Abstract
The Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV-2) has been recently found responsible for the pandemic outbreak of a novel coronavirus disease (COVID-19). In this work, a novel approach based on deep learning is proposed for identifying precursors of small active RNA molecules named microRNA (miRNA) in the genome of the novel coronavirus. Viral miRNA-like molecules have shown to modulate the host transcriptome during the infection progression, thus their identification is crucial for helping the diagnosis or medical treatment of the disease. The existence of the mature miRNAs derived from computationally predicted miRNA precursors (pre-miRNAs) in the novel coronavirus was validated with small RNA-seq data from SARS-CoV-2-infected human cells. The results demonstrate that computational models can provide accurate and useful predictions of pre-miRNAs in the SARS-CoV-2 genome, underscoring the relevance of machine learning in the response to a global sanitary emergency. Moreover, the interpretability of our model shed light on the molecular mechanisms underlying the viral infection, thus contributing to the fight against the COVID-19 pandemic and the fast development of new treatments. Our study shows how recent advances in machine learning can be used, effectively, in response to public health emergencies. The approach developed in this work could be of great help in future similar emergencies to accelerate the understanding of the singularities of any viral agent and for the development of novel therapies. Data and source code available at: https://sourceforge.net/projects/sourcesinc/files/aicovid/.
Keywords: COVID-19, Deep learning, Computational prediction, Pre-miRNAs
1. Introduction
MicroRNAs (miRNAs) are a special type of small non-coding RNA of 22 nucleotides in length that can be found in plants, metazoans and viruses. MiRNAs participate in gene regulation influencing diverse biological processes such as development, proliferation, cell differentiation and metabolism across different cell types (Bartel, 2004, Gurtan and Sharp, 2013). They also play important roles in disease development and progression. MiRNAS are processed from long intermediates, known as miRNA precursors (pre-miRNAs). In animals, the specificity and function of miRNAs are determined by the nucleotides 2 to 7 of the mature region of the miRNAs. The quantification of the dynamic abundance of specific miRNAs can assist in diagnosis, prognosis prediction and therapeutic assessment. Notably, host miRNAs have been recently associated with antiviral defense mechanisms triggered by a coronavirus, and the activity of miRNAs derived from viral genomes has also been proved (Guzzi et al., 2020, Li and Zou, 2019).
The Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV-2) is the agent responsible for the recent pandemic outbreak of a novel coronavirus disease (COVID-19). SARS-CoV-2 is a positive-single stranded RNA virus with a genome of 30 kb. The discovery of miRNAs in the novel virus is of paramount importance in the context of the current worldwide sanitary crisis, especially for contributing to the improvement of diagnostic and treatment strategies (Huan et al., 2015, Ivashchenko et al., 2020). Biochemical identification of novel miRNAs is hampered by the requirements for wet/biological experimental setup, which can be expensive and time-consuming especially in the case of complete genomes (Li et al., 2009). This difficulty led to the development of several computational approaches for predicting miRNAs and their precursors based on genomic information (Allmer and Yousef, 2012, Bugnon et al., 2021, Stegmayer et al., 2019).
Machine learning (ML) approaches for pre-miRNA prediction involve training a binary classifier following a supervised learning strategy using well-known pre-miRNA sequences (deposited in miRBase1 (Kozomara et al., 2018) as the positive class, for identifying genomic regions with the highest chance of being miRNA precursors. The pre-miRNAs adopt a very well-known RNA secondary structure during biogenesis, named stem-loop or hairpin, which allowed the development of feature extraction algorithms for their identification. This secondary structure typically exhibits a few internal loops or asymmetric bulges. A large amount of hairpin-like structures can be found in a genome, most of which do not behave as pre-miRNAs. Thus, their correct identification is still a big challenge (Bugnon et al., 2021).
The computational detection of pre-miRNAs additionally suffers from a big challenge: the presence of very high class-imbalance, which has important consequences on the learning process producing classifiers with very poor predictive accuracy for the minority class. The class-imbalance problem has been largely recognized as an important issue in ML (Haixiang et al., 2017). It occurs when there are significantly lower training examples of one class in comparison to the other one. Most ML algorithms work well with balanced data sets, although imbalanced data in a supervised classifier can produce a model completely biased towards the majority class, with very low performance on the minority one, and generating many false positives (Bugnon et al., 2020).
In the particular problem of pre-miRNAs prediction, the big challenge here is that there are only tens or hundreds of well-known pre-miRNAs (the positive class), versus millions of unknown (unlabeled) sequences across the rest of the genome, most of which are really negative class albeit including yet unknown hidden pre-miRNAs. For example, the Anopheles gambiae genome has only 66 well-known pre-miRNAs, but more than 4 million hairpin-like sequences, thus giving an imbalance of 1:60,000 (Bugnon et al., 2019). In the case of viruses, for example, the value of imbalance ranges from 1:30 approximately in the bovine leukemia virus, 1:130 in the Epstein-Barr virus, and up to 1:400 in the Herpes virus of turkeys, which has only 8 known pre-miRNAs and a genome of 159 kb.
In the last decade, a growing number of strategies have been proposed for tackling the computational detection of pre-miRNAs and overcome the mentioned challenges. The first models were based on transcriptomics as input data, using heuristics with a limited capacity to detect miRNA precursors with low similarity to the reference set (Wei et al., 2014). To overcome this limitation, different ML approaches appeared (Stegmayer et al., 2019), for example based on random forest (Jiang et al., 2007). These methods use features extracted from typical properties of known pre-miRNAs, i.e the number of loops in a sequence, the average length of the sequence, the minimum free energy when folding the secondary structure (MFE), among many others (Liu et al., 2012, Yones et al., 2015). More recently, deep learning (DL) models have been developed for this task, not requiring any feature engineering since they can automatically extract motifs (patterns of nucleotides) from a set of homologous sequences, and being able also to efficiently handle the large class imbalance. This kind of model can be fed with the predicted secondary structure of a sequence coded into a matrix, and the primary sequence information, easily represented with a one-hot encoding matrix (Tang & Sun, 2019). Very recent reviews on a large number and many types of methods for the discovery of novel pre-miRNAs, in several species, have clearly shown that the best methods for prediction are those based on DL (Bugnon et al., 2021). A comprehensive comparison with experimental results in the human genome has shown that, for the highest class imbalance (1:5,000), DL models have the highest performance (), clearly outperforming other methods based on random forest () and classical support vector machines () (Stegmayer et al., 2019).
In the context of the actual sanitary emergency worldwide, many recent articles indicate ML as the methods that can be employed for battling the COVID-19 by integrating and analyzing heterogeneous types of medical information. A survey on the state-of-the-art of AI and big data for the COVID-19 pandemic emphasizes their speed and importance in responding to the coronavirus outbreak trying to prevent its severe effects (Pham et al., 2020). For example, in Babukarthik et al. (2020) a Genetic Deep Learning Convolutional Neural Network was proposed to classify COVID-19 and normal chest RX images. A comprehensive review of DL approaches based on images for COVID-19 detection has compared several methods, finding that the best ones can reach almost 90% accuracy (Alakus & Turkoglu, 2020). Thus, ML has shown to have the potential to aid in rapid evaluation of medical data, for differentiation of COVID-19 findings from other clinical entities with DL and images (Harmon et al., 2020).
Additionally to methods based on images, several ML methods have helped fighting the virus, for example with Extreme Learning Machines for drugs recommendation and for the estimation of Remdesivir drug behavior on the patient’s treatments; with Long/Short Term Memory models for classifying the best treatment method and for the estimation of cardiac involvement caused by the viral infection; with Generative Adversarial Network for visualization and detection of new human coronaviruses, and for the probability estimation of the process of viral gastrointestinal infection (Jamshidi et al., 2020). Koppu et al. (2020) employed Principal Component Analysis for the extraction of features, which are used within a Deep Belief Network for disease prediction. All of these studies coincide in how ML techniques could help in speeding up research and assisting in the current COVID-19 crisis, and furthermore, how using ML could be a huge advantage in combating various similar viruses in the future.
Differently from the approaches reviewed, and in front of the full lack of knowledge about this novel virus nature and behavior, we propose here the first pipeline to discover new pre-miRNAs in SARS-CoV-2 with deep learning. We believe that deciphering the potential activity of novel miRNAs encoded within the viral genome with ML and hijacking the human transcriptome could help to advance the frontiers of actual strategies for diagnosis and therapeutics. Thus, we have developed a novel ML approach specifically designed for finding pre-miRNAs in the SARS-CoV-2 genome. The pipeline proposed allows the prediction from the full raw genome of the virus, without pre-processing, with deep learning. We present here our proposal in detail, the comparison of several alternative methods for the pipeline, and the results obtained. It has to be highlighted that, remarkably, some candidate pre-miRNAs that were computationally predicted by our pipeline were actually experimentally validated with small RNA-seq data from SARS-CoV-2 infected human cells.
This paper is organized as follows. Section 2 explains in detail the ML-based pipeline designed for finding novel pre-miRNAs in the SARS-CoV-2 genome and the ML models used in this work. In Section 3 the data sets used in this study and the feature extraction process are explained. Section 4 shows the results obtained and their discussion. Finally, the conclusions of this work can be found in Section 5.
2. Identifying novel pre-miRNAs in SARS-CoV-2
2.1. Processing pipeline
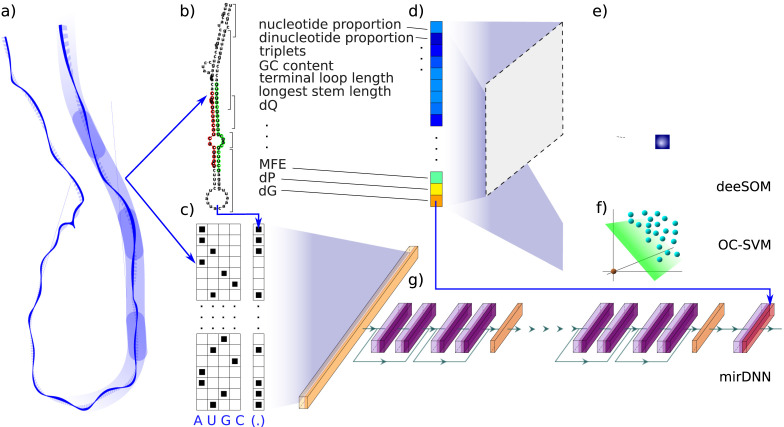
The approach developed based on ML for finding pre-miRNAs within the novel coronavirus genome is shown in Fig. 1. In the first step, the complete genome of the SARS-CoV-2 is cut into small sequences of a fixed length. This genome pre-processing step is crucial because it has a strong influence on the subsequent steps and the final results. For example, with respect to the cutting window length, if it is set arbitrarily, relevant sequences can be lost. If a too-short window length is used, a sequence with hairpin structure could be cut in half, leading to loss of structural features. If a too-long window length is used, many hairpins can be captured inside the same sequence, thus structural features become more complex and much more difficult to recognize by the classifier. These issues were discussed properly in previous works (Bugnon et al., 2019, Yones et al., 2015). Thus, to prevent these adverse influences and to ensure that no important sequences are lost nor inappropriately trimmed, the genome is cut into overlapped segments longer than the mean length of the pre-miRNAs of interest for the species under processing (in this case, viruses). The length of the cutting window has to be configured to define the maximum size that the stem-loops will have (this way shorter stems can also be identified). A stem-loop is a sequence that, once predicted its secondary structure, fulfills certain conditions such as minimum energy released when folding, unpaired nucleotides at the middle (the loop) and a minimum length in the remaining paired nucleotides (the stem). The window must be long enough to correctly include a complete hairpin, as well as to take into account the neighborhood of any possible hairpin when estimating the secondary structure. This is very important since the results of estimating a secondary structure can be greatly affected by the neighborhood of the sequences.
Fig. 1.
Processing pipeline based in machine learning for finding potential pre-miRNAs in the genome of the Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV-2), responsible for the novel coronavirus disease (COVID-19): (a) Genome cut into sequences, (b) Prediction of secondary structure and feature extraction, (c) One-hot encoding of the nucleotides (A, U, G, C) and matches (‘()’) or mismatches (‘.’) in the secondary structure, (d) Input feature vector, (e) The deeSOM classifier, (f) The OC-SVM classifier, (g) The mirDNN model.
The second step consists in the prediction of the secondary structure resulting from the folding of the sequences obtained in the previous windowing and cutting step. This is classically done with the RNAfold tool (Hofacker, 2003), an algorithm that uses dynamic programming for finding the secondary structure which minimizes the energy released. Then, simple representations are used to extract the main features of pre-miRNAs, which are based on the inherent characteristics of the sequences and the secondary structure of these types of molecules. Some typical features are, for example, the nucleotides and dinucleotides proportion, the matching triplets, the GC content, the length of the sequence, the MFE, the frequency of occurrence of certain pairs of nucleotides, among many others. A large number of studies indicate that local sequence features as well as secondary structure are very important for pre-miRNAs identification (Allmer and Yousef, 2012, Li et al., 2009, Liu et al., 2012). The candidate sequences, their secondary structures and the set of extracted features are then used as inputs to a ML classifier, specifically designed for pre-miRNA prediction. These classifiers provide scores according to the likelihood of each RNA sequence of being a pre-miRNA. For this study, we have selected three ML methods: a classical model as baseline and two top-performing and very recently published proposals based on deep learning. These methods have already been validated individually with benchmark data from well-known pre-miRNAs in humans (Bugnon et al., 2021, Yones et al., 2021). Finally, the best model was used for the pre-miRNA predictions in SARS-CoV-2.
2.2. Machine learning models for pre-miRNAs prediction
The ML methods must be trained for identifying RNA sequences highly likely to be miRNA precursors (Stegmayer et al., 2019). Among all possible supervised classifiers, support vector machines (SVM) have been the first and most widely applied algorithm for pre-miRNAs prediction (Xue et al., 2005). A classical supervised approach needs both positive (real well-known pre-miRNA) and negative sequences. In this study, a more recent approach was used, which employs only the positive labeled data for building a classification frontier: the one-class SVM (OC-SVM). It has been shown that this approach outperforms standard two-classes SVM in pre-miRNA prediction because it is capable of learning a decision frontier only from the well-known pre-miRNAs, avoiding the large class imbalance issue (Yousef et al., 2010). Thus, the OC-SVM was trained with features from known viral pre-miRNAs only from miRBase. Then, the fitted model was used on the sequences extracted from the SARS-CoV-2 full-genome.
The second method was the deeSOM model (Bugnon et al., 2020), which consists of several hierarchical layers with self-organizing maps (SOMs). This model has already proven to be very suited to the pre-miRNA prediction task (Bugnon et al., 2021). This model has an ensemble of unsupervised SOMs that are used in parallel at the first level. The unlabeled samples are provided as input data by splitting them among the members of the ensemble, which also receive the full set of positive class cases. This allows to reduce the imbalance at each SOM in the ensemble, each one learning a different unlabeled subspace. At each SOM layer, pre-miRNA neurons are identified as those having, at least, one positive class sample. Only the sequences that are in pre-miRNA neurons pass to the next level. At each level, the map size of each SOM layer is automatically determined by an adaptive algorithm, depending on the number of sequences that arrive from the previous layer. This changes the distribution of samples on each layer, allowing a further depuration of pre-miRNA candidates. Therefore, several deep layers are added with this self-size-adjusting method, until only known pre-miRNA samples remain at the last map. The best candidate sequences are identified as the ones in the pre-miRNA neurons of the last levels. Thus, this model was trained with the features of known pre-miRNAs sequences from other viruses (positive-class samples), 1 million of negative sequences from the human genome, and the features of all the sequences extracted from the full-genome of the novel coronavirus (marked as unlabeled). The largest possible number of structural features available in literature (Yones et al., 2015) were extracted.
Finally, the third method was mirDNN (Yones et al., 2021), a convolutional neural network based on a residual network. This model is trained directly with raw RNA sequences, their corresponding predicted secondary structure and MFE. Thus, the input is represented as a one-hot-encoding tensor of shape , being the maximum sequence length. Each row of the tensor represents the four possible ribonucleotides A, U, G, C and each column represents a position in the sequence. The tensor size is fixed and completed with zero-padding for sequences shorter than . The secondary structure is represented as a tensor of shape , where the value of each element indicates the type of match with the opposite nucleotide. These two tensors are concatenated over the first dimension to form a tensor of shape , which is the input of the model. The first layer of this network is a one-dimensional convolution, followed by stacked identity blocks (He et al., 2016) and pooling layers. The identity blocks allow the model to auto-define the number of convolutional layers needed during training, avoiding optimization of this critical hyperparameter. Each block is composed of two activation functions, two batch normalization layers, and two convolutions. The result is summed up to the input of the next identity block, which helps back-propagate the training error, allowing the addition of convolution layers without bothering the training of the model. After the identity blocks, a pooling layer is used to reduce the length of the sequence by 2. After several of these stages, another tensor is obtained, which is converted into a one-dimensional vector that then passes through activation and batch normalization layers. Then, the input sequence stability, calculated as -MFE/length of the sequence, is appended in order to form a new tensor that feeds a fully connected layer that generates the corresponding output score. For training this model, the focal loss (FL) function (Lin et al., 2020) has been used in order to tackle the high class-imbalance. Usually, when the negative examples (the majority class) are forwarded in the network, they generate an error to be back-propagated through the model whose sum is much larger than the contribution of the (few) positive examples. Thus, the model is heavily biased towards the negative class, meanwhile the positive class is not properly learned. In order to overcome this problem, the FL function can be used to reduce the weight given to the examples easily classified, and increase the weight of the most difficult samples. Therefore, in an imbalanced escenario, the model errors for both the minority (in this case, the positive) class and the unlabeled near the positive class increase in importance to a higher extent than the most obvious negative samples, driving the learning of the network. The mirDNN was trained using the known viral pre-miRNAs as positive class and 1 million of negative sequences from the human genome. After training, the complete genome of the SARS-CoV-2 virus was used for prediction.
3. Data preparation and performance measures
The whole sequence of the novel coronavirus genome was obtained from NCBI Reference Sequence NC_045512.2: Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1. The SARS-CoV-2 genome was segmented with HextractoR (Yones et al., 2020) into 600 nt long fragments with 500 nt overlaps, in order to avoid missing any important structure. The following parameters were used: single loop trimming, minimum sequence length 60, minimum number of base-pairs that must form a sequence 16, and final trimming optimizing the minimum free energy normalized by the sequence length (NMFE). The secondary structure of each segment obtained from the complete genome was predicted by using RNAfold (Hofacker, 2003) with default setting obtaining 597 hairpins in total, which were considered as unlabeled samples. The positive labeled samples (known pre-miRNAs) were downloaded from miRBase v22, retrieving 569 pre-miRNAs of viruses. A total of 73 structural features from the folded sequences of the virus genome and the well-known viruses pre-miRNAs were extracted with miRNAfe (Yones et al., 2015) as in Bugnon et al. (2019), and normalized with z-score. The included features are: length of the sequence, MFE, cumulative size of internal loops found in the secondary structure, number of loops, absolute and relative GC content, among many others (detailed information in Supplementary Material).
For the validation of the computational predictions on the SARS-CoV-2 premiRNAs, expression profiling by high throughput small RNA sequencing (RNA-seq) of the epithelial lung cancer cell line Calu-3 mock and infected with SARS-CoV-2 (USA-WA1/2020) were obtained from NCBI (Reference series GSE148729, made public on May 4th 2020). To identify human genes and cellular pathways influenced by SARS-CoV-2 infection, Singh et al. performed genome-wide measurements of different aspects of gene expression at the bulk and single-cell level. Calu-3 cell line was mock-treated or infected with SARS-CoV-2 and harvested at different time points. Gene expression profiling was determined using bulk and single-cell polyA RNA-seq, small RNA-seq, and total RNA-seq. Alignment files of small RNA-seq samples against SARS-CoV-2 genome were downloaded from NCBI repository. More details on the biological experiment and sequencing technologies used can be found in Singh et al. (2020).
In order to compare the ML methods for the same task of pre-miRNA prediction in virus, they have been tested in a cross-validation scheme with a dataset including all the known pre-miRNAs from viruses (positive samples), and different amounts of hairpin-like sequences (negative samples) from the human genome. The performance was assessed with
where , and are the number of true positives, false positives and false negatives, respectively. The sensitivity or recall () measures how good a classification method is for recognizing (and not missing) the TPs of the problem. The precision () measures the relation between TPs and FPs. In a realistic scenario for practical applications, precision is very important in imbalanced datasets because FPs can be many more than the TPs. Thus, considering the characteristics of the classification problem under study, it is important to take into account both sensitivity and precision. Therefore, was used as a global comparative measure.
Performance curves for the ML methods were drawn using the precision vs recall curve (PRC) plot, since this representation is preferred to assess binary classifiers with imbalanced data. For high imbalances, a classifier can reach a good performance in terms of specificity, but can perform poorly in providing good quality candidates, with a large amount of false positives. Instead, PRC plots can provide a more clear assessment of performance due to the fact that they evaluate the fraction of TPs among the total positive predictions.
4. Results
4.1. Performance of the ML methods with known pre-miRNAS from viruses
Several imbalance ratios (IR) were used to test the models in conditions close to the most common IRs expected in the publicly available viral genomes: 1:50, 1:100, and 1:200. The different IRs were created by maintaining always the same number of positives and varying the number of corresponding negative sequences.
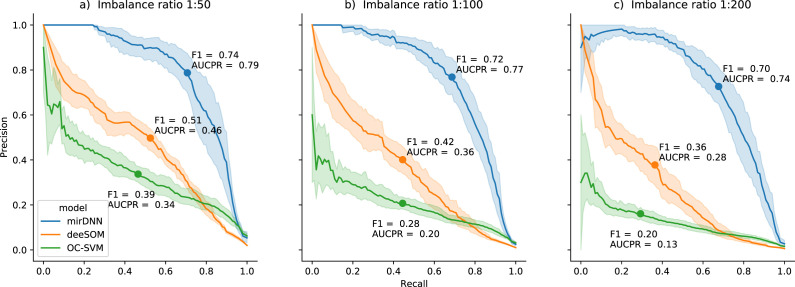
The PRCs of a 10-fold cross-validation of the models presented in Section 2.B, for the different imbalance ratios, are shown in Fig. 2. Each sub-plot in the Figure shows the PRC for each method at a particular IR, the maximum point over it and the area under the precision–recall curve (AUCPR). Results at the IR 1:50, depicted in Fig. 2.(a), indicate that mirDNN reached the best performance with maximum and . The second best method was deeSOM, with and . Then OC-SVM had and , close to deeSOM performance and reaching better results at high recall (and very low precision). For IRs of 1:100 and 1:200, the same general behavior was observed, where AUCPR of deeSOM and OC-SVM was affected by the imbalance (Fig. 2.b and Fig. 2.c, respectively). From the analysis of all three imbalanced situations, it can be seen that the optimal operation point, where is maximum, remains almost the same for mirDNN, with losses between 3%–5% for different imbalances. However, decreases for deeSOM (13%–15%) and especially OC-SVM (30%) as imbalance rises. This study has shown that methods using traditional features, such as OC-SVM and deeSOM, have close performance at low IR and they can be severely affected by imbalance. Also, it has shown that the mirDNN model can provide good accuracy independently of the imbalance.
Fig. 2.
Precision–recall curves for each model and imbalance ratio (a) 1:50, (b)1:100, (c)1:200. Bold line is the mean value, and the shaded area is its standard deviation from cross-validation results. Maximum and AUCPR are indicated for each curve.
4.2. Interpretable predictions of pre-miRNAs in SARS-CoV-2 with deep learning
The identification of potential pre-miRNAs encoded in the SARS-CoV-2 genome was performed with mirDNN, due to its best performance in the comparison of the previous subsection. For each candidate sequence, the method gives a score indicating whether it is a good miRNA precursor candidate (score close to 1) or not (score close to 0). In the case of mirDNN, the activation level of the pre-miRNA output neuron is used as a score.
The experimental validation of these computational predictions involved exploring the read profiles in the regions of the virus genome covered by the predicted precursors in the alignments of small RNA-seq samples (NCBI GSE148729 as described in Section 3). In this analysis, three of the top 5% candidates to pre-miRNAs found by the pipeline proposed in this work have had significant expression in the infected cells and a valid secondary structure in the hairpin (Merino et al., 2020).
The mirDNN model and its scores for the pre-miRNAs candidates with enough expression in infected cells were analyzed in detail. The aim was to find out which parts of those sequences were important for the deep model to give a high score. The importance level of each nucleotide of the sequence for the prediction task was measured as follows. First, each input sequence was evaluated with the already trained mirDNN. This way the prediction output for each input sequence can be considered as the reference score. Then, each nucleotide was masked, one-by-one, by converting the corresponding column to an all-zeros vector. Finally, each masked version of the input sequence was evaluated with the mirDNN, and the difference between the new score and the reference score was used as a measure of the importance level of each nucleotide. This is an important advantage of the mirDNN model over other models: the importance of each individual nucleotide on the output can be characterized, allowing the interpretability of the results. In the following, 2 examples of the attention provided by the deep model to the sequences that have had enough expression are analyzed in detail.
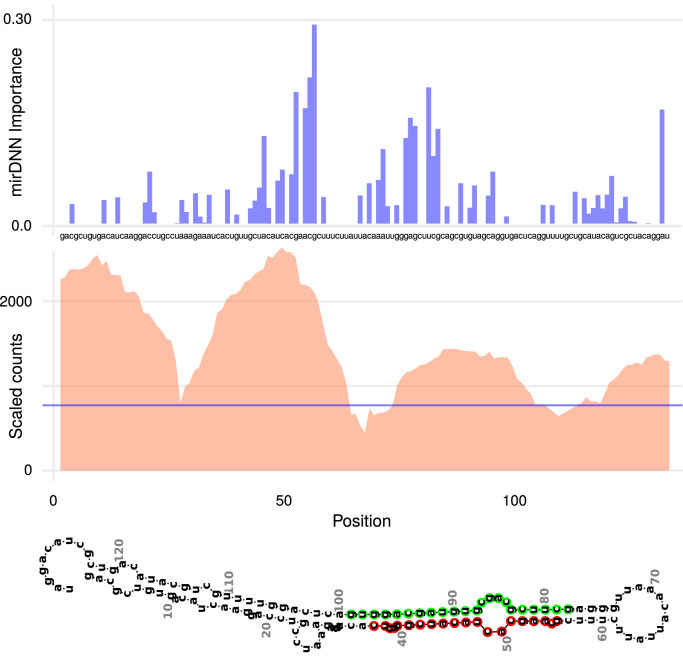
The first example of the coincidence between the importance given by the deep model to the candidate sequence, and the experimental reads, is shown in Fig. 3 for the sars_cov2_26601-27201_stem-392-524 sequence. In the top of this Figure, it is shown the importance that mirDNN has assigned to each nucleotide in the sequence. The vertical axis on the left shows the importance levels of each nucleotide, while the horizontal axis represents the nucleotides position within the sequence. It can be seen here that the deep model has given the highest importance to two sections in the middle of the sequence. Fig. 3 (middle) shows the corresponding experimental reads average (two biological replicates) for this sequence after 24 h upon infection with the virus, with a horizontal blue line indicating the average reads count for the complete genome. It is extremely interesting to see high expression in more than one section of the sequence. There is a high coincidence between the deep model attention zones and the regions of the sequence with high experimental reads. In particular, the highest peak of reads around the position 50 is also the point of maximum importance for the deep model. The zones with high reads, close to the central loop, can be used afterwards for the determination of the mature position. The estimated mature portion is the final miRNA molecule which has a biological function. Fig. 3 (bottom) shows the secondary structure corresponding to this pre-miRNA, with the two mature miRNAs marked in red and green, respectively, according to the importance given by mirDNN and confirmed with the experimental reads.
Fig. 3.
Interpretable prediction of the sars_cov2_26601-27201_stem-392-524 candidate sequence to pre-miRNA in SARS-CoV-2. (top) Importance given by mirDNN to each nucleotide in the sequence. (middle) Experimental reads for this sequence, average of two replicates of the biological experiment. Horizontal blue line indicating the average reads count for the complete genome. (bottom) Corresponding secondary structure predicted using RNAfold, in this case with two possible mature miRNAs marked in red and green, respectively.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
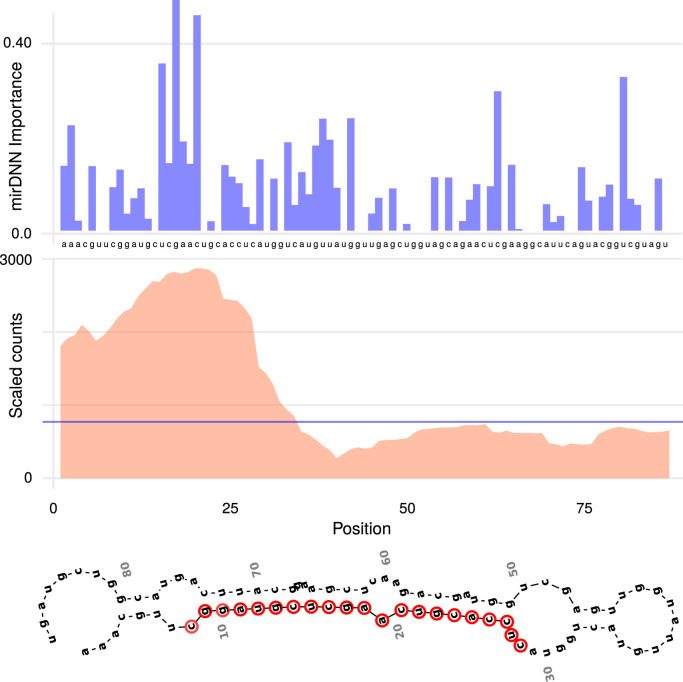
The second example is shown in Fig. 4 for the sars_cov2_101-701_stem-379-465 candidate to pre-miRNA. The top of the Figure presents the importance that mirDNN has assigned to each nucleotide in the sequence. It can be seen here that the deep model has given more importance to the zone around the first 25 nucleotides of the sequence. Fig. 4 (middle) shows the corresponding reads for this sequence. The expression of this predicted pre-miRNA is clearly seen here according to the notable increase of the reads in the infected tissue at 24 h upon infection. Remarkably, the region pointed by the deep model shows high overlap with the portion of the predicted pre-miRNA exhibiting a dynamic production of small RNAs during the virus infection, detected by high-throughput sequencing. There is a high coincidence between most parts of the deep model attention (particularly at the beginning) and the zones of the sequence with high experimental reads. Finally, Fig. 4 (bottom) shows the secondary structure corresponding to this pre-miRNA, with its mature miRNA marked in red, which was determined as in the other case according to the importance given by mirDNN and confirmed with the experimental reads.
Fig. 4.
Interpretable prediction of the sars_cov2_101-701_stem-379-465 candidate sequence to pre-miRNA in SARS-CoV-2. (top) Importance given by mirDNN to each nucleotide in the sequence. (middle) Experimental reads for this sequence, average of two replicates of the biological experiment. Horizontal blue line indicating the average reads count for the complete genome. (bottom) Corresponding secondary structure predicted using RNAfold, with the mature miRNA marked in red.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The next step, once the most likely mature miRNA derived from the pre-miRNA has been determined, it is to predict the corresponding target genes for each of the newly discovered mature miRNAs. After the analysis of the differentially expressed genes and their corresponding functional enrichment, a biological hypothesis could be proposed regarding their regulatory function during the novel coronavirus infection. This will likely contribute to the understanding about how these multiple miRNA-like molecules predicted from the SARS-CoV-2 genome may modulate the host transcriptome upon infection, hopefully helping in the design of innovative strategies for diagnosis and treatment of COVID-19.
5. Discussion
It is important to highlight the advantages of ML models like the ones presented here with respect to the algorithms that would normally be used in this scenario. Generally, the first step when having a new genome for which there is no previous information is to align it against genomes of already known closely-related species, in such a way as to find coding and non-coding areas that are highly similar. To this end, the biological community mainly uses sequence homology search, such as BLAST.2 However, this search approach has two major disadvantages. The first one is that finding a similar sequence by alignment does not necessarily guarantee a relationship with its molecular and biological functions. The second one is that it has a very limited capacity to detect miRNA sequences and precursors with low similarity to the reference set. Sequence homology search does not use any additional information from the sequence, such as features (length, stems number, GC content, number of base-pairs, number of nucleotides in the stem region, etc.), while ML models can capture and learn from them. These disadvantages lead to a loss of generalizability in the search for candidate sequences that have similar molecular functions, but differ substantially in their sequences. Hence, ML algorithms emerged here as an advantageous choice to overcome these limitations, identifying candidates by using features automatically learnt from the training set with deep learning, and classifying according to the values of the features and, moreover, their interactions.
The DL approach proposed here has two main advantages over classical methods: generalization capability and automatic explainability of the predicted sequences. The first one, by automatically extracting inherent characteristics of the data, the model can find pre-miRNAs that are rare and their sequences are different to those of well-known in other species. The second advantage has to do with the capability of analyzing why an algorithm has given a certain score to a sequence. Instead of the match score given by search or other ML methods, our deep model can provide a detailed analysis, nucleotide by nucleotide, for making better global decisions. This allows us to assess the attention given by the model to a certain region of each sequence, in order to determine its most important part. This way, differently from classical ML methods, the possibility of the existence of a biological function in the best-ranked candidate sequences can be hypothesized to be validated and tested with more directed wet-lab trials.
6. Conclusions
In this work we presented a novel approach based on machine learning to uncover potential miRNA precursors hidden in the genome of the SARS-CoV-2, the agent of COVID-19. The pipeline proposed allowed the prediction from the full raw genome of the virus, without pre-processing. Several ML methods were compared inside the pipeline for prediction, and a deep model has shown to be the best one for predicting novel pre-miRNAs. This model is a deep convolutional neural network based on residual learning, which is trained directly with raw RNA sequences, and their corresponding secondary structure. For each input sequence, the model gives a score indicating whether it is a good miRNA precursor candidate or not, and explains the decision providing the importance of each nucleotide in the input sequence. After the prediction, the candidate sequences computationally predicted were also experimentally validated with small RNA-seq data from SARS-CoV-2 infected human cells. In this analysis, 3 of the top 5% candidates to pre-miRNAs found by the pipeline proposed in this work have had significant expression in the infected cells and a valid secondary structure in the hairpin. Therefore, the approach developed allows finding novel pre-miRNAs upon the release of the viral genome sequence, even in the absence of associated transcriptomics of infected cells. This could be of great help in future similar emergencies to accelerate the understanding of the singularities of any viral agent and for the development of novel therapies.
The ML approach proposed here has two main advantages over classical methods: generalization capability and interpretability for the analysis of the predicted sequences. The pipeline allows the automatic extraction of inherent features from the raw sequences, finding shared features more than just exact nucleotides. The deep learning method can perform a very detailed analysis of a sequence, nucleotide by nucleotide, in order to determine its active region with potential biological function. The results demonstrate the usefulness of DL in the context of the current sanitary crisis worldwide. Immediately after genome sequencing, high-confidence pre-miRNAs candidates can be identified to speed up wet-lab trials, and help the fight against the pandemic and the fast development of new treatments. Furthermore, this approach can be replicated to other harmful RNA viruses that have not been fully characterized yet.
CRediT authorship contribution statement
L.A. Bugnon: Software and bioinformatics tools, Writing – review & editing. J. Raad: Software and bioinformatics tools, Writing – review & editing. G.A. Merino: Expression, Data curation, Analysis, Writing – review & editing. C. Yones: Software and bioinformatics tools, Writing – review & editing. F. Ariel: Discussion, Writing – review & editing. D.H. Milone: Conceptualization, Methodology, Figures, Writing – review & editing. G. Stegmayer: Conceptualization, Methodology, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors would like to thank Emanuel Wyler and Prof. M. Landthaler for providing the alignments of the small RNA-seq data of SARS-CoV-1/2 infected human cells, which were used for validating our predictions. We thank NVIDIA Corporation for the donation of the GPUs used for this project. This work was supported by ANPCyT, Argentina (PICT 2018 3384 and PICT 2018 2905) and UNER, Argentina (PID 2019 6204).
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.mlwa.2021.100150.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Structural features from the folded sequences of the virus genome and the well-known viruses pre-miRNAs used for training the ML models.
References
- Alakus T.B., Turkoglu I. Comparison of deep learning approaches to predict covid-19 infection. Chaos, Solitons & Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmer J., Yousef M. Computational methods for ab initio detection of microRNAs. Frontiers in Genetics. 2012;3:209–215. doi: 10.3389/fgene.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babukarthik R.G., Adiga V.A.K., Sambasivam G., Chandramohan D., Amudhavel J. Prediction of covid-19 using genetic deep learning convolutional neural network (gdcnn) IEEE Access. 2020;8:177647–177666. doi: 10.1109/ACCESS.2020.3025164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bugnon L.A., Yones C., Milone D.H., Stegmayer G. Deep neural architectures for highly imbalanced data in bioinformatics. IEEE Transactions on Neural Networks and Learning Systems. 2020;31:2857–2867. doi: 10.1109/tnnls.2019.2914471. [DOI] [PubMed] [Google Scholar]
- Bugnon L.A., Yones C., Milone D.H., Stegmayer G. Genome-wide discovery of pre-miRNAs: comparison of recent approaches based on machine learning. Briefings in Bioinformatics. 2021;22:1–15. doi: 10.1093/bib/bbaa184. [DOI] [PubMed] [Google Scholar]
- Bugnon L., Yones C., Raad J., Milone D., Stegmayer G. Genome-wide hairpins datasets of animals and plants for novel miRNA prediction. Data in Brief. 2019;25 doi: 10.1016/j.dib.2019.104209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtan A.M., Sharp P.A. The role of miRNAs in regulating gene expression networks. Journal of Molecular Biology. 2013;425:3582–3600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. Master regulator analysis of the SARS-CoV-2/human interactome. Journal of Clinical Medicine. 2020;9:982–990. doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haixiang G., Yijing L., Shang J., Mingyun G., Yuanyue H., Bing G. Learning from class-imbalanced data: Review of methods and applications. Expert Systems with Applications. 2017;73:220–239. doi: 10.1016/j.eswa.2016.12.035. [DOI] [Google Scholar]
- Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., Yang D., Myronenko A., Anderson V., Amalou A., Blain M., Kassin M., Long D., Varble N., Walker S.M., Bagci U., Ierardi A.M., Stellato E., Plensich G.G.…Turkbey B. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nature Communications. 2020;11 doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. 2016 IEEE conference on computer vision and pattern recognition (CVPR) 2016. Deep residual learning for image recognition. [DOI] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Research. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T., Rong J., Liu C., Zhang X., Tanriverdi K., Joehanes R., Chen B.H., Murabito J.M., Yao C., Courchesne P., Munson P.J., O’Donnell C.J., Cox N., Johnson A.D., Larson M.G., Levy D., Freedman J.E. Genome-wide identification of microRNA expression quantitative trait loci. Nature Communications. 2015;6:6601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashchenko A., Rakhmetullina A., Aisina D. 2020. How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV, and MERS-CoV. Research Square COVID-19 Preprints. [DOI] [Google Scholar]
- Jamshidi M., Lalbakhsh A., Talla J., Peroutka Z., Hadjilooei F., Lalbakhsh P., Jamshidi M., Spada L.L., Mirmozafari M., Dehghani M., Sabet A., Roshani S., Roshani S., Bayat-Makou N., Mohamadzade B., Malek Z., Jamshidi A., Kiani S., Hashemi-Dezaki H., Mohyuddin W. Artificial intelligence and COVID-19: Deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/access.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Wu H., Wang W., Ma W., Sun X., Lu Z. Mipred: classification of real and pseudo microrna precursors using random forest prediction model with combined features. Nucleic Acids Research. 2007;35:W339–W344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppu S., Maddikunta P.K.R., Srivastava G. Deep learning disease prediction model for use with intelligent robots. Computers and Electrical Engineering. 2020;87 doi: 10.1016/j.compeleceng.2020.106765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Research. 2018;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xu J., Yang D., Tan X., Wang H. Computational approaches for microRNA studies: a review. Mammalian Genome. 2009;21:1–12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- Li X., Zou X. An overview of RNA virus-encoded micrornas. ExRNA. 2019;1:1–10. doi: 10.1186/s41544-019-0037-6. [DOI] [Google Scholar]
- Lin T.-Y., Goyal P., Girshick R., He K., Dollar P. Focal loss for dense object detection. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2020;42:318–327. doi: 10.1109/tpami.2018.2858826. [DOI] [PubMed] [Google Scholar]
- Liu B., Li J., Cairns M.J. Identifying miRNAs, targets and functions. Briefings in Bioinformatics. 2012;15:1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino G.A., Raad J., Bugnon L.A., Yones C., Kamenetzky L., Claus J., Ariel F., Milone D.H., Stegmayer G. Novel SARS-CoV-2 encoded small RNAs in the passage to humans. Bioinformatics. 2020;11 doi: 10.1093/bioinformatics/btaa1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Q.-V., Nguyen D.C., Huynh-The T., Hwang W.-J., Pathirana P.N. Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: A survey on the state-of-the-arts. IEEE Access. 2020;8:130820–130839. doi: 10.1109/access.2020.3009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Reports. 2020;32 doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmayer G., Persia L.E.D., Rubiolo M., Gerard M., Pividori M., Yones C., Bugnon L.A., Rodriguez T., Raad J., Milone D.H. Predicting novel microrna: a comprehensive comparison of machine learning approaches. Briefings in Bioinformatics. 2019;20:1607–1620. doi: 10.1093/bib/bby037. [DOI] [PubMed] [Google Scholar]
- Tang X., Sun Y. Fast and accurate microRNA search using CNN. BMC Bioinformatics. 2019;20:S23. doi: 10.1186/s12859-019-3279-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Liao M., Gao Y., Ji R., He Z., Zou Q. Improved and promising identification of human micrornas by incorporating a high-quality negative set. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2014;11:192–201. doi: 10.1109/TCBB.2013.146. [DOI] [PubMed] [Google Scholar]
- Xue C., Li F., He T., Liu G.-P., Li Y., Zhang X. Classification of real and pseudo microrna precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yones C., Macchiaroli N., Kamenetzky L., Stegmayer G., Milone D. 2020. HextractoR: an r package for automatic extraction of hairpins from genome-wide data. bioRxiv. [DOI] [Google Scholar]
- Yones C., Raad J., Bugnon L., Milone D., Stegmayer G. High precision in microrna prediction: a novel genome-wide approach based on convolutional deep residual networks. Computers in Biology and Medicine. 2021;134 doi: 10.1016/j.compbiomed.2021.104448. [DOI] [PubMed] [Google Scholar]
- Yones C.A., Stegmayer G., Kamenetzky L., Milone D.H. miRNAfe: A comprehensive tool for feature extraction in microRNA prediction. Biosystems. 2015;138:1–5. doi: 10.1016/j.biosystems.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Yousef M., Najami N., Khalifav W. A comparison study between one-class and two-class machine learning for microrna target detection. Journal of Biomedical Science and Engineering. 2010;3:247–252. doi: 10.4236/jbise.2010.33033. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural features from the folded sequences of the virus genome and the well-known viruses pre-miRNAs used for training the ML models.