Abstract
Background
To fully enhance the feature extraction capabilities of deep learning models, so as to accurately diagnose coronavirus disease 2019 (COVID-19) based on chest CT images, a densely connected attention network (DenseANet) was constructed by utilizing the self-attention mechanism in deep learning.
Methods
During the construction of the DenseANet, we not only densely connected attention features within and between the feature extraction blocks with the same scale, but also densely connected attention features with different scales at the end of the deep model, thereby further enhancing the high-order features. In this way, as the depth of the deep model increases, the spatial attention features generated by different layers can be densely connected and gradually transferred to deeper layers. The DenseANet takes CT images of the lung fields segmented by an improved U-Net as inputs and outputs the probability of the patients suffering from COVID-19.
Results
Compared with exiting attention networks, DenseANet can maximize the utilization of self-attention features at different depths in the model. A five-fold cross-validation experiment was performed on a dataset containing 2993 CT scans of 2121 patients, and experiments showed that the DenseANet can effectively locate the lung lesions of patients infected with SARS-CoV-2, and distinguish COVID-19, common pneumonia and normal controls with an average of 96.06% Acc and 0.989 AUC.
Conclusions
The DenseANet we proposed can generate strong attention features and achieve the best diagnosis results. In addition, the proposed method of densely connecting attention features can be easily extended to other advanced deep learning methods to improve their performance in related tasks.
Keywords: Deep learning, Diagnose COVID-19, Chest CT, Self-attention mechanism, Attention features
1. Introduction
Since outbreak in December 2019, coronavirus disease (COVID-19) has spread rapidly around the world [[1], [2], [3]]. As of August 11, 2021, there have been 204,644,849 confirmed cases worldwide, among which 4323,139 people have died [4]. Compared with Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS), although COVID-19 has a lower mortality rate, it spreads more widely, causing more deaths [5]. The average incubation period of SARS-CoV-2 infection is 5.2 days, and during this time, patients will have flu-like symptoms such as fever and cough [6]. The SARS-CoV-2 virus-specific Reverse Transcription Polymerase Chain Reaction (RTPCR) is used clinically as the gold standard for the detection of COVID-19 [7], but RTPCR detection also has many limitations. On the one hand, the RTPCR detection may take up to two days to complete, and in order to reduce the false-negative results of patients, multiple tests need to be performed continuously, which will greatly reduce the efficiency of diagnosing COVID-19. On the other hand, RTPCR detection can only give qualitative analysis to patients, which is not conducive to the treatment of COVID-19 patients. In addition, the study [8] found that the positive rate of RTPCR assay for throat swab samples was 59%, and a number of any external factors may affect RTPCR testing results, including sampling operations, specimen source (upper or lower respiratory tract), sampling timing (different periods of the disease development) and performance of detection kits. Under such circumstances, there is an urgent need for supplementary methods to quickly and accurately diagnose COVID-19 with quantitative analysis.
Computed tomography (CT) examination, as a common method for detecting lung diseases, not only has a faster turnaround time than molecular diagnostic tests performed in standard laboratories, but also can provide more detailed imaging information. Besides, CT examination is more suitable for quantitative measurement of lesion size and lung involvement. Now, researchers find that the lungs of patients infected with SARS-CoV-2 often show consistent CT imaging manifestations, including glass opacities (GGO), multifocal patchy consolidation, and/or interstitial changes with a peripheral distribution [9]. Relying on these CT imaging characteristics, experienced radiologists can accurately identify COVID-19 through chest CT. In addition, chest CT examination has the advantages of fast detection speed and low cost, so it can be used as a supplementary method to detect COVID-19. Therefore, compared with RTPCR detection, chest CT examination is not only faster, but also can give a quantitative analysis of the patient's lung infection, which is more helpful to the patient's follow-up treatment and prognosis. Fang et al. [10] found that the sensitivity of chest CT was greater than that of RTPCR (98% vs 71%, respectively), indicating that CT is very useful in the early detection of suspected COVID-19 cases. However, it takes a lot of time for radiologists to screen CT slices from the entire 3D CT layer by layer to find the lesions. When faced with a large number of CT images, radiologists are prone to fatigue and will reduce the efficiency of diagnosis. So, there is an urgent need to develop an automatic CT-based method to assist radiologists in diagnosing COVID-19.
In recent years, deep learning methods have been widely used in the field of medical image analysis, and have achieved beyond human performance in the classification and segmentation of lesions [[11], [12], [13], [14], [15], [16], [17]]. The chest CT imaging manifestation of COVID-19 patient has strong intra-class variability, and is highly similar to the imaging manifestation of other common pneumonia, as shown in Fig. 1 . So, traditional machine learning methods based on manually extracted features are difficult to extract sufficiently fine features to distinguish COVID-19 from other common pneumonia. While deep learning methods can overcome the limitations of manually extracting features, and can automatically extract distinguishable features from CT images according to task requirements. Therefore, most recent researches [5,6,[18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]] for diagnosing COVID-19 adopt deep learning methods.
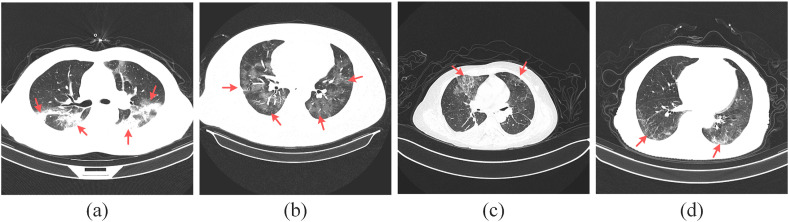
Fig. 1.
Chest CT images of the patients.
In Fig. 1, Fig. 1(a) and Fig. 1(b) are COVID-19, Fig. 1(c) and Fig. 1(d) are common pneumonia, and the red arrows point to the lung lesions. From Fig. 1, we can see that the chest CT images of patients with COVID-19 and common pneumonia are similar, which brings challenges to the research of detecting COVID-19 through CT images.
In the literature [6], the authors used a 2D convolutional neural network (CNN) to segment lung fields and lesions in chest CT images, and input the segmented lesions into a 3D CNN to detect COVID-19 patients. The results showed that their method can distinguish COVID-19 from common pneumonia and healthy controls with an accuracy of 92.49% and an AUC of 0.9797. Wu et al. [20] proposed a weakly-supervised deep active learning framework called COVID-AL to diagnose COVID-19 with CT scans, and experimental results showed that, with only 30% of the labeled data, the COVID-AL achieved over 95% accuracy of the deep learning method using the whole dataset. Chikontwe et al. [21] proposed an attention-based end-to-end weakly supervised framework for the rapid diagnosis of COVID-19 and achieved an overall accuracy of 98.6% and an AUC of 98.4%. Carvalho et al. [22] first proposed a convolutional neural network architecture to extract features from CT images, and then optimized the hyperparameters of the network using a tree Parzen estimator to choose the best parameters. Results showed that their model achieved an AUC of 0.987. Fang et al. [23] proposes a deep classification network model of COVID-19 based on convolution and deconvolution local enhancement. Experimental results showed their method achieved 0.98 sensitivity and 0.97 precision. Li et al. [24] proposed a 3D deep learning model called COVNet to detect COVID-19, and achieved an AUC of 0.96. Mei et al. [25] combined chest CT with clinical symptoms, exposure history and laboratory testing to established an artificial intelligence (AI) model for rapid diagnosis of COVID-19 patients through a 2D CNN and a multilayer perceptron (MLP). In a test set of 279 patients, the AI system achieved an AUC of 0.92. Through analysis, most of the above studies used 2D CT slices to diagnose COVID-19.
Singh et al. [30] used multi-objective differential evolution (MODE) to tune their proposed CNN model, and extensive results showed that their method can classify the chest CT images with a good accuracy. Wang et al. [31] developed a deep learning algorithm to diagnose COVID-19 using chest images, and achieved a total accuracy of 89.5% with a specificity of 0.88 and sensitivity of 0.87. Wang et al. [32] proposed a fully automatic deep learning system for diagnosing COVID-19, and obtained an AUC of 0.87. In the literature [33], a series of artificial intelligence algorithms based on deep learning were used to detect COVID-19, and the experimental results fully demonstrated the superiority of deep learning algorithms in the diagnosis of COVID-19.
In the literature [5], the authors found that there is an imbalance in the size distribution of the infected lung areas between COVID-19 patients and common pneumonia patients. So, they proposed a dual sampling strategy to alleviate the imbalance, and used an attention network to automatically diagnose COVID-19 patients. Results showed they obtained an AUC of 0.944 on the test set of 2057 patients. Xu et al. [34] first used a 3D deep learning model to segment the candidate infection regions, and used a location-attention classification model to classify COVID-19 and viral pneumonia. The results showed that their method achieved an overall accuracy of 86.7%. Li et al. [35] proposed the COVID-CT-DenseNet by using attention mechanism, and achieved 0.82 accuracy. In addition, their method can help overcome the problem of limited training data when using deep learning methods to diagnose COVID-19. Wang et al. [36] proposed a Prior-Attention Residual Learning method to diagnose COVID-19, which is an extension of the attention residual learning [37]. In the literature [36], the author trained two separate 3D networks by using chest CT images, where one branch was used to generate attention maps of lesions to guide the other branch to diagnose COVID-19 more accurately. Experimental results showed that the attention framework they proposed can significantly improve the performance of screening COVID-19. It can be seen from the above studies that the attention mechanism has been widely used in the detection of COVID-19 based on chest CT, but there is still a problem of not effectively using attention features. In literatures [5,36], attention features are only generated inside the feature extraction block, not between blocks or between features on different scales, resulting in incomplete focus areas learned by the model.
Although deep learning methods are widely used in all aspects of medical image analysis, there are still many challenges in using chest CT images to detect COVID-19 through deep learning methods. First of all, because chest CT is 3-dimensional (3D), and limited by computer memory and calculation speed, it is difficult to design a sufficiently deep 3D deep learning model to fully utilize 3D CT information through a 3D CNN. However, the ability of the shallow model to extract image features is weak, resulting in poor diagnosis effects of the shallow model in diagnosing COVID-19. Second, because there are a large number of non-lesion areas in the lungs of patients, the method of detecting COVID-19 in actual clinical practice is based on the accurate segmentation of the lesions.
In this paper, in order to design a deep learning model with strong feature extraction capabilities and eliminate lesion segmentation process, a 2D CNN-based densely connected attention network (DenseANet) was constructed. Our proposed DenseANet takes 2D chest CT slices as input and can realize accurate diagnosis of COVID-19 without the need to finely segment lesion areas. Although the 2D CNN may lose part of the spatial information compared to the 3D CNN, it still contains enough features that can be used to distinguish different types of pneumonia. The radiologists also examine the 2D slices when reading the CT, and then combine the results of each slice to make the final diagnosis. Therefore, only the 2D slice of the CT image is sufficient to detect COVID-19. In literatures [19,25], the authors also constructed a 2D CNN model by using 2D slice of the CT image to diagnose COVID-19. In addition, the literature [38] showed that the model constructed using 2D CNNs can prevent overfitting compared to the model constructed using 3D CNNs. Besides, compared with the 3D CNN, the 2D CNN can greatly reduce the number of model parameters. That is, in the same computing environment, we can use 2D CNN to design a deeper network and extract higher-order features that are more conducive to classification.
In order to avoid losing the image characteristics of the lung lesions of COVID-19 patients, we only segmented the lung fields, and used the segmented lung CT images as the input of DenseANet. Inspired by the design idea of DenseNet [39], on the basis of our previous work [40], we generated a series of attention features in the construction of the deep model, and densely connected these attention features to enhance the feature extraction capabilities of the model. In summary, our contributions have the following three aspects:
-
1)
We proposed a densely connected attention block (DAB) as the basic feature extraction block of CNN, this novel densely connected form of attention features can enhance the model's ability to extract features of different pneumonia types, thereby improving the model's diagnostic effect.
-
2)
The novel DAB block was introduced into U-Net, and an Attention U-Net model (A-U-Net) for semantic segmentation was constructed to achieve accurate segmentation of lung fields. We performed a five-fold cross-validation on a segmentation dataset containing 750 2D CT slices of 150 COVID-19 patients, and the results showed that the average Dice coefficient of A-U-Net for lung field segmentation was 0.980, and the average pixel accuracy (PA) was 0.987.
-
3)
A densely connected attention network (DenseANet) was constructed based on the DAB, which can maximize the use of attention features at different depths and scales. The proposed DenseANet takes the CT image of the segmented lung parenchymal region as input and can achieve accurate detection of COVID-19. We performed a five-fold cross-validation on a dataset containing 860 CT scans of 534 COVID-19 patients, and obtained an average of 96.06% Acc and 0.989 AUC. The results show that the DenseANet can improve the effect of diagnosing COVID-19 with only a few additional parameters.
2. Materials and methods
2.1. Materials
The dataset used in our research comes from China National Center for Bioinformation (CNCB). The dataset of the CT images and clinical metadata are constructed from cohorts from the China Consortium of Chest CT Image Investigation (CC-CCII), which consists of Sun Yat-sen Memorial Hospital and Third Affiliated Hospital of Sun Yat-sen University, The first Affiliated Hospital of Anhui Medical University, West China Hospital, Nanjing Renmin Hospital, Yichang Central People's Hospital, Renmin Hospital of Wuhan University. All CT images are classified into novel coronavirus pneumonia (NCP) due to SARS-CoV-2 virus infection, common pneumonia (CP) and normal controls (Normal). In the dataset, NCP annotation was given when a patient had pneumonia with a confirmed reverse-transcriptase-PCR, and CP includes viral pneumonia, bacterial pneumonia and mycoplasma pneumonia.
The dataset provides the locations of lesions in patients with NCP and CP. At the same time, we took 15 consecutive 2D slices from the whole CT of each healthy normal person in the middle position. Our experiment is based on these 2D slices, that is, only 2D slices are actually used to train and test the constructed DenseANet. The detailed data distribution of the dataset was shown in Table 1 . Besides, we also downloaded a dataset for lung field segmentation from CNCB. The segmentation dataset contains 750 chest CT 2D slices of 150 cases and the masks corresponding to the lung field regions.
Table 1.
The data distribution.
| Types | Num. of patients | Num. of CT scans | Num. of 2D slices |
|---|---|---|---|
| CP | 738 | 1055 | 36894 |
| NCP | 534 | 860 | 21872 |
| Normal | 849 | 1078 | 16170 |
| Total | 2121 | 2993 | 74936 |
2.2. The overall workflow
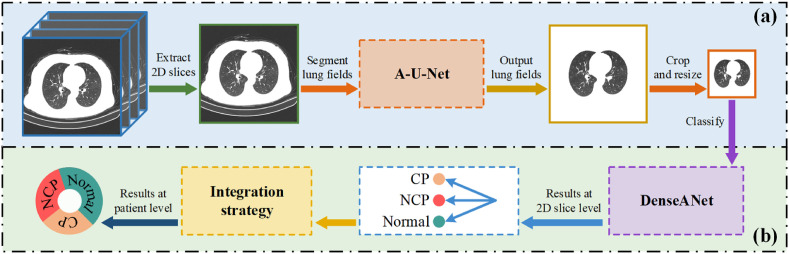
In this paper, based on our previous work [40], firstly, a densely connected attention block (DAB) was proposed. Secondly, we replaced the convolutional layer in U-Net with the DAB to construct an Attention U-Net segmentation model (A-U-Net) to segment lung fields in chest CT images. Finally, the DAB blocks were used to construct the DenseANet, which can utilize 2D CT images of segmented lung field regions to detect COVID-19. Since the DenseANet outputs 2D slice-level detection results, we also explored methods of generating patient-level diagnosis results using 2D results through mean strategy and voting strategy. Fig. 2 showed the overall workflow for diagnosing COVID-19 in our work.
Fig. 2.
The overall workflow for diagnosing COVID-19.
Fig. 2(a) represents data processing, and its main purpose is to generate lung CT images that only contain lung parenchymal regions. Fig. 2(b) represents the diagnosis process of COVID-19. The A-U-Net for segmentation and DenseANet for classification in Fig. 2 will be described in detail later.
2.3. Densely connected attention block
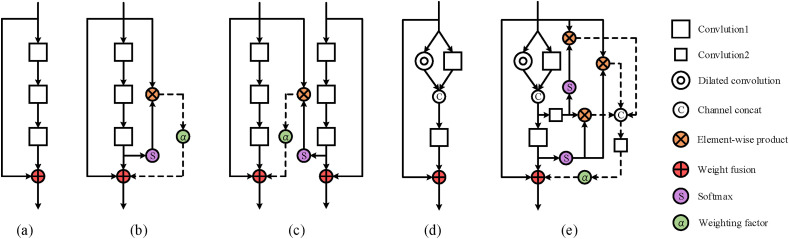
Since having excellent classification performance, ResNet [41] has been widely used in various tasks of image processing. ResNet is a residual structure that can use skip connections to alleviate the problem of gradient disappearance. Using the residual structure, researchers can design deeper deep learning models to improve the performance of the model on corresponding tasks. The residual attention block is based on the feature extraction block in the original ResNet, and has gradually derived a variety of forms. Fig. 3 shows some structures of residual attention blocks.
Fig. 3.
The residual attention blocks.
Fig. 3(a) is the feature extraction block in ResNet, and it does not contain any attention features. Fig. 3(b) is the attention residual learning block proposed in the literature [37], which generates attention features through higher-level features inside the block, and the generated attention features act on the output of the block. Therefore, the attention residual learning block is a kind of self-attention mechanism. Fig. 3(c) is the Prior-Attention block proposed in the literature [36], which provides attention features to the backbone network through a branch network, so the Prior-Attention block is not a self-attention mechanism. Fig. 3(d) is the feature extraction block proposed in our previous work [40]. It was verified in the literature [40] that the ability of Fig. 3(d) to extract features is better than that of Fig. 3(a). Therefore, we introduced the self-attention mechanism to the structure shown in Fig. 3(d) and constructed the DAB (Fig. 3(e)), where the expansion coefficient of dilated convolution was set to 2. In the DAB, a convolution kernel with a size of 3 × 3 was used to extract features, and a convolution kernel with a size of 1 × 1 was used to change the number of feature maps. In this way, attention feature maps at different depths can be merged.
Different from the attention residual learning block (Fig. 3(b)), all internal higher-level features in the DAB (Fig. 3(e)) will correct the low-level features in the spatial domain to generate attention features. These attention features in DAB are first spliced in the channel domain to obtain dense attention features f, then f is subjected to a 1 × 1 convolution to perform feature selection to obtain selected feature maps, and finally the selected feature maps are weighted and added to the output of this DAB. The mathematical expressions of above blocks in Fig. 3 are as follows:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Equations (1), (2), (3), (4), (5) correspond to the mathematical expressions of Fig. 3(a)–3(e), where y represents the output of the residual attention block, the function represents the mapping of the backbone path learned by the stacked layers in a block. The function represents the mapping of the branch path learned by the stacked layer in the Prior-Attention module, and the function represents the mapping of the main path learned by the stacked layer in Fig. 3(d). represents the result obtained through channel splicing in Fig. 3(d), and the weight coefficient α reflects the importance of the attention feature in the feature extraction block. represents the convolution operation with a size of 1 × 1 convolution kernel, and the function indicates that the feature map is spliced in the channel dimension. represents the softmax operation in the spatial domain, as shown in Eq. (7).
| (7) |
where represents the feature map with a size of w×h and a channel number of n.
2.4. A-U-Net
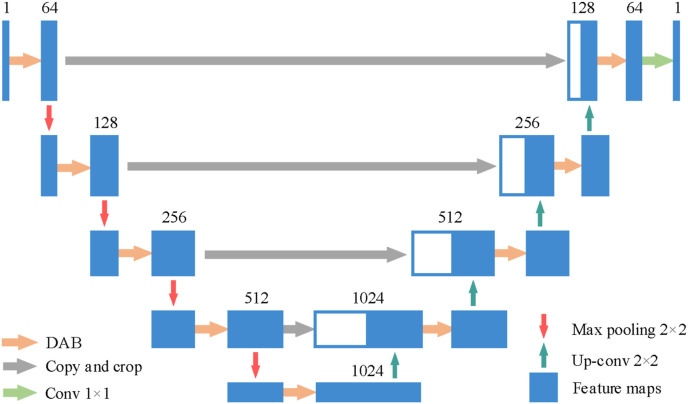
Nowadays, researchers have proposed various semantic segmentation models, including U-Net [42], FCN [43], RDUNET [44], SegNet [45] and DeepLabv3+ [46], UNet++ [47] etc. Since the skip connection in U-Net can combine low-resolution information and high-resolution information in a model, U-Net is conducive to accurate segmentation of object edges. On the basis of U-Net, using the DAB proposed in section 2.3, we constructed an A-U-Net to segment the lung field regions of the chest CT images. The structure of A-U-Net was shown in Fig. 4 .
Fig. 4.
The structure of the A-U-Net.
The number above the feature maps represented by the blue rectangular blocks in Fig. 4 represents the number of feature maps in different deep layers. We replaced the two convolutional layers in the original U-Net with the DAB on the same scale, and added the dropout [48] operation to DAB (drop rate = 0.85). Table 2 showed the detailed structure of the A-U-Net.
Table 2.
The detailed structure of the A-U-Net.
| Layer name | Output size | Layer name | Output size |
|---|---|---|---|
| Input | 512 × 512 × 1 | Up-conv | 64 × 64 × 512 |
| DAB | 512 × 512 × 64 | DAB | 64 × 64 × 512 |
| Max pooling | 256 × 256 × 64 | Up-conv | 128 × 128 × 256 |
| DAB | 256 × 256 × 128 | DAB | 128 × 128 × 256 |
| Max pooling | 128 × 128 × 128 | Up-conv | 256 × 256 × 128 |
| DAB | 128 × 128 × 256 | DAB | 256 × 256 × 128 |
| Max pooling | 64 × 64 × 256 | Up-conv | 512 × 512 × 64 |
| DAB | 64 × 64 × 512 | DAB | 512 × 512 × 64 |
| Max pooling | 32 × 32 × 512 | Conv | 512 × 512 × 1 |
| DAB | 32 × 32 × 1024 |
In the A-U-Net, except for the activation function of the last layer which is softmax, others are ReLU, which is defined in Eq. (8), and the softmax activation function is in the form of channel domain, which is defined in Eq. (9).
| (8) |
| (9) |
2.5. DenseANet
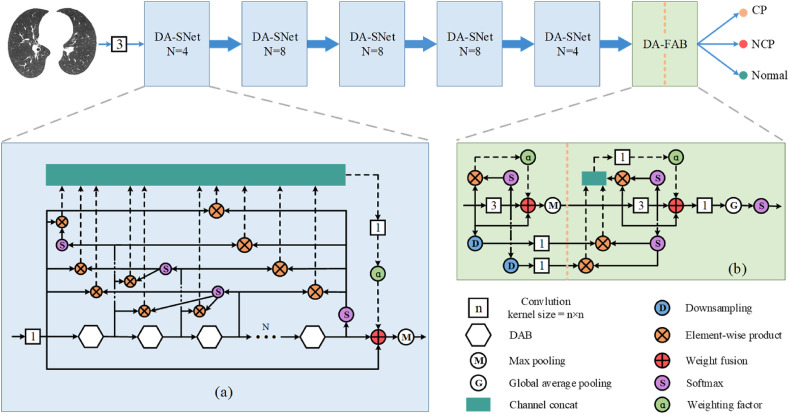
The overall framework of the DenseANet was shown in Fig. 5 . The inputs of the DenseANet are 2D CT images of segmented lung field regions, which are first operated by a 3 × 3 convolution kernel to extract primary features. After that, these primary features are sequentially passed through five densely connected attention sub-networks (DA-SNets) with different scales and a densely connected attention feature aggregation block (DA-FAB) to finally get the outputs of the DenseANet. The structures of the DA-SNet and DA-FAB were shown in Fig. 5(a) and (b), respectively.
Fig. 5.
The framework of the DenseANet.
As shown in Fig. 5(a), the backbone of the DA-SNet is in the form of residual structure, which is composed of N DAB stacked. The sizes of all attention feature maps inside a DA-SNet are the same, and the size of the feature maps are halved after a DA-SNet. Inside the DA-SNet, after the output of the later DAB passes through the softmax in the spatial domain, it is conducted element-wise product with the outputs of all previous DABs to generate attention features. All these attention features will be superimposed in the channel dimension. Finally, the Densely connected Attention Features (DAFs) are obtained. Then, these DAFs are selected through the 1 × 1 convolution kernel operation, and then the selected DAFs are weighted and added to the output of the last DAB. There are two ways to determine the weight coefficients α. One way to determine α is to be learned by the network as learnable parameters, and the other way is to be manually specified as hyperparameters. How to choose and determine α will be analyzed in the experimental part.
Generally speaking, the more abstract the features extracted in the deeper layers of the network, the more specific the location of lung lesions can be located. Therefore, in order to make full use of advanced abstract features, we added the DA-FAB after the output of the last DA-SNet, as shown in Fig. 5(b). The biggest difference between DA-FAB and DA-SNet is that the attention features densely connected in DA-FAB are of different scales.
In Fig. 5(b), the features of the previous layer are reduced to the same size as the latter features by downsampling, and then the down sampled features are selected through the 1 × 1 convolution kernel operation, and finally the selected down sampled features are conducted element-wise product with the latter features to generate attention features. These attention features are first spliced with the later attention features in the channel domain to generate DAFs, and then DAFs are selected through a 1 × 1 convolution kernel operation, and then the selected DAFs are weighted and finally added to the output of the second 3 × 3 convolution operation. In Fig. 5(b), the last 1 × 1 convolution layer will reduce the number of feature maps to the same as the number of categories, playing the role of a fully connected layer. Then, the output of DenseANet is obtained after global average pooling and softmax in the channel domain. The detailed structure of the entire DenseANet was shown in Table 3 .
Table 3.
The structure of the DenseANet.
| Layer name | Num. of DAB | Output size |
|---|---|---|
| Input | – | 224 × 224 × 1 |
| Convolution | – | 224 × 224 × 16 |
| DA-SNet | 4 | 112 × 112 × 16 |
| DA-SNet | 8 | 56 × 56 × 32 |
| DA-SNet | 8 | 28 × 28 × 64 |
| DA-SNet | 8 | 14 × 14 × 128 |
| DA-SNet | 4 | 7 × 7 × 256 |
| DA-FAB | – | 3 |
As can be seen from Table 3, the number of convolution layers in the backbone network of DenseANet is approximately the same as the number of convolution layers in ResNet101 [41], therefore, we will compare DenseANet with ResNet101 and its variants in the following experiment part.
In addition, to accurately detect COVID-19 patients, the diagnosis results need to be obtained at the patient-level rather than the single 2D slice-level of the chest CT. Therefore, after obtaining the detection results of all 2D slices of a patient, an integration strategy needs to be adopted to gather the detection results of all 2D slices together to generate patient-level diagnosis result. Here, we adopted two integrated strategies to generate patient-level diagnosis results. One is the average strategy, which averages the results of all 2D slices as the patient's diagnosis result, and the integrated strategy was given by Eq. (10). The other is the voting strategy, that is, the most frequently occurring result among all the 2D slice results of one patient is selected as the patient's diagnosis result, and this strategy was given by Eq. (11). Assuming that the detection results of 2D slices belong to one-hot form, represents the detection results of 2D slices, s represents the number of 2D slices in the chest CT.
| (10) |
| (11) |
| (12) |
where represents the value that appears most frequently in the set . In the experimental part, we conducted an experimental comparison of these two integrated strategies, and selected the strategy that has the best effect in diagnosing COVID-19 at the patient-level as the final integrated strategy.
3. Results
In this paper, in order to verify our DenseANet, a five-fold cross-validation was performed on a dataset containing 2993 chest CT scans of 2121 patients. The purpose of DenseANet is to accurately diagnose NCP, CP and Normal, therefore, the DenseANet is essentially a three-classification prediction model.
3.1. Evaluation metrics
In order to evaluate the segmentation performance of A-U-Net, we used Dice coefficient (Dice) and pixel accuracy (PA) as evaluation metrics. And to evaluate the classification performance of these models, Accuracy (Acc), Sensitivity (Se), Specificity (Sp), Precision (Pr), F1 score (F1) and area under the receiver operating characteristic curve (AUC) were used as evaluation metrics. The above metrics were defined as follows:
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
where TP, TN, FP, FN represent the number of true positive, true negative, false positive, false negative, respectively. is the indicator function which returns 1 if the argument is true and 0 otherwise. represents the nonlinear mapping of DenseANet from inputs to outputs, and , , , represent positive samples, negative samples, the number of positive samples and the number of negative samples, respectively. It should be noted that, unless otherwise specified, all evaluation metrics of a certain category are obtained by calculating this category and the other two categories.
3.2. Experimental analysis of the A-U-Net
The dataset used for lung field segmentation is composed of five 2D slices of each patient's chest CT, so 150 patients contain a total of 750 2D slices. The resolution of the input images to the A-U-Net model is 512 × 512, before entering the model, we first standardized the data according to Eq. (20).
| (20) |
where is the mean value of the image, and is the standard deviation of the input image.
The implement of the entire A-U-Net is based on the TensorFlow [49], and dual NVIDIA GTX1080ti GPUs (11 GB) were used to train and test the segmentation model. We used the “Xavier” algorithm to initialize A-U-Net, and used the Adam algorithm to update the parameters of A-U-Net. The initial learning rate was set to 3 × 10−4, the batch size was set to 8, and the epochs was set to 100. It should be pointed out that in the A-U-Net, we set the weight coefficient in DAB to 1, and used the cross-entropy loss function to train the A-U-Net model. The cross-entropy (CE) loss function was defined as follows:
| (21) |
where m represents the number of samples in a mini-batch, n represents the number of categories, represents ground truth (one-hot), represents the output of the A-U-Net, represents the input of the A-U-Net, and θ represents all parameters of the A-U-Net.
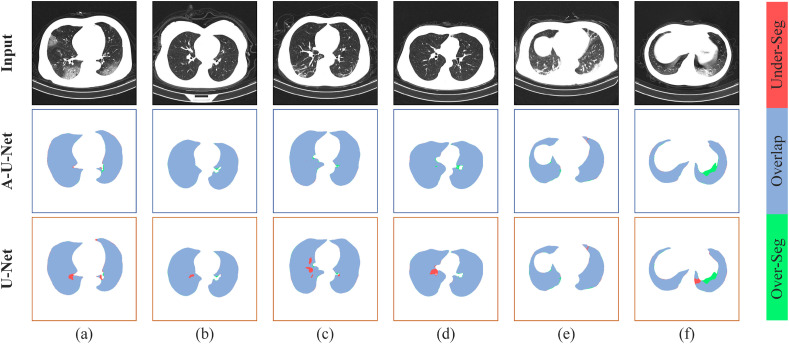
When using the A-U-Net to segment the lung field, in order to prevent data leakage, we divided the dataset at the patient-level. After the five-fold cross-validation, the average Dice coefficient of the segmentation results obtained by A-U-Net is 0.980 and the average pixel accuracy is 0.987, while the average Dice coefficient of the segmentation result obtained by original U-Net is 0.971 and the average PA is 0.980. Fig. 6 showed some segmented results of lung fields obtained by U-Net and A-U-Net.
Fig. 6.
Segmentation results of lung field. The top row showed the segmentation results of A-U-Net and the bottom row showed the segmentation results of U-Net. Blue area showed the overlap between the predicted result and the true result, red area showed the under-segmentation area and green area showed the over-segmentation area.
In Fig. 6, Fig. 6(a)–6(d) showed the segmentation results of lung fields under normal conditions, Fig. 6(e) and Fig. 6(f) showed the segmentation results under extreme conditions. It can be seen from Fig. 6 that even in extreme cases, A-U-Net can accurately segment lung field regions in chest CT. From the comparison of the segmentation results of U-Net and A-U-Net, we can see that A-U-Net can ensure the integrity of the segmented lung parenchyma, that is, the segmentation result of A-U-Net has a higher recall. Besides, the overall segmentation effect of A-U-Net is better than U-Net, which not only provides a reliable basis for utilizing CT images of lung field areas to diagnose Covid-19, but also proves the effectiveness of the DAB we proposed.
3.3. Experimental analysis of the DenseANet
3.3.1. Experiment setups and data preprocessing
In the process of analyzing the dataset, we found that the areas of the segmented lung fields are concentrated in the middle of the image, and the resolution of the CT images in the dataset is 512 × 512. Therefore, in order to reduce computational load, we crop the segmented images to a size of 448 × 448 at the center, and then downsample them to the resolution of 224 × 224. It should be noted that mixup method [50] was used for data enhancement. Specifically, we randomly mixed two images () in a certain ratio to generate a mixed image (), and mixed the corresponding labels () with one-hot form to obtain the mixed label () of , as shown in Eq. (22) and Eq. (23). Actually, the DenseANet was trained through and .
| (22) |
| (23) |
where λ obeys the beta distribution with parameters (β, β) in a batch, in our experiment, we set β = 2.
We performed a five-fold cross-validation on the patient-level, and randomly flip the input images left and right to enhance data during training. The “Xavier” algorithm was used to initialize the DenseANet, the Adam algorithm was used to update parameters of the DenseANet. The initial learning rate was set to 1 × 10−4, the batch size was set to 64, the epoch was set to 100, and the cross-entropy loss function was used to train the DenseANet. The entire DenseANet was built based on the TensorFlow deep learning framework, and dual NVIDIA GTX1080ti GPUs (11 GB) were used for model training and testing. Noted that the parameter size of the DenseANet model is 361 MB, and the diagnosis result of a 2D slice can be obtained in 0.04 s on the above-mentioned experimental platform.
3.3.2. Weight coefficient α
There are weight coefficients in both DAB and DA-SNet (between DABs), the weight coefficients in DAB are defined as , and the weight coefficients between DABs in the DA-SNet are defined as . In the literature [37], is obtained by training the network as a learnable parameter, the range of which is between (0, 2). In our experiment, we also tried the method in the literature [37] to automatically learn and through training the network. But we found that if and were used as learnable parameters, as the number of model training increases, and did not converge, which made the training results of the entire model poor. So, in our experiment, we took and as hyperparameters to train the constructed DenseANet model.
Specifically, given , firstly, was fixed to 0, was initialized to 0, and an ablation experiment was performed for every 0.1. When the accuracy of the model on the validation set reaches the highest, the at this time is selected as the final . Secondly, was fixed to , was initialized to 0, and another ablation experiment was performed for every 0.1. When the accuracy of the model on the validation set reaches the highest, the at this time is selected as the final . After a lot of experiments, was finally determined to 1.2 and was determined to 1.0, respectively. After a five-fold cross-validation, we found that the results in patient-level generated by the average strategy are better than that generated by the voting strategy in any classification metrics. Therefore, all patient-level results in the following were generated by the average strategy.
3.3.3. Experimental results
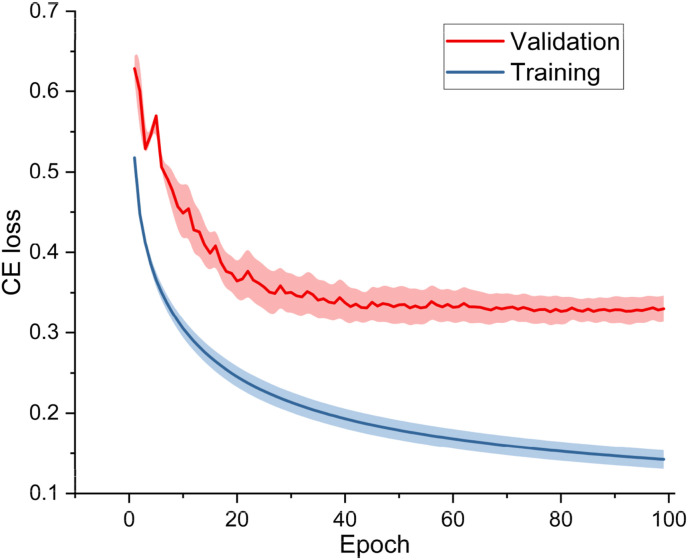
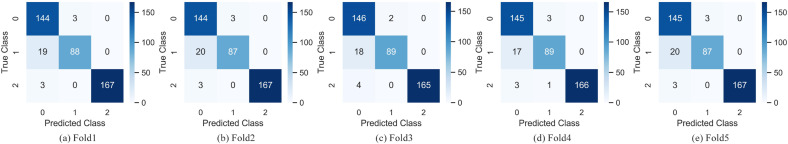
Since the number of convolution operations in the backbone network of the DenseANet is roughly the same as the number of that in ResNet101, and the backbone networks of the two networks are all residual structures, so, we introduced the structure in Fig. 3(b) into ResNet101 to construct a ResNet101+ARL model for comparing. At the same time, we combined ARL with the backbone network (Baseline) we proposed in the literature [40] to construct a Baseline+ARL model. In the experiments, we used ResNet101, ResNet101+ARL, Baseline, Baseline+ARL and DenseANet five networks to conduct five-fold cross-validations. The training/validation curves and confusion matrixes of the DenseANet in the five-fold cross-validation experiment were shown in Fig. 7 and Fig. 8 , respectively.
Fig. 7.
The training and validation curves of DenseANet.
Fig. 8.
The confusion matrixes of the DenseANet. 0 represents CP, 1 represents NCP and 2 represents Normal.
In order to quantitatively analyze the DenseANet, we showed all results with a 95% confidence interval of the above five models at the 2D slice-level and the patient-level in Table 4 .
Table 4.
Detailed results (%) with a 95% confidence interval of five models. The numbers in bold black font represent the best classification result in the corresponding category. Note that the p-values are calculated between DenseANet and Baseline+ARL.
| Models | Category | Acc | AUC | Se | Sp | Pr | F1 | |
|---|---|---|---|---|---|---|---|---|
| Results of 2D slices | ResNet101 | CP | 91.84 ± 1.21 | 97.64 ± 0.47 | 91.84 ± 1.05 | 91.85 ± 1.23 | 92.00 ± 1.37 | 91.72 ± 0.95 |
| NCP | 92.63 ± 0.92 | 97.80 ± 0.43 | 88.04 ± 1.58 | 94.54 ± 0.74 | 86.98 ± 1.74 | 87.31 ± 1.49 | ||
| Normal | 98.89 ± 0.27 | 99.71 ± 0.11 | 96.59 ± 0.37 | 99.47 ± 0.19 | 97.89 ± 0.55 | 97.03 ± 0.46 | ||
| ResNet101+ARL | CP | 92.99 ± 1.04 | 97.96 ± 0.35 | 91.72 ± 1.70 | 94.28 ± 0.76 | 94.25 ± 0.62 | 92.66 ± 0.82 | |
| NCP | 93.63 ± 1.19 | 98.01 ± 0.23 | 92.39 ± 1.21 | 94.14 ± 0.45 | 86.73 ± 1.29 | 89.37 ± 1.66 | ||
| Normal | 99.08 ± 0.21 | 99.47 ± 0.17 | 96.33 ± 0.85 | 99.77 ± 0.08 | 99.06 ± 0.16 | 97.48 ± 0.58 | ||
| Baseline | CP | 94.07 ± 0.87 | 98.28 ± 0.28 | 97.09 ± 0.65 | 90.99 ± 1.58 | 91.67 ± 1.07 | 94.03 ± 0.89 | |
| NCP | 94.90 ± 0.49 | 98.23 ± 0.31 | 86.61 ± 1.18 | 98.34 ± 0.31 | 95.59 ± 0.65 | 90.65 ± 1.07 | ||
| Normal | 98.95 ± 0.22 | 99.63 ± 0.14 | 96.81 ± 0.64 | 99.49 ± 0.18 | 97.95 ± 0.52 | 97.14 ± 0.41 | ||
| Baseline+ARL | CP | 93.50 ± 0.89 | 98.35 ± 0.20 | 92.08 ± 1.22 | 94.95 ± 0.53 | 94.91 ± 0.54 | 93.27 ± 0.73 | |
| NCP | 94.04 ± 0.72 | 98.40 ± 0.21 | 92.96 ± 0.97 | 94.48 ± 0.62 | 87.48 ± 1.17 | 90.04 ± 1.25 | ||
| Normal | 99.03 ± 0.14 | 99.73 ± 0.07 | 96.78 ± 0.52 | 99.60 ± 0.29 | 98.39 ± 0.23 | 97.35 ± 0.46 | ||
| DenseANet | CP | 94.81 ± 0.39 | 98.38 ± 0.24 | 98.25 ± 0.21 | 91.02 ± 0.85 | 93.07 ± 0.71 | 94.66 ± 0.56 | |
| NCP | 95.67 ± 0.32 | 98.61 ± 0.12 | 87.19 ± 1.68 | 98.61 ± 0.33 | 95.83 ± 0.65 | 91.41 ± 0.71 | ||
| Normal | 98.87 ± 0.16 | 99.57 ± 0.14 | 95.44 ± 0.81 | 99.74 ± 0.13 | 99.76 ± 0.14 | 97.53 ± 0.39 | ||
| p-value | CP | 0.003 | 0.742 | 6.35e-7 | 1.08e-4 | 0.002 | 0.007 | |
| NCP | 0.007 | 0.461 | 2.56e-5 | 5.38e-7 | 1.08e-7 | 0.012 | ||
| Normal |
0.477 |
0.215 |
0.035 |
0.269 |
0.014 |
0.083 |
||
| Results of patients | ResNet101 | CP | 91.05 ± 1.48 | 97.07 ± 0.52 | 90.55 ± 1.17 | 91.32 ± 1.19 | 84.65 ± 2.09 | 87.25 ± 2.17 |
| NCP | 92.77 ± 1.09 | 97.63 ± 0.64 | 83.43 ± 1.88 | 96.60 ± 0.63 | 90.97 ± 1.32 | 86.91 ± 2.06 | ||
| Normal | 98.28 ± 0.33 | 99.56 ± 0.13 | 97.63 ± 0.52 | 98.65 ± 0.38 | 97.63 ± 0.44 | 97.47 ± 0.42 | ||
| ResNet101+ARL | CP | 92.08 ± 0.96 | 97.55 ± 0.51 | 91.54 ± 1.29 | 92.37 ± 1.24 | 86.38 ± 2.17 | 88.65 ± 1.85 | |
| NCP | 93.80 ± 0.84 | 97.90 ± 0.32 | 86.39 ± 1.14 | 96.84 ± 0.58 | 91.82 ± 1.17 | 88.92 ± 1.75 | ||
| Normal | 98.28 ± 0.27 | 99.71 ± 0.15 | 97.16 ± 0.43 | 98.92 ± 0.39 | 98.09 ± 0.36 | 97.41 ± 0.49 | ||
| Baseline | CP | 92.08 ± 1.21 | 98.12 ± 0.35 | 96.02 ± 0.61 | 90.00 ± 1.23 | 83.55 ± 1.75 | 89.15 ± 1.25 | |
| NCP | 93.63 ± 1.05 | 98.43 ± 0.47 | 80.47 ± 2.03 | 99.03 ± 0.20 | 97.14 ± 0.49 | 87.96 ± 1.19 | ||
| Normal | 98.45 ± 0.24 | 99.53 ± 0.28 | 97.63 ± 0.34 | 98.92 ± 0.14 | 98.10 ± 0.38 | 97.64 ± 0.42 | ||
| Baseline+ARL | CP | 93.12 ± 0.71 | 97.80 ± 0.46 | 90.55 ± 0.91 | 94.47 ± 0.90 | 89.66 ± 1.26 | 90.04 ± 1.08 | |
| NCP | 93.80 ± 0.94 | 98.08 ± 0.32 | 88.17 ± 0.97 | 96.12 ± 0.57 | 90.30 ± 1.19 | 89.13 ± 1.12 | ||
| Normal | 98.62 ± 0.33 | 99.64 ± 0.09 | 98.58 ± 0.21 | 98.65 ± 0.17 | 97.65 ± 0.32 | 97.91 ± 0.63 | ||
| DenseANet | CP | 94.11 ± 0.25 | 98.81 ± 0.17 | 98.10 ± 0.33 | 91.99 ± 0.32 | 86.68 ± 0.50 | 92.03 ± 0.32 | |
| NCP | 94.86 ± 0.33 | 98.13 ± 0.11 | 82.40 ± 1.28 | 99.06 ± 0.24 | 96.71 ± 0.82 | 88.98 ± 0.76 | ||
| Normal | 99.20 ± 0.14 | 99.74 ± 0.08 | 98.00 ± 0.36 | 100.00 ± 0.00 | 100.00 ± 0.00 | 98.99 ± 0.18 | ||
| p-value | CP | 0.012 | 0.003 | 4.95e-7 | 1.75e-4 | 0.001 | 0.003 | |
| NCP | 0.004 | 0.098 | 9.17e-6 | 1.68e-5 | 9.22e-8 | 0.218 | ||
| Normal | 0.021 | 0.273 | 0.047 | 0.013 | 0.004 | 0.016 |
From Table 4, we can see that the classification effect of the DenseANet is significantly better than that of the other four models, indicating that the densely connected attention mechanism is effective. From Table 4, we can also see that the classification effect of Baseline is better than ResNet101, which is consistent with the conclusion of our previous work [40]. From the results of the Baseline, Baseline+ARL and DenseANet models, it can be seen that as the number of attention features in the model increases, the better the classification effect of the mode. This shows that using more attention features can improve the model's ability to classify chest CT images with different disease types, which is consistent with our starting point for designing the DenseANet model.
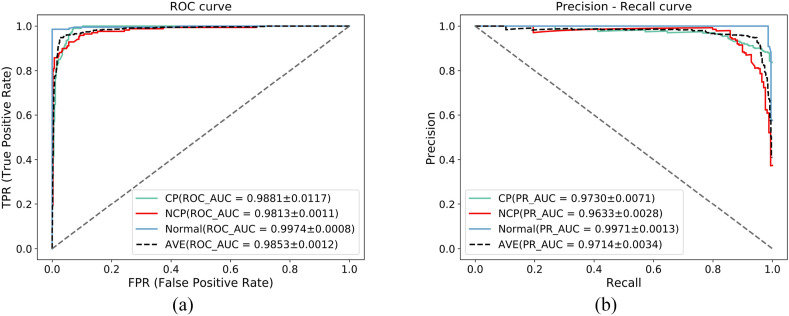
In addition, we also showed the average receiver operating characteristic (ROC) curves and precision-recall (PR) curves of the five-fold cross-validation of the DenseANet, as shown in Fig. 9 .
Fig. 9.
AUC curves and PR curves.
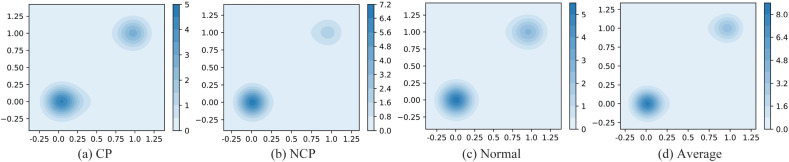
In order to further demonstrate the classification effect of the DenseANet on the test set, we plotted the kernel density estimation (KDE) diagram of the test results, as shown in Fig. 10 . Fig. 10(a) represents the KDE of the prediction results of the CP, Fig. 10 (b) represents the KDE of the prediction results of the NCP, Fig. 10(c) represents the KDE of the prediction results of the Normal, and Fig. 10(d) represents the KDE of the prediction results of the average type.
Fig. 10.
KDE of the probability values output by the DenseANet.
From Fig. 10 we can see that the output probability of the DenseANet are mostly concentrated around 0 or 1, indicating that DenseANet can accurately distinguish various types of pneumonia patients with a high degree of confidence.
In addition, well known the distinction between normal controls and any kind of pneumonia is relatively easy, while the distinction between novel coronavirus pneumonia (NCP) and common pneumonia (CP) is not. In order to further verify the performance of DenseANet, we also calculated various evaluation metrics between CP and NCP. Since the output of the model is the three-classification probability, we first excluded the Normal samples in the test set, and got the preliminary result R 1 (three-classification probability) through the model. Then the two-classification results (R 2) of CP and NCP was obtained by Eq. (25), and the detailed results of the two-classification between CP and NCP was shown in Table 5 .
| (24) |
| (25) |
Table 5.
Diagnosis results (%) of the DenseANet on the CP and NCP. The numbers in bold black font represent the best classification results. Note that the p-values are calculated between DenseANet and Baseline+ARL.
| Models | Acc | AUC | Se | Sp | Pr | F1 | |
|---|---|---|---|---|---|---|---|
| Results of 2D slices | ResNet101 | 88.65 ± 1.95 | 95.50 ± 0.61 | 93.03 ± 0.93 | 83.43 ± 1.47 | 86.98 ± 1.98 | 89.75 ± 1.35 |
| ResNet101+ARL | 90.00 ± 1.01 | 96.06 ± 0.52 | 93.03 ± 0.89 | 86.39 ± 1.93 | 89.05 ± 1.24 | 90.91 ± 1.16 | |
| Baseline | 90.00 ± 1.13 | 97.13 ± 0.49 | 98.01 ± 0.30 | 80.47 ± 2.64 | 85.65 ± 2.47 | 91.24 ± 0.95 | |
| Baseline+ARL | 90.54 ± 1.37 | 96.75 ± 0.56 | 92.04 ± 1.02 | 88.76 ± 1.53 | 90.69 ± 1.17 | 91.19 ± 1.09 | |
| DenseANet | 91.28 ± 0.87 | 97.24 ± 0.41 | 98.18 ± 0.24 | 82.33 ± 1.97 | 86.85 ± 1.47 | 92.03 ± 0.69 | |
|
p-value |
0.014 |
0.034 |
1.68e-5 |
8.61e-7 |
2.38e-4 |
0.002 |
|
| Results of patients | ResNet101 | 90.88 ± 1.28 | 97.02 ± 0.49 | 92.45 ± 0.82 | 88.15 ± 1.17 | 93.08 ± 0.88 | 92.58 ± 0.75 |
| ResNet101+ARL | 92.13 ± 0.89 | 97.41 ± 0.47 | 91.97 ± 0.97 | 92.43 ± 1.02 | 95.45 ± 0.47 | 93.81 ± 0.62 | |
| Baseline | 93.70 ± 0.77 | 97.72 ± 0.56 | 97.78 ± 0.66 | 86.67 ± 1.34 | 92.67 ± 0.91 | 95.07 ± 0.74 | |
| Baseline+ARL | 92.73 ± 0.93 | 97.98 ± 0.38 | 92.56 ± 1.04 | 93.04 ± 0.85 | 95.82 ± 0.58 | 93.94 ± 0.98 | |
| DenseANet | 94.67 ± 0.55 | 98.41 ± 0.27 | 98.31 ± 0.27 | 87.15 ± 1.16 | 93.47 ± 0.62 | 95.43 ± 0.38 | |
| p-value | 4e-4 | 0.015 | 1.33e-10 | 4.75e-7 | 0.001 | 0.002 |
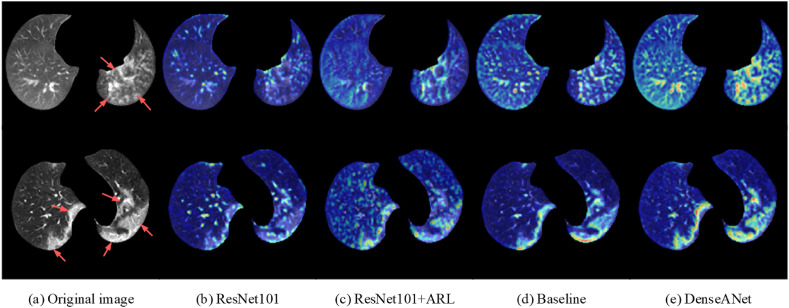
In addition, in order to show the classification effect of the DenseANet more intuitively and make the diagnosis results of the DenseANet more convincing, we used class activation mapping (CAM) [51] to visualize the attention regions in the lung fields, as shown in Fig. 11 .
Fig. 11.
Visualization of the attention regions.
In Fig. 11, Fig. 11(a) are original chest CT images, Fig. 11(b)–11(e) are images obtained by superimposing CAMs of each deep model on corresponding CT images, and red arrows point to the area of lung lesions. The top row represents CP and the bottom row represents NCP, respectively. In Fig. 11(b)–11(e), the redder the color on the image indicates that the model payed more attention to this area, that is, the model considers this area to be a lesion area worthy of attention.
From the CAM maps, we can see that the DenseANet can locate lung lesions more accurately than the other four models, so DenseANet can diagnose COVID-19 with a higher accuracy. From the comparison of Fig. 11(b) and (c) or from the comparison of Fig. 11(d) and (e), we can see that the introduction of the self-attention mechanism can indeed enhance the model's ability to locate lung lesions, leading the model to make more accurate judgments.
4. Discussions
4.1. Analysis of the DenseANet architecture
The core of A-U-Net for lung field segmentation and DenseANet for diagnosing COVID-19 are all the densely connected form of attention features. The obvious difference between our proposed method and existing attention models is the deep fusion of attention features at different depths and scales. From the structure of the model, DenseANet can obtain more potential discriminative representations by densely connecting the attention features of different depths and different scales. Compared with the self-attention model in the literature [37], DenseANet contains more attention features of different levels, both in the number and types of attention features. Due to the ability to use more attention features, compared with the other four models, DenseANet has the best classification accuracy in diagnosing COVID-19. More importantly, the form of densely connected attention features we proposed is a general paradigm that can be easily integrated into various existing deep learning models.
4.2. Result analysis of the DenseANet
To prove the advantages of our proposed DenseANet, we listed results of some models on the same data set in Table 4. From Table 4, we can see that the attention mechanism can indeed improve the classification ability of the model, which can support the argument in literature [37]. From the comparison of DenseANet and Baseline+ARL model, we can see that densely connected attention features can improve the performance of a model more than common attention features, which further prove the advantage of our proposed dense connection of attention features. So, from the whole results in Table 4, we can see that the densely connected form of the attention features at different depths and different scales is the reason for the higher performance of the proposed DenseANet.
Fig. 10 showed the statistical distribution of the output results of the DenseANet model on the test set, and it can be seen that DenseANet can output the correct diagnosis result with a higher degree of confidence. At the same time, in order to reveal the feature representation ability of the DenseANet, we used CAMs to visualize attention regions in the lung field. The visualized results showed that the DenseANet can accurately locate the location of lung lesions, and then made accurate classification judgments, thus supporting the excellent performance of the DenseANet to a certain extent.
Besides, to further demonstrate the effectiveness of our DenseANet in diagnosing COVID-19, we compared the DenseANet with COVID-AL [20], COVIDNet-CT [28] and COVID-CT-DenseNet [35] based on the same dataset. Table 6 showed the experimental results of the above four models at the patient level.
Table 6.
Diagnosis results (%) of the DenseANet, COVID-AL, COVID-CT-DenseNet and COVIDNet-CT. The numbers in bold black font represent the best classification result in the corresponding category. And the numbers in italic font indicate that the p-value (between DenseANet and other corresponding models) is greater than 0.05.
| Models | Category | Acc | AUC | Se | Sp | Pr | F1 |
|---|---|---|---|---|---|---|---|
| COVID-AL | CP | 92.27 ± 0.43 | 95.66 ± 0.49 | 95.12 ± 0.61 | 90.74 ± 0.71 | 84.59 ± 1.03 | 89.54 ± 0.55 |
| NCP | 92.93 ± 0.41 | 97.39 ± 0.33 | 78.47 ± 1.55 | 97.79 ± 0.01 | 92.29 ± 0.12 | 84.81 ± 0.96 | |
| Normal | 96.70 ± 0.32 | 98.73 ± 0.28 | 95.17 ± 0.55 | 97.72 ± 0.47 | 96.54 ± 0.68 | 95.85 ± 0.39 | |
| COVID-CT-DenseNet | CP | 93.21 ± 0.56 | 97.46 ± 0.31 | 96.48 ± 0.62 | 91.47 ± 0.71 | 85.79 ± 1.10 | 90.82 ± 0.75 |
| NCP | 93.82 ± 0.50 | 97.19 ± 0.23 | 80.34 ± 1.56 | 98.36 ± 0.16 | 94.28 ± 0.58 | 86.75 ± 1.11 | |
| Normal | 97.69 ± 0.34 | 98.85 ± 0.13 | 96.35 ± 0.55 | 98.58 ± 0.39 | 97.85 ± 0.57 | 97.09 ± 0.42 | |
| COVIDNet-CT | CP | 93.54 ± 0.51 | 96.53 ± 0.49 | 97.16 ± 0.81 | 91.61 ± 0.58 | 86.13 ± 0.93 | 91.29 ± 0.66 |
| NCP | 94.15 ± 0.85 | 97.39 ± 0.33 | 80.84 ± 2.44 | 98.67 ± 0.38 | 95.39 ± 1.30 | 87.50 ± 1.74 | |
| Normal | 98.40 ± 0.59 | 98.73 ± 0.28 | 97.05 ± 0.79 | 99.29 ± 0.47 | 98.92 ± 0.72 | 97.98 ± 0.74 | |
| DenseANet | CP | 94.11 ± 0.25 | 98.81 ± 0.17 | 98.10 ± 0.33 | 91.99 ± 0.32 | 86.68 ± 0.50 | 92.03 ± 0.32 |
| NCP | 94.86 ± 0.33 | 98.13 ± 0.11 | 82.40 ± 1.28 | 99.06 ± 0.24 | 96.71 ± 0.82 | 88.98 ± 0.76 | |
| Normal | 99.20 ± 0.14 | 99.74 ± 0.08 | 98.00 ± 0.36 | 100.00 ± 0.00 | 100.00 ± 0.00 | 98.99 ± 0.18 |
It can be seen from Table 6 that DenseANet has achieved the best results in each category, which fully proved the superiority of our method. In addition, in order to better prove the effectiveness and the generalization of the DenseANet in diagnosing COVID-19, we tested the previously trained DenseANet and the other models on the COVID-CT-Dataset [52], which contains 349 COVID-19 CT images and 463 non-COVID-19 CT images. The best result of each model was shown in Table 7 .
Table 7.
Diagnosis results of the DenseANet on the COVID-CT-Dataset. The numbers in bold black font represent the best classification results.
| Models | Acc | AUC | Se | Sp | Pr | F1 |
|---|---|---|---|---|---|---|
| ResNet101 | 0.8633 | 0.9274 | 0.8797 | 0.8510 | 0.8165 | 0.8469 |
| ResNet101+ARL | 0.8793 | 0.9416 | 0.8968 | 0.8661 | 0.8347 | 0.8646 |
| Baseline | 0.8903 | 0.9473 | 0.9083 | 0.8769 | 0.8476 | 0.8769 |
| Baseline + ARL | 0.8941 | 0.9607 | 0.9112 | 0.8812 | 0.8525 | 0.8809 |
| COVID-AL | 0.8670 | 0.9318 | 0.8739 | 0.8618 | 0.8266 | 0.8496 |
| COVID-CT-DenseNet | 0.8879 | 0.9475 | 0.9026 | 0.8769 | 0.8468 | 0.8738 |
| COVIDNet-CT | 0.8916 | 0.9514 | 0.9083 | 0.8791 | 0.8499 | 0.8781 |
| DenseANet | 0.9027 | 0.9564 | 0.9226 | 0.8877 | 0.8610 | 0.8907 |
It can be seen from Table 7 that even if we performed cross-testing between two completely different data sets, DenseANet still has a higher accuracy than other models in identifying COVID-19, which can indicate that DenseANet has strong generalization ability and universal applicability.
In our whole experiment, we also adopted the method in the literature [6] to establish a 3D classification model, but the classification effect is poor. Through further observation of the visualization method, we found that the 3D model payed more attention to the edge of the lung fields, rather than the whole lung fields. The reason for the above problem is that there is not sufficient data to train the model, since only part of the data in the literature [6] can be obtained. Therefore, the amount of patient-level data available to us is small, which makes it impossible to train a 3D prediction model with excellent classification effect.
4.3. Disadvantages of the DenseANet
Regarding the determination of the weight coefficient within the feature extraction block (DAB) and the weight coefficient between the DABs, in our experiment, we regarded and as two independent variables, without considering the relationship between and . In addition, the weight coefficients and in different depths and stages in the DenseANet should not be completely the same. However, there are 39 weight coefficients in the entire DenseANet model, if these parameters are used as different hyperparameters to be manually specified and determined through experiments, it is also unrealistic. Therefore, we treated the hyperparameters inside the DAB as the same, and treat the hyperparameters between the DABs as the same. From a methodology point of view, the biggest limitation of our proposed method is to manually determine the weight coefficients of the attention features. And from the perspective of diagnosing COVID-19, the biggest limitation of our proposed method is the inability to use complete 3D CT information.
4.4. The future work
Considering the method of determining and , a better approach is to use a grid search algorithm to comprehensively consider the relationship between and . However, the grid search algorithm will greatly increase the training times. In the case of a step size of 0.1, the grid search algorithm will require 400 retraining of the model, which greatly increases the computational cost and is unrealistic. In future work, we will search a more effective method to determine and more reasonably. In order to further solve the problem of determining and in different depth of the DenseANet model, we can try to divide the network into three different parts, namely the primary feature extraction part, the intermediate feature extraction part and the advanced feature extraction part. Under this experimental configuration, the and in these three parts will be determined separately, so that the model only has 6 different weight coefficients. We believe that this approach will make the model's diagnosis results more accurate. In addition, we further plan to track the CT images and treatment plans of patients during the treatment phase to carry out researches on evaluating the treatment effects of COVID-19 patients.
5. Conclusions
During the COVID-19 pandemic, the sooner patients with COVID-19 are identified, not only can the patients be treated in time, but also the transmission risk of COVID-19 can be effectively reduced. At present, although RTPCR detection is the gold standard for detecting COVID-19, since has long detection cycle and the disadvantage of can not give a quantitative analysis of patients, RTPCR detection will also be limited in the diagnosis of COVID-19. In this paper, a densely connected attention network (DenseANet) was constructed by utilizing chest CT images to accurately diagnose COVID-19. All experimental results show that our DenseANet has a better effect in diagnosing CIVID-19 than some existing models. The cross-validation experiment between different data sets show that the DenseANet model has a high robustness, which can provide solid reliability for the actual clinical application of the DenseANet model.
In clinic, the proposed DenseANet model can be used as a supplementary method for RTPCR detection. Especially in the current situation of Delta variant (a new variant of COVID-19) with high infection rate and long incubation period spreading, and in the case of long time of RTPCR detection and the false negatives that may easily exist in one RTPCR detection, our proposed DenseANet method can provide the quantitative analysis results of the examinee in time through lung CT images, which is helpful for the screening cases with COVID-19 in the early stage of the patients.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported in part by the Fundamental Research Funds for the Central Universities (China), in part by the National Natural Science Foundation of China under Grant 62171261, 81671848 and 81371635, in part by the Key Research and Development Project of Shandong Province under Grant 2019GGX101022.
References
- 1.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi H., Han X., Jiang N., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song F., Shi N., Shan F., et al. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1):210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/
- 5.Ouyang X., Huo J., Xia L., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imag. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 6.Zhang K., Liu X., Shen J., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zu Z., Jiang M., Xu P., et al. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296(2):15–25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai T., Yang Z., Hou H., et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):32–40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung M., Bernheim A., Mei X., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y., Zhang H., Xie J., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):115–117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue P., Dong E., Ji H. Lung 4D CT image registration based on high-order markov random field. IEEE Trans. Med. Imag. 2020;39(4):910–921. doi: 10.1109/TMI.2019.2937458. [DOI] [PubMed] [Google Scholar]
- 12.Lou M., Wang R., Qi Y., et al. MGBN: convolutional neural networks for automated benign and malignant breast masses classification. Multimed. Tool. Appl. 2021;81(17):26731–26750. [Google Scholar]
- 13.Fu Y., Xue P., Ren M., et al. Harmony loss for unbalanced prediction. IEEE J. Biomed. Health Inform. Early Acc. 2021 doi: 10.1109/JBHI.2021.3094578. [DOI] [PubMed] [Google Scholar]
- 14.Fu Y., Xue P., Li N., et al. Fusion of 3D lung CT and serum biomarkers for diagnosis of multiple pathological types on pulmonary nodules. Comput. Methods Progr. Biomed. 2021;210 doi: 10.1016/j.cmpb.2021.106381. [DOI] [PubMed] [Google Scholar]
- 15.Fabijańska A. Automatic segmentation of corneal endothelial cells from microscopy images. Biomed. Signal Process Contr. 2019;47:145–158. [Google Scholar]
- 16.Forestier G., Wemmert C. Semi-supervised learning using multiple clusterings with limited labeled data. Inf. Sci. 2016;361:48–65. [Google Scholar]
- 17.Liu Z., Hauskrecht M. Clinical time series prediction: toward a hierarchical dynamical system framework. Artif. Intell. Med. 2015;65(1):5–18. doi: 10.1016/j.artmed.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang H., Xia L., Yan F., et al. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imag. 2020;39(8):2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 19.Bai H., Wang R., Xiong Z., et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296(2):156–165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X., Chen C., Zhong M., et al. COVID-AL: the diagnosis of COVID-19 with deep active learning. Med. Image Anal. 2021;68 doi: 10.1016/j.media.2020.101913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chikontwe P., Luna M., Kang M., et al. Dual attention multiple instance learning with unsupervised complementary loss for COVID-19 screening. Med. Image Anal. 2021;72 doi: 10.1016/j.media.2021.102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho E., Silva E., Araújo F., et al. An approach to the classification of COVID-19 based on CT scans using convolutional features and genetic algorithms. Comput. Biol. Med. 2021 doi: 10.1016/j.compbiomed.2021.104744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang L., Wang X. COVID-19 deep classification network based on convolution and deconvolution local enhancement. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Qin L., Xu Y., et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):65–71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei X., Lee H.-C., Diao K-y, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Z., Wei B., Hong Y., et al. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imag. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Deng X., Fu Q., et al. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imag. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 28.Gunraj H., Wang L., Wong A. COVIDNet-CT: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest CT images. Front. Med. 2020;7 doi: 10.3389/fmed.2020.608525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia G., Lam H.-K., Xu Y. Classification of COVID-19 chest X-Ray and CT images using a type of dynamic CNN modification method. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh D., Kumar V., Vaishali, et al. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., Kang B., Ma J., et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19) Eur. Radiol. 2021;31:6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X., Jiang X., Ma C., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S., Zha Y., Li W., et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmon S., Sanford T., Turkbey B. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Zhang J., Li B., et al. COVID-19 diagnosis on CT scan images using a generative adversarial network and concatenated feature pyramid network with an attention mechanism. Med. Phys. 2021 doi: 10.1002/mp.15044. Early Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J., Bao Y., Wen Y., et al. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imag. 2020;39(8):2572–2583. doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Xie Y., Xia Y., et al. Attention residual learning for skin lesion classification. IEEE Trans. Med. Imag. 2019;38(9):2092–2103. doi: 10.1109/TMI.2019.2893944. [DOI] [PubMed] [Google Scholar]
- 38.Liu L., Dou Q., Chen H., et al. Multi-task deep model with margin ranking loss for lung nodule analysis. IEEE Trans. Med. Imag. 2020;39(3):718–728. doi: 10.1109/TMI.2019.2934577. [DOI] [PubMed] [Google Scholar]
- 39.Huang G., Liu Z., Maaten L., et al. Densely connected convolutional networks. IEEE Conf Comput Vis Pattern Recogn. 2017:2261–2269. [Google Scholar]
- 40.Fu Y., Xue P., Ji H., et al. Deep model with Siamese network for viable and necrotic tumor regions assessment in osteosarcoma. Med. Phys. 2020;47(10):4895–4905. doi: 10.1002/mp.14397. [DOI] [PubMed] [Google Scholar]
- 41.He K., Zhang X., Ren S., et al. Deep residual learning for image recognition. IEEE Conf Comput Vis Pattern Recogn. 2016:770–778. doi: 10.1109/CVPR.2016.90. [DOI] [Google Scholar]
- 42.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention (MICCAI), 9351. 10.1007/978-3-319-24574-4_28. [DOI]
- 43.Shelhamer E., Long J., Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(4):640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 44.Devalla S., Renukanand P., Sreedhar B., et al. DRUNET: a dilated-residual U-Net deep learning network to segment optic nerve head tissues in optical coherence tomography images. Biomed. Opt Express. 2018;9(7):3244–3265. doi: 10.1364/BOE.9.003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badrinarayanan V., Kendall A., Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 46.Liang C., Yukun Z., Papandreou G., et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. Computer vision - ECCV. 15th European conference. Proc. Lect. Notes Comput. Sci. 2018:833–851. [Google Scholar]
- 47.Zhou Z., Siddiquee M., Tajbakhsh N., et al. UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imag. 2020;39(6):1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinton G., Srivastava N., Krizhevsky A., et al. 2012. Improving Neural Networks by Preventing Co-adaptation of Feature Detectors. arXiv: 1207. [Google Scholar]
- 49.Abadi M., Barham P., Chen J., et al. 2016. Tensorflow: A System for Large-Scale Machine Learning. arXiv:1605. [Google Scholar]
- 50.Zhang H., Cisse M., Dauphin Y., et al. 2017. Mixup: beyond Empirical Risk Minimization. arXiv: 1710. [Google Scholar]
- 51.Zhou B., Khosla A., Lapedriza A., et al. Learning deep features for discriminative localization. IEEE Conf Comput Vis Pattern Recogn. 2016:2921–2929. [Google Scholar]
- 52.Yang X., He X., Zhao J., et al. 2020. COVID-CT-Dataset: A CT Image Dataset about COVID-19. arXiv:2003.13865vol. 3. [Google Scholar]