Abstract
Gastric cancer (GC) is one of the most common malignancies, with an ever-increasing incidence and high mortality rate. Chromobox4 (CBX4), also named hPC2, is a small ubiquitin-related modifier (SUMO) E3 ligase. Previous studies have found that high CBX4 expression is associated with tumor size, pathologic differentiation and decreased patient survival in hepatocellular carcinoma (HCC). However, the expression and prognostic value of CBX4 in GC have not been clarified. In our study, ONCOMINE, UALCAN, Kaplan-Meier Plotter, cBioPortal, DAVID 6.8 and TIMER were utilized. RT-PCR, immunohistochemistry (IHC), Western blot, CCK-8 assay, cell apoptosis assay, cell cycle assay were used to further verify in GC tissue samples or cell line. The transcriptional and protein level of CBX4 in GC tissues was found significantly elevated and a significant association between the expression of CBX4 and clinicopathological parameters was found in GC patients. Low expression of CBX4 in GC patients were correlated with a significantly improved prognosis. The functions of CBX4 are primarily related to the stem cell pluripotency signaling pathway, Hippo signaling pathway, HTLV-I infection, Notch signaling pathway, and N-glycan biosynthesis. Our results may provide novel insights for the selection of therapeutic targets and prognostic biomarkers for GC.
Keywords: Bioinformatics analysis, Chromobox4 (CBX4), Gastric cancer, Prognostic biomarker, Therapeutic target
Introduction
Gastric cancer (GC) is one of the most common malignant gastrointestinal cancers in the world.1, 2, 3, 4, 5 Both the mortality and morbidity of GC rank second among those of malignant tumors.1 Although considerable advances in both diagnosis and adjuvant therapy have been made, the pathogenesis of GC is still not completely clear.
Polycomb group (PcG) complexes are epigenetic regulatory complexes, and their dysregulation is associated with many types of cancer.6, 7, 8 Chromobox (CBX) family proteins are canonical components of PcG complexes that regulate tumorigenesis and the progression of many cancers, including hepatocellular carcinoma (HCC), by inhibiting the cell differentiation and self-renewal of cancer stem cells.9,10 CBX4, also named hPC2, is a small ubiquitin-related modifier (SUMO) E3 ligase. Previous studies found that CBX4 was overexpressed in clinical tissues and multiple HCC cell lines. High CBX4 expression was associated with tumor size, pathologic differentiation and decreased patient survival.11 In addition, CBX4 is involved in growth control.12 Taken together, these findings indicate that CBX4 may play a role in human cancers. However, its specific function in the tumorigenesis and progression of GC has not been documented.
To explore the role of CBX4 in the tumorigenesis and progression of GC, we analyzed the expression and mutation of CBX4 and its correlations with clinical parameters in GC patients. Furthermore, we also analyzed mutations in CBX4 and the predicted functions and pathways enriched in CBX4 and its 50 most frequently altered neighboring genes.
Materials and methods
ONCOMINE
ONCOMINE (www.oncomine.org) is a comprehensive online cancer microarray database that provides reliable data from genome-wide expression analysis of RNA or DNA sequences.13 In this study, we used the ONCOMINE database to evaluate the transcriptional expression of CBX4 in GC tissues and corresponding normal tissues. The significance thresholds were set as a P value of 0.01, a fold change of 1.5, a gene rank in the top 10% and a data type of mRNA. Student's t-test was utilized to analyze differences in the transcriptional expression of CBX4 between GC and normal tissues.
UALCAN
UALCAN (http://ualcan.path.uab.edu) is an integrated website that facilitates analysis of level 3 RNA sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) database. It can be used to analyze the relative transcriptional expression of potential genes of interest in cancer tissues and normal control tissues and the association of transcriptional expression with relative clinicopathologic parameters.14 In our study, the mRNA expression of CBX4 in GC tissue and its associations with clinicopathologic parameters were obtained by using the “Expression Analysis” module in UALCAN. Student's t-test was used to analyze differences in transcriptional expression, and the P value cutoff was 0.01.
Kaplan-Meier plotter
Kaplan-Meier plotter (http://kmplot.com/analysis/) is a website from which data regarding the association of gene expression with the survival of patients with ovarian cancer, GC, breast cancer, lung cancer and liver cancer can be easily accessed.15, 16, 17, 18 Cancer patients were divided into high and low expression groups based on median mRNA expression values obtained from Kaplan-Meier plotter and validated by Kaplan-Meier survival curves. Information about the number of at-risk cases, median mRNA expression levels, HRs, 95% CIs and P-values were found at the Kaplan-Meier plotter webpage. A statistically significant difference was indicated by a P value < 0.05. In our study, we used Kaplan-Meier plotter to analyze the prognostic value of CBX4 mRNA expression in GC.
cBioPortal
cBioPortal (www.cbioportal.org), an open-access web resource, facilitates the exploration, visualization, and analysis of multidimensional cancer genomics data.19 Genetic alterations were obtained from cBioPortal based on data from TCGA. In our study, 412 GC (TCGA, provisional) cases were analyzed. Mutations and putative copy-number alterations from GISTIC and mRNA expression z-scores (RNA Seq V2 RSEM) were obtained using a z-score threshold of ±1.8.
STRING
STRING (https://string-db.org/) is a comprehensive online database that aims to collect, score and integrate all publicly available information regarding protein–protein interactions (PPIs) and to supplement these data with computational predictions of potential functions.20 STRING was utilized to conduct a PPI network analysis of CBX4 to discover its interactions with other proteins.
DAVID 6.8
DAVID 6.8 (https://david.ncifcrf.gov/home.jsp) is a comprehensive and functional annotation website that helps investigators better clarify the biological functions of submitted genes.21 In our study, Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis ofCBX4 and closely related neighboring genes was conducted with DAVID 6.8. The results of GO enrichment analysis included information regarding biological processes (BPs), cellular components (CCs) and molecular functions (MFs).
TIMER
TIMER (https://cistrome.shinyapps.io/timer/) is a online resource help researchers to evaluate the infiltration of different immune cells and their clinical impact.22 In our study, we utilized “Gene module” to evaluate the association of CBX4 expression level and the infiltration of immune cells, and “Survival module” to assess the correlation among clinical outcome and the infiltration of immune cells and CBX4 expression.
Tissue samples
Four pairs of GC tissue specimens and corresponding normal peripheral tissue (more than 5 cm from tumor verge) from GC patients were collected by the Renji hospital (Shanghai, China) and were well-documented with clinical information. Tissue specimens were confirmed by pathological examination.
RNA extraction and RT-PCR
Briefly, total RNA was isolated from GC tissues and corresponding normal peripheral tissues using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized using the Prime Script 1st Strand cDNA Synthesis Kit (Takara D6110A). RT-PCR was performed with SYBR Green premix Ex Taq (TaKaRa, RR420A). The expression level of genes was measured using the comparative Ct method. The primers used to amplify the indicated genes are shown in Supplementary Materials and Methods.
Western blot
The whole cell lysates were isolated using RIPA buffer (Beyotime P0013C). Equal quantities of protein were resolved on 12% SDS-PAGE and transferred onto PVDF membranes (Millipore, ISEQ00010). The membranes were blocked using 5% non-fat milk, and were incubated with primary antibodies anti-CBX4 (1:500 Abcam, ab242149), anti-BMI-1 (1:500 Abcam, ab126783) and GADPH (1:5000 BioTNTabcam, A20120A0701) followed by horseradish peroxidase-conjugated secondary antibody (1:5000) 2 h. Protein bands were visualized using enhanced chemiluminescence (ECL) reagents (GE, 5978613).
Immunohistochemistry
Paraffifin-embedded sections were deparaffifinized and rehydrated. Endogenous peroxidase activity was blocked by incubation in 0.3% hydrogen peroxide for 30 min, then the slides were microwaved and boiled in 10 mM citrate buffer (pH 6.0) for antigen retrieval. Nonspecific antigens were blocked by incubation in sheep serum for 30 min. Slides were incubated overnight at 4°C with anti-CBX4 (1:100, Abcam, ab242149) and anti-BMI-1 (1:100, Abcam, ab126783).
CBX4 knockdown using siRNAs
Three CBX4 small interfering RNAs (siRNAs) and negative control siRNAs were purchased from Shanghai Genepharma RNAi Company (Shanghai, China). Each siRNA was transfected into gastric cancer cells MKN74 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The volume ratio of Lipofectamine 2000 and siRNA was 1:1. At 48 h after transfection, the treated cells were harvested and the silencing of CBX4 was confirmed by qRT-PCR and Western blot.
Cell proliferation CCK-8 assay
Gastric cancer cells MKN74, transfected with CBX4-siRNA and negative control siRNAs for 48 h, were detached using trypsin (S330JV, Shanghai Basal Media Technologies Co. LTD) and incubated in a 96-well plate (Corning Inc, New York, USA) at a density of 2 × 103 cells in 100 μl. After culturing for 48 h, the supernatant was removed and cell growth was detected using Cell Counting Kit-8 Kit (C0037, Beyotime, China) according to the manufacturer's instructions. Absorbance was measured at 450 nm using a microplate reader. All experiments were performed in triplicate and repeated at least three times.
Cell apoptosis assay
For the apoptosis experiment, after 48 h of transfection, cells were cultured in 37 °C incubator for 48 h. The fluorescein isothiocyanate Annexin V Apoptosis Detection kit (C1062S, Beyotime, China) was used. Briefly, the cells were collected and centrifuged at 2000×g for 5 min. Then the cells resuspended in 500 μl binding buffer, supplemented with 5 μl Annexin V and 10 μl propidium iodide (PI), for 15 min of dark treatment at the room temperature. The flow cytometry (FACS Calibur, BD Bioscience, USA) was utilized to analyze the samples.
Cell cycle assay
For cell cycle assay, 48 h after transfection, the cells were fixed in 70% ethanol at 4 °C in the refrigerator for 12–24 h. After that, cells were treated with staining solution, which containing 50 µg/ml propidium iodide (PI) (Biolegend, California, USA) and 1 mg/ml RNase A (IBD LSRⅡ, San Jose, CA, USA). Each experiment was repeated three times.
Statistical analyses
We utilized Cox regression analysis to assess the correlation of patient survival with mRNA expression of CBX4 by SPSS software version 20.0. First, we imputed missing covariates with methods similar to the methods of White.23 In addition, influence of mRNA expression of CBX4 and clinical parameters on survival of GC patients was assessed by univariate Cox regression and it was followed by a filter which reserved those with P ≤ 0.1 for subsequent analysis. Finally, we further analyzed correlation of patient survival with mRNA expression of CBX4 with multivariate Cox regression which was adjusted for other parameters (e.g., tumor grades and individual cancer stages) similar to the methods of Hou.24 Statically significant difference was considered when a P value < 0.05.
Results
Overexpression of CBX4 in patients with GC
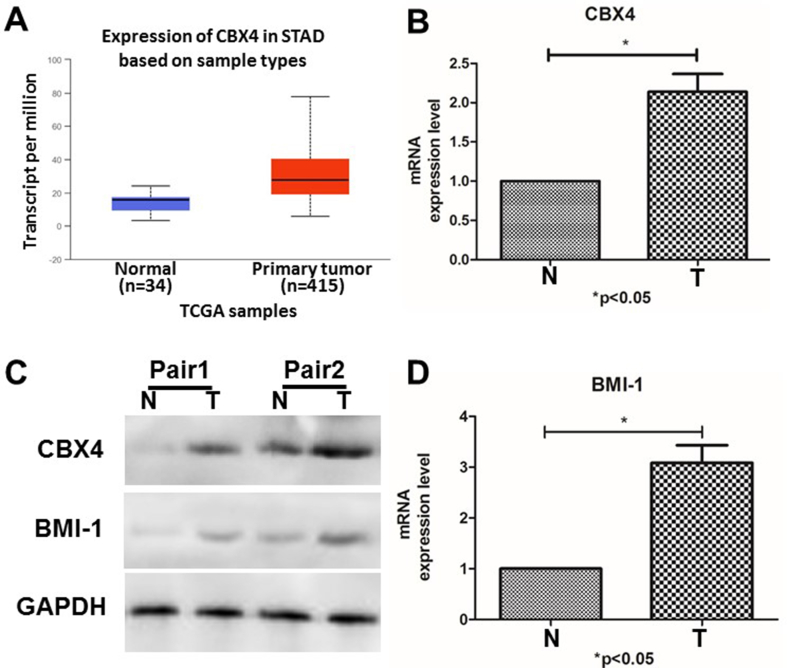
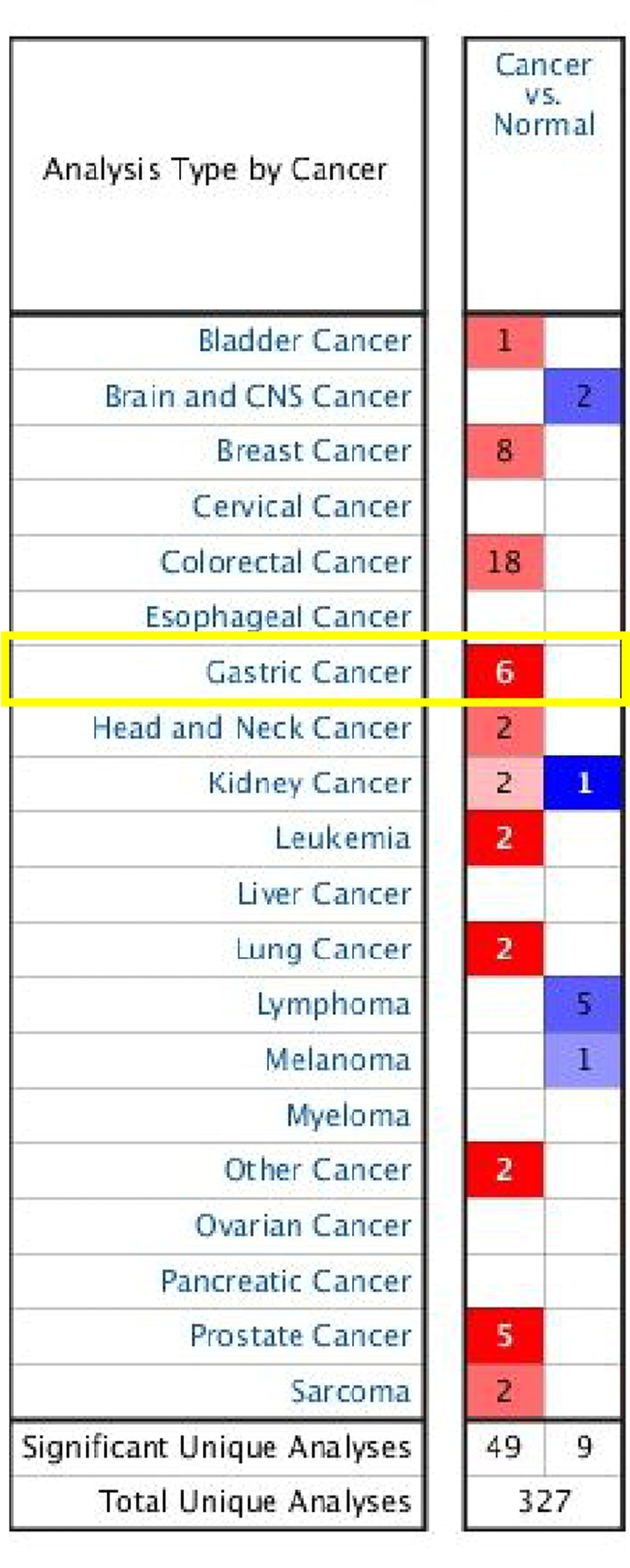
To explore the mRNA and protein expression of CBX4 in GC patients, we used the ONCOMINE database and UALCAN for data retrieval. We first utilized the ONCOMINE database to evaluate the mRNA expression of CBX4 in 20 types of cancers and normal samples. As shown in Supplemental Figure 1, the mRNA expression of CBX4 was significantly upregulated in GC tissues compared to normal tissues. Next, we used UALCAN to further evaluate the mRNA expression pattern of CBX4. As presented in Fig. 1A, significantly higher mRNA expression of CBX4 was observed in GC tissues than in normal tissues (P < 0.05).
Figure 1.
The expression of CBX4 in GC tissues (T) and corresponding adjacent normal gastric tissues (N). (A) CBX4 mRNA expression was increased in primary GC tissues compared to normal tissues. (UALCAN)∗∗∗ P < 0.001. (B) The mRNA expression of CBX4 was elevated in GC tissues compared to normal tissues. (C) The protein expression of CBX4 and BMI-1 was increased significantly in GC tissues compared to normal tissues (pair 1 and pair 2). (D) The mRNA expression of BMI-1 was increased in GC tissues compared to normal tissues.
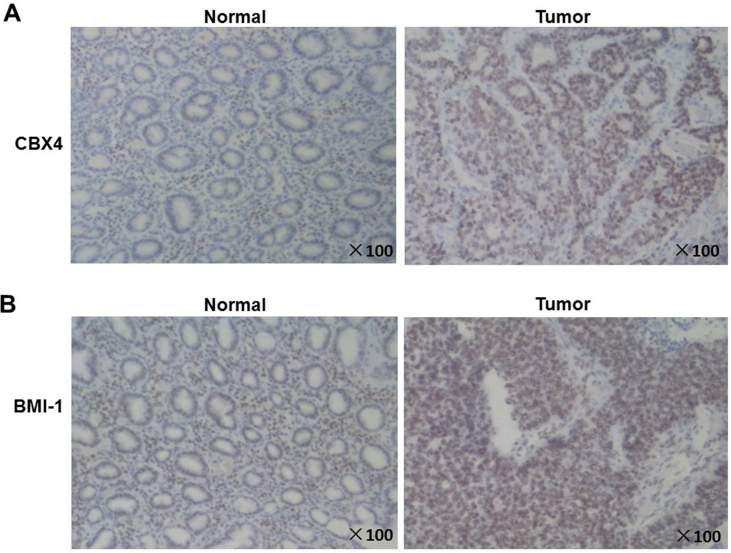
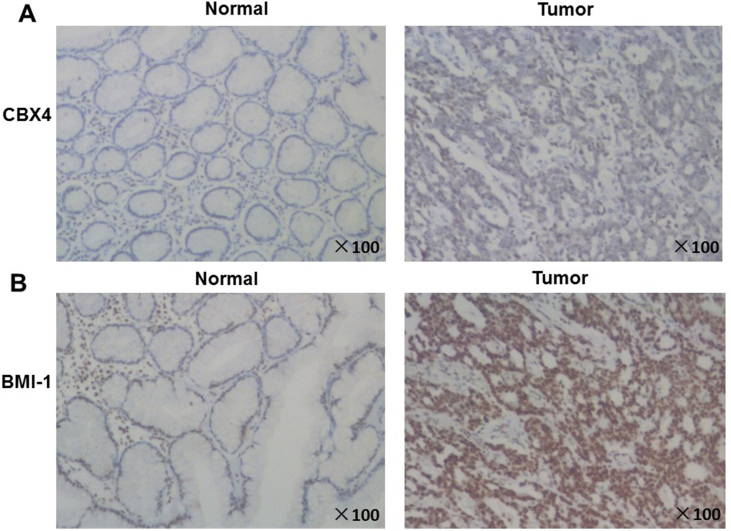
To verify these results, we detected the expression of CBX4 at mRNA level by RT-PCR and protein level by Western blot in four pairs of GC tissue specimens and corresponding normal peripheral tissue from GC patients. As for mRNA level, it increased to 2.139875225 folds in tumor tissues, compared with adjacent tissues (P = 0.0154, 95% CI -1.863 to −0.4137) (Fig. 1B). For protein level, the expression also increased significantly in gastric cancer (Fig. 1C and Fig. S2). To explore more, we did immunohistochemistry (IHC) in both tumor tissues and adjacent tissues from three GC patients. As shown in Fig. 2 and Supplementary materials (Fig. 3), in all the GC patients, CBX4 was located in nucleus; It was strongly expressed in tumor tissues, but low in adjacent tissues.
Figure 2.
Representative immunohistochemistry images of CBX4 and BMI-1 in GC tissues (T) and corresponding adjacent normal gastric tissues (N). (A) CBX4 was located in nucleus and it was strongly expressed in tumor tissues compared to adjacent normal tissues. (B) BMI-1 was located in nucleus and it was strongly expressed in tumor tissues compared to adjacent normal tissues. Expression trend of BMI-1 in GC was consistent with that of CBX4. Therefore, it was further confirmed that BMI-1 may be involved in the mechanisms of CBX4 in GC.
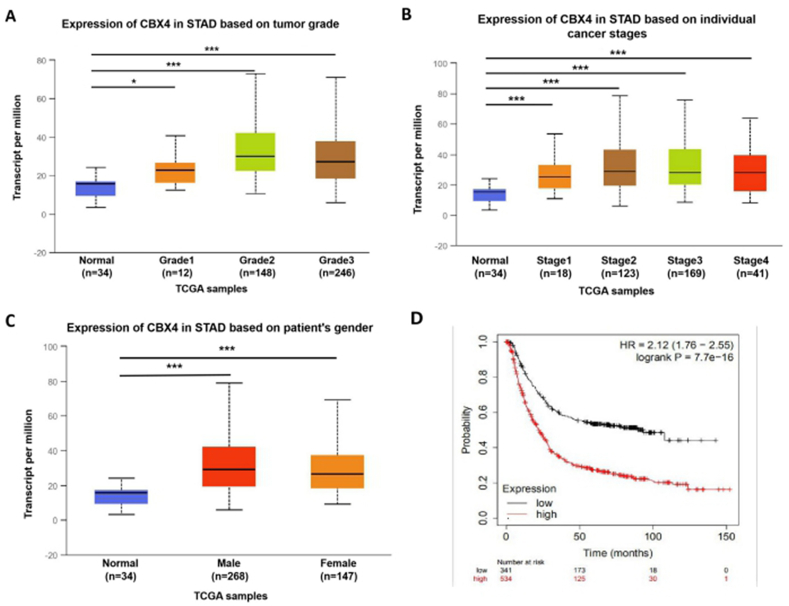
Figure 3.
Relationships between the expression of CBX4 and clinicopathological parameters in GC patients. (A) CBX4 expression was significantly associated with tumor grade, and as tumor grade increased, the expression of CBX4 tended to increase. The highest expression of CBX4 was discovered in grade 2 tumors. ∗P < 0.05, ∗∗∗P < 0.001. (B) CBX4 expression was remarkably related to individual cancer stage in GC patients, and patients with cancer at a more advanced stage tended to express higher levels of CBX4. The highest CBX4 expression was found in patients with stage 3 cancer. ∗∗∗P < 0.001. (C) There was no association between the expression of CBX4 and patient's gender. (D) Higher CBX4 expression was significantly correlated with a shorter overall survival (OS) in GC patients.
In summary, our results illustrated that transcriptional and protein expression of CBX4 was increased in patients with GC.
Relationships between the expression of CBX4 and clinicopathological parameters in patients with GC
After the mRNA and protein expression of CBX4 were discovered to be upregulated in patients with GC, UALCAN was used to analyze the association of CBX4 expression with the following clinicopathological parameters of GC patients: tumor grade, individual cancer stage and patient's gender. As shown in Fig. 3A, the expression of CBX4 was significantly associated with tumor grade, and as tumor grade increased, the expression of CBX4 tended to increase. The highest CBX4 expression was found in grade 2 tumors. Similarly, as shown in Fig. 3B, the expression of CBX4 was remarkably correlated with patients' individual cancer stage, and patients with cancer in a more advanced stage tended to express higher levels of CBX4. The highest CBX4 expression was found in patients with stage 3 cancer. The expression of CBX4 in stage 3 cancer seemed to be higher than that in stage 4 cancer, which may be attributed to the smaller sample size (41 GC patients had stage 4 GC). As for patient's gender, there was no association between the expression of CBX4 and patient's gender (Fig. 3C). In brief, the results above showed that the expression of CBX4 was remarkably related to clinicopathological parameters in patients with GC.
Furthermore, the prognostic value of CBX4 mRNA expression in patients with GC was analyzed by Kaplan-Meier plotter. We explored the association between CBX4 mRNA expression and the prognosis of patients with GC. As shown in Fig. 3D, higher CBX4 expression (HR = 2.12, 95% CI: 1.76–2.55, and P = 7.7e-16) was significantly correlated with shorter overall survival (OS) in patients with GC. These results suggested that the expression of CBX4 is remarkably related to GC patient prognosis and that CBX4 might be developed as a useful biomarker to predict GC patient survival.
Genetic mutation, co-expression and interaction analyses of CBX4 in patients with GC
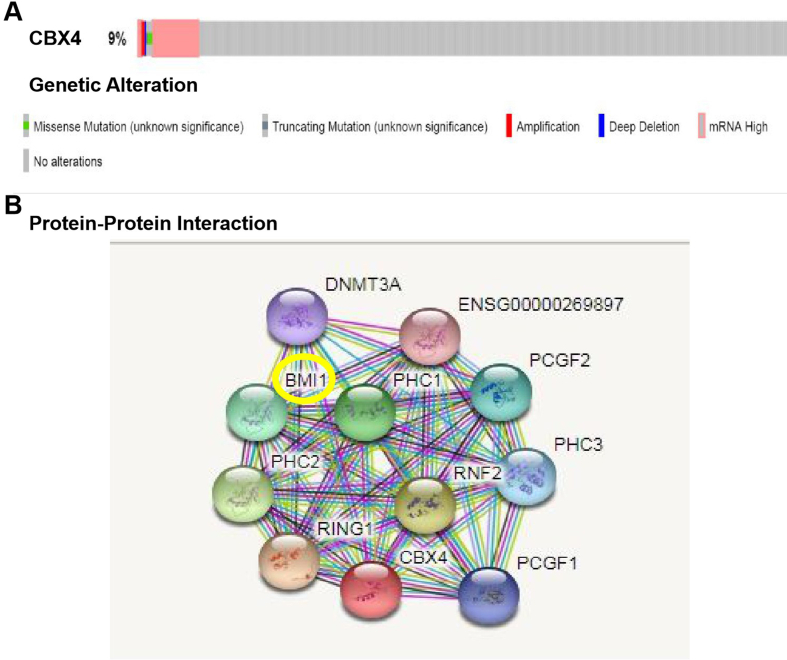
We comprehensively analyzed the molecular characteristics of CBX4. A provisional TCGA dataset was used to analyze the genetic alteration of CBX4. As shown in Fig. 4A, CBX4 was altered in 9% of the queried GC samples. Enhanced mRNA expression was the most common alteration in these samples.
Figure 4.
Genetic alteration of CBX4 and analysis of CBX4 interactions in GC patients. (A) CBX4 was altered in 9% of the queried GC samples. Enhanced mRNA expression was the most common alteration in these GC samples. (B) Several nodes (11) and several edges (54) were included in the PPI network. The function of CBX4 was associated with negative regulation of the G0 to G1 transition and histone ubiquitination.
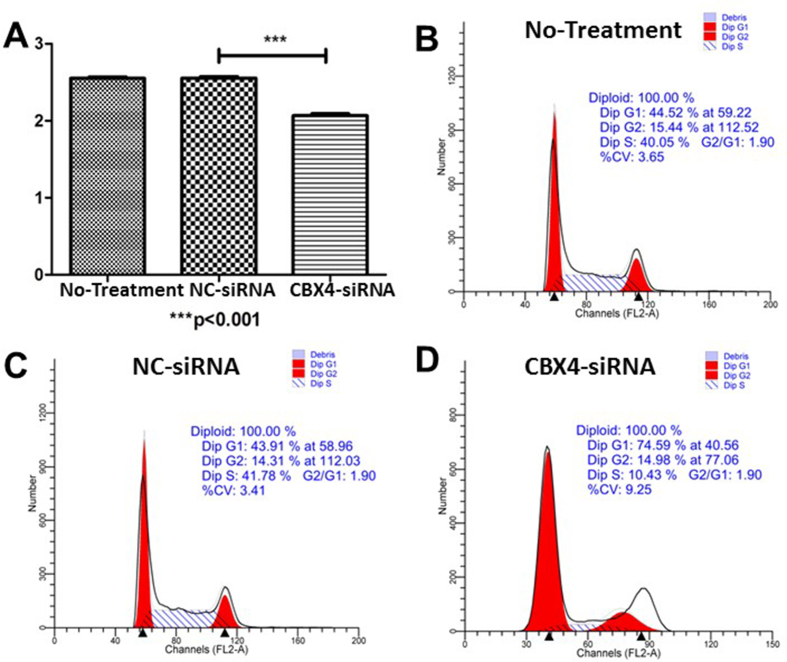
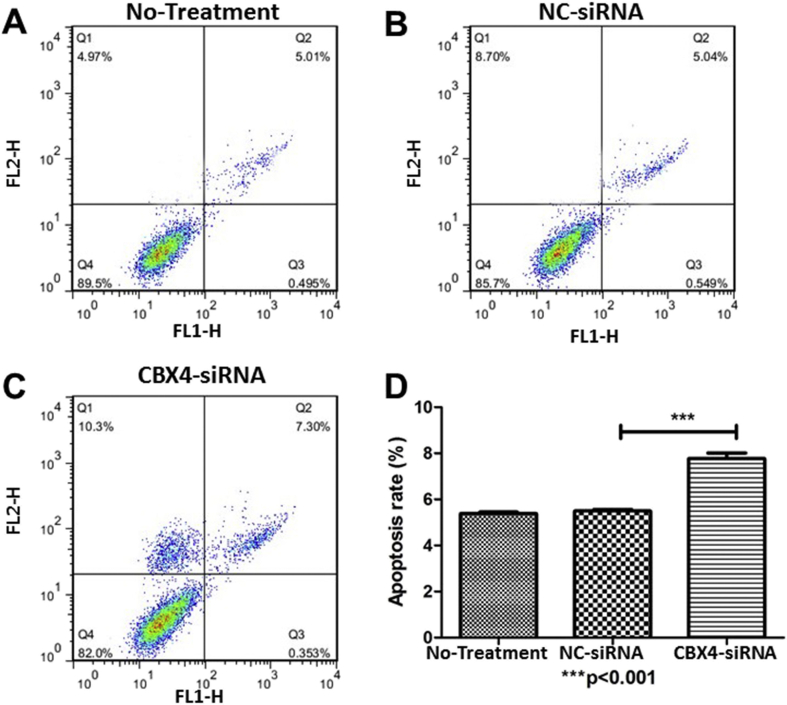
Then, we did in vitro experiments to explore the function of CBX4 in gastric cancer. After knocking down the expression of CBX4 by small interfering RNA in gastric cancer cell line MKN74, we detected cell viability, cell cycle and apoptosis. Our results showed that CBX4 siRNA transfection could inhibit cell viability (Fig. 5A), arrest the cell cycle in the G1 phase (Fig. 5B–D), and increase apoptosis rate (Fig. 6) in MKN74 cell line.
Figure 5.
The results of cell proliferation CCK8 assay and cell cycle assay. (A) Cell viability was inhibited after CBX4-siRNA transfection in MKN74. (B–D) The cell cycle of gastric cancer cell MKN74 was blocked in the G1 phase after CBX4-siRNA transfection.
Figure 6.
The results of cell apoptosis assay. (A–D) The number of apoptotic cells increased after CBX4-siRNA transfection.
In order to further explore the mechanism of CBX4 involved in gastric cancer, we performed PPI network analysis of CBX4 with STRING to investigate its potential interactions with other proteins. As expected, several nodes (11) and several edges (54) were included in the PPI network (Fig. 4B). According to the STRING analysis and literatures, we focused our eyes on B cell specific moloney murine leukemia virus integration site 1 (BMI-1). We detected BMI-1 expression at mRNA level by RT-PCR and protein level by Western blot in four pairs of GC tissue specimens and corresponding normal peripheral tissue from GC patients, together with CBX4. Our results suggested that, BMI-1 mRNA expression increased to 2.139875225 folds in tumor tissues, compared with adjacent tissues (p = 0.0154, 95% CI -1.863 to −0.4137) (Fig. 1B). And at protein level, the expression of BMI-1 also increased significantly in gastric cancer (Fig. 1C and Fig. S2) and IHC showed that BMI-1 was also located in nucleus in GC samples (Figure 1, Figure 2B). All those results were consistent with CBX4.
Functional enrichment analysis of CBX4 in patients with GC
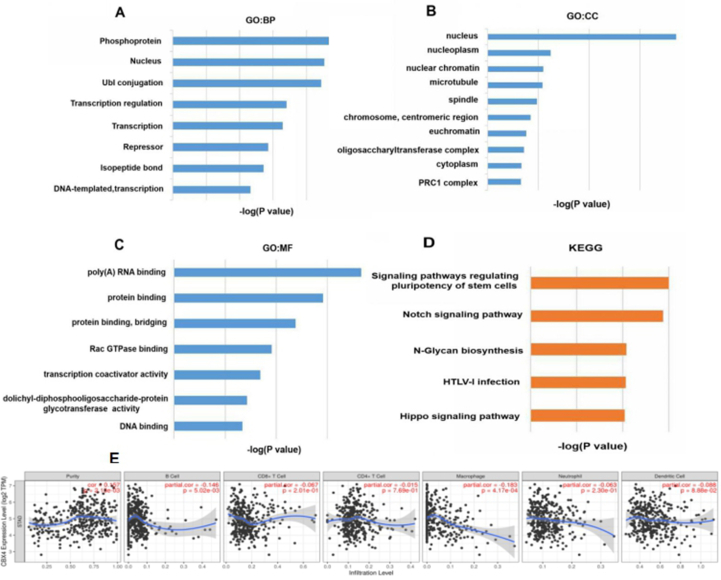
Metascape and DAVID 6.8 were used to analyze the functions of CBX4 and its neighboring genes. Fig. 7 shows the top 10 most highly enriched GO terms revealed using DAVID 6.8. As shown in Fig. 7A–C, among the 10 most highly enriched functions in the BP category, the phosphoprotein, nucleus, Ubl conjugation, transcription regulation and transcription BPs were remarkably enriched in CBX4 in GC. PRC1 complex, cytoplasm, oligosaccharyl transferase complex, euchromatin chromosome, centromeric region, spindle, microtubule, nuclear chromatin, nucleoplasm, and nucleus were the ten CCs most highly enriched in CBX4. Among MFs, CBX4 and its neighboring genes were mainly enriched in poly(A) RNA binding and protein binding. We also conducted KEGG pathway analysis, which revealed that the following 5 pathways were significantly enriched in CBX4 in GC: the stem cell pluripotency signaling pathway, Hippo signaling pathway, HTLV-I infection pathway, Notch signaling pathway, and N-glycan biosynthesis pathway (Fig. 7D). As for immune cell infiltration of CBX4 in GC patients. The TIMER database was used to explore the correlation between CBX4 and immune cell infiltration. We discovered that CBX4 expression was negatively associated with the infiltration of B cells (Cor = −0.146, P = 5.02E-3) and macrophages (Cor = −0.183, P = 4.17E-4) (Fig. 5E). We also assessed association of CBX4 with immune cell infiltration (Fig. 7E).
Figure 7.
Predicted functions and pathways enriched in CBX4 and its 100 most frequently altered neighboring genes in GC patients (DAVID 6.8) and immune cell infiltration of CBX4 in GC patients (TIMER). (A) Among the 10 most highly enriched functions in the BP category, the phosphoprotein, nucleus, Ubl conjugation, transcription regulation and transcription BPs were remarkably enriched in CBX4 in GC. (B) PRC1 complex, cytoplasm, oligosaccharyl transferase complex, euchromatin chromosome, centromeric region, spindle, microtubule, nuclear chromatin, nucleoplasm, and nucleus were the ten CCs most highly enriched in CBX4. (C) Among MFs, CBX4 and its neighboring genes were mainly enriched in poly(A) RNA binding and protein binding. (D) As for KEGG pathway analysis, the following 5 pathways were significantly enriched in CBX4 in GC: the stem cell pluripotency signaling pathway, Hippo signaling pathway, HTLV-I infection pathway, Notch signaling pathway, and N-glycan biosynthesis pathway. (E) CBX4 expression was negatively associated with the infiltration of B cells and macrophages.
Discussion
CBX4, a member of the CBX family, is a relatively unique PcG protein due to its SUMO E3 ligase activity, through which CBX4 plays significant roles in biological functions by influencing numerous key proteins, such as HIPK2 and CtBP.7,25 The role of CBX4 in cancers was first identified in hepatocellular cancer. Wang et al11 showed that knockdown of CBX4 resulted in inhibition of in vitro tumor growth. More recently, Li et al26 showed that CBX4 promoted the angiogenesis of hepatocellular cancer by regulating the proliferation, invasion and migration of tumor cells. However, the role of CBX4 in human GC has rarely been investigated.
In the present study, We first investigated the expression of CBX4. We discovered that CBX4 was upregulated in GC tissues compared with normal tissues. And the expression of CBX4 increased as the cancer progressed. Low CBX4 expression was significantly correlated with a more favorable OS in GC patients. Higher expression of CBX4 was associated with clinical parameters including tumor size, pathologic differentiation, shorter OS and RFS.27,28 What's more, the expression of CBX4 was remarkably associated with individual cancer stage and tumor grade in patients with GC. Furthermore, genetic alteration plays a significant role in tumorigenesis and the progression in GC. We explored the molecular characteristics of CBX4 in GC, and high CBX4 mutation rate (9%) was found. Enhanced CBX4 mRNA expression was the most common alteration. These results suggested that the expression of CBX4 is remarkably related to GC patient prognosis and that CBX4 might be developed as a useful biomarker to predict GC patient survival.
Then, we explored the predicted functions and pathways enriched in CBX4 and its neighboring genes. CBX4 is a specific PcG protein involved in tumor occurrence and cell cycle regulation. CBX4 expression varies in different types of cancer and has diverse biological functions. It is thought to be a cell cycle promoting gene with proliferative activity and can be up-regulated and exhibits pro-tumour effect by activating the HIF-1α signalling pathway in osteosarcoma.6,29,30 In our in vitro experiments, we found that knock-down of CBX4 expression by small interfering RNA could inhibit cell viability, arrest the cell cycle in the G1 phase, and increase apoptosis rate. Therefore, the expression level of CBX4 is associated with the malignant phenotype of gastric cancer cells.
Next, STRING was used to analyze PPI network of CBX4 to investigate its potential interactions with other proteins. According to the STRING analysis and literatures, BMI was noticed. BMI-1 is a polycomb ring finger oncogene which plays a crucial role in cell growth, metastasis and stem cell self-renewal.27,31, 32, 33, 34, 35 It has been reported that BMI-1 is a potential therapeutic target for glioma.14 Clinical studies revealed that BMI-1 expression was negatively correlated with survival of patient with colon cancer. In our study, BMI-1 was also detected together with CBX4 in GC tissues. Both its expression and location were consistent with those of CBX4. So it was further suggested that BMI-1 may be involved in the mechanisms of CBX4 in GC.
We then used GO enrichment analysis and KEGG pathway enrichment analysis to explore the function of CBX4. As expected, we discovered that the functions of these genes are primarily related to phosphoproteins, the nucleus, Ubl conjugation, transcription regulation, stem cell pluripotency signaling pathways, the Hippo signaling pathway, HTLV-I infection, the Notch signaling pathway and N-glycan biosynthesis. These data demonstrate that CBX4, which is overexpressed in GC, is a potential therapeutic drug target. Recently, CBX4 are widely acknowledged in all metazoans for its roles in lots of biological processes, such as cell cycle control, developmental controls, cell fate decisions, and maintenance of pluripotency and self-renewal in embryonic stem cells (ESCs) (Table 1). Furthermore, Li et al found that CBX4 enhanced angiogenesis of hepatocellular carcinoma by sumoylating of HIF-1α protein,26 which confirmed HIF-1α to be a new substrate for the SUMO E3 ligase activity of CBX4. We also sought to correlation between the expression of CBX4 and the infiltration of the six immune cell types. In this study, we found a significant correlation between the expression of CBX4 and the infiltration of the six immune cell types, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells, indicating that CBX4 is not only as prognostic indicators, but may also reflect immune status.
Table 1.
Pathways associated with CBX4.
Conclusion
In the present study, we analyzed the expression and mutation of CBX4 and its association with clinical parameters in patients with GC. Moreover, we explored the predicted functions and pathways enriched in CBX4 and its neighboring genes. The results of our study showed that CBX4 was overexpressed at the mRNA and protein levels in GC. A significant association between the expression of CBX4 and pathological stage was found in GC patients. Low transcriptional levels of CBX4 in GC patients were correlated with a significantly improved prognosis. The functions of CBX4 are primarily related to the stem cell pluripotency signaling pathway, Hippo signaling pathway, HTLV-I infection, Notch signaling pathway, and N-glycan biosynthesis. Taken together, these data demonstrate that CBX4, which is overexpressed in GC, is a potential therapeutic drug target. We hope our results provide new insights to assist in the design of new anticancer therapeutic drugs, to help clinicians choose proper drugs for their patients with GC, and to identify prognostic biomarkers to more accurately predict the survival of GC patients.
Conflict of Interests
All authors declare that there are no conflicts of interest related to the contents of this article.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.08.010.
Contributor Information
Zhijun Cao, Email: caozj_renji@163.com.
Shuliang Zhao, Email: shuliangzhao@126.com.
Funding
This project was supported by the National Natural Science Foundation of China [grant number 81972655], the Program for Young Eastern Scholars at the Shanghai Institutions of Higher Learning [grant number QD2016004], the Shanghai Science and Technology Commission Research Project [grant number 14441903103] and Shanghai Municipal Key Clinic Specialty.
Abbreviations
- GC
gastric cancer
- PcG
polycomb group
- CBX
chromobox
- TCGA
The Cancer Genome Atlas
- PPI
protein-protein interaction
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- BP
biological process
- CC
cellular component
- MF
molecular function
- OS
overall survival
Appendix A. Supplementary data
The following are the Supplementary data to this article:
1. Transcriptional expression of CBX4 in 20 different types of cancer (ONCOMINE database). The mRNA expression of CBX4 was significantly upregulated in GC tissues compared to normal tissues.
2. The protein expression of CBX4 and BMI-1in GC tissues and normal tissues (pair 3 and pair 4).
3. Immunohistochemistry images of CBX4 and BMI-1in GC tissues and normal gastric tissues in another pair.
4. The mRNA and protein expression of CBX4-siRNA in gastric cancer cell MKN74: Three different CBX4-siRNA were used and compared,; CBX4-siRNA2 was selected for the other in vitro experiments.
figs1.
figs2.

figs3.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Sitarz R., Skierucha M., Mielko J. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Z., Wu Y., Yang J., Yang D., Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7) doi: 10.1177/1010428317714626. e1010428317714626. [DOI] [PubMed] [Google Scholar]
- 4.Chon S.H., Berlth F., Plum P.S. Gastric cancer treatment in the world: Germany. Transl Gastroenterol Hepatol. 2017;2 doi: 10.21037/tgh.2017.05.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H.S., Kim W.H., Kwak Y. Molecular testing for gastrointestinal cancer. J Pathol Transl Med. 2017;51(2):103–121. doi: 10.4132/jptm.2017.01.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aloia L., Di Stefano B., Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140(12):2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 7.Müller J., Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19(2):150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Wang W., Qin J.J., Voruganti S., Nag S., Zhou J., Zhang R. Polycomb Group (PcG) proteins and human cancers: multifaceted functions and therapeutic implications. Med Res Rev. 2015;35(6):1220–1267. doi: 10.1002/med.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klauke K., Radulović V., Broekhuis M. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15(4):353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 10.Ma R.G., Zhang Y., Sun T.T., Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15(5):412–428. doi: 10.1631/jzus.B1400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B., Tang J., Liao D. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S684–S692. doi: 10.1245/s10434-013-3171-7. [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Lin C., Liu W. ncRNA- and Pc2 methylation dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes D.R., Yu J., Shanker K. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekar D.S., Bashel B., Balasubramanya S.A. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szász A.M., Lánczky A., Nagy Á. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Györffy B., Lanczky A., Eklund A.C. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 17.Gyorffy B., Lánczky A., Szállási Z. Implementing an online tool for genome-wide validation of survival associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 18.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Gable A.L., Lyon D. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Li T., Fan J., Wang B. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White I.R., Royston P. Imputing missing covariate values for the Cox model. Stat Med. 2009;28(15):1982–1998. doi: 10.1002/sim.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou X., He X., Wang K. Genome-wide network-based analysis of colorectal cancer identifies novel prognostic factors and an integrative prognostic index. Cell Physiol Biochem. 2018;49(5):1703–1716. doi: 10.1159/000493614. [DOI] [PubMed] [Google Scholar]
- 25.Richly H., Aloia L., Di Croce L. Roles of the polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2(9) doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Xu Y., Long X.D. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25(1):118–131. doi: 10.1016/j.ccr.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Zheng C., Li J., Wang Q. MicroRNA-195 functions as a tumor suppressor by inhibiting CBX4 in hepatocellular carcinoma. Oncol Rep. 2015;33(3):1115–1122. doi: 10.3892/or.2015.3734. [DOI] [PubMed] [Google Scholar]
- 28.Strand M.S., Lockhart A.C., Fields R.C. Genetics of gastric cancer. Surg Clin North Am. 2017;97(2):345–370. doi: 10.1016/j.suc.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Yang J., Cheng D., Zhu B., Zhou S., Ying T., Yang Q. Chromobox homolog 4 is positively correlated to tumor growth, survival and activation of HIF-1alpha signaling in human osteosarcoma under normoxic condition. J Cancer. 2016;7(4):427–435. doi: 10.7150/jca.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagey M.H., Melhuish T.A., Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113(1):127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 31.Ismail I.H., Gagne J.P., Caron M.C. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40(12):5497–5510. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X., Leung S.Y., Yuen S.T. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14(8):3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho J.Y., Lim J.Y., Cheong J.H. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mardaryev A.N., Liu B., Rapisarda V. Cbx4 maintains the epithe-lial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol. 2016;212(1):77–89. doi: 10.1083/jcb.201506065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao H.K., Xu Y., Li J. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis. 2015;6(3):e1689. doi: 10.1038/cddis.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]