Abstract
What is known about this topic?
The emerging H5Ny lineages of the avian influenza virus (AIV) with genomic reassortments have posed a continuous threat to animals and human beings. Since the first case of avian influenza A (H5N6) virus infection in 2014, the World Health Organization has reported a total of 38 cases by August 6, 2021.
What is added by this report?
A total of 5 new cases of H5N6 that occurred from May 2021 to July 2021 in Sichuan Province, China were reported in this study. Epidemiological and laboratory information of the five cases were investigated. The genomic analysis of the H5N6 genomes showed the features of AIV genomic reassortments and key residue substitutions.
What are the implications for public health practice?
The emergence of human cases infected by AIV H5Ny lineages through time demonstrates a risk of the persistence and evolution of the virus to trigger sporadic outbreaks and even pandemics, which need continuous surveillance.
Keywords: H5N6, AIV, influenza
As a highly contagious virus, the avian influenza virus (AIV) circulates among birds as its reservoir host (1). The spillover to human beings and other animal species occurs frequently, leading to cross-species infection that may trigger mild outbreaks and even pandemics (2). In recent years, AIV H5Ny lineages (i.e., H5N1, H5N2, H5N6, and H5N8) have proved the capacity for zoonotic spread and genomic reassortments amongst this viral group and thus pose a severe threat to animals and human beings (3–4). Among these AIVs, H5N6 was first detected in 1975 (5), and the first reported case of human infection with a novel H5N6 was dated in 2014 (6–7). By August 2021, the World Health Organization (WHO) reported a total of 38 laboratory-confirmed cases of human infection with influenza A(H5N6) virus, including 21 deaths. This year, 10 sporadic human infections have been recorded in Sichuan Province, Anhui Province, Guangxi Zhuang Autonomous Region, and Chongqing Municipality (8).
INVESTIGATION AND RESULTS
Herein, we report the 5 cases infected by AIV H5N6 in Sichuan Province, China in 2021. These 5 independent cases occurred in 5 different districts or counties from 4 cities (Figure 1A, i.e., Jinjiang District of Chengdu City, Kaijiang County and Xuanhan County of Dazhou City, Bazhou District of Bazhong City, and Nanxi District of Yibin City). All four cities are located in the east of Sichuan Province, China (Figure 1A). A case was sampled by nasopharyngeal swabs and sent to the laboratory for quantitative polymerase chain reaction (qPCR) testing. The positive results will be sequenced and sent to China CDC for virus isolation. When a case was found, the local CDC and the municipal and/or provincial CDCs form two or three levels of investigation groups to carry out the epidemiological investigation of avian influenza cases. The timeline and investigation group for each case were shown in Table 1.
Figure 1.
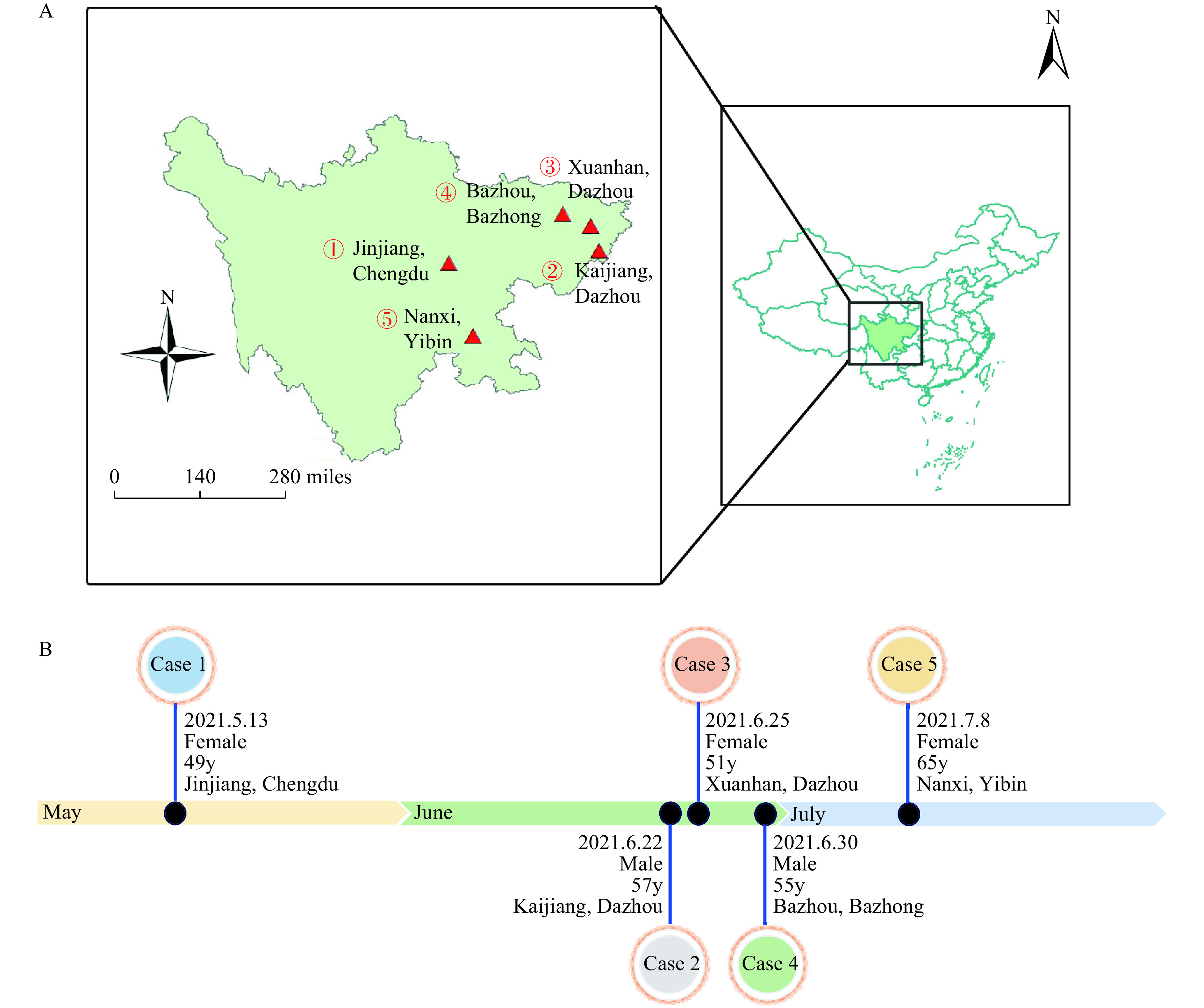
Temporal and spatial distribution of human infections with avian influenza A(H5N6) viruses in Sichuan Province, China, May to July 2021. (A) Geographical distribution of H5N6 virus infection among humans*; (B) The timeline and the basic demographic information of the H5N6 human cases.
Abbreviation: y=years old. * The sequence number of case-patients in place is based on the disease onset date.
Table 1. Demography and epidemiological profiles of the 5 cases in Sichuan.
| Case No. | Region | Age of case/years old | Date of onset | Date of hospitalization | Date of laboratory confirmation | Date of visited LPM | Contact with dead poultry | Consumption of dead poultry | Date of used Tamiflu | Outcome | Date of Investigation | Investigation group |
| Abbreviation: LPM=live poultry market. | ||||||||||||
| 1 | Jinjiang, Chengdu | 49 | 2021-05-13 | 2021-05-16 | 2021-05-26 | 2021-05-05 | Unknown | No | 2021-05-27 | Death | 2021-05-26 | Sichuan, Chengdu and Jinjiang CDC |
| 2 | Kaijiang, Dazhou | 57 | 2021-06-22 | 2021-07-05 | 2021-07-08 | 2021-06-13 | Yes | Yes | 2021-07-08 | Death | 2021-07-08 | Sichuan, Dazhou and Kaijiang CDC |
| 3 | Xuanhan, Dazhou | 51 | 2021-06-25 | 2021-07-02 | 2021-07-07 | 2021-06-04 | Yes | Unknown | Not used | Death | 2021-07-06 | Sichuan, Dazhou and Xuanhan CDC |
| 4 | Bazhou, Bazhong | 55 | 2021-06-30 | 2021-07-04 | 2021-07-06 | Unknown | Yes | No | 2021-07-05 | Recovered | 2021-07-05 | Sichuan, Bazhong and Bazhou CDC |
| 5 | Nanxi, Yibin | 65 | 2021-07-08 | 2021-07-13 | 2021-07-20 | Unknown | Yes | Yes | 2021-07-19 | Recovered | 2021-07-18 | Yibin and Nanxi CDC |
Patient 1 in Jinjiang District of Chengdu City was a 49-year-old (y) female who developed headache and nasal stuffiness on May 13. On May 26, the respiratory tract sample was tested for influenza viruses in Huaxi Hospital and was found to be H5N6 influenza virus positive, and on the next day, Chengdu Municipal CDC and Sichuan Provincial CDC confirmed the laboratory test as positive for H5N6 (Figure 1B and Table 1). The patient (Patient 2) from Kaijiang County of Dazhou City, who was a 57y male, had symptom onset on June 22 with cough and asthma. The patient was laboratory confirmed as H5N6 infection by Sichuan CDC on July 8. Patient 3 was a 51y female, from Xuanhan County of Dazhou City, that had a headache on June 25. Through qPCR testing of alveolar lavage fluid specimens, the case was laboratory diagnosed as H5N6 infection by Sichuan CDC on July 7. Patient 4 was a 55y male in Bazhou District of Bazhong City. He developed initial symptoms of fever on June 30 and was laboratory confirmed as H5N6 infection on July 6. Patient 5, female, 65y, from Nanxi District of Yibin City had symptom onset on July 8. The pharyngeal swab from Patient 5 was laboratory tested as H5N6 positive on July 20.
The epidemiological investigation found that all 5 patients had been exposed to live poultry before their disease onset (Table 1). Patient 1 bought duck from a poultry market on May 5, i.e., 8 days before the symptom onset. Patients 2, 3, 4, and 5 lived in rural areas and raised chickens, ducks, and gooses in their backyard for self-consumption. Patients 2 and 3 had visited the live poultry markets (LPMs) and purchased baby poult before symptom onset. Patients 4 and 5 had no history of visiting LPMs 4 weeks before their illness onset. The poultry farmed by Patient 5 were usually sent to the adjacent pond for breeding where wild birds (predominantly, white cranes) were frequently observed. There were dead poultry that were farmed by Patients 2, 3, 4, and 5, and they had contact with or ate these dead poultry before their symptom onset (Table 1).
In light of laboratory testing of samples collected from the poultry market contacted by Patient 1, H5N6 were qPCR detected positive. The poultry feeding settings of Patient 2–5 were tested, with positive results in the environment samples related to all four patients (Supplementary Table S1, available in http://weekly.chinacdc.cn/).
Table S1. Poultry environment investigation and testing.
| Case No. | Region | Source of poultry | Poultry species | Specify the upstream supplier | Poultry species of upstream supplier | qPCR of poultry environment | qPCR of upstream supplier environment | qPCR of living environment |
| *ND: Not done. | ||||||||
| 1 | Jinjiang, Chengdu | Market | Chicken, duck | Chenghua district market | Chicken, duck | + | + | + |
| 2 | Kaijiang, Dazhou | Domestic | Chicken, duck | Luo's chicken and duck farm | Chicken, duck | + | + | + |
| 3 | Xuanhan, Dazhou | Domestic | Chicken, duck | Yunchengzhai chicken farm | Chicken, duck | + | + | + |
| 4 | Bazhou, Bazhong | Domestic | Chicken, duck, goose | Not purchased in 2021 | Not purchased in 2021 | + | ND* | + |
| 5 | Nanxi, Yibin | Domestic | Chicken | Private vendors in Wangjia town | Chicken | + | ND | – |
All five patients had a history of contact with birds, so the following measures were taken. 1) Ten days of health monitoring was implemented of close contacts and possible exposed personnel of all patients. 2) Poultry and environmental disposal were carried out in affected areas. The patients’ families and the patients’ neighbors’ families carried out poultry culling and disposal, and thoroughly disinfected bird-related environments. 3) Strengthened management of poultry breeding sites and live poultry trading markets. This involved comprehensive rectification, cleaning, and disinfection of the live poultry trading market in the district and county. 4) Influenza-like illness (ILI) surveillance was strengthened in affected districts and counties. For outpatient and emergency patients who met the definition of ILI and patients with severe acute respiratory tract infection, the history of poultry exposure was inquired for and respiratory tract samples were collected for testing. During the strengthened surveillance period, the positive rate of ILI had a mild upward trend, but these ILI were all influenza B viruses. Influenza A viruses had not been isolated (except for one case of H9N2) in all of Sichuan Province, and there was no A (subtype not determined). 5) Strengthened health education by carrying out publicity and education on avian influenza prevention and control knowledge throughout the province, improving the disease prevention awareness of professional people, and reducing the risk of human infection with avian influenza virus.
The full genome of the virus from Patient 3 (A/Sichuan/06681/2021(H5N6), Dazhou/2021) and 6 gene segments except for PB2 and PA of the virus from Patient 4 (A/SiChuan-Bazhong/1/2021(H5N6), Bazhong/2021) were successfully sequenced. The nucleotide sequences similarity of 8 segments of Dazhou/2021 were analyzed with the online Basic Local Alignment Search Tool (BLAST) (Table 2), and the PB2 segment was found to have similarity with chicken H3N2 strain from Guangxi, China (96%) in 2014; PB1 and PA genes were 97%–98% similarity to that of H5N6 strains from poultry environment samples in Guangdong in 2017–2018 season. NA and NP segments were highly similarity to human infecting H5N6 strains from Anhui Province in 2020 (99%). The MP segment has a high similarity with chicken H5N8 strain from Kostroma, Russia in 2020 and the NS segment was highly similar to chicken H3N2 strain from Ganzhou, China in 2016. Interestingly, for strain Bazhong/2021 in our study, only the NS segment has a different closest stain i.e., H4N2 and H3N2 from Jiangxi, China in 2016, compared to the NS of Dazhou/2021 (Table 2). The maximum likelihood (ML) phylogenies tree [root at A/Goose/Guangdong/1/96(H5N1)] was generated by RAxML (version 8.2) program (9). The genomes of Dazhou/2021 and Bazhong/2021 have a similarity >99% for all the 6 available segments. The molecular phylogenetic analysis showed that the HA genes of both Dazhou/2021 and Bazhong/2021 belong to the 2.3.4.4b and have the highest homology with A/chicken/Omsk/0112/2020 (A/H5N8) from Omsk Russia ( Figure 2). The genomes of Dazhou/2021 and Bazhong/2021 have a similarity >99% for all the 6 available segments. The molecular phylogenetic analysis showed that the HA genes of both Dazhou/2021 and Bazhong/2021 belong to the 2.3.4.4b and have the highest homology with A/chicken/Omsk/0112/2020 (A/H5N8) from Omsk Russia ( Figure 2).
Table 2. Similarity of 8 segments of the viruses analyzed by online basic local alignment search tool.
| Segments | Bazhong/2021 Identity/Length (%) | Dazhou/2021 Identity/Length (%) | Collection location |
| PB2 | No sequence detected | A/chicken/Guangxi/165C7/2014 (A/H3N2) | Guangxi, China |
| 2,263/2,342 (96%) | |||
| PB1 | A/Env/Guangdong/zhanjiang/C18277136/2018-04-02 (A/H5N6) | A/Env/Guangdong/zhanjiang/C18277136/2018-04-02 (A/H5N6) | Guangdong, China |
| 2,304/2,344 (98%) | 2,310/2,345 (98%) | ||
| PA | No sequence detected | A/Env/Guangdong/zhanjiang/C17277346/2017-12-05 (A/H5N6) | Guangdong, China |
| 2,184/2,233 (97%) | |||
| HA | A/chicken/Omsk/0112/2020 (A/H5N8) | A/chicken/Omsk/0112/2020 (A/H5N8) | Omsk, Russia |
| 1,766/1,776 (99%) | 1,760/1,772 (99%) | ||
| NP | A/Anhui/2021-00011/2020 (A/H5N6) | A/Anhui/2021-00011/2020 (A/H5N6) | Anhui, China |
| 1,552/1,565 (99%) | 1,555/1,564 (99%) | ||
| NA | A/Anhui/2021-00011/2020 (A/H5N6) | A/Anhui/2021-00011/2020 (A/H5N6) | Anhui, China |
| 1,422/1,432 (99%) | 1,423/1,431 (99%) | ||
| MP | A/chicken/Kostroma/304-10/2020 (A/H5N8) | A/chicken/Kostroma/304-10/2020 (A/H5N8) | Kostroma, Russia |
| 1,025/1,027 (99%) | 1,026/1,027 (99%) | ||
| NS | A/Environment/Jiangxi/47054/2016 (A/H4N2) | A/chicken/Ganzhou/GZ43/2016 (A/H3N2) | Jiangxi, China |
| 870/891 (97%) | 873/890 (98%) |
Figure 2.
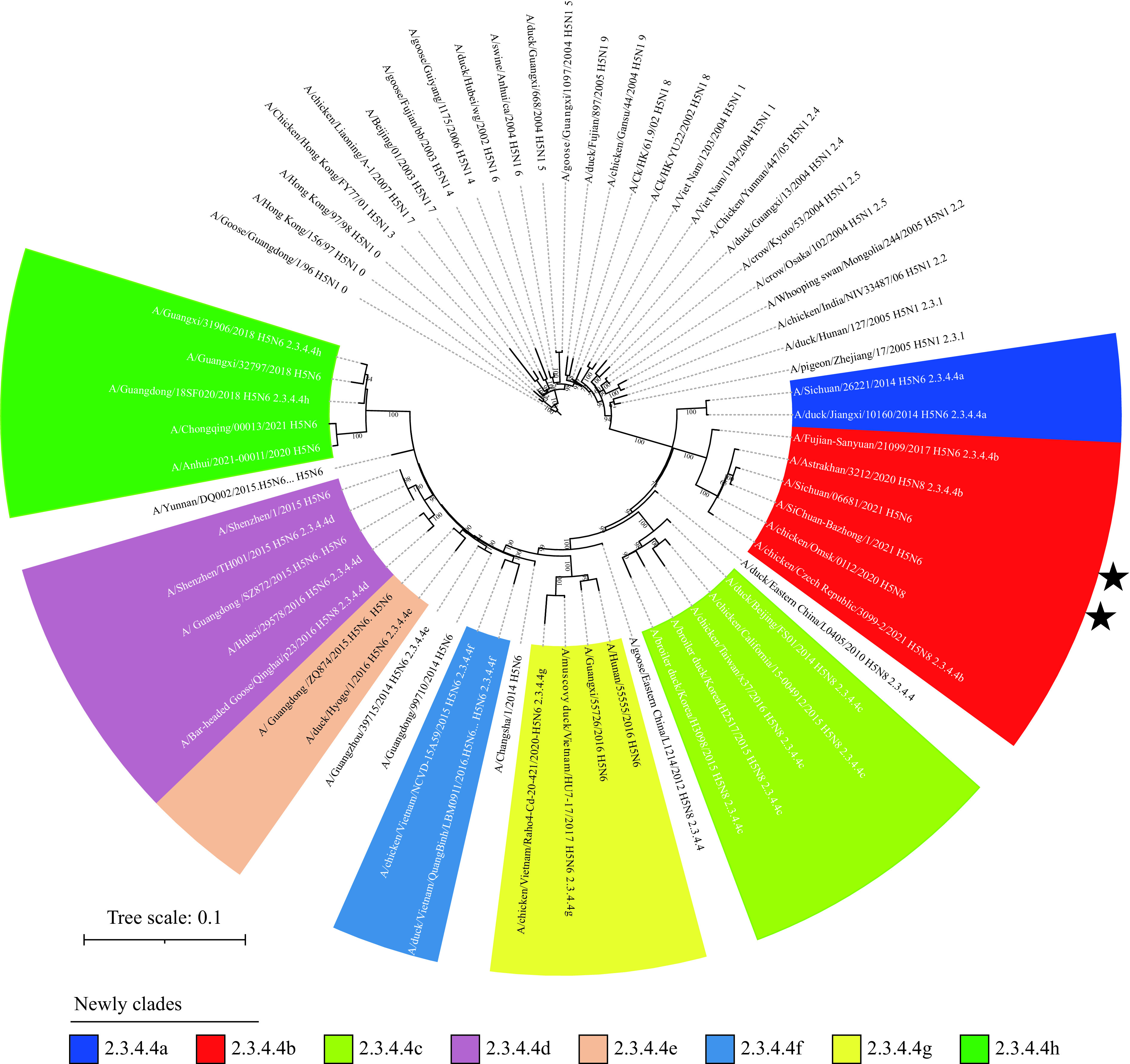
Maximum likelihood phylogenetic relationships of H5 viruses’ hemagglutinin (HA) genes.
Note: Phylogenetic included most of H5 clades (names were showed at leaves tip) and rooted to A/Goose/Guangdong/1/96(H5N1), constructed with 1,000 bootstrap replicates (above branches). The two stars indicate the strains of this study.
The molecular characterization of the Dazhou/2021 and Bazhong/2021 strains were analyzed and related key sites in all H5N6 were analyzed at the same time (Supplementary Table S2, available in http://weekly.chinacdc.cn/). The cleavage site of HA protein possessed a multiple basic amino acids motif (LREKRRKR↓G), which indicated high pathogenicity to chickens. The amino acid of the NS1 protein of these 2 strains at position 92 was aspartic acid, However, most H5N6 strains (n=1,329) have the 92E mutation; this D92E mutation has been correlated with increased virulence and/or cytokine resistance (10). NA protein was deleted at the stalk region (position 59–69), and no oseltamivir-associated resistance mutations in amino acid residues were found. Host-specific related sites, such as receptor binding Q226L of HA fragment and E627K of PB2 fragment were not found, which indicated that both Dazhou/2021 and Bazhong/2021 strains still possess features of avian origin.
Table S2. The molecular characteristics of the H5N6 influenza viruses isolated from Sichuan Province.
| Genes | AA position | Bazhong/2021 | Dazhou/2021 | All H5N6 * | Phenotypic effect |
| Note: Bold is consistent with the results of this study.
* Only show top 5, if the candidates are more than 5. | |||||
| HA | N158D | N | N |
N 1803 S 5 D 4 |
The substitution at residue 158 leads to a loss of glycan chain modification in the 150-loop, which avoids the potential steric hindrance for binding human receptors. |
| Q226L | Q | Q | Q 1813 | Critical for binding the α-2,6-linked receptor and enabling transmission in mammals | |
| Cleavage site | LREKRRKRG | LREKRRKRG | LRERRRKRG 1388
LREKRRKRG 305 SRERRRKRG 47 LKERRRKRG 29 QRETRG 10 |
Virulence increases in chickens | |
| NA | H274Y | H | H |
H 1744 Y 2 |
Reduces the susceptibility of neuraminidase inhibitors |
| PB2 | T271A | T | T |
T 1648 V 37 M 4 I 1 |
Enhances viral replication in mammalian cells in vitro |
| Q591K | Q | Q |
Q 1693 H 1 |
Increases pathogenicity in mice | |
| E627K | E | E |
E 1673 K 18 V 2 |
Associates with increased virulence of AIVs in mammals | |
| D701N | D | D |
D 1689 N 3 |
Altered virulence in mice | |
| NS1 | P212S | P | P |
P 1681 L 367 |
Promotes viral replication in mice |
| D92E | D | D | E 1329
D 355 G 1 |
Correlated with increased virulence and/or cytokine resistance | |
| M2 | L26F, V27A, A30T, S31N, G34E | L-V-A-S-G | L-V-A-S-G |
L-V-A-S-G1455 L-V-A-N-G 165 L-A-A-S-G 16 L-I-A-S-G 10 L-G-A-N-G 6 |
Antiviral amantadine resistance |
DISCUSSION
The AIV H5N6 was first reported as a low-pathogenic AIV (LPAIV) decades ago (5). Since the strain a/gs/gd/1/96 (H5N1) appeared in the 1990s, outbreaks of highly pathogenic AIV (HPAI) have occurred frequently. Before 2010 there was no evidence of reassortment of the H5N1 viruses with NA subtypes other than N1 (11). However, H5 reassorted with different specific NA subtypes, termed H5N2, H5N3, H5N5, H5N6, H5N8 after 2010 (12). These new virus subtypes emerged through multiple genetic reassortments of different subtype viruses within resident domestic and wild bird populations and continued to circulate in domestic poultry populations, leading to wider geographical spread and raising great concern based on their pandemic potential (13). With the cross-infection of wild birds and poultry, the epidemic of viruses in poultry has accelerated. To date, a total of 38 laboratory-confirmed cases of human infection with influenza A (H5N6) virus, including 21 deaths. Our study found that all the 5 patients had been exposed to live poultry before their disease onset. Given this, more research, enhanced surveillance, including genomic epidemiology, is necessary on wild birds, poultry, and the environment.
The novel clade 2.3.4.4 H5N6 that was first reported in 2013, then reported in Laos and Vietnam in 2014/2015, with evidence of sustained transmission and further geographical spread in both countries. Later on, a series of poultry outbreaks in Japan, Myanmar, and the Republic of Korea have been found to be related by H5N6. By 2017, it began spreading to some European countries such as Greece, Germany, the Netherlands, and Switzerland (14). Due to the genetic diversity of the virus, the World Health Organization (WHO) subdivided 2.3.4.4 into a–h clades, a total of 8 new clades. Even if it is so finely divided, recent serological studies have found that the vaccine strains recommended by WHO cannot fully cover the emerging strains in these clades (15). Thus, the balance between viral evolution and the capacity of the candidate vaccine protection needs to be timely updated which will reduce infection events and mortality in humans and animals.
Fortunately, there is no evidence of sustainable human-to-human transmission of H5N6, considering that all cases in this study were independent and have no epidemiological correlation. A one-health approach needs to be strengthened to trace the source of infection in time, block the cross-species spread of the virus, reduce risks, and protect people’s health.
Acknowledgments
Ms. Jie Zhang, Mr. Idrissa Kamara, and Dr. Chuansong Quan.
Funding Statement
Supported by the National Natural Science Foundation of China (No.82041033), Sichuan Science and Technology Program (2020FYS0015; 2020FYS0017). W.J.L. supported by the Excellent Young Scientist Program of the NSFC (No. 81822040)
Contributor Information
William J. Liu, Email: liujun@ivdc.chinacdc.cn.
Xingyu Zhou, Email: 17203409@qq.com.
Huiping Yang, Email: lydia_ping106@163.com.
References
- 1.Gao GF Influenza and the live poultry trade. Science. 2014;344(6181):235. doi: 10.1126/science.1254664. [DOI] [PubMed] [Google Scholar]
- 2.Liu WJ, Wu Y, Bi YH, Shi WF, Wang DY, Shi Y, et al Emerging HxNy influenza a viruses. Cold Spring Harb Perspect Med. 2020:a038406. doi: 10.1101/cshperspect.a038406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi WF, Gao GF Emerging H5N8 avian influenza viruses. Science. 2021;372(6544):784–6. doi: 10.1126/science.abg6302. [DOI] [PubMed] [Google Scholar]
- 4.Quan CS, Wang QL, Zhang J, Zhao M, Dai QG, Huang T, et al Avian influenza A viruses among occupationally exposed populations, China, 2014-2016. Emerg Infect Dis. 2019;25(12):2215–25. doi: 10.3201/eid2512.190261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi YH, Kun M, Shi WF, Liu D, Yu XL, Gao ZM, et al Two novel reassortants of avian influenza A (H5N6) virus in China. J Gen Virol. 2015;96(Pt 5):975-981. doi: 10.1099/vir.0.000056. [DOI] [PubMed] [Google Scholar]
- 6.Zhang RS, Chen TM, Ou XH, Liu RC, Yang Y, Ye W, et al Clinical, epidemiological and virological characteristics of the first detected human case of avian influenza A(H5N6) virus. Infect Genet Evol. 2016;40:236–42. doi: 10.1016/j.meegid.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan M, Gao RB, Lv Q, Huang SH, Zhou ZH, Yang L, et al Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect. 2016;72(1):52–9. doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 8.WHO. Avian influenza weekly update number 804. https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20210806.pdf?Status=Master&sfvrsn=5f006f99_42. [2021-8-6].
- 9.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MY, Wang BH Homology modeling and examination of the effect of the D92E mutation on the H5N1 nonstructural protein NS1 effector domain. J Mol Model. 2007;13(12):1237–44. doi: 10.1007/s00894-007-0245-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Liu XL, Yan JH, Liu WJ, Gao GF Interspecies transmission and host restriction of avian H5N1 influenza virus. Sci China C Life Sci. 2009;52(5):428–38. doi: 10.1007/s11427-009-0062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith GJD, Donis RO Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. Influenza Other Respir Viruses. 2015;9(5):271–6. doi: 10.1111/irv.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi YH, Li J, Li SQ, Fu GH, Jin T, Zhang C, et al Dominant subtype switch in avian influenza viruses during 2016-2019 in China. Nat Commun. 2020;11(1):5909. doi: 10.1038/s41467-020-19671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi YH, Chen QJ, Wang QL, Chen JJ, Jin T, Wong G, et al Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20(6):810–21. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Chen JH, Li X, Xu LY, Xie SM, Jia WX Health threats from increased antigenicity changes in H5N6-dominant subtypes, 2020 China. J Infect. 2021;83(2):e9–11. doi: 10.1016/j.jinf.2021.06.013. [DOI] [PubMed] [Google Scholar]


