Abstract
Cyclin-dependent kinase (CDK)-activating kinases (CAKs) carry out essential activating phosphorylations of CDKs such as Cdc2 and Cdk2. The catalytic subunit of mammalian CAK, MO15/Cdk7, also functions as a subunit of the general transcription factor TFIIH. However, these functions are split in budding yeast, where Kin28p functions as the kinase subunit of TFIIH and Cak1p functions as a CAK. We show that Kin28p, which is itself a CDK, also contains a site of activating phosphorylation on Thr-162. The kinase activity of a T162A mutant of Kin28p is reduced by ∼75 to 80% compared to that of wild-type Kin28p. Moreover, cells containing kin28T162A and a conditional allele of TFB3 (the ortholog of the mammalian MAT1 protein, an assembly factor for MO15 and cyclin H) are severely compromised and display a significant further reduction in Kin28p activity. This finding provides in vivo support for the previous biochemical observation that MO15-cyclin H complexes can be activated either by activating phosphorylation of MO15 or by binding to MAT1. Finally, we show that Kin28p is no longer phosphorylated on Thr-162 following inactivation of Cak1p in vivo, that Cak1p can phosphorylate Kin28p on Thr-162 in vitro, and that this phosphorylation stimulates the CTD kinase activity of Kin28p. Thus, Kin28p joins Cdc28p, the major cell cycle Cdk in budding yeast, as a physiological Cak1p substrate. These findings indicate that although MO15 and Cak1p constitute different forms of CAK, both control the cell cycle and the phosphorylation of the C-terminal domain of the large subunit of RNA polymerase II by TFIIH.
The eukaryotic cell division cycle is regulated by a series of kinase activities that increase and diminish with periodicity. These kinases, which belong to the cyclin-dependent kinase (CDK) family, are themselves positively regulated by cyclin binding partners and activating phosphorylations and are negatively regulated by inhibitory binding proteins and inhibitory phosphorylations (for general reviews, see references 36, 46, and 59). Full kinase activity of most CDKs requires activating phosphorylations of a threonine located in a flexible region termed the T loop. Cdc2 and Cdk2 both require this activating phosphorylation for functional activity in vivo and in vitro (14, 26, 27, 57, 60). Other CDKs, including Cdk4 (33), Cdk6 (30), and Cdk7 (23, 35), are also activated by similar phosphorylations. Crystallographic studies of Cdk2 suggest that this phosphorylation may help organize an acidic patch and thereby enhance protein substrate binding at the Cdk2 active site (52; reviewed in reference 47). In addition to stimulating substrate binding, activating phosphorylations may also stabilize interactions between certain CDKs and their cyclin partners (12, 14, 26, 43).
In mammals, activating phosphorylations of Cdc2 (11, 60), Cdk2 (21, 50, 58), Cdk4 (44), and Cdk6 (30) are mediated by a CDK-activating kinase (CAK). Purification of CAK activity from starfish and Xenopus revealed that CAK contains the protein kinase MO15 (21, 50, 58), which is also called Cdk7 (23). The activity of MO15 requires binding of its cyclin partner, cyclin H, to form a dimeric complex (23, 41) and either an activating phosphorylation (on threonine-170 in human MO15) (23, 35, 43) or binding of the assembly factor MAT1 to form a trimeric complex (13, 22, 63). In addition to their roles as a CAK, MO15, cyclin H, and MAT1 are subunits of the RNA polymerase II (Pol II) basal transcription factor TFIIH (1, 51, 54, 55). MO15 contained in TFIIH phosphorylates the heptapeptide repeat located in the carboxy-terminal domain (CTD) of the large subunit of RNA Pol II, stimulating transcriptional elongation (2, 40; reviewed in references 9, 42, and 49).
The Saccharomyces cerevisiae ortholog of MO15, Kin28p (56), is a subunit of yeast TFIIH but does not display CAK activity (7). Originally identified because of its homology with Cdc28p, Kin28p is essential (56), and mutants show diminished CTD phosphorylation and impaired RNA Pol II transcription (7, 66). In addition to Kin28p (20), the yeast orthologs of cyclin H and MAT1, Ccl1p (67) and Tfb3p (19) respectively, are both subunits of yeast TFIIH. Consistent with these observations, kin28 (7, 66), ccl1 (65), and tfb3 (17) mutant strains fail to display cell cycle arrest morphologies, and are all severely deficient in RNA Pol II transcription.
Biochemical purification and characterization of Cak1p, the CAK enzyme from yeast (15, 32, 64), showed that it is active as a monomer, inactive toward the CTD of RNA Pol II (31), the physiological CAK of Cdc28p (32, 64), and not a subunit of TFIIH. In contrast with kin28 mutants, cak1 mutants are blocked in cell cycle progression (32, 64). The genetically simpler yeast, therefore, has two separate gene products for CAK and TFIIH functions, whereas higher eukaryotes use MO15 and cyclin H for both.
Like MO15, Kin28p contains a potential site of activating phosphorylation. In order to characterize the function and regulation of Kin28p, we examined the properties of kin28 strains lacking this activating threonine in vivo. Kin28pT162A has significantly reduced kinase activity and displays a very strong phenotype in a tfb3-ts strain background, thus supporting the dual regulation of MO15 by activating phosphorylation and MAT1 binding suggested by experiments in vitro. In contrast to a recent report (16), activating phosphorylation of Kin28p is not essential for Kin28p function or cell viability in the presence of wild-type TFB3. Finally, we provide evidence that Cak1p regulates the activating phosphorylation of Kin28p in vivo, phosphorylates Kin28p on this site in vitro, and thereby activates Kin28p. Together, these experiments link the functions of Cak1p and Kin28p.
MATERIALS AND METHODS
Plasmids and strains.
Unless otherwise noted, all yeast strains were derived from YMW1 (MATα ade2-1 ade3-22 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100). Specific plasmids and genotypes of strains in this study are listed in Tables 1 and 2. Temperature-sensitive tfb3 strains (rig2-ts [17]) were crossed with YJK1744, transformed with pGK13 or pGK36, and grown on 5-fluoro-orotic acid (FOA) to derive YJK1844, YJK1845, YJK1848, and YJK1849.
TABLE 1.
Plasmids
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pAF21 | lacZ in YEplac112GAL | A. Fluegge |
| pGK11 | KIN28 in YCplac22 | This work |
| pGK12 | KIN28 in YEplac112 | This work |
| pGK13 | KIN28(HA) in YCplac22 | 7 |
| pGK14 | KIN28(HA) in YEplac112 | This work |
| pGK21 | KIN28 TRP1 (pSf19) | 7 |
| pGK22 | kin28-ts16 in YCplac22 | 7 |
| pGK33 | kin28(HA)-ts16 in YCplac22 | This work |
| pGK36 | KIN28(HA)T162A in YCplac22 | This work |
| pGK37 | KIN28(HA)T162A in YEplac112 | This work |
| pGK39 | KIN28(HA)AF in YCplac22 | This work |
| pGK40 | KIN28(HA)AF in YEplac112 | This work |
| pGK41 | KIN28(HA)AF/T162A in YCplac22 | This work |
| pGK42 | KIN28(HA)AF/T162A in YEplac112 | This work |
| pGK44 | KIN28(HA)T162S in YEplac112 | This work |
| pGK45 | KIN28(HA)T162E in YEplac112 | This work |
| pGK47 | KIN28(HA)T162D in YEplac112 | This work |
| pJK1 | KIN28(HA) in YCplac33 | This work |
| pJK12 | KIN28(HA)T162A in YCplac33 | This work |
| pJK25 | kin28(HA)-ts16T162A in YCplac22 | This work |
| pMS454 | KIN28(HA)D147N in YCplac22 | This work |
| pMS455 | KIN28(HA)T162S in YCplac22 | This work |
| pMS456 | KIN28(HA)D147N in YEplac112 | This work |
| pMS457 | KIN28(HA)T162S in YEplac112 | This work |
| YCplac22 | TRP1 CEN | 25 |
| YCplac33 | URA3 CEN | 25 |
| YEplac112 | TRP1 2μm | 25 |
| SB74 | cak1-22 CEN LEU2 | 32 |
| SB76 | CAK1 CEN LEU2 | 32 |
TABLE 2.
Strains
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| YGL16 | kin28Δ::LEU2 [pGK12] | This work |
| YGL17 | kin28Δ::LEU2 [pGK14] | This work |
| YGL20 | kin28Δ::LEU2 [pGK11] | This work |
| YGL24 | kin28Δ::LEU2 [pGK21] | This work |
| YGL26 | kin28Δ::LEU2 [pGK13] | This work |
| YGL42 | kin28Δ::LEU2 [pGK36] | This work |
| YGL43 | kin28Δ::LEU2 [pGK37] | This work |
| YGL44 | kin28Δ::LEU2 [pGK40] | This work |
| YGL45 | kin28Δ::LEU2 [pGK40] | This work |
| YGL47 | kin28Δ::LEU2 [pGK41] | This work |
| YGL48 | kin28Δ::LEU2 [pGK42] | This work |
| YGL62 | kin28Δ::LEU2 [pMS457] | This work |
| YGL68 | kin28Δ::LEU2 [pGK47] | This work |
| YGL66 | kin28Δ::LEU2 [pGK45] | This work |
| YJK1599 | cak1Δ::HIS3 kin28::LEU2 [pGK22-1, SB74] | This work |
| YJK1600 | cak1Δ::HIS3 kin28::LEU2 [pGK22-1, SB74] | This work |
| YJK1610 | cak1Δ::HIS3 [pGK13, SB74] | This work |
| YJK1614 | cak1Δ::HIS3 [pGK13, SB76] | This work |
| YJK1624 | cak1Δ::HIS3 [pGK14, SB76] | This work |
| YJK1625 | cak1Δ::HIS3 [pGK14, SB74] | This work |
| YJK1744 | kin28Δ::LEU2 [pJK1] | This work |
| YJK1747 | kin28Δ::LEU2 [pGK13, pAF21] | This work |
| YJK1749 | kin28Δ::LEU2 [pGK36, pAF21] | This work |
| YJK1755 | kin28Δ::LEU2 [pJK12, pMS454] | This work |
| YJK1756 | kin28Δ::LEU2 [pJK12, pMS456] | This work |
| YJK1767 | kin28Δ::LEU2 [pJK13, pMS454] | This work |
| YJK1768 | kin28Δ::LEU2 [pJK1, pMS456] | This work |
| YJK1773 | MATa bar1Δ kin28Δ::LEU2 [pGK13] | This work |
| YJK1828 | rig2-ts14 (GF2214 strain background) | 17 |
| YJK1829 | rig2-ts23 (GF2217 strain background) | 17 |
| YJK1844 | kin28Δ::LEU2 rig2-ts14 [pGK13] | This work |
| YJK1845 | kin28Δ::LEU2 rig2-ts14 [pGK36] | This work |
| YJK1848 | kin28Δ::LEU2 rig2-ts23 [pGK13] | This work |
| YJK1849 | kin28Δ::LEU2 rig2-ts23 [pGK36] | This work |
| YJK1869 | kin28Δ::LEU2 [pJK1, YCplac22] | This work |
| YJK1870 | kin28Δ::LEU2 [pJK12, YCplac22] | This work |
| SY143 | cak1::HIS3 [SB74] | 32 |
| SY162 | cak1::HIS3 [SB76] | 62 |
KIN28 disruption and constructs were derived from plasmids described previously (7). All non-temperature-sensitive point mutants were generated by PCR of a KIN28-HA plasmid (pGK13) by using oligonucleotides that introduce restriction sites for diagnostic purposes. For the ts/T162A allele, kin28-ts16 (pGK33) was used as a template for amplification. Sense primers for point mutants are as follows (altered codons are underlined, and diagnostic restriction sites are in parentheses): T162A, CCCACATGAGATACTGGCAAGTAACGTCGTAACAA (BsrI); AF, GTTGGTGAGGGTGCTTTTGCGGTTGTTTACTTGGG (eliminates RsaI); D147N, CTGATGGCCAGATAAAAGTCGCGAATTTCGGTCTAGCAAGGG (NruI); T162S, CCCCACATGAGATACTCTCGAGTAACGTCGTAACAAG (XhoI); and T162D and T162E, GCCCCACATGAGATACTCGAG/TTCGAACGTCGTAACAAGATG (BstBI and XhoI, respectively). All mutants were sequenced in their entirety and cloned into vector plasmids (25) by using PstI and HindIII restriction sites.
CAK1 constructs were described previously (32).
The β-galactosidase reporter plasmid contained lacZ after a GAL1/GAL10 promoter. The cloning strategy for the GAL1/GAL10 promoter-containing vector was described previously (34); lacZ was cloned by using BamHI and HindIII cloning sites in the vector.
Recombinant baculoviruses expressing FLAG-Kin28p and FLAG-Ccl1p were prepared and purified via the FLAG tags as described previously (28). FLAG-Kin28pT162A was produced by site-directed in vitro mutagenesis with oligonucleotide CCACATGAGATACTGGCAAGTAACGTCGTAACA. The mutation was verified by DNA sequencing. Staining of protein gels indicated that the preparations of FLAG-Kin28p and FLAG-Ccl1p were homogeneous (28) and that the preparations of monomeric FLAG-Kin28p were ∼1 to 5% pure. As indicated below, despite these differences in purity, approximately equal amounts of Kin28p were used as substrates for Cak1p.
Buffers.
The 1× protease inhibitor mix (PI) contained 10 μg each of leupeptin, chymostatin, and pepstatin (Chemicon) per ml. EB is 80 mM β-glycerophosphate (pH 7.3)–20 mM EGTA–15 mM MgCl2–10 mM dithiothreitol (DTT)–1 mg of ovalbumin per ml–1× PI; 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer is 0.5 M Tris (pH 8.0)–10% SDS–20 mM EDTA–250 mM DTT–50% glycerol. Plates and media were made as described previously (5).
Preparation of lysates.
Nondenaturing lysates were prepared by using a modified protocol previously applied for extracting TFIIH from yeast (18). Exponentially growing cultures were harvested by filtration, washed with deionized water, and transferred into 1.5-ml Microfuge tubes (0.2 to 0.4 g of pellets per tube), with addition of 0.8 g of acid-washed glass beads (0.5-mm diameter; Sigma) and lysis buffer [150 mM Tris acetate (pH 7.5), 300 mM (NH4)2SO4, 1 mM EDTA, 1 mM spermidine, 1 mM DTT, 10% glycerol, and 1× PI] to fill the remaining volume. Samples were lysed by seven 1-min pulses on a bead beater (Mini-Beadbeater-8; Biospec Products) followed by 1-min incubations in a −10°C ice-NaCl slurry. Glass beads and debris were removed by centrifugation at 15,000 rpm in a Microfuge at 4°C for 10 min. The supernatant was clarified by centrifugation at 70,000 rpm for 30 min at 4°C in the TLA 100.2 rotor in a Beckman Optima ultracentrifuge. Crude extracts were aliquoted, frozen in liquid nitrogen, and stored at −80°C. Typical extract concentrations were 4 to 7 mg/ml.
Denaturing lysates for Fig. 1B were prepared as follows: 100 to 200 μl of cell pellets was homogenized as described above in a bead beater for 7 min in an equal volume of SDS-PAGE sample buffer and acid-washed glass beads, boiled for 5 min, and clarified at 15,000 rpm in a Microfuge.
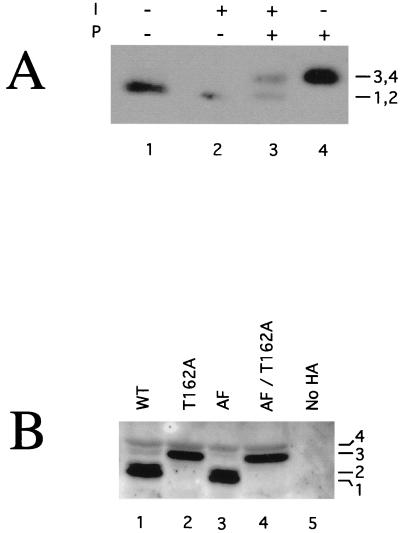
FIG. 1.
Kin28p is a phosphoprotein. (A) Phosphatase treatment. Kin28p-HA was immunoprecipitated from native yeast lysates and incubated with lambda phosphatase (P) and/or phosphatase inhibitor (I). Samples were subject to SDS-PAGE followed by immunoblotting. (B) Electrophoretic mobilities of Kin28p point mutants. Strains carrying a kin28 deletion covered by plasmids containing HA epitope-tagged Kin28p point mutants were harvested during exponential growth, lysed directly into SDS-PAGE sample buffer, and analyzed by immunoblotting. AF denotes a T18A/Y19F double mutant. “No HA” indicates that a strain lacks HA-tagged proteins. Immunoblots produced four Kin28p species (see also Fig. 2A). WT, wild type
Immunoblotting.
SDS–12.5% polyacrylamide gels were transferred to polyvinylidene difluoride membranes (Millipore) by using either a tank or semidry blotting transfer system. Membranes were blocked for at least 1 h with TBST Blotto (10 mM Tris [pH 8.0], 150 mM NaCl, 0.1% Tween, 5% milk powder), incubated with primary antibodies for at least 1 h, washed with TBST (TBST Blotto without milk powder) five times for 5 min each, incubated for 1 to 2 h with secondary antibodies, washed with TBST five times for 5 min each, washed with TBS (TBST without Tween 20), incubated with chemiluminescence reagents (Pierce), and exposed to film.
One-dimensional (1-D) antihemagglutinin (anti-HA) blots were probed with either 12CA5 antibody (10 μg/ml) or rabbit anti-HA antibodies (80 ng/ml) (Santa Cruz). 2-D blots were probed with rabbit anti-HA antibodies (50 ng/ml). FLAG immunoblotting was conducted with 40 ng of polyclonal rabbit anti-FLAG (Santa Cruz) per ml followed by 160 ng of horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Pierce) per ml.
Immunoprecipitation.
Twenty microliters of protein A-agarose beads (Gibco BRL) was used per immunoprecipitation. The beads were washed seven times with 800 μl of EB containing 1% Nonidet P-40 (NP-40), incubated for a minimum of 1 h at 4°C with 10 μg of 12CA5 antibody per immunoprecipitation, washed seven times with 800 μl of EB containing 1% NP-40, incubated for at least 1 h with 400 μg of yeast lysate diluted into 4 volumes of EB containing 1% NP-40, washed three times with 300 μl of EB containing 1% NP-40, and then washed three times with 300 μl of EB. Final pellets were resuspended to a total volume of 100 μl and either used in experiments or stored at −80°C following freezing in liquid nitrogen.
Phosphatase treatments.
Kin28p immunoprecipitates were prepared as described above, with EB replaced by HB (EB containing 20 mM HEPES [pH 7.3] instead of β-glycerophosphate). Immunoprecipitates were incubated at 37°C for 45 min in 1× lambda phosphatase buffer (New England Biolabs)–1× PI–1 mM phenylmethylsulfonyl fluoride with various combinations of 800 U of lambda phosphatase (New England Biolabs) and 20 mM sodium pyrophosphate.
2-D electrophoresis.
Nondenaturing yeast lysates were made by using yeast extract buffer (100 mM NaCl, 20 mM Tris [pH 7.5], 10 mM EDTA, 2 mM EGTA, 5% glycerol, 1× PI) according to the procedure described above. Fifty microliters of nondenaturing yeast lysate was treated with 10 U of protease-free DNase and 25 μg of protease-free RNase A (both from Worthington Biochemical Corp.) and 5 mM MgCl2 on ice for 30 min, followed by incubation at 4°C for 15 min. Proteins were precipitated overnight with 9 volumes of acetone at −20°C, pelleted at 8,000 rpm for 15 min in a Microfuge, and air dried to translucence. The dried pellets were dissolved in 100 μl of IEF sample buffer (9 M urea [American Bioanalytical], 65 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 65 mM DTT, 5% Resolyte 4-8 [BDH Biochemicals]), and 10 μl was loaded into 8-mm tube gels containing 5% acrylamide premix (derived from a 37.5:1 acrylamide-bisacrylamide mix), 9.25 M deionized urea solution, 27 mM CHAPS, 2.8% Resolyte 4-8, and 2.8% Resolyte 5-7 (BDH Biochemicals). Gels were focused for 4.5 h at 400 V with 20 mM NaOH catholyte and 10 mM phosphoric acid anolyte. The gels were extruded into equilibration buffer (77 mM Tris buffer [pH 6.8], 3.1% SDS, 32 mM DTT), incubated for 5 min, and subjected to SDS-PAGE and immunoblotting as described above.
Kinase assays. (i) CTD kinase assay.
CTD kinase assays were performed essentially as described previously (7). Briefly, 10 μl of immunoprecipitate was incubated in the presence of 3.0 μCi of [γ-32P]ATP, 0.375 μM ATP, and 4 μg of CTD peptide [(YSPTSPS)4] in a total volume of 16 μl (filled with EB) at room temperature. Assays were terminated after 15 min by adding 4 μl of 5× sample buffer, and the mixtures were loaded onto SDS–12.5% polyacrylamide gels. The gels were Coomassie blue stained, dried, and visualized by using phosphorimaging (Molecular Imager GS-250; Bio-Rad) and autoradiography.
(ii) Phosphorylation of CDKs.
Purified glutathione S-transferase–Cak1p (7.5 ng) was incubated with 10 ng of Cdk2, ∼30 ng of FLAG-Kin28p–Ccl1p, or ∼50 ng of FLAG-Kin28pT162A in the presence of 5 μCi of [γ-32P]ATP, 10 μM ATP, and 20 mM MgCl2 in EB (final volume, 16 μl; EB was as described above except that 10× PI mix was used). The reactions were terminated after 30 min at room temperature by adding 7 μl of 5× SDS-PAGE sample buffer, and the products were run on SDS–10% PAGE and analyzed by phosphorimaging and autoradiography.
Expression assays.
Northern blotting was conducted as follows. Cultures were grown to exponential phase at 30°C in yeast extract-peptone-dextrose (YPD) and reinoculated into YPD containing 0.9 M NaCl. RNA was extracted from intact yeast cells by a hot-phenol-chloroform protocol (5), and duplicate samples were run in formaldehyde-containing agarose (5) and transferred to a GeneScreen membrane (Dupont). RNA was UV-cross-linked to the membrane at 1,200 mJ/cm2. Labeled probe was prepared from a PstI/BglII fragment of GPD1 (the pUCGPD1 plasmid was a gift from Michael Gustin, Rice University [4]). Results were visualized and quantitated by phosphorimager analysis.
Galactose promoter induction assays were conducted as follows. Cells were grown to exponential phase in CM-Trp-Ura (5) with raffinose at 30°C, and expression of the GAL1/GAL10 promoter-linked lacZ reporter was induced by addition of 30% galactose to 2%. One-milliliter samples were harvested by centrifugation in Microfuge tubes. Cells were permeabilized by resuspending them in 500 μl of ZF1 buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 38 mM 2-mercaptoethanol, pH 7.0), adding 10 μl of 0.1% SDS and 20 μl of chloroform, vortexing, and incubating at 32°C for 5 min. Assays were conducted by adding 100 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml), incubating at 32°C for various intervals, stopping the reaction by adding 500 μl of Na2CO3, pelleting the cells, and measuring the optical density at 420 nm (OD420) of the supernatant. Because the assays were only approximately linear with respect to substrate incubation times and OD, the substrate incubation times were equal within each time point.
Cell synchronization.
A MATa bar1Δ strain (YJK1773) was grown in CM-Trp (5) at 30°C. When the OD600 reached ∼0.6, three 50-ml cultures were harvested. One was saved for the asynchronous sample; the other two were reinoculated into YPD containing 50 μg of benomyl (Dupont) per ml. Cultures were arrested for 2.5 h in YPD containing 100 ng of α-factor per ml. Cultures were harvested by filtration, washed twice with at least 100 ml of CM-Trp, and resuspended in prewarmed CM-Trp. Samples (50 ml) were harvested at 15-min intervals following release from α-factor. Benomyl-arrested cells were collected after 3.75 and 4.75 h of incubation. Each harvested sample was washed once with deionized water, frozen in liquid nitrogen, and stored at −80°C until lysis.
At each time point, 400-μl culture samples were preserved with 100 μl of 37% formaldehyde, mixed, and refrigerated. At the end of the time course, time point labels on each tube were covered to prevent any potential bias, samples were sonicated for 20 s, and bud indices were counted.
RESULTS
Kin28p is phosphorylated on threonine 162.
In the course of our studies, we noticed that Kin28p resolved into two to four species on SDS-PAGE (unless stated otherwise, all Kin28p used in this work contains a C-terminal influenza virus HA epitope tag [7]). To test whether Kin28p, like most other CDKs, is a phosphoprotein, we treated Kin28p immunoprecipitates with lambda phosphatase and monitored the effect on electrophoretic mobility by immunoblotting. Treatment with phosphatase converted the immunoprecipitated Kin28p to a slower-migrating species (Fig. 1A, compare lane 4 with lane 1). Inclusion of a phosphatase inhibitor partially blocked this effect (lane 3), indicating that the higher-mobility form of Kin28p is due to phosphorylation.
To examine some potential sites of phosphorylation, we constructed mutant alleles of KIN28 that substituted nonphosphorylatable residues at positions homologous to the sites of regulatory phosphorylation in other CDKs. Our point mutants included T162A (corresponding to the site of activating phosphorylation in Cdk2, Thr-160), T17A/Y18F (corresponding to sites of inhibitory phosphorylation at Thr-14 and Tyr-15 in Cdk2 and hereafter called AF), and mutants that contained all three substitutions (AF/T162A). We prepared extracts from strains containing each of the four mutants as well as a non-HA-tagged control and compared the electrophoretic mobilities of Kin28p by immunoblotting. In high-resolution gels, wild-type Kin28p resolved into four species (Fig. 1B, lane 1). The T162A mutant produced only the two lower-mobility species (lane 2). The AF mutant was indistinguishable from the wild type (lanes 1 and 3), and the AF/T162A triple mutant was indistinguishable from the T162A single mutant (lanes 4 and 2). Thus, Kin28p appears to be phosphorylated on Thr-162. We have never observed evidence for phosphorylation on Thr-17 or Tyr-18, including by immunoblotting with an antiphosphotyrosine antiserum (data not shown). Bands 1-2 and 3-4 in Fig. 1B correspond to the two bands observed in Fig. 2A. We do not fully understand why the resolution of identical samples varies from gel to gel.
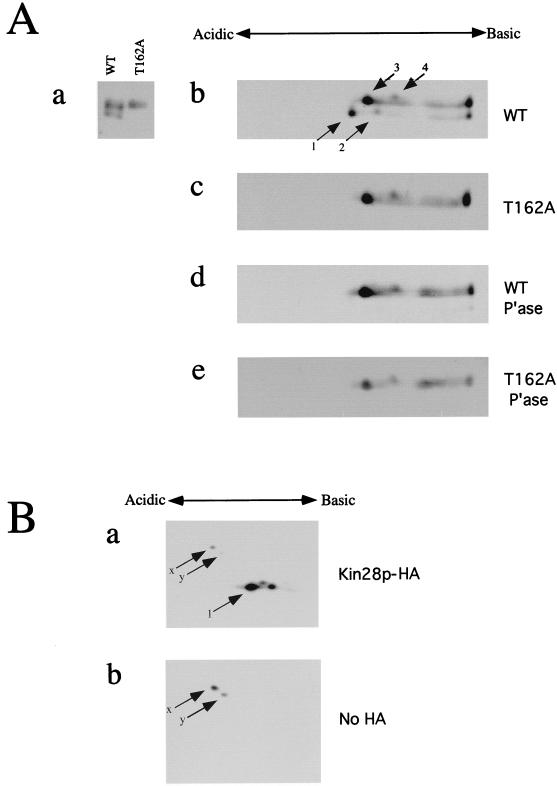
FIG. 2.
2-D gel analysis of Kin28p. (A) Protein samples containing Kin28p alleles were subjected to isoelectric focusing with a 1:1 mixture of pH 4 to 8 and pH 5 to 7 carrier ampholytes, run on SDS–12.5% polyacrylamide gels in the second dimension, and immunoblotted. To aid in identifying spots, lanes containing Kin28p and Kin28pT162A samples were run alongside tube gels in the second dimension (a). Kin28p alleles tested included wild-type (WT) Kin28p (b), Kin28pT162A (c), phosphatase (P’ase)-treated wild-type Kin28p (d), and phosphatase-treated Kin28pT162A (e). (B) To rule out the possibility of a cross-reacting species in these immunoblots, extracts from cells expressing tagged (a) or untagged (b) Kin28p were subjected to isoelectric focusing with pH 4 to 8 carrier ampholytes and processed as described for panel A.
To further analyze Kin28p phosphorylation, we resolved Kin28p by using 2-D electrophoresis. Phosphorylated Kin28p species should be more acidic than the unphosphorylated forms. Wild-type, mutant, and phosphatase-treated forms of Kin28p were focused by using a mixture of ampholytes that produced high resolution in the pH 5 to 7 range; 1-D SDS-PAGE lanes run in parallel allowed us to assign spots on the 2-D gels to bands in the vertical axis (Fig. 2A, panel a). Wild-type Kin28p produced four species (Fig. 2A, panel b), consistent with the four bands observed in Fig. 1B. Because not all of the Kin28p entered the first-dimension gel, streaks leading up to spots at the extreme basic end are apparent. The amount of this material was highly variable among experiments. To test whether any spots are attributable to nonspecific cross-reactivity of our antibody, we performed immunoblotting with extracts from strains containing or lacking HA-tagged Kin28p that had been resolved by isoelectric focusing (Fig. 2B, panels a and b, respectively). Spots 1 to 4 were completely absent from the non-HA sample (b), although cross-reacting species (spots x and y) in both blots confirmed that focusing and loading levels were similar. The T162A mutants produced only spots 3 and 4 (Fig. 2A, panel c), as did the phosphatase-treated samples (Fig. 2A, panels d and e). The AF mutant produced a pattern identical to that of the wild type (data not shown), suggesting that Kin28p is not modified at these sites. As expected, spots 3 and 4 produced by the T162A mutant or by phosphatase treatment of the wild-type Kin28p were shifted toward the basic pole compared with spots 1 and 2. We conclude that spots 1 and 2 correspond to the Thr-162-phosphorylated forms of spots 3 and 4, respectively.
We were surprised to observe two spots following phosphatase treatment of wild-type Kin28p or Kin28pT162A (Fig. 2A) or of the T18A/Y19F double mutant (data not shown). Because we observed corresponding faint bands on 1-D gels and consistently observed them in isoelectric focusing, we inferred that Kin28p is posttranslationally modified by something other than phosphorylation that shifts the isoelectric point by about twice the charge of a single phosphorylation. It is unclear which of the two species present following phosphatase treatment is the unmodified form, and it is difficult to derive a second species by conceptual proteolysis of a few residues from either end of Kin28p. A similar modification may have been observed previously with an untagged form of Kin28p (20). Whatever the nature of this modification, it appears to occur independently of Thr-162 phosphorylation.
Biochemical activity of Kin28p point mutants.
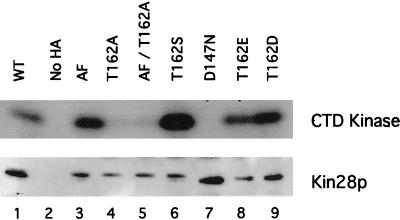
As part of our characterization of the biochemical properties of Kin28p, we tested whether any of the Kin28p point mutants affected kinase activity, expecting that the Kin28pT162A mutants would be severely compromised, as are analogous mutants of Cdc2 and Cdk2. We performed CTD kinase assays and anti-HA immunoblotting on immunoprecipitates of wild-type Kin28p and Kin28p point mutants expressed from multicopy plasmids (Fig. 3). Wild-type Kin28p showed strong CTD kinase activity (upper panel, lane 1), as did the Kin28pAF mutant (lane 3). In contrast, Kin28pT162A mutants had significantly lower kinase activity (lanes 4 and 5). Full kinase activity was present in mutants containing substituted serine (T162S) (lane 6) or acidic residues (T162D/T162E) (lanes 8 and 9) designed to mimic a constitutively phosphorylated Thr-162. Immunoprecipitates from a strain containing untagged Kin28p displayed no activity (lane 2), as did immunoprecipitates of a mutant designed to lack catalytic activity (D147N) (lane 7). Corresponding immunoblots demonstrated comparable loadings and indicated that the Kin28pT162E and Kin28pT162D mutants ran with the faster mobility observed for phosphorylated Kin28p (Fig. 3, lower panel, lanes 8 and 9). Subsequent experiments in which Kin28p was expressed from low-copy-number plasmids produced essentially identical results (data not shown), and phosphorimager quantification normalized to protein loading indicated that Kin28pAF and Kin28pT162S had the same kinase activities as wild-type Kin28p; Kin28pT162A, however, was only 20 to 25% as active. These results indicate that phosphorylation of Thr-162, although not essential for kinase activity, substantially increases Kin28p activity.
FIG. 3.
CTD kinase activity of Kin28p mutants. Kin28p was immunoprecipitated from extracts of strains expressing wild-type (WT) Kin28p or the indicated point mutants and assayed for CTD kinase activity. The portion of the gel containing the CTD peptide was processed for autoradiography (upper panel), while the portion containing Kin28p was immunoblotted with anti-HA antibodies (lower panel).
Activating phosphorylation of Thr-162 is not essential in vivo.
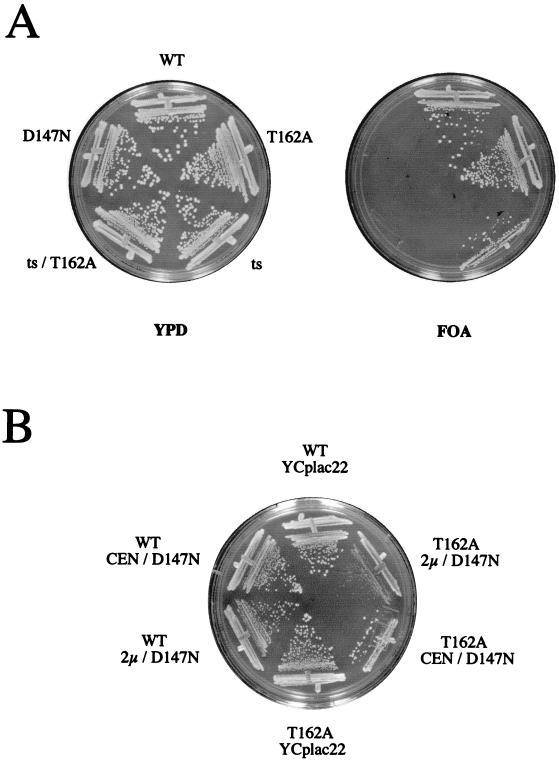
The substantial reduction of kinase activity observed in the T162A mutant, combined with the fact that KIN28 is an essential gene, led us to expect that strains containing only the T162A allele would be inviable. To test this prediction, we disrupted KIN28 in a diploid strain and attempted to isolate haploid progeny that were rescued by KIN28 or kin28T162A expressed from a low-copy-number plasmid. To our surprise, kin28T162A fully rescued the kin28 disruption, as did the AF, AF/T162A, T162S, T162E, and T162D mutants (data not shown); kin28D147N strains were inviable (see Fig. 5A). All of the viable strains containing the indicated KIN28 alleles and lacking a chromosomal copy of KIN28 were isolated at the expected frequencies.
FIG. 5.
Phenotype of a kin28T162A strain. (A) kin28Δ cells containing a URA3-marked wild-type (WT) KIN28 plasmid (pJK1) and TRP1-marked plasmids containing either KIN28 (pGK13), kin28T162A (pGK36), kin28-ts16 (pGK33), kin28D147N (pMS454), or kin28-tsT162A (pJK25) were plated on FOA-containing plates to select for loss of the URA3 marked plasmid. (B) kin28Δ cells containing KIN28 (YJK1869, YJK1767, and YJK1768) or kin28T162A (YJK1870, YJK1755, and YJK1756) and either an empty vector (YCplac22), kin28D147N on a low-copy-number plasmid (pMS454), or kin28D147N on a high-copy-number plasmid (pMS456) were plated on CM-Trp and grown at 37°C.
We therefore sought to determine whether T162A strains showed gross phenotypes that could be uncovered by simple selective conditions or assays. We found no differences in the survival of KIN28 or kin28T162A strains upon growth at various temperatures, heat shock, osmotic shock, UV exposure, cadmium exposure, or carbon starvation (data not shown). To test for subtle growth defects, a coculture experiment in which genetically marked KIN28 and kin28T162A strains were mixed and maintained in exponential phase for over 2 weeks was performed. We were unable to distinguish survival of the KIN28 and kin28T162A strains under these conditions (data not shown).
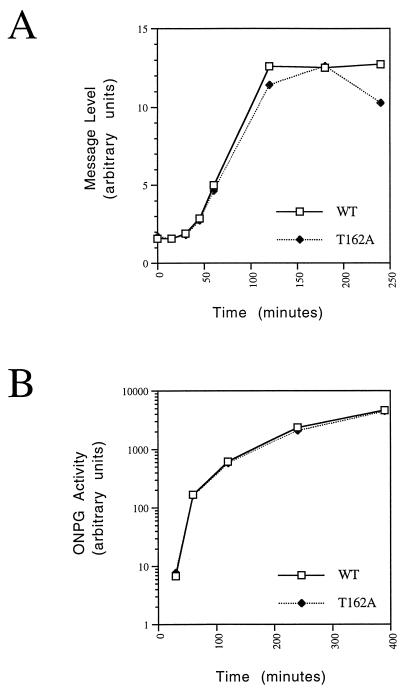
We next determined whether Thr-162 phosphorylation was necessary for high-level transcriptional induction. We first compared the induction kinetics of GPD1, a gene involved in survival following osmotic stress (4). GPD1 encodes an enzyme involved in glycerol synthesis; following hyperosmotic stress, yeast accumulates glycerol to counteract extracellular osmotic pressure (4, 39). Cultures of KIN28 and kin28T162A strains were transferred into medium containing 0.9 M NaCl and grown. GPD1 transcripts were quantitated by Northern blot analysis. KIN28 and kin28T162A cells showed very similar GPD1 induction kinetics (Fig. 4A). Comparable kinetics were also seen for HSP104, a heat shock gene also induced during osmotic stress (data not shown). We next compared the induction kinetics of a lacZ reporter gene from a galactose-inducible promoter following the transition from raffinose- to galactose-containing medium. Expression of β-galactosidase activity was virtually identical for the two strains (Fig. 4B). These findings were surprising, since they indicated that fine tuning of TFIIH activity via Kin28p probably plays little role during the transcriptional cycle.
FIG. 4.
Thr-162 phosphorylation of Kin28p is not essential for transcriptional induction. (A) Osmotic stress. Cells containing KIN28 (wild type [WT]) or kin28T162A (strains YGK26 and YGK42, respectively) were inoculated into YPD containing 0.9 M NaCl and maintained. RNA was extracted at various times after induction, and transcripts were analyzed by Northern blotting with a probe derived from GPD1. Signal intensities were quantitated by phosphorimaging. (B) Galactose induction. KIN28 or kin28T162A cells containing the galactose-inducible lacZ reporter plasmid pAF21 (strains YJK1747 and YJK1749) and growing in raffinose-containing medium were collected at various times following induction of the galactose promoter by addition of 30% galactose to a final concentration of 2%. β-Galactosidase activity was measured with o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate.
Finally, we tested whether the T162A mutant interacted genetically with RNA Pol II temperature-sensitive mutants. Since the CTD of the large subunit of RNA Pol II (Rpb1p) is the presumed target for the essential kinase activity of Kin28p, we reasoned that the lowered kinase activity of Kin28pT162A might enhance the temperature sensitivity of an rpb1-cs allele in which the CTD has been truncated (37) or of a structural mutant allele, rpb1-1 (48). We observed no enhancement of temperature sensitivity for either strain (data not shown).
Role of Thr-162 phosphorylation in vivo.
Our observations that Kin28p kinase activity is substantially reduced in T162A mutants led us to explore whether Thr-162 phosphorylation might be essential when Kin28p is otherwise compromised or limiting.
We first tested whether a temperature-sensitive allele, kin28-ts16 (7), would have increased thermosensitivity if we introduced a T162A point mutation (kin28-tsT162A). We constructed a kin28Δ strain containing both a URA3-marked KIN28 and a TRP1-marked kin28-tsT162A plasmid. Cells were plated on FOA to select against the wild-type plasmid. Although cells containing a TRP1-marked KIN28 gene survived on FOA at any temperature, both catalytically inactive (kin28D147N) and kin28-tsT162A plasmids were unable to rescue viability (Fig. 5A), indicating that both mutants are nonfunctional. To corroborate this result, we introduced kin28-tsT162A into a diploid strain in which one allele of KIN28 was disrupted with LEU2. No leucine prototrophs were recovered at 23°C; in contrast, Leu− cells contained the kin28-tsT162A plasmid, indicating that the inviability of kin28 disruptants was not due to plasmid loss or toxicity during sporulation (data not shown).
We hypothesized that if the kinase activity of Kin28pT162A was compromised compared with that of the wild-type, cells might be hypersensitive to overexpression of a kinase-inactive kin28 mutant, which could compete with Kin28p for activating pathways, components necessary for Kin28p function (such as TFIIH), or substrates. We tested the viability of kin28Δ strains containing low-copy KIN28 or kin28T162A combined with high- or low-copy kin28D147N. We observed that kin28T162A strains were impaired for growth in a dose-dependent manner when Kin28pD147N was simultaneously expressed (Fig. 5B) and were inviable when Kin28pD147N was expressed from a high-copy-number plasmid at 37°C (Fig. 5B). In contrast, strains containing the wild-type allele were not sensitive to overexpression of kin28D147N at any temperature.
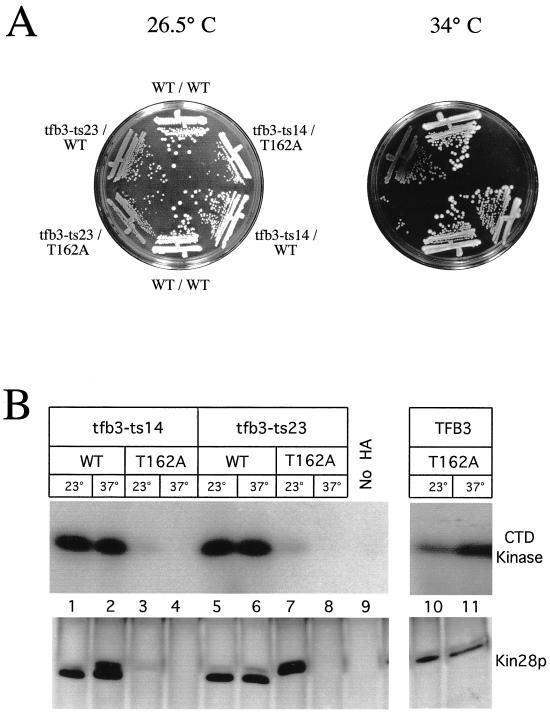
The higher eukaryotic Kin28p ortholog, MO15, can be activated in a phosphorylation-independent manner by binding MAT1 (13, 22, 63), an assembly factor that is also a subunit of TFIIH. The yeast ortholog of MAT1 is TFB3, an essential gene whose product is a subunit of TFIIH (17, 19). We tested whether kin28T162A could enhance the effects of a tfb3-ts mutation. We reasoned that if Kin28p-Ccl1p complexes can be activated either by Thr-162 phosphorylation or by association with Tfb3p, then the function of the Kin28pT162A mutant might be severely attenuated in a tfb3-ts strain. We constructed tfb3-ts strains containing either KIN28 or kin28T162A. Two different tfb3-ts alleles (tfb3-ts14 and tfb3-ts23) (17) were barely temperature sensitive in our strain background (Fig. 6A). However, when the tfb3-ts alleles were combined with kin28T162A, these strains were critically impaired for growth at 34°C (Fig. 6A).
FIG. 6.
Genetic interaction between kin28ST162A and tfb3-ts. (A) Strains containing TFB3 or one of two temperature-sensitive alleles of tfb3 and either KIN28 or kin28T162A were plated and grown at 26.5 and 34°C. Cells containing kin28T162A alone showed no growth defect even at 37°C (data not shown). (B) The strains described for panel A as well as a kin28T162A strain were grown at either 23 or 37°C for 4 h. Extracts were prepared, and Kin28p immunoprecipitates were assayed for CTD kinase activity (top panel) or immunoblotted for Kin28p (bottom panel). The apparently greater Kin28pT162A activity seen in this figure compared to Fig. 3 reflects the longer exposure used in this experiment; Kin28pT162A activity is still reduced by 75 to 80% compared to that of wild-type (WT) Kin28p.
To further investigate the cause of the growth defect in tfb3-ts kin28T162A strains, we prepared extracts from cells at permissive and restrictive temperatures, immunoprecipitated Kin28p, and performed CTD kinase assays and immunoblotting (Fig. 6B). We saw little effect of either tfb3-ts allele on wild-type Kin28p kinase activity at the restrictive temperature (Fig. 6B, lanes 1, 2, 5, and 6). In contrast, strains containing kin28T162A had substantially less or no kinase activity at the restrictive temperature (lanes 3, 4, 7, and 8). The amount of Kin28pT162A was reduced in both strains at 37°C and in the tfb3-ts14 strain even at 23°C (lanes 3, 4, and 8). Importantly, the tfb3-ts23 strain had normal levels of Kin28pT162A but reduced Kin28p activity at 23°C, indicating that Tfb3p activates Kin28p in the absence of Thr-162 phosphorylation (lane 7). Wild-type Kin28p retained approximately 50% of its activity in the tfb3-ts strains (data not shown).
Our results indicate that a role for the activating threonine can be unmasked if Kin28p function is otherwise compromised, because of either a feeble ts allele, competition with an inactive mutant, or reduction of Tfb3p function.
Kin28p phosphorylation and activity are invariant throughout the cell cycle.
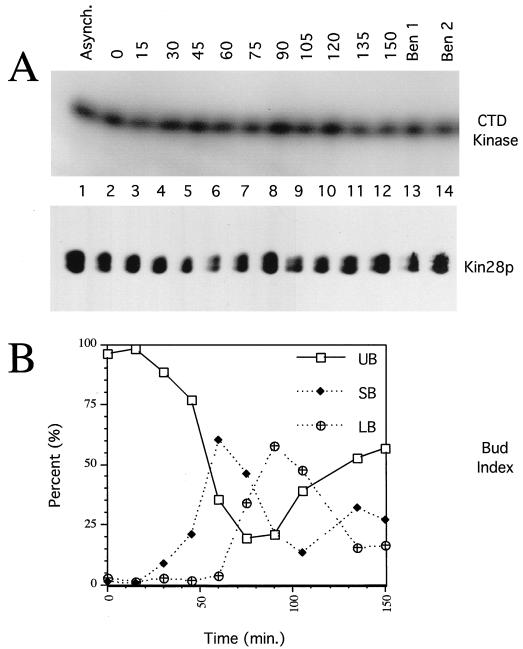
The activity of MO15 in mammalian TFIIH was recently shown to be reduced during mitosis as part of a pathway in which transcription is repressed during mitosis (3, 38). This effect is due to both an increase in Ser-164 phosphorylation (which inhibits phosphorylation of the CTD by MO15) and a decrease in Thr-170 phosphorylation (3). We therefore determined whether Kin28p activity or its phosphorylation on Thr-162 varies during the cell cycle. (Kin28p lacks a phosphorylatable residue corresponding to Ser-164 in MO15.) Kin28p was immunoprecipitated from extracts derived from synchronized cells, assayed for CTD kinase activity, and immunoblotted. Neither Thr-162 phosphorylation nor kinase activity varied significantly during the cell cycle (Fig. 7A) or in extracts from asynchronous or mitotically arrested cells (Fig. 7A, lanes 1, 13, and 14). We conclude that Kin28p activity and phosphorylation are not regulated in a cell cycle-dependent manner.
FIG. 7.
Kin28p level, activity, and phosphorylation state are constant during the cell cycle. (A) Exponentially growing KIN28 cells (YJK1773) were arrested with α-factor, washed, and released into fresh medium at 30°C. Extracts were prepared at 15-min intervals, and Kin28p immunoprecipitates were assayed for CTD kinase activity (top panel) or immunoblotted for Kin28p (bottom panel). Extracts of asynchronous cells (Asynch.) and cells arrested in mitosis with benomyl for 3.75 h (Ben 1) and 4.75 h (Ben 2) were also analyzed. (B) Bud indices of the cultures in panel A. UB, SB, and LB, unbudded, small-budded, and large-budded cells, respectively.
Kin28p is phosphorylated by Cak1p in vivo and in vitro.
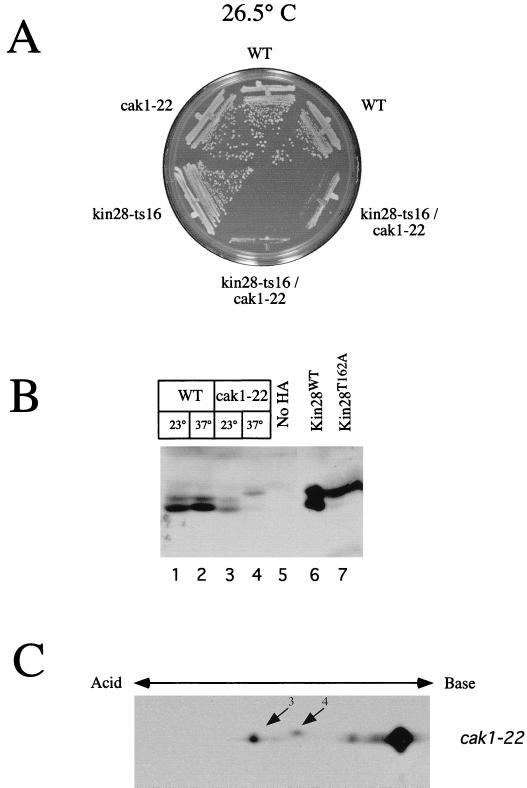
Cak1p phosphorylates Cdc28p in vivo on a site (Thr-169) equivalent to Thr-162 in Kin28p (32, 64). To investigate the relationship between Cak1p and Kin28p, we isolated cak1-22 kin28-ts16 double mutants and examined them for synthetic interactions. We found that cak1-22 kin28-ts16 strains had increased temperature sensitivity compared with strains containing the single mutations and grew poorly even at 26.5°C (Fig. 8A). Presumably, the already-compromised protein made by kin28-ts16 is rendered nonfunctional in the absence of phosphorylation by Cak1p, similar to the inviability of strains containing kin28-tsT162A (Fig. 5A).
FIG. 8.
CAK1 regulates the phosphorylation of Kin28p. (A) Genetic interaction between cak1-22 and kin28-ts16. The indicated wild-type (WT) and mutant strains (clockwise from top, YMW2, SY162, YJK1599, YJK1600, YGK24, and SY143) were plated and incubated at 26.5°C. Two isolates of wild-type and double-mutant strains are shown. (B) Kin28p is hypophosphorylated in a cak1-22 strain. Extracts were prepared from CAK1 and cak1-22 strains (YJK1610 and YJK1614, respectively) grown at 23 or 37°C for 6 h, and Kin28p immunoprecipitates were immunoblotted. (C) Extracts of a cak1-22 strain (YJK1625) grown at 37°C for 6 h were analyzed by 2-D gel electrophoresis as described for Fig. 2. Spots 3 and 4 correspond to the same Kin28p spots in Fig. 2.
To investigate further the nature of this genetic interaction, we prepared extracts of cak1-22 strains and examined the pattern of phosphorylation of wild-type Kin28p following SDS-PAGE. At the restrictive temperature for cak1-22, we found that Kin28p was hypophosphorylated in a manner similar to that for T162A mutants (Fig. 8B, lanes 4 and 7). (Although Espinoza et al. [16] did not observe a change in the electrophoretic pattern of Kin28p in a cak1-22 strain, they used a shorter incubation at the restrictive temperature. They found that Kin28p was rapidly dephosphorylated in strains containing other CAK1 alleles.) We performed 2-D isoelectric focusing to confirm that the shift in phosphorylation at the restrictive temperature corresponded to the elimination of a single phosphorylation site. Kin28p derived from the cak1-22 strain at the restrictive temperature produced only two species (Fig. 8C), which correspond to spots 3 and 4 on 2-D gels of Kin28p from a wild-type strain (Fig. 2B). This pattern of phosphorylation is identical to that produced by Kin28pT162A, indicating that Cak1p regulates the phosphorylation of Kin28p on Thr-162 in vivo. Kin28p from a wild-type strain at 37°C resolved into the same four species observed previously (data not shown). This hypophosphorylation of Kin28p is probably not an indirect consequence of the cell cycle block of cak1-22 cells at the restrictive temperature, since the extent of Thr-162 phosphorylation does not vary during the cell cycle (Fig. 7A). We also consider unlikely the possibility that Kin28p hypophosphorylation in the cak1-22 strain is an indirect effect of lowered Cdc28p activity, since we did not observe a similar hypophosphorylation in a cdc28-1 strain (data not shown).
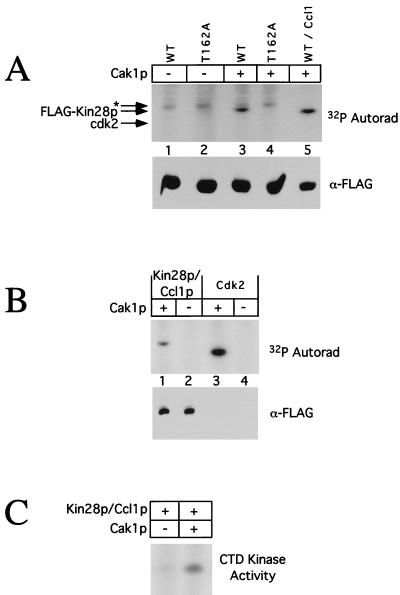
We tested directly whether Cak1p could phosphorylate Kin28p expressed in baculovirus-infected insect cells and isolated via a FLAG tag on Kin28p. Cak1p could phosphorylate both monomeric and Ccl1p-bound forms of Kin28p (Fig. 9A, lanes 3 and 5, and B, lane 1). No Kin28p phosphorylation occurred in the absence of Cak1p (Fig. 9A, lanes 1 and 2, and B, lane 2) or when a Kin28pT162A substrate was used (Fig. 9A, lane 4). For reference, phosphorylation of monomeric Cdk2, a known excellent Cak1p substrate, is shown (Fig. 9B, lanes 3 and 4). The bottom panels in Fig. 9A and B show that comparable amounts of Kin28p were used in the various assays. We could not phosphorylate Kin28p by using Cdk2-cyclin A, which can phosphorylate the equivalent site in the MO15 subunit of human TFIIH (22, 43), or by using MO15-cyclin H (data not shown). The specificity of this reaction is further indicated by our inability to phosphorylate another CDK component of the transcriptional apparatus, Srb10p (37), by using Cak1p (data not shown). Finally, Fig. 9C shows that the CTD kinase activity of Kin28p-Ccl1p complexes was increased sevenfold following incubation with Cak1p, confirming the importance of this phosphorylation for full Kin28p activity.
FIG. 9.
Cak1p phosphorylates Kin28p on Thr-162 in vitro. (A) Purified Cak1p was incubated with FLAG-Kin28p (lane 3), FLAG-Kin28pT162A (lane 4), or FLAG-Kin28pD147A–Ccl1p complexes (lane 5) in the presence of [γ-32P]ATP. As controls for autophosphorylation, FLAG-Kin28p and FLAG-Kin28pT162A were incubated in the absence of Cak1p (lanes 1 and 2). Phosphorylated proteins were detected by autoradiography (Autorad) following SDS-PAGE (upper panel). Note that the monomeric Kin28p samples (lanes 1 to 4) were only ∼1 to 5% pure (estimated by gel staining), resulting in significant background phosphorylation; the asterisk denotes a nonspecific species. The relative Kin28p levels in the kinase assays were determined by immunoblotting with antibodies to the FLAG tag (lower panel). WT, wild type. (B) FLAG-Kin28pD147A–Ccl1p complexes (lanes 1 and 2) or Cdk2 (lanes 3 and 4) was incubated with (lanes 1 and 3) or without (lanes 2 and 4) purified Cak1p in the presence of [γ-32P]ATP. Phosphorylated proteins were detected by autoradiography following SDS-PAGE (upper panel), and the relative Kin28p levels in the kinase assays were determined by immunoblotting with antibodies to the FLAG tag (lower panel). Cdk2 is not visible because it is not FLAG tagged. (C) Phosphorylation of Kin28p by Cak1p increases its CTD kinase activity. FLAG-Kin28p–Ccl1p complexes were incubated with (lane 2) or without (lane 1) purified Cak1p and assayed for CTD kinase activity. Phosphorimager quantitation showed that the CTD kinase activity of Kin28p increased sevenfold following incubation with Cak1p.
DISCUSSION
Phosphorylation of Kin28p.
Our goals in these experiments were to characterize the in vivo functions of Kin28p posttranslational regulation, to establish whether the modes of regulation of MO15 activity apply to Kin28p, and to clarify the relationship between Cak1p and Kin28p. We have shown that Kin28p, like many other CDKs, is phosphorylated in vivo on an activating residue (Thr-162) within its T loop. A Kin28pT162A mutant had significantly reduced activity in vitro and reduced function in vivo. In contrast to the situation in mammalian cells (38), both the extent of this phosphorylation and Kin28p activity were constant during the cell cycle, suggesting that the CTD kinase in yeast TFIIH is not a target for the regulation of transcription during mitosis.
Dual regulation of Kin28p.
Like most CDKs (27, 30, 33, 53, 60), Kin28p requires phosphorylation for full activity. Unlike Cdc28p (7) in S. cerevisiae and Cdc2 in Schizosaccharomyces pombe (14, 26), where analogous activating phosphorylation site mutants are nonfunctional, kin28T162A is functional in vivo. Like MO15, Kin28p is positively regulated both by phosphorylation and by binding to an assembly factor. In the case of human MO15, the MO15-cyclin H complex can be activated by phosphorylation on Thr-170. In contrast, the trimeric MO15-cyclin H-MAT1 complex is active whether or not Thr-170 is phosphorylated. Our results indicate that this model, which is based on experiments in vitro, also applies in vivo. A strain carrying kin28T162A or a temperature-sensitive allele of TFB3 grows well under normal conditions. However, a kin28T162A tfb3-ts strain is severely compromised at all temperatures. This genetic interaction was confirmed at the biochemical level: Kin28p activity in the double-mutant strains was greatly reduced compared to that in a wild-type strain or in a strain containing either single mutation.
Kin28pT162A appears to be destabilized in strains defective for TFB3. Both tfb3-ts strains had reduced levels of Kin28pT162A at the restrictive temperature, and the tfb3-14 strain had much less Kin28pT162A than a wild-type strain even at the permissive temperature. In contrast, the amount of wild-type Kin28p was unaffected by the status of TFB3, suggesting that either Thr-162 phosphorylation or association with Tfb3p can stabilize Kin28p. The stability of Kin28p may depend on its association with its cyclin partner, Ccl1p, resulting in the rapid elimination of free Kin28p. Activating phosphorylations of CDKs, in addition to increasing kinase activity, may also stabilize the CDK-cyclin complex (12). Thr-170 phosphorylation of MO15, for example, stabilizes MO15-cyclin H complexes (41, 43). Furthermore, MAT1 (the homolog of Tfb3p) stabilizes the MO15-cyclin H complex in the absence of an activating phosphorylation (13, 43). The instability of Kin28p can be further inferred from the observation that temperature-sensitive alleles of CDC37 are synthetically lethal with kin28-ts alleles (reference 66 and our unpublished results). A number of newly translated CDKs, including Cdc28p and Cdk4, require stabilization by the Cdc37p chaperone (24, 61).
Relationship between Cak1p and Kin28p.
The data presented here establish Kin28p as the second physiological substrate for the yeast CAK, Cak1p. Cak1p can phosphorylate activating sites in a number of CDKs, including Cdc28p and a variety of mammalian CDKs (15, 31, 32, 64), and was a logical candidate for the activating kinase acting on the equivalent site in Kin28p. We showed that Cak1p could phosphorylate wild-type Kin28p but not Kin28pT162A in vitro and that Thr-162 phosphorylation of Kin28p was eliminated upon inactivation of Cak1p in vivo. In contrast, inactivation of Kin28p does not affect Cak1p activity (data not shown). In addition, we and others (66) have shown that mutations in KIN28 and CAK1 display synthetic lethal interactions.
The phosphorylation of Kin28p by Cak1p is highly specific. Cak1p was unable to phosphorylate MO15 (a Kin28p ortholog), and MO15 (another CAK) was unable to phosphorylate Kin28p (data not shown). Although MO15-cyclin H is a substrate for Cdk2-cyclin A (22, 43), Kin28p was not (data not shown). Furthermore, we observed no genetic interaction between kin28-ts16 and cdc28-1 alleles (data not shown) or any changes in Kin28p phosphorylation in a cdc28-ts strain under restrictive growth conditions (data not shown).
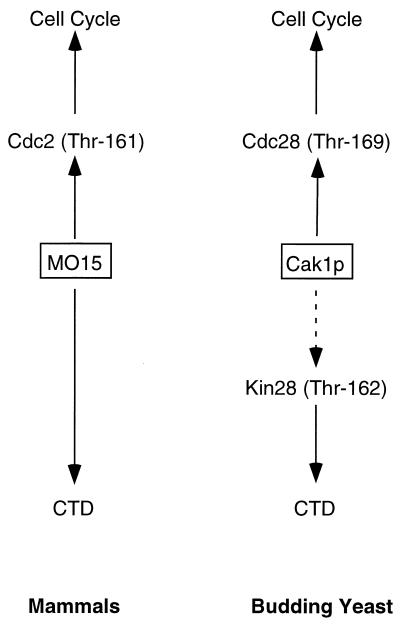
A highly mutated allele of CDC28 that is functional but whose protein product cannot be phosphorylated by Cak1p has been isolated (8). A strain carrying this allele can grow in the absence of CAK1, but not as well as when CAK1 is present. These findings revealed a nonessential role for Cak1p in addition to its essential function as an activating kinase for Cdc28p. A plausible nonessential function is the activating phosphorylation of Kin28p. That our work should bring together Cak1p and Kin28p is ironic, since these two proteins reflect the evolutionary separation of the CAK and TFIIH functions performed by MO15 in other species. Although Cak1p is a very different CAK from MO15, the close connection of each to both the cell cycle and the basal transcription apparatus is intriguing (Fig. 10). Both control the cell cycle via phosphorylation of the major CDKs in their respective species. MO15 directly controls transcription as a subunit of TFIIH by phosphorylating the CTD of RNA Pol II, whereas Cak1p modulates TFIIH activity by phosphorylating Kin28p. Both yeast and mammalian cells can therefore coordinately affect both the cell cycle and CTD phosphorylation through a single CAK.
FIG. 10.
Dual regulation of transcription and the cell division cycle by CAKs. Both mammalian CAK (MO15) and yeast Cak1p affect transcription (via CTD phosphorylation) and cell division (via CDK phosphorylation). The dotted line denotes a nonessential phosphorylation.
A relationship similar to that of Cak1p and Kin28p may also exist in S. pombe. S. pombe appears to contain two CAKs. One, the kinase Mcs6 (also called Mop1/Crk1), is similar to MO15 in sequence and has both CAK activity and CTD kinase activity in vitro (6, 10, 45). The gene for the second, csk1, was isolated as a multicopy suppresser of a mutation of mcs2 (encoding the fission yeast cyclin H homolog) (45). Csk1 is distantly related to Cak1p. Mcs2-associated kinase activity is reduced in a csk1Δ strain (45), and Csk1 can phosphorylate and activate Mcs6 in vitro and in vivo (29). Whether organisms such as humans also contain a Cak1p-like or Csk1-like CAK capable of phosphorylating MO15 remains an open question.
Espinoza et al. (16) reported related results after the completion of this work. They found that Kin28p is phosphorylated on Thr-162, that Kin28p activity is reduced in the absence of this phosphorylation, and that Cak1p is probably the kinase that phosphorylates Kin28p on Thr-162. Our results differ in two notable aspects. First, Espinoza et al. concluded that Kin28pT162A was nonfunctional in that it could not rescue a temperature-sensitive allele of KIN28 at the nonpermissive temperature. In contrast, we observed little phenotypic effect of expressing kin28T162A at wild-type levels as the sole Kin28p in the cell. Both studies were carried out in the W303 strain background. We are confident that our strains contained kin28T162A, since the lack of Thr-162 phosphorylation was detectable by SDS-PAGE. Moreover, sequencing of our kin28T162A allele indicated the absence of any unintended mutations. These different conclusions may reflect differences in experimental design. It is possible, for example, that the activity of Kin28pT162A was compromised in the kin28-3 strain used by Espinoza et al., particularly at the nonpermissive temperature for kin28-3. In fact, we observed that Kin28pT162A functions very poorly in a strain overexpressing a catalytically inactive form of Kin28p, Kin28pD147N (Fig. 5B), a situation somewhat similar to that employed by Espinoza et al. (This comparison is imperfect, however, since we expressed Kin28pD147N from the KIN28 promoter on a high-copy-number plasmid, resulting in ∼three to fivefold overexpression of Kin28p compared to that with a low-copy-number plasmid.) Their observation that the activity of Kin28p (wild type or mutant) expressed from a heterologous promotor on a plasmid was surprisingly low (their data not shown) strengthens the possibility that the presence of inactivated Kin28-3p could compromise Kin28pT162A, presumably via competition for Ccl1p and/or Tfb3p. Whatever the explanation for the different conclusions, we feel that the direct approach taken in this study indicates that Kin28pT162A is fully functional as the only Kin28p in the cell. In a separate experiment, Espinoza et al. found that a strain with CAK1 deleted was viable (see also reference 8), even though Kin28p lacked activating phosphorylation, supporting our conclusion that Thr-162 phosphorylation is not essential for Kin28p function. Activating phosphorylation also appears to be nonessential for the Kin28p homolog in S. pombe, Mcs6 (29).
A second difference between our results and those of Espinoza et al. (16) concerns the requirements for Kin28p phosphorylation by Cak1p. We found that Cak1p could phosphorylate Kin28p approximately equally well whether the Kin28p was free or bound to Ccl1p, whereas they observed only weak phosphorylation of monomeric Kin28p, moderate phosphorylation of Kin28p bound to Ccl1p, and strong phosphorylation of Kin28p in the presence of both Ccl1p and Tfb3p. These results may also reflect differences in experimental design. Espinoza et al. examined Kin28p phosphorylation following coinfection of insect cells with recombinant baculoviruses expressing Kin28p, Ccl1p, Tfb3p, Cak1p, and Cdc37p, whereas we examined phosphorylation by using isolated proteins. Free Kin28p may be rapidly dephosphorylated by an insect cell phosphatase. An intriguing second possibility is that high expression of the Cdc37p protein kinase chaperone both aids in the proper folding of Kin28p and interferes with phosphorylation by Cak1p. Further work will be required to resolve this difference.
ACKNOWLEDGMENTS
Many technical aspects of this work depended on the help of our coworkers, including Janet Burton, Beth Egan, Deb Enke, Karen Ross, Zach Pitluk, Joyce Wall, and the other members of the Solomon lab. We are especially grateful for the patience of Jackie Vogel and Kate Long, who helped navigate the flatlands of isoelectric focusing. Critical reagents were graciously provided by Gerard Faye (rig2-ts strains), Mike Gustin (GPD1 plasmids), Ann Sutton (cak1 plasmids and strains), Peter Novick (HSP104 probe), Henrik Dohlman (bar1 disruption plasmid), and Amy Fluegge (pAF21 reporter plasmid). For their sharing of equipment and materials, we thank Ken Williams, Bill Konigsberg, Peter Lengyel, Peter Novick, Sandy Wolin, and the members of their labs. David Stern, Mike Snyder, and David Gonda are gratefully acknowledged for advice and reagents.
This work was supported by a long-term fellowship from the Swiss National Science Foundation (to P.K.), the National Institutes of Health (grants GM34365 to R.A.Y. and GM47830 to M.J.S.), and the Searle Scholars Program/The Chicago Community Trust (M.J.S.). M.J.S. is a Leukemia Society of America Scholar.
Footnotes
J.K. dedicates this paper to Sara Laimon.
REFERENCES
- 1.Adamczewski J P, Rossignol M, Tassan J-P, Nigg E A, Moncollin V, Egly J-M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Akoulitchev S, Mäkelä T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 3.Akoulitchev S, Reinberg D. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 1998;12:3541–3550. doi: 10.1101/gad.12.22.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. J. Boston, Mass: Wiley & Sons, Inc.; 1995. [Google Scholar]
- 6.Buck V, Russell P, Millar J B A. Identification of a cdk-activating kinase in fission yeast. EMBO J. 1995;14:6173–6183. doi: 10.1002/j.1460-2075.1995.tb00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross F R, Levine K. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol Cell Biol. 1998;18:2923–2931. doi: 10.1128/mcb.18.5.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 10.Damagnez V, Mäkelä T P, Cottarel G. Schizosaccharomyces pombe Mop1-Mcs2 is related to mammalian CAK. EMBO J. 1995;14:6164–6172. doi: 10.1002/j.1460-2075.1995.tb00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai D, Wessling H C, Fisher R P, Morgan D O. The effect of phosphorylation by CAK on cyclin binding by CDC2 and CDK2. Mol Cell Biol. 1995;15:345–350. doi: 10.1128/mcb.15.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devault A, Martinez A-M, Fesquet D, Labbé J-C, Morin N, Tassan J-P, Nigg E A, Cavadore J-C, Dorée M. MAT1 (‘menage à trois’), a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995;14:5027–5036. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducommun B, Brambilla P, Félix M-A, Franza Jr B R, Karsenti E, Draetta G. cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 1991;10:3311–3319. doi: 10.1002/j.1460-2075.1991.tb04895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinoza F H, Farrell A, Erdjument-Bromage H, Tempst P, Morgan D O. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 16.Espinoza F H, Farrell A, Nourse J L, Chamberlin H M, Gileadi O, Morgan D O. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faye G, Simon M, Valay J G, Fesquet D, Facca C. Rig2, a RING finger protein that interacts with the Kin28/Ccl1 CTD kinase in yeast. Mol Gen Genet. 1997;255:460–466. doi: 10.1007/s004380050518. [DOI] [PubMed] [Google Scholar]
- 18.Feaver W J, Gileadi O, Kornberg R D. Purification and characterization of yeast RNA polymerase II transcription factor b. J Biol Chem. 1991;266:19000–19005. [PubMed] [Google Scholar]
- 19.Feaver W J, Henry N L, Wang Z, Wu X, Svejstrup J Q, Bushnell D A, Friedberg E C, Kornberg R D. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. J Biol Chem. 1997;272:19319–19327. doi: 10.1074/jbc.272.31.19319. [DOI] [PubMed] [Google Scholar]
- 20.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 21.Fesquet D, Labbé J-C, Derancourt J, Capony J-P, Galas S, Girard F, Lorca T, Shuttleworth J, Dorée M, Cavadore J-C. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 23.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 24.Gerber M R, Farrell A, Deshaies R J, Herskowitz I, Morgan D O. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc Natl Acad Sci USA. 1995;92:4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 26.Gould K L, Moreno S, Owen D J, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu Y, Rosenblatt J, Morgan D O. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengartner C J, Meyer V E, Liao S M, Wilson C J, Koh S S, Young R A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–54. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 29.Hermand D, Pihlak A, Westerling T, Damagnez V, Vandenhaute J, Cottarel G, Mäkelä R P. Fission yeast Csk1 is a CAK-activating kinase (CAKAK) EMBO J. 1998;17:7230–7238. doi: 10.1093/emboj/17.24.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iavarone A, Massagué J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 31.Kaldis P, Russo A A, Chou H S, Pavletich N P, Solomon M J. Human and yeast cdk-activating kinases (CAKs) display distinct substrate specificities. Mol Biol Cell. 1998;9:2545–2560. doi: 10.1091/mbc.9.9.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaldis P, Sutton A, Solomon M J. The cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 33.Kato J-Y, Matsuoka M, Storm D K, Sherr C J. Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol Cell Biol. 1994;14:2713–2721. doi: 10.1128/mcb.14.4.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolman C J, Toth J, Gonda D K. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labbé J-C, Martinez A-M, Fesquet D, Capony J-P, Darbon J-M, Derancourt J, Devault A, Morin N, Cavadore J-C, Dorée M. p40MO15 associates with a p36 subunit and requires both nuclear translation and Thr176 phosphorylation to generate cdk-activating kinase activity in Xenopus oocytes. EMBO J. 1994;13:5155–5164. doi: 10.1002/j.1460-2075.1994.tb06845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lees E. Cyclin dependent kinase regulation. Curr Opin Cell Biol. 1995;7:773–780. doi: 10.1016/0955-0674(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 37.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 38.Long J J, Leresche A, Kriwacki R W, Gottesfeld J M. Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol Cell Biol. 1998;18:1467–1476. doi: 10.1128/mcb.18.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mager W H, Varela J C S. Osmostress response of the yeast Saccharomyces. Mol Microbiol. 1993;10:253–258. [PubMed] [Google Scholar]
- 40.Mäkelä T P, Parvin J D, Kim J, Huber L J, Sharp P A, Weinberg R A. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mäkelä T P, Tassan J-P, Nigg E A, Frutiger S, Hughes G J, Weinberg R A. A cyclin associated with the CDK-activating kinase MO15. Nature. 1994;371:254–257. doi: 10.1038/371254a0. [DOI] [PubMed] [Google Scholar]
- 42.Maldonado E, Reinberg D. News on initiation and elongation of transcription by RNA polymerase II. Curr Opin Cell Biol. 1995;7:352–361. doi: 10.1016/0955-0674(95)80090-5. [DOI] [PubMed] [Google Scholar]
- 43.Martinez A-M, Afshar M, Martin F, Cavadore J-C, Labbé J-C, Dorée M. Dual phosphorylation of the T-loop in cdk7: its role in controlling cyclin H binding and CAK activity. EMBO J. 1997;16:343–354. doi: 10.1093/emboj/16.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuoka M, Kato J-Y, Fisher R P, Morgan D O, Sherr C J. Activation of cyclin-dependent kinase 4 (Cdk4) by mouse MO15-associated kinase. Mol Cell Biol. 1994;14:7265–7275. doi: 10.1128/mcb.14.11.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molz L, Beach D. Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J. 1993;12:1723–1732. doi: 10.1002/j.1460-2075.1993.tb05817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 47.Morgan D O. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–772. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- 48.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 50.Poon R Y C, Yamashita K, Adamczewski J P, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J-P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J-M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 52.Russo A A, Jeffrey P D, Pavletich N P. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 53.Santos R C, Waters N C, Creasy C L, Bergman L W. Structure-function relationship of the yeast cyclin-dependent kinase Pho85. Mol Cell Biol. 1995;15:5482–5491. doi: 10.1128/mcb.15.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serizawa H, Mäkelä T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 55.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 56.Simon M, Seraphin B, Faye G. Kin28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986;5:2697–2701. doi: 10.1002/j.1460-2075.1986.tb04553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solomon M J. The function(s) of CAK, the p34cdc2 activating kinase. Trends Biochem Sci. 1994;19:496–500. doi: 10.1016/0968-0004(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 58.Solomon M J, Harper J W, Shuttleworth J. CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J. 1993;12:3133–3142. doi: 10.1002/j.1460-2075.1993.tb05982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solomon M J, Kaldis P. Regulation of cdks by phosphorylation. In: Pagano M, editor. Results and problems in cell differentiation. 22. Cell cycle control. Heidelberg, Germany: Springer; 1998. pp. 79–109. [DOI] [PubMed] [Google Scholar]
- 60.Solomon M J, Lee T, Kirschner M W. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stepanova L, Leng X, Parker S B, Harper J W. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 62.Sutton A, Freiman R. The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics. 1997;147:57–71. doi: 10.1093/genetics/147.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tassan J-P, Jaquenoud M, Fry A M, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thuret J-Y, Valay J-G, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 65.Valay J-G, Dubois M-F, Bensaude O, Faye G. Ccl1, a cyclin associated with protein kinase Kin28, controls the phosphorylation of RNA polymerase II largest subunit and mRNA transcription. C R Acad Sci. 1996;319:183–189. [PubMed] [Google Scholar]
- 66.Valay J-G, Simon M, Dubois M-F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 67.Valay J G, Simon M, Faye G. The Kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]