Abstract
Infections caused by Gram-negative bacteria can be challenging to treat due to the outer membrane permeability barrier and the increasing emergence of antibiotic resistance. During infection, Gram-negative pathogens must acquire iron, an essential nutrient, in the host. Many Gram-negative bacteria utilize sophisticated iron acquisition machineries based on siderophores, small molecules that bind iron with high affinity. In this review, we provide an overview of siderophore-mediated iron acquisition in Enterobacteriaceae and show how these systems provide a foundation for the conceptualization and development of approaches to prevent and/or treat bacterial infections. Differences between the siderophore-based iron uptake machineries of pathogenic Enterobacteriaceae and commensal microbes may lead to the development of selective “Trojan-horse” antimicrobials and immunization strategies that will not harm the host microbiota.
Keywords: Enterobacteriaceae, Gram-negative bacteria, iron, microcins, siderophores
The Enterobacteriaceae comprise a large family of Gram-negative bacteria that belong to the phylum Proteobacteria. Many Enterobacteriaceae are commonly found in the mammalian gastrointestinal tract as components of the gut microbiota, a large and diverse community of microbes that play fundamental roles in host physiology. In healthy individuals, the gut microbiota primarily comprises commensal anaerobic bacteria of the phyla Bacteroidetes and Firmicutes [1], whereas Proteobacteria comprise only a small fraction (eg, Escherichia coli constitutes <1%) [2]. Even so, commensal E coli strains biosynthesize molecules crucial for host homeostasis such as B-complex vitamins and vitamin K [3].
Escherichia coli, a prominent species of Enterobacteriaceae, is represented not only by gut commensals but also by pathogens and pathobionts (ie, bacteria that can become pathogenic under certain circumstances) [4]. Another member of the Enterobacteriaceae family, Salmonella enterica, is a pathogen that represents one of the leading causes of infectious diarrhea worldwide [5]. Many Salmonella serovars including S enterica serovar Typhimurium (S Typhimurium) induce intestinal inflammation, which enables the pathogen to compete with the gut microbiota and establish a niche in the inflamed gut [6]. Several studies have demonstrated that intestinal inflammation induces the loss of obligate anaerobes (ie, Bacteroidetes and Firmicutes) and favors a bloom of Proteobacteria [7, 8]. For example, uropathogenic E coli and adherent-invasive E coli are able to thrive in the inflamed intestine [7]. Finally, some Enterobacteriaceae can disseminate from the gut and cause diseases including urinary tract infection, meningitis, and sepsis. Altogether, pathogenic Enterobacteriaceae are responsible for substantial morbidity and mortality worldwide [9].
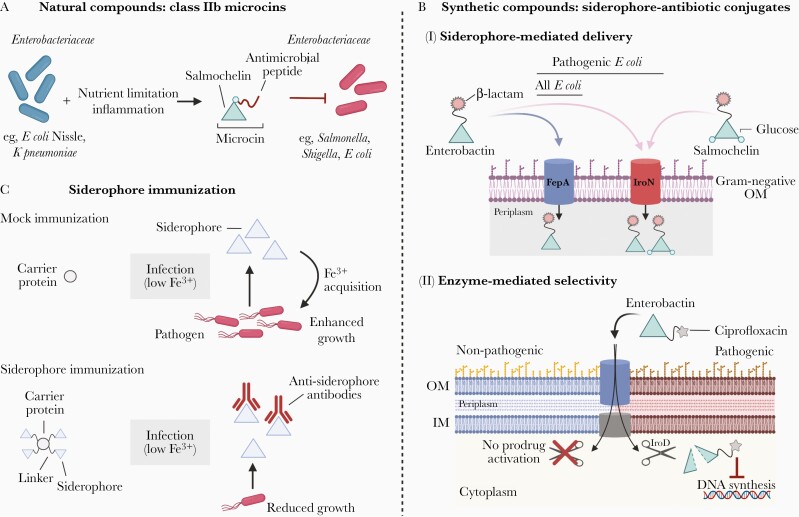
Treatment of infections caused by Enterobacteriaceae and other Gram-negative bacterial pathogens is inherently difficult because these organisms have a semipermeable outer membrane that serves as a barrier and blocks cellular entry of many antibiotics. Moreover, the overuse of antibiotics has led to increased multidrug resistance in Gram-negative pathogens [10]. In general, treatment of Gram-negative bacterial infections relies on broad-spectrum antibiotics that disrupt the microbiota and can lead to life-threatening secondary infections such as those caused by Clostridioides difficile (formerly known as Clostridium difficile) [11]. In this review, we discuss potential antimicrobial strategies that are inspired by mechanisms of iron acquisition in Gram-negative bacteria and by the iron-withholding innate immune response (Figure 1). In addition, such strategies could provide antibiotics that are more selective towards pathogens and reduce perturbation of the microbiota.
Figure 1.
Overview of siderophore-based antimicrobial strategies to target Enterobacteriaceae. A, Upon nutrient limitation, certain members of the family Enterobacteriaceae such as Escherichia coli Nissle 1917 and Klebsiella pneumoniae secrete small antibacterial peptides known as microcins. Class IIb microcins are siderophore-antibiotic conjugates (SACs) characterized by the following: (1) a conserved C-terminal domain that carries a salmochelin-like siderophore that mediates cellular uptake through catecholate siderophore receptors (eg, FepA and IroN, as shown in B); and (2) an N-terminal domain that exerts antibacterial activity. B, Siderophore-mediated delivery of antimicrobials can be mimicked by synthetic SACs. β-lactam antibiotics linked to the siderophores enterobactin or salmochelin achieve selectivity based on the siderophore’s receptor expression in the outer membrane (OM) of E coli (I). In E coli, enterobactin can be internalized by both FepA and IroN, whereas salmochelin uptake requires IroN, which is predominantly expressed by pathogens. Thus, salmochelin-antibiotic conjugates are more selective towards pathogens. Differences in bacterial enzyme expression are another way to achieve targeted antibacterial activity (II). The synthetic enterobactin-ciprofloxacin conjugate is transported through FepA in all E coli strains, although it is inactive unless this prodrug’s siderophore moiety is hydrolyzed in the cytoplasm by the esterase IroD, which is primarily found in pathogenic strains. Thus, the enterobactin-ciprofloxacin conjugate exerts its antibiotic activity only on E coli strains that express the IroD esterase. C, Siderophores can be leveraged for immunization if they are conjugated to a carrier protein that acts as an adjuvant. The resulting anti-siderophore antibodies confer protection against pathogen outgrowth in mouse models of intestinal Salmonella infection and in models of urinary tract infection with E coli.
IRON ACQUISITION IN GRAM-NEGATIVE BACTERIA: SIDEROPHORES
Iron (Fe) is the most abundant transition metal in the human body. Because excess iron is toxic, iron homeostasis in humans is tightly controlled by regulators, iron transport systems, and iron storage proteins. Most iron in the human body is found in intracellular spaces, where it is predominantly bound to the iron-storage protein ferritin, to hemoglobin, and to other proteins that use iron as a cofactor. In extracellular spaces, most iron is associated with high-affinity iron-binding proteins such as the iron-transport protein transferrin and, during inflammation, lactoferrin [12]. For almost all characterized bacteria, iron is a cofactor for many enzymes including those associated with proliferation and host colonization [13], and most bacteria require iron concentrations of 10–8 to 10–6 M to survive [14]. During infection, the host further limits iron availability as a strategy to inhibit bacterial replication. The host defense mechanisms that limit the availability of iron and other essential nutrients to pathogens are collectively termed nutritional immunity [15].
To overcome the iron limitation imposed by the innate immune response, bacteria use several strategies, including scavenging iron in the ferric (Fe3+) and ferrous (Fe2+) forms as well as heme. Under aerobic conditions, such as those encountered in the inflamed gut, Fe3+ predominates. Upon encountering iron-limited environments, Enterobacteriaceae biosynthesize and secrete small Fe3+-chelating molecules termed siderophores [11]. These molecules bind Fe3+ with affinities that can exceed those of host Fe3+-binding proteins such as transferrin and lactoferrin [16], which allows the siderophore producer to scavenge Fe3+. One such siderophore is enterobactin, which is produced by commensal and pathogenic Enterobacteriaceae alike, including E coli, Klebsiella pneumoniae, and Salmonella [17]. To limit enterobactin-mediated iron acquisition by Enterobacteriaceae, the host secretes the antimicrobial protein lipocalin-2 (also known as neutrophil gelatinase-associated lipocalin [NGAL] and as 24p3), which sequesters Fe3+-bound enterobactin and thus inhibits the growth of microbes that rely on this siderophore (eg, many commensal Enterobacteriaceae) [18]. In contrast, many pathogenic Enterobacteriaceae evade this host response by secreting additional siderophores with different chemical structures that cannot be captured by lipocalin-2. For example, Salmonella, Klebsiella, and a number of pathogenic E coli produce salmochelins, glucosylated derivatives of enterobactin. The sugar moieties of salmochelins are bulky and hydrophilic, which impede capture of these molecules by lipocalin-2.
In addition to competing with the host for iron, bacteria can compete with one another for this nutrient. In general, microbial competition occurs by 2 modes: exploitative competition or interference competition. Exploitative competition encompasses microbes competing for a common nutrient, whereas interference competition involves microbes secreting molecules to target their competitors [19]. Both modes of competition can occur between members of the Enterobacteriaceae when these bacteria compete for iron. An example of exploitative competition occurs in the inflamed gut, where the production of multiple siderophores enables the probiotic bacterium E coli Nissle 1917 to successfully outcompete S Typhimurium for acquiring iron [20]. Interference competition can occur when a bacterium secretes antimicrobial molecules that target the siderophore uptake machineries of its bacterial competitors. This particular mode of interference competition is often referred to as a “Trojan-horse” strategy, because it involves disguising a toxic payload as a beneficial molecule.
NATURAL TROJAN-HORSE ANTIBIOTICS: CLASS IIB MICROCINS
An example of a natural Trojan-horse strategy is when a bacterium biosynthesizes and deploys a siderophore-antibiotic conjugate (SAC); the antibacterial moiety is disguised by being attached to a beneficial iron chelator (the siderophore) that is recognized by the cognate siderophore acquisition system on a recipient bacterium [21]. Sideromycins and class IIb microcins are 2 types of natural SACs deployed by Gram-negative bacteria [21]. Although these SACs have different chemical structures and mechanisms of antibacterial activity, they exemplify the same design principle of linking a toxic cargo to a siderophore. Broadly, microcins are ribosomally synthesized antimicrobial peptides that are produced by various members of the Enterobacteriaceae [21]. In particular, the class IIb subfamily of microcins represent SACs where the ribosomal peptide is posttranslationally modified at its C-terminus with a salmochelin moiety (Figure 1a). This remarkable structural feature was first reported in 2004 during studies of MccE492, a microcin produced by K pneumoniae RYC492 [22]. In vitro studies demonstrated that MccE492 exerts potent antimicrobial activity (minimum inhibitory concentration = 40–150 nM) against phylogenetically related bacterial strains and can inhibit the growth of E coli, Salmonella, Klebsiella, Citrobacter, and Shigella [23]. Subsequent to these findings, the siderophore modification has been predicted for other microcins including MccM and MccH47, which are produced by a few E coli strains, including uropathogenic E coli CFT073 and probiotic E coli Nissle 1917 [23, 24].
In vitro, the class IIb microcins target susceptible Enterobacteriaceae and have been proposed to be important mediators of competition among different species and strains of this family [23]. In vivo, a role for MccM and MccH47 in the competition among Enterobacteriaceae has been shown in the inflamed gut (ie, during colitis), where hypoferremia is observed [6]. Indeed, microcin-producing strains have a competitive advantage in such iron-deplete environments, where commensal and pathogenic Enterobacteriaceae produce siderophores and compete for iron. Moreover, consistent with the prediction that microcins only target Enterobacteriaceae, microcin production had no broad effects on the gut microbiota composition [6]. These in vivo results should motivate additional investigations of the antimicrobial activity of MccM and MccH47, including fundamental studies of cellular uptake and target engagement. Based on our current knowledge, one could foresee a potential application of class IIb microcins or microcin-inspired molecules as antimicrobials that limit the expansion of some enteric pathogens and pathobionts in environments that are otherwise favorable to their growth, without perturbing the gut microbiota at large. At present, investigations of the therapeutic potential of class IIb microcins are hampered by limited availability of the peptides. These natural products are difficult to isolate and purify from cultures of the producer organisms in quantities that are needed for fundamental and clinical investigations. Alternative methods of production such as heterologous expression or chemical or chemoenzymatic synthesis may hold promise and warrant consideration.
NONNATURAL TROJAN-HORSE ANTIBIOTICS: SYNTHETIC SIDEROPHORE-ANTIBIOTIC CONJUGATES
Over several decades, synthetic chemists in academia and industry have designed and synthesized SACs where a native siderophore or siderophore-like molecule is linked to a clinically approved antibiotic such as a β-lactam or fluoroquinolone. In early studies, a primary objective was to hijack siderophore uptake machinery to overcome outer membrane permeability in Gram-negative bacteria [25]. More recently, synthetic SACs have gained interest for their potential as narrow-spectrum antibiotics that target specific strains, species, or group of species on the basis of siderophore receptor expression [26–29]. This synthetic work presents several design challenges because both the siderophore and the antibiotic moieties must be modified and connected together such that neither loses its function. Two noteworthy examples of synthetic SACs include an artificial siderophore modified with aminopenicillins, which exhibited potent antipseudomonal activity relative to the parent antibiotics [30], and a mycobactin-artemisinin conjugate that kills Mycobacterium tuberculosis and Plasmodium falciparum [31].
In 2019, cefiderocol was approved by the US Food and Drug Administration for the treatment of complicated urinary tract infections [32]. It was also evaluated in clinical trials for the treatment of Gram-negative bacterial infections, including bloodstream infection and pneumonia. Cefiderocol is described as a siderophore-cephalosporin conjugate; however, the “siderophore” portion is a simplified scaffold containing a monochlorinated catechol moiety reminiscent of the catechols found in many bacterial siderophores including enterobactin and salmochelins. Cefiderocol is reported to overcome 3 primary resistance mechanisms of Gram-negative bacteria to β-lactams—porin channel alterations, β-lactam inactivation, and efflux pump overproduction [32]. Furthermore, cefiderocol is stable against all classes of β-lactamase enzymes, including carbapenemases that are the predominant mechanism of β-lactam resistance in many bacterial pathogens that cause urinary tract infections [32]. Nonetheless, cefiderocol followed an accelerated clinical development pathway, and further clinical data are needed to inform its wide use in the clinic [33]. Future studies should also address the microbial transport of cefiderocol and whether its administration perturbs the gut microbiota.
Several recent investigations of synthetic SACs that target Enterobacteriaceae illustrated the potential for using siderophores in narrow-spectrum antibacterial approaches [27, 29]. One series of SACs that harbor a β-lactam (ampicillin or amoxicillin) linked to enterobactin or salmochelin by a polyethylene glycol linker provided proof-of-concept evidence for targeting pathogenicity (Figure 1b) [27, 28]. All E coli encode for the biosynthesis and transport of enterobactin, whereas only a subset of Enterobacteriaceae (mostly pathogens, including K pneumoniae, S Typhimurium, and uropathogenic E coli) harbor the iroA gene cluster (iroBCDEN), which encodes machinery for salmochelin biosynthesis, transport, and processing. All non-pathogenic and pathogenic E coli express FepA, the outer membrane receptor for enterobactin, and all such tested strains were susceptible to enterobactin-β-lactam conjugates. In contrast, salmochelin-β-lactam conjugates harboring diglucosylated enterobactin provided strain-selective targeting based on the expression of IroN, the outer membrane receptor for salmochelin. Moreover, these siderophore-β-lactams exhibited up to 1000-fold improved antibacterial activity against E coli compared with the unmodified β-lactam, and these SACs killed E coli in the presence of other bacteria, including the commensal Lactobacillus rhamnosus [28]. Overall, these studies of enterobactin- and salmochelin-β-lactam conjugates support the notion that the activity profile of a clinically approved, broad-spectrum antibiotic can be tuned by siderophore modification, and they indicate that siderophores provide a means to target pathogenicity.
Subsequently, a report of an enterobactin-ciprofloxacin conjugate uncovered the potential for leveraging a pathogen-associated enzyme for targeted antibacterial activity (Figure 1c) [29]. An antibacterial activity assay screen against 4 E coli strains indicated that the enterobactin-ciprofloxacin conjugate provided antibacterial activity comparable to ciprofloxacin, but only against E coli strains that harbor the iroA gene cluster. Further investigation demonstrated that the enterobactin-ciprofloxacin conjugate was transported into the E coli cytoplasm by the enterobactin transport machinery (FepA and the inner membrane transport system FepCDG). It is remarkable that IroD, an iroA gene cluster-encoded cytoplasmic esterase that can hydrolyze enterobactin and salmochelins, was required for the antibacterial activity of the enterobactin-ciprofloxacin conjugate. Hence, the siderophore modification generated an inactive prodrug that guided the antibiotic into the cytoplasm, whereafter intracellular processing by IroD afforded an active deoxyribonucleic acid gyrase/topoisomerase inhibitor. Looking ahead, such SACs may have the ability to limit the growth of some enteric pathogens (eg, uropathogenic E coli, Salmonella) and pathobionts (eg, adherent-invasive E coli) that express the iroA gene cluster without perturbing the gut microbiota at large.
SIDEROPHORE-BASED IMMUNIZATION STRATEGIES
Another promising strategy for designing nontraditional antibacterial therapies focuses on targeting components of bacterial iron acquisition systems for vaccine development. Some studies have targeted iron receptors (eg, for siderophores, heme) for immunization against Gram-negative bacteria, including uropathogenic E coli [34–36]. This approach has had some success but also some limitations because immunization against only a subset of iron receptors present in uropathogenic E coli induced a sustained and robust antibody response [34–36]. An alternative immunization strategy would be to use siderophores as the antigens (Figure 1d). Because siderophores are small molecules that are not inherently immunogenic, they need to be conjugated to immunogenic carrier proteins. The promise of this approach was demonstrated by an early study showing that injection of the Vibrio cholerae siderophore vibriobactin conjugated to either ovalbumin or bovine serum albumin induced the production of immunoglobulin G (IgG) that binds to vibriobactin in immunized mice [37]. However, whether this response conferred protection during V cholerae infection was not reported.
Recently, 2 independent studies using murine models of infection showed that immunization with siderophores conjugated to carrier proteins conferred protection in response to infections caused by enteric pathogens [38, 39]. In one study, immunization of mice with the siderophores yersiniabactin and aerobactin, each conjugated to bovine serum albumin, conferred protection against uropathogenic E coli [38]. In the other study, immunization of mice with enterobactin conjugated to the cholera toxin B subunit resulted in the production of mucosal immunoglobulin A (IgA) that binds enterobactin and salmochelin and reduced intestinal colonization and systemic dissemination of S Typhimurium [39]. Moreover, analysis of the gut microbiota revealed that reduction of S Typhimurium colonization in the inflamed gut was accompanied by expansion of Lactobacillus spp, beneficial commensal organisms that thrive in similar locales as Enterobacteriaceae [39].
OUTLOOK
Siderophores have captivated interest at the interface of basic science and medicine for decades, resulting in consideration of these molecules and their derivatives for translational applications that include the prevention and treatment of microbial infections. The examples highlighted in this review illustrate recent advances in this field and provide a foundation for further investigations at the interface of chemistry, biology, and medicine that address siderophore-based strategies to target bacterial pathogens. We believe that targeting siderophores and siderophore acquisition systems will provide nontraditional, narrow-spectrum approaches for preventing and treating microbial infections. For these approaches to be successful, a fundamental understanding of the chemistry and biology of the siderophore system being targeted is necessary. In particular, increasing our knowledge of siderophore-mediated iron acquisition in vivo, including the production and uptake of siderophores by commensals and pathogens in different niches, is an important area for future research.
Along these lines, commensal bacteria also require iron, and 2 recent reports show that enterobactin utilization is more widespread than initially appreciated [40, 41]. Rothia mucilaginosa, a commensal bacterium of the oral cavity that can become pathogenic, biosynthesizes enterobactin [40]. Bacteroides thetaiotaomicron, a gut commensal, lacks the biosynthetic machinery for enterobactin and salmochelin, but it expresses a receptor that can import these siderophores when they are produced by neighboring microbes, which enables the bacterium to colonize the inflamed gut [41]. Further studies, including those that leverage metagenomics and new genetic tools to manipulate commensal microbes, are needed to assess how widespread this phenomenon is across the microbiota. In addition, it will be informative to evaluate whether such commensal organisms are susceptible to natural and synthetic SACs and to siderophore-based immunization.
Despite the promise of siderophore-based antimicrobial approaches, some concerns have been raised over the years. First, many pathogens produce and utilize multiple different siderophores in the host. For example, uropathogenic E coli CFT073 produces enterobactin, salmochelin, yersiniabactin, and aerobactin. It is thus possible that such a pathogen could evolve to evade siderophore-mediated antibiotic delivery by particular SACs (eg, enterobactin-antibiotic conjugates) through decreasing the expression of the targeted biosynthetic pathways and uptake receptors (eg, enterobactin-related machinery), while also increasing the expression of other iron-uptake mechanisms. To counter such a scenario, a combination of antibiotics conjugated to different siderophores could be used. Likewise, from the standpoint of developing effective immunization strategies, the optimal approach to prevent infection with such a pathogen requires immunization studies that evaluate combinations of different siderophores conjugated to immunogenic carrier proteins. Because siderophores have complex structures that are assembled by multistep biosynthetic pathways, it is difficult for a mutation to result in the production of a new siderophore. Although pathogens can acquire new siderophore gene clusters via horizontal gene transfer, a limited repertoire of these molecules have been identified in Gram-negative bacteria that cause human disease. Looking beyond siderophore uptake systems, efforts to elucidate mechanisms of siderophore biosynthesis and excretion in diverse bacterial pathogens may uncover new targets for the development of narrow-spectrum antibiotics.
CONCLUSIONS
Many exciting fundamental and translational lines of research exist for harnessing the iron-uptake machinery of Enterobacteriaceae to achieve new antibacterial strategies. Moving forward, these efforts may be broadly applicable to combat infections caused by other bacterial and fungal pathogens.
Notes
Acknowledgments. The figure was created with BioRender.com. We thank Sean-Paul Nuccio for critical reading of the manuscript.
Financial support. M. R. and E. M. N. siderophore immunization work is funded by Public Health Service Grant AI114625. M. R. is also funded by Public Health Service Grants AI126277 and AI145325, by the Chiba University-UCSD Center for Mucosal Immunology, Allergy, and Vaccines, and by an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. E. M. N. siderophore conjugate work is funded by a Professor Amar G. Bose Research Grant. E. M. N. microcin work is funded by a Pilot Project Grant from the MIT Center for Environmental Health Sciences (NIH P30-ES002109). R. R. G. is partly supported by a fellowship from the Crohn’s and Colitis Foundation.
Supplement sponsorship. This work is part of a supplement sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health (NIH) and the Centers for Disease Control and Prevention (CDC).
Potential conflicts of interest. M. R. and E. M. N. hold patents related to this work. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010; 90:859–904. [DOI] [PubMed] [Google Scholar]
- 2.Winter SE, Bäumler AJ. Why related bacterial species bloom simultaneously in the gut: principles underlying the ‘Like will to like’ concept. Cell Microbiol 2014; 16:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blount ZD. The unexhausted potential of E. coli. eLife 2015; 4:e05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol 2004; 2:123–40. [DOI] [PubMed] [Google Scholar]
- 5.Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis 2001; 32:263–9. [DOI] [PubMed] [Google Scholar]
- 6.Sassone-Corsi M, Nuccio SP, Liu H, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016; 540:280–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winter SE, Winter MG, Xavier MN, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013; 339:708–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol 2008; 16:107–14. [DOI] [PubMed] [Google Scholar]
- 9.Bennett JE, Dolin R, Blaser MJ. Douglas and Bennett’s Principles and Practice of Infectious Diseases. 8th Ed. Philadelphia, PA: Elsevier; 2014. [Google Scholar]
- 10.Reardon S. Antibiotic resistance sweeping developing world. Nature 2014; 509:141–2. [DOI] [PubMed] [Google Scholar]
- 11.Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nat Rev Gastroenterol Hepatol 2016; 13:206–16. [DOI] [PubMed] [Google Scholar]
- 12.Masson PL, Heremans JF, Schonne E. Lactoferrin, an iron-binding protein in neutrophilic leukocytes. J Exp Med 1969; 130:643–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol 2006; 2:132–8. [DOI] [PubMed] [Google Scholar]
- 14.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe 2013; 13:509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg ED. Nutritional immunity. Host’s attempt to withold iron from microbial invaders. JAMA 1975; 231:39–41. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths E. Iron in biological systems. In: Bullen J, Griffiths E (eds). Iron and Infection—Molecular, Physiological and Clinical Aspects. Chichester, UK: Wiley-Interscience; 1999: pp 1–25. [Google Scholar]
- 17.O’Brien IG, Gibson F. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim Biophys Acta 1970; 215:393–402. [DOI] [PubMed] [Google Scholar]
- 18.Flo TH, Smith KD, Sato S, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004; 432:917–21. [DOI] [PubMed] [Google Scholar]
- 19.Stubbendieck RM, Straight PD. Multifaceted interfaces of bacterial competition. J Bacteriol 2016; 198:2145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deriu E, Liu JZ, Pezeshki M, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 2013; 14:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wencewicz TA, Miller MJ. Sideromycins as pathogen-targeted antibiotics. Topics in Medicinal Chemistry. New York, NY: Springer-Verlag; 2018: pp 151–83. [Google Scholar]
- 22.Thomas X, Destoumieux-Garzón D, Peduzzi J, et al. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J Biol Chem 2004; 279:28233–42. [DOI] [PubMed] [Google Scholar]
- 23.Baquero F, Moreno F. The microcins. FEMS Microbiol Lett 1984; 23:117–124. [Google Scholar]
- 24.Vassiliadis G, Destoumieux-Garzón D, Lombard C, Rebuffat S, Peduzzi J. Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob Agents Chemother 2010; 54:288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MJ, McKee JA, Minnick AA, Dolence EK. The design, synthesis and study of siderophore-antibiotic conjugates. Siderophore mediated drug transport. Biol Met 1991; 4:62–9. [DOI] [PubMed] [Google Scholar]
- 26.Zheng T, Bullock JL, Nolan EM. Siderophore-mediated cargo delivery to the cytoplasm of Escherichia coli and Pseudomonas aeruginosa: syntheses of monofunctionalized enterobactin scaffolds and evaluation of enterobactin-cargo conjugate uptake. J Am Chem Soc 2012; 134:18388–400. [DOI] [PubMed] [Google Scholar]
- 27.Zheng T, Nolan EM. Enterobactin-mediated delivery of β-lactam antibiotics enhances antibacterial activity against pathogenic Escherichia coli. J Am Chem Soc 2014; 136:9677–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chairatana P, Zheng T, Nolan EM. Targeting virulence: salmochelin modification tunes the antibacterial activity spectrum of β-lactams for pathogen-selective killing of Escherichia coli. Chem Sci 2015; 6:4458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann W, Sassone-Corsi M, Raffatellu M, Nolan EM. Esterase-catalyzed siderophore hydrolysis activates an enterobactin-ciprofloxacin conjugate and confers targeted antibacterial activity. J Am Chem Soc 2018; 140:5193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji C, Miller PA, Miller MJ. Iron transport-mediated drug delivery: practical syntheses and in vitro antibacterial studies of tris-catecholate siderophore-aminopenicillin conjugates reveals selectively potent antipseudomonal activity. J Am Chem Soc 2012; 134:9898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller MJ, Walz AJ, Zhu H, et al. Design, synthesis, and study of a mycobactin-artemisinin conjugate that has selective and potent activity against tuberculosis and malaria. J Am Chem Soc 2011; 133:2076–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jean SS, Hsueh SC, Lee WS, Hsueh PR. Cefiderocol: a promising antibiotic against multidrug-resistant gram-negative bacteria. Expert Rev Anti Infect Ther 2019; 17:307–9. [DOI] [PubMed] [Google Scholar]
- 33.Shields RK. Case commentary: the need for cefiderocol is clear, but are the supporting clinical data? Antimicrob Agents Chemother. 2020; 64: e00059-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brumbaugh AR, Smith SN, Mobley HL. Immunization with the yersiniabactin receptor, FyuA, protects against pyelonephritis in a murine model of urinary tract infection. Infect Immun 2013; 81:3309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alteri CJ, Hagan EC, Sivick KE, Smith SN, Mobley HL. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog 2009; 5:e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forsyth VS, Himpsl SD, Smith SN, et al. Optimization of an experimental vaccine to prevent Escherichia coli urinary tract infection. mBio 2020; 11:e00555-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergeron RJ, Bharti N, Singh S, McManis JS, Wiegand J, Green LG. Vibriobactin antibodies: a vaccine strategy. J Med Chem 2009; 52:3801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mike LA, Smith SN, Sumner CA, Eaton KA, Mobley HL. Siderophore vaccine conjugates protect against uropathogenic Escherichia coli urinary tract infection. Proc Natl Acad Sci U S A 2016; 113:13468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassone-Corsi M, Chairatana P, Zheng T, et al. Siderophore-based immunization strategy to inhibit growth of enteric pathogens. Proc Natl Acad Sci U S A 2016; 113:13462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uranga C, Arroyo P, Duggan BM, Gerwick WH, Edlund A. Commensal oral Rothia mucilaginosa produces enterobactin – a metal chelating siderophore. mSystems 2020; 5:e00161-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu W, Winter MG, Spiga L, et al. Xenosiderophore utilization promotes Bacteroides thetaiotaomicron resilience during colitis. Cell Host Microbe 2020; 27:376–88.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]