Abstract
The adenovirus E1A gene can act as an oncogene or a tumor suppressor, with the latter effect generally arising from the induction of apoptosis or the repression of genes that provide oncogenic growth stimuli (e.g., HER-2/c-erbB2/neu) or increased metastatic invasiveness (e.g., metalloproteases). In this study, coexpression of E1A and p50E4F, a cellular transcription factor whose DNA binding activity is stimulated by E1A, suppressed colony formation by NIH 3T3 cells and transformation of primary rat embryo fibroblasts but had no observed effect in the absence of E1A. Domains in p50E4F required for stimulation of the adenovirus E4 promoter were required for the suppressive effect, indicating a transcriptional mechanism. In serum-containing media, retroviral expression of p50E4F in E1A13S/ras-transformed NIH 3T3 fibroblasts had little effect on subconfluent cultures but accelerated a decline in viability after the cultures reached confluence. Cell death occurred by both apoptosis and necrosis, with the predominance of each process determined by culture conditions. In serum-free media, p50E4F accelerated E1A-induced apoptosis. The results suggest that p50E4F sensitizes cells to signals or conditions that cause cell death.
Ectopic expression of the adenovirus E1A oncogene can cause a variety of phenotypic changes that range from immortalization and oncogenic cotransformation to the induction of apoptosis or the suppression of tumorigenic or metastatic growth. The diverse and, at times, contradictory nature of these changes reflects the number and complexity of cellular regulatory pathways that are targeted by E1A. Promotion of oncogenic growth is achieved mainly through the interaction of E1A with products of the retinoblastoma tumor suppressor gene family, Rb, p107, and p130, and the transcriptional coactivators p300 and CBP (for reviews see references 17, 18, 51, and 52). E1A interaction with Rb family proteins abrogates control of E2F transcription factors and the expression of E2F-regulated genes required for DNA synthesis or cell cycle progression (15, 41, 53, 75), whereas E1A interaction with p300/CBP functionally inactivates other factors that use p300/CBP to stimulate the transcription of cyclin-dependent kinase (CDK) inhibitors p15 and p21 (12, 42, 68) and/or otherwise maintain cells in a nonproliferative or differentiated state (8, 19, 30, 36, 37, 57, 67, 70, 73, 78). Additional mechanisms have also been proposed for E1A deregulation of the CDK inhibitors p21 and p27 through other pathways (1, 21, 47).
In various circumstances, these same interactions can also adversely affect cell growth and survival. For example, in a variety of breast and ovarian carcinoma cell lines, the interaction of E1A with p300/CBP is responsible for E1A-mediated repression of the HER-2/c-erbB2/neu oncogene and thereby suppresses tumor formation by those cell lines in rodents (4, 5, 76). In primary fibroblasts, E1A interaction with Rb family proteins and possibly p300/CBP leads to the stabilization of p53 and induction of p53-dependent apoptosis following serum withdrawal (7, 16, 27, 46, 74). Recent evidence indicates that E1A stimulates the expression of the p19ARF tumor suppressor (16), whose interaction with Mdm-2 and p53 inhibits p53 ubiquitination and degradation (34, 35, 55). Moreover, E1A-induced apoptosis was significantly reduced in ARF-null mouse embryo fibroblasts (MEFs), supporting the contention that cellular transformation by nuclear oncogenes such as E1A or c-myc initially requires abrogation of the ARF-p53 pathway, either by mutation or by introduction of a cooperating oncogene (e.g., one encoding activated Ras, E1B 19K, or Bcl-2) (13, 43, 45, 80). It should be noted, however, that E1A-induced apoptosis was not completely eliminated in ARF-null MEFs, indicating the involvement of p19ARF-independent mechanisms as well.
Other distinct oncogenic and tumor suppressor properties have also been attributed to E1A. Although perhaps more subtle in effect, they include both positive and negative modulation of E1A’s own immortalization and transformation capacities (3, 6, 9, 26) as well as repression of tumorigenesis and metastatic growth by a variety of tumor cell lines (3, 22, 23, 44, 56, 77). Although the mechanisms responsible for many of these effects have not been fully established, a number of them require regions in the C-terminal half of E1A and do not appear to involve a direct interaction with Rb family proteins or p300/CBP. Thus, a number of regulatory systems that influence E1A-induced phenotypic changes remain to be determined.
A primary function of E1A is to regulate transcription. One of the mechanisms utilized by E1A to transactivate the adenovirus E4 promoter involves the differential regulation of two related transcription factors, p120E4F and a proteolytically derived amino-terminal fragment of p120E4F termed p50E4F. The two factors have the same DNA binding domain (DBD) but exert opposite effects on the E4 promoter: p120E4F represses E4 transcription, whereas p50E4F stimulates it. During adenovirus infection, E1A induces the independent phosphorylation of both factors, causing the down-regulation of p120E4F DNA binding activity and stimulation of p50E4F DNA binding activity, which thereby contributes to activation of the E4 promoter (20).
We recently showed that ectopic expression of p120E4F in NIH 3T3 fibroblasts led to the stabilization of CDK inhibitors p21 and p27 and inhibited colony formation due to cell cycle arrest near the G1/S transition (21). Expression of either E1A or activated ras alleviated p120E4F-induced growth suppression through stimulation of cyclin D1 expression by ras or by direct interaction with Rb and down-regulation of p120E4F by E1A. Due to their coregulation by E1A, we surmised that both p120E4F and p50E4F might comprise a regulatory system involved in cell cycle checkpoint control. However, p50E4F had no apparent effect on nontransformed NIH 3T3 fibroblasts.
In this report, we show that ectopic expression of p50E4F can suppress the cotransformation of NIH 3T3 cells and primary rat embryo fibroblasts (REFs) by E1A and activated ras but, consistent with past results, has no effect in the absence of E1A. Suppression requires p50E4F domains that are required for regulation of the E4 promoter, indicating that the effect is due to p50E4F-regulated transcription. In serum-containing media, ectopic p50E4F expression increased the onset of cell death in confluent cultures of NIH 3T3 cells that coexpress E1A and activated ras, while no effect was seen in subconfluent cultures. Cell death occurred predominantly by necrosis (accidental cell death) when the medium was not frequently changed and predominantly by apoptosis (programmed cell death) in cultures with fresh media. Moreover, p50E4F expression increased the onset of apoptosis in subconfluent E1A-expressing cells upon serum withdrawal. We infer from these results that p50E4F does not directly induce cell death per se but rather sensitizes cells to whatever death signals are triggered by existent conditions.
MATERIALS AND METHODS
Plasmids.
pCMVs-E4F262 contains E4F residues 1 to 262 fused to the S-peptide sequence (Novagen) and expressed from the immediate-early cytomegalovirus (CMV) promoter in vector pCMV5 (20). pCMVs-E4FN1, pCMVs-E4FN2, pCMVs-E4FN5, pCMVs-E4FC1, pCMVs-E4FC2, and pCMVs-E4FC3 contain N-terminal and C-terminal truncations of the E4F262 sequence, as described below, subcloned into pCMV5 from previously described pCITE-E4F constructs (63). pCMV-E4FN1(fs) contains a 1-bp deletion at the beginning of the E4FN1 sequence. pβA-E1A(12S) and pβA-E1A(13S) contain the E1A(12S) and E1A(13S) cDNAs, respectively, cloned into pβA-Pr-neo (28, 73); E1A expression is driven by the human β-actin promoter, and the neomycin resistance (Neor) gene is driven by the simian virus 40 (SV40) promoter. pSP72-ras contains an SV40 promoter-driven T24 H-ras oncogene (64). pCMV-NF-IL6 and pCMV-ATF-2 express human cDNAs for NF-IL6 and ATF-2, respectively (20). pCMV-E1B19K (a gift from Eileen White, Rutgers University, Piscataway, N.J.) expresses the E1B19K cDNA (59). pE4-CAT1 contains the adenovirus E4 promoter (−224 to +32) fused to the bacterial chloramphenicol acetyltransferase (CAT) gene (20). The proviral vector pEQG1Na contains the Neor gene cloned between Moloney murine leukemia virus 5′ and 3′ long terminal repeats (54); the pEQ backbone contains the SV40 origin. pEQG1-E4F262-tkneo expresses E4F residues 1 to 262 from the Moloney murine leukemia virus 5′ long terminal repeat and the Neor gene from an internal herpes simplex virus thymidine kinase gene promoter. Retroviral replication and packaging functions were provided by the amphotropic helper plasmid pEQPAM3 (54). pEQG1Na and pEQPAM3 were a gift from Elio Vanin, St. Jude Children’s Research Hospital, Memphis, Tenn., with permission from Genetic Therapy, Inc.
Cell culture.
Primary fibroblasts and all cell lines were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum unless otherwise noted. NIH 3T3 cell lines expressing activated H-ras (3T3/ras) contain pSP72-ras and either pβA-Pr-neo or pRMM (PGK-HYGRO) (a gift from Paul Ney) for G418 or hygromycin B resistance, respectively. E1A12S/3T3 cell lines were created by using pβA-E1A(12S) and selected for G418 resistance. To create E1A13S/ras cell lines, hygromycin B-resistant (Hygror) 3T3/ras cells were transfected with pβA-E1A(13S) and selected in medium containing G418 (0.4 mg/ml) and hygromycin (0.8 mg/ml). E1A and Ras protein levels in isolated colonies were determined by Western blotting.
Colony formation and transformation assays.
NIH 3T3 fibroblasts (a gift from Martine Roussel, St. Jude Children’s Research Hospital) were transfected by the DNA-calcium phosphate coprecipitation method as previously described (21). Transfected cells were split 1:10 the next day and grown in medium containing G418 (0.4 mg/ml) for 24 days; the medium was replaced every 3 to 5 days. Colonies were stained after 24 days with 1× crystal violet stain (Sigma) in phosphate-buffered saline (PBS) and scored visually. Primary REFs from 8.5-day-old Fisher rat embryos were a generous gift from Gerard Zambetti (St. Jude Children’s Research Hospital) or obtained from BioWhittaker. Passage 3 REFs were transfected by the DNA-calcium phosphate coprecipitation method; 105 cells were transfected by the DNA-calcium phosphate coprecipitation method; 105 cells were transfected with a total of 20 μg of DNA containing 1.25 μg of each plasmid (as specified) and denatured salmon sperm DNA. Cells were fed every 3 to 5 days with Dulbecco’s modified Eagle’s medium containing 5% fetal bovine serum and stained after 28 days with 1× magic stain (200× magic stain contains 3 g of crystal violet and 0.8 g of ammonium oxalate in 100 ml of 20% ethanol) in PBS. Transformed foci were scored over a light box.
Retrovirus production and infection.
Proviral vectors pEQG1Na and pEQG1-E4F262-tkneo were cotransfected with pEQPAM3 into Cos 7 cells by electroporation (54). The medium was changed the following morning; viral supernatants were collected 72 h later, filtered, and frozen in aliquots at −80°C until use. Viral titers (CFU per milliliter) were measured by selection of G418-resistant colonies after NIH 3T3 cells were infected with serial dilutions of the supernatants. For bulk infections, cells underwent two consecutive 6-h incubations with viral supernatants containing 10 μg of Polybrene per ml, each at a multiplicity of infection (MOI) of 5 to 10 CFU/cell. An equal volume of fresh medium was added for an additional 12 h after the second infection, after which the cultures were trypsinized, counted, and plated at the densities indicated.
Cell viability, cell cycle, and apoptosis assays.
Infected cells were plated at 2.0 × 104 cells/well in 17-mm-diameter wells or 104 cells/well in 35-mm-diameter wells. In serum deprivation experiments, infected cells were washed and refed with serum-free medium 2 days after plating. At the indicated times, attached cells (harvested with trypsin) and floating cells (in the culture supernatant) were collected by centrifugation and resuspended in PBS containing 10 μg of soybean trypsin inhibitor (Boehringer Mannheim) per ml. Cell viability was determined by trypan blue dye exclusion. Cell cycle distribution and DNA content were determined by fluorescence-activated cell sorting (FACS) of propidium iodide-stained nuclei (PI-FACS) as previously described (38). The extent of apoptosis and necrosis was determined by FACS analysis after double staining with annexin V–R-phycoerythrin conjugate (annexin V-PE) and 7-aminoactinomycin D (7-AAD) as recommended by the supplier (PharMingen).
CAT assays.
Transfections were performed in 35-mm-diameter wells by using the Lipofectamine reagent (Life Technologies); 105 NIH 3T3 cells were plated in each well 24 h prior to transfection. Each well was transfected with a total of 2.0 μg of DNA containing 250 ng of pE4-CAT1, 0 to 1,000 ng of pCMV-E1A(13S), 0 to 500 ng of pCMVs-E4F262 or -E4F truncation mutant, 250 ng of pCMV-E1B19K, and pCMV4 as carrier. After 40 h, cells were washed in PBS and lysed by addition of 0.5 ml of 0.25 M Tris-HCl (pH 8.0)–0.04% Nonidet P-40 to each well. Cell extracts were directly assayed for CAT activity in microcentrifuge tubes (20) and normalized for protein concentration in 96-well plates. All transfections were performed in duplicate, and each specific set was repeated 3 to 12 times. The amounts of acetylated and unacetylated [14C]chloramphenicol were quantitated by PhosphorImager (Molecular Dynamics) analysis.
Western blots.
Whole-cell or membrane fraction lysates were prepared from NIH 3T3 cell lines or transfected NIH 3T3 and HeLa cells, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western blotting using standard procedures (29). E4F-N4-specific polyclonal antiserum was previously described (20). Monoclonal and polyclonal antibodies against E1A and Ras proteins were from Santa Cruz Biotechnologies. Immune complexes were detected using SuperSignal Ultra enhanced chemiluminescence reagents (Pierce).
Gel shift assays.
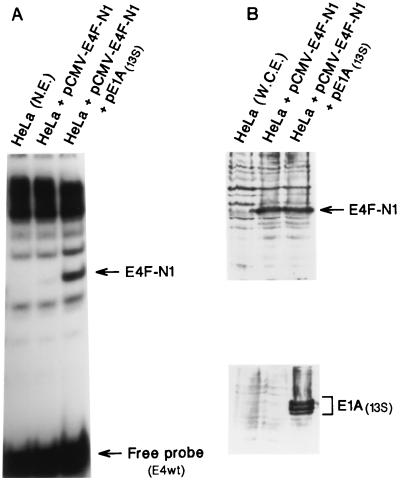
Nuclear extracts from HeLa cells transfected with pCMVs-E4FN1 were prepared 48 h posttransfection, applied to a 1-ml heparin-agarose column in buffer containing 0.1 M KCl, washed with buffer containing 0.25 M KCl, and eluted with buffer containing 0.4 M KCl as described previously (61). E4F-N1 DNA binding activity was bound to 32P-labeled E4wt (wild-type E4) probe and assayed under standard E4F gel shift conditions (62).
RESULTS
E4F262 suppresses cotransformation by E1A and activated ras.
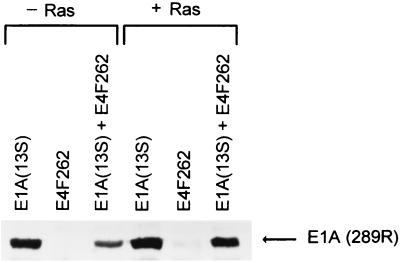
We previously demonstrated that a polypeptide containing the first 262 amino acids encoded by the E4F cDNA (E4F262) is functionally equivalent to endogenous p50E4F in terms of (i) DNA binding specificity and stability and (ii) transcriptional activation (20, 63); E4F residues 84 to 262 are sufficient for E1A-regulated DNA binding activity (Fig. 1). To assess the ability of p50E4F to influence E1A-mediated transformation, NIH 3T3 fibroblasts were cotransfected with an E4F262 expression construct in combination with separate constructs that express one of the two major E1A transcripts, E1A(13S) or E1A(12S), and activated ras and then assayed for colony formation (Table 1). E4F262 expression in the absence of E1A (with or without activated ras) had no significant effect on colony formation, consistent with our previous report that the growth and viability of NIH 3T3 cell lines expressing E4F262 or E4F262 and activated ras were the same as for the parental line (21). In contrast, coexpression of E4F262 with either E1A(13S) or E1A(12S) and activated ras reduced the number of G418-resistant colonies nearly 10-fold relative to control transfections expressing E1A and ras. Moreover, coexpression of two other transcription factors that recognize the E4F binding site (ATF-2, NF-IL6) with E1A(12S) and activated ras did not reduce colony numbers relative to controls, indicating that the suppressive effect was E4F262 specific. As a further control, Western blot analysis showed that E4F262 did not significantly reduce expression from pβA-E1A(13S) in NIH 3T3 cells (Fig. 2).
FIG. 1.
E1A stimulates the DNA binding activity of the E4F-N1 protein. (A) Gel shift analysis of E4F-N1 (E4F residues 84 to 262) DNA binding activity in extracts of HeLa cells transiently transfected with pCMVs-E4FN1 with or without pCMV-E1A(13S). Nuclear extracts (N.E.) were prepared 48 h posttransfection and step fractionated on heparin-agarose. (B) Western blots of whole-cell extracts (W.C.E.) from the transfected cells. E4F-N4-specific antiserum was used to detect E4F-N1 protein (top); monoclonal antibody M73 was used to detect E1A protein (bottom).
TABLE 1.
Effect of E4F262 expression on NIH 3T3 colony formation
| Expression construct | Coexpressed oncogene(s) | Fold change in colony no. (SD)a |
|---|---|---|
| pCMV5 | 1.00 | |
| E4F262b | 1.16 (0.06) | |
| pCMV5 | T24 ras | 1.00 |
| E4F262 | T24 ras | 0.95 (0.24) |
| pCMV5 | E1A(13S) + T24 ras | 1.00 |
| E4F262 | E1A(13S) + T24 ras | 0.23 (0.02) |
| pCMV5 | E1A(12S) + T24 ras | 1.00 |
| E4F262 | E1A(12S) + T24 ras | 0.11 (0.04) |
| E4F-N1 | E1A(12S) + T24 ras | 0.08 (0.07) |
| E4F-N2 | E1A(12S) + T24 ras | 0.40 (0.08) |
| E4F-C1 | E1A(12S) + T24 ras | 0.70 (0.31) |
| E4F-C2 | E1A(12S) + T24 ras | 0.93 (0.12) |
| NF-IL6 | E1A(12S) + T24 ras | 0.77 (0.23) |
| ATF-2 | E1A(12S) + T24 ras | 1.19 (0.31) |
Calculated as number of G418-resistant colonies from transfection containing pCMV construct expressing E4F262, E4F262 mutant, NF-IL6, or ATF-2/number of G418-resistant colonies from pCMV5 control transfection. For each set of transfections containing a pCMV-expressed transcription factor, parallel transfections containing pCMV5 and the indicated coexpressed oncogene(s) were used as controls. Values were derived from two to three independent experiments, each performed in duplicate or triplicate. pCMV5 control transfections typically produced 30 to 50 colonies in the absence of E1A and 50 to 150 colonies in the presence of E1A. A colony diameter of ≥3 mm was used as a criterion of unabated growth.
Each transfection of 5 × 105 NIH 3T3 fibroblasts contained 1.0 μg of pCMV5 or the indicated pCMV5-transcription factor construct, 1.0 μg of pβA-Pr-neo, 1.0 μg each of pCMV-E1A(13S), pCMV-E1A(12S), or pSP72-ras (T24 ras), as indicated, and pBluescript plasmid, for a total of 20.0 μg of DNA.
FIG. 2.
E4F262 does not inhibit E1A expression from pβA-E1A(13S). Shown are Western blots of lysates from NIH 3T3 cells transfected with E1A expression plasmid pβA-E1A(13S), p50E4F expression plasmid pCMVs-E4F262, or pβA-E1A(13S) plus pCMVs-E4F262; a duplicate set of transfections contained the activated ras expression plasmid pSP72-ras (+ Ras). Polyclonal antibodies against the E1A(13S) product were used to detected E1A protein [E1A (289R)].
The effect of E4F262 on E1A-mediated transformation was also assessed by coexpressing E4F262 with E1A(13S) or E1A(12S) and activated ras in primary REFs and then assaying for the formation of transformed foci (Table 2). Consistent with many past studies, coexpression of E1A(13S) or E1A(12S) with activated ras readily produced transformed foci; expression of E1A(13S), E1A(12S), activated ras, or E4F262 alone did not. Coexpression of E4F262 with E1A(13S) and activated ras reduced the number of transformed foci nearly 20-fold. Coexpression of E4F262 with E1A(12S) and activated ras was somewhat less effective but still consistently reduced the number of transformed foci threefold. As controls, coexpression of NF-IL6 or an E4F262 mutant containing a frameshift at residue 84 did not reduce the number of transformed foci. Thus, the results from two independent systems indicate that E4F262 overexpression can specifically suppress the growth of E1A-transformed cells.
TABLE 2.
Effect of E4F262 expression of E1A transformation of primary REFs
| Expression construct | Coexpressed oncogene(s) | Fold change in no. of REF foci (SD)a |
|---|---|---|
| pCMV5 | E1A(13S) + T24 ras | 1.00 |
| E4F262b | E1A(13S) + T24 ras | 0.05 (0.01) |
| E4F262 | E1A(12S) + T24 ras | 0.35 (0.05) |
| E4F-N1 | E1A(12S) + T24 ras | 0.37 (0.03) |
| E4F-N2 | E1A(12S) + T24 ras | 0.90 (0.04) |
| E4F-N5 | E1A(12S) + T24 ras | 0.86 (0.01) |
| E4F-C3 | E1A(12S) + T24 ras | 0.89 (0.04) |
| E4F-N1(fs) | E1A(12S) + T24 ras | 1.18 (0.07) |
| NF-IL6 | E1A(12S) + T24 ras | 1.13 (0.02) |
Calculated as number of transformed foci from transfection containing pCMV construct expressing E4F262, E4F262 mutant, NF-IL6, or ATF-2/number of foci from parallel pCMV5 control transfections. Values were derived from two independent experiments, each performed in duplicate or triplicate. pCMV5 control transfections containing E1A and T24 ras expression constructs typically produced 100 to 200 transformed foci per plate. pCMV5 or pCMVs-E4F262 transfections that did not contain E1A and T24 ras expression constructs produced zero to four foci per plate. A focus diameter of ≥2 mm was used as the criterion for transformation.
Each transfection of 105 passage 3 REFs contained 1.25 μg of pCMV5 or the indicated pCMV-transcription factor construct, 1.25 μg of pCMV-E1A(13S) or pCMV-E1A(12S), 1.25 μg of pSP72-ras (T24 ras), and sonciated salmon sperm DNA, for a total of 20.0 μg of DNA.
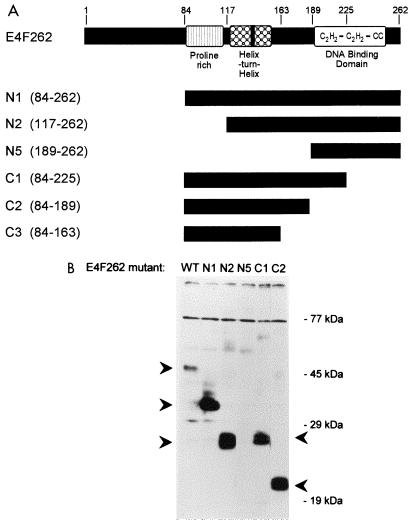
Domains in E4F262 required for transformation suppression.
Previous experiments indicated that the p50E4F DBD is located between residues 189 and 262 (63). To determine if the DBD or other functional domains are required for transformation suppression, we tested a series of expression constructs containing N- and C-terminal truncations of the E4F262 sequence (Fig. 3) for the ability to suppress the growth of E1A(12S)/ras-transformed NIH 3T3 cells and REFs. In the NIH 3T3 colony formation assay (Table 1), deletion of N-terminal residues 1 to 83 (N1 truncation) had no effect on colony suppression, whereas further deletion of residues 1 to 116 (N2 truncation) greatly reduced the suppressive effect. C-terminal truncations were introduced into the E4F-N1 construct and tested as described above. Deletion of residues 226 to 262 (C1 truncation), which covers half of the DBD, abolished the suppressive effect, as did deletion of residues 190 to 262 (C2 truncation), which covers the entire DBD.
FIG. 3.
E4F262 truncation mutants. (A) Schematic depiction of E4F262, N-terminal deletion mutants (N1, N2, and N5), and C-terminal deletion mutants (C1, C2, and C3) used in this study; E4F262 residues that remain in each truncation mutant are indicated by black bars and denoted in parentheses. (B) Western blot of lysates from NIH 3T3 cells transfected with pCMVs constructs expressing wild-type (WT) E4F262 and the truncation mutants. E4F-N4-specific antiserum was used to detect E4F proteins (marked by arrowheads); E4F-N5 and E4F-C3 (not shown) proteins do not contain the epitope recognized by this antiserum.
In the REF focus formation assay (Table 2), the (N1 truncation) again had no effect, whereas further deletion of residues 1 to 116 and 1 to 188 (N2 and N5 truncations) abolished the suppression of transformed foci. On the C-terminal end, deletion of residues 164 to 262 (C3 truncation), which covers the entire DBD, also abolished the suppressive effect in REFs. The results from both assays indicate that residues in the DBD and a separate upstream region with an N-terminal boundary between residues 84 and 116 are required for transformation suppression.
Domains in E4F262 required for transcriptional regulation.
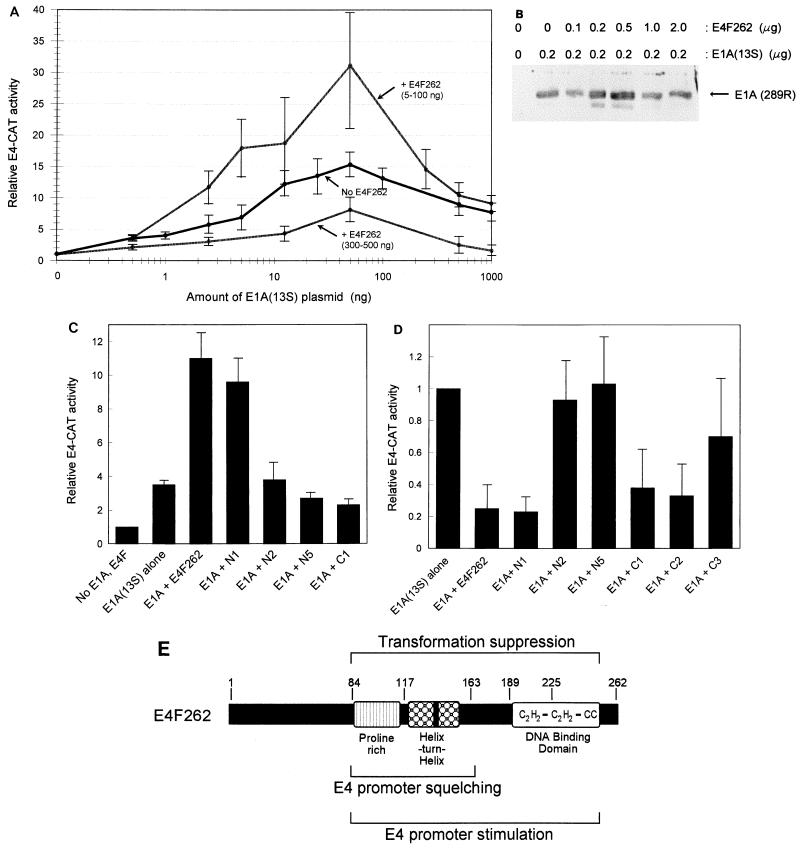
To determine if the regions in p50E4F that were required for transformation suppression correlated with those regions responsible for transcriptional regulation, E4F262 and the truncation mutants were transiently coexpressed with E1A(13S) in NIH 3T3 fibroblasts and analyzed for the ability to regulate the expression of an E4 promoter-driven CAT reporter construct (E4-CAT). E1A(13S) stimulated E4-CAT activity in a biphasic manner, with activation starting at low E1A(13S) levels, increasing to a peak at intermediate E1A(13S) levels, and then decreasing at high E1A(13S) levels, presumably due to promoter squelching (Fig. 4A). This type of response curve is consistent with many past studies, although peak activity levels in NIH 3T3 cells (ca. 15-fold) were significantly lower than those that we typically observed in HeLa cells (ca. 80-fold) (20).
FIG. 4.
Identification of regions in p50E4F that are required for regulation of the E4 promoter. (A) Stimulation and squelching of the E4 promoter by E4F262 and E1A(13S) in NIH 3T3 cells. The relative CAT activity from pE4-CAT1 was determined after cotransfection with increasing amounts of pCMV-E1A(13S) (0.5 to 1,000 ng) and pCMVs-E4F262 (5 to 500 ng). CMV promoter concentrations were kept at a constant level in all transfections by addition of pCMV4. CAT activities normalized for protein concentration, and internal control activities (when included) were divided by the basal CAT activity obtained in the absence of E1A and E4F262 to calculate relative E4-CAT activity. Each individual data point was derived from 3 to 7 independent experiments performed in duplicate; all data points were derived from a total of 12 independent experiments. Standard deviations are shown by thin error bars. Each data point showing p50E4F-mediated stimulation [+ E4F262 (5-100 ng)] or squelching [+ E4F262 (300-500 ng)] was derived from the highest CAT activity level obtained within the specified concentration range of pCMVs-E4F262 for each individual amount of pCMV-E1A(13S). (B) E4F262 does not significantly alter pCMV-E1A(13S) expression. Shown are Western blot of lysates from NIH 3T3 cells cotransfected with 0.2 μg of pCMV-E1A(13S) and the specified amounts of pCMVs-E4F262. E1A protein [E1A (289R)] was detected by polyclonal antibodies against the E1A(13S) product. (C) Regions of p50E4F required for stimulation of the E4 promoter. Relative E4-CAT activities were determined after cotransfection of 5 ng of pCMV-E1A(13S) and 30 ng of pCMVs constructs expressing E4F262 or E4F262 truncation mutants (N1, N2, N5, and C1). Values for each mutant were derived from two to four independent experiments. (D) Regions of p50E4F required for squelching of the E4 promoter. Relative E4-CAT activities were determined after cotransfection of 500 ng of pCMV-E1A(13S) and 500 ng of pCMVs constructs expressing E4F262 or E4F262 truncation mutants (N1, N2, N5, C1, C2, and C3). Values for each mutant were derived from two to six individual experiments. (E) Schematic depiction of p50E4F domains that are required for suppression of E1A-mediated transformation and regulation of the adenovirus E4 promoter.
Expression of E4F262 in the absence of E1A(13S) had no significant effect on E4-CAT activity. However, when increasing amounts of E4F262 were coexpressed with E1A(13S), relatively low amounts of E4F262 stimulated E4-CAT activity 1.5- to 3-fold at suboptimal and peak E1A levels but had little or no effect at high E1A levels (Fig. 4A). In contrast, coexpression of high amounts of E4F262 repressed or squelched E4-CAT activity at all E1A levels (Fig. 4A). pCMV-E1A(13S) expression did not significantly change when cotransfected with increasing amounts of pCMVs-E4F262 (Fig. 4B).
Over the course of many experiments, the amounts of E4F262 and E1A(13S) that produced maximal stimulation of E4-CAT activity varied, ranging between 5 and 100 ng for each expression plasmid. In addition, it was not uncommon for the E4F262 stimulatory effect to be more pronounced at low, suboptimal E1A levels. To compensate for this variability, the transfection experiments were performed in a matrix format where increasing amounts of E4F262 or E4F truncation mutants were cotransfected with increasing amounts of E1A(13S), to ensure that any effect on E4-CAT activity would be detected. Under conditions in which E4F262 stimulated E4-CAT activity, maximal stimulation by the N1 mutant was only slightly (10 to 20%) lower than that by E4F262 (Fig. 4C). In contrast, no stimulation was observed with the N2 and N5 mutants, setting the N-terminal boundary of a domain required for E4 promoter stimulation between residues 84 and 116. The C1 mutant was also unable to stimulate E4-CAT activity, indicating that the DBD was required.
The N- and C-terminal deletion mutants were also tested at higher amounts for the ability to squelch E4-CAT activation by E1A(13S) (Fig. 4D). Under these conditions, the N1 mutant reduced E4-CAT activity to the same extent as E4F262, whereas the N2 and N5 mutants had no effect. C-terminal mutants C1 and C2 also squelched E4-CAT activity similarly to E4F262, whereas the C3 mutant had a variable but reduced effect, setting the C-terminal boundary of the domain required for squelching between residues 163 and 190; the variability of the C3 mutant suggests the C-terminal boundary is close to residue 163. Taken together, these results identify a central region in p50E4F, approximately between residues 84 and 163, that is necessary and sufficient for squelching of the E4 promoter and is required along with the p50E4F DBD for stimulation of the E4 promoter. The same regions are required for suppression of E1A-mediated transformation (Fig. 4E).
E4F262 stimulates cell death in confluent cultures of E1A-expressing cells.
We examined the nature of E4F262 growth suppression by measuring cell growth and viability over a 2-week period after infecting parental NIH 3T3, 3T3/ras, or E1A13S/ras cells with a replication-defective retrovirus that expresses E4F262 (rvE4F262) or a control virus (rvG1Na) (Fig. 5A). Cultures were infected by consecutive applications of viral supernatants at an MOI of 5 to 10 CFU/cell to ensure that ≥90% of the cells were infected. Four independently derived E1A13S/ras cell lines were tested individually or as a pool, and no significant differences were observed between them (data not shown); results of experiments using the pooled cell lines are presented.
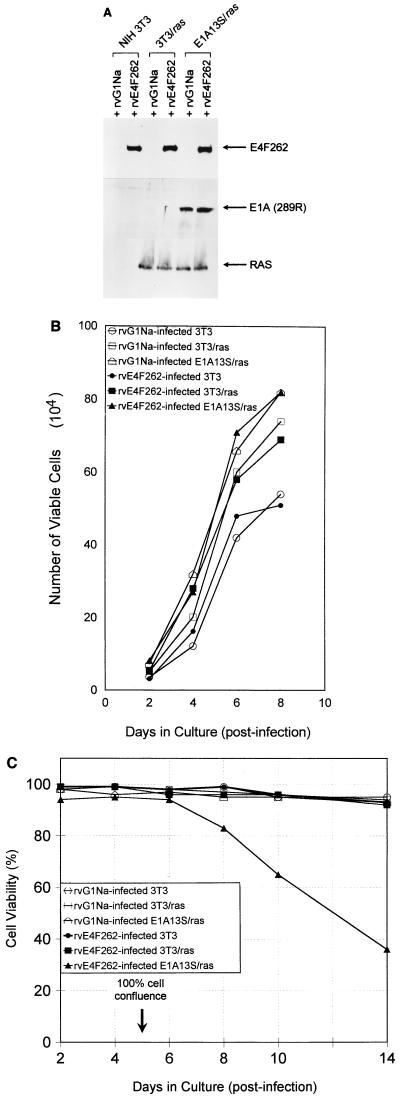
FIG. 5.
Retroviral expression of E4F262 reduces cell viability in confluent cultures of E1A13S/ras cells. (A) Western blots of lysates from NIH 3T3, 3T3/ras, and E1A13S/ras cells infected with rvE4F262 or control retrovirus rvG1Na. Polyclonal antibodies were used to detect E4F262 and E1A(289R) proteins in total cell lysates; rat monoclonal immunoglobulin G was used to detect Ras proteins in lysates of membrane-enriched fractions. Lysates were harvested 96 h postinfection. (B) NIH 3T3, 3T3/ras, and E1A13S/ras cells were infected with rvE4F262 or rvG1Na (MOI = 5 to 10 CFU/cell) for 24 h and grown in 35-mm-diameter wells after replating at 104 cells/well. Cell counts were performed after trypan blue staining at the indicated times. Growth curves were performed in triplicate, with a standard deviation of ≤6% between triplicates for all points. Cell viability was ≥95% at all points. NIH 3T3 cells were a single parental line; 3T3/ras cells were a pool of one Neor- and one Hygror NIH 3T3 cell lines that both express activated ras; E1A13S/ras cells were a pool of four Neor and Hygror NIH 3T3 cell lines that express the E1A(13S) cDNA and activated ras. (C) rvE4F262- or rvG1Na-infected NIH 3T3, 3T3/ras, and E1A13S/ras cells were replated in 17-mm-diameter wells at 2.0 × 104 cells/well and received fresh medium every 48 h. Cell viability was determined by trypan blue exclusion at the indicated times. Viability curves were performed in duplicate in two independent experiments, with a standard deviation of ≤9% between duplicates for all points in both experiments. Cell confluence was determined by visual inspection.
We observed no differences in growth rates or viability between exponentially growing cultures of rvE4F262- and control-infected cells (Fig. 5B). Also, PI-FACS revealed no differences in cell cycle distribution (data not shown). However, starting 24 to 48 h after the infected cultures reached cell confluence, the viability of rvE4F262-infected E1A13S/ras cells decreased from 94 to 35% over an 8-day period whereas the viability of control-infected E1A13S/ras cells remained above 90% (Fig. 5C). In contrast, the viability of cells not expressing E1A (parental NIH 3T3 and 3T3/ras) did not change with rvE4F262 infection over the same time period. After 2 weeks, cell viability gradually declined in all of the cultures (not shown).
Having observed a decrease in viability only in confluent rvE4F262-infected cultures, we assessed several parameters that could potentially affect the growth of cells in colony and focus formation assays, i.e., high-density growth, time in culture, and exhaustion of medium (Fig. 6A). One day after infection, rvE4F262- and control-infected E1A13S/ras cells were plated at two densities (104 cells/35-mm-diameter well and 2.0 × 104 cells/17-mm-diameter well), and viability was assessed over a 2-week period. For each set of cultures, half of the wells received fresh medium every 2 days and the other half were not replenished. Independent of the original cell density, viability began to decrease only after cell confluence was reached; length of time in culture did not correlate with loss of viability. Replenishment of the culture medium delayed the onset of cell death, particularly in cultures plated at the lower cell density (medium in cultures plated at the higher cell density usually became acidic within 24 h of replenishment after the culture exceeded 100% confluence). These data indicate that both cell confluence and medium exhaustion play a role in triggering the death of p50E4F-expressing E1A/ras cells.
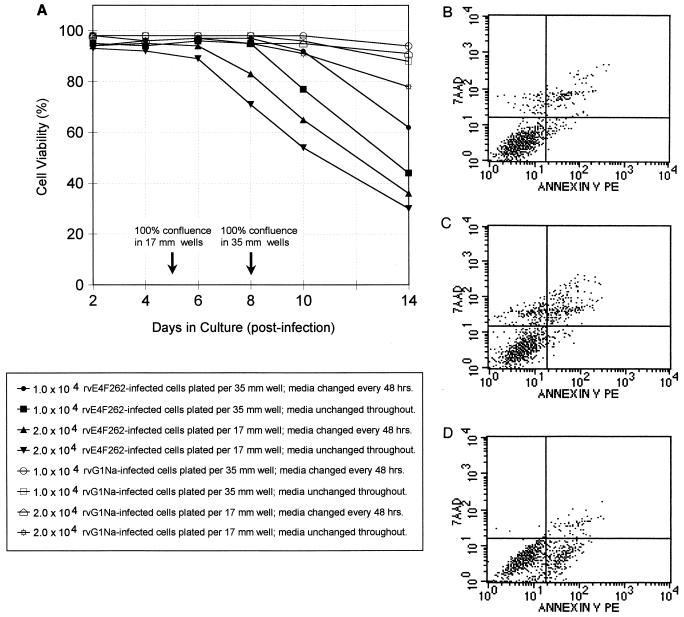
FIG. 6.
Culture conditions influence the onset and mechanism of cell death in rvE4F262-infected E1A13S/ras cells. (A) Quadruplicate cultures of rvE4F262- or rvG1Na-infected E1A13S/ras cells were replated at two cell densities, 104 cells/35-mm-diameter well and 2.0 × 104 cells/17-mm-diameter well. Half of each set of cultures received fresh medium every 48 h; the medium was unchanged in the other half. Average cell viability from duplicate wells was determined by trypan blue exclusion at the indicated times; standard deviations were ≤10% between duplicates at all points. Two independent experiments were performed, and the results from one representative experiment are shown. (B to D) Quadruplicate sets of rvE4F262-infected E1A13S/ras cells were replated at 2.0 × 104 cell/17-mm-diameter well with fresh medium every 48 h (B), 104 cells/35-mm-diameter well with no fresh media after plating (C) and 104 cells/35-mm-diameter well with fresh medium every 24 h (D). Cells were stained with annexin V-PE and 7-AAD and analyzed by FACS approximately 48 h after reaching confluence. Representative histograms from each quadruplicate set are shown. Viable cells not undergoing cell death do not bind annexin V and exclude 7-AAD (lower left quadrant); cells in early apoptosis bind annexin V but still exclude 7-AAD (lower right quadrant); cells in early necrosis do not bind annexin V but do not exclude 7-AAD (upper right quadrant); cells in late apoptosis or necrosis and dead cells bind annexin V and do not exclude 7-AAD (upper right quadrant). Quadrant boundaries were set by using rvG1Na-infected E1A13S/ras cells and uninfected NIH 3T3 cells.
Initially, microscopic examination and PI-FACS analysis were inconclusive as to whether confluent rvE4F262-infected E1A13S/ras cultures were dying by apoptosis or necrosis (data not shown). However, FACS analysis of cells stained with annexin V and the vital dye 7-AAD gave consistent results regarding the mechanism of cell death. During the early stages of apoptosis, cells begin to lose membrane asymmetry and annexin V can bind to phosphotidylserine that has translocated to the outer surface of the plasma membrane (48, 72). Thus, early apoptotic cells stain positive for annexin V and negative for vital dyes, whereas cells during early necrosis stain annexin V negative and vital dye positive prior to cell rupture; late apoptotic, late necrotic, and dead cells have lost membrane integrity and stain positive for both (65, 66, 71).
Quadruplicate sets of rvE4F262-infected E1A13S/ras cells were grown in three culture conditions. One set was plated at 2.0 × 104 cells/17-mm-diameter well and received fresh medium every 2 days; the second set was plated at 104 cells/35-mm-diameter well and was not replenished with fresh medium after plating; the third set was plated at 104 cells/35-mm-diameter well and received fresh medium every 24 h to ensure against medium depletion or acidification. Cells were harvested 48 h after reaching confluence and analyzed by FACS after staining with annexin V and 7-AAD (Fig. 6B to D). The higher-density cultures (Fig. 6B) were 82 to 86% viable (as judged by vital dye exclusion), with 4 to 7% of the cells showing evidence of early necrosis (upper left quadrant), 3 to 5% showing evidence of early apoptosis (lower right quadrant), and 8 to 12% dead (upper right quadrant). The lower-density cultures that were not replenished (Fig. 6C) were 69 to 82% viable (as judged by vital dye exclusion), with 7 to 13% of cells showing evidence of early necrosis, 1 to 2% showing evidence of early apoptosis, and 9 to 15% dead. In contrast, the lower-density cultures that were replenished every 24 h (D) were still 92 to 95% viable (as judged by vital dye exclusion), but with 12 to 15% of cells showing evidence of early apoptosis, 0 to 1% showing evidence of early necrosis, and 4 to 7% dead. Control-infected E1A13S/ras cells grown under the three culture conditions were all ≥94% viable, and the number of cells in either the upper left (necrosis) or lower right (apoptosis) quadrant was ≤2% of the total population (data not shown). These data indicate that p50E4F accelerates cell death regardless of which process is triggered but may not be a trigger itself.
E4F262 stimulates E1A-induced apoptosis.
Having demonstrated that E4F262 could stimulate cell death under conditions that approximated those in colony and transformed focus formation assays (i.e., high density cell growth in 10% serum), we next determined the effect of E4F262 in NIH 3T3 cells under conditions typically used to study E1A-induced apoptosis. Two E1A(12S)-expressing NIH 3T3 cell lines, one E1A13S/ras cell line, 3T3/ras cells, and parental NIH 3T3 cells were infected with rvE4F262 or control virus and plated at low density (104 cells/35-mm-diameter well). Two days later, the infected cells were serum deprived for 24 and 48 h, and the percentage of apoptotic cells with a subdiploid DNA content was determined by PI-FACS analysis; the average percentages of apoptotic cells from E1A-expressing cells and parental cells are presented with representative histograms (Fig. 7).
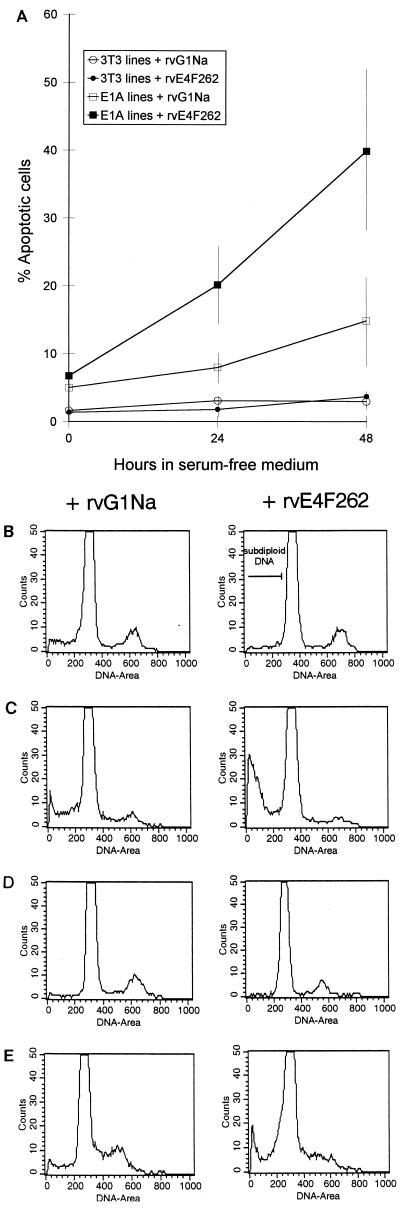
FIG. 7.
E4F262 expression increases E1A-induced apoptosis in serum starved NIH 3T3 cells. After infection by rvE4F262 or control virus (rvG1Na), two independent E1A12S/3T3 cell lines, one E1A13S/ras cell line, one 3T3/ras cell line, and parental NIH 3T3 cells were replated at 104 cells/ 35-mm-diameter well, cultured for 48 h, and then shifted to serum-free medium for 24 and 48 h. At each time point, the degree of apoptosis was determined by the percentage of cells with a subdiploid DNA content as measured by PI-FACS analysis. (A) Average percentages of apoptotic cells from E1A-expressing and non-E1A-expressing (3T3) cell lines infected with rvE4F262 or rvG1Na. Standard deviations are indicated by thin vertical lines. (B to E) Representative DNA histograms of rvE4F262- or rvG1Na-infected NIH 3T3 (B), E1A12S/3T3 (C), 3T3/ras (D), and E1A13S/ras (E) cells after 24 h in serum-free medium. The position of the subdiploid DNA peak is indicated by a bar in the upper right panel.
Although NIH 3T3 cells do not express p19ARF (58), a modest increase in apoptosis was still observed in control-infected E1A-expressing cells, similar to the attenuated induction of apoptosis seen in ARF−/− MEFs (13). This occurred in both E1A13S/ras cells and E1A12S/3T3 cells, confirming a result previously seen in REF52 cells that coexpression of activated ras does not completely protect against E1A-induced apoptosis (46). Importantly, the percentage of apoptotic cells tripled in rvE4F262-infected E1A-expressing cells when compared to control-infected E1A-expressing cells. By contrast, in NIH 3T3 and 3T3/ras cells, there was little or no indication of apoptosis with either rvE4F262 or the control virus. Moreover, no effect was observed when the cultures were maintained in 10% serum (data not shown). Thus, again, E4F262 significantly increased the onset of apoptosis when expressed in the presence of an activator (E1A) and triggering stimuli (E1A and serum depletion).
DISCUSSION
Here we have shown that ectopic expression of p50E4F, a cellular transcription factor whose DNA binding activity is stimulated by E1A, can suppress the growth of primary and established REFs transformed by E1A and activated ras. The expression of p50E4F by itself or with activated ras did not suppress colony formation, strongly suggesting that the suppressive effect required specific stimulation of p50E4F DNA binding activity by E1A, although it might also reflect a more general effect caused by oncogenic, hyperproliferative growth. Clearly, p50E4F’s ability to function as a transcription factor is required for growth suppression, as shown by the negative impact of deletions within the C-terminal DBD and the central region required for E4 promoter stimulation. The requirement for both regions indicates that the suppressive effect is not simply due to squelching or competitive binding to promoter elements. This inference is also supported by observations in our laboratory that the levels of E4F262 expression achieved by the transfection conditions used in the colony suppression assay or by infection with rvE4F262 are considerably (10- to 100-fold on a per-cell basis) lower than that achieved by the transfection conditions used for E4 promoter squelching (60). Thus, regardless of whether it is controlled directly or indirectly by E1A, transcriptional regulation by p50E4F is likely responsible for the suppressive effect in the colony and transformed focus formation assays.
p50E4F is generated by proteolysis of p120E4F, a low-abundance zinc finger protein that is ubiquitously expressed in all tissues (20, 61). We previously demonstrated that ectopic expression of p120E4F will also suppress colony formation but, unlike p50E4F, does so only in the absence of E1A or activated ras (21). Thus, growth suppression by both E4F factors initially appeared to correlate with their differential regulation by E1A (E1A down-regulates p120E4F DNA binding activity and up-regulates p50E4F DNA binding activity) and suggested the possibility that they regulate growth through the same pathway. However, p120E4F suppresses cell growth by inducing cell cycle arrest through stabilization of CDK inhibitors p21WAF1 and p27KIP1 (21), whereas the results in Fig. 5 to 7 clearly show that p50E4F affects cell viability, not cell cycle progression, and therefore operates through a fundamentally different pathway.
In the retrovirus-infected cultures, three parameters that most affected p50E4F-stimulated cell death were E1A expression, cell confluence, and medium replenishment. The fact that rvE4F262 potentiated cell death only in E1A-expressing cells is consistent with a requirement for E1A-induced activation of p50E4F. However, the lack of an effect in subconfluent, exponentially growing cultures showed that p50E4F activation by itself is not sufficient, although it is conceivable that an effect on cell viability would have become evident in longer-term cultures. Instead, cell death was evident only in confluent cultures and was further increased in the absence of medium replenishment, suggesting that cell confluence, or perhaps more specifically the growth of cells in a confluent culture, and medium exhaustion created the triggering events and p50E4F sensitized cells to them or accelerated their response. The same triggering events would also have been present in the colony and focus formation assays, given the high-density cell growth and potential for medium exhaustion that occurs in these assays.
FACS analysis of confluent rvE4F262-infected E1A13S/ras cultures stained with annexin V-PE and 7-AAD demonstrated evidence of early apoptosis (annexin V positive, 7-AAD negative) and early necrosis (annexin V negative, 7-AAD positive), indicating cell death occurred through both processes. Under extreme culture conditions, cell death occurred almost exclusively through one or the other process, indicating that E4F262 expression potentiated either process independently of the other. The increase in cell death (Fig. 6A) and the predominance of necrosis (Fig. 6C), in nonreplenished cultures is consistent with the induction of necrosis by an ischemic condition, e.g., medium acidification or nutrient depletion caused by high-density cell growth. The more gradual onset of cell death (Fig. 6A) and predominance of apoptosis (Fig. 6D) in cultures frequently replenished with serum-containing media is consistent with the induction of suspension-induced apoptosis (anoikis), a process that occurs when E1A/ras-transformed cells lose integrin-mediated matrix adhesion (24, 25, 50). Notably, both are p53-independent processes.
Under most culture conditions, however, both forms of cell death were observed, with the percentage of necrotic cells usually being higher than the percentage of those undergoing apoptosis. This suggests that rvE4F262-infected cells may be more prone to necrotic cell death, although it might also reflect the relative ease at which each process is triggered in these cultures instead of some inherent predisposition of the cell. The variable occurrence of both apoptosis and necrosis, and the mimicry of some characteristics of apoptosis (e.g., nuclear fragmentation and DNA degradation) that can occur in slowly accumulating dead cells (10, 11), also likely accounted for the inconsistent or inconclusive results initially obtained by microscopic examination and PI-FACS analysis.
A less complicated result was obtained with the induction of apoptosis in E1A-expressing cells by serum withdrawal. In primary fibroblasts, the majority of the apoptotic response induced by E1A and serum withdrawal is p53 dependent (2, 13, 74), occurring through ARF-dependent and independent pathways (16). However, in our experiments, the apoptotic response was solely ARF independent, as the NIH 3T3 cells used here do not express p19ARF (58), and thus was attenuated in comparison to the level of induction normally observed in wild-type primary fibroblasts. Regardless, E4F262 expression clearly potentiated the induction of apoptosis in NIH 3T3 cells (Fig. 7), indicating its effect lies outside of the p19ARF pathway. Recent evidence suggests that the ARF-independent pathways utilized by E1A may overlap those involved in DNA damage-induced apoptosis (16). It will be of interest to determine if p50E4F can also potentiate the response to DNA-damaging agents.
Regardless of the actual trigger, the ability of p50E4F to potentiate both apoptosis and necrosis suggests that at least one of its target genes must be involved in both processes. A critical mechanism in both apoptosis and necrosis involves in both processes. A critical mechanism in both apoptosis and necrosis involves a change in mitochondrial membrane permeability, called the mitochondrial permeability transition (reviewed in references 32, 39, 40, and 79). Perhaps those genes whose products regulate the mitochondrial permeability transition (e.g., Bcl-2 family members), or some other mitochondrial function, would be good candidates for p50E4F targets (14, 31, 33, 49, 69). Ultimately, identification of bona fide p50E4F target genes will be required to understand how p50E4F influences cell death and its physiological significance.
ACKNOWLEDGMENTS
We thank Ruby Tharp and Kristen Rothammer for excellent technical assistance, Mike Cook and Lynn Martinek of the Duke Comprehensive Cancer Center Flow Cytometry Facility for FACS analysis, Paul Ney for pRMM, Elio Vanin for pEQG1Na and pEQPAM3, Eileen White for pCMV-E1B19K, and Gerard Zambetti for pSP72-ras and primary REF cells. R.J.R. also thanks Scott Hiebert and Joseph R. Nevins for their encouragement and support.
This work was supported in part by National Institutes of Health grant R01GM51299 (R.J.R.), National Cancer Institute Cancer Center Support (core) grant 5P30CA21765 (St. Jude Children’s Research Hospital), and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Alevizopoulos K, Catarin B, Vlach J, Amati B. A novel function of adenovirus E1A is required to overcome growth arrest by the CDK2 inhibitor p27(Kip1) EMBO J. 1998;17:5987–5997. doi: 10.1093/emboj/17.20.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attardi L D, Lowe S W, Brugarolas J, Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd J M, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Hung M C. Involvement of co-activator p300 in the transcriptional regulation of the HER-2/neu gene. J Biol Chem. 1997;272:6101–6104. doi: 10.1074/jbc.272.10.6101. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Yu D, Chinnadurai G, Karunagaran D, Hung M C. Mapping of adenovirus 5 E1A domains responsible for suppression of neu-mediated transformation via transcriptional repression of neu. Oncogene. 1997;14:1965–1971. doi: 10.1038/sj.onc.1201030. [DOI] [PubMed] [Google Scholar]
- 6.Chinnadurai G. Adenovirus E1a as a tumor-suppressor gene. Oncogene. 1992;7:1255–1258. [PubMed] [Google Scholar]
- 7.Chiou S K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condorelli G, Giordano A. Synergistic role of E1A-binding proteins and tissue-specific factors in differentiation. J Cell Biochem. 1997;67:423–431. doi: 10.1002/(sici)1097-4644(19971215)67:4<423::aid-jcb1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Cone R D, Grodzicker T, Jaramillo M. A retrovirus expressing the 12S adenoviral E1A gene product can immortalize epithelial cells from a broad range of rat tissues. Mol Biol Cell. 1998;8:1036–1044. doi: 10.1128/mcb.8.3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darzynkiewicz Z, Bedner E, Traganos F, Murakami T. Critical aspects in the analysis of apoptosis and necrosis. Hum Cell. 1998;11:3–12. [PubMed] [Google Scholar]
- 11.Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami T, Traganos F. Cytometry in cell necrobiology: analysis of apoptosis and accidental cell death (necrosis) Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- 12.Datto M B, Hu P P, Kowalik T F, Yingling J, Wang X F. The viral oncoprotein E1A blocks transforming growth factor beta-mediated induction of p21/WAF1/Cip1 and p15/INK4B. Mol Cell Biol. 1997;17:2030–2037. doi: 10.1128/mcb.17.4.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 14.Decaudin D, Geley S, Hirsch T, Catedo M, Marchetti P, Macho A, Kofler R, Kroemer G. Bcl-2 and Bcl-XL antagonize the mitochrondrial dysfunction preceding nuclear apoptosis induced by chemotherapeutic agents. Cancer Res. 1997;57:62–67. [PubMed] [Google Scholar]
- 15.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis-and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Stancina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 19.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 20.Fernandes E R, Rooney R J. The adenovirus E1A-regulated transcription factor E4F is generated from the human homolog of nuclear factor φAP3. Mol Cell Biol. 1997;17:1890–1903. doi: 10.1128/mcb.17.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes E R, Zhang J Y, Rooney R J. Adenovirus E1A-regulated transcription factor p120E4F inhibits cell growth and induces the stabilization of the cdk inhibitor p21WAF1. Mol Cell Biol. 1998;18:459–467. doi: 10.1128/mcb.18.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frisch S M. Reversal of malignancy by the adenovirus E1a gene. Mutat Res. 1996;350:261–266. doi: 10.1016/0027-5107(95)00103-4. [DOI] [PubMed] [Google Scholar]
- 23.Frisch S M, Dolter K E. Adenovirus E1a-mediated tumor suppression by a c-erbB-2/neu-independent mechanism. Cancer Res. 1995;55:5551–5555. [PubMed] [Google Scholar]
- 24.Frisch S M, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frisch S M, Ruoslahti E. Integrins and anoikis. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- 26.Gopalakrishnan S, Douglas J L, Quinlan M P. Immortalization of primary epithelial cells by E1A 12S requires late, second exon-encoded functions in addition to complex formation with pRB and p300. Cell Growth Differ. 1997;8:541–551. [PubMed] [Google Scholar]
- 27.Grossman S R, Perez M, Kuang A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 28.Gunning P, Leavitt J, Muscat G, Ng S-Y, Kedes L. A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci USA. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 30.Hasegawa K, Meyers M B, Kitsis R N. Transcriptional coactivator p300 stimulates cell type-specific gene expression in cardiac myocytes. J Biol Chem. 1997;272:20049–20054. doi: 10.1074/jbc.272.32.20049. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch T, Decaudin D, Susin S A, Marchetti P, Larochette N, Resche-Rigon M, Kroemer G. PK11195, a ligand of the mitochondrial benzodiazepine receptor, facilitates the induction of apoptosis and reverses Bcl-2-mediated cytoprotection. Exp Cell Res. 1998;241:426–434. doi: 10.1006/excr.1998.4084. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch T, Marchetti P, Susin S A, Dallaporta B, Zamzami N, Marzo I, Geuskens M, Kroemer G. The apoptosis-necrosis paradox. Apoptogenic proteases activated after mitochrondrial permeability transition determine the mode of cell death. Oncogene. 1997;15:1753–1581. doi: 10.1038/sj.onc.1201324. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch T, Marzo I, Kroemer G. Role of the mitochrondrial permeability transition pore in apoptosis. Biosci Rep. 1997;17:67–76. doi: 10.1023/a:1027339418683. [DOI] [PubMed] [Google Scholar]
- 34.Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki H, Song J, Eckner R, Ugai H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama K K. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirshenbaum L A, Schneider M D. Adenovirus E1A represses cardiac gene transcription and reactivates DNA synthesis in ventricular myocytes, via alternative pocket protein- and p300-binding domains. J Biol Chem. 1995;270:7791–7794. doi: 10.1074/jbc.270.14.7791. [DOI] [PubMed] [Google Scholar]
- 38.Krishan A. A rapid flow cytometric analysis of the mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroemer G, Dallaporta B, Resche-Rigon M. The mitochrondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol. 1998;60:619–42. doi: 10.1146/annurev.physiol.60.1.619. :619–642. [DOI] [PubMed] [Google Scholar]
- 40.Lemasters J J, Nieminen A L, Qian T, Trost L C, Elmore S P, Nishimura Y, Crowe R A, Cascio W E, Bradham C A, Brenner D A, Herman B. The mitochrondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 41.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 43.Lin H J, Eviner V, Prendergast G C, White E. Activated H-ras rescues E1A-induced apoptosis and cooperates with E1A to overcome p53-dependent growth arrest. Mol Cell Biol. 1995;15:4536–4544. doi: 10.1128/mcb.15.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linder S, Popowicz P, Svensson C, Marshall M, Bondesson M, Akusjarvi G. Enhanced invasive properties of rat embryo fibroblasts transformed by adenovirus E1A mutants with deletions in the carboxy-terminal exon. Oncogene. 1992;7:439–443. [PubMed] [Google Scholar]
- 45.Lowe S W, Jacks T, Housman D E, Ruley H E. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci USA. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 47.Mal A, Poon R Y, Howe P H, Toyoshima H, Hunter T, Harter M L. Inactivation of p27Kep1 by the viral E1A oncoprotein in TGFbeta-treated cells. Nature. 1996;380:262–265. doi: 10.1038/380262a0. [DOI] [PubMed] [Google Scholar]
- 48.Martin S J, Reutelingsperger C P, McGahon A J, Rader J A, van Schie R C, LaFace D M, Green D R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–155. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marzo I, Brenner C, Zamzami N, Jurgensmeier J M, Susin S A, Vieira H L, Prevost M C, Xie Z, Matsuyama S, Reed J C, Kroemer G. Bax and adenine nucleotide translocator cooperate in the mitochrondiral control of apoptosis. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 50.McGill G, Shimamura A, Bates R C, Savage R E, Fisher D E. Loss of matrix adhesion triggers rapid transformation-selective apoptosis in fibroblasts. J Cell Biol. 1997;138:901–911. doi: 10.1083/jcb.138.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nevins J R. Adenovirus E1A: transcription regulation and alteration of cell growth control. Curr Top Microbiol Immunol. 1995;199:25–32. doi: 10.1007/978-3-642-79586-2_2. [DOI] [PubMed] [Google Scholar]
- 52.Nevins J R, Leone G, DeGregori J, Jakoi L. Role of the Rb/E2F pathway in cell growth control. J Cell Physiol. 1997;173:233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 53.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Persons D A, Mehaffey M G, Kaleko M, Nienhuis A W, Vanin E F. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol Dis. 1998;24:167–182. doi: 10.1006/bcmd.1998.0184. [DOI] [PubMed] [Google Scholar]
- 55.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 56.Pozzatti R, McCormack M, Thompson M A, Khoury G. The E1a gene of adenovirus type 2 reduces the metastatic potential of ras-transformed rat embryo cells. Mol Biol Cell. 1988;8:2984–2988. doi: 10.1128/mcb.8.7.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1994;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 59.Rao I, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. . (Erratum, 89:9974.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rooney, R. J. Unpublished data.
- 61.Rooney R J, Daniels R R, Jenkins N A, Gilbert D J, Rothammer K, Morris S W, Higgs D R, Copeland N G. Chromosomal location and tissue expression of the gene encoding the adenovirus E1A-regulated transcription factor E4F in humans and mice. Mamm Genome. 1998;9:320–323. doi: 10.1007/s003359900758. [DOI] [PubMed] [Google Scholar]
- 62.Rooney R J, Raychaudhuri P, Nevins J R. E4F and ATF, two transcription factors that recognize the same site, can be distinguished both physically and functionally: a role for E4F in E1A trans activation. Mol Cell Biol. 1990;10:5138–5149. doi: 10.1128/mcb.10.10.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rooney R J, Rothammer K, Fernandes E R. Mutational analysis of p50E4F suggests that DNA binding activity is mediated through an alternative structure in a zinc finger domain that is regulated by phosphorylation. Nucleic Acids Res. 1998;26:1681–1688. doi: 10.1093/nar/26.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos E, Tronick S R, Aaronson S A, Pulcianni S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB and Harvey-MSV transforming genes. Nature. 1982;298:343–347. doi: 10.1038/298343a0. . (Abstract.) [DOI] [PubMed] [Google Scholar]
- 65.Schmid I, Krall W J, Uittenbogaart C H, Braun J, Giorgi J V. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13:204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 66.Schmid I, Uittenbogaart C H, Giorgi J V. Sensitive method for measuring apoptosis and cell surface phenotype in human thymocytes by flow cytometry. Cytometry. 1994;15:12–20. doi: 10.1002/cyto.990150104. [DOI] [PubMed] [Google Scholar]
- 67.Shisler J, Duerksen-Hughes P, Herminston T M, Wold W S, Gooding L R. Induction of susceptibility to tumor necrosis factor by E1A is dependent on binding to either p300 or p105-Rb and induction of DNA synthesis. J Virol. 1996;70:68–77. doi: 10.1128/jvi.70.1.68-77.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Somasundaram K, el-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 69.Susin S A, Zamzami N, Castedo M, Hirsch T, Marchetti P, Macho A, Daugas E, Geuskens M, Kroemer G. Bcl-2 inhibits the mitochrondrial release of an apoptogenic protease. J Exp Med. 1996;184:1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor D A, Kraus V B, Schwarz J J, Olsen E N, Kraus W E. E1A-mediated inhibition of myogenesis correlates with a direct physical interaction of E1A12S and basic helix-loop-helix proteins. Mol Cell Biol. 1993;13:4714–4727. doi: 10.1128/mcb.13.8.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Engeland M, Nieland L J, Ramaekers F C, Schutte B, Reutelingsperger C P. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31:1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 72.Verems I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 73.Weigel R J, Devoto S H, Nevins J R. Adenovirus 12S E1A gene represses differentiation of F9 tetratocarcinoma cells. Proc Natl Acad Sci USA. 1990;87:9878–9882. doi: 10.1073/pnas.87.24.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White E. Regulation of p53-dependent apoptosis by E1A and E1B. Curr Top Microbiol Immunol. 1995;199:34–58. [PubMed] [Google Scholar]
- 75.Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins J R, Williams R S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu D, Scorsone K, Hung M C. Adenovirus type 5 E1A gene products act as transformation suppressors of the neu oncogenes. Mol Biol Cell. 1991;11:1745–1750. doi: 10.1128/mcb.11.3.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu D, Shi D, Scanlon M, Hung M C. Reexpression of neu-encoded oncoprotein counteracts the tumor-suppressing but not the metastasis-suppressing function of E1A. Cancer Res. 1993;53:5784–5790. [PubMed] [Google Scholar]
- 78.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 79.Zamzami N, Hirsch T, Dallaporta B, Petit P X, Kroemer G. Mitochondrial implication in accidental and programmed cell death: apoptosis and necrosis. J Bioenerg Biomembr. 1997;29:185–193. doi: 10.1023/a:1022694131572. [DOI] [PubMed] [Google Scholar]
- 80.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]