Abstract
The work of Reddy et al. (S. A. Reddy, J. A. Huang, and W. S. Liao, J. Biol. Chem. 272:29167–29173, 1997) reveals that phosphatidylinositol 3-kinase (PI3K) plays a role in transducing a signal from the occupied interleukin-1 (IL-1) receptor to nuclear factor κB (NF-κB), but the underlying mechanism remains to be determined. We have found that IL-1 stimulates interaction of the IL-1 receptor accessory protein with the p85 regulatory subunit of PI3K, leading to the activation of the p110 catalytic subunit. Specific PI3K inhibitors strongly inhibit both PI3K activation and NF-κB-dependent gene expression but have no effect on the IL-1-stimulated degradation of IκBα, the nuclear translocation of NF-κB, or the ability of NF-κB to bind to DNA. In contrast, PI3K inhibitors block the IL-1-stimulated phosphorylation of NF-κB itself, especially the p65/RelA subunit. Furthermore, by using a fusion protein containing the p65/RelA transactivation domain, we found that overexpression of the p110 catalytic subunit of PI3K induces p65/RelA-mediated transactivation and that the specific PI3K inhibitor LY294,002 represses this process. Additionally, the expression of a constitutively activated form of either p110 or the PI3K-activated protein kinase Akt also induces p65/RelA-mediated transactivation. Therefore, IL-1 stimulates the PI3K-dependent phosphorylation and transactivation of NF-κB, a process quite distinct from the liberation of NF-κB from its cytoplasmic inhibitor IκB.
Interleukin-1 (IL-1), a proinflammatory cytokine, mediates numerous host responses (14). Although much is known about the mechanisms involved in IL-1-dependent signaling, much remains to be elucidated. IL-1 induces the rapid activation of the latent transcription factor nuclear factor κB (NF-κB) (3, 30, 31). The term NF-κB refers to a group of binary complexes of proteins with related promoter-binding and transactivation activities. The prototypical NF-κB complex consists of a p65-p50 heterodimer (46). p65/RelA, RelB, and c-Rel stimulate transcription, whereas p50 and p52 serve primarily to bind to DNA (25). Activation of NF-κB by IL-1, tumor necrosis factor alpha (TNF-α), H2O2, and phorbol-12-myristate-13-acetate is accompanied by increased phosphorylation of the p65/RelA subunit (7, 29). The activity of NF-κB is regulated by IκBs, which sequester NF-κB in the cytosol. Upon activation of signaling, IκB is phosphorylated and degraded, allowing NF-κB to enter the nucleus and bind to DNA (1, 41, 43, 46). The activation of NF-κB by IL-1 occurs through a discrete set of molecules recruited by the activated IL-1 receptor (IL-1R) complex, which includes IL-1R type I and the IL-1R accessory protein (IL-1R AcP) (17, 18, 22, 49).
A recent study indicates that phosphatidylinositol 3-kinase (PI3K) is a downstream effector of IL-1 signaling, involved in liberating NF-κB from IκB (34). PI3K consists of catalytic (p110) and regulatory (p85) subunits. The SH2 domains of p85 interact with the phosphotyrosine YXXM motifs of several activated cytokine and growth factor receptors (11, 19). p85 activates p110 by bringing it into contact with p110 lipid substrates at the cell membrane. The phosphorylated lipid products are secondary messengers, activating protein kinases such as Akt, also known as protein kinase B, and certain isoforms of protein kinase C (44). Recent work reveals that the p110α and -γ subunits of PI3K can also phosphorylate the p85 adapter protein and possibly other target proteins directly (9).
At present, it is unclear how PI3K and its downstream effectors feed into a signal transduction cascade that leads to the activation of NF-κB (6, 13, 15, 20, 26, 34, 39, 47, 52). However, a recent study shows that the activation of an NF-κB-dependent reporter gene by TNF-α or IL-1 is blocked by the phosphatidylcholine-specific phospholipase C inhibitor D609 or by the protein kinase C inhibitor R031-8220 (6). Moreover, IL-1-induced IκB degradation, NF-κB nuclear translocation, and DNA binding are not affected by these inhibitors, indicating that the phosphorylation and degradation of IκB are not sufficient for IL-1-induced, NF-κB-dependent transcription (6). In addition, other studies have shown that the transcriptional activity of NF-κB is regulated independently of IκB. For example, IκB-associated protein kinase A is involved in phosphorylating the p65/RelA subunit of NF-κB, allowing it to bind to the transcriptional coactivator CREB-binding protein/p300 (16, 33, 50, 51). Additionally, TNF-α was shown to mediate the transactivation of p65/RelA, which was in turn blocked by inhibitors of p38 and mitogen-activated protein kinases (45). Most recently, the activation by TNF-α of NF-κB-dependent transcription was shown to be mediated through phosphorylation of p65/RelA on serine 529 (47). These studies provide evidence for a second signaling pathway, induced by IL-1 and TNF-α, that is activated in parallel to the cascade leading to IκB degradation.
Our results indicate that IL-1 stimulates PI3K activity by causing the p85 regulatory subunit to bind to a specific region of the cytoplasmic domain of IL-1R AcP. PI3K then activates a pathway that parallels but is separate from IκB degradation, leading to the phosphorylation of p65/RelA, which is required for its full activity as a transcriptional activator.
MATERIALS AND METHODS
Cell culture and treatment with cytokines and inhibitors.
Recombinant human IL-1β was provided by the National Cancer Institute. LY294,002 and wortmannin were obtained from the Alexis Corporation. Polyclonal anti-p85, anti-Gal4 DNA binding domain, anti-glycogen synthase kinase 3 (anti-GSK-3), anti-phospho-c-Jun NH2-terminal kinase (anti-phospho-JNK1), anti-JNK1, and anti-IκBα were from Santa Cruz Biotechnology. Monoclonal anti-PY20, directed against phosphotyrosine, and anti-IL-1R AcP were from Transduction Laboratories. Polyclonal anti-phospho-GSK-3α (serine 21) antibody was obtained from New England Biolabs. Polyclonal anti-p65/RelA, anti-p50, and anti-c-Rel antibodies were the kind gift of Warner C. Greene. The plasmids encoding recombinant IL-1R AcP–glutathione S-transferase (GST) fusion proteins were the kind gift of Grace Ju. Protein A-Sepharose and glutathione-agarose beads were from Pharmacia. Phosphatidylinositol was from Sigma. The human hepatoma cell line HepG2, from the American Type Culture Collection, was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin G (100 μg/ml), and streptomycin (100 μg/ml). For all experiments, unless otherwise indicated, cells at 80% confluence on 100-mm-diameter dishes were incubated with specific PI3K inhibitors (20 μM LY294,002 or 100 nM wortmannin) for 30 min at 37°C prior to stimulation with IL-1β (1 ng/ml) at 37°C for the times indicated below. All of the results shown are typical of at least three independent experiments.
Immunoblotting and immunoprecipitation.
Cells were washed once with phosphate-buffered saline and lysed for 30 min at 4°C in 1 ml of 0.5% Nonidet P-40 lysis buffer as described previously (48). Cellular debris was removed by centrifugation at 16,000 × g for 15 min. For immunoblotting, cell extracts were fractionated directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Immunoblot analysis was performed with various primary antibodies, which were visualized with horseradish peroxidase-coupled goat anti-rabbit or anti-mouse immunoglobulins, by using the enhanced chemiluminescence Western blotting detection system (Amersham). For immunoprecipitations, cell extracts were incubated with 1 μl of primary antibody for 4 h followed by incubation for 1 h with 50 μl of protein A-Sepharose beads (20% suspension). The beads were washed three times with lysis buffer, and samples were analyzed by SDS-PAGE and autoradiography.
Analysis of interactions with an IL-1R AcP–GST fusion protein.
The full length IL-1R AcP cytoplasmic domain and C-terminal deletions were expressed as GST fusion proteins in bacteria and purified after sonication at 4°C in 0.5% Nonidet P-40 lysis buffer as described previously (21). Cellular debris was removed by centrifugation at 16,000 × g for 5 min. IL-1R AcP–GST fusion proteins were isolated with glutathione-agarose beads. HepG2 cells, either left unstimulated or stimulated with IL-1β for 0.5 min, were lysed for 1 min by sonication on ice with a Fisher model 300 sonic dismembrator at setting 35 in detergent-free lysis buffer as described previously (48). Cellular debris was removed by centrifugation at 16,000 × g for 15 min. IL-1R AcP–GST beads were incubated with rocking for 1 h at 4°C with the HepG2 cell extracts. Following the binding reaction, the beads were washed three times with GST fusion protein lysis buffer and the bound proteins were analyzed by SDS-PAGE, followed by immunoblotting with anti-p85.
PI3K assay.
Cells were washed once with phosphate-buffered saline and lysed for 30 min at 4°C in 1 ml of 1% Triton X-100 lysis buffer as described previously (36). Cellular debris was removed by centrifugation at 16,000 × g for 15 min at 4°C. PI3K activity was measured as described previously (36). Briefly, immunoprecipitation was performed with anti-PY20. Equal amounts of protein were incubated with 1 μg of anti-PY20 for 4 h followed a 1-h incubation with 50 μl of protein A-Sepharose (20% suspension). The bound beads were subjected to several washes with ice-cold buffers and were resuspended in kinase buffer containing phosphatidylinositol (4,5)P2 [PI(4,5)P2; 0.2 mg/ml], 10 μCi of [γ-32P]ATP, and 20 mM MgCl2 for 10 min. The reactions were terminated by adding chloroform–methanol–12 M HCl (50:100:1), and the products were extracted with chloroform, washed with methanol–1 M HCl (1:1), and freeze-dried. The products were resuspended in 15 μl of chloroform, resolved by thin-layer chromatography in chloroform-methanol-ammonium hydroxide-water (86:76:10:14), and visualized by autoradiography.
Northern transfers.
The cells were stimulated with IL-1β for 4 h. Total RNA was isolated with TRIzol reagent (Gibco BRL). RNA was fractionated by electrophoresis in a formaldehyde gel and transferred to Hybond-N (Amersham), a positively charged nylon membrane, according to the manufacturer’s directions. Probes from IL-8 and glyceraldehyde-3-phosphate dehydrogenase cDNAs were made with a random priming kit from Amersham. Probe hybridization and washing were performed according to procedures specified by Amersham, and signals were visualized by autoradiography.
Transfection and reporter assay.
The NF-κB-dependent E-selectin–luciferase reporter plasmid pElam[−143]-luc was a kind gift from Paul DiCorleto, Cleveland Clinic Foundation. This reporter plasmid has the ATF site deleted from the E-selectin promoter and contains three NF-κB sites. For the reporter gene assay, 2 × 105 HepG2 cells were transfected with Lipofectin (Gibco BRL) together with 1 μg of pElam[−143]-luc and 1 μg of pSV2-β-gal. In one experiment, 2 × 105 HepG2 cells were transfected with Lipofectin together with 1 μg of pElam[−143]-luc, 1 μg of pSV2-β-gal, and 1 μg of either a vector or a dominant negative mutant of Akt, which contains the first 159 amino acids of Akt, including the pleckstrin homology domain, and an additional group of highly conserved 40 amino acids (12), kindly provided by Julian Downward. After 48 h, the cells were harvested. Where indicated, the cells were incubated with LY294,002 or wortmannin prior to stimulation with IL-1β for 4 h. Luciferase or β-galactosidase (β-Gal) activities were determined with the luciferase assay system (Promega) or with chemiluminescent reagents (Promega), respectively. Luciferase activity was normalized to β-Gal activity to control for transfection efficiency. The viability of each transfected cell population was measured by trypan blue exclusion at the time of harvesting.
Gel mobility and supershift assays.
For electrophoretic mobility shift assays (EMSAs), HepG2 cells were incubated with LY294,002 or wortmannin prior to stimulation with IL-1β for 30 min. The NF-κB binding site (5′-GAGCAGAGGGAAATTCCGTAACTT-3′) from the IP10 gene was used as a probe. Briefly, complementary oligonucleotides, end labeled with polynucleotide kinase and [γ-32P]ATP, were annealed by slow cooling. Approximately 20,000 cpm of probe was used per reaction mixture. Nuclear and cytoplasmic extracts were prepared in binding reaction buffer as described previously (5). The binding reaction was carried out at room temperature for 30 min in a total volume of 20 μl. The DNA–NF-κB complexes were separated on 5% polyacrylamide gels by electrophoresis in low-ionic-strength Tris-borate-EDTA buffer. The gels were dried, and the labeled complexes were visualized by autoradiography. For supershifts, approximately 1 μg of polyclonal antibody against any of the NF-κB subunits p65/RelA, p50, and c-Rel was added to the binding reactions after 15 min, and the incubations were continued for 15 min more.
Cell labeling and immunoprecipitation of phosphorylated p65/RelA.
HepG2 cells in 100-mm plates were preincubated for 30 min in serum-free medium lacking phosphate. This medium was replaced with the same medium, containing [32P]orthophosphate (100 μCi/ml), and the incubation was continued for 4 h. Where indicated, cells were incubated with LY294,002 prior to stimulation with IL-1β for 5 min. The cells were washed once with phosphate-buffered saline and lysed for 30 min at 4°C in 1 ml of 1% Triton X-100 lysis buffer as described previously (7). Cellular debris was removed by centrifugation at 16,000 × g for 15 min at 4°C. Immunoprecipitation was performed with a polyclonal antibody against p65/RelA, which pulls down a complex of p65/RelA, p50 NF-κB, and IκB. Equal amounts of protein were incubated overnight with 1-μg portions of the antibody, followed by a 1-h incubation with 50 μl of protein A-Sepharose (20% suspension). The bound beads were washed with ice-cold solutions as described previously (7). The beads were resuspended in SDS-PAGE sample buffer and analyzed by electrophoresis. The gel was dried, and labeled proteins were visualized by autoradiography.
Transactivation of the p65/RelA fusion protein.
Mammalian expression plasmids, containing full-length and truncated versions of the transactivation domain of the p65/RelA subunit of NF-κB fused to the DNA binding domain of the transcription factor Gal4 (8), were a kind gift from Bryan R. Cullen. The C terminus of p65/RelA has a potent transactivating domain of 135 amino acids (2, 37, 38), which can be divided into three subdomains extending from amino acids 416 to 458 (domain I), 458 to 521 (domain II), and 508 to 550 (domain III) (8, 28, 40). The plasmid pGal4-RelA contains all three transactivating domains, pGal4-RelA 458-550 contains domains II and III, and pGal4-RelA 508-550 contains only domain III (8). The reporter plasmid pGal4-luc (pFR-Luc), obtained from Stratagene, contains the luciferase gene controlled by the Gal4 upstream activating sequence. Cells were transfected with 0.5 μg of pGal4-luc (pFR-luc) and 1 μg of a RelA/Gal4 construct alone or with 1 μg of one of the following: a construct expressing the p110 subunit of PI3K, kindly provided by Warren Liao; constructs expressing constitutively activated p110 (rCD2p110) or kinase-dead p110 (rCD2p110 R/P), kindly provided by Doreen Cantrell and Karin Reif (35); or constructs expressing constitutively activated Akt (gag Akt) or kinase-dead Akt (Akt K179A), kindly provided by Julian Downward, as described previously (10). After 48 h, the cells were harvested and, where indicated, incubated with LY294,002 prior to stimulation with IL-1β for 4 h. Luciferase or β-Gal activities were determined with the luciferase assay system (Promega) or with chemiluminescent reagents (Promega), respectively. Luciferase activity was normalized to β-Gal activity to control for transfection efficiency. The expression of the Gal4-RelA fusion proteins was monitored at the time of harvesting by Western analysis of cell extracts with Gal4 DNA binding domain polyclonal antibody.
RESULTS
IL-1 induces the association of the p85 subunit of PI3K with a specific region of the cytoplasmic domain of the IL-1R AcP.
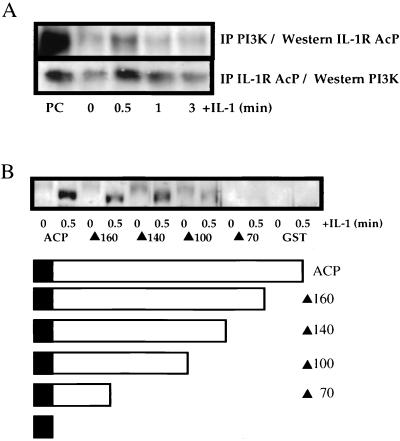
IL-1 induces the formation of a ternary complex with the IL-1R type I subunit and IL-1R AcP which, in turn, recruits several cytoplasmic signal-transducing proteins. To assess recruitment of the p85 subunit of PI3K to this complex, antibodies against p85 and IL-1R AcP were used to coprecipitate the associated proteins. IL-1 stimulation of HepG2 cells induced binding of IL-1R AcP to p85, as determined by coimmunoprecipitation of IL-1R AcP with an antibody against p85 and of p85 with an antibody against IL-R AcP (Fig. 1A). The association was maximal at 0.5 min, and little association was evident 3 min after IL-1 treatment. The interaction of IL-1R AcP with p85 was better defined by the use of GST fusion proteins containing either the cytoplasmic domain of IL-1R AcP or a series of C-terminal deletions. Extracts of HepG2 cells that were either left unstimulated or stimulated with IL-1 for 0.5 min were incubated with the recombinant GST fusion proteins (Fig. 1B). p85 bound to the full-length IL-1R AcP cytoplasmic domain (188 amino acids) as well as to mutants with deletions removing 28 or 48 amino acids from the C terminus, respectively. However, deletion of 88 amino acids from the C terminus abolished the interaction.
FIG. 1.
IL-1 induces the association of IL-1R AcP with p85. (A) HepG2 cells were stimulated with IL-1β (1 ng/ml) as indicated, and cell lysates were prepared. Polyclonal antibodies against either p85 or IL-1R AcP were used to coimmunoprecipitate the associated proteins. After separation by SDS-PAGE, the immunoprecipitates were immunoblotted with either anti-IL-1R AcP or anti-p85. PC, positive control (human endothelial cell extract). (B) Beads carrying GST fusion proteins with full-length and C-terminally deleted forms of the IL-1R AcP cytoplasmic domain (diagrammed) were bound to proteins in extracts of HepG2 cells. Following binding, the beads were washed, and the bound proteins were analyzed by SDS-PAGE, followed by immunoblotting with anti-p85. ACP, AcP. Mutant proteins are designated by a solid triangle followed by the number of amino acids remaining after deletion; e.g., ▴160 resulted from the deletion of 28 amino acids from the full-length protein (188 amino acids).
IL-1 stimulates the activation of PI3K, which is blocked by specific inhibitors.
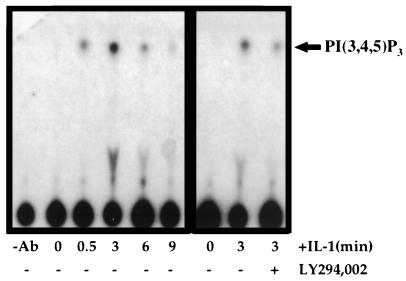
Extracts of IL-1-treated cells were assayed for PI3K activity by using phosphatidylinositol as a substrate. Treatment of HepG2 cells with IL-1 resulted in the rapid elevation of PI3K activity (Fig. 2, left). PI3K activity was detected after only 30 s, with maximal activity 3 min after IL-1 stimulation, followed by a rapid decline (Fig. 2, left). Pretreatment of the cells with 20 μM LY294,002 (Fig. 2, right), a specific PI3K inhibitor, or with 100 nM wortmannin (data not shown) dramatically reduced IL-1-stimulated PI3K activity, by ∼80 to 90% at 3 min.
FIG. 2.
PI3K activity is stimulated by IL-1 and inhibited by LY294,002. Where indicated, HepG2 cells were incubated with 20 μM LY294,002 for 30 min prior to stimulation with IL-1β for the indicated time periods. PI3K activity was measured as the phosphorylation of phosphatidylinositol(4,5)P2 yielding phosphatidylinositol(3,4,5)P3 [PI(3,4,5)P3]. −Ab, no anti-PY20.
LY294,002 inhibits IL-1-stimulated, NF-κB-dependent gene expression.
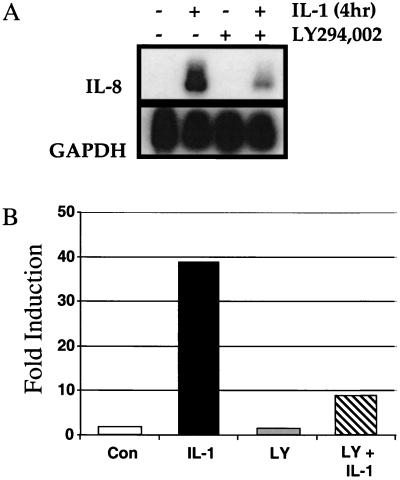
IL-8 is a chemotactic cytokine for neutrophils and lymphocytes. IL-8 expression is induced in response to various proinflammatory stimuli, including IL-1. Many of the cis elements regulating its expression have been identified, including binding sites for NF-κB (23, 24, 42). As IL-1-induced IL-8 expression is partially NF-κB dependent, we next investigated the effect of inhibiting IL-1-induced PI3K activity on IL-8 expression. Pretreatment of HepG2 cells with 20 μM LY294,002 reduced IL-1-stimulated IL-8 mRNA expression by ∼70 to 80% as determined by Northern analysis (Fig. 3A). LY294,002 pretreatment also caused an ∼70 to 80% decline in IL-1-stimulated luciferase expression from the transiently transfected, NF-κB-dependent reporter plasmid pElam[−143]-luc (Fig. 3B). Pretreatment with wortmannin (100 nM) also led to an ∼70 to 80% decline in IL-1-stimulated reporter gene expression (data not shown). As a previous study indicated that the degradation of IκB is insufficient for IL-1-induced NF-κB-dependent transcription (6), we investigated how PI3K might regulate this process.
FIG. 3.
NF-κB-dependent gene expression is stimulated by IL-1 and inhibited by LY294,002. Where indicated, HepG2 cells were incubated with 20 μM LY294,002 for 30 min prior to stimulation with IL-1β for 4 h. (A) Cells were lysed and analyzed for the expression of IL-8 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs by the Northern procedure. (B) HepG2 cells were transiently transfected with the NF-κB-dependent reporter plasmid pElam[−143]-luc and analyzed 48 h later. Where indicated, the cells were incubated with 20 μM LY294,002 (LY) for 30 min prior to stimulation with IL-1β for 4 h. Luciferase activity was normalized for transfection efficiency. Data are expressed as fold induction, the ratio between the expression level in the experimental and the level in the untreated control (Con) cells.
LY294,002 inhibits an NF-κB activation pathway distinct from IκBα degradation, NF-κB nuclear translocation, and DNA binding.
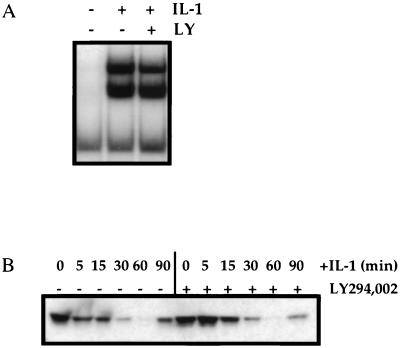
Despite the dramatic decrease in NF-κB-induced gene expression, pretreatment with LY294,002 had no effect on the IL-1-stimulated degradation of IκBα or on the nuclear translocation and DNA binding of NF-κB itself. Pretreatment with either LY294,002 (Fig. 4A). or wortmannin (data not shown) had no substantial effect on the ability of NF-κB, prepared from nuclear extracts of IL-1-stimulated HepG2 cells, to bind to a radiolabeled consensus κB oligonucleotide, as demonstrated by EMSA (Fig. 4A). Additionally, pretreatment with LY294,002 had no substantial effect on the degradation of IκBα, as demonstrated by Western analysis of IL-1-stimulated HepG2 cell extracts following IL-1 stimulation (Fig. 4B), or on the ability of p65/RelA to translocate from the cytoplasm to the nucleus, as measured by p65/RelA immunofluorescence in HepG2 cells 30 min after IL-1 stimulation (data not shown). The degradation of IκBβ, which did not occur until much later, was also not affected by LY294,002 pretreatment (data not shown). The lack of effect of LY294,002 on IκBα degradation is consistent with the EMSA results.
FIG. 4.
LY294,002 inhibits an NF-κB activation pathway distinct from IκBα degradation. Where indicated, HepG2 cells were incubated with 20 μM LY294,002 (LY) for 30 min prior to stimulation with IL-1β for 30 min for EMSA or for the indicated times for IκBα analysis. (A) EMSA for NF-κB. (B) Immunoblot analysis of IκBα.
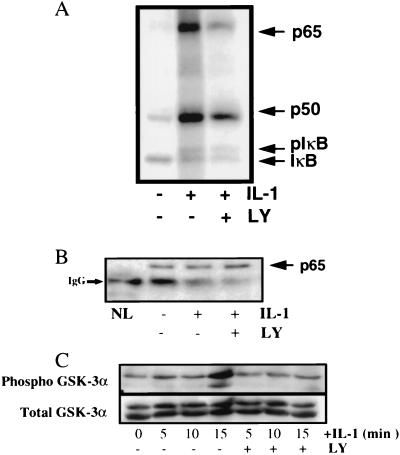
LY294,002 inhibits IL-1-induced phosphorylation of the p65/RelA subunit of NF-κB.
Cytokine-mediated phosphorylation of NF-κB subunits has been demonstrated previously (7) and might be involved in regulating NF-κB activity. Therefore, we investigated how treatment with LY294,002 affects NF-κB itself. The subunit composition of the two NF-κB EMSA complexes induced by IL-1 in HepG2 cell extracts (Fig. 4A) was investigated with antibodies to p65/RelA, p50, and c-Rel NF-κB in supershift experiments. The faster-migrating complex was a heterodimer of p65/RelA and p50, while the slower-migrating complex was possibly either a homodimer of p65/RelA or a heterodimer of p65/RelA with a different NF-κB subunit (Fig. 5). To investigate the effect of LY294,002 on IL-1-stimulated NF-κB phosphorylation, we used an antibody to the p65/RelA subunit known to precipitate a complex containing p65/RelA, p50, and IκB (7). Phosphorylated proteins labeled for 5 min in vivo following IL-1 stimulation of HepG2 cells were prepared either from untreated cells or from cells pretreated with 20 μM LY294,002. IL-1 stimulation induced the phosphorylation of p65/RelA, p50, and IκB (Fig. 6A). Pretreatment with LY294,002 led to a 90% decrease in the IL-1-stimulated phosphorylation of p65/RelA, with less of an effect on the phosphorylation of the p50 NF-κB subunit, which was decreased by ∼50% (Fig. 6A). The identity of the phosphorylated proteins was confirmed by Western analysis with specific antibodies to p65/RelA (Fig. 6B) as well as to p50 and IκB (data not shown). Although the decrease in phosphorylation of the p65/RelA subunit was greater than that of the p50 subunit, inhibition of PI3K did decrease the phosphorylation of both subunits. However, the p50 and p52 NF-κB subunits primarily serve to bind DNA, whereas the p65/RelA, RelB, and c-Rel subunits, containing C-terminal transactivation domains, are the main transcriptionally active members of the NF-κB family (22). These C-terminal transactivation domains are thought to be regulated, at least in part, through phosphorylation events (8), probably allowing recruitment of various transcription activators. Additionally, IL-1 stimulated the phosphorylation of a known PI3K- and Akt-dependent target, glycogen synthase kinase GSK-3α, and LY294,002 pretreatment blocked this phosphorylation (Fig. 6C). GSK-3α, a ubiquitously expressed serine/threonine kinase whose activity is inhibited by phosphorylation of serine 21, is a downstream element in the PI3K and Akt cell survival pathway (32). Pretreatment with LY294,002, however, did not block IL-1-induced phosphorylation of JNK1, a protein target that is phosphorylated in response to IL-1 but is not a target of the PI3K and Akt phosphorylation cascade (data not shown).
FIG. 5.
IL-1 stimulates the formation of a p65-p50 heterodimer and a second p65/RelA complex in HepG2 cells. Cells were either left untreated or stimulated with IL-1β for 30 min. The cells were lysed, and the supershift analysis was performed with the antibodies indicated. p65/? is either a homodimer of p65/RelA or a heterodimer of p65/RelA with another NF-κB family member.
FIG. 6.
LY294,002 inhibits NF-κB phosphorylation. HepG2 cells were preincubated in phosphate-free medium containing [32P]orthophosphate (100 μCi/ml). Where indicated, the cells were incubated with 20 μM LY294,002 (LY) for 30 min prior to stimulation with IL-1β for 5 min (A and B) or for the indicated times (C). Cell lysis and immunoprecipitation of the phosphorylated NF-κB complex were performed as described in Materials and Methods. (A) Analysis of phosphorylated NF-κB proteins. pIκB, phosphorylated IκB. (B) Western analysis of phosphorylated p65/RelA proteins. IgG, immunoglobulin G antibody heavy chain; NL, no cell lysate. (C) Analysis of phosphorylated phospho-GSK-3α (serine 21-phosphorylated GSK-3α).
The PI3K pathway regulates the activity of NF-κB.
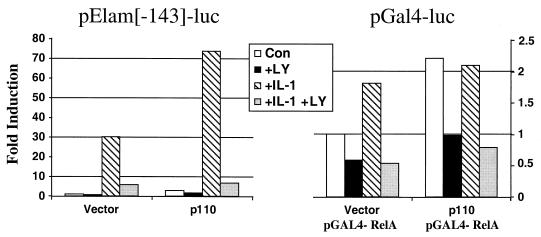
The C terminus of p65/RelA has a potent transactivating domain of 135 amino acids (2, 37). Cytokine-dependent phosphorylation of this domain may facilitate transactivation. We have shown that IL-1-stimulated PI3K regulates NF-κB-dependent promoter expression independently of the IκB degradation–NF-κB liberation pathway and that inhibition of PI3K decreases the IL-1-induced phosphorylation of the p65/RelA subunit of NF-κB substantially. To investigate this effect further, we examined the effect of overexpressing the p110 subunit of PI3K on the NF-κB-dependent promoter pElam[−143]-luc. Transient cotransfection of p110 led to a two- to threefold induction of NF-κB-dependent luciferase activity compared to that in vector-transfected cells, and this increase was inhibited by LY294,002 (Fig. 7, left). However, in cells overexpressing p110 and stimulated with IL-1, the NF-κB-dependent promoter was activated synergistically, and this activation was inhibited by LY294,002 (Fig. 7, left). We next evaluated the effect of PI3K activation on p65/RelA-dependent transactivation, independently of IκB degradation, with Gal4-dependent one-hybrid analysis of the transcriptional activation of p65/RelA. In this system, a plasmid encoding a hybrid transcription factor containing the transactivation domain of p65/RelA fused to the DNA binding domain of the Gal4 transcription factor (8) is cotransfected with a reporter construct containing the luciferase gene under the control of the Gal4 upstream activating sequence. These fusion proteins are regulated independently of IκB (2, 8, 37). The effects of IL-1 alone and IL-1 plus LY294,002 were evaluated. Stimulation with IL-1 led to a modest but reproducible increase in luciferase activity over that observed in untreated control cells, and 20 μM LY294,002 inhibited both the high basal activation of the p65/RelA fusion protein in untreated cells and the modest increase caused by IL-1 stimulation (Fig. 7, right). The effect of overexpressing the p110 catalytic subunit of PI3K was also examined. Transient cotransfection with p110 led to a two- to threefold induction of p65/RelA-dependent luciferase activity compared to that in the vector-transfected control, and this increase was inhibited by LY294,002 (Fig. 7, right). Additionally, stimulation with IL-1 was not able to activate promoter expression in cells overexpressing p110 (Fig. 7, right).
FIG. 7.
Involvement of the PI3K pathway in regulating p65/RelA transactivation. (Left) HepG2 cells were cotransfected transiently with the NF-κB-dependent reporter plasmid pElam[−143]-luc and either an empty vector or a plasmid expressing the p110 catalytic subunit of PI3K. The cells were harvested 48 h after transfection. Where indicated, the cells were incubated with 20 μM LY294,002 (LY) for 30 min prior to stimulation with IL-1β for 4 h. Luciferase activity was normalized for transfection efficiency. Data are expressed as fold induction, the ratio between the expression level in the experimental cells and the level in vector-transfected untreated control cells (Con). (Right) HepG2 cells were cotransfected transiently with the pGal4-luc reporter plasmid, pGal4-RelA (expressing the p65-Gal4 fusion protein), and either the vector or a plasmid expressing p110; the cells were harvested 48 h after transfection. Where indicated, the cells were incubated with 20 μM LY294,002 for 30 min prior to stimulation with IL-1β for 4 h. Luciferase activity was normalized for transfection efficiency. The data are expressed as fold induction, the ratio between the expression level in the experimental and the level in vector-transfected untreated control cells (Con).
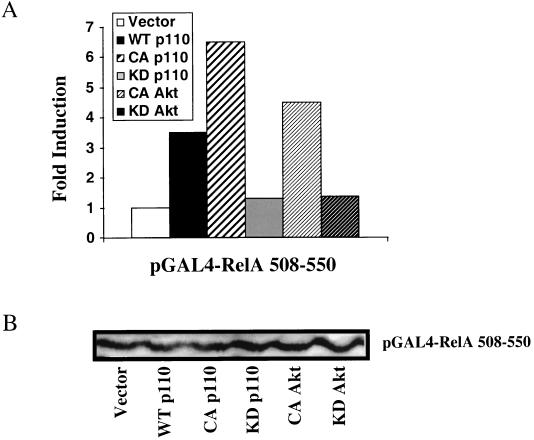
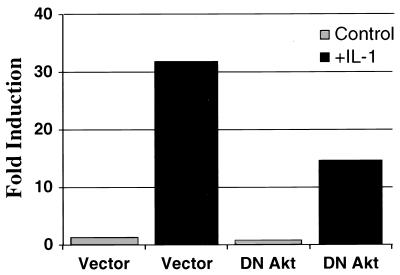
Recently, activation by TNF-α of NF-κB-dependent transcription was shown to be mediated through phosphorylation of p65/RelA on serine 529 (47). Since this phosphorylated serine lies in transactivation domain III, we investigated the ability of constitutively activated constructs of the p110 subunit of PI3K and its downstream effector Akt to transactivate p65/RelA through domain III, using the Gal4 one-hybrid system. Transient cotransfection of constitutively activated p110 and Akt, but not of their kinase-dead counterparts, led to a marked induction of luciferase activity driven by pGal4-RelA 508-550 compared to that in the vector control (Fig. 8A). This induction of promoter activity by the activated p110 and Akt was due to increasing the transcriptional potency of the Gal4-RelA fusion protein rather than to increasing the total amount of the fusion protein (Fig. 8B). Finally, overexpressed dominant negative Akt inhibited the ability of IL-1 to stimulate the NF-κB-dependent promoter construct pElam[−143]-luc by ∼50 to 60% (Fig. 9). This construct did not block IL-1 stimulation of an AP1-luciferase reporter construct (twofold induction; data not shown). The effects of the transfected p110 and Akt proteins are indeed due to transcriptional regulation and not to alterations in cell survival and cell death, since the viability of the various transfected cell populations did not differ from that of cells transfected with the vector control, as measured by trypan blue exclusion (data not shown).
FIG. 8.
Constitutively activated p110 and Akt regulate p65/RelA transactivation. HepG2 cells were cotransfected with the pGal4-luc reporter plasmid, pGal4-RelA 508-550, and either a vector or plasmids expressing wild-type (WT) p110, constitutively activated (CA) p110 or Akt, or their kinase-dead (KD) derivatives. The cells were harvested 48 h after transfection. (A) Luciferase activity was normalized for transfection efficiency. Data are expressed as fold induction, the ratio between the expression level in the experimental cells and the level in vector-transfected control cells. (B) Immunoblot analysis of the Gal4-RelA 508-550 fusion protein.
FIG. 9.
Dominant negative Akt blocks IL-1 signaling to an NFκB-dependent promoter. HepG2 cells were transiently cotransfected with the NFκB-dependent reporter plasmid pElam[−143]-luc, together with either a vector or a construct expressing a dominant negative (DN) derivative of Akt. The cells were analyzed 48 h later, with a 4-h stimulation with IL-1β where indicated. Luciferase activity was normalized for transfection efficiency. The data are expressed as fold induction, the ratio of the expression level in the experimental cells and the level in the vector-transfected untreated control cells.
DISCUSSION
We have analyzed the protein-protein interactions responsible for the IL-1-dependent activation of PI3K and how IL-1-stimulated PI3K leads to the activation of NF-κB. IL-1 stimulates interaction of the p85 subunit of PI3K with the cytoplasmic domain of IL-1R AcP, and we have defined a specific region of this domain that is necessary for the interaction. Additionally, we have shown that the IL-1-stimulated recruitment and activation of PI3K initiate a pathway of NF-κB activation that is distinct from IκB degradation, NF-κB nuclear translocation, and DNA binding. In contrast to the lack of effect of inhibiting PI3K on IκB degradation or NF-κB DNA binding, pretreatment of HepG2 cells with LY294,002 caused a dramatic inhibition of the IL-1-stimulated phosphorylation of p65/RelA and the ability of a p65/RelA-Gal4 hybrid factor to activate transcription. Indeed, using a Gal4 one-hybrid reporter system that is independent of IκB, we established a clear role for PI3K and its downstream effector Akt in modulating the transactivation potential of p65/RelA.
We do not know if components other than IL-1R AcP are necessary to recruit p85 to the activated IL-1R complex. A previous study has shown by coimmunoprecipitation that p85 interacts with the type I IL-1R (34). We have also seen an interaction between p85 and the IL-1 type I receptor (data not shown), but due to low expression in HepG2 cells, quantitation is difficult. Together with our study, the results of Reddy et al. (34) indicate that both receptor subunits may be necessary to recruit PI3K to the receptor complex upon stimulation with IL-1. We are currently investigating the role of tyrosine phosphorylation of IL-1R AcP in recruiting and activating PI3K to the activated receptor in HepG2 cells.
Inhibition of IL-1-stimulated PI3K activity by pretreatment with LY294,002 or wortmannin also causes a dramatic loss of NF-κB-dependent gene expression, consistent with the results of Reddy et al. (34). However, unlike the previous results and despite the dramatic decrease in NF-κB-induced gene expression, in our study pretreatment with LY294,002 or wortmannin had no effect on the IL-1-stimulated degradation of IκBα or the nuclear translocation or DNA binding of NF-κB itself in HepG2 cells. Reddy et al. (34) reported the loss of the ability of IL-1-activated NF-κB to bind to DNA following pretreatment with wortmannin, but IκBα degradation following IL-1 stimulation was not measured. At this time, we do not have a clear explanation for the differences in our results. Perhaps wortmannin, the only drug used in the previous study, has nonspecific effects on other proteins, such as the DNA-dependent protein kinase. At high concentrations, wortmannin has been shown to affect IκB degradation in other systems (4). However, our results are consistent with the results of Bergmann et al. (6), who found that inhibitors of phosphatidylcholine-specific phospholipase C and protein kinase C blocked IL-1- and TNF-α-induced, NF-κB-dependent gene expression without affecting cytokine-induced IκB degradation or the nuclear translocation or DNA binding of NF-κB.
The activation of NF-κB by cytokines and other stimuli has been shown to be accompanied by phosphorylation of the p65/RelA subunit in its C-terminal transactivation domain (7, 29). IκB-associated protein kinase A was previously shown to be involved in phosphorylating p65/RelA in this domain, allowing it to bind to the transcriptional coactivator CREB-binding protein/p300 (16, 33, 50, 51), and p65/RelA kinase activity was found to be associated with the IκB kinase (27). Most recently, the mutation to alanine of serine 529 in the C-terminal p65/RelA transactivation domain was shown to block the ability of p65/RelA to activate transcription in response to TNF-α without affecting p65/RelA nuclear translocation or DNA binding (47). These studies, as well as our data, support the hypothesis that cytokine-dependent phosphorylation of p65/RelA is required for its activation as a transcription factor. It will be of interest to determine how the phosphorylation of p65/RelA is regulated by different stimuli, and it is possible that the differential regulation of transcription may be determined by differential phosphorylation in response to different signals. At this time we have not determined whether p65/RelA must be dissociated from the IκB complex in order for phosphorylation to occur or whether the p65/RelA kinase is inducible or constitutive. How the p65/RelA kinase is regulated will be elucidated only after it has been identified.
The PI3K pathway seems to cooperate with a separate IL-1-induced signal transduction pathway to activate NF-κB-dependent transcription fully. Overexpression of the PI3K subunit p110 leads only to a 2- to 3-fold induction of an NF-κB-dependent reporter construct but causes a synergistic (IL-1 alone yielded 30-fold, p110 alone yielded 2- to 3-fold, and IL-1 plus p110 yielded 74-fold) induction of NF-κB-dependent expression in response to IL-1. These results, together with the lack of effect of PI3K inhibition on IL-1-induced IκB degradation, suggest that the PI3K pathway leading to p65/RelA phosphorylation and transactivation also requires IκB degradation and NF-κB liberation to activate NF-κB-dependent transcription.
By using an IκB-independent reporter system that requires only the transactivation of p65/RelA to activate transcription, we demonstrated that PI3K and its downstream effector Akt may mediate this transactivation in response to IL-1. Overexpression of the p110 catalytic subunit of PI3K led to a two- to threefold induction of p65/RelA transactivation-dependent luciferase activity compared to the activity in vector-transfected cells, and this activation was not enhanced by IL-1 treatment. LY294,002 is able to inhibit the high basal activity as well as the activation by either IL-1 or p110, consistent with our data showing that PI3K regulates the activation of p65/RelA (Fig. 7). Additionally, activated Akt induces p65/RelA activation, and a dominant negative derivative of Akt blocks the ability of IL-1 to signal to an NF-κB-dependent promoter. In summary, we provide evidence that IL-1 stimulates the recruitment of PI3K to the receptor and its subsequent activation, initiating a pathway of NF-κB activation that is distinct from IκB degradation and the liberation of NF-κB. This pathway is required for IL-1-stimulated transcriptional activation by NF-κB. The PI3K-dependent activation of NF-κB involves the phosphorylation and transactivation of p65/RelA, probably through signals transduced through Akt. The downstream effectors of PI3K and Akt required for the transactivation of p65/RelA remain to be identified.
ACKNOWLEDGMENTS
We thank Doreen A. Cantrell, Bryan R. Cullen, Paul DiCorleto, Julian Downward, Warner C. Greene, Grace Ju, Warren S.-L. Liao, and Karin Reif for the reagents used in this work.
REFERENCES
- 1.Baeuerle P A, Henkel T. Function and activation of NF κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 2.Ballard D W, Dixon E P, Peffer N J, Bogerd H, Doerre S, Greene W C. The p65 kD DNA binding subunit of the human NFκB complex functions as a potent transcriptional activator and a target for repression by the v-Rel oncoprotein. Proc Natl Acad Sci USA. 1992;89:1875–1879. doi: 10.1073/pnas.89.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes P J, Karin M. Nuclear factor-κB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 4.Basu S, Rosenzweig K R, Youmell M, Price B D. The DNA-dependent protein kinase participates in the activation of NFκB following DNA damage. Biochem Biophys Res Commun. 1998;247:79–83. doi: 10.1006/bbrc.1998.8741. [DOI] [PubMed] [Google Scholar]
- 5.Beg A A, Finco T S, Nantermet P V, Baldwin A S., Jr Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism for NF-κB activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann M, Hart L, Lindsay M, Barnes P J, Newton R. IκBα degradation and nuclear factor-B DNA binding are insufficient for interleukin-1β and tumor necrosis factor-α-induced B-dependent transcription. J Biol Chem. 1998;273:6607–6610. doi: 10.1074/jbc.273.12.6607. [DOI] [PubMed] [Google Scholar]
- 7.Bird T A, Schooley K, Dower S K, Hagen H, Virca G D. Activation of nuclear transcription factor NFκB by interleukin-1 is accompanied by casein kinase II-mediated phosphorylation of the p65 subunit. J Biol Chem. 1997;272:32606–32612. doi: 10.1074/jbc.272.51.32606. [DOI] [PubMed] [Google Scholar]
- 8.Blair W S, Bogerd H P, Madore S J, Cullen B R. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bondeva T, Pirola L, Bulgarelli-Leva G, Rubio I, Wetzker R, Wymann M P. Bifurcation of lipid and protein kinase signals of PI3K gamma to the protein kinases PKB and MAPK. Science. 1998;282:293–296. doi: 10.1126/science.282.5387.293. [DOI] [PubMed] [Google Scholar]
- 10.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter C L, Cantley L C. Phosphoinositide kinases. Curr Opin Cell Biol. 1996;8:153–157. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 12.Datta K, Franke T F, Chan T O, Makris A, Yang S-I, Kaplan D R, Morrison D K, Golemis E A, Tsichlis P N. AH/PH domain-mediated interaction between Akt molecules and its potential role in Akt regulation. Mol Cell Biol. 1995;15:2304–2310. doi: 10.1128/mcb.15.4.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz-Meco M T, Dominguez I, Sanz L, Dent P, Lozano J, Municio M M, Berra E, Hay R T, Sturgill T W, Moscat J. Zeta PKC induces phosphorylation and inactivation of IκB-alpha in vitro. EMBO J. 1994;13:2842–2848. doi: 10.1002/j.1460-2075.1994.tb06578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinarello C A. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 15.Genot E M, Parker P J, Cantrell D A. Analysis of the role of protein kinase C-alpha, -epsilon, and -zeta in T cell activation. J Biol Chem. 1995;270:9833–9839. doi: 10.1074/jbc.270.17.9833. [DOI] [PubMed] [Google Scholar]
- 16.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenfeder S A, Nunes P, Kwee L, Labow M, Chizzonite R A, Ju G. Molecular cloning and characterization of a second subunit of the interleukin 1 receptor complex. J Biol Chem. 1995;270:13757–13765. doi: 10.1074/jbc.270.23.13757. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Gao X, Li S, Cao Z. Recruitment of IRAK to the interleukin-1 receptor complex requires interleukin-1 receptor accessory protein. Proc Natl Acad Sci USA. 1997;94:12829–12832. doi: 10.1073/pnas.94.24.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- 20.Janosch P, Schellerer M, Seitz T, Reim P, Eulitz M, Brielmeier M, Kolch W, Sedivy J M, Mischak H. Characterization of IκB kinases. I κB-alpha is not phosphorylated by Raf-1 or protein kinase C isozymes, but is a casein kinase II substrate. J Biol Chem. 1996;271:13868–13874. doi: 10.1074/jbc.271.23.13868. [DOI] [PubMed] [Google Scholar]
- 21.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 22.Korherr C, Hofmeister R, Wesche H, Falk W. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. Eur J Immunol. 1997;27:262–267. doi: 10.1002/eji.1830270139. [DOI] [PubMed] [Google Scholar]
- 23.Kunsch C, Lang R K, Rosen C A, Shannon M F. Synergistic transcriptional activation of IL-8 gene by NF-κB p65 and NF-IL6. Immunology. 1994;153:153–164. [PubMed] [Google Scholar]
- 24.Kunsch C, Rosen C A. NF-κB subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liou H C, Baltimore D. Regulation of NF-κB/rel transcription factor and I kappa B inhibitor system. Curr Opin Cell Biol. 1993;5:477–487. doi: 10.1016/0955-0674(93)90014-h. [DOI] [PubMed] [Google Scholar]
- 26.Lozano J, Berra E, Municio M M, Diaz-Meco M T, Dominguez I, Sanz L, Moscat J. Protein kinase C zeta isoform is critical for κB-dependent promoter activation by sphingomyelinase. J Biol Chem. 1994;269:19200–19202. [PubMed] [Google Scholar]
- 27.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 28.Moore P A, Ruben S M, Rosen C A. Conservation of transcriptional activation functions of the NF-κB p50 and p65 subunits in mammalian cells and Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1666–1674. doi: 10.1128/mcb.13.3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naumann M, Scheidereit C. Activation of NF-κB in vivo is regulated by multiple phosphorylations. EMBO J. 1994;13:4597–4607. doi: 10.1002/j.1460-2075.1994.tb06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Neill L A J. Towards an understanding of the signal transduction pathways for interleukin 1. Biochim Biophys Acta. 1995;1266:31–44. doi: 10.1016/0167-4889(94)00217-3. [DOI] [PubMed] [Google Scholar]
- 31.O’Neill L A J. Molecular mechanisms underlying the actions of the pro-inflammatory cytokine interleukin 1. Biochem Soc Trans. 1997;25:295–302. doi: 10.1042/bst0250295. [DOI] [PubMed] [Google Scholar]
- 32.Pap M, Cooper G M. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-kinase/Akt cell survival pathway. J Biol Chem. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 33.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 34.Reddy S A, Huang J H, Liao W S. Phosphatidylinositol 3-kinase in interleukin 1 signaling. J Biol Chem. 1997;272:29167–29173. doi: 10.1074/jbc.272.46.29167. [DOI] [PubMed] [Google Scholar]
- 35.Reif K, Burgering B M, Cantrell D A. Phosphatidylinositol 3-kinase links the interleukin-2 receptor to protein kinase B and p70 S6 kinase. J Biol Chem. 1997;272:14426–14433. doi: 10.1074/jbc.272.22.14426. [DOI] [PubMed] [Google Scholar]
- 36.Royal I, Park M. Hepatocyte growth factor-induced scatter of Madin-Darby canine kidney cells requires phosphatidylinositol 3-kinase. J Biol Chem. 1995;270:27780–27787. doi: 10.1074/jbc.270.46.27780. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz M L, Baeuerle P A. The p65 subunit is responsible for the strong transcription activating potential of NF κB. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz M L, dos Santos Silva M A, Baeuerle P A. Transactivation domain 2 (TA2) of p65 NF κB. J Biol Chem. 1995;270:15576–15584. doi: 10.1074/jbc.270.26.15576. [DOI] [PubMed] [Google Scholar]
- 39.Schutze S, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Kronke M. TNF activated NF κB by phosphatidylcholine-specific phospholipase C-induced “acidic” sphingomyelin breakdown. Cell. 1992;71:765–776. doi: 10.1016/0092-8674(92)90553-o. [DOI] [PubMed] [Google Scholar]
- 40.Seiple K O, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 42.Stein B, Baldwin A S., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 44.Toker A, Meyer M, Reddy K K, Falck J R, Aneja R, Aneja S, Parra A, Burns D J, Ballas L M, Cantley L C. Activation of protein kinase C family members by the novel polyphosphoinositides PtdIns-3,4-P2 and PtdIns-3,4,5-P3. J Biol Chem. 1994;269:32358–32367. [PubMed] [Google Scholar]
- 45.Vander Berghe W, Plaisance S, Boone E, De Bosscher K, Schmitz M L, Fiers W, Haegeman G. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor κB p65 transactivation mediated by tumor necrosis factor. J Biol Chem. 1998;273:3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 46.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF κB/I κB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 47.Wang D, Baldwin A S. Activation of nuclear factor κB-dependent transcription by tumor necrosis factor-alpha is mediated through phosphorylation of RelA/p65 on serine 529. J Biol Chem. 1998;273:29411–29416. doi: 10.1074/jbc.273.45.29411. [DOI] [PubMed] [Google Scholar]
- 48.Welch P J, Wang J Y J. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 49.Wesche H, Korherr C, Kracht M, Falk W, Resch K, Martin M U. The interleukin-1 receptor accessory protein (IL-1RAcP) is essential for IL-1-induced activation of interleukin-1 receptor-associated kinase (IRAK) and stress-activated protein kinases (SAP kinases) J Biol Chem. 1997;272:7727–7731. doi: 10.1074/jbc.272.12.7727. [DOI] [PubMed] [Google Scholar]
- 50.Zhong H, Suyang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF κB is regulated by the IκB-associated PKA subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 52.Zumbansen M, Stoffel W. Tumor necrosis factor alpha activates NF κB in acid sphingomyelinase-deficient mouse embryonic fibroblasts. J Biol Chem. 1997;272:10904–10909. doi: 10.1074/jbc.272.16.10904. [DOI] [PubMed] [Google Scholar]