Abstract
P58IPK is a tetratricopeptide repeat-containing cochaperone that is involved in stress-activated cellular pathways and that inhibits the activity of protein kinase PKR, a primary mediator of the antiviral and antiproliferative properties of interferon. To gain better insight into the molecular actions of P58IPK, we generated NIH 3T3 cell lines expressing either wild-type P58IPK or a P58IPK deletion mutant, ΔTPR6, that does not bind to or inhibit PKR. When treated with double-stranded RNA (dsRNA), ΔTPR6-expressing cells exhibited a significant increase in eukaryotic initiation factor 2α phosphorylation and NF-κB activation, indicating a functional PKR. In contrast, both of these PKR-dependent events were blocked by the overexpression of wild-type P58IPK. In addition, the P58IPK cell line, but not the ΔTPR6 cell line, was resistant to dsRNA-induced apoptosis. Together, these findings demonstrate that P58IPK regulates dsRNA signaling pathways by inhibiting multiple PKR-dependent functions. In contrast, both the P58IPK and ΔTPR6 cell lines were resistant to tumor necrosis factor alpha-induced apoptosis, suggesting that P58IPK may function as a more general suppressor of programmed cell death independently of its PKR-inhibitory properties. In accordance with this hypothesis, although PKR remained active in ΔTPR6-expressing cells, the ΔTPR6 cell line displayed a transformed phenotype and was tumorigenic in nude mice. Thus, the antiapoptotic function of P58IPK may be an important factor in its ability to malignantly transform cells.
P58IPK, a member of the tetratricopeptide repeat (TPR) family of proteins (31), is a cellular inhibitor of the interferon-induced protein kinase PKR. This property of P58IPK has been exploited by influenza virus, which recruits P58IPK to repress PKR-mediated eukaryotic initiation factor 2α (eIF-2α) phosphorylation (32, 33), thereby enabling influenza virus to evade the host antiviral response by maintaining a high level of protein synthesis. P58IPK also has growth-regulatory properties in the absence of virus infection, and the overexpression of P58IPK results in malignant transformation (5). Although the mechanism by which this occurs has not been defined, one possibility is that P58IPK transforms cells by interfering with PKR-regulated pathways. There is a well-established correlation among PKR inhibition, reduced eIF-2α phosphorylation, and malignant transformation (4, 6, 13, 27, 37). The stimulation of mRNA translation initiation rates, which occurs in response to decreased eIF-2α phosphorylation, may result in an increase in the translation of normally poorly translated mRNAs, such as those encoding growth factors or oncogenes. In addition, decreased eIF-2α phosphorylation may contribute to the suppression of apoptosis (44), consistent with numerous reports that have implicated PKR as a mediator of programmed cell death pathways (3, 12, 30, 56, 57).
Studies examining the regulatory pathways governing P58IPK function have yielded additional insight into the cellular activities of this protein. Under “normal” physiological conditions, P58IPK is present in an inactive complex with one or more regulatory proteins. These proteins include the molecular chaperone Hsp40 (35) and a novel protein referred to as P52rIPK (for regulator of the inhibitor of protein kinase) (17). Several types of stimuli, including influenza virus infection and heat shock, promote the disruption of the Hsp40–P58IPK complex and the activation of P58IPK (36). The same stimuli trigger an association between P58IPK and the ATPase domain of Hsp70. P58IPK shares structural similarities with other Hsp70-interactive proteins, such as the cochaperones Hip, Hop, and Cyp40, all of which contain TPR domains that are required for stable Hsp70 interaction (42). The carboxyl terminus of P58IPK contains a J domain, and like other J-domain proteins, such as Hsp40, Hdj-2, and auxilin (8), P58IPK stimulates the ATPase activity of Hsp70. Thus, P58IPK also functions as a cochaperone protein. Interestingly, the second regulator of P58IPK, P52rIPK, contains homology to a segment of the molecular chaperone Hsp90, which is essential for the function of a variety of steroid hormone receptors, transcription factors, and protein kinases (39). P58IPK, through its interaction with Hsp40, Hsp70, and an Hsp90-related protein, therefore appears to be intimately tied to the molecular chaperone machinery and to cellular stress response pathways.
Given the emerging relationship between the molecular chaperone machinery and apoptosis (23, 24, 45), and the numerous reports of the apoptotic functions of PKR (reviewed in reference 49), we sought to examine the role of P58IPK as an inhibitor of the programmed cell death response and to determine whether this property could contribute to the ability of P58IPK to malignantly transform cells. To this end, we generated NIH 3T3 cell lines expressing either wild-type P58IPK or a P58IPK deletion mutant, ΔTPR6, that does not bind to PKR or inhibit its ability to phosphorylate eIF-2α (18, 50), thus enabling us to evaluate whether P58IPK also acts outside PKR-dependent pathways. We found that overexpression of wild-type P58IPK inhibited the double-stranded RNA (dsRNA)-induced phosphorylation of eIF-2α and the activation of NF-κB, whereas these PKR-dependent functions remained active in ΔTPR6-expressing cells. Moreover, the P58IPK cell line, but not the ΔTPR6 cell line, was resistant to dsRNA-induced apoptosis. In contrast, both the P58IPK and ΔTPR6 cell lines were resistant to apoptosis induced by tumor necrosis factor alpha (TNF-α). Significantly, both cell lines exhibited a transformed phenotype and induced tumor formation in nude mice. Thus, our findings suggest that inhibition of PKR is not a requirement for P58IPK-induced transformation and that suppression of apoptosis may be a primary mechanism by which P58IPK malignantly transforms cells. A model depicting the role of P58IPK as a regulator of apoptosis is presented.
MATERIALS AND METHODS
Construction of P58IPK and ΔTPR6 cell lines.
Murine NIH 3T3 fibroblasts (American Type Culture Collection, Manassas, Va.) were grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). To generate stable cell lines, plasmid DNA consisting of wild-type P58IPK/pcDNAI/NEO (31), ΔTPR6/pcDNAI/NEO (50), or pcDNAI/NEO (Invitrogen, Carlsbad, Calif.), was introduced into monolayer cell cultures (20 μg of DNA/1.3 × 106 cells) by calcium phosphate transfection (2). Following a 20-h transfection period, the DNA mix was removed and replaced with fresh medium. This medium was removed after an additional 24-h incubation and replaced by medium supplemented with 600 μg of G418 (Geneticin; Life Technologies, Inc., Gaithersburg, Md.) per ml. After 9 days of drug selection, G418-resistant cells were trypsinized and individual clones were harvested. Clonal cell lines were maintained in medium containing 400 μg of G418 per ml.
Preparation of polyclonal antiserum to P58IPK.
Purification of glutathione S-transferase (GST)-P58IPK fusion protein was performed as described previously (31). For the generation of a polyclonal antiserum to P58IPK, New Zealand White rabbits were immunized with 100 μg of GST-P58IPK in 5 ml of incomplete Freund’s adjuvant containing 100 μg of N-acetylmuramyl-l-alanyl-d-isoglutamine. Two subsequent boosts of 100 μg of GST-P58IPK in incomplete Freund’s adjuvant were administered at monthly intervals.
Immunoblot analysis.
Monolayer cell lines or dissociated tumor cells were washed twice with ice-cold Hanks’ balanced salt solution and lysed in Triton lysis buffer (10 mM Tris hydrochloride [pH 7.5], 50 mM KCl, 1 mM dithiothreitol, 2 mM MgCl2, aprotinin [100 μg/ml], 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, polypeptides were transferred to nitrocellulose membranes (54) and detected with the P58IPK monoclonal antibody 2F8 (5) or with the P58IPK polyclonal antibody described above.
eIF-2α phosphorylation analysis.
The state of eIF-2α phosphorylation in cultured cells was determined by vertical-slab isoelectric focusing and immunoblotting. Cell lysates were prepared essentially as described previously (40), but without BPA-1000 treatment. The 10,000 × g supernatant (20 μg of protein) was subjected directly to vertical-slab isoelectric focusing (without prior immunoprecipitation of eIF-2α), and the resolved proteins were transferred to nitrocellulose membranes. After transfer, blots were blocked in phosphate-buffered saline (PBS) containing 10% (wt/vol) nonfat dried milk and 0.2% Tween 20. All incubations with antibody against eIF-2α were performed in PBS containing 0.1 to 0.2% Tween 20. Autoradiographs were analyzed by scanning laser densitometry to determine the ratio of phosphorylated to unphosphorylated eIF-2α in each sample. A change in the ratio in response to poly(I · C) is considered indicative of PKR activity.
Electrophoretic mobility shift assay (EMSA) for NF-κB activation.
Cells (80% confluent in 10-cm-diameter dishes) were incubated in serum-free medium for 18 h, after which they were stimulated with either poly(rI · rC) (100 μg/ml) or TNF-α (10 ng/ml; Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Whole-cell extracts were prepared as described previously (19), with the exception that the 15,000 × g supernatant material was not subjected to dialysis. To assay for activation of NF-κB, 5 μg of cell extract was incubated with 0.2 ng of a 32P-labeled oligonucleotide encoding positive regulatory domain II (PRDII) in binding buffer [20 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 0.6 mM EDTA, 1.0 μg of poly(dI · dC) per ml] for 20 min at room temperature. Protein-DNA complexes were resolved by native 5% polyacrylamide gel electrophoresis in 0.5× TBE (1× TBE is 90 mM Tris, 90 mM boric acid, and 2 mM EDTA). The gels were dried, and NF-κB–DNA complexes were visualized by autoradiography.
Apoptosis assay.
Cells (5 × 105/sample) were treated with poly(I · C) (1, 10, or 100 μg/ml) together with actinomycin D (50 ng/ml) for 16 h. Apoptosis-induced DNA fragmentation was detected by the in situ labeling of DNA strand breaks with fluorescein dUTP and terminal deoxynucleotidyltransferase as described by the manufacturer (Boehringer Mannheim). Fluorescein incorporation was analyzed with a FACStar flow cytometer (Immunocytometry Systems; Becton Dickinson, Mountain View, Calif.). Alternatively, cells were treated with TNF-α (0.2 to 1.0 ng/ml) together with actinomycin D (50 ng/ml) for 16 h and analyzed for cell viability by trypan blue dye exclusion or for apoptosis-induced DNA laddering as described previously (12).
Cell growth assays.
The growth rate of exponential-phase cells was measured by seeding 2 × 104 cells in DMEM supplemented with 10% FBS and 400 μg of G418 per ml. The culture medium was changed every 3 days, and the doubling time was determined by counting cells at 2-day intervals. To determine cloning efficiency, 104 cells were suspended in medium containing 0.35% agarose and overlaid onto medium containing 0.5% agarose in 35-mm-diameter plates. Colonies were then counted 2 to 4 weeks after plating. Percent cloning efficiency is defined as the number of colonies present 2 to 4 weeks after plating, divided by the number of cells plated, multiplied by 100.
Injection of nude mice and propagation of tumor cell lines.
Four- to six-week-old athymic mice (BALB/c nu/nu; Taconic Farms, Germantown, N.Y.), housed in a specific-pathogen-free environment, were injected subcutaneously in the right inguinal area with 2 × 106 cells in 500 μl of DMEM. Tumors were extracted from euthanized mice under aseptic conditions, washed four times in PBS to remove excess blood, fat, and necrotic tissue, and minced. The tumor pieces were then added to DMEM containing 0.5% collagenase-dispase (Sigma Chemical Co., St. Louis, Mo.) and were homogenized with a Dounce vessel, and the homogenate was incubated for 2 h at 37°C with gentle stirring. Dissociated cells were washed once with complete DMEM and seeded into plates for the generation of tumor cell lines.
RESULTS
A P58IPK protein that lacks the sixth TPR motif does not inhibit phosphorylation of eIF-2α.
We reported previously on the construction and characterization of P58IPK deletion mutants to determine the regions of P58IPK required to inhibit PKR activity (50). Using this approach, we demonstrated that a P58IPK mutant lacking the sixth TPR motif, ΔTPR6, was unable to bind to PKR (18) or to inhibit PKR-mediated eIF-2α phosphorylation in vitro (50). This characteristic of ΔTPR6 enabled us to generate a cell line that would allow us to evaluate the ability of P58IPK to function outside PKR-dependent pathways. To generate stable cell lines, NIH 3T3 cells were transfected with a pcDNAI/NEO expression construct encoding the ΔTPR6 protein. Control cell lines were generated in a similar fashion by using a pcDNAI/NEO construct encoding the wild-type bovine P58IPK (31) or by using the pcDNAI/NEO vector alone (NEO). After selection for G418 resistance, multiple clones from each transfection were analyzed by immunoblotting for ΔTPR6 or P58IPK protein production. Using a P58IPK polyclonal antibody that detects bovine P58IPK but does not recognize the endogenous mouse protein, we confirmed that the bovine P58IPK or ΔTPR6 protein was produced in each of the clones examined (Fig. 1).
FIG. 1.
Bovine P58IPK or ΔTPR6 is efficiently produced in stably transfected NIH 3T3 cell lines. Immunoblot analysis of P58IPK and ΔTPR6 protein production was performed. Protein extracts (100 μg) from wild-type P58IPK (P58IPK-1 and P58IPK-2) and ΔTPR6 (ΔTPR6-1 and ΔTPR6-2) cell lines were analyzed with a polyclonal antibody that recognizes bovine P58IPK (lane 1, Madin-Darby bovine kidney cells) but not the endogenous P58IPK protein present in murine cells (lane 2, NEO). Since the deletion in ΔTPR6 is small, the ΔTPR6 protein migrates at approximately the same position as P58IPK in the 12% acrylamide gel shown. Gradient gels (10 to 20% acrylamide) were therefore used to distinguish between the 58-kDa wild-type P58IPK and the 54-kDa ΔTPR6 protein (not shown).
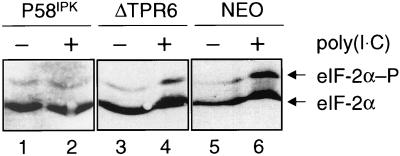
Using a variety of biochemical and genetic techniques, we have previously demonstrated that ΔTPR6 fails to interact with PKR (18), disrupt the formation of PKR dimers (48), or inhibit PKR activity (50). To confirm these results in stable cell lines, we used isoelectric focusing to resolve the phosphorylated and unphosphorylated forms of eIF-2α in the wild-type P58IPK, ΔTPR6, and NEO cell lines. In this assay, a change in the ratio of phosphorylated to unphosphorylated eIF-2α in response to poly(I · C) is considered indicative of PKR activity. When treated with poly(I · C) to activate PKR, the P58IPK cell line exhibited only a 1.3-fold increase in the ratio of phosphorylated to unphosphorylated eIF-2α (Fig. 2; compare lane 2 to lane 1), indicating that PKR activity was inhibited by the overexpression of wild-type P58IPK. In contrast, the ΔTPR6 cell line exhibited a 4.5-fold increase in the ratio of phosphorylated to unphosphorylated eIF-2α (Fig. 2; compare lane 4 to lane 3), demonstrating that PKR is active in ΔTPR6-expressing cells. A 4.5-fold change in the ratio of phosphorylated to unphosphorylated eIF-2α was also observed in NEO cells. Thus, consistent with our earlier in vitro studies, a P58IPK mutant that lacks the sixth TPR motif is also unable to block PKR-mediated eIF-2α phosphorylation in vivo.
FIG. 2.
PKR phosphorylates eIF-2α in ΔTPR6-expressing cells. An analysis of the steady-state level of eIF-2α phosphorylation in cultured cell lines was performed. Extracts were prepared from P58IPK-1, ΔTPR6-1, or NEO cells in mid-log phase, and 20 μg of each extract was subjected to vertical-slab isoelectric focusing. The resolved proteins were transferred to a nitrocellulose membrane, and the blot was probed with an eIF-2α monoclonal antibody. The ratio of phosphorylated to unphosphorylated eIF-2α in each lane was determined by scanning laser densitometry. A change in the ratio in response to treatment with poly(I · C) is indicative of PKR activity. The ratio of phosphorylated to unphosphorylated eIF-2α increased 1.3-fold in the P58IPK cell line and 4.5-fold in the ΔTPR6 and NEO cell lines. Phosphorylated (P) and unphosphorylated forms of eIF-2α are indicated by arrows.
P58IPK, but not ΔTPR6, inhibits the dsRNA-induced activation of NF-κB.
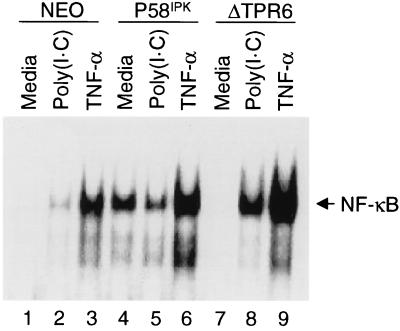
Given the multiple functions of PKR, we next sought to evaluate the ability of P58IPK to inhibit additional PKR-dependent pathways. PKR is essential for the dsRNA-induced activation of the transcriptional regulator NF-κB (28, 34, 55). We therefore began by examining whether the ability of dsRNA to induce activation of NF-κB was altered in the P58IPK or ΔTPR6 cell line. An EMSA was used to detect NF-κB DNA-binding activity as a measure of NF-κB activation. We found that when NEO cells were treated with poly(I · C), there was an increase in NF-κB DNA-binding activity (Fig. 3; compare lanes 1 and 2). Similarly, in ΔTPR6-expressing cells, poly(I · C) treatment also resulted in activation of NF-κB, but to an even greater extent than that observed in NEO cells, indicating a functional dsRNA signaling pathway (Fig. 3, lanes 7 and 8). In contrast, poly(I · C) treatment did not appear to increase NF-κB activation in P58IPK-overexpressing cells (Fig. 3, lanes 4 and 5). This observation suggests that P58IPK not only inhibits PKR’s ability to phosphorylate eIF-2α but also can block the ability of PKR to activate NF-κB. It should be noted, however, that the effect of poly(I · C) on NF-κB activation in P58IPK-overexpressing cells may be somewhat obscured, since P58IPK-overexpressing cells exhibited a high basal level of NF-κB activation even in the absence of poly(I · C) (Fig. 3; compare lane 1 to lane 4). This effect was observed in multiple P58IPK cell lines (data not shown) and may be similar to the increase in NF-κB-dependent gene expression that also occurs in response to the expression of oncogenic forms of Ras, Raf-1, or Neu (ErbB-2/HER2) (14–16).
FIG. 3.
P58IPK, but not ΔTPR6, inhibits the dsRNA-induced activation of NF-κB. NF-κB DNA-binding activities in NEO, P58IPK-1, and ΔTPR6-1 cell lines are shown. Cells were treated with medium alone, medium containing poly(I · C) (100 μg/ml), or medium containing TNF-α (10 ng/ml). NF-κB activation was detected in an EMSA using a 32P-labeled PRDII oligonucleotide probe. NF-κB–PRDII complexes are indicated by the arrow.
As an alternative method of activating NF-κB, we treated each of the cell lines with TNF-α. In contrast to the effect observed with poly(I · C), treatment with TNF-α resulted in a marked increase in NF-κB DNA-binding activity in both the ΔTPR6 and P58IPK cell lines (Fig. 3). This result demonstrates that activation of NF-κB, in response to an inducer other than dsRNA, is not impaired in P58IPK-overexpressing cells. Moreover, the ability of P58IPK to inhibit NF-κB activation is specific for dsRNA signaling pathways. These findings are consistent with those of previous studies, which found that the elimination of PKR (through gene knockout, targeted mRNA ablation, or expression of a transdominant negative PKR mutant) impairs the ability of poly(I · C) to induce NF-κB activation but has no effect on the ability of TNF-α to activate NF-κB (12, 34, 44, 55). Again, this effect was observed in multiple P58IPK and ΔTPR6 cell lines (data not shown).
P58IPK, but not ΔTPR6, mediates resistance to dsRNA-induced apoptosis.
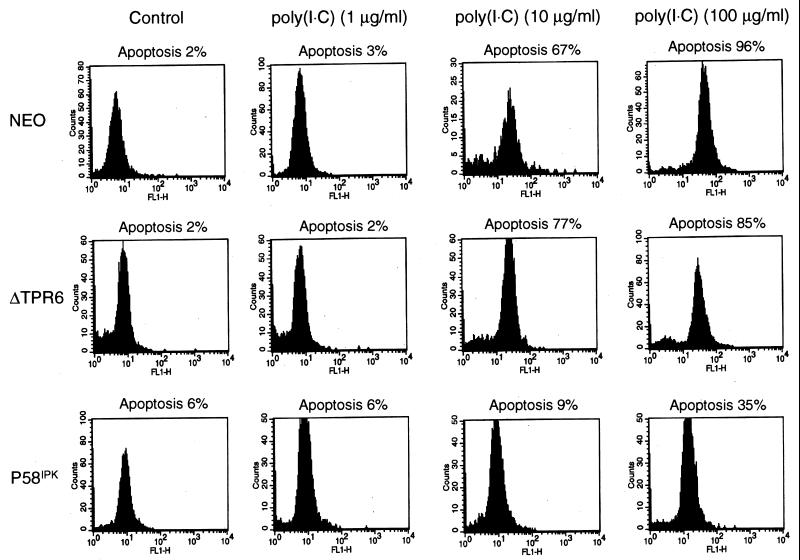
Since PKR has been implicated as a mediator of the apoptotic pathway that is activated in cells upon treatment with dsRNA (3, 12, 44), we examined the ability of dsRNA to induce apoptosis in the NEO, ΔTPR6, and P58IPK cell lines. For these experiments, cells were treated with poly(I · C), and apoptosis-induced DNA fragmentation was detected by the in situ labeling of DNA strand breaks with fluorescein dUTP and terminal deoxynucleotidyltransferase. We found that treatment with poly(I · C) induced apoptosis in a dose-dependent manner in both the NEO and ΔTPR6 cell lines. When ΔTPR6-expressing cells were treated with poly(I · C) at a concentration of 10 μg/ml, 77% of the cells underwent apoptosis, and 85% of the cells became apoptotic at a poly(I · C) concentration of 100 μg/ml (Fig. 4). Thus, ΔTPR6 does not inhibit the ability of PKR to mediate dsRNA-induced apoptosis. In contrast, only 9% of P58IPK-overexpressing cells underwent apoptosis when treated with poly(I · C) at a concentration of 10 μg/ml. Even at a 10-fold higher concentration of poly(I · C), only 35% of P58IPK-overexpressing cells underwent apoptosis. These observations were consistent in multiple clonal cell lines and further extend the role of P58IPK as a regulator of multiple PKR-dependent functions.
FIG. 4.
P58IPK, but not ΔTPR6, mediates resistance to dsRNA-induced apoptosis. An analysis of dsRNA-induced apoptosis in NEO, ΔTPR6-1, and P58IPK-1 cell lines was performed. Cells were treated with poly(I · C) (1, 10, or 100 μg/ml) for 16 h. Apoptosis-induced DNA fragmentation was detected by the labeling of DNA strand breaks with fluorescein dUTP and deoxynucleotidyltransferase. The percentage of apoptotic cells was quantified by flow cytometry as described in Materials and Methods.
P58IPK and ΔTPR6 cell lines are resistant to TNF-α-induced apoptosis.
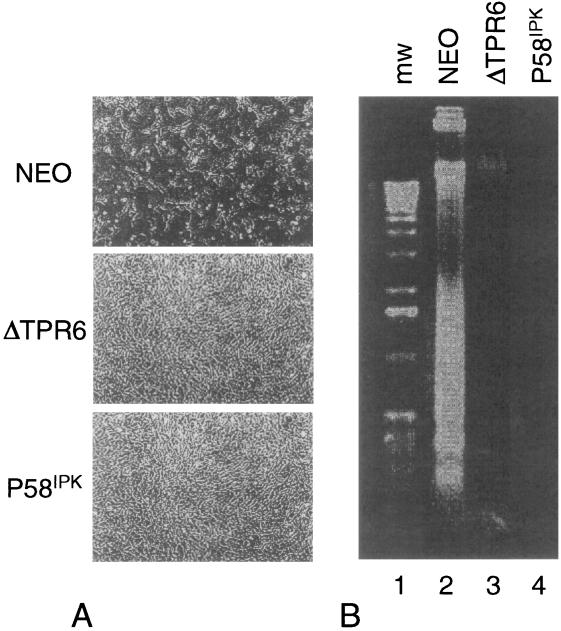
To extend these observations, and to determine if P58IPK might function as a more general apoptotic inhibitor, we examined the ability of TNF-α to induce apoptosis in the P58IPK and ΔTPR6 cell lines. Microscopic examination revealed that in response to TNF-α, only 20% of NEO cells remained viable, whereas the P58IPK and ΔTPR6 cell lines each maintained 75% cell viability (Fig. 5A). The loss of viability in NEO cells was confirmed to be due to apoptosis by the presence of extensive DNA laddering (Fig. 5B), a characteristic apoptotic signature of DNA cleavage into oligonucleosome-sized fragments. In contrast, even at a TNF-α concentration of 1.0 ng/ml, DNA laddering was not detected in either the P58IPK or the ΔTPR6 cell line. It should be noted that since both cell lines showed activation of NF-κB in response to TNF-α (Fig. 3), resistance to TNF-α-induced apoptosis did not reflect an inability to respond to this cytokine. Moreover, the recent report that PKR0/0 cells exhibit no defect in the apoptotic response to TNF-α (1) indicates that the ability of P58IPK (and ΔTPR6) to mediate resistance to TNF-α-induced apoptosis is not mediated through inhibition of PKR. Rather, it appears that P58IPK may function to prevent cells from undergoing programmed cell death in response to a variety of inducers, establishing a novel role for P58IPK as an inhibitor of apoptosis.
FIG. 5.
P58IPK and ΔTPR6 mediate resistance to TNF-α-induced apoptosis. The NEO, ΔTPR6-1, and P58IPK-1 cell lines were treated with TNF-α for 16 h and examined for cell viability and apoptosis-induced DNA fragmentation as described in Materials and Methods. (A) Morphologic characteristics of cell monolayers 16 h post-TNF-α treatment. (B) Electrophoretic analysis of DNA prepared from TNF-α-treated cells.
ΔTPR6-expressing cells exhibit a transformed phenotype and are tumorigenic in nude mice.
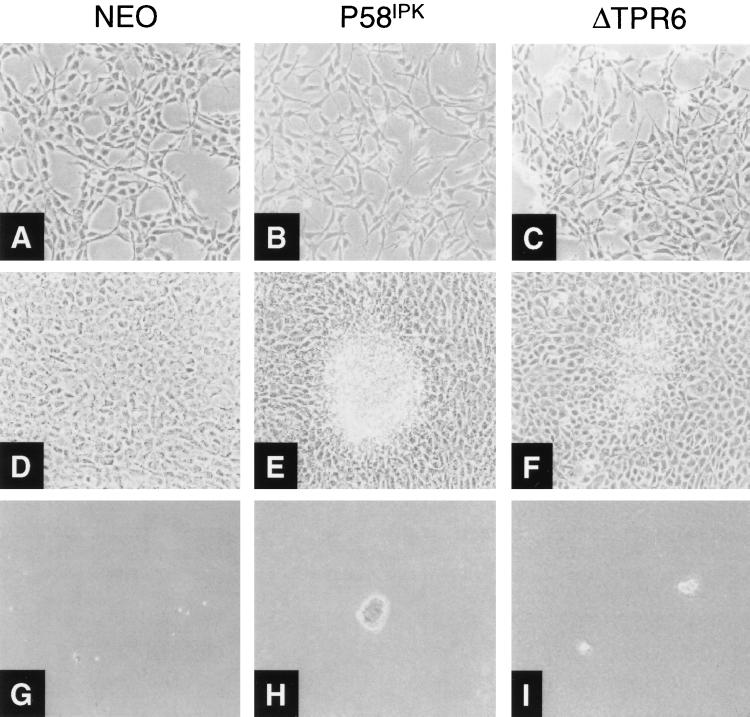
Our observation that ΔTPR6 was unable to inhibit PKR-dependent functions but acted as a suppressor of TNF-α-induced apoptosis prompted us to examine whether ΔTPR6 retained additional growth-regulatory properties. Microscopic examination revealed that, like the P58IPK cell line, ΔTPR6-expressing cells exhibited spindle-shaped morphology and increased refractivity (Fig. 6B and C). Consistent with these morphological changes, transformed foci readily formed on top of ΔTPR6 cell monolayers (Fig. 6F), and ΔTPR6-expressing cells grew faster and to a higher saturation density than the NEO cell line (Table 1). Most significantly, the ΔTPR6 cell line formed colonies in soft agar, indicating anchorage-independent growth (Fig. 6I). We noted, however, that ΔTPR6-expressing cells were approximately twofold less efficient at forming colonies in soft agar than cells overexpressing wild-type P58IPK (Table 1). Thus, although ΔTPR6-expressing cells exhibit the characteristics of a transformed phenotype, this phenotype is less pronounced than that observed in the wild-type P58IPK cell line.
FIG. 6.
ΔTPR6-expressing cells exhibit a transformed phenotype. Morphologic and growth characteristics of the NEO, P58IPK-1, and ΔTPR6-1 cell lines are shown. Cell lines were plated at 2 × 104 cells per 100-mm-diameter dish in DMEM containing 10% FBS and 400 μg of G418 per ml. (A through C) Morphologic characteristics of mid-log-phase cells. In contrast to NEO cells, P58IPK and ΔTPR6 cell lines exhibited spindle-shaped morphology and increased refractivity. (D through F) Cells maintained in culture 4 days after they reached confluency, demonstrating transformed foci on P58IPK and ΔTPR6 cell monolayers. (G through I) Growth in soft agar. Anchorage-independent growth was observed in P58IPK and ΔTPR6 cell lines. Magnification, ×100.
TABLE 1.
Growth properties and tumorigenicity of P58IPK and ΔTPR6 cell lines
| Clone | Growth propertya
|
Tumorigenicity
|
|||
|---|---|---|---|---|---|
| Doubling time (h) | Saturation density (106 cells) | Cloning efficiency (%) | Tumor forma-tionb | Latency (days)c | |
| NEO | 32.7 ± 0.4 | 1.4 ± 0.1 | 0 | 0/5 | |
| P58IPK-1 | 21.9 ± 0.2 | 4.9 ± 0.3 | 15.2 ± 3.8 | 5/5 | 14–16 |
| P58IPK-2 | 23.3 ± 1.3 | 3.1 ± 0.4 | 13.3 ± 5.3 | 2/2 | 14–16 |
| ΔTPR6-1 | 22.8 ± 0.9 | 4.8 ± 0.2 | 3.1 ± 0.4 | 5/5 | 29–35 |
| ΔTPR6-2 | 25.7 ± 0.2 | 3.3 ± 0.4 | 7.0 ± 0.1 | 5/5 | 29–51 |
| ΔTPR6-1 (TCL)d | ND | ND | ND | 6/6 | 4 |
| ΔTPR6-2 (TCL) | ND | ND | ND | 6/6 | 4 |
Assays to determine growth properties of P58IPK and ΔTPR6 cell lines are described in Materials and Methods. Saturation density is the number of cells present 4 days after cells become confluent. Cloning efficiency (%) is the number of colonies present in soft agar 2 to 4 weeks after plating, divided by the number of cells plated, multiplied by 100. Each value represents the average from two experiments. ND, not determined.
Number of animals with tumors of ≥2 mm per number of animals inoculated.
Number of days required to produce tumors of ≥2 mm.
TCL, tumor cell line.
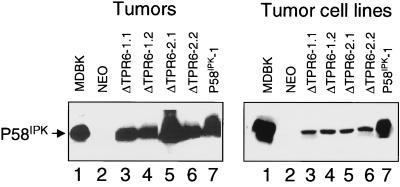
As a further test of the transforming properties of ΔTPR6, we examined whether ΔTPR6-expressing cells could form tumors in nude mice. For these studies, BALB/c nu/nu mice were injected in the right inguinal area with 2 × 106 ΔTPR6-expressing cells per animal. As controls, additional animals were injected with P58IPK or NEO cells. As observed previously (5), mice injected with P58IPK-overexpressing cells developed tumors approximately 2 weeks after injection (Table 1). In accord with their transformed phenotype, mice injected with ΔTPR6-expressing cells also developed tumors, although with a longer latency period. No tumors were observed in mice injected with NEO cells. To verify that the tumors produced either ΔTPR6 or the wild-type bovine P58IPK protein, cellular extracts were prepared from tumor homogenates and analyzed by immunoblotting. In each case, a high level of ΔTPR6 or P58IPK protein production was observed (Fig. 7, left panel).
FIG. 7.
Bovine P58IPK or ΔTPR6 is produced in tumors and tumor cell lines. Immunoblot analysis of P58IPK and ΔTPR6 protein production was performed. (Left) Four tumors from mice injected with ΔTPR6 cell lines (two from mice injected with ΔTPR6-1-expressing cells, designated ΔTPR6-1.1 and ΔTPR6-1.2, and two from mice injected with ΔTPR6-2-expressing cells, designated ΔTPR6-2.1 and ΔTPR6-2.2) and one tumor from a mouse injected with the P58IPK-1 cell line (P58IPK-1) were analyzed by using the anti-P58IPK monoclonal antibody, 2F8 (5). (Right) Cell lines derived from ΔTPR6-expressing and P58IPK-overexpressing tumors were analyzed by immunoblotting using the P58IPK polyclonal antibody. In each panel, cell extracts prepared from MDBK cells were used as a source of bovine P58IPK protein to serve as a positive control (lane 1). Cell extracts prepared from the NEO cell line were analyzed in parallel as a negative control (lane 2).
To confirm that P58IPK or ΔTPR6 was responsible for inducing tumor formation, we generated tumor-derived cell lines by culturing excised tumors and selecting for G418-resistant cells. When analyzed by immunoblotting, cell lines derived from P58IPK tumors showed a high level of bovine P58IPK protein production. However, a comparatively low level of the ΔTPR6 protein was produced in cell lines derived from ΔTPR6 tumors (Fig. 7, right panel). This low level of ΔTPR6 may reflect an instability of the ΔTPR6 protein or a downregulation of ΔTPR6 mRNA expression during cell culture. Nevertheless, cell lines derived from ΔTPR6 tumors were capable of inducing rapid tumor formation when injected into a second round of nude mice (Table 1). Together, these results demonstrate that cells expressing a P58IPK mutant that lacks the ability to inhibit PKR are tumorigenic in nude mice, indicating that inhibition of PKR is not an absolute requirement for P58IPK-induced malignant transformation. Rather, it appears that P58IPK also functions in a PKR-independent manner to malignantly transform cells.
DISCUSSION
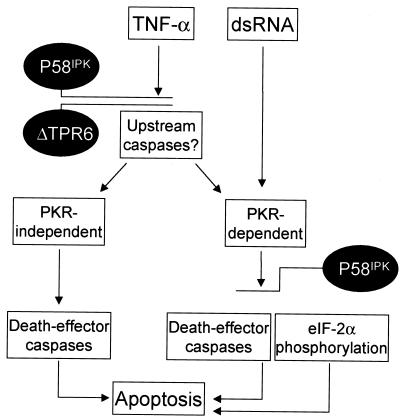
In the present report, we have established that P58IPK, originally identified as a cellular inhibitor of PKR, clearly has biological properties independent of the PKR pathway. We have found that P58IPK is an antiapoptotic protein, able to mediate inhibition of programmed cell death in response to both TNF-α and dsRNA signaling. Utilization of cell lines expressing the P58IPK deletion mutant ΔTPR6 has allowed us to further elucidate the complex molecular pathways involving the P58IPK cochaperone. In our model, we suggest that P58IPK functions through at least two separate pathways to mediate resistance to apoptosis (Fig. 8). In response to dsRNA, wild-type P58IPK, but not ΔTPR6, can inhibit PKR-mediated NF-κB activation and eIF-2α phosphorylation. These events lead to a repression of apoptosis through pathways that may involve death effector caspases (3) or deregulation of the translation of mRNAs encoding proapoptotic proteins (44). In contrast to dsRNA-mediated events, both wild-type P58IPK and the ΔTPR6 mutant can inhibit apoptosis in response to TNF-α. Based on studies with PKR-null mice (1, 12) and the work presented in this report, it is probable that TNF-α signals primarily through PKR-independent pathways, although one cannot completely rule out a minor PKR involvement. It is tempting to speculate, therefore, that P58IPK and ΔTPR6 suppress apoptosis by blocking an event upstream of PKR. For example, P58IPK may disrupt the recruitment or activation of an upstream caspase, such as caspase 8 (FLICE), which is at the apex of the TNF-α-mediated apoptotic cascade (11). This step in the apoptotic pathway is also a key point for inhibition by a variety of viral antiapoptotic proteins, collectively referred to as FLIPs (FLICE-inhibitory proteins) (7, 53). Together, our results indicate that the ability of P58IPK to suppress apoptosis may be a primary factor in P58IPK-induced malignant transformation. In addition, the ability of wild-type P58IPK to also inhibit PKR may account for the more-pronounced transformed phenotype observed in P58IPK-overexpressing cells. Compared with the ΔTPR6 cell lines, P58IPK-overexpressing cells exhibited a higher cloning efficiency and faster tumor formation in nude mice.
FIG. 8.
Model of P58IPK suppression of apoptosis. PKR has been implicated as an essential component of the apoptotic pathway that is induced by dsRNA, and inhibition of PKR by overexpression of P58IPK results in resistance to dsRNA-induced apoptosis. Consistent with the inability of ΔTPR6 to inhibit PKR-dependent functions, ΔTPR6-expressing cells undergo apoptosis in response to dsRNA treatment. In contrast, both the P58IPK and ΔTPR6 cell lines were resistant to TNF-α-induced apoptosis. We propose that P58IPK (and ΔTPR6) can also act in a PKR-independent fashion to regulate the TNF-α pathway. This regulation most likely occurs at a point in the pathway that is upstream of PKR. Suppression of apoptosis may be a primary mechanism by which overexpression of P58IPK induces malignant transformation. See Discussion for additional details.
The ability of P58IPK to suppress apoptosis suggests that there may be parallels between P58IPK and other antiapoptotic proto-oncogenes, such as Bcl-2. Interestingly, Bcl-2 blocks both influenza virus-induced and PKR-mediated apoptosis (22, 30), but in contrast to P58IPK, it does not affect the ability of PKR to inhibit translation. Thus, Bcl-2 is most likely exerting its action downstream of PKR, at or before the point where the PKR-dependent and -independent pathways converge into a common (or core) cell death pathway. Intriguingly, the recently described cochaperone activities of P58IPK may provide additional insight into the mechanism by which P58IPK suppresses apoptosis. The ability of P58IPK to bind to and regulate Hsp70 may provide a link between P58IPK, the cellular chaperone machinery, and the programmed cell death response. This may be analogous to the link that has been proposed for the antiapoptotic protein BAG-1. BAG-1 interacts with and enhances the activity of Bcl-2, and like P58IPK, BAG-1 interacts with the ATPase domain of Hsp70 (23, 24, 45). Thus, the cochaperone activities of P58IPK may facilitate the interaction of P58IPK, and possibly Hsp70, with one or more essential components of the apoptotic pathway.
By functioning as a regulator of apoptosis, P58IPK may be important not only in cellular antiproliferative pathways but also in the cellular response to influenza virus infection. We demonstrated previously that influenza virus recruits P58IPK to downregulate PKR, thereby ensuring the efficient synthesis of viral proteins. Intriguingly, influenza virus infection induces apoptosis (22, 46), and there is evidence for the involvement of PKR in this process (47). Therefore, the activation of P58IPK by influenza virus may also be a mechanism to inhibit or delay PKR-mediated apoptosis, thereby providing sufficient time for viral replication to occur. The ability to inhibit apoptosis is proving to be a general theme among many viruses, and a variety of viral antiapoptotic proteins have been identified (reviewed in references 20, 38, and 51). In addition to the inhibition of FLICE, discussed above, these viral proteins may either mimic the function of cellular antiapoptotic proteins or target various activators of apoptosis for inhibition. For instance, the E1B protein of adenovirus (25) and the KSbcl-2 protein of human herpesvirus 8 (9) are functional homologues of the cellular antiapoptotic protein Bcl-2. Alternatively, the papillomavirus E6 protein blocks apoptosis by targeting p53 for proteolysis (41), and the cowpox virus CrmA protein is a specific inhibitor of caspases (43, 52). The relationship between viral inhibition of PKR and apoptosis is also becoming apparent (49). Infection with a vaccinia virus mutant that lacks the E3L gene, which encodes a dsRNA-binding protein that is a potent inhibitor of PKR activation, triggers an apoptotic response that is not observed in cells infected with the wild-type virus (26, 29). There is also evidence that herpes simplex virus may utilize its γ134.5 gene product to counteract both the translational regulatory and apoptotic activities of PKR. It appears that the γ134.5 gene product interacts with and directs a cellular type-1 protein phosphatase to reverse PKR-mediated eIF-2α phosphorylation (21). This prevents the shutoff of protein synthesis, an event that is associated with neuronal apoptosis (10). Influenza virus, rather than encoding a viral gene product to inhibit apoptosis, may instead recruit P58IPK. Thus, P58IPK may represent a novel example of a cellular antiapoptotic protein being recruited by a virus to avoid the host’s programmed cell death response.
ACKNOWLEDGMENTS
We thank Marjorie Domenowske for help in figure preparation and Dagma Daniel for administrative support.
This investigation was supported by Public Health Service grants AI 22646, AI 41629, and RR 00166 from the National Institutes of Health to M.G.K. and AI 34039 to B.R.G.W. N.M.T. was supported by a Public Health Service National Research Service Award, T32 GM07270, from the National Institute of General Medical Sciences. M.G. is supported by the Helen Hay Whitney Foundation.
REFERENCES
- 1.Abraham N, Stojdl D F, Duncan P I, Méthot N, Ishii T, Dubé M, Vanderhyden B C, Atkins H L, Gray D A, McBurney M W, Koromilas A E, Brown E G, Sonenberg N, Bell J C. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 3.Balachandran S, Kim C N, Yeh W-C, Mak T W, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J Biol Chem. 1995;270:17423–17428. doi: 10.1074/jbc.270.29.17423. [DOI] [PubMed] [Google Scholar]
- 5.Barber G N, Thompson S, Lee T G, Strom T, Jagus R, Darveau A, Katze M G. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G-H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham M E, Caplan A J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng E H Y, Nicholas J, Bellows D S, Hayward G S, Guo H G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou J, Roizman B. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen G M. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Der S D, Yang Y-L, Weissman C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donzé O, Jagus R, Koromilas A E, Hershey J W B, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finco T S, Baldwin A S., Jr κB site-dependent induction of gene expression by diverse inducers of nuclear factor κB requires Raf-1. J Biol Chem. 1993;268:17676–17679. [PubMed] [Google Scholar]
- 15.Galang C K, García-Ramírez J J, Solski P A, Westwick J K, Der C J, Neznanov N N, Oshima R G H C A. Oncogenic Neu/Erb-2 increases Ets, AP-1, and NF-κB-dependent gene expression, and inhibiting Ets activation blocks Neu-mediated cellular transformation. J Biol Chem. 1996;271:7992–7998. doi: 10.1074/jbc.271.14.7992. [DOI] [PubMed] [Google Scholar]
- 16.Galang C K, Der C J, Hauser C A. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene. 1994;9:2913–2921. [PubMed] [Google Scholar]
- 17.Gale M, Jr, Blakely C M, Hopkins D A, Melville M W, Wambach M, Romano P R, Katze M G. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol. 1998;18:859–871. doi: 10.1128/mcb.18.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale M, Jr, Tan S-L, Wambach M, Katze M G. Interaction of the interferon-induced PKR protein kinase with inhibitory proteins P58IPK and vaccinia virus K3L is mediated by unique domains: implications for kinase regulation. Mol Cell Biol. 1996;16:4172–4181. doi: 10.1128/mcb.16.8.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque S J, Williams B R G. Identification and characterization of an interferon (IFN)-stimulated response element-IFN-stimulated gene factor 3-independent signaling pathway for IFN-α. J Biol Chem. 1994;269:19523–19529. [PubMed] [Google Scholar]
- 20.Hardwick J M. Virus-induced apoptosis. Adv Pharmacol. 1997;41:295–336. doi: 10.1016/s1054-3589(08)61063-7. [DOI] [PubMed] [Google Scholar]
- 21.He B, Gross M, Roizman B. The γ34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinshaw V S, Olsen C W, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol. 1994;68:3667–3673. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höhfeld J. Regulation of the heat shock cognate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol Chem. 1998;379:269–274. [PubMed] [Google Scholar]
- 24.Höhfeld J, Jentsch S. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang D C S, Cory S, Strasser A. Bcl-1, Bcl-x(1) and adenovirus EIB19k are functionally equivalent in their ability to inhibit cell death. Oncogene. 1997;14:405–414. doi: 10.1038/sj.onc.1200848. [DOI] [PubMed] [Google Scholar]
- 26.Kibler K V, Shors T, Perkins K B, Zeman C C, Banaszak M P, Biesterfeldt J, Langland J O, Jacobs B L. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J Virol. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 28.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 30.Lee S B, Rodríguez D, Rodríguez J R, Esteban M. The apoptosis pathway triggered by the interferon-induced protein kinase PKR requires the third basic domain, initiates upstream of Bcl-2, and involves ICE-like proteases. Virology. 1997;231:81–88. doi: 10.1006/viro.1997.8494. [DOI] [PubMed] [Google Scholar]
- 31.Lee T G, Tang N, Thompson S, Miller J, Katze M G. The 58,000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee T G, Tomita J, Hovanessian A G, Katze M G. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci USA. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee T G, Tomita J, Hovanessian A G, Katze M G. Characterization and regulation of the 58,000-dalton cellular inhibitor of the interferon-induced, dsRNA-activated protein kinase. J Biol Chem. 1992;267:14238–14243. [PubMed] [Google Scholar]
- 34.Maran A, Maitra R K, Kumar A, Dong B, Xiao W, Li G, Williams B R G, Torrence P F, Silverman R H. Blockage of NF-κB signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- 35.Melville M W, Hansen W J, Freeman B C, Welch W J, Katze M G. The molecular chaperone hsp40 regulates the activity of P58IPK, the cellular inhibitor of PKR. Proc Natl Acad Sci USA. 1997;94:97–102. doi: 10.1073/pnas.94.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melville M W, Tan S-L, Wambach M, Song J, Morimoto R I, Katze M G. The cellular inhibitor of the PKR protein kinase, P58IPK, is an influenza virus-activated co-chaperone that modulates heat shock protein to activity. J Biol Chem. 1999;274:3797–3803. doi: 10.1074/jbc.274.6.3797. [DOI] [PubMed] [Google Scholar]
- 37.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien V. Viruses and apoptosis. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 39.Pratt W B. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 40.Savinova O, Jagus R. Use of vertical slab isoelectric focusing and immunoblotting to evaluate steady-state phosphorylation of eIF-2α in cultured cells. Methods: Companion. Methods Enzymol. 1997;11:419–425. doi: 10.1006/meth.1996.0438. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner M, Werness B A, Huibregstse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 42.Smith D F. Sequence motifs shared between chaperone components participating in the assembly of progesterone receptor complexes. Biol Chem. 1998;379:283–288. doi: 10.1515/bchm.1998.379.3.283. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava S P, Kumar K U, Kaufman R J. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 45.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J Gen Virol. 1993;74:2347–2355. doi: 10.1099/0022-1317-74-11-2347. [DOI] [PubMed] [Google Scholar]
- 47.Takizawa T, Ohashi K, Nakanishi Y. Possible involvement of double-stranded RNA-activated protein kinase in cell death by influenza virus infection. J Virol. 1996;70:8128–8132. doi: 10.1128/jvi.70.11.8128-8132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan S-L, Gale M J, Jr, Katze M G. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan, S.-L., and M. G. Katze. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J. Interferon Cytokine Res., in press. [DOI] [PubMed]
- 50.Tang N M, Ho C Y, Katze M G. The 58-kDa cellular inhibitor of the double stranded RNA-dependent protein kinase requires the tetratricopeptide repeat 6 and DnaJ motifs to stimulate protein synthesis in vivo. J Biol Chem. 1996;271:28660–28666. doi: 10.1074/jbc.271.45.28660. [DOI] [PubMed] [Google Scholar]
- 51.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tewari M, Telford W G, Miller R A, Dixit V M. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J Biol Chem. 1995;270:22705–22708. doi: 10.1074/jbc.270.39.22705. [DOI] [PubMed] [Google Scholar]
- 53.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J-L, Schroter M, Scaffidi C, Krammer P H, Peter M E. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 54.Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y-L, Reis L F L, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams B R G, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeung M C, Lau A S. Tumor suppressor p53 as a component of the tumor necrosis factor-induced, protein kinase PKR-mediated apoptotic pathway in human promonocytic U937 cells. J Biol Chem. 1998;273:25198–25202. doi: 10.1074/jbc.273.39.25198. [DOI] [PubMed] [Google Scholar]
- 57.Yeung M C, Liu J, Lau A S. An essential role for the interferon-inducible, double-stranded RNA-activated protein kinase PKR in the tumor necrosis factor-induced apoptosis in U937 cells. Proc Natl Acad Sci USA. 1996;93:12451–12455. doi: 10.1073/pnas.93.22.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]