2, 0.1% Triton X-100) and were assayed for kinase activity as described elsewhere (28) by using 40S ribosomal protein S6 as a substrate. Proteins were resolved by SDS-PAGE. 32P-labelled S6 proteins were detected by autoradiography or quantitated with a PhosphorImager (Molecular Dynamics). Expression of myc epitope-tagged p70s6k was revealed by Western blot analysis using the myc epitope-specific antibody 9E10.
Transfections and CAT assay.
Kit225 cells were deprived of IL-2 as indicated prior to transfection, and 15 × 106 cells were transfected by electroporation with the amounts of DNA indicated in the figure legends. Electroporation was carried out with a Gene Pulser (Bio-Rad) set at 320 V and 960 μF. After transfection, cells were replaced in culture in the absence or presence of 20 ng of IL-2/ml for 16 to 18 h prior to lysis. Cells were lysed in buffer containing 10 mM Tris (pH 8.0), 1 mM EDTA, 150 mM NaCl, and 0.65% NP-40. Samples were then assayed for CAT activity by the radioisotope method (12).
Western blotting.
Cells (107 per ml of lysis buffer) were lysed in buffer containing 25 mM HEPES (pH 7.4), 75 mM NaCl, 10 mM NaF, 1% NP-40, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of chymostatin/ml, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 1 mM Na3VO3. Proteins were concentrated by precipitation with 1.5 volumes of acetone. Proteins from 5 × 106 cells were separated by SDS-PAGE using the following gel conditions: for E2F and pocket proteins, 7.5% acrylamide–0.2% bis, and for S6kinase, 10% acrylamide–0.16% bis. Proteins were transferred to polyvinylidene difluoride membranes and detected by Western blot analysis with antibodies as indicated in the figures by using the ECL detection system (Amersham).
Dual staining for BrdU and myc-tagged p70s6k.
Quiesced Kit225 cells were transfected and stimulated with IL-2. They were then labelled overnight with bromodeoxyuridine (BrdU). Following labelling they were harvested, fixed in 1% paraformaldehyde, and treated with 2 M HCl–0.5% Triton X-100. myc-tagged p70s6k was detected with a biotinylated 9E10 antibody (1.7 μg/ml) and revealed with avidin-conjugated Tricolour (Caltag). BrdU was detected with directly conjugated fluorescein isothiocyanate-labelled antibody (Boehringer Mannheim). Samples were analyzed with a Becton Dickinson fluorescence-activated cell sorter (FACS).
RESULTS
IL-2 and PI 3-kinase control E2F activity by rapamycin-sensitive pathways.
IL-2 regulates the transcriptional activity of E2Fs by PI 3-kinase-mediated signals (5). To monitor E2F transcriptional activity in T cells, we used E2ACAT, a reporter plasmid containing two E2F binding sites upstream of the CAT gene (26, 31, 33). The IL-2-dependent T-cell line Kit225 was used in these transfection experiments because these cells are dependent on IL-2 for mitosis but not for survival. Kit225 cells are like normal peripheral blood-derived T lymphoblasts in that they proliferate in medium supplemented with serum and IL-2 (40). In the absence of serum, the cells apoptose, whereas in the absence of IL-2, they arrest in G1. Importantly, after IL-2 deprivation and G1 arrest, Kit225 cells remain IL-2 responsive for cell growth following transient transfection. The selectivity and sensitivity of E2ACAT for detecting transcriptionally active E2Fs in IL-2-activated Kit225 cells have been described previously (5).
Kit225 cells were deprived of IL-2 for 24 h, transfected with E2ACAT, and stimulated with IL-2. E2F activity is low in IL-2-deprived Kit225 cells, but it can be induced by IL-2 (Fig. 1A). These data also show that expression of CD2p110, a membrane-targeted catalytic subunit of PI 3-kinase that constitutively induces accumulation of PI(3,4)P2 and PI(3,4,5)P3 in vivo, activates E2F transcriptional activity in the absence of IL-2 (Fig. 1A). An E2CAT construct containing the −85-to-+40 portion of the E2A promoter with mutations in the E2F binding sites [E2CAT(E2F−)] was not induced by either IL-2 or PI 3-kinase (Fig. 1A). Thus, IL-2 and PI 3-kinase activation of the E2ACAT reporter is dependent on the integrity of E2F binding sites.
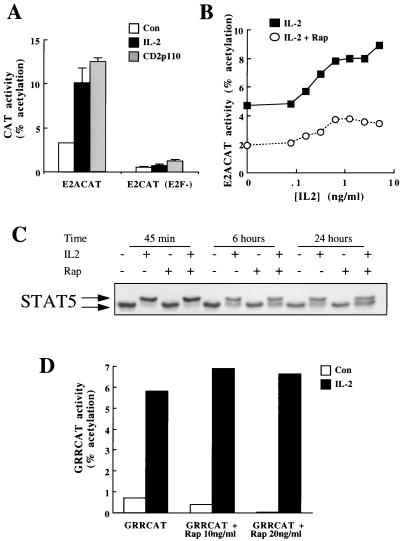
FIG. 1.
Rapamycin inhibits IL-2 activation of E2ACAT. (A) Quiescent Kit225 cells (1.5 × 107 per sample) were cotransfected with E2ACAT or the mutated reporter [E2CAT(E2F−)] and either empty vector or CD2p110 (active PI 3-kinase) (20 μg). After 2 h, cells transfected with empty vector were stimulated with IL-2 (20 ng/ml). Open bars, control (Con); solid bars, IL-2; shaded bars, CD2p110. After 22 h, samples were harvested and lysed, and CAT activity was measured. (B) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with E2ACAT (20 μg). Cells were treated with 20 ng of rapamycin/ml (open circles) or left untreated (solid squares) for 20 min prior to stimulation with various doses of IL-2 as indicated. After 18 h, samples were harvested and assayed for CAT activity. (C) Kit225 cells (2 × 106 per ml; 5 ml per sample) were pretreated with rapamycin (20 ng/ml) and incubated with IL-2 (20 ng/ml) as indicated for 45 min, 6 h, and 24 h. Total cell lysates were generated and resolved by SDS-PAGE, and Western blotting was performed. Protein was detected by using antibodies specific for STAT5. (D) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with GRRCAT (20 μg). Cells were pretreated with rapamycin (20 ng/ml) for 20 min prior to stimulation with IL-2 (20 ng/ml) as indicated. After 18 h, samples were harvested and assayed for CAT activity.
In T cells, PI 3-kinase initiates responses controlled by the rapamycin target, mTOR. To determine whether rapamycin-controlled signaling pathways impinge on IL-2 and PI 3-kinase regulation of E2Fs, we investigated the effects of rapamycin on IL-2 and PI 3-kinase E2ACAT responses. Kit225 cells were deprived of IL-2 for 24 h, transfected with E2ACAT, allowed to recover for 2 h, and pretreated for 20 min with rapamycin prior to IL-2 addition. After 18 h, cells were lysed and E2ACAT activity was measured. IL-2 induction of E2ACAT shows a maximal response at 5 to 10 ng/ml, with a half-maximal response at 0.2 ng/ml (Fig. 1B). These levels of IL-2 correspond to those that occupy the high-affinity IL-2 receptor and induce proliferation. The data in Fig. 1B show the effects of rapamycin on an IL-2 dose response for E2F activation. These results show that rapamycin treatment suppresses IL-2 activation of E2ACAT.
To investigate the specificity of rapamycin inhibition of E2ACAT, we examined the effects of this drug on IL-2-induced phosphorylation and transcriptional activation of STAT5. IL-2 induces rapid and sustained tyrosine and serine phosphorylation of STAT5B, which results in nuclear translocation, DNA binding, and activation of this transcription factor (2). Quiescent Kit225 cells were pretreated with rapamycin for 20 min and stimulated with IL-2 for the times indicated. STAT5B hyperphosphorylation was monitored by examining its electrophoretic mobility in SDS-PAGE, with the upper band corresponding with phosphorylated STAT5B. The results in Fig. 1C show that 20 ng of rapamycin/ml did not prevent IL-2 induction of STAT5 hyperphosphorylation. STAT5 transcriptional activity was assessed by using GRRCAT, a reporter gene with five STAT5 binding sites upstream of the CAT gene. Kit225 cells were IL-2 deprived for 24 h, transfected with GRRCAT, and pretreated with rapamycin prior to stimulation with IL-2. Rapamycin does not affect STAT5 transcriptional activity in IL-2-activated cells (Fig. 1D) and is thus not a general inhibitor of IL-2-activated transcription.
Rapamycin did not completely abrogate E2F activation by IL-2 (Fig. 1B). From nine experiments, rapamycin inhibition of IL-2-induced E2ACAT activity was consistently 50%. Hence, rapamycin signaling pathways are clearly involved in E2F regulation, but there is also a rapamycin-resistant component of E2F responses to IL-2. A rapamycin dose-response curve confirms this observation (Fig. 2A). Figure 2A shows that doses from 5 to 20 ng of rapamycin/ml cause a reduction of approximately 50% in the E2F response. The data also show that rapamycin inhibited the E2ACAT activity induced in cells expressing constitutively active PI 3-kinase. Like IL-2-induced E2ACAT activity, PI 3-kinase-induced E2ACAT activity is inhibited only partially, approximately 50%, by rapamycin (Fig. 2A).
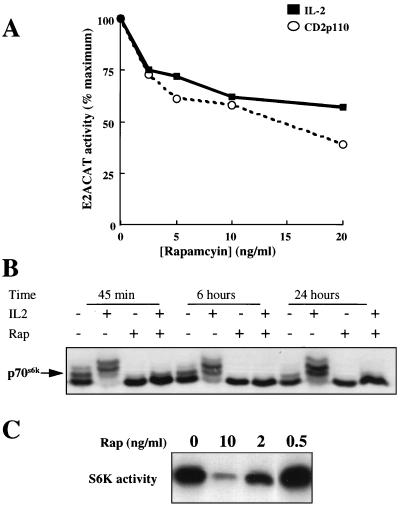
FIG. 2.
Rapamycin effects on E2ACAT and p70s6k. (A) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with E2ACAT (20 μg) and with either 20 μg of empty vector (solid squares) or pEF rCD2p110 (active PI 3-kinase) (open circles). Cells were left for 4 h. They were pretreated with rapamycin (20 ng/ml), and the cells transfected with empty vector were stimulated with IL-2 (20 ng/ml). After 18 h, samples were harvested and assayed for CAT activity. Data are expressed as percentages of maximum activity, with 100% representing 15% ± 3% acetylation for IL-2 and 24% ± 4% acetylation for cells cotransfected with CD2p110. (B) Kit225 cells (106 per ml; 5 ml per sample) were pretreated with rapamycin (20 ng/ml) and incubated with IL-2 (20 ng/ml) as indicated for 45 min, 6 h, and 24 h. Total cell lysates were generated and resolved by SDS-PAGE, and Western blotting was performed. Protein was detected by using antibodies specific for S6 kinase. (C) Kit225 cells (1.5 × 107 per sample) were transfected with an expression vector for myc epitope-tagged p70s6k. Cells were left overnight and treated for 20 min with rapamycin at the doses indicated. Cells were lysed, the expressed p70s6k was immunoprecipitated with myc tag-specific antibody, and kinase activity was measured. S6 substrate phosphorylation for S6 kinase assays was analyzed by autoradiography.
IL-2 activates p70s6k by a rapamycin-dependent pathway (34). To confirm the effectiveness of rapamycin in the E2F activity experiments, a series of parallel experiments was performed to examine the ability of the drug to regulate p70s6k activity in Kit225 cells. The activity of endogenous p70s6k is regulated by multiple phosphorylation events that can be monitored by the reduced electrophoretic mobility of this enzyme in SDS-PAGE gels. The data in Fig. 2B show that in quiescent Kit225 cells, p70s6k migrates predominantly as a doublet, whereas in IL-2-activated cells, four discrete phosphoforms of the enzyme can be readily discerned. In rapamycin-treated cells, p70s6k migrates as a single band corresponding to the hypophosphorylated forms of the enzyme. The data in Fig. 2B show that the inhibitory effects of the macrolide on p70s6k activity are sustained at 6 and 24 h following IL-2 stimulation. Hence rapamycin was effective at blocking p70s6k signaling pathways throughout the duration of the E2F functional activity assays. Rapamycin inhibition of p70s6k phosphorylation correlates with an inhibition of p70s6k activity, as judged from in vitro kinase assays using 40S ribosomal protein S6 as a substrate. These results show that the rapamycin-resistant component of E2F responses to IL-2 is not due to ineffectiveness of the drug and that this pathway must bifurcate above p70s6k.
Rapamycin regulates E2F activity but not the expression or DNA binding of E2F proteins.
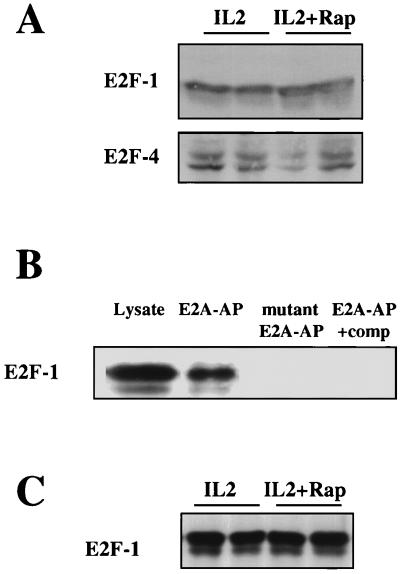
One mechanism by which rapamycin could block E2F activity is to downregulate cellular levels of E2Fs. However, Western blot analyses of total lysates from Kit225 cells treated with IL-2 or IL-2 plus rapamycin showed no effect of rapamycin on the expression of E2F-1 and E2F-4 (Fig. 3A), the main E2Fs found in T cells (30). To measure the effects of rapamycin on E2F DNA binding, biotinylated oligonucleotides corresponding to the E2F binding sequence in E2ACAT were used to affinity purify E2F complexes from cells activated with IL-2 in the presence or absence of rapamycin. E2F-1 Western blot analysis showed that E2A oligonucleotides can effectively affinity purify E2Fs from cell lysates (Fig. 3B); E2F-1 binding was lost when an oligonucleotide with two point mutations in each of the DNA binding sites in the E2A oligonucleotide was used in the affinity purifications or when cell lysates were preincubated with unbiotinylated competitor oligonucleotide. Figure 3C shows that pretreatment of Kit225 cells with rapamycin prior to stimulation with IL-2 for 20 h had no effect on E2F DNA binding. This suggests that rapamycin inhibits the transcriptional activity of DNA-bound E2F complexes, not their expression or DNA binding.
FIG. 3.
Rapamycin and E2F. (A) Kit225 cells (106 per ml; 5 ml per sample) were pretreated with rapamycin (Rap) (20 ng/ml) and incubated with IL-2 (20 ng/ml) as indicated for 20 h. Total cell lysates were generated and resolved by SDS-PAGE, and Western blotting was performed. Protein was detected by using specific antibodies for E2F-1 and E2F-4. (B) Kit225 cells (106 per ml; 10 ml per sample) were lysed, and DNA affinity precipitations were performed with biotinylated oligonucleotides containing E2A sequence (E2A-AP), with a mutant E2A biotinylated oligonucleotide (two point mutations in each E2F binding site) (mutant E2A-AP) or with biotinylated oligonucleotides containing E2A sequence in the presence of a 10-fold excess of unbiotinylated E2F binding oligonucleotide as a competitor (E2A-AP+comp). Samples were analyzed by SDS-PAGE and Western blotting. Protein was detected with E2F-1-specific antibodies. (C) Kit225 cells (106 per ml; 10 ml per sample) were pretreated with rapamycin (20 ng/ml) and stimulated with IL-2 (20 ng/ml) for 20 h. Samples were lysed, and DNA affinity precipitations were performed with biotinylated oligonucleotides containing E2A sequence. Samples were analyzed by SDS-PAGE and Western blotting. Protein was detected with E2F-1-specific antibodies.
Rapamycin inhibits pocket protein phosphorylation.
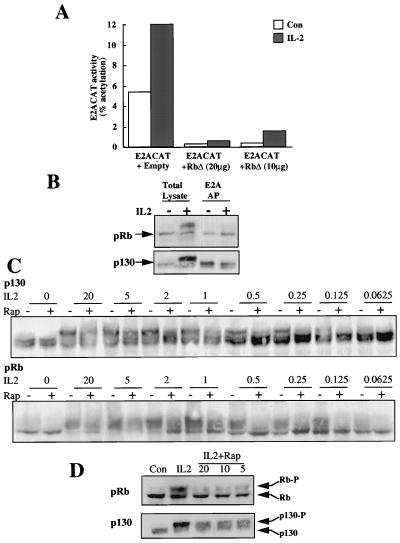
One key mechanism to control E2F transcriptional activity is the formation of E2F pocket protein complexes, a process controlled by pocket protein phosphorylation. When Kit225 cells are transfected with E2ACAT and an Rb construct (pRbΔ) in which the p34cdc2 phosphorylation sites are mutated (13), E2F transcriptional activity is lost (Fig. 4A). In quiescent T lymphocytes, hypophosphorylated forms of the pocket binding proteins Rb and p130 predominate. The phosphorylation of pocket proteins was monitored by electrophoretic mobility in SDS-PAGE gels. In these biochemical analyses, we studied IL-2 responses in Kit225 cells and also in human peripheral blood-derived T lymphocytes. The data from peripheral blood lymphocytes are shown because of the physiological relevance of these cells as the target for the in vivo actions of rapamycin; the same result was observed in Kit225 cells. Figure 4B demonstrates that hyperphosphorylation of Rb and p130 is induced by IL-2, as judged by their reduced electrophoretic mobilities following IL-2 treatment (Fig. 4B). Furthermore, the data in Fig. 4B show that E2F-DNA protein complexes contain only the fast-migrating, hypophosphorylated forms of the pocket proteins p130 and pRb. The slower electrophoretic mobility forms of pRb and p130, corresponding to the hyperphosphorylated pocket proteins, are unable to bind to the E2F complexes, although they are readily detectable in total cell lysates.
FIG. 4.
Rapamycin and pocket proteins. (A) Quiescent Kit225 cells (1.5 × 107 per sample) were cotransfected with E2ACAT reporter with a mammalian expression vector for a form of pRb in which the cdc2 phosphorylation sites had been mutated (pRbΔ) or with empty vector as a control. After 4 h, the cells were left untreated (Con) or stimulated with IL-2 (20 ng/ml). Eighteen hours later, the cells were harvested, lysed, and assayed for CAT. (B) Peripheral blood lymphocytes (106 per ml; 10 ml per sample) were incubated with IL-2 (20 ng/ml) as indicated for 20 h. Samples were lysed, and DNA affinity precipitations (AP) were performed with biotinylated oligonucleotides containing E2A sequence. Samples were analyzed by SDS-PAGE and Western blotting. Protein was detected with specific pRb and p130 antibodies. (C) Peripheral blood lymphocytes (106 per ml; 5 ml per sample) were pretreated with rapamycin (20 ng/ml) and stimulated with IL-2 at the doses indicated (in nanograms per milliliter) for 20 h. Total cell lysates were generated and resolved by SDS-PAGE, and Western blotting was performed. Protein was detected by using specific antibodies for pRb and p130. (D) Peripheral blood lymphocytes (106 per ml; 5 ml per sample) were pretreated with various doses of rapamycin as indicated (in nanograms per milliliter) and stimulated with IL-2 (20 ng/ml) for 20 h. Total cell lysates were generated and resolved by SDS-PAGE, and Western blotting was performed. Protein was detected with specific antibodies for pRb and p130. Con, control.
To explore whether the inhibitory effects of rapamycin on E2F transcriptional activity could be explained by effects of rapamycin on pocket protein phosphorylation, we analyzed the phosphorylation of pRb and p130 in rapamycin-treated T cells. The data shown in Fig. 4C are for peripheral blood-derived T lymphocytes, but indistinguishable results were seen in Kit225 cells. In the experiment shown in Fig. 4C, cells were activated with a range of concentrations of IL-2. For each dose of IL-2, the effect of 20 ng of rapamycin/ml on the induction of pocket protein phosphorylation was monitored. The data show that IL-2 induces the hyperphosphorylation of pRb and p130 at cytokine concentrations coinciding with those that induce E2F transcriptional activity and cell cycle progression. There is inhibition of the hyperphosphorylation of both Rb and p130 in the presence of rapamycin. This inhibition is suppressed at high concentrations of IL-2. It is most marked in cells activated by low concentrations of cytokine that give suboptimal induction of pocket protein phosphorylation. The data in Fig. 4D show a rapamycin dose response for the inhibition of Rb and p130 hyperphosphorylation. The effects of rapamycin are seen in cells treated with 5 ng of rapamycin/ml, and increasing the drug dose to 20 ng/ml has no further inhibitory effect on pocket protein phosphorylation. These inhibitory effects of rapamycin on the phosphorylation of pocket proteins could explain why this drug can antagonize E2F transcriptional activity. The failure of rapamycin to totally suppress IL-2-induced Rb and p130 phosphorylations is consistent with the failure of rapamycin to totally abrogate E2F transcriptional activity. These results are also fully consistent with the fact that rapamycin blocks T-cell proliferation by delaying G1 transit times rather than by causing an absolute mitotic block (43).
p70s6k regulates E2Fs in Kit225 cells.
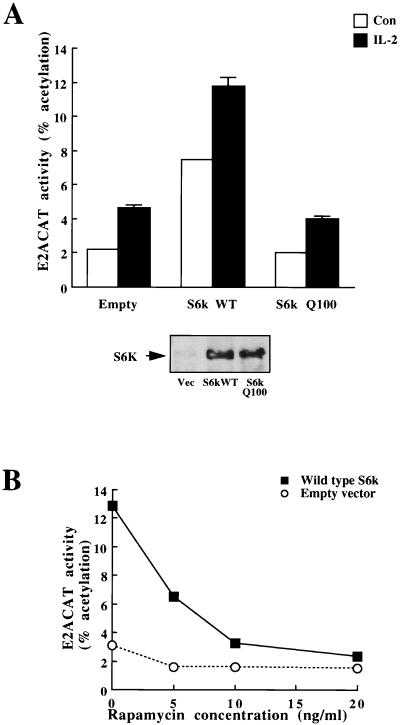
IL-2 activates p70s6k in a PI 3-kinase- and rapamycin-dependent pathway (34). To explore the role of p70s6k in E2F regulation in Kit225 cells, we examined the effects of overexpression of p70s6k on IL-2 activation of E2ACAT. The results show that overexpression of a myc epitope-tagged wild-type p70s6k raised basal levels of E2ACAT and augmented the effects of IL-2 (Fig. 5A). This response required p70s6k catalytic activity, as it was not seen in cells expressing comparable levels of p70s6kQ100, a catalytically inactive variant in which the critical lysine residue in the ATP binding site of the kinase is mutated (Fig. 5A). The effect of expression of wild-type p70s6k on the upregulation of E2ACAT activity was abrogated by rapamycin (Fig. 5B), whereas IL-2–E2F responses were only 50% inhibited (Fig. 2A), consistent with the hypothesis that rapamycin-resistant pathways from IL-2 and PI 3-kinase bifurcate above p70s6k.
FIG. 5.
p70s6k increases E2F transcriptional activity. (A) (Top) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with E2ACAT (20 μg) and with 10 μg each of either empty vector, wild-type p70s6k (S6k WT), or p70s6kQ100 (Q100) (kinase-dead S6k). Cells were left for 4 h and were either stimulated with 20 ng of IL-2/ml (solid bars) or left unstimulated (open bars) (Con). After 18 h, samples were harvested and assayed for CAT activity. (Bottom) In parallel experiments, Kit225 cells (1.5 × 107 per sample) were transfected with an expression vector for myc epitope-tagged S6k constructs. Cells were left overnight and were lysed, and the expressed S6k was immunoprecipitated with myc tag-specific antibody and separated by SDS-PAGE. Transfected myc-tagged S6k was detected with myc tag-specific antibody (9E10). (B) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with E2ACAT (20 μg) and with 20 μg of wild-type p70s6k or empty vector. Cells were left for 4 h. They were then treated with various doses of rapamycin. After 18 h, samples were harvested and assayed for CAT activity.
The rapamycin-resistant p70s6k mutant, p70s6kD3E-E389, regulates E2F activity in Kit225 cells.
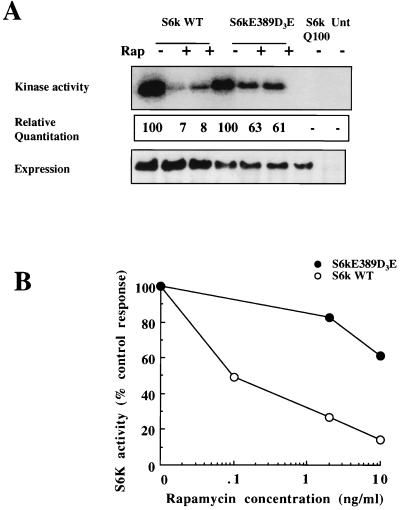
p70s6kD3E-E389 is a mutant in which threonine 389, the main target of rapamycin-induced p70s6k inactivation, has been replaced by a glutamic acid and the four serine or threonine phosphorylation sites in the autoinhibitory domain have been changed to either aspartic or glutamic acid (19). This mutant shows rapamycin-insensitive catalytic activity in kidney 293 cells (19, 46). It retains 20% of serum-stimulated activity compared to the 2% residual S6 kinase activity seen in wild-type p70s6k when cells are treated with 20 nM rapamycin (equivalent to 18.3 ng/ml) (19). Similarly, p70s6kD3E-E389 retains 50% of its catalytic activity in rapamycin- and insulin-treated 293 cells compared to the 5% residual activity present in wild-type enzyme isolated from rapamycin- and insulin-treated cells (46). The rapamycin sensitivity of p70s6kD3E-E389 catalytic activity in T cells has not been determined. Figure 6A shows the kinase activity, relative quantitation, and expression in IL-2-activated Kit225 cells of wild-type p70s6k, p70s6kD3E-E389, and p70s6kQ100. Robust S6 kinase activity was detected in immunoprecipitates of both wild-type p70s6k and p70s6kD3E-E389, whereas p70s6kQ100 was catalytically inactive. When cells were treated with 10 or 20 ng of rapamycin/ml for 20 min, p70s6kD3E-E389 retained more than 60% catalytic activity in rapamycin-treated cells, compared to the 5 to 10% activity of the wild-type enzyme under similar conditions. The clear resistance to rapamycin inhibition of p70s6kD3E-E389 is also shown by a comparison of the rapamycin dose response for inhibition of p70s6kD3E-E389 compared to that of the wild-type kinase (Fig. 6B).
FIG. 6.
p70s6kD3E-E389 is rapamycin resistant in T-cells. (A) Kit225 cells (1.5 × 107 per sample) were transfected with 30 μg each of empty vector, wild-type p70s6k (S6k WT), the rapamycin-resistant mutant p70s6kD3E-E389, or the kinase-dead mutant p70s6kQ100. Cells were left overnight and treated for 20 min with rapamycin (20 ng/ml). Cells were lysed, the expressed S6k was immunoprecipitated with myc tag-specific antibody, and kinase activity was measured. S6 substrate phosphorylation for S6 kinase assays was analyzed by autoradiography (top), and expression was measured by Western blot analysis (bottom). (Center) Incorporated radioactivity was measured with a PhosphorImager. Data are expressed relative to activity in the absence of rapamycin, taken as 100. (B) Kit225 cells (1.5 × 107 per sample) were transfected with 30 μg each of empty vector, wild-type p70s6k, p70s6kD3E-E389, or p70s6kQ100. Cells were left overnight and treated for 20 min with rapamycin at the doses indicated. Cells were lysed, the expressed S6k was immunoprecipitated with myc tag-specific antibody, and kinase activity was measured. S6 substrate-incorporated radioactivity was measured with a PhosphorImager. Data are expressed relative to activity in the absence of rapamycin, taken as 100%.
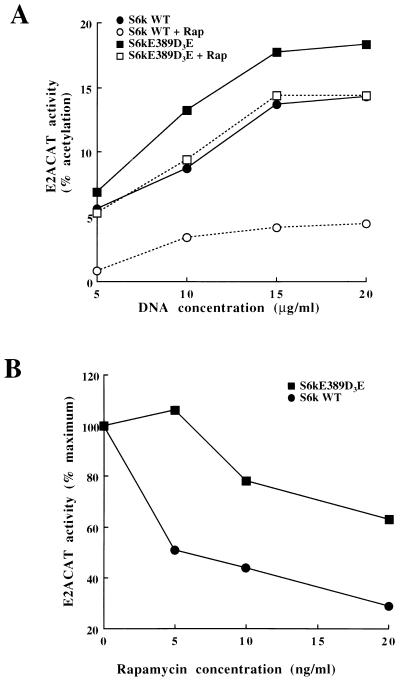
If p70s6k plays a role in the rapamycin-sensitive responses that regulate E2Fs, then the expression of a rapamycin-resistant mutant of p70s6k should rescue E2F transcriptional activity in rapamycin-treated cells. Figure 7A shows that expression of the rapamycin-resistant S6 kinase mutant (p70s6kD3E-E389) raises basal E2ACAT activity. The activation of E2ACAT stimulated by expression of the rapamycin-resistant mutant of p70s6k was strikingly less sensitive to rapamycin treatment than that for the wild-type p70s6k (Fig. 7A). It is clear that the E2F response to p70s6kD3E-E389 still retains some sensitivity to rapamycin treatment, but this is expected given that this mutant is not completely rapamycin resistant. The expression of p70s6kD3E-E389 also potentiated IL-2 induction of E2F (data not shown) and conferred a significant degree of rapamycin resistance to the E2F regulation by IL-2 (Fig. 7B). In data averaged from seven experiments, rapamycin caused approximately 55 to 60% inhibition of the E2F responses to IL-2 in cells expressing wild-type p70s6k, whereas in cells expressing the p70s6kD3E-E389 mutant, the inhibition was only 20 to 25%. Expression of p70s6kD3E-E389 can thus rescue E2F responses to IL-2 in rapamycin-treated cells. It does not fully rescue the E2F response, but this is explained by the partial rapamycin sensitivity of the p70s6kD3E-E389 catalytic function. Collectively, these experiments with p70s6k mutants indicate that p70s6k is the intermediate of the rapamycin-sensitive pathway from IL-2 and PI 3-kinase to E2Fs.
FIG. 7.
p70s6kD3E-E389 rescues rapamycin inhibition of E2F transcriptional activity. (A) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with E2ACAT (20 μg) and with wild-type (WT) p70s6k or the rapamycin-resistant mutant (p70s6kD3E-E389) at various concentrations. Cells were left for 4 h and then treated with rapamycin (Rap) (20 ng/ml). After 18 h, samples were harvested and assayed for CAT activity. (B) Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with E2ACAT (20 μg) and with wild-type (WT) p70s6k or the rapamycin-resistant mutant (p70s6kD3E-E389). Cells were left for 4 h. They were then pretreated for 20 min with various doses of rapamycin and were stimulated with IL-2 (20 ng/ml). After 18 h, samples were harvested and assayed for CAT activity.
The rapamycin-resistant p70s6k mutant, p70s6kD3E-E389, does not rescue rapamycin inhibition of cell cycle entry.
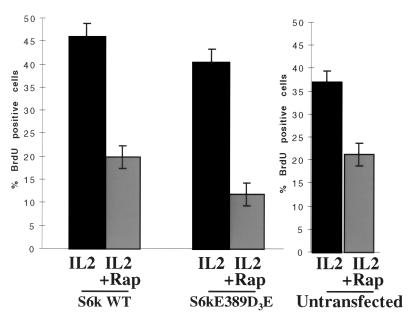
A rapamycin-resistant p70s6k was sufficient to rescue E2F transcriptional activity in rapamycin-treated T cells. However, even though the transcriptional activation of E2Fs is required, it is not sufficient to drive T-cell cycle progression in T cells (5). In addition, it is clear that mTOR has other downstream effectors distinct from p70s6k (10). We therefore investigated whether expression of a rapamycin-resistant p70s6k was sufficient to restore T-cell cycle progression in the presence of rapamycin or whether other rapamycin-sensitive targets may be involved in the IL-2-mediated response. To perform this experiment, we transfected cells with myc epitope-tagged wild-type p70s6k and the rapamycin-resistant mutant p70s6kD3E-E389. We then labelled the cells with BrdU and used antibodies to detect p70s6k expression and BrdU incorporation. The results show BrdU incorporation in Kit225 cells expressing either wild-type p70s6k or the rapamycin-resistant mutant p70s6kD3E-E389 (Fig. 8). In the presence of wild-type p70s6k, rapamycin had a dramatic effect on DNA incorporation, lowering the percentage of cells entering the cell cycle from about 40% to less than 20% of cells. In cells expressing p70s6kD3E-E389, DNA synthesis was inhibited to an equivalent extent, consistent with the observation that mTOR has other downstream effector targets involved in T-cell cycle progression in addition to p70s6k.
FIG. 8.
p70s6kD3E-E389 does not rescue rapamycin inhibition of T-cell cycle entry. Quiescent Kit225 cells (1.5 × 107 per sample) were transfected with wild-type (WT) p70s6k or p70s6kD3E-E389. Cells were left overnight. They were then pretreated for 20 min with rapamycin (Rap) (20 ng/ml) and stimulated with IL-2 (20 ng/ml). After 8 h, 10 μM BrdU was added to samples. Twelve hours later, cells were harvested and assayed for BrdU incorporation and protein expression by FACS.
DISCUSSION
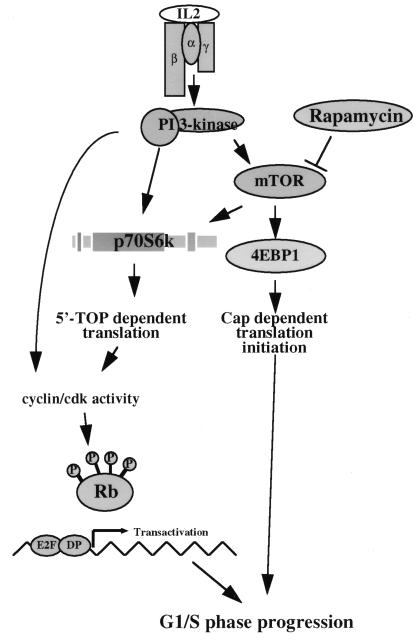
In T lymphocytes, the cytokine IL-2 controls the regulation of the transcriptional activity of E2Fs, a critical cell cycle event. IL-2 activation of E2Fs is controlled by PI 3-kinase (5), and one objective of the present study was to explore the PI 3-kinase signaling pathways involved in E2F regulation in T cells. We have shown previously that PI 3-kinase regulates the activity of the ribosomal S6 kinase, p70s6k, by a rapamycin-sensitive mechanism (34). The present results show that IL-2 and PI 3-kinase regulation of E2Fs is sensitive to inhibition with rapamycin. Rapamycin inhibits the catalytic activity of p70s6k, and the inhibitory effects of rapamycin on E2F transcriptional activity can be rescued by expression of a rapamycin-resistant S6kinase. However, a rapamycin-resistant p70s6k mutant does not rescue rapamycin inhibition of T-cell cycle entry. These results demonstrate that IL-2–PI 3-kinase and rapamycin pathways interact centrally at p70s6k, leading to E2F transcriptional activity, but that other pathways are also involved in cell cycle entry. We have illustrated these concepts schematically in Fig. 9.
FIG. 9.
IL-2 signaling to E2F and the cell cycle. Role of p70s6k in E2F transactivation in T cells. Our model places p70s6k upstream of translation events leading to E2F transcriptional activity but demonstrates that other rapamycin-sensitive signals are required for cell cycle progression.
Rapamycin is an immunosuppressant because it prolongs G1 transit times in T lymphocytes, thereby inhibiting T-cell clonal expansion. E2F transcriptional activity is a rate-limiting step for mitosis (15), and any influence of rapamycin on this process would have an impact on T-cell cycle G1 progression and would explain the ability of this drug to markedly prolong G1 transit time in T cells (43). The present results thus provide valuable insights about the cell cycle targets for the action of rapamycin in T cells. The ability of rapamycin to suppress E2F activity in T cells correlates with the ability of rapamycin to inhibit the accumulation of phosphorylated pocket proteins in T cells. Several studies have examined the effects of rapamycin on pocket protein phosphorylation in nonlymphoid cells, but the results have been discrepant (9, 14). There have been no previous reports about the effects of rapamycin on IL-2-regulated pocket protein phosphorylation in T cells, even though these cells are the pharmacologically relevant target for rapamycin. Herein we show that rapamycin inhibits IL-2-induced hyperphosphorylation of pRb and p130 in Kit225 and normal human peripheral blood-derived T cells. Rapamycin does not totally ablate the generation of hyperphosphorylated Rb and p130 in T cells, but the present results show that there is a significant accumulation of hypophosphorylated pocket proteins in rapamycin-treated T cells that would translate into an inhibition of E2F activity. The effect of rapamycin is most striking at limiting concentrations of cytokine, as seen in the IL-2 dose response for the induction of pRb and p130 phosphorylation. The fact that rapamycin regulates threshold responses rather than totally ablating them might explain the discrepancies in the literature regarding the effects of rapamycin on pocket protein phosphorylation. However, it is equally possible that rapamycin has different modes of action in controlling cell cycle progression in different cell types.
Rapamycin controls at least two distinct pathways that bifurcate at the level of mTOR: the activation of p70s6k and the regulation of the phosphorylation of 4EBP1 (44). To test the involvement of p70s6k in E2F regulation, we looked at the effects of expressing a rapamycin-resistant mutant of p70s6k on E2F responses to IL-2. The rationale for these experiments is that expression of a rapamycin-resistant mutant of p70s6k should rescue E2F transcriptional activity in rapamycin-treated cells if p70s6k plays a role in the rapamycin-sensitive responses that regulate E2Fs. Our data show that overexpression of wild-type p70s6k raises basal levels of E2F activity and potentiates E2F responses to IL-2. Moreover, expression of p70s6kD3E-E389, a rapamycin-resistant mutant of p70s6k, confers rapamycin resistance on the E2F response to IL-2. These results demonstrate that p70s6k is an important cellular target for the action of rapamycin in T cells and reveal that p70s6k plays a critical role in regulation of T-cell cycle progression, acting to link PI 3-kinase and the cell cycle machinery. The molecular basis for the effects of p70s6k on E2F are likely to be indirect, because p70s6k is thought to have a single cellular substrate, ribosomal protein S6. By controlling S6 phosphorylation, p70s6k regulates the translation of an essential subset of mRNAs that contain an oligopyrimidine tract at their transcriptional start site (19). These RNAs must include molecules that can regulate E2Fs. In preliminary experiments we observed that rapamycin prevents IL-2 induction of cyclins D2 and D3 (unpublished observation). These D-type cyclins form complexes with the kinases cdk4 or cdk6, which are responsible for the phosphorylation of pocket proteins. Accordingly, one prediction is that p70s6k controls E2F activity via regulation of D-type cyclins.
The present study has focused on characterization of the rapamycin-sensitive molecules involved in IL-2 and PI 3-kinase activation of E2F because this is an important control point for T-cell cycle progression. p70s6k mediates rapamycin effects on E2Fs, but additional rapamycin-controlled pathways are clearly involved in cell cycle entry. These could be mediated by the other well-characterized target for rapamycin, 4EBP1, which, when phosphorylated by mTOR (7), is released from eIF4E, allowing formation of the productive mRNA cap binding complex critical for mRNA translation. There is no discrepancy between the ability of a rapamycin-resistant p70s6k mutant to rescue rapamycin-suppressed E2F activity and its inability to restore T-cell cycle entry in rapamycin-treated cells because the activation of E2Fs is not sufficient for T-cell cycle progression (5). Cell cycle control in mammalian cells is a complicated process involving integration of a network of rapamycin-sensitive and -insensitive signaling pathways. There are multiple, diverse rapamycin-insensitive pathways controlling T-cell cycle progression. The present results reveal that there is similar complexity to the rapamycin-controlled events important for T-cell proliferation.
In summary, the present study brings together the biochemical processes linking the IL-2 receptor to the cell cycle with the study of the mechanisms of action of rapamycin, an important immunosuppressant. Rapamycin inhibits T-cell proliferation, and it is accordingly important to understand the biochemical events required for its mode of action in T lymphocytes. We show that targets for the action of rapamycin in T cells are signaling pathways that regulate E2F transcriptional activity. Rapamycin completely ablates the catalytic activity of p70s6k, and the inhibitory effects of rapamycin on E2F transcriptional activity can be rescued by a rapamycin-resistant p70s6k mutant, suggesting that IL-2–PI 3-kinase and rapamycin pathways interact centrally at p70s6k, leading to E2F transcriptional activity. The effects of rapamycin on E2F activity reveal a molecular mechanism for the immunosuppressive effects of rapamycin. They also reveal a role for p70s6k as a mediator of IL-2 and PI 3-kinase activation of E2F, a key event in the G1 phase of the cell cycle.
ACKNOWLEDGMENTS
This work was supported by Human Frontiers Science Program (grant RG 445/95), EC HCM Network CHRX-CT94-0537, and the Imperial Cancer Research Fund.
REFERENCES
- 1.Beadling C, Guschin D, Witthuhn B A, Ziemiecki A, Ihle J N, Kerr I M, Cantrell D A. Activation of JAK kinases and STAT proteins by interleukin-2 and interferon-α, but not the T cell antigen receptor, in human T lymphocytes. EMBO J. 1994;13:5605–5615. doi: 10.1002/j.1460-2075.1994.tb06898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beadling C, Ng J, Babbage J W, Cantrell D A. Interleukin-2 activation of STAT5 requires the convergent action of tyrosine kinases and a serine/threonine kinase distinct from the Raf-1/Erk2 MAP kinase pathway. EMBO J. 1996;15:1902–1913. [PMC free article] [PubMed] [Google Scholar]
- 3.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 4.Botz J, Zerfass-Thome K, Spitzovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Durr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan P, Babbage J W, Burgering B M T, Groner B, Reif K, Cantrell D A. Phosphatidylinositol 3-kinase controls E2F transcriptional activity in response to interleukin-2. Immunity. 1997;7:679–689. doi: 10.1016/s1074-7613(00)80388-x. [DOI] [PubMed] [Google Scholar]
- 6.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 7.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C J, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 8.Cantrell D A, Smith K A. The interleukin-2 T-cell system: a new cell growth model. Science. 1984;224:1312–1316. doi: 10.1126/science.6427923. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Knudsen E S, Wang J Y. The RB/p107/p130 phosphorylation pathway is not inhibited in rapamycin-induced G1-prolongation of NIH3T3 cells. Oncogene. 1996;13:1765–1771. [PubMed] [Google Scholar]
- 10.Dennis P B, Fumagalli S, Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 11.Fairhurst R M, Daeipour M, Amaral M C, Nel A E. Activation of mitogen-activated protein kinase/ERK-2 in phytohaemagglutinin in blasts by recombinant interleukin-2: contrasting features with CD3 activation. Immunology. 1993;79:112–118. [PMC free article] [PubMed] [Google Scholar]
- 12.Genot E M, Parker P J, Cantrell D A. Analysis of the role of the protein kinase Cα, ε, and ζ in T cell activation. J Biol Chem. 1995;270:9833–9839. doi: 10.1074/jbc.270.17.9833. [DOI] [PubMed] [Google Scholar]
- 13.Hamel P A, Gill R M, Phillips R A, Gallie B L. Regions controlling hyperphosphorylation and conformation of the retinoblastoma gene product are independent of domains required for transcriptional repression. Oncogene. 1992;7:693–701. [PubMed] [Google Scholar]
- 14.Hashemolhosseini S, Nagamine Y, Morley S J, Desrivières S, Mercep L, Ferrari S. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J Biol Chem. 1998;273:14424–14429. doi: 10.1074/jbc.273.23.14424. [DOI] [PubMed] [Google Scholar]
- 15.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 16.Hori T, Uchiyama T, Tsudo M, Umadome H, Ohno H, Fukuhara S, Kita K, Uchino H. Establishment of an interleukin-2-dependent human T cell line from a patient with T cell chronic lymphocytic leukemia who is not infected with human T cell leukemia/lymphoma virus. Blood. 1987;70:1069–1072. [PubMed] [Google Scholar]
- 17.Hou J, Schindler U, Henzel W J, Wong S C, McKnight S L. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity. 1995;2:321–329. doi: 10.1016/1074-7613(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 18.Izquierdo Pastor M, Reif K, Cantrell D. The regulation and function of p21ras during T-cell activation and growth. Immunol Today. 1995;16:159–164. doi: 10.1016/0167-5699(95)80134-0. [DOI] [PubMed] [Google Scholar]
- 19.Jefferies H B, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston J, Kawamura M, Kirken R, Chen Y, Blake T, Shibuya K, Ortaldo J, McVicar D, O’Shea J. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature. 1994;370:151–153. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 21.Johnston J A, Bacon C M, Finbloom D S, Rees R C, Kaplan D, Shibuya K, Ortaldo J R, Gupta S, Chen Y Q, Giri J D, O’Shea J. Tyrosine phosphorylation and activation of Stat5, Stat3, and Janus kinases by interleukin-2 and interleukin-15. Proc Natl Acad Sci USA. 1995;92:8705–8709. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karnitz L M, Burns L A, Sutor S L, Blenis J, Abraham R T. Interleukin-2 triggers a novel phosphatidylinositol 3-kinase-dependent MEK activation pathway. Mol Cell Biol. 1995;15:3049–3057. doi: 10.1128/mcb.15.6.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasame H, Papst P, Webb S, Keller G M, Johnson G L, Gelfand E W, Terada N. Targeted disruption of p70S6k defines its role in protein synthesis and rapamycin sensitivity. Proc Natl Acad Sci USA. 1998;95:5033–5038. doi: 10.1073/pnas.95.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J-X, Migone T-S, Tsang M, Friedmann M, Weatherbee J A, Zhou L, Yamauchi A, Bloom E T, Mietz J, John S, Leonard W J. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 25.Loeken M R, Brady J. The adenovirus EIIA enhancer. Analysis of regulatory sequences and changes in binding activity of ATF and EIIF following adenovirus infection. J Biol Chem. 1989;264:6572–6579. [PubMed] [Google Scholar]
- 26.Mann D J, Jones N C. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr Biol. 1996;6:474–483. doi: 10.1016/s0960-9822(02)00515-8. [DOI] [PubMed] [Google Scholar]
- 27.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- 28.Ming X-F, Burgering B M T, Wennström S, Claesson-Welsh L, Heldin C-H, Bos J L, Kozma S C, Thomas G. Activation of p70/p85 S6 kinase by a pathway independent of p21ras. Nature. 1994;371:426–429. doi: 10.1038/371426a0. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu Z-J, Oishi I, Silvennoinen O, Witthuhn B, Ihle J, Taniguchi T. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 30.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murthy S C, Bhat G P, Thimmappaya B. Adenovirus EIIA early promoter: transcriptional control elements and induction by the viral pre-early EIA gene, which appears to be sequence independent. Proc Natl Acad Sci USA. 1985;82:2230–2234. doi: 10.1073/pnas.82.8.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nourse J, Firpo E, Flanagan W M, Coats S, Polyak K, Lee M H, Massague J, Crabtree G R, Roberts J M. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature. 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 33.Ohtani K, Nevins J R. Functional properties of a Drosophila homolog of the E2F1 gene. Mol Cell Biol. 1994;14:1603–1612. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reif K, Burgering B M T, Cantrell D A. Phosphatidylinositol 3-kinase links the interleukin-2 receptor to protein kinase B and p70 S6 kinase. J Biol Chem. 1997;272:14426–14438. doi: 10.1074/jbc.272.22.14426. [DOI] [PubMed] [Google Scholar]
- 35.Reif K, Nobes C D, Thomas G, Hall A, Cantrell D A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- 36.Remillard B, Petrillo R, Maslinski W, Tsudo M, Strom T B, Cantley L, Varticovski L. Interleukin-2 receptor regulates activation of phosphatidylinositol 3-kinase. J Biol Chem. 1991;266:14167–14170. [PubMed] [Google Scholar]
- 37.Russell S, Johnston J, Noguchi M, Kawamura M, Bacon C, Friedman M, Berg M, McVicar D, Witthuhn B, Silvennoinen O, Goldman A, Schmalstieg F, Ihle J, O’Shea J, Leonard W. Interaction of IL-2R β and γ chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 38.Sabatini D M, Erdjumentbromage H, Lui M, Tempst P, Snyder S H. Raft1—a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast Tors. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 39.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y, Wang R, Sharma A, Gao C, Collins M, Penn L, Mills G B. Dissociation of cytokine signals for proliferation and apoptosis. J Immunol. 1997;159:5318–5328. [PubMed] [Google Scholar]
- 41.Shima H, Pende M, Chem Y, Fumagalli S, Thomas G, Kozma S C. Disruption of the p70S6k/p85S6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith K A, Cantrell D A. Interleukin 2 regulates its own receptors. Proc Natl Acad Sci USA. 1985;82:864–868. doi: 10.1073/pnas.82.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terada N, Takase K, Papst P, Nairn A, Gelfand E. Rapamycin inhibits ribosomal protein synthesis and induces G1 prolongation in mitogen activated T lymphocytes. J Immunol. 1995;155:3418–3426. [PubMed] [Google Scholar]
- 44.Thomas G, Hall M N. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 45.Turner B, Rapp U, App H, Greene M, Dobashi K, Reid J. Interleukin-2 induces tyrosine phosphorylation and activation of p72-74 Raf-1 kinase in a T cell line. Proc Natl Acad Sci USA. 1991;88:1227–1232. doi: 10.1073/pnas.88.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Von Manteuffel S R, Gingras A C, Ming X F, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the Frap-p70 S6 kinase pathway and is independent of mitogen-activated protein-kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witthuhn B, Silvennoinen O, Miura O, Lai K, Cwik C, Liu E, Ihle J. Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature. 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]