Abstract
Although it is widely accepted that human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are readily available, robustly reproducible, and physiologically appropriate human cells for clinical applications and research in the cardiovascular field, hiPSC-CMs cultured in vitro retain an immature metabolic phenotype that limits their application, and little is known about the underlying molecular mechanism controlling mitochondrial metabolic maturation during human induced pluripotent stem cells (hiPSCs ) differentiation into cardiomyocytes. In this study, we found that peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) played an important role in inducing mitochondrial biogenesis and establishing oxidative phosphorylation (OXPHOS) during the cardiac differentiation of hiPSCs. Knocking down PGC-1α by siRNA impaired mitochondrial respiration, while upregulating PGC-1α by ZLN005 promoted mitochondrial biosynthesis and function by regulating the expression of downstream genes involved in mitochondrial dynamics and oxidative metabolism in hiPSC-CMs. Furthermore, we found that estrogen-related receptor α (ERRα) was required for the induction of PGC-1α stimulatory effects in hiPSC-CMs. These findings provide key insights into the molecular control of mitochondrial metabolism during cardiac differentiation and may be used to generate more metabolically mature cardiomyocytes for application.
Keywords: Cardiac differentiation, hiPSCs, Mitochondrial metabolism, PGC-1α, Maturation
Introduction
Despite advances in medical technology and treatment, heart disease remains the leading cause of mortality worldwide, partially because of the lack of appropriate human cardiomyocyte sources for repairing injured hearts.1 With the rapid development and improvement of cardiac differentiation2,3 and enrichment techniques,4 highly purified contracting cardiomyocytes can be obtained from human induced pluripotent stem cell (hiPSCs), which can be reprogrammed from many somatic cell sources, thus avoiding many ethical issues and immune rejection; therefore, hiPSCs play key roles in cardiac regeneration.5 Because cardiomyocytes can be derived from them, hiPSCs hold tremendous potential for clinical application, heart development, disease modelling, drug discovery and toxicity screenings in the cardiovascular field.6
It has been established that the metabolic transition from glycolysis to oxidative phosphorylation (OXPHOS) is necessary for cellular differentiation.7 The successful transition of PSCs to cardiomyocytes requires a switch from glycolytic metabolism to mitochondrial OXPHOS to meet the increasing demands for energy8,9; however, the specific molecular mechanisms of this process remain unknown. Additionally, human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) in traditional culture medium differ from adult cardiomyocytes in terms of structure, electrophysiology, calcium handling, and metabolism, with greater similarity to foetal cardiomyocytes,10, 11, 12 which limits their application. To address these limitations, various approaches have been employed to facilitate hiPSC-CM maturity with varying degrees of success.13 While most studies have focused on characterizing cardiomyocyte morphological, electrical and contractile phenotypes, mitochondrial metabolism and regulatory mechanisms in the cardiac differentiation of hiPSCs are incompletely understood. It is particularly important to clarify the specific role and mechanism of energy metabolism in regulating cell fate to further regulate the fate of cells through modifications to cell energy metabolism.
Peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) was originally identified as a coregulator of PPARγ in mitochondrial-rich brown adipocytes.14 PGC-1α interacts with and potentiates the activity of a diverse set of transcription factors and nuclear receptors, including nuclear respiratory factor 1 (NRF1) and ERRα, which collectively control the expression of a large number of genes involved in energy metabolism and mitochondrial biogenesis.15,16 For this reason, PGC-1α has been identified as a master transcriptional regulator of cellular metabolism.17 During heart development, a major metabolic switch from glycolytic to oxidative energy production occurs shortly after birth.18 Several studies have demonstrated that PGC-1α is a crucial regulator of oxidative metabolism in cardiac development, especially because it is required for mitochondrial biosynthesis during perinatal cardiac maturation.19, 20, 21 Furthermore, PGC-1α is also essential for normal cardiac energy and metabolic capacity, and loss of PGC-1α results in a reduction in the expression of genes involved in OXPHOS and a decrease in cardiac mitochondrial activity.22,23 However, the roles and regulatory mechanisms of PGC-1α in regulating metabolism during the cardiac differentiation of hiPSCs are not completely understood. Given the central role of PGC-1α in regulating energetic metabolism and mitochondrial biogenesis, we hypothesize that PGC-1α plays an essential role in regulating metabolic maturation during cardiac differentiation.
In this study, we have confirmed that the enhancement of mitochondrial respiratory function after hiPSCs differentiated into hiPSC-CMs is positively correlated with the protein expression of PGC-1α and its downstream proteins. Knocking down PGC-1α by siRNA impairs mitochondrial respiration, while the small-molecule compound ZLN005 promotes PGC-1α expression and the maturation of hiPSC-CM mitochondrial metabolism. The possible mechanism of PGC-1α regulation of mitochondrial metabolism involves the regulated expression of proteins involved in mitochondrial dynamics and oxidative metabolism, especially ERRα, which combines with PGC-1α to form a coactivation complex to regulate the expression of downstream proteins. This research has preliminarily indicated the importance of PGC-1α in inducing oxidative metabolism and mitochondrial biogenesis during cardiac differentiation of hiPSCs and revealed its possible regulatory mechanisms. These findings lay the foundation for increasing mitochondrial metabolic maturation and enhancing the applications of hiPSC-CMs.
Material and methods
Cell culture and cardiac differentiation
Undifferentiated hiPSCs (kindly provided by Cellapy, China) were maintained on Matrigel (Corning, USA)-coated plates in feeder-free culture conditions with fresh PGM1 PSC culture medium (Cellapy, China) and subcultured with ethylenediaminetetraacetic acid (EDTA) (Cellapy, China) every 3–4 days. The schematic shown in Fig. 1A illustrates the hiPSC culture, differentiation, and dissociation timeline. Cardiac differentiation methods were adapted from previously published manuscripts.2,24 After 3 days the cells were 95%~100% confluent, referred to as day 0, and the medium was replaced with RPMI 1640 medium (Sigma, USA) plus B27 supplement minus insulin (Thermo Fisher Scientific, USA). For the first 48 h, 6 μM CHIR-99021 (Selleck, USA) was added to the basal medium. Then, the medium was replaced with basal medium for another 24 h. On days 3–5, 5 μM IWP2 (Selleck, USA), a WNT inhibitor, was added to the medium. On day 7 of differentiation, the medium was changed to RPMI 1640 plus B27 supplement with insulin (Thermo Fisher Scientific, USA) and incubated for 4–5 days, at which time, it was replaced with cardiac enrichment medium and incubated for 3–4 days. The cardiac enrichment medium used was glucose-free RPMI 1640 medium (Thermo Fisher Scientific, USA) supplemented with 4 mM sodium l-lactate (Sigma–Aldrich, USA). After this enrichment phase, the medium was switched to RPMI 1640 plus knockout serum replacement (KSR) (Thermo Fisher Scientific, USA). Between days 20 and 25 of differentiation, spontaneously beating cardiomyocytes were dissociated with TrypLE Express enzyme (Thermo Fisher Scientific, USA), centrifuged, resuspended, and re-plated onto Matrigel-coated plates for the appropriate experiments. All cultures were grown at 37 °C in 5% oxygen and 5% CO2 conditions.
Figure 1.
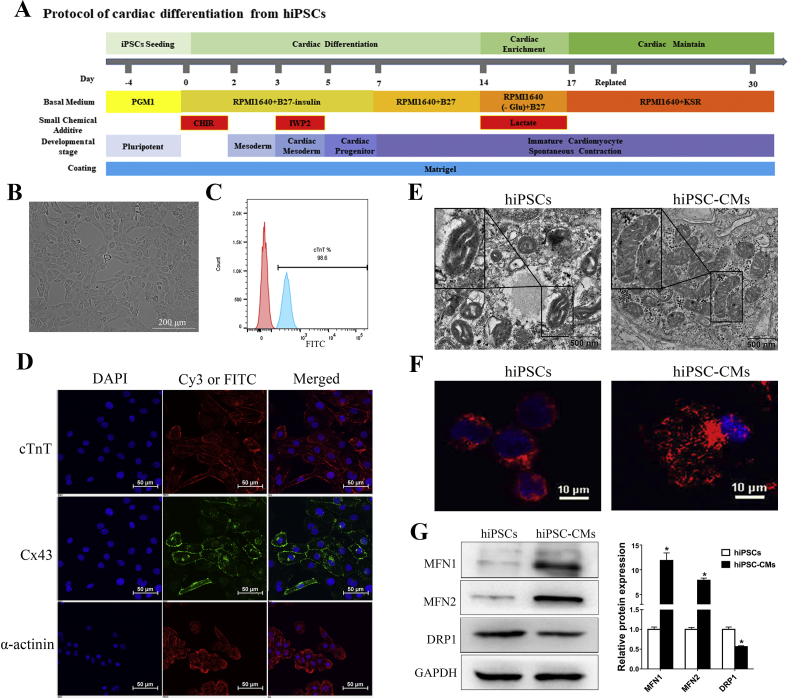
Mitochondrial morphology changes after the differentiation of hiPSCs into cardiomyocytes. (A) Schematic depicting the procedure for the generation of cardiomyocytes from hiPSCs via temporal modulation of Wnt/β-catenin signaling pathway, and metabolic purification of hiPSC-CMs. (B) The morphology of single cardiomyocytes from hiPSCs after metabolic purification and replating. (C) The efficiency of cardiac differentiation after metabolic purification was 98.6% measured by flow cytometry for cTnT positive on day 30 cells. (D) Immunofluorescence staining showing day 30-hiPSC-CMs expressing typical specific cardiomyocyte markers cTnT, α-actinin and Cx43. Nuclei are stained in blue with DAPI (blue). (E–F) Transmission electron microscopy (TEM) (E) and MitoTracker red (F) were used to investigate the mitochondrial morphology changes during differentiation. Note that in hiPSCs, mitochondria were round, contain poorly developed cristae networks, whereas in day 30-hiPSC-CMs, mitochondria were elongated, and contain dense cristae networks. (G) The expression of mitochondrial fusion proteins (MFN1 and MFN2) increased while mitochondrial fission protein DRP1 decreased after hiPSCs differentiated into cardiomyocytes. Compared with hiPSCs group, ∗P < 0.05.
Flow cytometry
HiPSCs and day 30-hiPSC-CMs were dissociated into single cells, washed with sterile DPBS and pelleted at 200×g for 5 min. Following fixation in 4% paraformaldehyde, the cells were washed twice with DPBS, pelleted and resuspended in 200 μl of DPBS. Fixed hiPSCs and hiPSC-CMs were incubated with fluorescently labelled anti-OCT4 and anti-cTnT antibodies (see Table S1) for 1 h at room temperature, respectively. The cells were then washed 3 times. Analysis was performed on BD FACSCanto II cytometer, and the results were analysed and plotted using FlowJo v10 software.
Immunofluorescence staining
HiPSCs and day 30-hiPSC-CMs were cultured on Matrigel-coated coverslips. The cells were washed with DPBS and fixed in 4% paraformaldehyde for 20 min. After 3 washes with DPBS, the cells were permeabilized with 0.5% Triton X-100 (Sigma–Aldrich, USA) for 15 min, blocked with 10% goat serum, and then incubated overnight with primary antibodies (anti-OCT4, anti-SOX2, anti-SSEA4, anti-TRA-1-60, anti-cTnT, anti-α-actinin, and anti-Cx43, see Table S1) at 4 °C. After 3 washes with DPBS, secondary antibodies conjugated to Cy3 or Alexa Fluor® 488 were incubated for 1 h at room temperature in the dark. After nuclei were stained with DAPI for 10 min, and the cells were washed 3 times with DPBS, micrographs were taken with a fluorescence microscope (BX51; Olympus).
Transmission electron microscopy
HiPSCs and day 30-hiPSC-CMs were immediately fixed with 4% glutaraldehyde solution for 2 h and then postfixed with 1% osmium tetroxide for 2 h at 4 °C. The fixed cells were rinsed with distilled water, dehydrated with an ethanol and methanol gradient, and embedded in epoxy resin. Subsequently, the samples were sectioned and contrast-stained for imaging. TEM images were acquired at random locations throughout the samples. Micrographs were taken with a transmission electron microscope (TEM; H-7500).
Mitochondrial staining
For mitochondrial imaging, the MitoTracker® red CMXRos (Thermo Fisher Scientific, USA) intracellular dye was used following the manufacturer's protocol. HiPSCs and day 30-hiPSC-CMs were incubated with 25 nM MitoTracker red for 30 min. Then, the nuclei were stained with Hoechst for 10 min. After washed with DPBS, the cells were imaged using light confocal microscopy (Nikon, Japan). The average fluorescence intensity was analysed using NIS-Elements Viewer software.
Transient transfection of siRNA
Cells were seeded onto 12-well plates and transfected with siRNA according to the manufacturer's instructions. Transfection of siRNA into day 27-hiPSC-CMs was performed utilizing RNAiMax reagent (Thermo Fisher Scientific, USA) at a final concentration of 100 nM. Protein collection was conducted 72 h after transfection. The siRNA sequences for PGC-1α were 5′-CUCGGAGCUUCUCAAAUAU tt-3′ and 5′-AUAUUUGAGAAGCUCCGAG tt -3’.
Seahorse XF24 metabolic flux analysis
The oxygen consumption rate (OCR) was determined using the Seahorse XF24 Extracellular Flux Analyzer (Agilent Technologies, USA). Undifferentiated hiPSCs were plated onto a Seahorse XF-24 cell culture plate 24 h before the analysis. Cardiomyocytes at differentiation day 25 were dissociated using TrypLE Express enzyme (Thermo Fisher Scientific, USA) and subsequently plated onto a Matrigel (1:50) precoated Seahorse XF-24 cell culture plate (Agilent Technologies, USA) at a density of 2.5 × 105 cells/well. After they resumed beating, the day 27-hiPSC-CMs were treated with a negative control, siRNA, DMSO control, ZLN005 or ZLN005 plus XCT790 for 3 days. Mitochondrial function was analysed using a XF Cell Mito Stress kit (Agilent Technologies, USA). One hour before the assay, the cells were washed once with XF assay medium (unbuffered DMEM + 10 mM glucose + 2 mM l-glutamine + 1 mM sodium pyruvate) and incubated in 500 μL base medium at 37 °C in a non-CO2 incubator. Three mitochondrial inhibitors—oligomycin (2.5 μM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP, 2.5 μM) and rotenone (0.5 μM) + antimycin A (0.5 μM)—were diluted in base medium and sequentially added into each well during the measurements. OCR (pmol/min) was measured according to the manufacturer's (Agilent Technologies) instructions. The results were normalized to 1 μg of protein as determined by a BCA protein assay kit (Beyotime Biotech, China).
Protein extraction and Western blot
The proteins were extracted from hiPSCs and day 30-hiPSC-CMs using RIPA reagent (Beyotime Biotech, China). Protein samples were mixed with 5 × buffer and boiled for 5 min before being loaded onto a 10% SDS-PAGE gel. After electrophoresis, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). According to markers, the membranes were cut into pieces and incubated in 5% non-fat milk in PBST for 1 h. The membranes were incubated overnight with primary antibodies (see Table S1) at 4 °C and then washed with PBST 3 times for 10 min each time. The membranes were incubated with the corresponding secondary antibody. Positive bands were detected by chemiluminescent reactions (Millipore, USA).
Quantitative real-time PCR
Total RNA preparations were obtained by using TRIzol reagent (TaKaRa, Japan) from hiPSCs and day 30-hiPSC-CMs, as described elsewhere. One microgram of total RNA was reverse transcribed to cDNA using the PrimeScript RT reagent kit (TaKaRa, Japan). Then, the resulting cDNA samples were amplified with gene-specific primers and a SYBR Green dye kit (TaKaRa, Japan). The forward and reverse primers for PCR are shown in Table S2. GAPDH served as an endogenous control. The relative amount of mRNA to endogenous control was calculated using the 2−ΔΔCT method.
Mitochondrial membrane potential assay
The mitochondrial membrane potential was measured using a mitochondrial membrane potential assay kit (JC-1, Beyotime Biotech, China) according to the manufacturer's instructions. HiPSCs and day 30-hiPSC-CMs were incubated with a JC-1 working solution for 30 min. After washing with DPBS, they were imaged using a light confocal microscope. The average fluorescence intensity was analysed using NIS-Elements Viewer software. The relative ratio between the monomeric (Em = 590 nm) and aggregate (Em = 525 nm) forms of JC-1 represents the change in the mitochondrial membrane potential. A high ratio indicates a high mitochondrial membrane potential.
Co-immunoprecipitation (co-IP) assay
Coimmunoprecipitation was conducted using a co-IP assay kit (Thermo Fisher Scientific, USA), and the total protein concentrations of the cell lysates were measured by BCA assay. An anti-PGC-1α antibody (see Table S1) was used to pull down PGC-1α, and an anti-IgG antibody was used as a negative control. The protein collected was analysed by Western blot analysis using an anti-ERRα antibody (see Table S1) to detect the co-combination between PGC-1α and ERRα.
Treatment of hiPSC-CMs with small-molecule compounds
Cardiomyocyte differentiation cultures were dissociated using TrypLE Express and re-plated onto plates at an appropriate density. HiPSC-CMs at differentiation day 27 were treated with the PGC-1α agonist ZLN005 (MCE) at different concentrations (0, 4, 8, and 16 μmol/L) for 72 h, and the optimal concentration was determined for the subsequent experiments. To evaluate the role of ERRα in PGC-1α-induced mitochondrial biogenesis, hiPSC-CMs were treated at differentiation day 27 with the following compounds for 3 days: (1) DMSO control, (2) ZLN005, and (3) ZLN005 + 10 μmol/L XCT790 (ERRα inhibitor, MCE);
Statistical analysis
Each experiment was repeated at least three times. All data are expressed as the means ± standard deviation (SD). Statistical signifcance was evaluated using either unpaired Student's t test or one-way analysis of variance (ANOVA). SPSS 17.0 software (SPSS Inc., Armonk, NY, USA) was used for the statistical analysis. For all analyses, a value of P < 0.05 was considered to be significant.
Results
Mitochondrial respiratory function increases after the differentiation of hiPSCs into cardiomyocytes
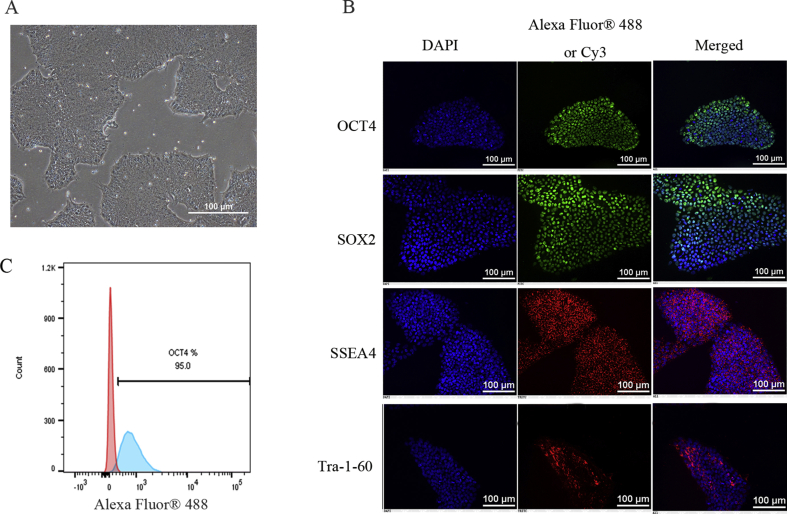
Because partially differentiated hiPSCs diminish cardiac differentiation efficiency, the hiPSCs that we employed were maintained in an undifferentiated state. They exhibited a uniform typical colony morphology (Fig. S1A) and expressed the pluripotent markers OCT4, SOX2, SSEA4, and TRA-1-60 (Fig. S1B). The pluripotent marker OCT4 was expressed in more than 95% of the cells, as assessed by flow cytometry (Fig. S1C). We used a modified monolayer cardiac differentiation protocol (Fig. 1A) to induce the hiPSCs to differentiate into cardiomyocytes. The spontaneous beating of cardiomyocytes was first visible at approximately day 8 (Movie S1), and the hiPSC-CMs displayed spontaneous rhythmic beating at approximately day 12 (Movie S2). After purification, the cardiomyocytes were digested into single cells and re-plated onto new plates (Fig. 1B). The flow analysis indicated that metabolic purification led to a purified population containing more than 95% cTnT-positive cells by day 30 (Fig. 1C). Immunofluorescence analysis confirmed that, on day 30-hiPSC-CMs were positively stained for the typical cardiomyocyte structural protein cardiac troponin T (cTnT), α-actinin and the cardiac-specific junction connexin 43 (Cx43) (Fig. 1D). We also observed well developed sarcomeres in day 30-hiPSC-CMs through transmission electron microscopy (TEM) (Fig. S2). On day 30, the cardiomyocytes were collected for functional analyses.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.gendis.2020.12.006
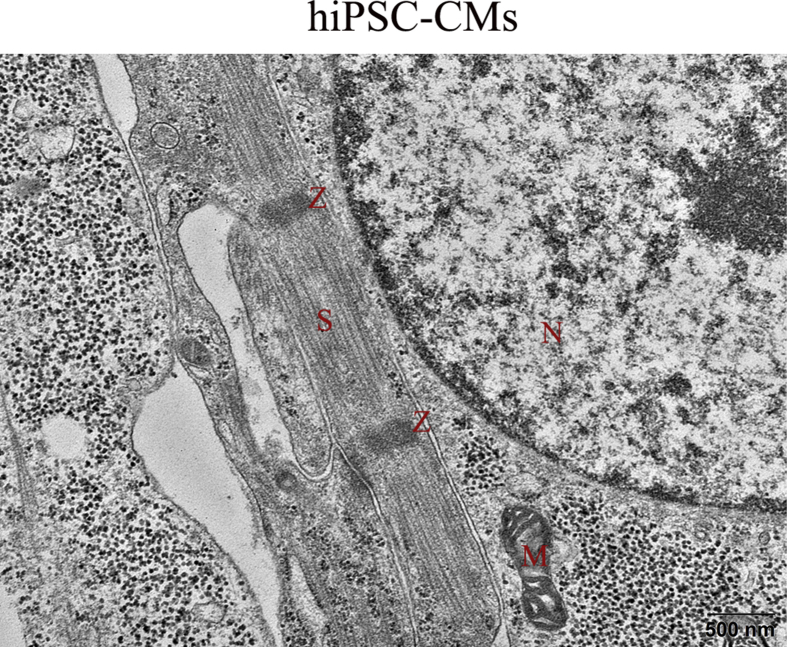
To observe changes in mitochondrial morphology during the cardiac differentiation of hiPSCs, the morphology of the mitochondria and expression of the proteins related to mitochondrial morphology were observed for the undifferentiated iPSCs and the derived cardiomyocytes. We used transmission electron microscopy to examine the mitochondrial ultrastructure. As expected, we observed some changes in the structure of mitochondria in the day 0-hiPSCs and the day30-hiPSC-CMs. The day30-hiPSC-CMs exhibited a few more mature elongated, large mitochondria with denser intramitochondrial cristae and compacted matrix that were observed in the day 0-hiPSCs, which had mostly immature round-shaped mitochondria with sparse cristae and an expanded matrix (Fig. 1E). Consistent with evidence of pro-fusion events in the day 30-hiPSC-CMs, images of the cellular distribution of the mitochondria identified with MitoTracker red revealed disparate mitochondrial morphologies in the day 0-hiPSCs and day 30-hiPSC-CMs, with the former containing isolated mitochondria with fragmented foci whereas the latter containing rod-shaped mitochondria (Fig. 1F). These results were consistent with previous findings showing substantial structural changes of mitochondria during cardiac differentiation.25,26 Mitochondrial morphology was associated with the expression of proteins involved in mitochondrial dynamics (fusion and fission). These changes in morphology were accompanied by significantly higher protein expression of the mitochondrial fusion proteins MFN1 and MFN2 and lower protein expression of the mitochondrial fission protein DRP1 in the cardiomyocytes than were observed in the undifferentiated hiPSCs (Fig. 1G).
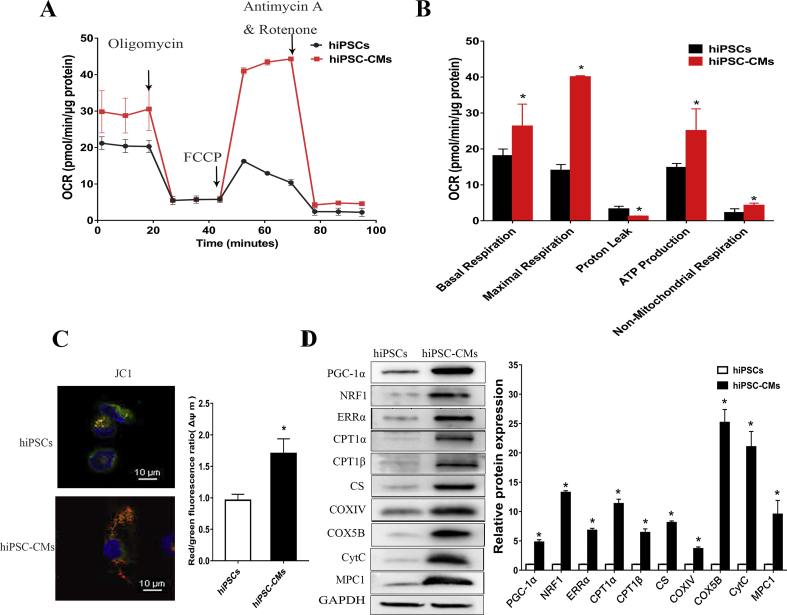
Cardiomyocyte metabolism is predominantly oxidative-based metabolism, which differs from the glycolysis-based metabolism evident in pluripotent stem cells.8 To compare the metabolic status of hiPSCs and differentiated cells, we measured the oxygen consumption rate (OCR) of the hiPSCs and day 30-hiPSC-CMs with a XF24 Extracellular Flux Analyzer, which indicated mitochondrial function. Fig. 2A shows representative traces of both hiPSCs and day 30-hiPSC-CMs. Basal respiration is usually controlled substantially by ATP turnover, partly by substrate oxidation and proton leak, and it was significantly increased in the hiPSC-CMs, as well as maximal respiration (Fig. 2B). In addition, day 30-hiPSC-CMs showed a trend for increased coupling to ATP synthesis when treated with the ATP synthase inhibitor oligomycin (Fig. 2B), indicating that hiPSC-CMs produce ATP mainly through OXPHOS. Non-mitochondrial respiration is mediated by various enzymes in the cell membrane and cytoplasm, which control detoxification and oxidation, and non-mitochondrial OCR also increased after the hiPSCs differentiated into cardiomyocytes (Fig. 2B). Furthermore, mitochondrial membrane potential (Δψm), which is a key indicator of the integrality of mitochondrial structure and function, was measured by fluorescence probe JC-1 assay. The results showed that Δψm increased after cardiac differentiation (Fig. 2C), and an increased Δψm could enhance the ATP generation capacity.
Figure 2.
Mitochondrial respiratory function increased after the differentiation of hiPSCs into cardiomyocytes. (A) Representative two OCR traces of day 0-hiPSCs and day 30-hiPSC-CMs respectively, in response to oligomycin, FCCP, and antimycin A. (B) OCR parameters representing mitochondrial function in day 0-hiPSCs and day 30-hiPSC-CMs. (C) The cellular ATP content of day 0-hiPSCs and day 30-hiPSC-CMs was measured using a bioluminescent assay system. (D) Mitochondrial membrane potential analysis was measured using a fluorescence probe JC-1 assay system in day 0-hiPSCs and day 30-hiPSC-CMs. The ratio of red/green fluorescence represented the level of Δψm. (E) The expression of PGC-1α and its downstream metabolic-related proteins increased after hiPSCs differentiate into cardiomyocytes. Protein bands were detected by Western blot analysis, and the proteins assessed were PGC-1α, NRF1, ERRα, CPT1α, CPT1β, CS, COXⅣ, COX5B, CytC and MPC1. Compared with hiPSCs group ∗P < 0.05.
To observe whether the metabolic changes observed after the hiPSCs differentiated into cardiomyocytes were related to PGC-1α, we assessed the protein expression of PGC-1α and its downstream oxidative metabolism-related proteins in day 0-hiPSCs and day 30-hiPSC-CMs by Western blot analysis. Western blot analysis revealed increased expression of PGC-1α and its downstream metabolic transcription factors NRF1 and ERRα in the hiPSC-derived cultures compared with that of the hiPSCs. In addition, the expression of carnitine palmitoyl transferase 1α and β (CPT1α and CPT1β), which regulate the entry of medium- and long-chain fatty acids into mitochondria, increased significantly. Citrate synthetase (CS), which is the first rate-limiting enzyme of the tricarboxylic acid (TCA) cycle, and the important proteins cytochrome c oxidase subunit Ⅳ and 5B (COXIV and COX5B) and cytochrome c (CytC) in the electronic respiratory chain (ETC) were also increased. Furthermore, the expression of the MPC1 protein, which mediates the transport of pyruvate into mitochondria increased (Fig. 2D). These results suggested that mitochondrial activity was positively correlated with the expression of PGC-1α and its downstream targets, which were involved in the TCA cycle, ETC, fatty acid oxidation and pyruvate transport, indicating that these mitochondrial functions might be induced by PGC-1α. These observations prompted us to investigate the role of PGC-1α in regulating the energy metabolism of cardiac differentiation.
PGC-1α plays an important role in cardiomyocyte mitochondrial respiratory function
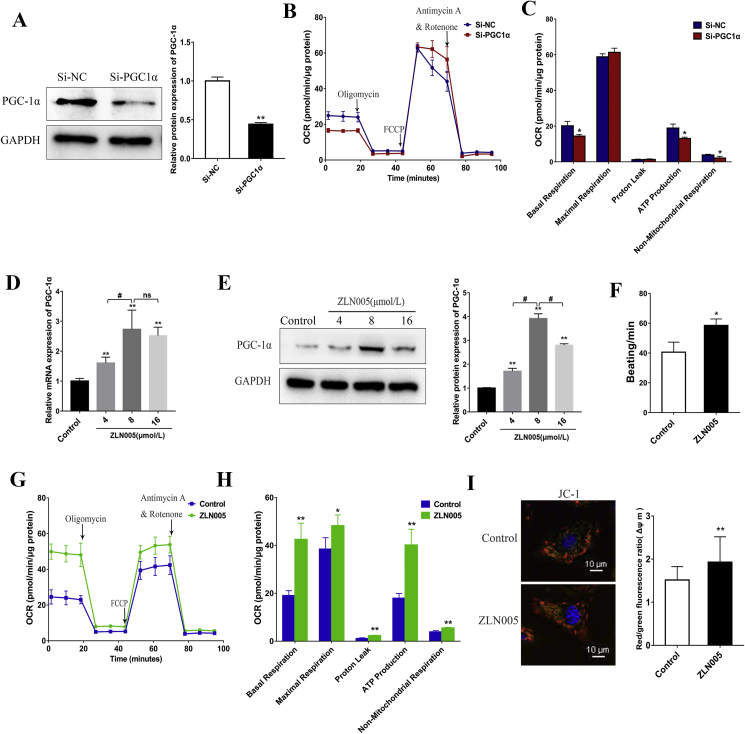
To evaluate the functional role of PGC-1α in the mitochondrial oxidative metabolism of hiPSC-CMs, we used siRNA to knock down PGC-1α expression. After transfection with PGC-1α siRNA, the expression of PGC-1α was assessed by western blot analysis. Protein level of PGC-1α was significantly suppressed by PGC-1α siRNA compared with that of NC siRNA (Fig. 3A). We then investigated the effect of the knockdown of PGC-1α on mitochondrial respiration function by the sequential addition of oligomycin, FCCP and antimycin A. We observed that the suppression of PGC-1α by siRNA added to the medium significantly decreased the OCRs, which are linked to basal respiration, ATP production, and non-mitochondrial respiration (Fig. 3B and C). The decreased values of these major mitochondrial oxidation parameters indicated a reduced mitochondrial oxidative capacity. The fact that PGC-1α-depleted cells functionally displayed impaired mitochondrial respiration, combined with the previous results, suggested that the enhancement of mitochondrial oxidative metabolism of the hiPSC-CMs might be induced by the increased expression of PGC-1α. Therefore, we further hypothesized that the upregulation of PGC-1α would increase mitochondrial respiration and promote the metabolic maturation of hiPSC-CMs.
Figure 3.
PGC-1α plays an important role in cardiomyocyte mitochondrial respiratory function. (A) Western blot analysis detected the expression of PGC1α in hiPSC-CMs treated with negative control siRNA (Si-NC) or PGC-1α siRNA (Si-PGC1α). (B) Representative two OCR traces of hiPSC-CMs treated with negative control siRNA (Si-NC) or PGC-1α siRNA (Si-PGC1α) for 3 days, respectively, in response to oligomycin, FCCP, and antimycin A. (C) OCR parameters representing mitochondrial function in PGC-1α siRNA-treated hiPSC-CMs were significantly reduced. (D–E) RT-qPCR (D) and Western blot analysis (E) of PGC-1α expression in hiPSC-CMs treated with ZLN005 at different concentrations (0, 4, 8, 16 μmol/L). 8 μmol/L ZLN005 could effectively upregulate the expression of PGC-1α. (F) Upregulating PGC-1α by ZLN005 (8 μmol/L) increased beating frequency of hiPSC-CMs. (G) Representative traces for control and 8 μmol/L ZLN005-treated hiPSC-CMs responding to oligomycin, FCCP, and rotenone and antimycin A. (H) OCR parameters representing mitochondrial function in 8 μmol/L ZLN005-treated hiPSC-CMs were significantly increased. (I) Mitochondrial membrane potential analysis was measured using a fluorescence probe JC-1 assay system in day 0-hiPSCs and day 30-hiPSC-CMs. The ratio of red/green fluorescence represented the level of Δψm. Compared with control group, ∗P < 0.05, ∗∗P < 0.01; ns, no significant difference.
To determine whether upregulation of PGC-1α promotes energy metabolism and maturation of the hiPSC-CMs, we employed the small-molecule compound ZLN005 to activate the expression of PGC-1α in the hiPSC-CMs. First, we evaluated the effect of the different concentrations of ZLN005 by treating day 27-hiPSC-CMs with 0, 4, 8 and 16 μmol/L ZLN005 for 3 days. The qRT-PCR and Western blot results showed that the mRNA and protein expression of PGC-1α was significantly increased after treatment with ZLN005, and the optimal activation concentration of ZLN005 was 8 μmol/L (Fig. 3D and E), which we used in subsequent experiments. After treating the hiPSC-CMs with 8 μmol/L ZLN005, we observed an increase in their beating frequency (Fig. 3F). We next investigated the modulation of oxidative metabolism, and the measurement of the oxygen consumption rate (OCR) in the hiPSC-CMs, as determined by extracellular flux analysis, showed that the upregulation of PGC-1α led to a robust and significant increase in multiple aspects of respiration measured, including the basal respiratory rate, maximum respiration rate and ATP-dependent respiration rate (Fig. 3G and H), demonstrating that PGC-1α strongly induced mitochondrial respiration. Consistent with this augmented mitochondrial respiratory function, the ZLN005-treated CMs had an overall higher ΔΨm than the control group (Fig. 3I).
The effects of upregulating PGC-1α on mitochondrial morphology of the hiPSC-CMs
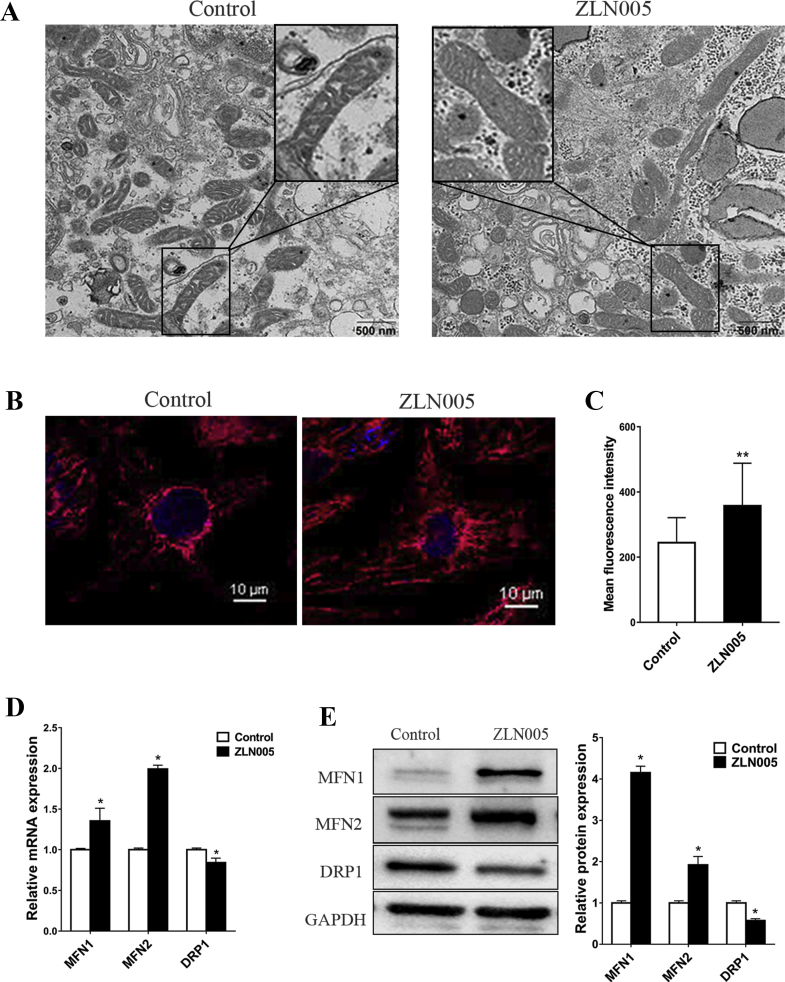
To examine the effect of ZLN005 on mitochondrial morphology in the hiPSC-CMs, the morphology of mitochondria and the expression of genes related to mitochondrial morphology in the control group and the ZLN005 group were compared. The distribution and morphology of the mitochondria in the ZLN005-treated group were different from those of the mitochondria in the control group. The mitochondria in the ZLN005-treated group were scattered throughout the cytoplasm and showed a widely interconnected filamentous network with denser intramitochondrial cristae matrix than the mitochondria in the control group, which were rod-shaped structures located around the nucleus and had few reticular connections (Fig. 4A and B). The mean fluorescence intensity of MitoTracker red was significantly higher in the ZLN005-treated cells than it was in the DMSO-treated cells, indicating an increased mitochondrial content in the hiPSC-CMs (Fig. 4C). We found that these changes in mitochondrial morphology and content were accompanied by a significantly higher mRNA expression level of MFN1 and MFN2 and a lower mRNA expression level of DRP1 (Fig. 4D). The Western blot results confirmed that ZLN005 promoted the protein expression of MFN1 and MFN2 but inhibited the protein expression of DRP1 compared with those expression levels in the control group (Fig. 4E). These results indicate that PGC-1α influences mitochondrial morphology by regulating the expression of mitochondrial fusion and fission genes.
Figure 4.
The effect of up-regulation of PGC-1α by ZLN005 on mitochondrial membrane potential and mitochondrial morphology. (A–C) Transmission electron microscopy (TEM) (A) and MitoTracker red (B) were used to investigate the mitochondrial morphology changes in DMSO-treated or ZLN005-treated hiPSC-CMs. (C) Mean fluorescence intensity of MitoTracker red in DMSO-treated or ZLN005-treated hiPSC-CMs. (D) RT-qPCR detected the effect of upregulating PGC-1α by ZLN005 on mRNA expression levels of MFN1, MFN2 and DRP1. (E) Western blot analysis detected the effect of upregulating PGC-1α by ZLN005 on protein expression levels of MFN1, MFN2 and DRP1. Compared with control group, ∗P < 0.05, ∗∗P < 0.01.
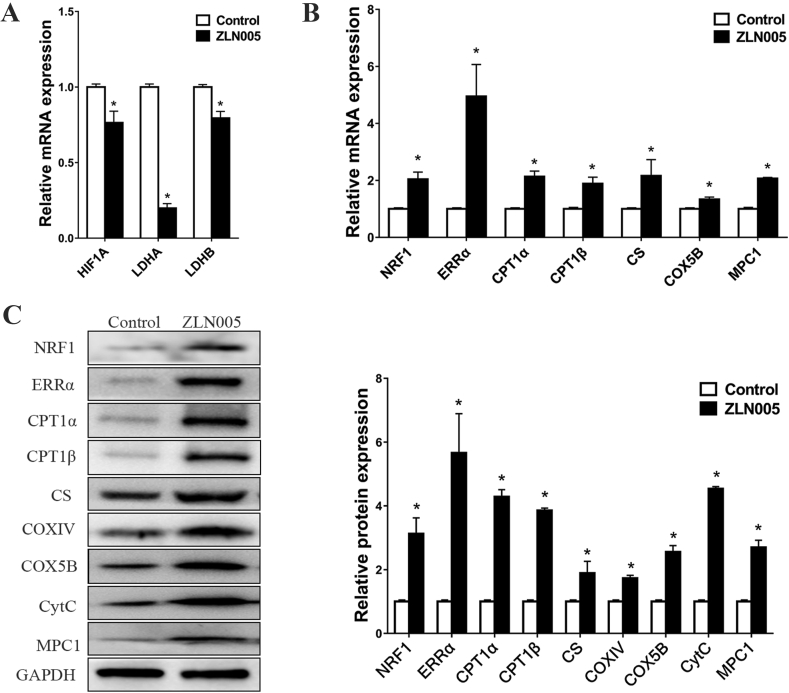
The effects of upregulating PGC-1α on the expression of downstream metabolism-related targets
PGC-1α is a central regulator of metabolism and promotes OXPHOS at the expense of glycolysis.27 To investigate the effects of PGC-1α on the expression of downstream metabolism-related targets, we analysed the expression of a series of metabolism-related genes involved in glycolysis (HIF-1α, LDHA, and LDHB), metabolic transcription factors (ERRα and NRF1), fatty acid oxidation (CPT1α and CPT1β), TCA cycle (CS), ETC (COX5B) and mitochondrial pyruvate transport (MPC1) in the hiPSC-CMs by treatment with ZLN005. On the one hand, the mRNA expression of HIF-1α, LDHA and LDHB was significantly decreased (Fig. 5A), indicating that the upregulation of PGC-1α could inhibit glycolysis. On the other hand, the upregulation of PGC-1α induced the expression of ERRα and NRF1, which regulate genes related to functions involving fatty acid metabolism and OXPHOS. Consistent with these results, multiple fatty acid β-oxidation and OXPHOS genes, including CPT1α, CPT1β, CS, COX5B and MPC1, were also significantly induced by PGC-1α (Fig. 5B), and the Western blot analysis results confirmed the stimulatory effects (Fig. 5C), suggesting that PGC-1α may induce the oxidative metabolism in hiPSC-CMs, a supposition paralleling our observations of hiPSCs differentiating into cardiomyocytes (Fig. 2). Additionally, among the genes we measured, PGC-1α had a robust stimulatory effect on ERRα mRNA and protein levels, suggesting that ERRα might play an important role in the oxidative metabolism induced by PGC-1α.
Figure 5.
The effects of upregulating PGC-1α on the expression of downstream metabolism-related targets. (A–B) RT-qPCR detected the effect of upregulating PGC-1α by ZLN005 on mRNA expression levels of glycolysis-related genes (A) and oxidative metabolism-related genes (B). (C) Western blot analysis detected the effect of upregulating PGC-1α by ZLN005 on the expression levels of oxidative metabolism-related proteins. The proteins assessed were NRF1, ERRα, CPT1α, CPT1β, CS, COXⅣ, COX5B, CytC and MPC1. Compared with control group, ∗P < 0.05.
PGC-1α regulates energy metabolism by interacting with ERRα and regulating its expression
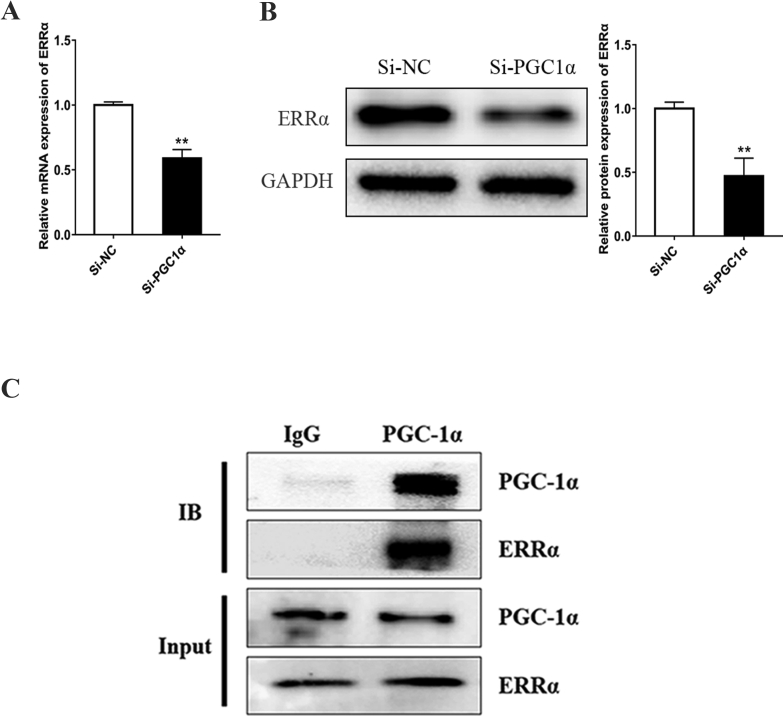
ERRα has been shown to promote mitochondrial biogenesis by promoting the transcription of genes involved in OXPHOS and other metabolic pathways.28 Among the metabolism-related proteins we detected, ERRα increased most obviously after PGC-1α was upregulated (Fig. 5B and C). We also found that PGC-1α knockdown by PGC-1α siRNA inhibited the mRNA and protein expression levels of ERRα (Fig. 6A and B). Taken together, these results have demonstrated a role for ERRα as an important effector of PGC-1α, and the expression and activity of ERRα are regulated by PGC-1α in the hiPSC-CMs, which is consistent with findings from previous research.29 In addition to inducing the expression of ERRα, PGC-1α interacts physically with ERRα, enabling it to activate transcription.30,31 To validate the presence of PGC-1α-ERRα complexes in the hiPSC-CMs, we confirmed the endogenous interaction between these two proteins using immunoprecipitation analysis. An interaction was observed between PGC-1α and ERRα in the hiPSC-CMs (Fig. 6C).
Figure 6.
ERRα is associated with PGC1α in hiPSC-CMs. (A–B) PGC-1α depletion by siRNA inhibited the expression of ERRα detected by qPCR (A) and Western blot analysis (B). Compared with negative control siRNA (Si-NC) group, ∗∗P < 0.01. (C) Co-IP experiments confirmed that PGC-1α interacts with ERRα in hiPSC-CMs. Immunoprecipition of PGC-1α was followed by immunoblotting using the indicated antibodies.
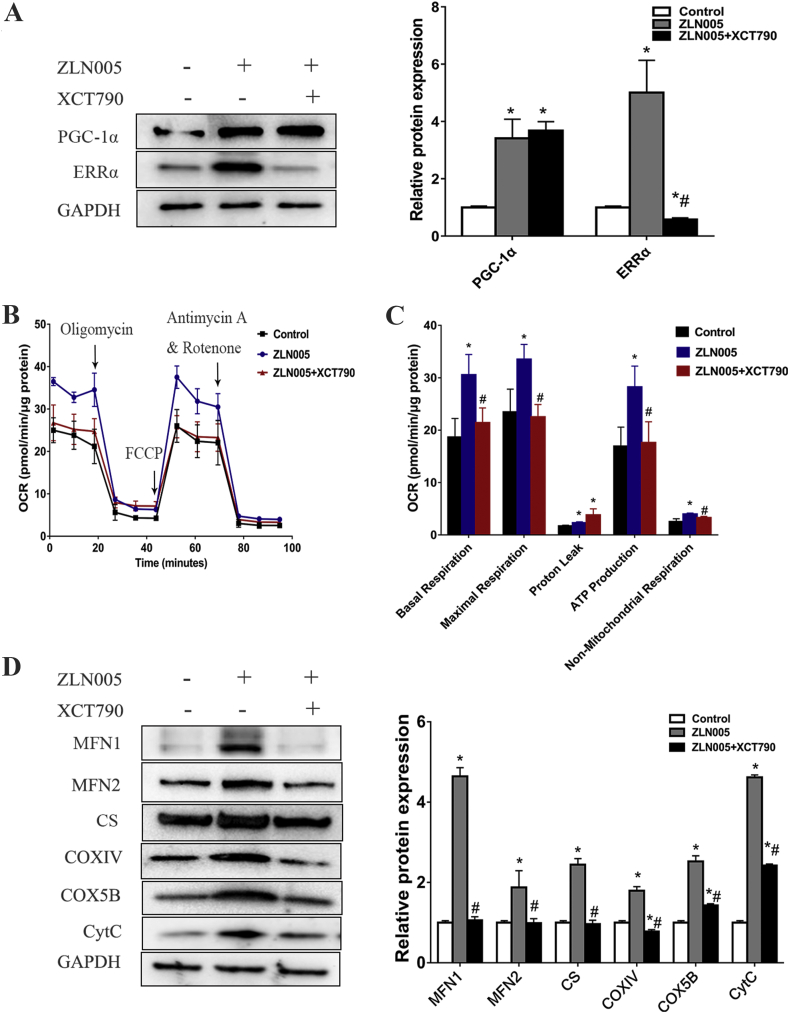
Because of the tight physical interaction between these two proteins, in combination with our data indicating that higher PGC-1α expression is associated with increased mitochondrial biosynthesis and function, we hypothesized that ERRα played an important role in PGC-1α stimulatory effects. To determine whether the stimulatory effects of PGC-1α on mitochondrial respiratory function were mediated through ERRα, we suppressed ERRα expression via XCT790 treatment in ZLN005-treated cells and measured the mitochondrial respiratory function and mitochondrial proteins with a Seahorse XF analyser and Western blot analysis, respectively. As shown in Fig. 7A, XCT790 significantly inhibited the expression of ERRα but had no effect on the expression of PGC-1α. Compared with those of the ZLN005 group, the OCRs of basal respiration, maximum respiration, ATP-associated respiration and non-mitochondrial respiration of the ZLN005 + XCT790 group were significantly reduced, and there was no difference compared with the levels in the control group (Fig. 7B and C), indicating that depletion of ERRα completely abrogated the PGC-1α-induced enhancement of mitochondrial respiration. Consistent with these results, depletion of ERRα also significantly abolished PGC-1α-induced elevation of mitochondrial protein expression, including factors of mitochondrial fusion (MFN1 and MFN2) and components of the TCA cycle (CS), and ETC (COXⅣ, COX5B and CYTC) (Fig. 7D). Taken together, these data indicated that the expression of ERRα was critical for the PGC-1α-mediated stimulation of mitochondrial oxidative metabolism in the hiPSC-CMs.
Figure 7.
ERRα is required for PGC-1α-mediated mitochondrial OXPHOS and mitochondrial biogenesis. (A) Western blot analysis detected the protein expression of PGC-1α and ERRα in hiPSC-CMs after ZLN005/XCT790 treatment. (B) OCR profiles of hPSC-CMs either pretreated with DMSO (control), ZLN005 or ZLN005+XCT790 in response to oligomycin, FCCP, and rotenone and antimycin using the Seahorse flux analyser. (C) OCR parameters representing mitochondrial function in DMSO (control), ZLN005 or ZLN005+XCT790 group. (D) Western blot analysis detected the protein expression in control, ZLN005 or ZLN005+XCT790 group, and the proteins assessed were MFN1, MFN2, CS, COXⅣ, COX5B, and CytC. Compared with control group, ∗P < 0.05; Compared with ZLN005 group, #P < 0.05.
Discussion
Despite encouraging achievements in the efficiency and application of cardiac differentiation of hiPSCs, hiPSC-CMs exhibit an immature phenotype and structurally and functionally resemble foetal cardiomyocytes.10, 11, 12 Numerous studies have been developed to promote cardiac maturation using genetic, chemical, and biomechanical approaches, which led to substantial improvements in hiPSC-CM maturity,13 such as long-term culture,32 three-dimensional (3D) tissue engineering,33 and electric stimulation34; however, few studies have been dedicated to a thorough investigation of the underlying molecular mechanism. In this study, we focus on oxidative metabolism as a critical rubric for determine the maturation state of hiPSC-CMs and have explored the molecular mechanism of metabolic maturation.
Mitochondrial and metabolic remodelling is a central characteristic of differentiation. The morphology of mitochondria changes during cell fate conversion. Undifferentiated PSCs contain immature mitochondria with poorly developed cristae that are located primarily in the perinuclear regions of the cytoplasm and a poorly developed mitochondrial network. As PSCs differentiate into cardiomyocytes, the granular mitochondria become more tubular and form net-like reticular networks containing elongated cristae throughout the cytoplasm.26 In our study, we have also observed similar morphological changes during the cardiac differentiation of hiPSCs. It has been demonstrated that mitochondrial morphology is critical for cardiomyocyte development and maturation, which is directly regulated by mitochondrial dynamics (fusion and fission).19,35 In this regard, in vivo and in vitro experiments demonstrated that ablation of the mitochondrial fusion proteins MFN1 and MFN2 in the embryonic mouse heart or gene trapping of MFN2 or optic atrophy 1 (OPA1) in mouse embryonic stem cells arrested mouse heart development and impaired the differentiation of embryonic stem cells into cardiomyocytes.25 Additionally, a previous study demonstrated that shifting the balance of mitochondrial morphology towards fusion by inhibiting DRP1 promoted the cardiac mesodermal differentiation of hiPSCs, which included a metabolic shift from glycolysis towards oxidative phosphorylation.26 These findings highlight the important role of mitochondrial dynamics in stem cell fate determination. Consistent with mitochondrial morphological changes, the cells underwent significant metabolic alterations, including enhanced mitochondrial respiration and increased mitochondrial membrane potential, resulting in a great difference in total ATP output. These results were consistent with previous studies demonstrating that hiPSCs produce ATP and maintain pluripotency mainly through anaerobic glycolysis, not mitochondrial OXPHOS as exhibited in differentiated cells.36,37
PGC-1α has been identified as a master transcriptional regulator of cellular metabolism, and its expression is crucial for oxidative metabolism in cardiac development.17,19, 20, 21 Metabolic processes such as the TCA cycle, OXPHOS and β-oxidation of fatty acids are undertaken in mitochondria and are inseparable from the contribution of mitochondrial proteins. The transfer of electrons along the ETC is coupled to the transport of protons across the inner mitochondrial membrane, which establishes the electrochemical gradient, also called the mitochondrial transmembrane potential, and drives ATP synthesis with the contribution of proteins in the ETC such as COXⅣ, COX5B and CytC. The TCA cycle begins with the entry of acetyl-CoA, which is generated by the oxidation of energy substrates, mainly fatty acids and glucose-pyruvate, in normal adult hearts.38 Activated fatty acids fatty acids, FA-CoAs, are converted to FA carnitines and transported into mitochondria by carnitine palmitoyl transferase 1 (CPT1, with three isoforms CPT1α, β and C), which is the rate-controlling step in the mitochondrial oxidation of long-chain fatty acids.39 Fatty acids are then repeatedly cleaved to produce NADH, FADH and acetyl-CoAs though mitochondrial β-oxidation, and these cleaved products are coupled to the TCA cycle and ETC to produce ATP.40 Furthermore, pyruvate produced from glucose by glycolysis is transported into mitochondria by the mitochondrial pyruvate carrier (MPC)41 and forms acetyl-CoA. The MPC is composed of MPC1 and MPC2, and it has been confirmed that MPC1 is essential for PGC-1α-induced mitochondrial respiration and biogenesis in renal cell carcinoma.42 Acetyl-CoA enters the TCA cycle and is catalysed by CS, which is the first rate-limiting enzyme in the TCA cycle. In our study, we found that the protein expression levels of PGC-1α and its downstream metabolic-related proteins were significantly increased in the hiPSC-CMs compared with their levels in the hiPSCs. Taken together, these results suggested that the increased respiratory capacity of the hiPSC-CMs was related to the increased expression of PGC-1α, and PGC-1α might enhance mitochondrial function by promoting the expression of downstream proteins involved in mitochondrial oxidative metabolism. These observations prompted us to further study the role of PGC-1α in regulating mitochondrial metabolism during cardiac differentiation. PGC-1α knockdown by siRNA damaged mitochondrial respiration, suggesting that PGC-1α plays an important role in mitochondrial oxidative metabolism in hiPSC-CMs, a finding consistent with previous results found during hESC differentiation into CMs.43
The structural and functional changes of mitochondria are critical components of maturation during cardiac differentiation. PGC-1α is known to be critical for mitochondrial biogenesis in various tissues and cell lines.17,44 A previous study found that PGC-1α induces the expression of MFN1 and MFN2 and also confirmed that PGC-lα directly regulates MFN1 gene transcription by coactivating ERRα at a conserved DNA element in H9c2 myotubes.19 Additionally, overexpression of PGC-1α reduced DRP1 expression by directly binding to its promoter and inhibited mitochondrial fission in H9c2 cells.45 In our study, we found that the upregulation of PGC-1α also promoted mitochondrial fusion in the hiPSC-CMs by promoting the expression of MFN1 and MFN1 while inhibiting the expression of DRP1. These morphological alterations contribute to the increased oxidative capacity in mature mitochondria. In addition to regulating mitochondrial dynamics, we found that the upregulation of PGC-1α by ZLN005 increased hiPSC-CM mitochondrial respiration and mitochondrial membrane potential, resulting in an increase in beating frequency.
PGC-1α is a metabolic regulator of cell fate, for example, it has been confirmed that PGC-1α promotes a coordinate metabolic shift toward mitochondrial utilization and protects against tumorigenesis by promoting mitochondrial-mediated apoptosis via ROS accumulation.46 Obviously, cardiac differentiation is significantly different from intestinal tumorigenesis, it's necessary to further explore the molecular mechanism by which PGC-1α promotes mitochondrial function and metabolic maturity in hiPSC-CMs. We examined the expression of a series of metabolism-related genes, and found that the upregulation of PGC-1α inhibited the expression of HIF-1α and its downstream targets LDHA and LDHB. HIF-1α signaling has been shown to control metabolic signalling during cardiac development and maturation in vivo,47 and inhibition of HIF-1α can promote the metabolic maturation of hiPSC-CMs in vitro.48 Additionally, we found that the expression of transcription factors (NRF1 and ERRα) and mitochondrial genes involved in the TCA cycle, OXPHOS, β-oxidation and pyruvate transport was induced by PGC-1α. Among the proteins we detected, ERRα, one of the key nuclear regulators of genes involved in mitochondrial function and biogenesis,49 was induced most notably by PGC-1α. ERRα can promote cardiac metabolic processes such as mitochondrial OXPHOS, fatty acid β-oxidation and mitochondrial biosynthesis.50 ERRα-knockout mice subjected to pressure overload developed the signatures of heart failure, such as chamber dilatation and reduced LV fractional shortening, and the ERRα target genes involved in energy substrate oxidation, ATP synthesis, and phosphate transfer were downregulated.51 Previous reports indicated that PGC-1α regulates ERRα at two levels: PGC-1α induces the expression of ERRα and interacts physically with ERRα, enabling it to activate transcription, while ERRα functions in PGC-1α-induced mitochondrial biogenesis.28, 29, 30 Considering these results, we confirmed that PGC-1α not only regulates the expression of ERRα but also forms a complex with ERRα in the hiPSC-CMs. Furthermore, depletion of ERRα compromised PGC-1α-induced elevation of mitochondrial respiratory function and mitochondrial protein expression, suggesting that ERRα is required for PGC-1α-mediated stimulation of mitochondrial oxidative metabolism in hiPSC-CMs. Our results showed that PGC-1α is a metabolic regulator of hiPSC-CMs and promotes mitochondrial oxidation metabolism mainly though ERRα.
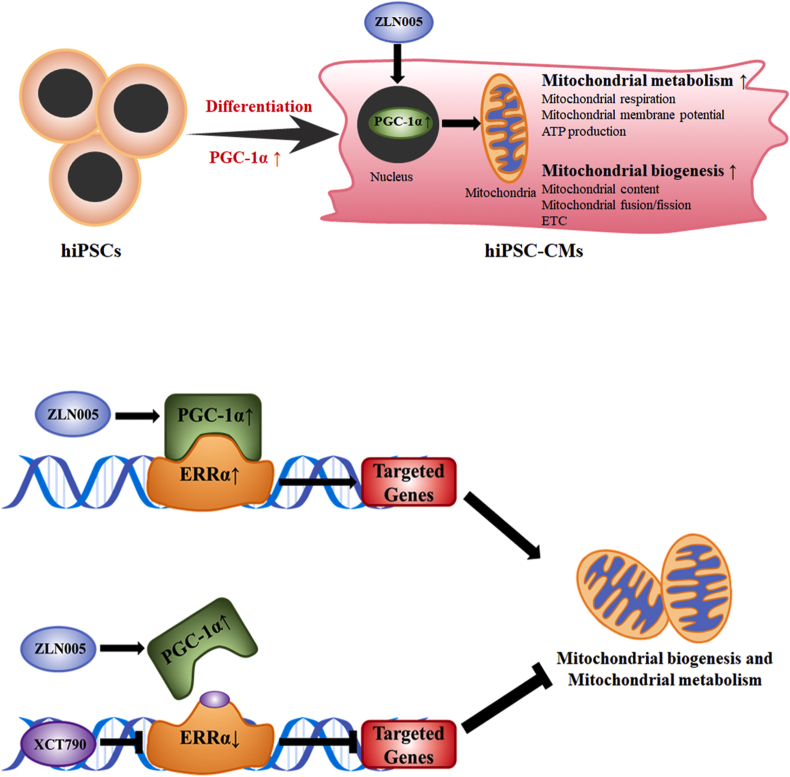
This study is based on hiPSC cardiac differentiation, and the results highlight the importance of PGC-1α in regulating mitochondrial metabolic maturation during hiPSC differentiation into cardiomyocytes. Considering these results, we propose a schematic representation of PGC-1α regulating mitochondrial metabolism and molecular mechanisms during cardiac differentiation and maturation (Fig. 8). After hiPSCs differentiate into hiPSC-CMs, mitochondrial biogenesis and metabolism are improved concomitantly with an increase in PGC-1α expression. Depletion of PGC-1α impairs mitochondrial respiration, while the promotion of PGC-1α expression by ZLN005 further promoted mitochondrial biosynthesis and function, resulting in enhanced mitochondrial respiration, a higher mitochondrial membrane potential, an increased number of mitochondria, improved mitochondrial fusion and increased expression of mitochondrial proteins. In particular, ERRα, which is not only regulated by PGC-1α but also interacts with it, is required for PGC-1α-induced mitochondrial biogenesis and metabolism in hiPSC-CMs. These results indicate that PGC-1α regulates mitochondrial biosynthesis and metabolism though ERRα, and ZLN005 might be used as an additive to hiPSC-CM medium to promote hiPSC-CM metabolic maturation.
Figure 8.
Schematic representation of PGC-1α regulates mitochondrial metabolism and molecular mechanism during cardiac differentiation and maturation. After hiPSCs differentiated into hiPSC-CMs, mitochondrial biogenesis and metabolism is improved, concomitantly with the increase of PGC-1α expression. Upregulating PGC-1α by ZLN005 further promote mitochondrial oxidation function though regulating its downstream proteins, particularly ERRα. Besides regulated by PGC-1α, ERRα interacts with it and is required in the PGC-1α-introduced mitochondrial biogenesis and metabolism in hiPSC-CMs.
In conclusions, our findings indicate that PGC-1α plays an essential role in regulating metabolic maturation during cardiac differentiation from hiPSCs and provide a better understanding of cardiac metabolism and its regulatory mechanism, further lay the foundation for facilitating the clinical application of hiPSC-CMs.
Authors contribution
Jing Zhu and Jie Tian conceived and designed the project. Qin Zhou and Hao Xu carried out the experiments and drafted the manuscript, Bin Tan contributed to edit the figures. Qin Yi performed statistical analyses. Liang Ye and Xinyuan Zhang carried out the sample collection. Liang Yan contributed to cells culture, reagent procurement and management.
Conflict of Interests
The authors declare that there are no competing interests associated with the manuscript.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.12.006.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 81670270, 81970244, 81700250].
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- 1.Katarzyna R. Adult stem cell therapy for cardiac repair in patients after acute myocardial infarction leading to ischemic heart failure: an overview of evidence from the recent clinical trials. Curr Cardiol Rev. 2017;13(3):223–231. doi: 10.2174/1573403X13666170502103833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lian X., Hsiao C., Wilson G. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breckwoldt K., Letuffe-Brenière D., Mannhardt I. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat Protoc. 2017;12(6):1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- 4.Ban K., Bae S., Yoon Y.S. Current strategies and challenges for purification of cardiomyocytes derived from human pluripotent stem cells. Theranostics. 2017;7(7):2067–2077. doi: 10.7150/thno.19427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreau A., Boutjdir M., Chahine M. Induced pluripotent stem-cell-derived cardiomyocytes: cardiac applications, opportunities, and challenges. Can J Physiol Pharmacol. 2017;95(10):1108–1116. doi: 10.1139/cjpp-2016-0726. [DOI] [PubMed] [Google Scholar]
- 6.Musunuru K., Sheikh F., Gupta R.M. Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: a scientific statement from the american heart association. Circ Genom Precis Med. 2018;11(1) doi: 10.1161/HCG.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cliff T.S., Dalton S. Metabolic switching and cell fate decisions: implications for pluripotency, reprogramming and development. Curr Opin Genet Dev. 2017;46:44–49. doi: 10.1016/j.gde.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaspar J.A., Doss M.X., Hengstler J.G., Cadenas C., Hescheler J., Sachinidis A. Unique metabolic features of stem cells, cardiomyocytes, and their progenitors. Circ Res. 2014;114(8):1346–1360. doi: 10.1161/CIRCRESAHA.113.302021. [DOI] [PubMed] [Google Scholar]
- 9.Chung S., Dzeja P.P., Faustino R.S., Perez-Terzic C., Behfar A., Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S60–S67. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Berg C.W., Okawa S., Chuva de Sousa Lopes S.M. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development. 2015;142(18):3231–3238. doi: 10.1242/dev.123810. [DOI] [PubMed] [Google Scholar]
- 11.Yang X., Pabon L., Murry C.E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114(3):511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keung W., Boheler K.R., Li R.A. Developmental cues for the maturation of metabolic, electrophysiological and calcium handling properties of human pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther. 2014;5(1) doi: 10.1186/scrt406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed R.E., Anzai T., Chanthra N., Uosaki H. A brief review of current maturation methods for human induced pluripotent stem cells-derived cardiomyocytes. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 15.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C., Lin J.D. PGC-1 coactivators in the control of energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2011;43(4):248–257. doi: 10.1093/abbs/gmr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villena J.A. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282(4):647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 18.Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56(2):130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 19.Martin O.J., Lai L., Soundarapandian M.M. A role for peroxisome proliferator-activated receptor gamma coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ Res. 2014;114(4):626–636. doi: 10.1161/CIRCRESAHA.114.302562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell L.K., Mansfield C.M., Lehman J.J. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94(4):525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 21.Lai L., Leone T.C., Zechner C. Transcriptional coactivators PGC-1 and PGC-l control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22(14):1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman J.J., Boudina S., Banke N.H. The transcriptional coactivator PGC-1α is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol. 2008;295(1):H185–H196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arany Z., He H., Lin J. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1(4):259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Sharma A., Li G., Rajarajan K., Hamaguchi R., Burridge P.W., Wu S.M. Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J Vis Exp. 2015;(97) doi: 10.3791/52628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasahara A., Cipolat S., Chen Y., Dorn G.W., Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342(6159):734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- 26.Hoque A., Sivakumaran P., Bond S.T. Mitochondrial fission protein Drp1 inhibition promotes cardiac mesodermal differentiation of human pluripotent stem cells. Cell Death Dis. 2018;4(1) doi: 10.1038/s41420-018-0042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo C., Widlund H.R., Puigserver P. PGC-1 coactivators: shepherding the mitochondribBiogenesis of tumors. Trends Cancer. 2016;2(10):619–631. doi: 10.1016/j.trecan.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber S.N., Emter R., Hock M.B. The estrogen-related receptor alpha (ERRalpha) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101(17):6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber S.N., Knutti D., Brogli K., Uhlmann T., Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRα) J Biol Chem. 2003;278(11):9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 30.Huss J.M., Kopp R.P., Kelly D.P. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277(43):40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 31.Gaillard S., Dwyer M.A., McDonnell D.P. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol Endocrinol. 2007;21(1):62–76. doi: 10.1210/me.2006-0179. [DOI] [PubMed] [Google Scholar]
- 32.Bhute V.J., Bao X., Dunn K.K. Metabolomics identifies metabolic markers of maturation in human pluripotent stem cell-derived cardiomyocytes. Theranostics. 2017;7(7):2078–2091. doi: 10.7150/thno.19390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correia C., Koshkin A., Duarte P. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol Bioeng. 2018;115(3):630–644. doi: 10.1002/bit.26504. [DOI] [PubMed] [Google Scholar]
- 34.Ronaldson-Bouchard K., Ma S.P., Yeager K. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556(7700):239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Chan D.C. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu W., Gaeta X., Sahakyan A. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell. 2016;19(4):476–490. doi: 10.1016/j.stem.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khacho M., Slack R.S. Mitochondrial activity in the regulation of stem cell self-renewal and differentiation. Curr Opin Cell Biol. 2017;49:1–8. doi: 10.1016/j.ceb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Stanley W.C., Recchia F.A., Lopaschuk G.D. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85(3):1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 39.Duran M. Physician’s Guide to the Laboratory Diagnosis of Metabolic Disease. Springer-Verlag Berlin Heidelberg; 2003. Disorders of mitochondrial fatty acid oxidation and ketone body handling; pp. 309–334. [Google Scholar]
- 40.Luo X., Cheng C., Tan Z. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017;16(1) doi: 10.1186/s12943-017-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzig S., Raemy E., Montessuit S. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337(6090):93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 42.Koh E., Kim Y.K., Shin D., Kim K.S. MPC1 is essential for PGC-1alpha-induced mitochondrial respiration and biogenesis. Biochem J. 2018;475(10):1687–1699. doi: 10.1042/BCJ20170967. [DOI] [PubMed] [Google Scholar]
- 43.Birket M.J., Casini S., Kosmidis G. PGC-1alpha and reactive oxygen species regulate human embryonic stem cell-derived cardiomyocyte function. Stem Cell Reports. 2013;1(6):560–574. doi: 10.1016/j.stemcr.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LeBleu V.S., O'Connell J.T., Gonzalez Herrera K.N. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding M., Feng N., Tang D. Melatonin prevents Drp1-mediated mitochondrial fission in diabetic hearts through SIRT1-PGC1alpha pathway. J Pineal Res. 2018;65(2) doi: 10.1111/jpi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Errico I., Salvatore L., Murzilli S. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc Natl Acad Sci U S A. 2011;108(16):6603–6608. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krishnan J., Ahuja P., Bodenmann S. Essential role of developmentally activated hypoxia-inducible factor 1alpha for cardiac morphogenesis and function. Circ Res. 2008;103(10):1139–1146. doi: 10.1161/01.RES.0000338613.89841.c1. [DOI] [PubMed] [Google Scholar]
- 48.Hu D., Linders A., Yamak A. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ Res. 2018;123(9):1066–1079. doi: 10.1161/CIRCRESAHA.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villena J.A., Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19(8):269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang T., McDonald C., Petrenko N.B. Estrogen-related receptor α (ERRα) and ERRγ are essential coordinators of cardiac metabolism and function. Mol Cell Biol. 2015;35(7):1281–1298. doi: 10.1128/MCB.01156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huss J.M., Imahashi K., Dufour C.R. The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6(1):25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.