ABSTRACT
Phytophthora diseases cause devastation to crops and native ecosystems worldwide. In New Zealand, Phytophthora agathidicida is threatening the survival of kauri, an endemic, culturally and ecologically important tree species. The current method for detecting P. agathidicida is a soil bating assay that is time-consuming and requires high levels of expertise to assess, thus limiting the analytical sample throughput. Here, we characterized the fatty acid methyl ester (FAME) profile of P. agathidicida. We also compared it with the FAME profile of P. cinnamomi and assessed the efficacy of FAME analysis as a diagnostic tool for detecting the pathogen in soil samples. In FAME analysis, the total fatty acid content is isolated from a sample and converted to FAMEs for analysis, a process that takes less than a day. Unique fatty acid acyl chains can serve as biomarkers for specific organisms. We detected 12 fatty acids in P. agathidicida, two of which (20:4ω6 and 20:5ω3) show promise as potential Phytophthora specific biomarkers. Collectively, these findings advance our fundamental understanding of P. agathidicida biology and provide a promising technique to increase the rate of sample processing and the speed of pathogen detection for P. agathidicida in soil.
Keywords: Phytophthora, Phytophthora agathidicida, diagnostics, fatty acid methyl ester analysis
The fatty acid methyl ester (FAME) profile of P. agathidicida was characterized and assessed for its potential use in detecting P. agathidicida in soil.
INTRODUCTION
Phytophthora agathidicida is a recently identified plant pathogen that is threatening New Zealand's native kauri trees (Agathis australis; Weir et al. 2015; Bellgard et al. 2016). It is the causative agent of kauri dieback disease, which has spread throughout most regions within the natural range of kauri (Bradshaw et al. 2020). Kauri trees are massive and can live for thousands of years. Because of this, there can be a latency period from infection to the expression of symptoms (Bradshaw et al. 2020). Thus, detection of P. agathidicida in soil, before the onset of disease symptoms, is critical for managing the spread of kauri dieback. Currently, there are limited tools available to control the spread of disease, and improved surveillance and diagnostics is an urgent priority for research (Bradshaw et al. 2020).
Phytophthora can spread in a variety of ways, including by water, root to root contact and movement of contaminated plant tissue or soil (Erwin and Ribeiro 1996). The lifecycle of Phytophthora species is commonly characterized by the production of vegetative mycelial growth and various spore types (Erwin and Ribeiro 1996). Zoospores are short-lived, motile spores that initiate infection in roots (Bradshaw et al. 2020). Oospores are thick-walled, dormant spores that can survive in soil for years (Bradshaw et al. 2020). Each lifecycle stage plays a critical role in disease spread. In New Zealand, there are at least 30 known species of Phytophthora that cause disease in agricultural, horticultural and indigenous forest settings (Scott and Williams 2014). For example, P. cinnamomi often co-occurs with P. agathidicida at sites of infected kauri (Waipara et al. 2013).
Currently the primary method used to detect P. agathidicida in soil is a baiting assay (Beever et al. 2010). Soil-baiting is a method that is used for detection and isolation of many different Phytophthora species (Erwin and Ribeiro 1996; O'Brien, Williams and Hardy 2009). This method is effective but has limitations; it is slow (2–3 weeks) and requires a high level of expertise to correctly identify the colony and spore morphology that distinguishes P. agathidicida from other Phytophthora species (Beever et al. 2010; Bradshaw et al. 2020). These limitations reduce sample throughput, which limits the capacity to monitor the geographic spread of P. agathidicida in a timely manner. More rapid DNA-based molecular diagnostic tools are in development, but so far have limited effectiveness for detecting P. agathidicida in soil samples (Than et al. 2013; McDougal et al. 2014; Winkworth et al. 2020).
Fatty acid methyl ester (FAME) analysis is potentially an alternative or complementary tool for the detection of organisms in soil. FAME analysis has been used extensively to characterize microbial community structure in soils and has also been applied as a diagnostic tool for the detection of specific organisms in environmental samples (Cavigelli, Robertson and Klug 1995; Drenovsky et al. 2004; Rastogi and Rajesh 2011; Yousef et al. 2012). In FAME analysis, total lipids are extracted from an organism or environmental sample, acyl chains are released as fatty acids and converted to their corresponding methyl esters with subsequent gas chromatography–mass spectrometry (GC–MS) analysis (Welch 1991; Drenovsky et al. 2004). Organisms differ in the types and quantities of acyl chains that are components of their lipids (White et al. 2002; Ehrhardt et al. 2010). Consequently, the presence of or varying ratios of specific FAMEs can indicate the presence and quantitative abundance of particular taxa in a sample, in which case the FAME or ratio of FAMEs may be considered a biomarker. For example, the neutral lipid 16:1ω5 is widely used as an indicator of arbuscular mycorrhizal fungi in soils (Olsson 1999). Specific to Phytophthora, it was shown that soils with P. sojae zoospores added showed increased levels of 18:2ω6, 20:4ω6 and 22:1ω6, indicating that increased ratios of these fatty acids to background fatty acids may potentially serve as a biomarker for the presence of P. sojae in soil (Yousef et al. 2012). Additionally, A significant advantage of a FAME approach, relative to soil baiting and PCR-based methods, is the potential to quantify the biomass of the pathogen in the sample. Finally, the characterization of FAMEs is relatively rapid; the process from a sample to quantitative detection of a biomarker can take less than a day.
Here, we present the results of FAME analysis of P. agathidicida across several key lifecycle stages, including mycelia, oospores and zoospores. Additionally, a comparison of FAME profiles of P. agathidicida and P. cinnamomi reveals that many fatty acids are conserved between the two species; no fatty acids unique to P. agathidicida were identified. However, elevated ratios of the long-chain polyunsaturated fatty acids 20:5ω3 and 20:4ω6 were observed in soil samples with P. agathidicida oospores added. This suggests that FAME analysis may be a useful diagnostic tool for the detection of Phytophthora species in soil.
MATERIALS AND METHODS
Culture conditions and mycelia production
Phytophthora agathidicida NZFS 3770 and P. cinnamomi NZFS 3910 (obtained from Scion, Rotorua, NZ) were maintained regularly on 10% clarified V8 (cV8) agar plates in the dark at 22°C. Mycelia is a common tissue source of lipids for FAME profiling of Phytophthora species (Larkin and Groves 2003; Duan, Riley and Jeffers 2013). Soil microbes may adjust the relative concentrations of membrane lipids to acclimate to different growth temperatures (Griffiths et al. 2003; Yousef et al. 2012). Therefore, we initially characterized the basic fatty acid profile of P. agathidicida mycelia at two different temperatures. We selected 16°C as a value similar to mean annual temperature in the host's range and 22°C as P. agathidicida shows optimal growth in vitro culture at this value. Mycelial mats for lipid extraction were grown in liquid 10% cV8 broth for 48 h at 16°C and 22°C in the dark. Mycelia were separated from the agar plug from which growth was initiated and washed in deionized water to remove residual cV8 broth.
Oospore production
Phytophthora agathidicida oospores were produced as described in Fairhurst et al. (Fairhurst, Deslippe and Gerth 2021). Briefly, three 3 mm agar plugs were taken from the leading edge of mycelial growth on agar plates and inoculated in 15 mL of 4% w/v carrot broth containing 12 µg/mL β-sitosterol and grown in the dark at 22°C for 2 weeks. The resulting mycelial mats were harvested and oospores were isolated by homogenization using a tissue homogenizer for 2 min followed by sonication for 1 min on ice. The homogenized mixture was sequentially filtered through 100 and 40 µm filters to separate the oospores from mycelial fragments. The concentration of the filtered oospore suspension (oospores/mL) was estimated using a disposable hemocytometer by averaging three separate counts.
Zoospore production
Phytophthora agathidicida zoospores were produced as described in Lacey et al. (2021). Briefly, mycelial mats were initially grown in 2% w/v carrot broth supplemented with 15 µg/mL β-sitosterol for 30 h. The mycelial mats were then washed with 2% w/v soil solution and incubated under light for 14 h. The soil solution was removed and the mycelial mats were washed with water. Zoospore release was induced by adding ice-cold water and incubating the mycelial mats at 4°C for 20 min. After sufficient zoospore release, the concentration of spores was estimated using disposable hemocytometers by averaging three separate spore counts.
Fatty acid extraction, conversion to FAMEs and GC-MS analysis
For FAME analyses, mycelia (∼200 mg) from P. agathidicida and P. cinnamomi were grown and prepared as described above. For P. agathidicida oospore and zoospore analyses, each spore type was produced as described above. A total of 250 000 zoospores or 100 000 oospores in 250 µL was used for lipid extraction. For the detection of oospores in soil, we collected five soil samples from forest and garden locations in Wellington, NZ, a region outside the native range of kauri and presumably free of P. agathidicida. Soil samples were collected with a trowel, which was sterilized with 70% ethanol between samples. Soil samples were sealed in plastic bags and placed on ice for transport to the laboratory. Samples were then freeze-dried and stored at –20°C. Laboratory grown oospores were added in 250 µL aliquots to 0.5 g sub-samples of soil sample A. The mixtures of soil and laboratory-grown oospores were subsequently subjected to lipid extraction and FAME production.
For all conditions, lipid extraction and conversion to FAMEs was carried out as described in Duan, Riley and Jeffers (2013) with slight modifications. Initially, fatty acids were released and saponified by the addition of 3.75 M NaOH dissolved in 50% MeOH(aq) (1 mL) to the sample and incubation at 80°C for 30 min with occasional mixing. Next, free fatty acids were methylated by addition of 62.5% 6 N HCl: 37.5% MeOH solution (2 mL) and incubation at 80°C for 10 min. Phase separation was then carried out by the addition of 1 mL of a 1:1 mixture of n-hexane and methyl-tert butyl ether with vigorous mixing by vortex for 30 s. After sufficient phase separation, the top organic phase was transferred to a new glass tube and washed with 3 mL of 0.3 M NaOH(aq). The top organic phase was again transferred to a new tube and concentrated under a stream of nitrogen gas. The remaining FAMEs were then resuspended in 150 µL n-hexane and transferred to 2 mL GC vials with inserts and processed immediately or stored at –20°C until analysis.
Samples were analysed on a Shimadzu GCMS-QP2010 Plus. The GC column used was a Restek RXI-5SilMS (30 m × 0.25 mm × 0.25 µm) and He was the carrier gas. An aliquot (2 µL) of the sample was injected (8:1 split ratio) at 260°C with a column flow rate of 1.09 mL/min and linear velocity of 39.6 cm/s. The GC conditions were as follows: 2 min hold at 150°C followed by a 10°C increase per min to 280°C with a final 10 min hold at 280°C. The MS transfer line temp was 260°C. MS analysis began after 4 min and ended at 25 min with 3 scans/s. Ions between 40 and 600 m/z were detected. The EI ion source operated at 70 eV ionization energy. Chromatograms were analysed using Shimadzu GCMSsolution Software. FAMEs were annotated using mass spectral matching to the National Institute for Standards and Technology (NIST) 2011 database. These annotations were confirmed based on a comparison of the retention time and fragmentation pattern with those of authentic FAME standards (Larodan). Fatty acids are named using standard nomenclature (fatty acid chain length: number of double bonds followed by an ω, finally the carbon number from the methyl terminus where the first double bond starts, e.g. linoleic acid ((9Z,12Z)-octadeca-9,12-dienoic acid) is 18:2ω6).
FAMEs were quantified in two ways. First, the relative amount of each P. agathidicida FAME was determined as a % of the total fatty acid content. To do this, the % of each FAME was determined by integrating the area under the peak representing each FAME on the chromatogram. The values were then converted to percentages with all identified peaks per sample combined to total 100%. Thus, each FAME is calculated as a fraction of 100% of the total FAMEs identified per sample. Second, we quantified molar amounts of 20:4ω6 and 20:5ω3 in soil samples using a 19:0 fatty acid standard as the internal standard. These two fatty acids were chosen for quantification due to the difference in ratio of 20:5ω3 to 20:4ω6 when comparing soil samples and cultured P. agathidicida. For use as an internal standard, 100 nmol of 19:0 fatty acid was added directly to soil samples prior to lipid extraction and conversion to FAMEs. For quantification, response factors for 20:4ω6, 20:5ω3 and 19:0 were first determined and relative response factors between 20:4ω6/19:0 and 20:5ω3/19:0 were subsequently determined (Dodds et al. 2005). For each condition, five biological replicates were performed, where each replicate represents a separate extraction, analysed on a separate day. Statistical analyses and graphical representations of the data were performed and generated using Prism GraphPad (Version 8.2.1).
Statistical analyses
In each experiment, treatments were applied to five biological replicates. To be considered a biological replicate, samples of Phytophthora life-cycle stages must have been isolated on different days. Replicate soil samples consisted of individual aliquots of a homogenized bulk soil sample. In all figures, errors bars indicate standard deviation of the five replicates per treatment. We used t-tests to determine the statistical significance of treatment effects. To account for multiple comparisons within experiments, we applied Bonferroni's correction to adjust P-values. All statistical analyses and graphical representations of the data were performed using Prism GraphPad (Version 8.2.1).
RESULTS
Comparison of FAME profiles of P. agathidicida and P. cinnamomi
A total of 12 fatty acids were regularly detected from P. agathidicida. The five most abundant fatty acids were 14:0, 16:0, 18:2ω6, 18:1ω9 and 20:5ω3, constituting greater than 75% of the total fatty acid content (Fig. 1). Comparing P. agathidicida mycelia grown at 16 and 22°C revealed slight changes in relative fatty acid amounts. Of the five most abundant fatty acids only 18:2ω6 was significantly increased at 22°C compared to 16°C (Table S1, Supporting Information). We also examined the FAME profile of P. cinnamomi mycelia at 16 and 22°C (Fig. 1). The FAME profiles of P. agathidicida and P. cinnamomi were largely similar. A total of 11 fatty acids were detected in P. cinnamomi, all of which overlapped with fatty acids present in P. agathidicida (Fig. 1). The five most abundant fatty acids were also consistent between both species at 16 and 22°C (Fig. 1). Of the five most abundant fatty acids 18:2ω6, 18:1ω9 and 20:5ω3 were significantly different between P. agathidicida and P. cinnamomi at both 16 and 22°C (Table S1, Supporting Information). Interestingly, the fatty acid 22:1ω9 was regularly present in P. agathidicida but not detectable from P. cinnamomi. However, previous studies have shown that this fatty acid is present in P. cinnamomi at low relative percentages (Duan, Riley and Jeffers 2013). In general, our fatty acid profile for P. cinnamomi was similar to a previously published report that spanned multiple isolates of this species (Duan, Riley and Jeffers 2013). Duan et al. detected fifteen fatty acids from P. cinnamomi. The four additional fatty acids detected in that study comprised less than 2% of the total fatty acid content.
Figure 1.
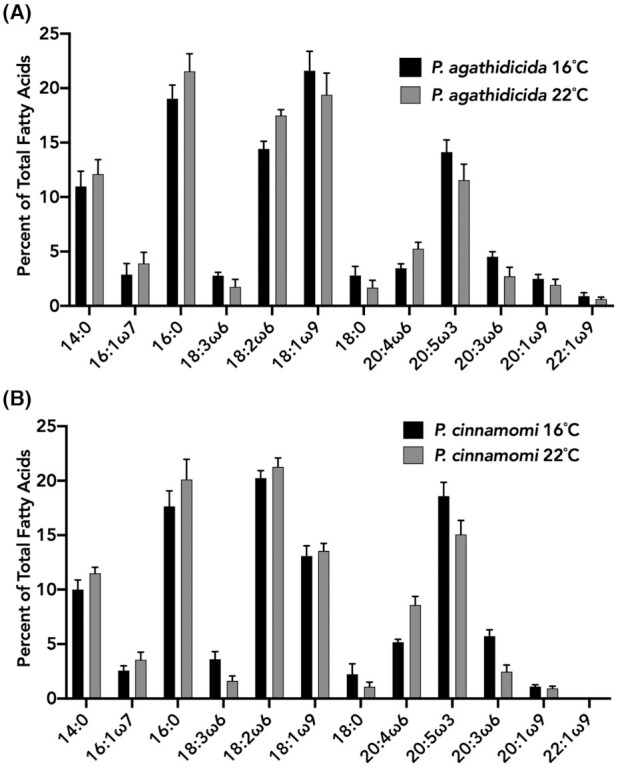
Fatty acid profile of P. agathidicida and P. cinnamomi at varying temperatures. FAMEs were produced and analysed from mycelia of P. agathidicida(A) and P. cinnamomi(B) grown at 16 and 22°C. The % of each fatty acid is the average relative % of five biological replicates. Error bars indicate standard deviation.
Comparison of FAME profiles of various P. agathidicida lifecycle stages
Environmental conditions can affect the relative abundances of oomycete life-cycle stages in soil (Erwin and Ribeiro 1996). We therefore compared the FAME profiles of P. agathidicida oospores, zoospores and mycelia (Fig. 2). Compared to mycelia, the FAME profiles of oospores and zoospores were less diverse in quality and quantity of acyl chains that were present. The five most abundant fatty acids (14:0, 16:0, 18:2ω6, 18:1ω9 and 20:5ω3) were the same for each lifecycle stage. Significant differences for each of these fatty acids between lifecycle stages can be found in Table S2 (Supporting Information). Each fatty acid that was detected from the mycelial samples was also detected in oospores; however, the long-chain unsaturated fatty acids were in lower quantities relative to shorter chain length fatty acids when compared with mycelia. In mycelia, three polyunsaturated, long-chain fatty acids (20:4ω6, 20:5ω3, 20:3ω6) and two mono-unsaturated, long-chain fatty acids (20:1ω9, 22:1ω9) were detected and all except 22:1ω9 constituted >1% of the total FAME profile. In oospores and zoospores, 22:1ω9 was not detected. In zoospores, 18:3ω6 and 20:1ω9 were also not detected. Overall, oospores and zoospores produced lower quantities of long-chain fatty acids. Different growth media are known to induce changes in the FAME profiles of Phytophthora species (Larkin and Groves 2003; Duan, Riley and Jeffers 2011), and may have contributed to the variation we observed among life-cycles stages. However, these effects are difficult to quantify since no single laboratory procedure exists to produce oospore and zoospores in P. agathidicida.
Figure 2.
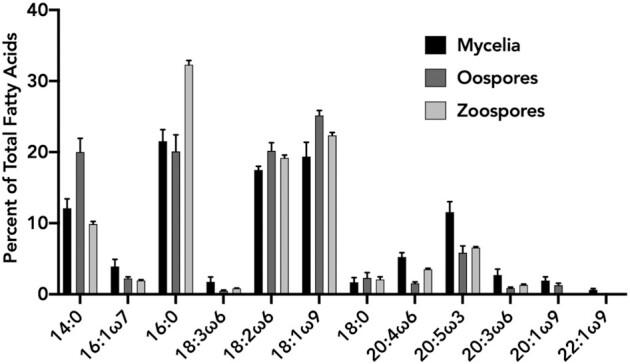
Fatty acid profile comparison of different P. agathidicida lifecycle stages. FAMEs were produced and analysed from P. agathidicida mycelia, oospores and zoospores. The % of each fatty acid is an average relative % of five biological replicates. Error bars indicate standard deviation.
Assessment of the FAME profile of soils with and without P. agathidicida oospores added
To determine if the identified acyl chains could serve as biomarkers for identifying P. agathidicida in soil, we first examined the fatty acid profile of five field-collected soil samples. In each soil sample, we characterized the 12 fatty acids produced by P. agathidicida. Of the 12 P. agathidicida fatty acids, eight were readily detectable in varying quantities across samples (Figure S1, Supporting Information). The addition of oospores to the soil revealed that no unique P. agathidicida fatty acids were detectable (Figure S2, Supporting Information). However, the relative % of the long-chain polyunsaturated fatty acid 20:5ω3, which was present in low quantities in soil (Figure S1, Supporting Information) and is relatively abundant in P. agathidicida (Fig. 1), increased linearly with increasing oospores added to soil (Figure S2, Supporting Information). In contrast, the % of 20:4ω6 remained relatively stable with increasing numbers of oospore added (Figure S2, Supporting Information). With this observation, we determined the molar quantity of 20:5ω3 and 20:4ω6 and examined the ratios of these fatty acids in each sample. The ratio of 20:5ω3 to 20:4ω6 in soil samples alone is <1 (Table 1). In P. agathidicida oospores, this ratio is >4 (Table 1). Thus, the presence of P. agathidicida in soil should lead to an increase in the ratio of 20:5ω3 to 20:4ω6 relative to P. agathidicida-free soil. As oospores were added to soil, the molar quantity of 20:5ω3 and 20:4ω6 both increased (Fig. 3 and Figure S2, Supporting Information). However, this change was greater for 20:5ω3 than for 20:4ω6 leading to a greater ratio of 20:5ω3 to 20:4ω6 as oospores increased (Table 1). In P. agathidicida-free soil, the 20:5ω3 to 20:4ω6 ratio was 0.92. This ratio shifted to 1.06 at the lowest concentration of added oospores (25 000), and increased to 1.4 at the highest concentration of added oospores (200 000). At 25 000, 100 000 and 200 000 added oospores, the ratio of 20:5ω3 to 20:4ω6 was significantly greater than that of soil alone (Table 3).
Table 1.
Ratios of 20:5ω3 to 20:4ω6 fatty acids in soil, soil with P. agathidicida oospores added, and P. agathidicida oospores alone.
| Soil alone | 25 k oospores in soil | 50 k oospores in soil | 100 k oospores in soil | 200 k oospores in soil | 100 k oospores alone | |
|---|---|---|---|---|---|---|
| 20:5ω3/20:4ω6 | 0.92 ± 0.30 | 1.06 ± 0.24* | 1.12 ± 0.34 | 1.41 ± 0.37** | 1.4 ± 0.44* | 4.05 ± 0.78** |
The ratios of 20:5ω3 to 20:4ω6 were determined by dividing the concentration of 20:5ω3 by 20:4ω6 for each condition across five biological replicates with the average and standard deviation shown. A single * indicates a statistical difference when compared with soil alone at P < 0.05, while ** indicates a statistical difference at P < 0.005.
Figure 3.
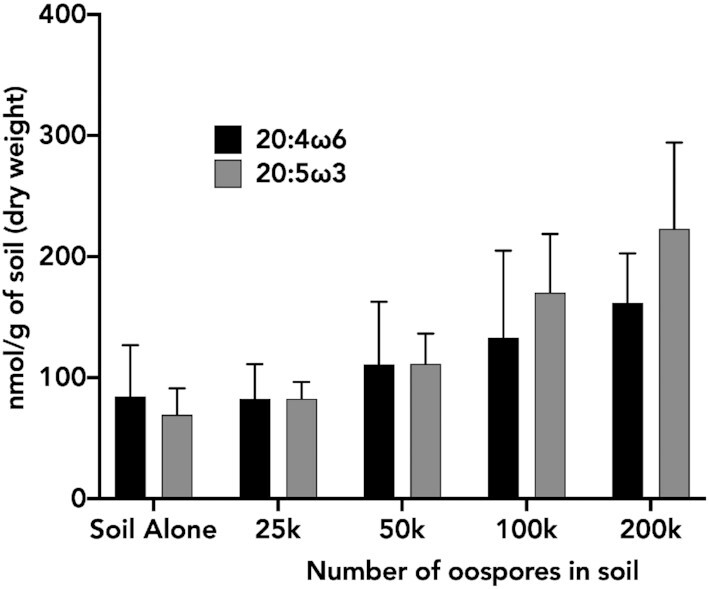
Quantification of 20:4ω6 and 20:5ω3 fatty acids in soil samples with P. agathidicida oospores added. Oospores were added at varying concentrations to 0.5 g of soil containing a 19:0 fatty acid internal standard. FAMEs were then produced and analysed from each sample, and the concentration of 20:4ω6 and 20:5ω3 was determined as nmol/g of soil. The values are an average of five biological replicates. Error bars indicate standard deviation.
DISCUSSION
Rapid and reliable diagnostics are essential when trying to manage diseases in both humans and plants. In the case of kauri dieback, tracking the spread of the P. agathidicida is particularly difficult due to the latent onset of disease symptoms (Bradshaw et al. 2020). In this study, we present the first FAME profile of P. agathidicida. We also assessed the potential of FAME analysis as a tool for detecting P. agathidicida in soil. A significant advantage of a FAME approach relative to soil baiting and PCR-based methods, is the potential to quantify the pathogen in the sample. Overall, the FAME profile of P. agathidicida is largely consistent with the FAME profiles of other Phytophthora species. The five most abundant P. agathidicida fatty acids (14:0, 16:0, 18:2ω6, 18:1ω9 and 20:5ω3) are also the five most abundant fatty acids in six other species of Phytophthora (Larkin and Groves 2003; Duan, Riley and Jeffers 2013). The similarity of P. agathidicida and other studied Phytophthora species is particularly interesting as P. agathidicida falls into the recently categorized (but as of yet understudied) Phytophthora clade five (Weir et al. 2015); our data suggest that lipid profiles may be conserved across different clades of Phytophthora.
While we identified no unique FAME biomarker for P. agathidicida, our data reveal that the addition of increasing amounts of P. agathidicida oospores to soil leads to an increase in the ratio of 20:5ω3 to 20:4ω6. Similar results have been observed with P. sojae where long-chain polyunsaturated fatty acids were detected above background soil levels when zoospores were added to soil (Yousef et al. 2012). Combined, this suggests that the ratio of 20:5ω3 to 20:4ω6 could be a useful biomarker indicating the presence or absence of Phytophthora species in forest soils. However, other oomycete genera may also influence the ratios of 20:5ω3 to 20:4ω6 in soil. For example, of the Plasmopara species examined by Spring and Haas, there was a high degree of variability in FAME profiles with only Plasmopara halstedii producing high levels of 20:5ω3 (Spring and Haas 2002). Pythium species also produce variable FAME profiles. Pythium aphanidermatum produces low levels of 20:5ω3, whereas Pythium irregulare and Pythium ultimum produce high levels of 20:5ω3 and 20:4ω6 at ratios close to 1:1 (Cheng et al. 1999; Spring and Haas 2002; Larkin and Groves 2003). Overall, of the Phytophthora species with FAME profiles reported in the literature, all produce high ratios of 20:5ω3 to 20:4ω6, while non-Phytophthora oomycetes present more variable FAME profiles. Thus, high 20:5ω3 to 20:4ω6 ratios are likely to indicate the presence of Phytophthora, but other oomycetes may also be detected.
The work presented here highlights the potential of the ratio of 20:5ω3 to 20:4ω6 to function as a tool for the detection of Phytophthora in soils. Nevertheless, further studies and analyses are needed to optimize FAME analysis as a diagnostic tool that can be effective in assessing field samples. Our results suggest that basic qualitative FAME analysis is not sufficient for differentiating P. agathidicida from other Phytophthora species and potentially other oomycetes. However, previous studies indicate that FAME analysis can be used to distinguish not only Phytophthora species but isolates within a species as well (Larkin and Groves 2003; Duan, Riley and Jeffers 2013). For example, cluster analysis based on FAME profiles was used to differentiate cultured P. cactorum, P. citrophthora, P. cinnamomi, P. cryptogea and P. nicotianae (Duan, Riley and Jeffers 2013). In another study, using similar techniques, individual isolates of cultured P. infestans were differentiated (Larkin and Groves 2003). This suggests that a more detailed characterization of P. agathidicida FAMEs may provide sufficient information for distinguishing P. agathidicida from other Phytophthora species. Additionally, utilizing extraction techniques that target specific groups of lipids such as phospholipids or neutral lipids may help to further enrich target fatty acids such as 20:5ω3 and reduce background signals from complex environmental samples like soil (Drenovsky et al. 2004). These techniques and analyses could lead to a determination of the threshold of detection for Phytophthora in infected soil samples.
Despite the current limitations of FAME analysis, detecting Phytophthora on a genus level and possibly other oomycete species is useful and can potentially be used in conjunction with soil bating assays or other molecular diagnostics for species confirmation. For example, large scale soil sampling and screening of samples via FAME analysis could be used to identify samples with 20:5ω3: 20:4ω6 >1. These samples could subsequently be subjected to the soil-baiting assay to confirm the presence of P. agathidicida. This is similar to the ‘funnel and filter’ model used for screening soils contaminated with P. ramorum (Smart et al. 2021). In that example, soils were pre-screened for the presence of Phytophthora using an immunosorbent assay. Soils positive for Phytophthora were then examined using qPCR to determine if P. ramorum was present.
In contrast to FAME analysis, several DNA based diagnostic tools have been explored for use in detecting P. agathidicida and other Phytophthora species. These include both conventional qPCR and loop-mediated isothermal amplification (LAMP) methods (Than et al. 2013; McDougal et al. 2014; Hansen et al. 2016; Winkworth et al. 2020). Recently, Winkworth et al. (2020) combined aspects of the existing P. agathidicida soil-baiting assay with LAMP to reduce the overall sample processing time. LAMP is a DNA amplification-based method that uses primers to amplify regions of the genome-specific to an organism of interest (Wong et al. 2018). Efficient amplification can lead to a detectable signal, such as color-change, that can often be assessed quickly and on-site spectrophotometrically. LAMP assays efficiently detected P. agathidicida immediately following baiting (Winkworth et al. 2020). This variation on standard soil-baiting effectively eliminated the final week of the assay, shortening the process to 2 weeks. It is unclear whether or not LAMP would be useful at directly testing soil samples for the presence of P. agathidicida.
Currently, our knowledge of the geographical spread of P. agathidicida is limited primarily to the presence of visible disease symptoms combined with soil-baiting assays (Bradshaw et al. 2020). More thorough knowledge of the full range of P. agathidicida could potentially help answer questions regarding disease resistance in kauri trees and environmental factors that lead to infection. The knowledge gap is mainly due to the lack of a quick, cost-effective and simple diagnostic tool. While effective, soil-baiting is not amenable to high-throughput, widespread testing. Both FAME analysis and DNA based molecular diagnostics are promising but have limitations. Further development of these techniques in conjunction with soil-baiting would help to expand our knowledge of the distribution of P. agathidicida in New Zealand enabling better management of kauri dieback disease.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Natascha Lewe for her assistance with fatty acid extractions and Mike Fairhurst for his assistance in isolating oospores.
Contributor Information
Randy F Lacey, School of Biological Sciences, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand; Centre for Biodiscovery, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand.
Blake A Sullivan-Hill, School of Biological Sciences, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand.
Julie R Deslippe, School of Biological Sciences, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand; Centre for Biodiversity and Restoration Ecology, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand.
Robert A Keyzers, Centre for Biodiscovery, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand; School of Chemical and Physical Sciences, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand; Centre for Biodiversity and Restoration Ecology, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand.
Monica L Gerth, School of Biological Sciences, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand; Centre for Biodiscovery, Victoria University of Wellington, PO Box 600 Wellington 6140, New Zealand.
FUNDING
This work was supported via strategic research funds (to MLG) from the School of Biological Sciences at Victoria University of Wellington and Seed and Scope funding (to JRD) from Ngā Rākau Taketake, part of the Bioheritage National Science Challenge.
Conflicts of Interest
None declared.
REFERENCES
- Beever RE, Bellgard SE, Dick MAet al. Detection of Phytophthora taxon Agathis (PTA). Landcare report LC0910/137. Ministry for Agriculture & Forestry, Biosecurity New Zealand, Wellington, New Zealand, 2010. [Google Scholar]
- Bellgard SE, Padamsee M, Probst CMet al. Visualizing the early infection of Agathis australis by Phytophthora agathidicida, using microscopy and fluorescent in situ hybridization. For Pathol. 2016;46:622–31. [Google Scholar]
- Bradshaw RE, Bellgard SE, Black Aet al. Phytophthora agathidicida: research progress, cultural perspectives and knowledge gaps in the control and management of kauri dieback in New Zealand. Plant Pathol. 2020;69:3–16. [Google Scholar]
- Cavigelli MA, Robertson GP, Klug MJ. Fatty acid methyl ester (FAME) profiles as measures of soil microbial community structure. Plant Soil. 1995;170:99–113. [Google Scholar]
- Cheng MH, Walker TH, Hulbert GJet al. Fungal production of eicosapentaenoic and arachidonic acids from industrial waste streams and crude soybean oil. Bioresour Technol. 1999;67:101–10. [Google Scholar]
- Dodds ED, McCoy MR, Rea LDet al. Gas chromatographic quantification of fatty acid methyl esters: flame ionization detection vs. electronImpact mass spectrometry. Lipids. 2005;40.419–28. [DOI] [PubMed] [Google Scholar]
- Drenovsky RE, Elliot GN, Graham KJet al. Comparison of phospholipid fatty acid (PLFA) and total soil fatty acid methyl esters (TSFAME) for characterizing soil microbial communities. Soil Biol Biochem. 2004;36:1793–800. [Google Scholar]
- Duan CH, Riley MB, Jeffers SN. Effects of growth medium, incubation temperature, and mycelium age on production of five major fatty acids by six species of Phytophthora. Arch Phytopathol Plant Protect. 2011;44:142–157. [Google Scholar]
- Duan CH, Riley MB, Jeffers SN. Evaluation of fatty acid methyl ester profile and amplified fragment length polymorphism analyses to distinguish five species of Phytophthora associated with ornamental plants. Arch Phytopathol Plant Protect. 2013;46:295–311. [Google Scholar]
- Ehrhardt CJ, Chu V, Brown Tet al. Use of fatty acid methyl ester profiles for discrimination of Bacillus cereus T-strain spores grown on different media. Appl Environ Microbiol. 2010;76:1902–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DC, Ribeiro OK. Phytophthora Diseases Worldwide. St. Paul, MN: APS Press, 1996. [Google Scholar]
- Fairhurst MJ, Deslippe JR, Gerth ML. A fluorescence-based viability assay for Phytophthora agathidicida oospores. PhytoFrontiers. 2021, In press. [Google Scholar]
- Griffiths RG, Dancer J, O'Neill Eet al. Effect of culture conditions on the lipid composition of Phytophthora infestans. New Phytol. 2003;158:337–44. [Google Scholar]
- Hansen ZR, Knaus BJ, Tabima JFet al. Loop-mediated isothermal amplification for detection of the tomato and potato late blight pathogen, Phytophthora infestans. J Appl Microbiol. 2016;120:1010–20. [DOI] [PubMed] [Google Scholar]
- Lacey RF, Fairhurst MJ, Daley KJet al. Assessing the effectiveness of oxathiapiprolin towards Phytophthora agathidicida, the causal agent of kauri dieback disease. bioRxiv. 2021. DOI: 10.1101/2021.03.10.434845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RP, Groves CL. Identification and characterization of isolates of Phytophthora infestans using fatty acid methyl ester (FAME) profiles. Plant Dis. 2003;87:1233–43. [DOI] [PubMed] [Google Scholar]
- McDougal RL, Bellgard SE, Scott PMet al. Comparison of a real-time PCR assay and a soil bioassay technique for detection of Phytophthora taxon Agathis from soil. MPI contract report 53789. Wellington, New Zealand: New Zealand Forestry Research Institute Ltd, 2014. [Google Scholar]
- O'Brien PA, Williams N, Hardy GES. Detecting Phytophthora. Crit Rev Microbiol. 2009;35:169–81. [DOI] [PubMed] [Google Scholar]
- Olsson PA. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiol Ecol. 1999;29:303–10. [Google Scholar]
- Rastogi G, Rajesh KS. Molecular techniques to assess microbial community structure, function, and dynamics in the environment. In: Ahmad I, Ahmad F, Pichtel J (eds), Microbes and Microbial Technology. New York, NY: Springer, 2011, 29–57. [Google Scholar]
- Scott PM, Williams NM. Phytophthora diseases in New Zealand forests. New Zeal J For. 2014;59:14–21. [Google Scholar]
- Smart A, Byrne J, Hammerschmidt Ret al. Evolving plant diagnostics during a pandemic. Plant Health Progr. 2021;22. DOI:10.1094/PHP-08-20-0074-MR. [Google Scholar]
- Spring O, Haas K. The fatty acid composition of Plasmopara halstedii and its taxonomic significance. Eur J Plant Pathol. 2002;108:263–7. [Google Scholar]
- Than DJ, Hughes KJD, Boonham Net al. A TaqMan real-time PCR assay for the detection of Phytophthora ‘taxon Agathis’ in soil, pathogen of Kauri in New Zealand. For Pathol. 2013;43. DOI:10.1111/efp.12034. [Google Scholar]
- Waipara NW, Hill S, Hill LMWet al. Surveillance methods to determine tree health, distribution of kauri dieback disease and associated pathogens. N Z Plant Protect. 2013;66.235–241. [Google Scholar]
- Weir BS, Paderes EP, Anand Net al. A taxonomic revision of Phytophthora Clade 5 including two new species, Phytophthora agathidicida and Phytophthora cocois. Phytotaxa. 2015;205:21–38. [Google Scholar]
- Welch DF. Applications of cellular fatty acid analysis. Clin Microbiol Rev. 1991;4:422–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DC, Lytle CA, Gan YDet al. Flash detection/identification of pathogens, bacterial spores and bioterrorism agent biomarkers from clinical and environmental matrices. J Microbiol Methods. 2002;48:139–47. [DOI] [PubMed] [Google Scholar]
- Winkworth RC, Nelson BCW, Bellgard SEet al. A LAMP at the end of the tunnel: a rapid, field deployable assay for the kauri dieback pathogen, Phytophthora agathidicida. PLoS ONE. 2020;15:e0224007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YP, Othman S, Lau YLet al. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J Appl Microbiol. 2018;124:626–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef LF, Wojno M, Dick WAet al. Lipid profiling of the soybean pathogen Phytophthora sojae using Fatty Acid Methyl Esters (FAMEs). Fung Biol. 2012;116:613–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


