Abstract
Objectives
SF3B1 mutations are the most common mutations in myelodysplastic syndromes (MDS). The International Working Group for the Prognosis of MDS (IWG-PM) recently proposed SF3B1-mutant MDS (SF3B1-mut-MDS) as a distinct disease subtype. We evaluated the spectrum and molecular landscape of SF3B1-mutated myeloid disorders and assessed the prognostication in MDS harboring SF3B1 mutations (MDS-SF3B1).
Methods
Cases were selected by retrospective review. Clinical course and laboratory and clinical findings were collected by chart review. SF3B1-mut-MDS was classified following IWG-PM criteria.
Results
SF3B1 mutations were identified in 75 of 955 patients, encompassing a full spectrum of myeloid disorders. In MDS-SF3B1, Revised International Prognostic Scoring System (IPSS-R) score greater than 3 and transcription factor (TF) comutations were adverse prognostic markers by both univariate and multivariate analyses. We confirmed the favorable outcome of IWG-PM-defined SF3B1-mut-MDS. Interestingly, it did not show sharp prognostic differentiation within MDS-SF3B1.
Conclusions
SF3B1 mutations occur in the full spectrum of myeloid disorders. We independently validated the favorable prognostication of IWG-PM-defined SF3B1-mut-MDS. However it may not provide sharp prognostication within MDS-SF3B1 where IPSS-R and TF comutations were prognostic-informative. Larger cohort studies are warranted to verify these findings and refine MDS-SF3B1 prognostication.
Keywords: SF3B1, Next-generation sequencing, Myeloid disorders, Transcription factor, Myelodysplastic syndrome
Key Points.
SF3B1 mutations occur in the full spectrum of myeloid disorders, ranging from clonal cytopenia with undetermined significance to acute myeloid leukemia.
Revised International Prognostic Scoring System score greater than 3 and presence of transcription factor comutations are adverse prognostic markers in myelodysplastic syndromes (MDS) harboring SF3B1 mutations (MDS-SF3B1).
We validated the favorable clinical outcome of SF3B1-mutant MDS as defined by the International Working Group for the Prognosis of MDS; however, this designation, by its proposed exclusion criteria, may not provide sharp prognostication within MDS-SF3B1.
The messenger RNA (mRNA) spliceosome is composed of a complex of small nuclear ribonucleoproteins (snRNP) U1, U2, U4, U5, and U6 and plays critical roles in regulating mRNA splicing and subsequent protein expression. It is assembled through binding of U1 snRNP to the 5′ splice site of the pre-mRNA, recruitment of U2snRNP to the 3′ splicing site at the intron–exon junction guided by the branch region sequence, and joining of the tri-snRNP U4/U5/U6.1-3 SF3B1, an essential protein in the U2snRNP complex, facilitates U2snRNP binding at the branch point near the 3′ splicing site. The critical role of SF3B1 and other spliceosome components (SRSF2, U2AF1, and ZRSR2) in hematopoiesis was initially recognized from the discovery of their frequent mutations in myelodysplasia by whole-exome sequencing studies.4-6 SF3B1 mutants were shown to alter U2snRNP function by promoting alternative branch-point usage and induction of cryptic 3′ splice site selection, thereby generating aberrantly spliced mRNA transcripts subject to nonsense-mediated decay and downregulation of target transcripts and protein expression.7,8 Morphologically, mutations in SF3B1 show a strong association with ring sideroblasts (RS) in myelodysplastic syndromes (MDS) with RS (MDS-RS) and MDS/myeloproliferative neoplasm with RS and thrombocytosis (MDS/MPN-RS-T).5,6,9 In MDS harboring SF3B1 mutations (MDS-SF3B1), aberrant splicing was seen in genes involved in heme metabolism and iron homeostasis, such as the iron transporter ABCB7, whose downregulation led to impaired erythroid differentiation, accumulation of mitochondrial ferritin, and presentation of RS.10-16 Besides MDS-RS and MDS/MPN-RS-T, SF3B1 mutations have also been reported in other myeloid neoplasms such as MPN17 and acute myeloid leukemia (AML),18 as well as chronic lymphocytic leukemia4 and solid tumors,19-22 albeit at lower frequencies. However, the molecular landscape of the full spectrum of SF3B1-mutated myeloid disorders has not been thoroughly characterized because most published studies focus on one specific myeloid neoplasm (eg, MDS or MDS/MPN-RS-T).
Furthermore, the majority of studies, although not all, reported an independent association of SF3B1 mutations with favorable clinical outcomes in MDS.5,9,23-28 The International Working Group for the Prognosis of MDS (IWG-PM) recently proposed SF3B1-mutant MDS (SF3B1-mut-MDS) as a distinct disease subtype,29 based on cumulative data supporting somatic SF3B1 mutation as a disease-defining genetic abnormality, demonstrated by the following characteristics: (1) it commonly represents a founding genetic lesion, (2) it is a major determinant of disease phenotype characterized by erythroid dysplasia with RS and ineffective erythropoiesis, (3) it showed independent prognostic value on survival and risk of AML progression in the majority of studies, and (4) it may predict response to specific agents such as transforming growth factor-β ligand trap luspatercept.5,9,23-28,30 The proposed SF3B1-mut-MDS diagnostic criteria included (1) cytopenia by standard hematologic values; (2) somatic SF3B1 mutation; (3) morphologic dysplasia with or without RS; (4) bone marrow blasts less than 5% and peripheral blood blasts less than 1%; and (5) not meeting World Health Organization (WHO) criteria for MDS with isolated del(5q), MDS/MPN, and MPN. Additional exclusion criteria included adverse genetic markers of monosomy 7, inv(3) or abnormalities of chromosome 3q26, complex karyotype (3 or more alterations), and comutation in RUNX1 and/or EZH2. Previous studies in MDS have also described mutations in FLT3, NPM1, NRAS, PTPN11, WT1, IDH1, and IDH2 as type 1 mutations, given their association with AML progression, and mutations in RUNX1, GATA2, ZRSR2, TP53, STAG2, ASXL1, KRAS, and TET2 as type 2 mutations with enrichment in high-grade MDS.31,32 Our study aimed to evaluate the spectrum of SF3B1 mutation-harboring myeloid disorders, to analyze their molecular landscape and associated clinical and laboratory findings, and to further explore the prognostication in MDS-SF3B1, including the newly proposed classification of SF3B1-mut-MDS by the IWG-PM and prognostic impacts of comutations.
Methods
Case Selection and Data Collection
Following Mayo Clinic institutional review board approval, we retrospectively screened Mayo Clinic cases that had a targeted 35-gene OncoHeme next-generation sequencing (NGS) panel performed for diagnosed or suspected hematologic neoplasms by December 2016. Cases that had SF3B1 mutations and bone marrow examination were selected. The bone marrow pathologic diagnoses were verified by at least 2 hematopathologists and were based on the 2016 revision of the WHO classification.33 Patients’ clinical courses and pertinent laboratory, pathologic, and clinical findings were collected by chart review.
Cytogenetic Analysis
Fresh bone marrow aspirate samples were cultured and harvested following standard cytogenetic methods, and chromosome preparations were stained using GTL banding with trypsin and Leishman stain. A total of 20 metaphases were analyzed and reviewed for each sample when available.
NGS Analysis
NGS testing was performed using a targeted, myeloid neoplasm–focused OncoHeme panel that interrogates 35 genes, including epigenetic modifiers ASXL1, DNMT3A, TET2, IDH1, IDH2, EZH2, and WT1; splicing factors SF3B1, SRSF2, U2AF1, and ZRSR2; transcription factors (TF) BCOR, CEBPA, ETV6, GATA1, GATA2, NOTCH1, and RUNX1; signaling and kinase factors BRAF, CALR, CBL, CSF3R, FLT3, JAK2, KIT, KRAS, MPL, MYD88, NRAS, and PTPN11; tumor suppressors TP53 and PHF6; molecular chaperone NPM1; telomerase reverse transcriptase TERT; and the functionally poorly defined SETBP1. DNA was extracted from bone marrow and peripheral blood using Qiagen EZI. The 200-ng sheared DNA was target-enriched with a custom hybridization-capture reagent (SureSelectXT; Agilent) and sequenced on the MiSeq or HiSeq platforms (Illumina) at the Mayo Clinic Clinical Genome Sequencing Laboratory. The read length was 151 bp and 101 bp on the MiSeq and HiSeq platforms, respectively. Sequencing data were processed through a custom bioinformatics pipeline, Mayo NGS Workbench, using CLC Bio Genomics Server v6.0 (Qiagen) for alignment and variant calling. The aligned BAM files were further processed through an in-house developed breakpointSearch tool for large insertion and deletion detection. BAM files of all variant calls were manually reviewed in the genome browser Alamut Visual (Interactive Biosoftware) for confirmation. The limit of detection of the NGS assay is 5% with a minimum 250× depth of coverage. More than 95% of tested regions had greater than 1,000× depth of coverage in the clinical assay. Genetic variants were curated and annotated in the molecular hematopathology laboratory following the American College of Medical Genetics and Genomics 5-tier system.34
Statistical Analysis
Numerical variables were presented by median and range; categorical variables were described by count and percentage in each category. The correlation between SF3B1 variant allele fraction (VAF) percentage and RS percentage were evaluated using the Pearson correlation test. We used the Wilcoxon rank sum or Mann-Whitney U test to compare the median hemoglobin level among different SF3B1 mutations and other numerical and ordinal variables. The Fisher exact test was performed to determine the significance of associations between categorical variables. Overall survival (OS) was defined as the time from the date of diagnosis to the date of death (for patients who were deceased) or last follow-up (for patients who were censored). Progression-free survival (PFS) was defined as the time from the date of diagnosis to the date of leukemic transformation (for patients transformed to AML) or last follow-up (for censored patients). The Kaplan-Meier survival curve and the Cox proportional hazards regression model (log-rank test, both univariate and multivariate analysis) were used to identify the potential impact of different variables on OS and PFS. Statistical significance was based on P < .05. Statistical analyses for this study were performed by using JMP Pro software version 14 (SAS Institute).
Results
SF3B1 Mutation Distribution in Myeloid Disorders
Of the 955 patients who had undergone NGS testing for diagnosed or suspected hematologic neoplasms, we identified 75 patients (8%) harboring an SF3B1 mutation. The mutations were all missense mutations, and the most common ones were K700E (n = 38, 51%), K666R/T/N/Q (n = 15, 20%), H662D/Q/Y (n = 6, 8%), and R625C (n = 5, 7%). Other less frequent mutations included E622D, Y623C, Q659R, K741N, G742D, and D781G. Patients’ demographics and clinical and laboratory findings are summarized in Table 1. SF3B1 mutations were detected across the full spectrum of myeloid disorders and comprised 7% (3/42) of clonal cytopenia of uncertain significance (CCUS), 19% (40/206) of MDS, 14% (12/89) of MDS/MPN, 4% (8/202) of MPN, 5% (11/209) of AML, and 11% (1/9) of systemic mastocytosis (SM) cases in the entire study cohort of 955 cases. SF3B1 mutations were not detected in 121 cytopenia or cytosis cases without evidence of myeloid disorders, 16 cases of aplastic anemia or paroxysmal nocturnal hematuria, or other miscellaneous cases including 18 with low-grade B-cell lymphoproliferative disorder, 14 with plasma cell proliferative disorder, and 10 with B-lymphoblastic lymphoma/leukemia. The disease distribution within the 75 SF3B1 mutation-positive cases was as follows: 4% CCUS (3/75), 53% MDS (40/75), 16% MDS/MPN (12/75), 11% MPN (8/75), 15% AML (11/75), and 1% SM (1%, 1/75) (Table 1).
Table 1.
Demographic, Clinical, Pathologic, and Genetic Characteristics of SF3B1-Mutated Myeloid Disorders
| Total | CCUS | MDS | MDS/MPN | MPN | AML | SM | |
|---|---|---|---|---|---|---|---|
| Patients, No. (%) | 75 | 3 (4) | 40 (53) | 12 (16) | 8 (11) | 11(15) | 1 (1) |
| Female sex, No. | 25 | 0 | 15 | 3 | 2 | 4 | 1 |
| Male sex, No. | 50 | 3 | 25 | 9 | 6 | 7 | 0 |
| Age, median (range), y | 73 (35-89) | 73 (72-76) | 74 (47-85) | 73 (53-89) | 71 (57-80) | 69 (35-85) | 68 |
| Progression to AML, No. | 5 | 0 | 5 | 0 | 0 | NA | 0 |
| Death, No. | 25 | 1 | 11 | 5 | 2 | 6 | 0 |
| Hemoglobin, median (range), g/dL | NA | 9.3 (7.5-9.5) | 8.7 (6.0-12.6) | 9.0 (6.1-11.4) | 9.7 (7.1-17.6) | 8.1 (6.5-11.6) | 11.8 |
| RDW, median (range), % | NA | 15.6 (14.7-16.9) | 18.4 (14.1-23.4) | 22.6 (16.9-22.8) | 19.9 (17.2-25.6) | 19.3 (15.7-26.9) | 21.3 |
| WBC, median (range), ×109/L | NA | 3.5 (3.0-33.7) | 4.5 (1.2-103.2) | 7.1 (2.5-23.0) | 10.8 (3.5-41.7) | 2.7 (0.2-27.5) | 11.2 |
| Platelet count, median (range), ×109/L | NA | 63 (38-73) | 199 (18-632) | 435 (156-791) | 191 (32-664) | 79 (21-274) | 32 |
| Ring sideroblasts, median (range), % | NA | 0 (0) | 16 (0-90) | 50 (15-75) | 0 (0) | 15 (0-30) | 0 |
| SF3B1 VAF, median (range), % | NA | 26 (6-27) | 38 (6-48) | 36 (13-48) | 34 (6-48) | 35 (14-46) | 5 |
| SF3B1 mutation position | |||||||
| p.E622 | 1 | 1 | |||||
| p.Y623 | 1 | 1 | 1 | ||||
| p.R625 | 5 | 4 | 1 | ||||
| p.N626 | 1 | 1 | |||||
| p.Q659 | 1 | 1 | |||||
| p.H662 | 6 | 1 | 2 | 2 | 3 | ||
| p.K666 | 15 | 6 | 4 | 3 | 1 | 1 | |
| p.K700 | 38 | 1 | 23 | 6 | 3 | 5 | |
| p.K741 | 1 | 1 | |||||
| p.G742 | 1 | 1 | |||||
| p.D781 | 1 | 1 | 1 | ||||
| No. of comutations, median (range) | NA | 0 (0-1) | 1 (0-5) | 1 (0-3) | 3 (1-5) | 2 (1-5) | 0 |
| Abnormal karyotype, No. | 27 | 0 | 8 | 6 | 5 | 8 | 0 |
AML, acute myeloid leukemia; CCUS, clonal cytopenia with undetermined significance; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; NA, not applicable; RDW, red cell distribution of width; SM, systemic mastocytosis; VAF, variant allele frequency.
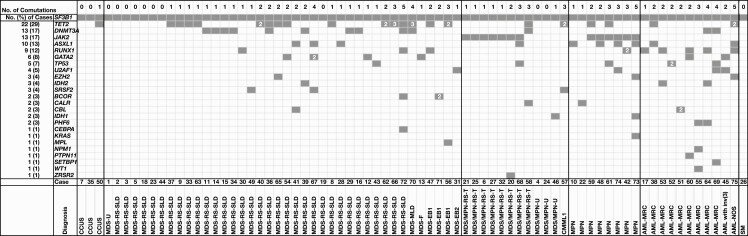
We then focused our study on the 75 SF3B1-mutated cases of myeloid disorders Figure 1. Of the 40 cases with MDS-SF3B1, 83% (33/40) were MDS-RS, including 23 MDS-RS–single lineage dysplasia (SLD) and 10 MDS-RS–multilineage dysplasia (MLD). Only 10% (4/40) were high-grade MDS with excess blasts (MDS-EB) including 3 MDS-EB1 and 1 MDS-EB2. The other 3 MDS cases were MDS-MLD, MDS with fibrosis, and MDS unclassifiable. Of the 12 patients with SF3B1-mut-MDS/MPN, 67% (8/12) had MDS/MPN-RS-T, besides 1 with chronic myelomonocytic leukemia and 3 with MDS/MPN unclassifiable. The 8 cases of MPN included 1 primary myelofibrosis, 1 polycythemia vera (PV), 1 post-PV myelofibrosis, 3 post–essential thrombocythemia myelofibrosis, and 2 MPN unclassifiable. Of the 11 patients with AML, 82% (9/11) had AML with myelodysplasia-related changes (AML-MRC), of whom 6 had a prior history of MDS. The remaining 2 had AML with inv(3)(q21q26.2) and AML not otherwise specified. The SM case was mast cell leukemia.
Figure 1.
Summary of molecular landscape and pathology diagnoses in 75 patients with SF3B1-mutated myeloid disorders. The information of each patient is present in a column. Black boxes indicate the presence of mutations. The numbers in the box indicate the number of mutations of the corresponding gene. AML, acute myeloid leukemia; AML-MRC, acute myeloid leukemia with myelodysplasia-related changes; AML-NOS, acute myeloid leukemia not otherwise specified; CCUS, clonal cytopenia with undetermined significance; CMML1, chronic myelomonocytic leukemia 1; MDS-EB, myelodysplastic syndrome with excess blasts; MDS-F, myelodysplastic syndrome with fibrosis; MDS-MPN-RS-T, myelodysplastic/myeloproliferative neoplasm with ring sideroblasts and thrombocytosis; MDS/MPN-U, myelodysplastic/myeloproliferative neoplasm, unclassifiable; MDS-RS-MLD, myelodysplastic syndrome with ring sideroblasts and multilineage dysplasia; MDS-RS-SLD, myelodysplastic syndrome with ring sideroblasts and single lineage dysplasia; MDS-U, myelodysplastic syndrome, unclassifiable; MPN, myeloproliferative neoplasm; SM, systemic mastocytosis.
SF3B1 Mutations and Association With Laboratory Findings
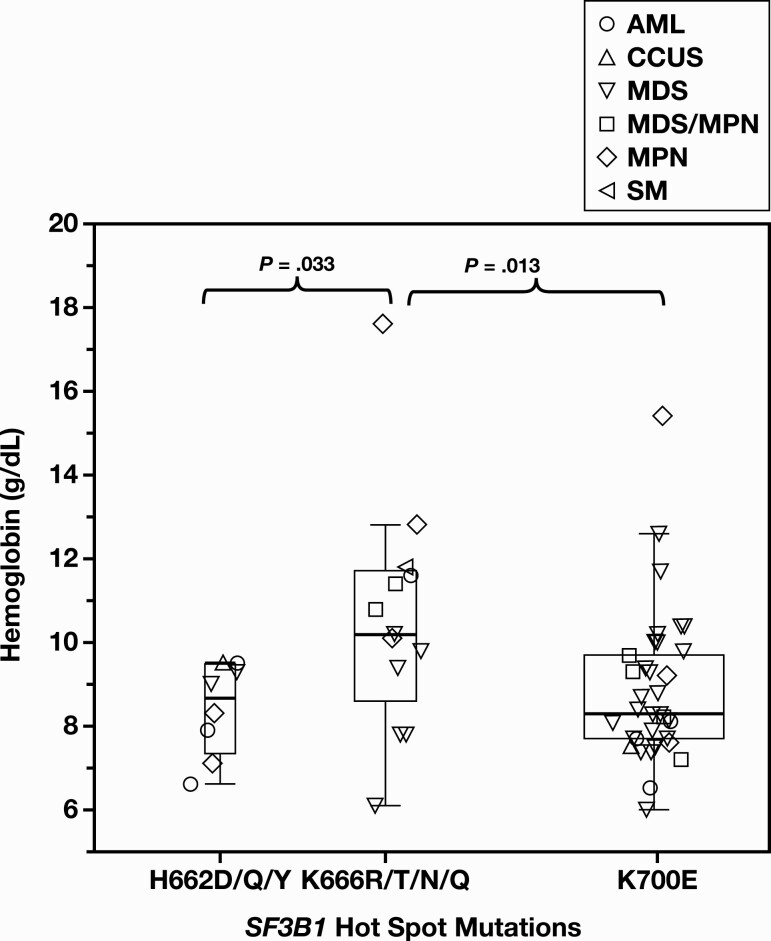
The VAF of SF3B1 mutations showed no statistically significant differences among various myeloid disorders (P > .05). It also did not demonstrate statistically significant associations with patient age, sex, hemoglobin level, RBC distribution width, neutrophil count, platelet count, number of comutations, or cytogenetic abnormalities. The distributions of SF3B1 mutations were similar among various myeloid disorders, with K700, K666, and H662 consistently being the mutational hotspots (Table 1). Interestingly, among the 3 hotspots, cases harboring K666 (R/T/N/Q) mutations exhibited higher hemoglobin levels than those seen in K700 (P = .13) or H662 (P = .033) mutations Figure 2.
Figure 2.
Association between SF3B1 mutational hotspots and hemoglobin levels. A whisker plot of the hemoglobin levels of 3 SF3B1 mutational hotspots K700, K666, and H662, showing range, interquartile range, and median of the hemoglobin levels. Different label shapes mark the specific disease categories of the cases.
Of 59 cases with adequate bone marrow aspirate that had Prussian blue iron stain performed, 55 (93%) showed the presence of RS, seen in 0 of 1 CCUS, 36 of 38 MDS, 12 of 12 MDS/MPN, 1 of 2 MPN, and 6 of 6 AML cases. The 36 MDS cases comprised 23 MDS-RS-SLD and 10 MDS-RS-MLD with 5% to 90% RS and 3 MDS-EB1 with 4% to 8% RS. All 12 MDS/MPN cases (including 8 MDS/MPN-RS-T) had 15% to 75% RS. All 6 AML cases with RS (range, 5%-30%) were AML-MRC, and 5 had a prior history of MDS. Four cases lacked RS (Supplementary Table 1; all supplemental material can be found at American Journal of Clinical Pathology online), including one with CCUS (case 35), one with MDS-MLD (case 70), one with MDS-EB2 (case 31), and one with MPN (PV, case 59), with the PV case showing absence of storage iron. No statistically significant numerical correlation between SF3B1 mutational VAF percentage and RS percentage was observed (Pearson correlation coefficient, 0.25; P = .09). In addition, no statistically significant association between RS percentage and Revised International Prognostic Scoring System (IPSS-R) risk categories was observed (median [range]: 10% [0%-30%] vs 17% [0%-90%]; P = .09, IPSS-R greater than 3 vs 3 or lower).
Molecular Landscape of Comutations in SF3B1-Mutated Myeloid Disorders
A total of 123 comutations were detected in 26 genes in 62 of 75 SF3B1-mutated cases of myeloid disorders, involving TET2, ASXL1, DNMT3A, EZH2, IDH1, and WT1 in epigenetic modifiers; RUNX1, GATA2, BCOR, and CEBPA in TFs; JAK2, CBL, CALR, KIT, KRAS, MPL, and PTPN11 in signaling and kinase factors; U2AF1, SRSF2, and ZRSR2 in splicing factors; and TP53 and PHF6 in tumor suppressors, in addition to NPM1 and SETBP1 (Figure 1). The most frequent comutations occurred in TET2 (22/75, 29%), DNMT3A (13/75, 17%), JAK2 (13/75, 17%), and ASXL1 (10/75, 13%). Mutations in these 4 genes accounted for 51% of the total number of comutations. Two or more TET2 mutations occurred in 7 of 22 cases. Although the JAK2 mutation was observed exclusively in MPN and MDS/MPN-RS-T, TET2, ASXL1, and DNMT3A mutations were present across different categories of myeloid disorders (Figure 1).
In terms of disease, only 1 of 3 CCUS cases showed one comutation in TET2. Of the 40 MDS-SF3B1 cases, comutations were seen in TET2 (n = 16); DNMT3A (n = 10); GATA2 (n = 4); ASXL1 (n = 3); RUNX1 (n = 3); BCOR (n = 2); and EZH2, TP53, U2AF1, IDH2, CBL, CEBPA, SRSF2, MPL, and TERT (n = 1 for each), in descending order of frequency. As for the 11 AML cases, comutations were detected in RUNX1 (n = 5); TET2 (n = 2); DNMT3A (n = 2); ASXL1 (n = 2); IDH2 (n = 2); GATA2 (n = 2); TP53 (n = 2); PHF6 (n = 2); U2AF1 (n = 2); and EZH2, CBL, NPM1, PTPN11, SETBP1, and WT1 (n = 1 for each). Type 1 mutations31 associated with AML progression were seen in IDH2, NPM1, PTPN11, and WT1 and were present mainly in AML rather than MDS cases (5/11 vs 1/40; P = .001). In the 12 MDS/MPN cases, comutations were present in JAK2 (n = 7); TET2 (n = 2); and CALR, ASXL1, TP53, IDH1, SRSF2, and ZRSR2 (n = 1 for each). Among the 8 MPN cases, comutations occurred in JAK2 (n = 6); ASXL1 (n = 4); TET2 (n = 2); and CALR, RUNX1, EZH2, TP53, U2AF1, CEBPA, IDH1, and KRAS (n = 1 for each). The number of comutations in the 75 SF3B1-mutated cases of myeloid disorders ranged from 0 to 5 with a median (range) of 0 (0-1) in CCUS, 1 (0-5) in MDS, 1 (0-3) in MDS/MPN, 3 (1-5) in MPN, and 2 (1-5) in AML (Figure 1). The numbers of comutations in MDS were lower than those in MPN (P = .015) and AML (P = .004). The MDS/MPN cases also had fewer comutations than those in AML (P = .037). Twenty-seven of the 75 SF3B1-mutated cases of myeloid disorders had chromosome abnormalities and did not show association with the numbers of comutations (P = .65) (Supplementary Table 1).
Prognostic Factors in MDS-SF3B1
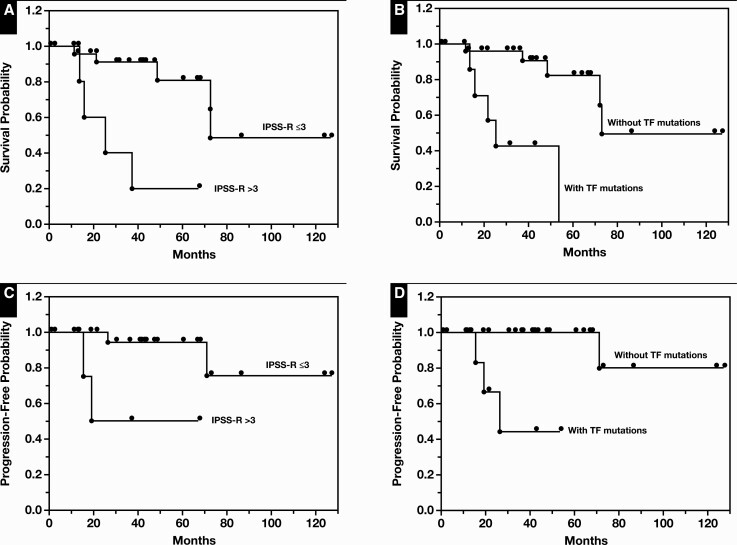
Among the 40 MDS-SF3B1 cases, 36 had available cytogenetics data. Their median (range) IPSS-R score was 2.3 (1-7). With a cutoff score of 3, the group with IPSS-R score greater than 3 (intermediate, high, or very high IPSS-R risk category, n = 8) showed an adverse outcome with a median OS of 25 months, in contrast to 73 months observed in the group with a score of 3 or lower (very low or low IPSS-R risk category, n = 28; P = .002) Figure 3A. In addition, during a median (range) follow-up time of 37 months (0.3-127 months), 4 of 40 patients with MDS-SF3B1 progressed to AML, including 1 with MDS-EB1, 1 with MDS with fibrosis, and 2 with MDS-RS-SLD Table 2. Median PFS for the 2 groups was 20 months vs not reached, respectively (P = .006, IPSS-R score greater than 3 vs 3 or lower) Figure 3C.
Figure 3.
Clinical impact of Revised International Prognostic Scoring System (IRSS-R) and presence of transcription factor (TF) comutations on overall survival (OS) and progression-free survival (PFS) in myelodysplastic syndrome (MDS) harboring SF3B1 mutations (MDS-SF3B1). A, OS in patients with IPSS-R scores greater than 3 vs 3 or lower (P = .002). B, OS in patients with TF comutations vs without TF mutations (P = .0003). C, PFS in patients with IPSS-R scores greater than 3 vs 3 or lower (P = .006). D, PFS in patients with TF comutations vs without TF mutations (P = .0001).
Table 2.
Genetic Findings in Patients With Myelodysplastic Syndrome Harboring SF3B1 Mutations With TF Comutations and/or Progression to AML
| Case No. | Age, y/Sex | Diagnosis | Chromosome Results | NGS Results | Time to AML, mo |
|---|---|---|---|---|---|
| 47 | 76/Ma | MDS-EB1 | 46,XY[20] | RUNX1:p.R174Qln (37%) | 26.4 |
| SF3B1: p.K666N (42%) | |||||
| TET2: p.Q1020a (41%) | |||||
| 13 | 79/Ma | MDS-F | 46,Y,t(X;8)(q22;q24.1)[12]/46,XY[8] | GATA2: p.S251Cfsa29 (38%) | 15.6 |
| SF3B1:p.K700E (46%) | |||||
| 54 | 76/Fa | MDS-RS-SLD | 46,XX,-2,-20,+2mar[2]/46,XX[18] | GATA2:p.L321F (6%) | 19.2 |
| SF3B1:p.K700E (10%) | |||||
| TET2: p.P1575Qfsa21 (10%) | |||||
| 67 | 80/Fa | MDS-RS-SLD | 46,XX[20] | ASXL1: p.G646Wfsa12 (34%) | NA |
| GATA2:p.A234Gfsa45 (32%) | |||||
| GATA2: p.T387_M388del (34%) | |||||
| SF3B1: p.K666N (48%) | |||||
| SRSF2: p.P95A (46%) | |||||
| 12 | 59/Ma | MDS-RS-MLD | 46,XY,+1,der(1;7)(q10;p10)[6]/47,idem,+8[14] | GATA2: p.K389_Q394del (23%) | NA |
| SF3B1: p.K700E (28%) | |||||
| 30 | 73/Ma | MDS-RS-SLD | ND | RUNX1:p.G172Q (40%)SF3B1: p.H662Q (41%) | NA |
| 72 | 77/Fa | MDS-RS-MLD | 46,XX,del(11)(q13q23)[3]/46,XX[17] | BCOR: p.N575Qfsa36 (34%) | NA |
| CEBPA: p.F31Gfsa72 (15%) | |||||
| DNMT3A: p.E774a (41%) | |||||
| RUNX1:p.R174Q (38%) | |||||
| SF3B1:p.K700E (39%) | |||||
| TET2: p.S657Hfsa43 (34%) | |||||
| 71 | 77/Ma | MDS-EB1 | 46,XY[20] | BCOR: p.Q1653Kfsa21 (41%) | NA |
| BCOR: p.R1661a (5%) | |||||
| SF3B1: p.R625C (38%) | |||||
| 3 | 68/M | MDS-RS-SLD | 46,XY[20] | SF3B1: p.K700E (38%) | 47.5 |
AML, acute myeloid leukemia; MDS-EB1, myelodysplastic syndrome with excess blasts-1; MDS-F, myelodysplastic syndrome with fibrosis; MDS-RS-MLD, myelodysplastic syndrome with ring sideroblasts–multilineage dysplasia; MDS-RS-SLD, myelodysplastic syndrome with ring sideroblasts–single lineage dysplasia; NGS, next-generation sequencing; NA, not applicable; ND, not determined; TF, transcription factor.
aCases harboring TF mutations.
In terms of mutations, SF3B1 mutational VAF and number of comutations showed no effect on OS or PFS (data not shown; P > .05). The most commonly comutated genes by pathway involved epigenetic modifiers (28/40; TET2, DNMT3A, ASXL1, EZH2, and IDH2) and TF (8/40; RUNX1, GATA2, BCOR, and CEBPA [coexisting with RUNX1 and BCOR]) (Table 2 and Supplementary Table 1). Comutations in other pathways were infrequent, including 2 in signaling and kinase factors (CBL, MPL), 2 in splicing factors (U2AF1, SRSF2), and one in tumor suppressor (TP53) (Figure 1). Although the presence of comutations in epigenetic modifiers showed no impact on clinical outcomes (data not shown; P > .05), the presence of TF comutations demonstrated inferior OS (median, 25 vs 73 months; P = .0003) Figure 3B and PFS (median, 26 months vs not reached; P = .0001) Figure 3D in comparison to MDS-SF3B1 cases without TF comutations.
Because the IWG-PM recently proposed SF3B1-mut-MDS as a distinct disease subtype with specified inclusion and exclusion criteria,29 we evaluated the prognostic impact of this new entity following the proposed diagnostic criteria in our study cohort with SF3B1-mut MDS. Among the 40 cases of MDS-SF3B1, 27 fulfilled the diagnostic criteria for IWG-PM–defined SF3B1-mut-MDS (Supplementary Table 1). Ten were excluded (IWG-PM–excluded MDS-SF3B1) according to the IWG-PM exclusion criteria, based on the presence of complex karyotype (cases 12, 43, and 54), RUNX1 mutation (cases 30 and 72), EZH2 mutation (case 65), and MDS-EB (cases 31, 47, 56, and 71). Three cases could not be classified because of a lack of cytogenetic data and failure to meet any exclusion criteria. The IWG-PM–defined SF3B1-mut-MDS cases demonstrated favorable clinical outcomes with a median OS of 73 months and unreached median PFS, similar to the data reported from the IWG-PM study.29 Interestingly, the IWG-PM–defined SF3B1-mut-MDS cases did not show statistically significant differences from the IWG-PM–excluded MDS-SF3B1 cases in either OS (median, 73 vs 54 months; P = .22, IWG-PM defined vs excluded) or PFS (median, both not reached; P = .41, IWG-PM defined vs excluded). Given the observed adverse impact of TF comutations, we explored the possible added prognostic impact of TF comutations in this context. By adding the presence of TF comutation into the exclusion criteria (RUNX1 already in the IWG-PM exclusion criteria, GATA2 and BCOR comutations added), statistically significant superior OS was seen in the refined IWG-PM–defined SF3B1-mut-MDS group in comparison to the refined IWG-PM–excluded MDS-SF3B1 group (median, not reached vs 37 months; P = .02, n = 25 vs n = 12). PFS stayed not reached in both groups (P = .08).
To further define the prognostic contribution of various factors to OS in MDS-SF3B1, we performed univariate and multivariate analyses using a Cox proportional hazards regression model Table 3. Univariate analyses of hazard ratio (HR) for OS included age, sex, hemoglobin level, neutrophil count, platelet count, bone marrow blast count, RS percentage, cytogenetics risk, number of comutations, SF3B1 mutation VAF, epigenetic modifier comutation status, TF comutation status, IPSS-R score, and IWG-PM–defined SF3B1-mut-MDS status. The presence of TF comutations and IPSS-R score greater than 3 were shown to be poor prognostic markers, with HRs of 9.67 (P = .003) and 7.88 (P = .008), respectively. Neutrophil count and RS percentage showed a minimal impact (HR, 1.04 [P = .012] and 0.96 [P = .04], respectively). In the multivariate analysis, IPSS-R score greater than 3 retained its clinical validity, showing an HR of 5.12 (P = .049), and the presence of TF comutations demonstrated independent prognostic value even after adjustment for IPSS-R (HR, 9.14; P = .04).
Table 3.
Cox Proportional Hazards Ratio Analyses of Variables in Myelodysplastic Syndrome Harboring SF3B1 Mutations Prognostic of Overall Survival
| Risk Factor | Hazard Ratio | 95% CI | P |
|---|---|---|---|
| Univariate analysis | |||
| Age, <70 vs ≥70 y | 1.80 | 0.38-8.56 | 0.46 |
| Sex, M/F | 1.08 | 0.29-4.03 | 0.91 |
| Hemoglobin, <10 vs ≥10 g/dLa | 3.16 | 0.92-14.69 | 0.095 |
| Neutrophil count, <0.8 vs ≥0.8 × 109/La | 1.04 | 1.01-1.08 | 0.012 |
| Platelet count, <50 vs 50~100 vs ≥100 × 109/La | 3.75 | 0.45-19.34 | 0.19 |
| Bone marrow blast counta | 0.91 | 0.05-5.07 | 0.92 |
| Ring sideroblasts, % | 0.96 | 0.92-1.00 | 0.04 |
| Cytogenetic risks, high/very high vs intermediate/low/very lowa | 4.72 | 0.53-42.40 | 0.17 |
| Number of comutations | 1.26 | 0.74-1.98 | 0.79 |
| SF3B1 VAF. %b | 0.72 | 0.09-8.80 | 0.78 |
| Epigenetic comutationsc | 0.72 | 0.18-2.89 | 0.64 |
| TF comutationsc | 9.67 | 2.23-41.94 | 0.003 |
| IPSS-R score, >3 vs ≤3 | 7.88 | 1.72-35.33 | 0.008 |
| SF3B1-mut-MDS by IWG-PMd | 0.46 | 0.13-1.61 | 0.23 |
| Multivariate analysis | |||
| TF comutationsc | 9.14 | 1.1-76.1 | 0.04 |
| IPSS-R score, >3 vs ≤3 | 5.12 | 1.0-26.1 | 0.049 |
| SF3B1-mut-MDS by IWG-PMd | 1.43 | 0.3-6.76 | 0.64 |
IPSS-R, Revised International Prognostic Scoring System; IWG-PM, International Working Group for the Prognosis of MDS; SF3B1-mut-MDS, SF3B1-mutant myelodysplastic syndrome; TF, transcription factor; VAF, variant allele fraction.
aIPSS-R categories.
bRange hazard ratio.
cPresent vs absent.
dYes vs no.
Discussion
In this study, we evaluated the distribution of SF3B1 mutations in 955 patients who had undergone NGS testing for hematologic neoplasms. We identified 75 cases harboring an SF3B1 mutation, spanning the full pathologic spectrum of myeloid disorders from CCUS to AML. Consistent with previous reports, SF3B1 mutations were highly associated with MDS-RS and MDS/MPN-RS-T (55%, 41/75), supporting a causal relationship between SF3B1 mutations and RS formation.5 Among the 40 MDS-SF3B1 cases, 90% were low-grade disease; only 4 (10%) had the high-grade diagnosis of MDS-EB, concordant with previous reports.29,35 The frequencies of SF3B1 mutations observed in AML and MPN were also on par with previous reports,5 supporting the representative nature of our study cohort. It was striking to see that 9 of 11 AML cases were AML-MRC, with 6 having evolved from a precedent MDS. This observation echoes the findings of Lindsley et al,18 who showed that SF3B1 mutation along with several other mutations in AML were more than 95% specific for secondary AML and conferred a worse clinical outcome. The diagnoses of the 3 CCUS cases were established by the presence of SF3B1 mutations in the absence of cytogenetic or definitive morphologic abnormalities.36,37SF3B1 mutation has been shown to be one of the most frequent mutations in clonal hematopoiesis of indeterminate potential (CHIP), a premalignant condition that occurs in elderly individuals without hematologic malignancies and confers an increased risk of subsequent development of hematologic malignancies.38-41 The observed wide distribution of SF3B1 mutations across the full spectrum of myeloid disorders, along with their frequent occurrence in CHIP, underscores the biology that SF3B1 mutations occur early in MDS development as founder mutations.42-44
SF3B1 mutation hotspots involved K700E, K666, and H662, similar to previous reports.5,6,9,12,45 We did not see disparate hotspot patterns among the various myeloid disorders. Interestingly, for the first time, an association between K666 mutations and higher hemoglobin levels was seen, suggesting that these mutations may exert less disruptive effects on erythropoiesis. Although some studies showed similar aberrant splicing patterns among mutants of the 3 hotspots, a recent study described a distinct missplicing pattern in K666N.8,46 Further studies are required to confirm this finding and understand its clinical significance.
The molecular landscape of SF3B1-mutated myeloid disorders showed unique but overlapping features (Figure 1). Of 123 comutations, TET2, DNMT3A, JAK2, and ASXL1 accounted for 51% of total comutations. The occurrence of epigenetic modifier DNMT3A, TET2, and ASXL1 (DTA) mutations across the broad spectrum of myeloid disorders was not surprising because these mutations have also been shown to be founder mutations in the early development of myeloid neoplasms. Similarly, these genes are the most frequently mutated genes in CHIP.32,38-42,47 Persistent DTA mutations in AML minimal residual disease monitoring also failed to predict disease relapse.48 The enrichment of type 1 mutations in AML was in keeping with their reported association with AML progression.31,32JAK2 comutations unsurprisingly and exclusively occurred in MPN and MDS/MPN-RS-T.49-54 The lower numbers of comutations seen in MDS and MDS/MPN are likely attributable to the enrichment of lower grade diseases innately associated with SF3B1 mutations (MDS-RS and MDS/MPN-RS-T). The higher numbers of comutations seen in AML and MPN reflect clonal heterogeneity and complexity during disease evolution.
SF3B1 mutations are the most common mutations in MDS, as seen in approximately 20% to 25% of cases. Consequently, MDS-SF3B1 comprises the largest MDS subgroup by genetic abnormalities. We explored the prognostication in this MDS subgroup. Significantly inferior outcomes for OS and PFS were seen in MDS-SF3B1 with IPSS-R scores greater than 3 (intermediate, high, and very high risk) in comparison to IPSS-R scores of 3 or lower (low or very low risk); this finding is concordant with that reported in the IWG-PM cohort, for which the favorable prognostic value of SF3B1 mutation was maintained only in the low or very low risk categories of MDS-SF3B1.29 In addition, in our study, the presence of TF comutations was associated with inferior OS and PFS. The prognostic value of IPSS-R and TF comutations in MDS-SF3B1 was confirmed in both univariate and multivariate analyses, and TF comutations were shown to be independent adverse prognostic markers even after adjustment for IPSS-R.
IWG-PM recently proposed SF3B1-mut-MDS as a distinct disease subtype with specific inclusion and exclusion criteria, and not all MDS-SF3B1 cases qualify as SF3B1-mut-MDS. In our independent cohort of MDS-SF3B1, we confirmed the favorable outcome of IWG-PM–defined SF3B1-mut-MDS with similar OS and PFS observed. We were interested in what a head-to-head outcome comparison in MDS-SF3B1 would reveal between the IWG-PM–defined SF3B1-mut-MDS and those excluded per IWG-PM exclusion criteria29 (IWG-PM–excluded MDS-SF3B1). Interestingly, we did not observe a statistically significant difference in OS (median, 73 vs 54 months; P = .22) or PFS (neither median reached; P = .41) among the 2 subgroups. When we explored prognostic refinement of the IWG-PM–defined SF3B1-mut-MDS in MDS-SF3B1 by adding TF comutations into the exclusion criteria (GATA2 and BCOR added, RUNX1 already in the IWG-PM exclusion criteria), statistically significant favorable OS was observed in the refined IWG-PM–defined SF3B1-mut-MDS group.
We cannot completely rule out that the lack of a demonstrable statistically significant prognostic differentiation among IWG-PM–defined SF3B1-mut-MDS, and IWG-PM–excluded MDS-SF3B1 may be related to the relatively small size of our MDS-SF3B1 cohort; however, it is worth noting that such an outcome comparison between the 2 subgroups of MDS-SF3B1 was not reported by the IWG-PM study.29 Furthermore, consistent with the IWG-PM study, we independently confirmed the adverse prognostic value of IPSS-R greater than 3 in MDS-SF3B1 and the favorable clinical outcome of IWG-PM–defined SF3B1-mut-MDS, with similar median OS and PFS in our study. These findings support an overall representative nature of our study cohort. The constellation of findings from our study and previous studies, including the strong association between SF3B1 mutations and RS, support recognition of SF3B1-mut-MDS as a distinct disease entity genetically, phenotypically, prognostically, and potentially therapeutically, as proposed by IWG-PM. However, based on its current diagnostic criteria, this designation may not provide sharp prognostication within MDS-SF3B1. This may not be surprising, as the IWG-PM exclusion criteria were based on previous reports and not all were fully validated in the IWG-PM cohort—for example, among the 3 excluding cytogenetic abnormalities monosomy 7, inv(3)/3q26 abnormalities, and complex karyotype, only monosomy 7 was fully confirmed.29 The IWG-PM diagnostic criteria also exclude the prognostically favorable MDS with del(5q) in which SF3B1 mutations were reported in approximately 20% of cases. In our study, IPSS-R outperformed the IWG-PM–defined SF3B1-mut-MDS designation in the prognostication of MDS-SF3B1. We also identified TF comutations as a novel risk marker with independent prognostic value in MDS-SF3B1, which may further refine the risk stratification of IWG-PM–defined SF3B1-mut-MDS. Because the TF gene RUNX1 is already included in the IWG-PM exclusion criteria, our data confirmed its validity as an exclusion criterion and suggested the possibly improved prognostic value of expanding the exclusion criteria to additional TF markers GATA2 and BCOR. Given our relatively small study cohort size, further evaluation in large, well-characterized patient cohorts is warranted to evaluate the clinical significance and biological basis of TF comutations and other informative prognostic markers in this patient cohort for improved prognostication, monitoring, and clinical management.
Biologically, it is conceivable that disturbed transcription activities secondary to TF mutations in RUNX1, GATA2, or BCOR may potentiate the deleterious effects from the aberrant transcript splicing of SF3B1 mutants and confer worse prognosis. Somatic mutations in RUNX1 were shown to be independent poor prognostic risk factors in MDS.25 Consistent with our findings, comutations of RUNX1 demonstrated negative prognostic value in MDS-SF3B1 in subsequent studies and were included in the IWG-PM SF3B1-mut-MDS exclusion criteria.23,24,29 As a master TF, balanced GATA2 expression is required for proper hematopoiesis, and its disruption may result in tumorigenesis. Somatic mutations in RUNX1 and GATA2 are considered type 2 mutations, given their association with progression from low- to high-risk MDS.31,32BCOR mutations have also been shown to be associated with inferior outcomes in MDS.55 These findings provide supporting evidence for our observation of TF comutation as a negative prognostic marker in MDS-SF3B1. One BCOR and RUNX1 comutated case (case 72) had a subclonal CEBPA mutation. Given its presence in a single case and subclonal nature, the prognostic impact of CEBPA was unclear in our cohort. Although germline mutations in RUNX1, GATA2, and CEBPA may occur in myeloid neoplasms with germline predisposition,33 and germline mutations in BCOR occur in oculofaciocardiodental syndrome,56 patients’ clinical and family histories, age, and sex and/or mutational VAF of the TF comutations all supported a somatic nature of these TF comutations.
In conclusion, we report the presence of SF3B1 mutations across the full spectrum of myeloid disorders, ranging from CCUS to AML. SF3B1 mutations are highly associated with RS and the disease types of MDS-RS, MDS/MPN-RS-T, and AML-MRC. In MDS-SF3B1, IPSS-R greater than 3 and TF comutations were shown to be adverse prognostic markers. We also independently validated the favorable outcome of the new entity SF3B1-mut-MDS as proposed by IWG-PM. However, the IWG-PM–defined SF3B1-mut-MDS designation may not provide a sharp prognostic differentiation within MDS-SF3B1 among those included and excluded for this new entity following the IWG-PM exclusion criteria. Pending further verification in larger cohort studies, the IWG-PM–proposed SF3B1-mut-MDS exclusion criteria may benefit from refinement by other informative markers such as TF comutations in GATA2 and BCOR besides RUNX1. Our data add to the growing body of evidence demonstrating the value of molecular analysis and incorporation of the pertinent prognostic markers into the MDS risk stratification scheme to further refine and improve personalized medicine.
Supplementary Material
References
- 1.Golas MM, Sander B, Will CL, et al. . Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980-984. [DOI] [PubMed] [Google Scholar]
- 2.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701-718. [DOI] [PubMed] [Google Scholar]
- 4.Rossi D, Bruscaggin A, Spina V, et al. . Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, Cazzola M, Boultwood J, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium . Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida K, Sanada M, Shiraishi Y, et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64-69. [DOI] [PubMed] [Google Scholar]
- 7.Alsafadi S, Houy A, Battistella A, et al. . Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darman RB, Seiler M, Agrawal AA, et al. . Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 2015;13:1033-1045. [DOI] [PubMed] [Google Scholar]
- 9.Malcovati L, Papaemmanuil E, Bowen DT, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium and of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative . Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeBoever C, Ghia EM, Shepard PJ, et al. . Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. Plos Comput Biol. 2015;11:e1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellagatti A, Armstrong RN, Steeples V, et al. . Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood. 2018;132:1225-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazzola M, Rossi M, Malcovati L; Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative . Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood. 2013;121:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikpour M, Scharenberg C, Liu A, et al. . The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia. 2013;27:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiozawa Y, Malcovati L, Gallì A, et al. . Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun. 2018;9:3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visconte V, Rogers HJ, Singh J, et al. . SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120:3173-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolatshad H, Pellagatti A, Liberante FG, et al. . Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia. 2016;30:2322-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boiocchi L, Hasserjian RP, Pozdnyakova O, et al. . Clinicopathological and molecular features of SF3B1-mutated myeloproliferative neoplasms. Hum Pathol. 2019;86:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Lindsley RC, Mar BG, Mazzola E, et al. . Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbour JW. Genomic, prognostic, and cell-signaling advances in uveal melanoma. Am Soc Clin Oncol Educ Book. 2013:388-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong Y, Krauthammer M, Halaban R. Rare SF3B1 R625 mutations in cutaneous melanoma. Melanoma Res. 2014;24:332-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis MJ, Ding L, Shen D, et al. . Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biankin AV, Waddell N, Kassahn KS, et al. ; Australian Pancreatic Cancer Genome Initiative . Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malcovati L, Karimi M, Papaemmanuil E, et al. . SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malcovati L, Papaemmanuil E, Ambaglio I, et al. . Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124:1513-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bejar R, Stevenson K, Abdel-Wahab O, et al. . Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haferlach T, Nagata Y, Grossmann V, et al. . Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patnaik MM, Tefferi A. Refractory anemia with ring sideroblasts and RARS with thrombocytosis. Am J Hematol. 2015;90:549-559. [DOI] [PubMed] [Google Scholar]
- 28.Damm F, Thol F, Kosmider O, et al. . SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26:1137-1140. [DOI] [PubMed] [Google Scholar]
- 29.Malcovati L, Stevenson K, Papaemmanuil E, et al. . SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood. 2020;136:157-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenaux P, Platzbecker U, Mufti GJ, et al. . Luspatercept in patients with lower-risk myelodysplastic syndromes. N Engl J Med. 2020;382:140-151. [DOI] [PubMed] [Google Scholar]
- 31.Makishima H, Yoshizato T, Yoshida K, et al. . Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa S. Genetics of MDS. Blood. 2019;133:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swerdlow SH, Campo E, Harris NL, et al. . WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Rev 4th ed. Lyon, France: IARC; 2017. [Google Scholar]
- 34.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makishima H, Visconte V, Sakaguchi H, et al. . Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steensma DP. The clinical challenge of idiopathic cytopenias of undetermined significance (ICUS) and clonal cytopenias of undetermined significance (CCUS). Curr Hematol Malig Rep. 2019;14:536-542. [DOI] [PubMed] [Google Scholar]
- 37.Kwok B, Hall JM, Witte JS, et al. . MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126:2355-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie M, Lu C, Wang J, et al. . Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaiswal S, Fontanillas P, Flannick J, et al. . Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genovese G, Kähler AK, Handsaker RE, et al. . Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371: 2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malcovati L, Gallì A, Travaglino E, et al. . Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129:3371-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steensma DP, Bejar R, Jaiswal S, et al. . Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsunawa M, Yamamoto R, Sanada M, et al. . Haploinsufficiency of Sf3b1 leads to compromised stem cell function but not to myelodysplasia. Leukemia. 2014;28:1844-1850. [DOI] [PubMed] [Google Scholar]
- 45.Broséus J, Alpermann T, Wulfert M, et al. ; MPN and MPNr-EuroNet (COST Action BM0902) . Age, JAK2(V617F) and SF3B1 mutations are the main predicting factors for survival in refractory anaemia with ring sideroblasts and marked thrombocytosis. Leukemia. 2013;27:1826-1831. [DOI] [PubMed] [Google Scholar]
- 46.Dalton WB, Helmenstine E, Pieterse L, et al. . The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 2020;4:1192-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corces-Zimmerman MR, Hong WJ, Weissman IL, et al. . Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111:2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. . Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378:1189-1199. [DOI] [PubMed] [Google Scholar]
- 49.Baxter EJ, Scott LM, Campbell PJ, et al. ; Cancer Genome Project . Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054-1061. [DOI] [PubMed] [Google Scholar]
- 50.Levine RL, Wadleigh M, Cools J, et al. . Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387-397. [DOI] [PubMed] [Google Scholar]
- 51.Kralovics R, Passamonti F, Buser AS, et al. . A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779-1790. [DOI] [PubMed] [Google Scholar]
- 52.James C, Ugo V, Le Couédic JP, et al. . A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144-1148. [DOI] [PubMed] [Google Scholar]
- 53.Malcovati L, Della Porta MG, Pietra D, et al. . Molecular and clinical features of refractory anemia with ringed sideroblasts associated with marked thrombocytosis. Blood. 2009;114:3538-3545. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt-Graeff AH, Teo SS, Olschewski M, et al. . JAK2V617F mutation status identifies subtypes of refractory anemia with ringed sideroblasts associated with marked thrombocytosis. Haematologica. 2008;93:34-40. [DOI] [PubMed] [Google Scholar]
- 55.Damm F, Chesnais V, Nagata Y, et al. . BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122:3169-3177. [DOI] [PubMed] [Google Scholar]
- 56.Davoody A, Chen IP, Nanda R, et al. . Oculofaciocardiodental syndrome: a rare case and review of the literature. Cleft Palate Craniofac J. 2012;49:e55-e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.