Abstract
The tissue-specific expression of major histocompatibility complex class I genes is determined by a series of upstream regulatory elements, many of which remain ill defined. We now report that a distal E-box element, located between bp −309 and −314 upstream of transcription initiation, acts as a cell type-specific enhancer of class I promoter activity. The class I E box is very active in a neuroblastoma cell line, CHP-126, but is relatively inactive in the HeLa epithelial cell line. The basic helix-loop-helix leucine zipper proteins upstream stimulatory factor 1 (USF1) and USF2 were shown to specifically recognize the class I E box, resulting in the activation of the downstream promoter. Fine mapping of USF1 and USF2 amino-terminal functional domains revealed differences in their abilities to activate the class I E box. Whereas USF1 contained only an extended activation domain, USF2 contained both an activation domain and a negative regulatory region. Surprisingly, the naturally occurring splice variant of USF2 lacking the exon 4 domain, U2ΔE4, acted as a dominant-negative regulator of USF-mediated activation of the class I promoter. This latter activity is in sharp contrast to the known ability of U2ΔE4 to activate the adenovirus major late promoter. Class I E-box function is correlated with the relative amount of U2ΔE4 in a cell, leading to the proposal that U2ΔE4 modulates class I E-box activity and may represent one mechanism to fine-tune class I expression in various tissues.
Class I molecules of the major histocompatibility complex (MHC) serve as receptors for endogenously generated peptides and as targets for cytotoxic T lymphocytes. Consistent with their role as sentinels for the immune response against malignant and virally infected cells, MHC class I genes are constitutively expressed on nearly all nucleated somatic cells. However, the level of class I gene expression varies considerably among different tissues (22, 37). The number of class I molecules expressed at the cell surface, in most instances, parallels the amount of class I RNA in any given tissue, indicating that class I gene expression is primarily regulated at the level of transcription (40). The precise mechanism(s) by which class I genes maintain tissue-appropriate levels of expression is not fully understood.
In general, gene transcription is determined by the action of multiple, distal regulatory elements which modulate a basal level of transcription directed by a core promoter region (12). Several studies in which class I transgenic mice were generated by using truncated class I promoter regions reported aberrant class I transgene expression that could be corrected by including additional upstream class I promoter sequences in the targeting vectors (9, 26, 36). Our studies and those of others indicate that the tissue-specific expression of MHC class I genes is, in part, determined by regulatory elements which reside upstream of the known proximal promoter sequences (Fig. 1). Despite these observations, the regulatory elements which reside upstream of the 5′ boundary of the proximal class I promoter (defined as the enhancer A element located approximately 200 bp upstream of the transcriptional start site in most class I genes) and their contribution to class I expression remain relatively unexplored.
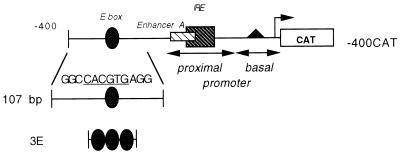
FIG. 1.
The MHC class I reporter construct -400CAT is shown with selected regulatory motifs indicated. The E box is located between bp −309 and −314 upstream of the point of transcription initiation. The proximal promoter extends from bp −203 to −68 and contains an enhancer, enhancer A, and an overlapping interferon response element (IRE). The basal promoter extends from bp −68 to +1 and includes the CCAAT box, promoter elements, and the site of transcription initiation. The derivation of the 107-bp and 3E oligonucleotide probes used in gel shift analysis is also shown; there is a detailed description in Materials and Methods. The core hexamer sequence of the 3E probe is shown.
Sequence comparison between the upstream regulatory region of a class I gene and known DNA binding motifs revealed the presence of a consensus E-box element upstream of the proximal promoter at bp −309 to −314. The E box is a highly conserved DNA element (CANNTG) recognized by the basic helix-loop-helix (bHLH) family of transcription factors. These proteins are characterized by bDNA binding and HLH protein-protein interaction domains (16). The various bHLH family members are segregated into three groups: (i) broadly expressed class A proteins (E12, E47, E2-2, and Daughterless), (ii) tissue-restricted class B proteins (MyoD, myogenin, muscle-specific regulatory factor 4, and Achaete-Scute protein), and (iii) the class C proteins (upstream stimulatory factor [USF], Mad, Max, Myc, activator protein 4, transcription factor E3 [TFE3], TFEB, TFII I, and sterol regulatory element-binding protein [SREBP]) which contain an additional leucine zipper (Z) protein-protein interaction motif (21). Precise DNA binding site selection by individual bHLH family members is determined both by the central dinucleotide contained in the core hexamer sequence and the flanking nucleotides (2, 6–8, 10, 13, 18, 28, 31, 32).
The bHLH-Z transcription factor, USF, was originally identified as a positive trans-acting factor which stimulated adenovirus major late promoter (AdMLP) activity by binding to its upstream activating element, a 12-bp element containing an E-box core (29, 34). Subsequently, two USF proteins were purified from HeLa cells, distinguished by their molecular sizes of 43 and 44 kDa and referred to as USF1 and USF2, respectively (35, 38, 39). USF proteins form homo- as well as heterodimers; both partners of the dimer contribute to DNA binding (5). USF1 or USF2 dimers are identical in their abilities to bind the 12-bp AdMLP upstream activating element (35, 37). A naturally occurring splice variant of USF2 lacking exon 4, U2ΔE4 (also called USF2b) has been described previously (23, 41). The relative ratio of wild-type, full-length (FL) USF2 to the alternatively spliced U2ΔE4 species varies among different cell types (41); however, the functional consequence, if any, of the U2ΔE4 splice variant is unknown.
In this report we demonstrate that (i) USF1 and USF2 are capable of enhancing class I promoter activity through an upstream E box; (ii) fine mapping of USF2 reveals the presence of both positive and negative regulatory domains, whereas USF1 has only trans-activating regions; and (iii) the U2ΔE4 splice variant of USF2 acts as a negative regulator of MHC class I E-box activity. Importantly, class I E-box activity was found to be cell type specific: it is active in neuroblastoma CHP-126 cells but not in HeLa epithelial cells. These cell types also differ in the relative abundance of U2ΔE4: CHP-126 cells contain relatively less U2ΔE4 than HeLa cells. Therefore, we propose that the cell type-specific activity of the E box is a reflection of the cellular U2ΔE4 content.
MATERIALS AND METHODS
Cell lines and cultivation.
Human HeLa epithelial cells and CHP-126 neuroblastoma cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 2 mM l-glutamine, HEPES (pH 7.2), gentamicin sulfate (10 μg/ml), and 10% fetal bovine serum and maintained in a humidified incubator at 37°C with 7% CO2.
Plasmids and cloning strategies.
The MHC class I promoter used in these studies derived from the swine class I gene PD1 (15). The -294CAT and -400CAT constructs were previously described (19). Control pSG5 and USF expression plasmids were also previously described (24), and the AdMLP E-box reporter construct U2E1bCAT and its control (E1bCAT) were provided by R. Roeder (Rockefeller University, New York, N.Y.) and described in reference 14. The class I E-box sequence (three copies) 5′-TCGAGGGCCACGTGAGGGGCCACGTGAGGGGCCACGTGT-3′ was subcloned into the XhoI/XbaI site in E1bCAT (directly upstream of the TATA box in the control construct E1bCAT) to generate 3E E1bCAT. The -342CAT construct was generated by PCR amplification with the oligonucleotide primers GGTTCTAGAGAAATCGCTGGG and GAGAAGCTTGAGCAGAGC and cloned into the XbaI/HindIII site of pSV3CAT as previously described (19). Wild-type and mutant class I E-box sequences were cloned into the XbaI site upstream of -294CAT to generate -294(WT E-box)CAT and -294(Mut E-box)CAT constructs, respectively. Oligonucleotide sequences for WT E box and Mut E box are CTAGATGGGCCACGTGAGGCACTGGAGACAT and CTAGATGGGCGGATCCAGGCACTGGAGACAT (the E-box hexamer sequence is shown in boldface type).
Transfections.
Transient transfections were performed by using a total of 20 μg of DNA; final concentrations were adjusted to 20 μg with pUC19 supercoiled DNA, as needed. Twenty-four hours prior to transfection 106 HeLa or CHP-126 cells were seeded in 100-mm-diameter tissue culture dishes. Transfections utilized standard calcium phosphate precipitation as previously described (19). The medium was replaced 24 h after transfection with fresh medium, and cells were harvested after an additional 24 h. Chloramphenicol acetyltransferase (CAT) activity was normalized to luciferase activity by cotransfecting an internal plasmid control, pSV2LUC. CAT activity was determined by using an AMBIS 4000 radioanalytic imaging detector. All CAT enzyme assays were measured in the linear range; control [14C]chloramphenicol values ranged between 20 to 80%, among the different experiments.
Gel shift mobility assays.
HeLa and CHP-126 whole-cell extracts (WCE) were prepared as previously described (42). The probes used in gel shift mobility assays included a 107-bp fragment encompassing the DdeI fragment from the −398 to −291 base pairs (relative to the point of initiation) of the class I gene PD1 or class I E-box oligonucleotides. Sense sequences for the double-stranded oligonucleotides used in gel shift analysis were as follows: for the class I E box (3E), 5′-TCGAGGGCCACGTGAGGGGCCACGTGAGGGGCCACGTGT-3′ (the E-box hexamer sequence is underlined), and for the nonspecific (NS) probe, 5′-AGCTTCATCGTCCCATCCTGACTGAGG-3′. For gel shift mobility assays, 4.5 μg of WCE was added to 1.5 fmol of end-labeled probe. The extract, probe, and specific antibodies were combined and incubated on ice for 30 min. Specific antibodies were obtained from Santa Cruz Biotechnology. The binding buffer consisted of 12 mM HEPES (pH 7.9), 10% glycerol, 5 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, 50 μg of bovine serum albumin per ml, 0.5 mM EDTA, 0.05% Nonidet P-40, and 3 μg of poly(dG-dC). DNA-protein complexes were separated from the unbound free probe by electrophoresis through a nondenaturing 4% acrylamide gel in a 0.5× Tris-borate-EDTA running buffer run at 160 V.
Western blotting.
USF proteins present in CHP-126 and HeLa WCE (150 μg) were analyzed by sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis (PAGE) followed by electrophoretic transfer to nitrocellulose membranes. Membranes were blocked in Blotto A (5% milk, 10 mM Tris-HCl [pH 8.0], 150 mM NaCl) for 12 to 16 h at 4°C. Subsequently, an antiserum directed against either USF1 or USF2 (Santa Cruz Biotechnology) was added and incubated in Blotto A–0.05% Tween 20 for 60 min at room temperature. Blots were washed twice in Tris-buffered saline (10 mM Tris-HCl [pH 8.0], 150 mM NaCl)–0.05% Tween 20. A sufficient amount of a secondary antibody (anti-rabbit immunoglobulin G horseradish peroxidase-conjugated antibody; Santa Cruz Biotechnology) was added to Blotto A–0.05% Tween 20 and incubated for a further 60 min. Blots were then extensively washed in Tris-buffered saline–0.05% Tween 20; specific proteins were detected by chemiluminescence with SuperSignal substrate (Pierce).
RESULTS
USF proteins bind the class I E-box element.
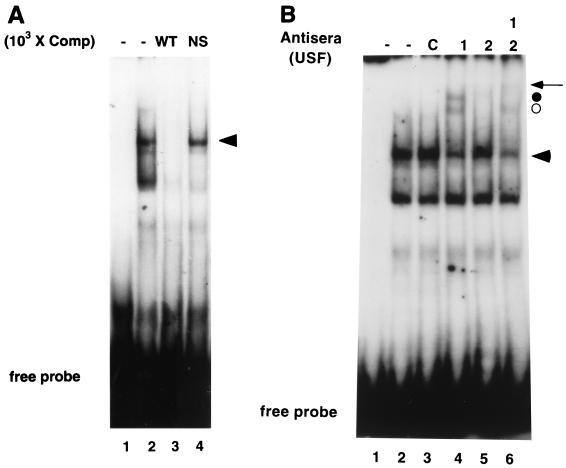
The MHC class I gene PD1 (15) contains a canonical E box (CACGTG) at bp −309 to −314, relative to the major start site of transcription (Fig. 1). The central CG dinucleotide, found in the class I E box, has been associated with binding by bHLH-Z proteins such as Myc, Max, and USF (3, 11). As a first step to determine whether this sequence contributes to MHC class I gene expression we performed gel shift analyses by using the extended class I E box, consisting of 12 bp, as a probe. HeLa WCE generated two major complexes with a double-stranded oligonucleotide probe containing three tandem class I E-box elements (Fig. 1 and 2A, lane 2). The slower-mobility complex was specific for the E box, since unlabeled wild-type oligonucleotide competed this complex, whereas an irrelevant oligonucleotide did not (Fig. 2A, lanes 3 and 4). The faster-mobility complex was not specific, since it was competed by both oligonucleotides.
FIG. 2.
The MHC class I E box forms complexes with USF proteins. The 3E oligonucleotide was end labeled, and DNA binding assay mixtures were incubated for 30 min at 4°C prior to the separation of bound and free probe in native 4% PAGE. Each lane contains 4.5 μg of HeLa WCE and competitor oligonucleotides or antisera (1 μg), as indicated, in a total volume of 20 μl. (A) A specific complex (indicated by the arrowhead) was shown to be bound to the class I E box. Lane 1, probe alone; lane 2, HeLa WCE; lanes 3 and 4, HeLa WCE (WT) and a 1,000-fold excess of self and nonspecific (NS) oligonucleotide competitors, respectively. (B) Antibody supershift examination of the 3E specific complex (represented by the arrowhead). Lane 1, probe alone; lane 2, HeLa WCE; lane 3, HeLa WCE and control (C) antisera (directed against the p50 subunit of NF-κB); lanes 4 to 6, antisera against USF1, USF2, and USF1 plus USF2, respectively, were added to DNA binding assay mixtures. Anti-USF1 antisera generated two supershifted complexes (indicated by the circles). Antisera to USF2 generated only one supershifted complex (closed circle). Combining anti-USF1 and anti-USF2 antisera together resulted in a further supershift of the USF2 containing complex (arrow).
We next determined the identity of factors that specifically bind to the class I E box by adding to the DNA binding reactions antisera to known E-box binding proteins. Antisera recognizing bHLH family members, such as Myc, Max, and SREBP-1 and antisera against unrelated transcription factors, such as activating transcription factor 2, c-jun, or the p50 subunit of NF-κB, had no effect on either complex formation or complex mobility (Fig. 2B, lane 3, and data not shown). In marked contrast, the addition of anti-USF1 antiserum generated two distinct supershifted complexes (Fig. 2B, lane 4). This observation is consistent with the known ability of USF1 to generate DNA binding complexes with distinct mobilities in gel shift analyses depending on whether the 43-kDa USF1 binds as a homodimer or as a heterodimer with the 44-kDa USF2. Therefore, the possible presence of USF2 in either of the two class I E-box complexes was assessed with anti-USF2 antiserum. The addition of anti-USF2 antiserum alone generated a faint supershifted complex (Fig. 2B, lane 5). The addition of both USF1 and USF2 antisera together had no effect on the appearance of the more rapidly migrating supershifted complex (Fig. 2B, lane 6), whereas the faint slower-migrating complex was further supershifted (Fig. 2B, lane 6). Anti-USF2 appears to preferentially eliminate, not supershift, USF2-containing complexes. These results are consistent with the interpretation that the rapidly migrating supershifted complex is generated by homodimers of USF1, while the slower-migrating supershifted complex contains heterodimers composed of USF1 and USF2. A complex of USF1 bound to the class I E box in a 107-bp DNA fragment of the native promoter was also observed in supershift assays (data not shown). Taken together, these data demonstrate that USF1 homodimers and USF1-USF2 heterodimers associate with the class I E box and are the primary bHLH proteins in HeLa cells to do so.
USF activates the class I promoter.
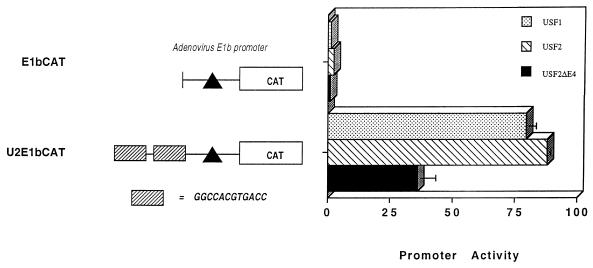
Since USF binds the class I E box in vitro, the ability of USF to regulate class I expression in vivo was examined. HeLa cells were cotransfected with an MHC class I reporter construct (-400CAT), which contains the E box (Fig. 1), together with either USF1 or USF2 expression construct. Ectopically expressed USF1 and USF2 each activated -400CAT three- to fivefold (Fig. 3). This activation was mediated through the E box, since the introduction of the class I E box upstream of a heterologous E1b promoter (3E E1bCAT) rendered it responsive to exogenous USF1 and USF2 (Table 1); in the absence of an upstream E box, the E1b promoter was unresponsive. These data indicate that the class I E box is a target for USF regulation.
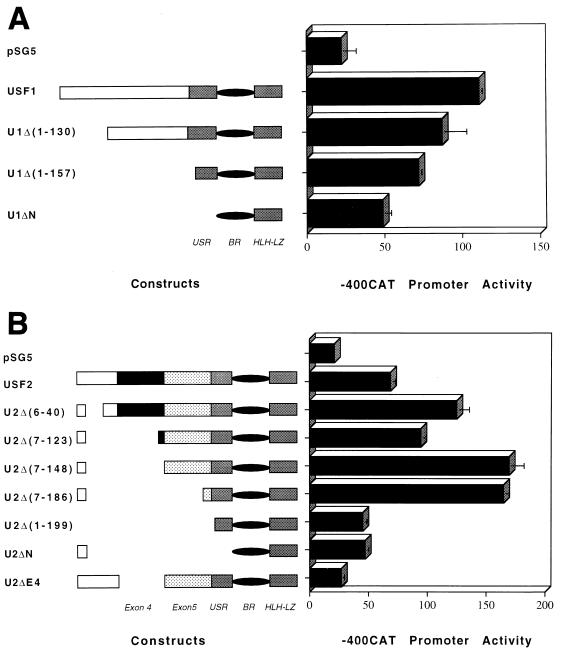
FIG. 3.
USF1 and USF2 activate the class I promoter. HeLa cells were cotransfected with 10 μg of -400CAT class I reporter construct (Fig. 1) and 3 μg of the vector control, pSG5, or the indicated USF expression plasmids. Representative maps of the expressed USF sequences and deleted regions are given on the left-hand side of the figure: upstream regulatory region (USR), binding region (BR), and HLH-Z interaction domain (HLH-LZ). Numbers in parentheses refer to deleted amino acids. Basal -400CAT activity (cotransfected with pSG5) is shown at the top of each panel. (A) The ability of USF1 and derivative truncations to trans-activate MHC class I promoter expression. (B) The ability of USF2 and truncated derivatives to activate class I expression. Data are expressed as relative percentages of acetylation normalized to the transfection control, pSV2LUC. Error bars indicate standard errors.
TABLE 1.
Class I E box confers USF inducibility on a heterologous promoter
| Construct | % Acetylation (± SE)a with:
|
||
|---|---|---|---|
| PSG5 | USF1 | USF2 | |
| 1.3 ± 0.6 | 2.4 ± 1.1 | 1.5 ± 0.8 | |
| 2.0 ± 0.2 | 62.6 ± 4.6 | 67.3 ± 4.1 | |
HeLa epithelial cells were transfected with control E1bCAT or 3E E1bCAT reporter construct and either control pSG5, USF1, or USF2 expression construct. Promoter activity was determined and expressed as percent acetylation. Percent acetylation values were normalized to that of cotransfected internal control plasmid pSV2LUC.
Although USF1 and USF2 are highly homologous in their carboxy termini (which contain the DNA-binding and dimerization domains), their amino termini (which contain the activation domains) are quite divergent (39). Therefore, the effect of various amino-terminal deletions on the ability of USF1 or USF2 to activate class I promoter activity was examined (Fig. 3). The truncation of the amino terminus of USF1 resulted in successively decreasing levels of activation of the class I promoter, consistent with an extended activation domain within this segment (Fig. 3A). Interestingly, a construct that expressed only the dimerization and DNA-binding domains (U1ΔN) still activated the promoter approximately twofold, indicating the presence of weak activation signals within the carboxy terminus of USF1.
A completely different pattern was observed for USF2. The removal of sequences between residues 6 and 186 significantly increased the ability of the USF2 variants to activate the class I promoter (Fig. 3B). These observations indicate that an inhibitory autoregulatory domain exists in the USF2 amino terminus. The further deletion of residues 186 to 199 resulted in a dramatic loss of activity, indicating the presence of an activation domain in this segment. As in the case of USF1, residual activity remained in constructs containing only the carboxy terminus (U2ΔN and U2Δ[1-199]), suggesting that a minor activation domain is present in this region.
U2ΔE4 binds to the class I E box but fails to activate the class I promoter.
In most tissues, normal in vivo alternative splicing gives rise to a variant of USF2 from which exon 4 has been deleted (U2ΔE4). However, U2ΔE4 still contains intact DNA-binding and dimerization domains as well as the activation domains encoded by exons 5 and 6 (USR). Surprisingly, the U2ΔE4 variant was unable to activate the class I promoter (Fig. 3B). The failure to activate the class I promoter does not reflect a global activation defect, since U2ΔE4 activates the AdMLP E box-containing construct, U2E1bCAT (Fig. 4).
FIG. 4.
U2ΔE4 activates an AdMLP E box-containing reporter construct. The AdMLP E-box construct U2E1bCAT contains two copies of the AdMLP E box upstream of the E1b TATA promoter. HeLa cells were cotransfected with 3 μg of control or USF expression vectors and 10 μg of reporter construct. Data are expressed as relative percentages of acetylation normalized to the transfection control, pSV2LUC. Error bars indicate standard errors.
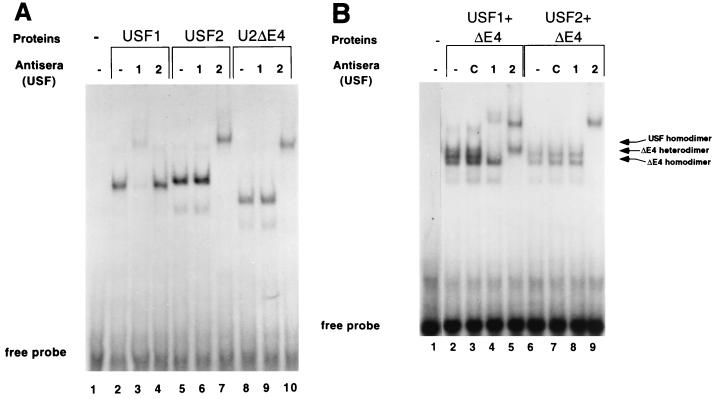
We considered the possibility that the failure of U2ΔE4 to activate the class I promoter is due to an inability to enter into functional complexes capable of binding the class I E box. However, in vitro-translated U2ΔE4 protein bound to the class I E box (Fig. 5). Individual USF proteins, translated in vitro, were combined with radiolabeled class I E-box probe, and the bound complexes were resolved in a nondenaturing acrylamide gel (Fig. 5A). USF2 complexes migrated slightly slower than USF1 complexes (Fig. 5A; compare lanes 2 and 5), consistent with the higher molecular weight of USF2. Notably, the splice variant U2ΔE4 also bound the class I E box as efficiently as either USF1 or USF2 (Fig. 5A, lane 8); the faster mobility of U2ΔE4 reflects its decreased molecular weight due to the loss of exon 4. The specificity of binding was determined by the addition of an anti-USF1 or anti-USF2 antiserum. Complexes containing in vitro-translated USF1 were supershifted by the anti-USF1 antiserum, whereas the anti-USF2 antiserum had no effect (Fig. 5A, lanes 3 and 4). Likewise, complexes containing in vitro-translated USF2 were supershifted by the anti-USF2 antiserum but not by the anti-USF1 antiserum (Fig. 5A, lanes 6 and 7). Complexes containing U2ΔE4 were recognized by the anti-USF2 antiserum, but not anti-USF1, in supershift analysis (Fig. 5A, lanes 8 to 10).
FIG. 5.
U2ΔE4 can bind the class I E box. Equivalent amounts of in vitro-translated USF proteins (determined by SDS-PAGE of 35S-labeled proteins; data not shown) were added to end-labeled class I E-box probe, and binding reactions were analyzed as described in the legend to Fig. 2 and in Materials and Methods. (A) Homodimer of U2ΔE4 can bind the class I E box. Lane 1, probe alone; lanes 2 to 4, USF1; lanes 5 to 7, USF2; lanes 8 to 10, U2ΔE4. Antisera against USF1 or USF2 were included to verify the identities of the translated proteins and demonstrate the specificity of the individual antisera. Lanes 3, 6, and 9 contain the anti-USF1 antiserum. Lanes 4, 7, and 10 contain the anti-USF2 antiserum. Specific supershifted complexes are present in lanes 3 (USF1), 7 (USF2), and 10 (U2ΔE4). Note the slower mobility of USF2 (due to its higher molecular weight) than that of USF1 and the faster mobility of U2ΔE4. (B) U2ΔE4 can generate class I E box-binding dimers with USF1 and USF2. U2ΔE4 was cotranslated with either USF1 or USF2, and the specific binding complexes generated were examined by antibody supershift analyses. Lane 1, probe alone; lanes 2 to 5, U2ΔE4 and USF1; lanes 6 to 9, U2ΔE4 and USF2. The antiserum against either USF2 or USF1 was added to distinguish homodimers from heterodimers. Control (C) antisera against the p50 subunit of NF-κB was added to the binding reaction mixtures shown in lanes 3 and 7. The antiserum against either USF1 or USF2 was added to the binding reaction mixtures shown in lanes 4 and 8 and 5 and 9, respectively.
Furthermore, the cotranslation of U2ΔE4 with either USF1 or USF2 generated heteromeric complexes capable of binding the class I E box (Fig. 5B). The cotranslation of USF1 and U2ΔE4 generated three major complexes in gel shift analyses (Fig. 5B, lane 2) that could be distinguished by their mobilities and through the addition of either the anti-USF1 or anti-USF2 antiserum. The slowest-migrating complex migrated to the same position in the gel as USF1 homodimers (data not shown). Also, this complex was supershifted by the anti-USF1 antiserum, but not by the anti-USF2 antiserum (Fig. 5B, lanes 4 and 5), indicating that it is composed only of USF1. In contrast, the fastest-migrating complex contained only U2ΔE4, since this complex was supershifted by the anti-USF2 antiserum, but not anti-USF1 (Fig. 5B; compare lanes 4 and 5). The intermediate complex was supershifted by both anti-USF1 and anti-USF2 antisera, indicating that it contained both USF1 and U2ΔE4. Although the antisera do not distinguish between USF2 and U2ΔE4, a comparison of the mobilities of complexes generated by cotranslated USF2 and U2ΔE4 (Fig. 5B, lanes 6 to 9) to those of USF1 and U2ΔE4 (Fig. 5B, lanes 2 to 5) indicated that USF2 and U2ΔE4 can also form stable class I E box binding complexes. These data indicate that defective-U2ΔE4 activation of the class I promoter is not due to its inability to enter into multimeric complexes or to bind DNA.
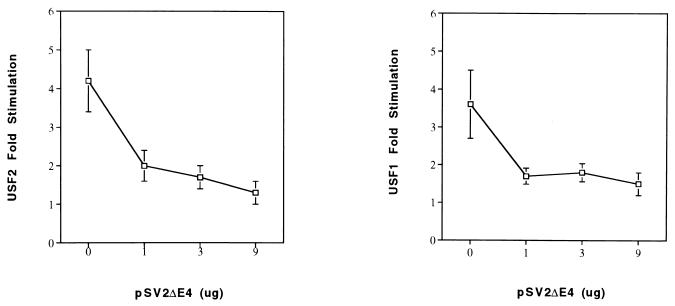
The ability of U2ΔE4 to dimerize with USF1 or USF2 and bind DNA, despite its failure to enhance class I promoter activity directly, suggested the possibility that it may function as a dominant-negative regulator of USF-mediated, class I E box-dependent enhancer activity. Many transcription factors are known to have their activities regulated by the production of sterile, alternatively spliced gene products which allow protein-protein interactions, and even DNA binding, but block trans activation (4, 17, 20, 27, 33). In order to determine whether U2ΔE4 inhibits USF activation of the class I promoter, the following experiment was conducted. The class I promoter construct -400CAT, which responds to USF, was transfected with constant amounts of either the USF2 or USF1 expression construct and increasing amounts of the U2ΔE4 construct. In the absence of U2ΔE4, both USF1 and USF2 activated class I promoter activity (Fig. 6). The addition of increasing amounts of U2ΔE4 resulted in an increased inhibition of USF1 and USF2 activation of -400CAT. At the highest concentrations, U2ΔE4 completely blocked the effects of USF1 and USF2. These data reveal a novel regulatory activity of the U2ΔE4 protein, in which it regulates activation by either USF1 or USF2. Thus, we propose that one of the in vivo functions of U2ΔE4 is to modulate USF activation of responsive genes, such as class I genes.
FIG. 6.
U2ΔE4 abrogates USF activation of MHC class I E-box activity. HeLa cells were cotransfected with 10 μg of -400CAT class I reporter construct (Fig. 1) and 3 μg of either USF1 (right panel) or USF2 (left panel) expression construct. Fold stimulation of the class I reporter, relative to the control and in the absence of added U2ΔE4, is indicated on the ordinate. Increasing amounts of the U2ΔE4 expression construct were included (shown on the abscissa as 1, 3, and 9 μg). All assays were normalized to the cotransfected internal control plasmid pSV2LUC.
The class I E box regulates promoter function in CHP-126 cells.
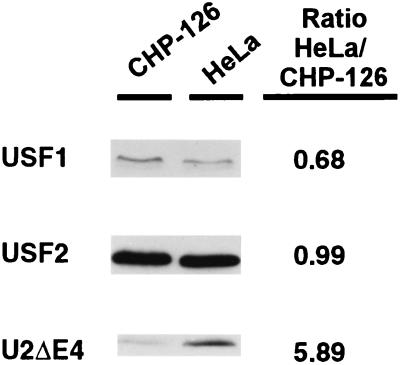
The above-mentioned findings suggest that the relative cellular concentrations of U2ΔE4 and FL USF2 may regulate the activity of the class I promoter in different tissues and cells. Therefore, we examined the U2ΔE4/FL ratio of two cell lines known to have markedly different patterns of class I expression: human HeLa epithelial cells, which express moderate levels of class I genes, and a human neuroblastoma line, CHP-126, which expresses very low levels of class I. Since U2ΔE4 appears to function as a dominant-negative regulator of class I expression, we expected it to be relatively more abundant in CHP-126 cells than in HeLa cells. Western analysis of the USF protein content of the two cell lines revealed that both contain similar amounts of USF1 and FL USF2 (Fig. 7). In contrast, the U2ΔE4/FL ratio was much lower in CHP-126 cells than in HeLa cells (Fig. 7). Inconsistent with the above prediction, the class I-expressing HeLa cell line contained sixfold more dominant-negative U2ΔE4 splice product than the CHP-126 line, which expresses barely detectable levels of class I genes (Fig. 7). This surprising result led us to compare the role of the E box in class I expression in these two cell lines.
FIG. 7.
CHP-126 neuroblastoma cell line contains less U2ΔE4 relative to HeLa epithelial cells. Approximately 150 μg of either CHP-126 or HeLa WCE was separated by SDS–12.5% PAGE and transferred to nitrocellulose membranes, and USF proteins were distinguished by Western blotting with antisera against USF1 and USF2. Densitometric values to quantitate the relative ratios of USF1 and USF2 to U2ΔE4 between the two cell lines are provided to the right.
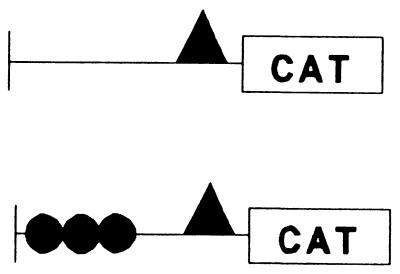
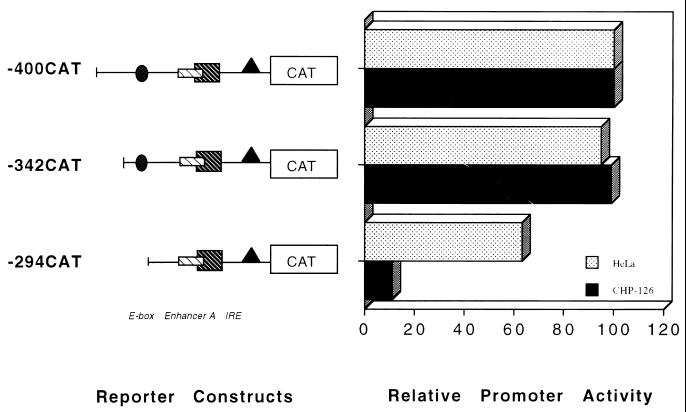
Consistent with the low level of endogenous class I expression in CHP-126 cells, an FL class I promoter construct containing 1,000 bp of upstream sequences transfected into CHP-126 cells is not expressed (30). The interval between bp −1000 and −400 contains a series of tissue-specific silencer elements which specifically represses class I expression in CHP-126 neuroblastoma cells (30). However, a promoter construct containing only the proximal 400 bp of upstream class I sequences is actively transcribed in both CHP-126 cells and HeLa cells (Fig. 8). The class I E box is contained within this 400-bp upstream segment (at bp −309 to −314), allowing an analysis of its relative role in class I promoter activity in both CHP-126 and HeLa cells. Removing the region of the promoter between positions −400 and −342 had no effect on class I promoter activity in either cell type (compare -400CAT to -342CAT [Fig. 8]). However, a truncation to position −294 (-294CAT), which removes the E box, reduced promoter activity 10-fold in CHP-126 cells, whereas only a modest reduction was observed in HeLa cells. These data demonstrate the presence of a strong tissue-specific enhancer element in the segment between positions −342 and −294 that functions preferentially in CHP-126 neuroblastoma cells, relative to HeLa cells. However, the activity of the enhancer in CHP-126 cells is revealed only in the truncated class I promoter construct which is completely dependent upon the E box for activity.
FIG. 8.
A region of the class I promoter containing the E box is active in the CHP-126 neuroblastoma cell line but is inactive in HeLa epithelial cells. HeLa epithelial cells and CHP-126 neuroblastoma cell lines were transfected with 10 μg of the indicated MHC class I reporter constructs, and promoter function was assessed. Note that the actual class I promoter activities in HeLa and CHP-126 cells were normalized to facilitate this comparison; class I expression in the CHP-126 cell line (although significant) is approximately 40-fold less than in HeLa cells.
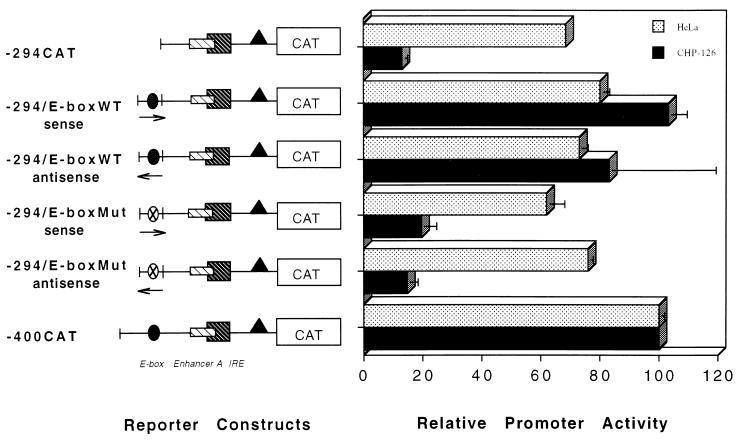
To determine whether the E box accounted for the strong CHP-126-specific enhancer activity present in this gene fragment, wild-type (WT) and mutant (Mut) class I E boxes (Fig. 9) were individually cloned upstream of -294CAT to generate the -294(WT) E-boxCAT and -294(Mut) E-boxCAT constructs, respectively. The mutant E box had no effect on basal class I promoter activity in either CHP-126 or HeLa cells (Fig. 9). In contrast, placement of the WT E box upstream of -294CAT (in either the sense or antisense orientation) stimulated its promoter activity in CHP-126 cells approximately fivefold, to a level equal to that of -400CAT. The E box had no significant effect on basal class I promoter activity in HeLa cells. However, as shown in Fig. 3 and 6, the overexpression of exogenous USF1 or USF2 does activate class I promoter activity in transiently transfected HeLa cells consistent with a titration of the dominant-negative effect of U2ΔE4. Taken together, these results support the proposal that U2ΔE4 functions as a dominant-negative regulator of class I E-box enhancer activity. In HeLa cells, the relative abundance of U2ΔE4 suppresses enhancer activity. In CHP-126 cells the lower U2ΔE4/FL ratio enables the E box to act as a strong enhancer of class I promoter activity in the absence of upstream tissue-specific silencers.
FIG. 9.
The ability of the class I E box to stimulate transcription is cell type specific. Wild-type (WT) or mutant (Mut) class I E boxes, in sense and antisense orientations, were cloned upstream of -294CAT (which lacks an E box). HeLa and CHP-126 cell lines were transfected with 10 μg of the indicated reporter construct, and promoter activity was determined.
DISCUSSION
Cell-surface MHC class I expression, in any given tissue, is largely determined at the level of transcription and consists of both tissue-specific and dynamic control mechanisms. Tissue-specific transcriptional mechanisms define a characteristic “set-point” level of gene expression (for example, class I expression is high in lymphoid tissue, moderate to low in most other tissues, and very low in the brain) and depends on a number of distinct positive and negative regulatory elements. Whether a common set of regulatory elements variably regulate gene expression in different tissues or different tissues utilize unique sets of regulatory elements to establish tissue-appropriate levels of expression is not clear. While the control mechanisms (regulatory elements and their cognate DNA binding factors) responsible for dynamic modulation of class I expression have been extensively examined, those elements responsible for the maintenance of tissue-appropriate levels of expression remain relatively uncharacterized.
We have identified a DNA sequence element, referred to as an E box, and have demonstrated that it is a cell type-specific regulator of class I promoter activity. The extended class I E-box element, G−6G−5C−4C−3A−2C−1G+1T+2G+3A+4G+5G+6, spans 12 bases. Although it contains a canonical binding site for bHLH-Z proteins, the class I E box shows a marked specificity in its interactions. USF proteins bind to the E box, either as in vitro-translated proteins or from WCE, whereas other bHLH-Z proteins, such as c-Myc and Max, do not (data not shown). Consistent with this discrimination of binding, only USF1 and USF2 activate the class I E box; c-Myc does not (data not shown). USF proteins are not known to be inducible, suggesting that the E box regulates the constitutive and not dynamic control of class I expression. The contribution of the E box to class I expression is determined by the ratio of USF1 and USF2 to the splice variant U2ΔE4. Both USF1 and USF2 can activate the class I promoter, whereas U2ΔE4 acts as a specific dominant-negative repressor of E-box function. All three USF proteins can enter into heteromeric complexes and appear to bind the class I E box with approximately equal affinities. Therefore, the ability of the E box to enhance class I expression, in the absence of ectopically expressed USF1 or USF2, is determined by the cellular ratio of endogenous U2ΔE4 splice variant to FL USF proteins. In neuroblastoma cells where there is relatively less U2ΔE4, the E box can function as a potent enhancer increasing class I promoter activity approximately fivefold. In contrast, in HeLa cells where U2ΔE4 is relatively more abundant, the E box does not function as a strong enhancer. However, E-box activity can be evoked in HeLa cells by overexpressing either USF1 or USF2 in a transient transfection, thereby disrupting the WT balance of U2ΔE4/FL USF protein and skewing in favor of FL USF dimers and an active E box.
USF1 and USF2 have both been shown to inhibit cell proliferation, raising the possibility that the observed effects on class I promoter activity may be an indirect result of cell growth effects (1, 25). This is unlikely because (i) the effects on the class I promoter depend on the presence of the E box and (ii) USF mutants, including U2ΔE4 and U2Δ(7-186), do not affect cell growth (25a) but differentially affect the class I promoter.
U2ΔE4 does not activate the MHC class I promoter, but it has been shown to directly activate a variety of natural and artificial promoters, including the AdMLP (24), the pyruvate kinase promoter (41), and a promoter containing two copies of the AdMLP E box upstream of the E1b TATA box (23). Although U2ΔE4 alone does not repress basal class I promoter activity in HeLa cells, it effectively inhibits the ability of either USF1 or USF2 to activate class I expression. These data indicate that U2ΔE4 acts as a dominant-negative regulator of USF-mediated activation of the class I promoter, presumably by forming heteromeric complexes with other USF proteins which can bind E-box elements but fail to activate. Importantly, U2ΔE4 directly activates a promoter construct consisting of three copies of the class I E box upstream of the E1b TATA box (data not shown). Therefore, the repressive effect of U2ΔE4 is not intrinsic to the class I E box. These findings support the hypothesis that the differential activities of U2ΔE4 and USF2 are either determined by promoter context or influenced by regulatory elements situated downstream of the E box. Differences in the AdMLP and class I core promoter regions may determine the trans-acting potential of U2ΔE4. Luo and Sawadogo demonstrated that a core promoter initiator-like sequence is required for optimal trans activation by U2ΔE4, as well as U2Δ(7-186) (24). Whereas U2ΔE4 and U2Δ(7-186) minimally activated U2E1bCAT (which contains only the E1b TATA box), the insertion of an initiator-like sequence downstream of the TATA box allowed U2ΔE4 and U2Δ(7-186) to activate almost to the level of USF2. However, whether U2ΔE4 or U2Δ(7-186) could act as a dominant repressor of USF2 activation of U2E1bCAT activity was not investigated in that study. Although U2ΔE4 did not activate class I promoter activity, U2Δ(7-186) was a very potent activator of class I expression. Therefore, the contribution of class I core promoter structures to the ability of USF to activate is unclear.
Exon 4 in U2ΔE4 contains at least two regulatory elements, as revealed by the fine mapping. One is a negative regulatory element between amino acids 123 and 148. After the deletion of this region, the resulting USF2 protein has an enhanced ability to activate class I promoter activity. The other element in exon 4 appears to be an activation domain. However, this domain does not directly activate but rather appears to inhibit the activity of a repression domain, located between amino acids 7 and 76. This is best appreciated by comparing the two constructs U2Δ(7-148) and U2ΔE4. U2Δ(7-148), which has the sequences between amino acids 7 and 76 as well as exon 4 deleted, is the most active of the USF2 constructs. Thus, exon 4 itself is not necessary for maximal activity. In contrast, U2ΔE4, which is devoid of only exon 4, has no activity. Since the only difference between these two mutants is the presence of amino-terminal residues 7 to 76, it is concluded that this region contains a suppressive autoregulatory domain which is blocked in the presence of exon 4.
Consistent with the model of USF-regulating tissue-specific class I expression, the class I E box functions as a cell type-specific enhancer. Both USF1 and USF2 are ubiquitously expressed: extracts from HeLa (adenocarcinoma), Jurkat (T lymphocyte), and CHP-126 (neuroblastoma) cells all give rise to similar complexes with E-box probes (Fig. 2 and data not shown). However, the relative ratios of USF homo- and heterodimers are known to vary in different cell lines (41). As detailed here and also reported by others, HeLa cells primarily express USF1-USF2 heterodimers and some USF1 homodimers (39). Whether the different compositions of USF dimers affects their abilities to activate different promoters has not been examined. However, it is likely that the relative level of U2ΔE4 in a cell determines the activity of the class I E box as an enhancer. The E box is active in the CHP-126 neuroblastoma cell line, where the relative abundance of U2ΔE4 is low, compared to HeLa cells, where the E box is much less active.
The finding that the E box is an active enhancer in CHP-126 neuroblastoma cells, whose class I expression is actively repressed by upstream silencers, would appear to be paradoxical. However, previous studies have demonstrated that these silencers are labile, and class I expression is readily induced in the face of infections and interferon (30, 42). Thus, it is tempting to speculate that the active E-box enhancer maintains the endogenous class I gene poised for active transcription, to allow the rapid development of an immune response.
In conclusion, the present studies suggest that the E box contributes to the tissue-specific regulation of class I gene expression, through the actions of USF1, USF2, and U2ΔE4. Together, these factors modulate enhancer activity, providing a mechanism to fine-tune the level of class I expression in different tissues.
ACKNOWLEDGMENTS
We gratefully acknowledge Carol Thiele, Stephen Straus, Barbara L. Rellahan, Shelby Berger, and Julie Brown for valuable suggestions and discussions. We thank Robert Roeder for providing the U2E1bCAT reporter construct.
S.J.H. was supported by the HHMI Summer Research Program.
REFERENCES
- 1.Aperlo C, Boulukos K E, Pognonec P. The basic region/helix-loop-helix/leucine repeat transcription factor USF interferes with Ras transformation. Eur J Biochem. 1996;241:249–253. doi: 10.1111/j.1432-1033.1996.0249t.x. [DOI] [PubMed] [Google Scholar]
- 2.Beckman H, Su L-K, Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer uE3 motif. Genes Dev. 1989;4:1730–1740. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 3.Bendall A J, Molloy P L. Base preferences for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodor J, Habener J F. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem. 1998;273:9544–9551. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
- 5.Bresnick E H, Felsenfeld G. The leucine zipper is necessary for stabilizing a dimer of the helix-loop-helix transcription factor USF but not for maintenance of an elongated conformation. J Biol Chem. 1994;269:21110–21116. [PubMed] [Google Scholar]
- 6.Cai M, Davis R W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 7.Carr C S, Sharp P A. A helix-loop-helix protein related to immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carthew R W, Chodosh L A, Sharp P A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985;43:439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain J W, Vasavada H A, Ganguly S, Weissman S M. Identification of cis sequences controlling efficient position-independent tissue-specific expression of human major histocompatibility complex class I genes in transgenic mice. Mol Cell Biol. 1991;11:3564–3572. doi: 10.1128/mcb.11.7.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corneliussen B, Thornell A, Hallberg B, Grundstrom T J. Helix-loop-helix transcriptional activators bind to a sequence in glucocorticoid response elements of retrovirus enhancers. J Virol. 1991;65:6084–6093. doi: 10.1128/jvi.65.11.6084-6093.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang C V, Dolde C, Gillison M L, Kato G J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das G, Hinkley C S, Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 13.Davis R L, Cheng P-F, Lassar A B, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 14.Du H, Roy A L, Roeder R G. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. EMBO J. 1993;12:501–511. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrlich R, Maguire J, Singer D. Identification of negative and positive regulatory elements associated with a class I major histocompatibility complex gene. Mol Cell Biol. 1988;8:695–703. doi: 10.1128/mcb.8.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferre-D’Amare A R, Pognonec P, Roeder R G, Burley S K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girardet C, Walker W H, Habener J F. An alternatively spliced polycistronic mRNA encoding cyclic adenosine 3′,5′-monophosphate (cAMP)-responsive transcription factor CREB (cAMP response element-binding protein) in human testis extinguishes expression of an internally translated inhibitor CREB isoform. Mol Endocrinol. 1996;10:879–891. doi: 10.1210/mend.10.7.8813728. [DOI] [PubMed] [Google Scholar]
- 18.Gregor P D, Sawadogo M, Roeder R G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 19.Howcroft T K, Richardson J C, Singer D S. MHC class I gene expression is negatively regulated by the proto-oncogene, c-jun. EMBO J. 1992;12:3163–3169. doi: 10.1002/j.1460-2075.1993.tb05985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones N. Transcriptional regulation by dimerization: two sides to an incestuous relationship. Cell. 1990;61:9–11. doi: 10.1016/0092-8674(90)90207-u. [DOI] [PubMed] [Google Scholar]
- 21.Kim J B, Spotts G D, Halvorsen Y-D, Shih H-M, Ellenberger T, Towle H C, Spiegelman B M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeBouteiller P. HLA class I chromosomal region, genes, and products: facts and questions. Crit Rev Immunol. 1994;14:89–129. doi: 10.1615/critrevimmunol.v14.i2.10. [DOI] [PubMed] [Google Scholar]
- 23.Lin Q, Luo X, Sawadogo M. Archaic structure of the gene encoding transcription factor USF. J Biol Chem. 1994;269:23894–23903. [PubMed] [Google Scholar]
- 24.Luo X, Sawadogo M. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X, Sawadogo M. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc Natl Acad Sci USA. 1996;93:1308–1313. doi: 10.1073/pnas.93.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Luo, X., and M. Sawadogo. Unpublished observation.
- 26.Maguire J E, Frels W I, Richardson J C, Weissman J D, Singer D S. In vivo function of regulatory DNA sequence elements of a major histocompatibility complex class I gene. Mol Cell Biol. 1992;12:3078–3086. doi: 10.1128/mcb.12.7.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991;251:33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- 28.Mellor J, Jiang W, Funk M, Rathjen J, Barnes C A, Hinz T, Hegemann J H, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto N G, Moncollin V, Chambon P. Specific interaction between a transcription factor and the upstream element of the adenovirus-2 major late promoter. EMBO J. 1985;4:3563–3570. doi: 10.1002/j.1460-2075.1985.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy C, Nikodem D, Howcroft K, Weissman J D, Singer D S. Active repression of major histocompatibility complex class I genes in a human neuroblastoma cell line. J Biol Chem. 1996;271:30992–30999. doi: 10.1074/jbc.271.48.30992. [DOI] [PubMed] [Google Scholar]
- 31.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 32.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 33.Oettgen P, Akbarali Y, Boltax J, Best J, Kunsch C, Libermann T A. Characterization of NERF, a novel transcription factor related to the Ets factor ELF-1. Mol Cell Biol. 1996;16:5091–5106. doi: 10.1128/mcb.16.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawadogo M, Roeder R G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 35.Sawadogo M, VanDykes M W, Gregor P D, Roeder R G. Multiple forms of the human gene-specific transcription factor USF. J Biol Chem. 1988;263:11985–11993. [PubMed] [Google Scholar]
- 36.Schmidt C M, Ehlenfeldt R G, Athanasiou M C, Duvick L A, Heinrichs H, David C S, Orr H T. Extraembryonic expression of the human MHC class I gene HLA-G in transgenic mice. Evidence for a positive regulatory region located 1 kilobase 5′ to the start site of transcription. J Immunol. 1993;151:2633–2645. [PubMed] [Google Scholar]
- 37.Singer D, Maguire J. Regulation of the expression of class I MHC genes. Crit Rev Immunol. 1990;10:235–257. [PubMed] [Google Scholar]
- 38.Sirito M, Walker S, Lin Q, Kozlowski M T, Klein W H, Sawadogo M. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 1992;2:231–240. [PMC free article] [PubMed] [Google Scholar]
- 39.Sirito M, Lin Q, Maity T, Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ting J P-Y, Baldwin A S. Regulation of MHC gene expression. Curr Biol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 41.Viollet B, Lefrancois-Martinez A M, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 42.Weissman J D, Singer D S. A complex regulatory DNA element associated with a major histocompatibility complex class I gene consists of both a silencer and an enhancer. Mol Cell Biol. 1991;11:4217–4227. doi: 10.1128/mcb.11.8.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]