Abstract
The discovery of novel pathogenic mechanisms engaged during bacterial infections requires the evolution of advanced techniques. Here, we evaluate the dual polarity matrix norharmane (NRM) to improve detection of bacterial lipid A (endotoxin), from host and vector tissues infected withFrancisella novicida (Fn). We evaluated NRM for improved detection and characterization of a wide range of lipids in both positive and negative polarities, including lipid A and phospholipids across a range of matrix-assisted laser desorption-ionization-coupled applications. NRM matrix improved the limit of detection (LOD) for monophosphoryl lipid A (MPLA) down to picogram level representing a 10-fold improvement of LOD versus 2,5-dihydroxybenzoic acid and 100-fold improvement of LOD versus 9-aminoacridine (9-AA). Improved LOD for lipid A subsequently facilitated detection of theFn lipid A major ion (m/z 1665) from extracts of infected mouse spleen and the temperature-modifiedFn lipid A atm/z 1637 from infectedDermacentor variabilis ticks. Finally, we simultaneously mapped bacterial phospholipid signatures within anFn-infected spleen along with an exclusively host-derived inositol-based phospholipid (m/z 933) demonstrating coprofiling of the host-pathogen interaction. Expanded use of NRM matrix in other infection models and endotoxin-targeting imaging experiments will improve our understanding of the lipid interactions at the host-pathogen interface.
Keywords: mass spectrometry imaging, host-pathogen interaction, endotoxin
This study presents a versatile tool for host-pathogen interaction studies by mass spectrometry imaging; we emphasize the improved detection of pathogen-specific lipids (endotoxin) achievable only with norharmane (NRM), an underutilized matrix substance facilitating simultaneous detection of host and pathogen lipids.
INTRODUCTION
Lipopolysaccharide (LPS) is the major component of the outer leaflet of the outer membrane of most Gram-negative bacteria. The membrane anchor region, lipid A, imparts endotoxin activity to LPS with specific lipid A structural configurations indicative of antimicrobial resistance (commonly terminal phosphate modifications) or local growth conditions (e.g. acyl shortening observed in growth at low temperature) (Gunn2001; Liet al.2012; Needham and Trent2013). Lipid A is substantially hydrophobic and is readily ionizable in the negative-ion mode due to the terminal phosphate moieties. Matrix-assisted laser desorption-ionization (MALDI) is commonly used for detection of diverse lipid A structures, but detection can be problematically matrix dependent. In order to study low abundance and exotic lipid A structures, improved detection methods are necessary (Heeren2015). Due to the relationship between lipid A structure and virulence,in situ description of lipid A and its structure is crucial (Gunn2001; Hajjaret al.2002; Hagaret al.2013; Pelletieret al.2013). Here we will describe an underutilized matrix for sensitive detection of lipid A including modified lipid A structures directly from infected vector and host extracts.
Norharmane (NRM; β-carboline, 9H-pyrido[3,4-b]indole]) is an indole alkaloid molecule commonly found in plants, including coffee and tobacco (Schmeltz and Hoffmann1977; Luxembourget al.2003; Wojtowiczet al.2015). NRM was first reported as a matrix substance for MALDI in 1999 where it was used to facilitate ionization of sialyl oligosaccharides (Yamagaki and Nakanishi1999; Cerrutiet al.2012). Following this initial report, the use of NRM as a MALDI matrix for work in negative-ion mode was systematically evaluated by Brownet al. in 2001, alongside common matrices such as 2,5-dihydroxybenzoic acid (DHB), sinapinic acid, ±-cyano-4-hydroxycinnamic acid (HCCA) and 9-nitroanthracene (Folch, Lees and Sloane Stanley1957; Bligh and Dyer1959; Brownet al.2001). This work identified NRM and a related molecule, harmane, as ideal matrices for analysis of hydrophobic molecules. The historical use of DHB, HCCA and 9-aminoacridine (9-AA) for lipid A and phospholipid characterization has resulted in an absence of well-defined uses for NRM, though a growing number of reports have appeared including the use of NRM as a matrix for laser-induced post-ionization, notated MALDI-2 (Soltwischet al.2015).
We have previously reported the use of NRM for mass spectrometry imaging (MSI) and 1D thin layer chromatography (TLC)-MALDI experiments (Nchoutmboubeet al.2013; Shireyet al.2013; Scottet al.2014). MSI is a technique used to characterize the spatial relationship of molecular targets to histological features, and MALDI is the most commonly used ionization method for MSI (Caprioli, Farmer and Gile1997; Stoeckliet al.2002; Cornettet al.2007; Schwamborn and Caprioli2010; Chaurand2012; Heeren2015). TLC-MALDI is a technique that couples traditional TLC separation with mass/charge identification by MALDI (Gusevet al.1995; Nicola, Gusev and Hercules1996; Fuchset al.2007). TLC-MALDI is another powerful tool for lipid profiling since it is rapid and offers improved lipid identification by virtue of the separation of lipid mixtures based on head group chemistry and it is especially useful for differential identification of isobaric species of phosphatidylcholine and phosphatidylethanolamine (PE) (Fuchset al.2009). Both techniques require a matrix capable of ionizing the molecular target(s) of interest, for example, lipids (van Hove, Smith and Heeren2010). Several matrices have been reported for lipid-targeting MSI, including DHB, 1,5-diaminonapthalene, HCCA and 9-AA) (Fuchs, Süss and Schiller2010; Zemiski Berryet al.2011; Cerrutiet al.2012). DHB is a widely used matrix for MALDI-MSI and applicable for lipid analysis in positive-ion mode, whereas 9-AA is used in negative-ion mode. 9-AA offers the advantage that the mass spectra consist largely of deprotonated or molecular ions, which simplifies lipid identification (Cerrutiet al.2012). Neither 9-AA nor DHB can serve as universal matrices for bacterial lipid analysis due to the need for large amounts of sample to produce mass spectra; 1 ng and 100 pg of lipid A are currently required, respectively. To highlight the relative lack of sensitivity, ∼109 colony-forming units (CFUs) of bacteria will yield 1 µg of LPS, only a fraction of which can be efficiently hydrolyzed and detected as the membrane anchor lipid A. Identifying an improved matrix, with near universal compatibility for a broader range of lipids in both positive- and negative-ion modes, including lipid A, would greatly benefit bacterial lipid research.
The first report of the use of NRM for MALDI analysis of lipid A was made by d'Hautevilleet al. (2002) to describe the activity of two lipid A biosynthesis genes (msbB1 andmsbB2) active inShigella flexneri. Prior to this study, lipid A was extracted from large-scale culture (>1L) and analyzed by MALDI using NRM matrix to assessin vitro-grown structures, but the sensitivity did not exist to analyze low inputin vivo-grown structures. To advance the study of lipid A structural modifications in primary clinical samples, improved detection limits are necessary for direct observation in tissue (Liet al.2012; O'Haraet al.2013; Pelletieret al.2013). This required the development of advanced extraction methodologies, as well as sensitive lipid A detection techniques. In 2005, a method for lipid A microextraction was reported by the Caroff group in which the authors achieved lipid A extraction from 10 mg of lyophilized bacteria followed by mass spectrometric analysis (Hamidiet al.2005). This report revolutionized the study of the lipid A structure-function relationship by making lipid A analysis from low-level bacterial cultures possible. Here, we present work combining these two advances to analyze thein vivo lipid A structure in both a mouse (host) and tick (vector) model ofFrancisella novicida (Fn) by extracting lipid A directly from burdened tissue.
Lipid A structure is influenced by a variety of factors including osmolarity, nutrient availability, presence of host/vector factors and growth temperature. The precise conformation of lipid A in any given condition is one of many components contributing to membrane integrity, permeability, topology and content.Fn lipid A, although primarily found with 18-carbon acyl chains at 37°C, is modified by 16-carbon acyl chains when grown at lower temperatures (to resemble the ambient conditions of a tick or the environment) (Gunn and Ernst2007; Shafferet al.2007; Liet al.2012; Needham and Trent2013). This shortening alters membrane permeability and resistance to antimicrobial agents and is likely a reflection ofFn lipid A structure in ticks (Liet al.2012). To date, the detection levels necessary to evaluate lipid A shorteningin vivo have not been identified. In this work, we present the findings of a lipid A extraction directly from whole hard-bodiedDermacentor variabilis (Dv) ticks infected withFn confirming the predictions made fromin vitro studies. Finally, by coupling the highly sensitive detection limit of NRM to MALDI-MSI, we can directly map phospholipids of bacterial origin (expected to be in low overall abundance versus host phospholipids) within infected host tissue, expanding the utility of lipid MSI studies to improve our understanding of bacterial pathogenic mechanisms. The results presented herein expand the fields of pathogenesis, general microbiology and lipid profiling by offering a versatile alternative matrix for lipid analysis.
MATERIALS AND METHODS
Ethics statement
All experiments were performed in accordance with the University of Maryland, Baltimore Institutional Animal Care and Use Committee protocol approval #0814005 in adherence with the Guide for the Care and Use of Animals (NIH), the Animal Welfare Act and applicable US Federal laws.
Matrices and solvents
9-AA, DHB and NRM were all purchased from Sigma-Aldrich (St. Louis, MO, USA). Solvent solutions were volumetric parts as follows: 1:1 E:W (ethanol:water), 1:2:0.8 C:M:W (chloroform:methanol:water), 1:1 C:M (chloroform:methanol), 2:1 C:M chloroform:methanol. Ethanol, methanol and chloroform were obtained from Sigma-Aldrich. Phosphate-buffered saline (PBS) and certified endotoxin-free water were sourced from Gibco (Grand Island, NY, USA). Matrix application was technique specific, as given.
Lipid A limit of detection
A commercial preparation of monophosphoryl lipid A (MPLA) fromSalmonella enterica serovar Minnesota (R595) was purchased from Sigma-Aldrich. For spot analysis, a concentrated stock solution of 1 mg mL−1 MPLA was made in 1:1 C:M. A 10-fold dilution series (1 µL−1 through 10 ng µL−1) was made in the same solvent. One microliter of each dilution in the series was spotted onto a stainless steel MALDI target plate, air-dried and followed by 1 µL of matrix (NRM: 20 mg mL−1 in 2:1 C:M, 9-AA 20 mg mL−1 in 2:1 C:M, DHB 40 mg mL−1 in 2:1 C:M, concentrations optimized for MPLA detection). MPLA spotted for the limit of detection (LOD) study was analyzed on a Bruker Daltonics (Billerica, MA, USA) solariX XR (MALDI FT-ICR,12T) calibrated to 1 ppm using sodium trifluoroacetic acid in negative-ion mode. LOD was defined as the quantity of MPLA spotted in the last detectable spot in the dilution series; minimum criterion for detection was defined as the presence of at least two isotopic peaks in addition to the corresponding, monoisotopic peak. LOD data were analyzed in DataAnalysis software (Bruker Daltonics) using the Sophisticated Numerical Annotation Procedure.
Bacterial strains and growth conditions
Francisella novicida (Fn) was grown in tryptic soy broth containing 0.1 g L−1 L-cysteine (TSBC) (Broth, Becton-Dickenson, Hunt Valley, MD, USA; L-cysteine, Sigma-Aldrich) in shaking liquid culture (225 RPM) to mid-log phase. Culture plates consisted of the same TSBC broth formulation with the addition of 1.5% (w/v) agar (Becton-Dickenson). For lipid A microextraction, 1 µl of liquid culture was harvested into a microfuge tube, pelleted (8000 ×g, 5 min) and supernatant was discarded. The remaining pellet was processed by the microextraction method described below. For rodent infections, an overnight shaking liquid culture (300 µL) was used to inoculate a large volume of fresh, warmed (37°C) TSBC (15 mL). These early-log phase subcultures (3 h culture) were prepared as follows for injection.
Mice, ticks and infection
Uninfected and infected solid organs (kidney, spleen) were collected from female C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME, USA), 6-8 weeks of age. Briefly, mice were housed in biosafety level 2 microisolator cages and provided food and waterad libitum. Infectious doses ofFn were prepared as follows: 1 mL of the 1:50 (v:v) large volume subculture was pelleted (3500 ×g, 5 min) and resuspended in PBS. This solution was diluted further in PBS to contain ∼300 CFUs in 50 µL (injection volume). Doses were administered subcutaneously, and control groups received sterile PBS injections. CFUs in duplicate 50 µL doses were assessed on TSBC agar plates (above). Mice were euthanized by carbon dioxide narcosis prior to tissue collection, followed by secondary thoracotomy. Spleens were collected for lipid A extraction 48 h post-infection along with uninfected, matching control tissue. Tissues were excised then snap-frozen by floating on foil in a pool of liquid nitrogen and stored at −80°C for sectioning or lipid A extraction.Dermacentor variabilis (Dv) ticks were kindly provided by Daniel Sonenshine (Department of Biological Sciences, Old Dominion University).Fn U112 (10 000-30 000 CFU) and PBS were injected into the emargination cavity of unfed male and females using pulled glass capillaries attached to a Nanoject II pump (Drummond Scientific, Broomall, PA, USA). Ticks were incubated overnight at 23°C with 95% humidity and then were washed with 3% hydrogen peroxide, sterile water and 70% ethanol in succession, dried and placed in a sterile 5 mL conical for processing (counting or microextraction).
Lipid A microextraction
Microextraction of lipid A from 1 mL of turbid shaking culture (in vitro), ticks (in vivo) and mouse spleen (in vivo) was performed as previously described with the following deviations: ticks or mouse spleens were extracted in a double volume of the initial extraction solution (800 µL total) starting with a tissue shredding step consisting of three 10-15 s full-speed pulses of the spleen in the extraction solution (Tissue Tearor Homogenizer, Cole Parmer, Vernon Hills, IL, USA) (Hamidiet al.2005). Briefly, 1 mL of mid-log phaseFn grown in TSBC was pelleted and supernatant was discarded. The pellet was extracted in 400 µL of a solution containing five parts of isobutyric acid: three parts of 1M ammonium hydroxide and heated at 100°C for 1 h followed by a 15-min incubation on ice and centrifugation at 2000 ×g for 15 min. Supernatant was collected and mixed in equal parts with water and then frozen and lyophilized. Contaminants were washed from the dried material by two rounds of methanol washes: 1 mL of methanol, vortexing and pelleting at 10 000 ×g for 5 min. The final product was reconstituted in 2:1 C:M (100 µL) along with 4-8 grains of Dowex ion exchange resin (Fisher Scientific, Pittsburgh, PA, USA), incubated with vortexing for at least 5 min. A portion of 1 mL of the extraction was spotted with 1 µL of NRM as above for MALDI analysis on a Bruker AutoFlex Speed in negative-ion mode calibrated with Agilent Tuning Mix (Santa Clara, CA, USA) and data were processed in flexAnalysis (Bruker Daltonics). All microextraction chemicals were obtained from Sigma-Aldrich unless otherwise noted.
Total lipid extraction
Total lipids were extracted fromFn (10 mL mid-log shaking culture at 37°C, pelleted) as previously described (Bligh and Dyer1959). Briefly, pellets were extracted on ice in 11.4 mL of the single-phase extraction solution—1:2:0.8 C:M:W—for 30 min with vigorous agitation (stirbar). Insoluble product was pelleted at 1000 ×g for 10 min at 4°C. The supernatant was converted into two phases by adding 3 mL of water and 3 mL chloroform, shaken vigorously until the solution turned cloudy (∼30 s to 1 min) and allowed to separate for 5 min at room temperature followed by a 15-min separation at 1500 ×g at room temperature. The organic phase was collected and dried under a stream of nitrogen. Aqueous phases were washed with 3 mL chloroform and separated as above to recover remaining lipids, processed as above and pooled with the first organic phase collection. Total lipids were reconstituted in 100 µL of 2:1 C:M solution. A portion of 12 µL of total lipid extract was spotted for 2D-TLC, as shown below.
Two-dimensional TLC-MALDI
2D-TLC-MALDI was performed onFn lipid extracts grown at 37°C in TSBC as described above. Lipid extracts were spotted onto aluminum-backed TLC silica gel 60 F254 plates (20 cm × 20 cm, EMD Chemicals Inc., Germany) that were prerun in an equilibrated chamber of 1:1 C:M in the direction of the first dimension and dried. Loaded TLC plates were run in an equilibrated chamber of 65:25:3.6:0.4 (v:v:v:v) chloroform:methanol:water:ammonium hydroxide, air dried, rotated 90° and separated in the second dimension for acyl complexity in an equilibrated chamber of 60:60:10 (v:v:v) toluene, pyridine and water. Solvent fronts were marked and lipid migration spots were determined using water exclusion (dried plates were sprayed with water to define lipid spot pattern). For MALDI analysis, the appropriate region of the TLC plate was excised (∼5 cm × 7.5 cm), spray coated with matrix (NRM matrix solution, 12 mg mL−1 in 2:1 C:M applied with a TLC sprayer) and scanned in negative-ion mode at 500 µm spatial resolution on a Bruker AutoFlex Speed; data were processed in flexImaging (Bruker Daltonics). Unless otherwise noted, all reagents were sourced from Sigma-Aldrich.
Tissue preparation and imaging
Tissue profiling experiments were performed on uninfected, unfixed, frozen mouse kidney and spleen. Twelve micron sections were cut using a ThermoFisher cryostat (Waltham, MA, USA), mounted onto a cold glass slide and incubated at 37°C until visibly dry. For spot profiling and MSI, matrices were applied with a Bruker ImagePrep (Billerica) device except where noted. Matrix crystal characterization was performed on 12 µm sections of mouse brain tissue, applied with a Suncollect automated pneumatic sprayer device (Sunchrom GmbH, Friedrichsdorf, Germany) as follows: DHB—18 layers (6 mg mL−1), 9-AA—13 layers (6 mg mL−1) and NRM—13 layers (6 mg mL−1). All matrices were solvated in 1:2:0.8 C:M:W for crystal sizing and description, and were applied to a level of comparably similar coverage. For the lipid-depleting experiment, methanol washing steps were performed as follows: 1 min wash in 70% methanol followed by a 1 min wash in 100% methanol, after which the sections were allowed to dry under ambient conditions and then prepared for imaging using the ImagePrep as above. Tissue spot profiling and washing MSI data were collected on a Synapt G1 from Waters (Milford, MA, USA) using MassLynx Software (Waters), calibrated with a polyethylene glycol mixture. Spectra were processed for image construction in BioMap 3.7.5.5 software (Novartis, Basel, Switzerland,www.maldi-msi.org). Simultaneous mapping tissue mapping experiments were prepared as described, with matrix deposition on the Sunchrom device and analysis on a Bruker solariX 12T MALDI-FTICR calibrated to 1 ppm in negative-ion mode using infused sodium trifluoroacetate clusters. Root-mean-square normalization was performed in flexImaging version 4.0 and ion identities were predicted in the Lipid Maps database (Lipid Maps Consortium,www.lipidmaps.org) along with the support of previously published lipid identities.
RESULTS AND DISCUSSION
Physiological range detection of lipid A made possible using NRM
We first sought to establish the detection limits for common MALDI matrices used for lipid A, making future analysis of primary samples possible. First, we evaluated the dynamic range and LOD of lipid A using NRM as a matrix. For this analysis, we used synthetically derived MPLA, a representative lipid A molecule derived from the LPS ofSalmonella enterica serovar Minnesota (Se). MPLA was 10-fold serially diluted from 1 µg µL−1 (1 µg spotted) through 10 pg µL−1 (10 pg spotted) followed by the addition of 1 µL of DHB, 9-AA or NRM (concentrations given in the Methods section). This analysis showed a two-log improvement of the LOD of MPLA with NRM compared to 9-AA and a log improvement compared to DHB. All three matrices resulted in detection of the singly charged lipid A in negative-ion mode, where the monoisotopic peak ism/z 1744 from 1000 pg of MPLA, the total amount in a single spot (Fig. S1, Supporting Information). MPLA was undetectable below 100 pg when spotted with DHB and 1000 pg when spotted with 9-AA. Only NRM yielded the monoisotopic peak and at least two additional isotopic peaks from 10 pg of MPLA (Fig. S1C, Supporting Information). To evaluate performance in a narrow range of concentration near and below LOD, signal-to-noise (S:N) ratios were calculated from triplicate samples (Table 1) diluted to an intermediate range bracketing 10 pg of MPLA. MPLA spotted at 12.5 and 6.3 pg total results in an average S:N ratios of 7.0 and 3.9, respectively. For the purposes of defining an endpoint value, S:N ≥ 3 (form/z 1744) was established as a detection cutoff. MPLA applied at 5.0 pg yielded an average S:N < 3 (2.5), establishing the refined LOD at 6.3 pg total material, the lowest concentration tested resulting in S:N ≥ 3.
Table 1.
MPLA detection performance in NRM matrix near LOD.
Below detection threshold of S:N < 3.0.
Low-level detection of lipid A is central to structural analysis from biological extracts, including infected tissues and primary clinical isolates. Given that 109-10 CFUs representing a small colony of bacteria on an agar plate will yield ∼10 mg of LPS (∼3-5 µg hydrolyzed lipid A,Escherichia coli for reference), this improved LOD for lipid A represents a detectable signal from six orders of magnitude fewer bacteria (Watsonet al.1977). Detection of lipid A from <103 CFUs puts detection by MALDI on a clinically relevant scale; for example, in a typical urinary tract infection, 104-5 CFU mL−1 are present (Schmiemannet al.2010; Kwonet al.2012). Therefore, we sought to detect lipid A directly from infected tissues.
For lipid A detection from biological samples, we extracted lipid A from two sources: cultures (in vitro) and infected mouse spleen (in vivo).Francisella novicida (Fn) lipid A is a tetraacylated, monoglycosylated structure and is readily detectable as a negative ion atm/z 1665 (Fig. 1A). Them/z 1637 species represents a shortening of one of the fatty acids by two carbon units (Fig. S2, Supporting Information). We infected mice subcutaneously withFn, harvested infected spleens (2.8 × 106 CFU mL−1, blood) and extracted lipid A using the microextraction method (Hamidiet al.2005).Fn lipid A was extracted from liquid culture using the Caroff isobutyric acid/ammonium hydroxide microextraction method, solvated in 2:1 chloroform:methanol (50 µL) from which 1 µL was spotted onto a MALDI target plate with 1 µL NRM matrix and analyzed using MALDI (Hamidiet al.2005). As expected, extractedFn lipid A is readily detectable (Fig. 1A) from a 1 mLin vitro culture (>108 CFU mL−1) as two major species,m/z 1665 andm/z 1637. The previously described structure of the larger molecule (m/z 1665) is given in Fig. S2 and is the dominant structure whenFn is grown at 37°C (Shafferet al.2007). Both of theFn lipid A products detectedin vitro were robustly detectedin vivo, and in similar relative abundances (Fig. 1B) to thein vitro lipid A profile; the dominant lipid A species (m/z 1665) along with the minor species at (m/z 1637) were both observedin vivo. These results represent the first lipid A analysis by MALDI-MS from directly extracted infected tissue, as opposed to lipid A extraction from expanded cultures,ex vivo. This approach will be a powerful tool to study the effect of host influences on bacterial lipid A structures.
Figure 1.
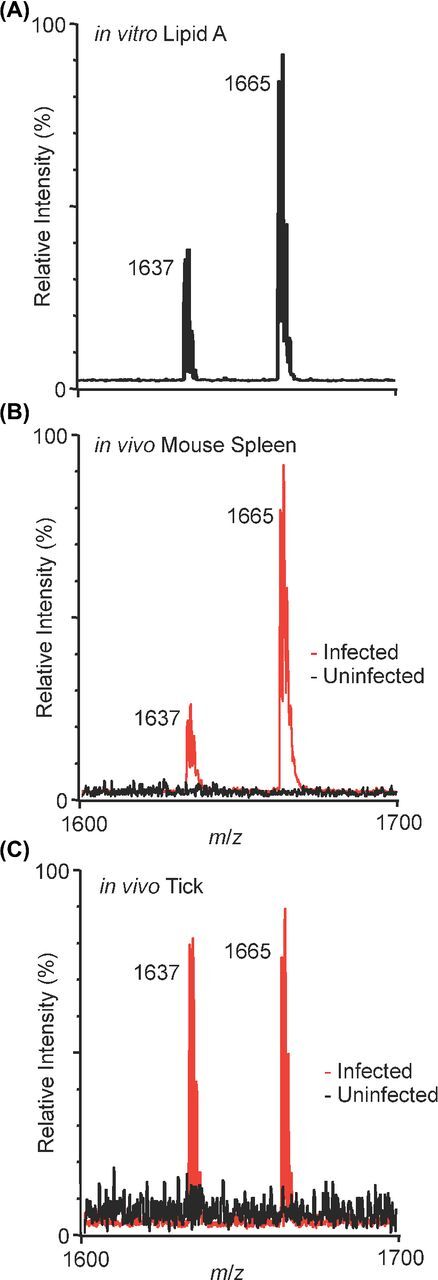
NRM matrix delivers high sensitivity for physiological-level lipid A detection. (A)In vitro-grownFn lipid A extract, 37°C, from 1 mL of culture. Two major peaks detected:m/z 1665 and 1637. Structure ofm/z 1665 given for reference in Fig. S2, them/z 1637 peak isFn lipid A lacking two carbons in one of the two amide-linked acyl chains. (B)In vivo-grownFn lipid A extract from a whole mouse spleen infected for 48 h, red trace. Uninfected, extracted spleen, black trace. Expected burden at 48 h post-infection ∼106-107 CFUs per spleen. (C)In vivo-grownFn lipid A extract from a whole hard-bodied tick infected for 48 h, red trace. Uninfected, extracted tick, black trace. Bacterial burden at 48 h post-infection 107 CFUs per tick in parallel replicates. (A-C) NRM matrix, negative-ion mode MALDI,m/z 1600-1700, total ion current (TIC), relative intensity.
Given the low input necessary for lipid A detection using the combination of NRM matrix and the Caroff microextraction method, we aimed to confirm the major lipid A ions ofFn growing in ticks, one of the arthropod vectors of multipleFrancisella subspecies (Nano2006). We posited that an intermediate (vector, tick) temperature-controlled structure ofFn lipid A should increase in relative abundance compared to the warm (host, mouse) temperature-controlled structure as demonstrated previously,in vitro (Liet al.2012).Dermacentor variabilis (Dv) ticks were injected withFn and maintained at 23°C for 48 h. Lipid A was extracted from whole ticks (1 × 107 CFUs per tick) by microextraction and spotted with NRM matrix for analysis. Compared to the relative abundance of the warm temperature structure ofFn lipid A (m/z 1665), we observed a nearly equal balance (Fig. 1C) with a previously described intermediate-length structure (m/z 1637) in the whole tick extracts (Shafferet al.2007; Li, Wang and Ernst2011). These results taken together support the hypothesis thatFn lipid A structure is modulated in response to local environmental cues such as growth at alternative temperatures or in an arthropod vector (Shafferet al.2007; Liet al.2012).
The LOD to detect lipid A was compared using NRM, 9-AA and DHB revealing a two-log improvement in LOD between 9-AA (100-fold) and NRM and a log improvement between DHB (10-fold) and NRM. The expansion of the working range of lipid A LOD, from routine analysis of nanogram quantities to routine analysis of picogram quantities, makes NRM a powerful tool for characterizing low-yield lipid A extractions from clinical or environmental samples, including bacteria isolated from biofilms on implanted devices or directly from infected wound sites. Overall, NRM will have wide applications for the detection and study of complex lipids.
Simultaneous bacterial lipid A and phospholipid profiling from lipid extracts
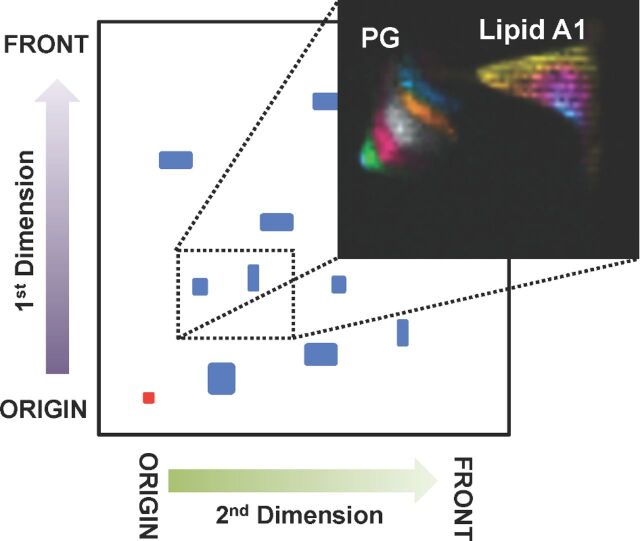
After growth at environmental (≤25°C) and mammalian (37°C) temperatures, bacteria rapidly remodel their membrane lipids to maintain proper membrane function and fluidity (Liet al.2012). Profiling these changes at both the phospholipid and lipid A levels, simultaneously, will yield a greater understanding of the relationship between temperature and global membrane remodeling. Harnessing the improved lipid A detection level conferred by NRM, we profiled both lipid A and phospholipids from aFn lipid extract using 2D-TLC coupled to MALDI (2D-TLC-MALDI). Due to the complexity of bacterial lipid preparations, we wanted to first resolve lipid classes and subsequently visualize the discrete acyl length variants of two majorFn lipid components: lipid A and phosphatidylglycerol (PG). For this analysis, we separated totalFn lipid extracts in a 2D-TLC format followed by MALDI-MSI to visualize discrete lipid bands. Figure2 illustrates the separation approach, as well as codetection of lipid A and several PG lipids. The previously observed improvements in LOD of lipid A from primary extracts using NRM was also apparent in the TLC silica plate format, with three lipid A variant structures detectable:m/z 1665, 1637 and 1609, a minor constituent at mammalian growth temperature (37°C) representing a further shortening of one of the fatty acids by two carbon units. The referenceFn lipid A structures (Fig. S2), their expected temperature-controlled abundance ratios and the major ions within the peaks have been previously described (Shafferet al.2007; Liet al.2012). TLC-MALDI is an established method for rapid phospholipid profiling. Here, we have demonstrated the use of a single matrix to profile both lipid A and phospholipids simultaneously from a single TLC-MALDI experiment. The approach can be readily translated to multiple infection model systems.
Figure 2.
Simultaneous analysis of both phospholipids and lipid A from 2D-TLC-MALDI (Fn). TOP: Cartoon depicting the 2D-TLC separation used. First dimension: polar separation, second dimension: apolar separation, red dot indicates spot origin, blue spots depict lipid separations. Dotted rectangle represents approximate area of large format TLC plate cut out for profiling by MALDI-MSI (inset). NRM matrix, negative-ion mode MALDI, 500 µm spatial resolution, relative intensity within color band. Lipid A band identities given in Fig. S2. PG spot colorized for uncharacterized isomer bands increasing by two carbons in length starting with PG 30:0 in the lower left (green) through PG 38:0 (blue).
The spleen is a dynamic secondary lymphoid organ responsible for clearance of compromised red blood cells in the red pulp and immune response and surveillance in the white pulp. During the course of an immune response, splenic architecture undergoes dramatic restructuring, forming germinal centers and follicles. To determine if NRM could be effective as a lipid matrix for MSI of the host response, naïve spleen sections were prepared with NRM matrix dissolved in a single-phase lipid extraction solution (1:2:0.8 chloroform:methanol:water) and spot analyzed in positive- and negative-ion modes (Cerrutiet al.2012; Scottet al.2014). We sought to establish a baseline profile of lipids in the spleen in both polarities, including those containing polyunsaturated fatty acids (PUFAs), such as arachidonic acid (AA), released from membrane phospholipids and required for the production of specific classes of inflammatory lipids. The most abundant ion detected in negative-ion mode wasm/z 885.6, an arachidonic acid-containing lipid phosphatidylinositol (PI) described in numerous other tissues (Fig. 3) (Murphy, Hankin and Barkley2008). Higher molecular weight lipid ions were also detected in the spleen, again highlighting increased NRM efficiency in higher mass ranges (Figs3 and S3, Supplemental Text). Together, these data highlight the dual polarity application of NRM for efficient lipid profiling experiments on tissue and widen the available methods to study lipid-mediated inflammation.
Figure 3.
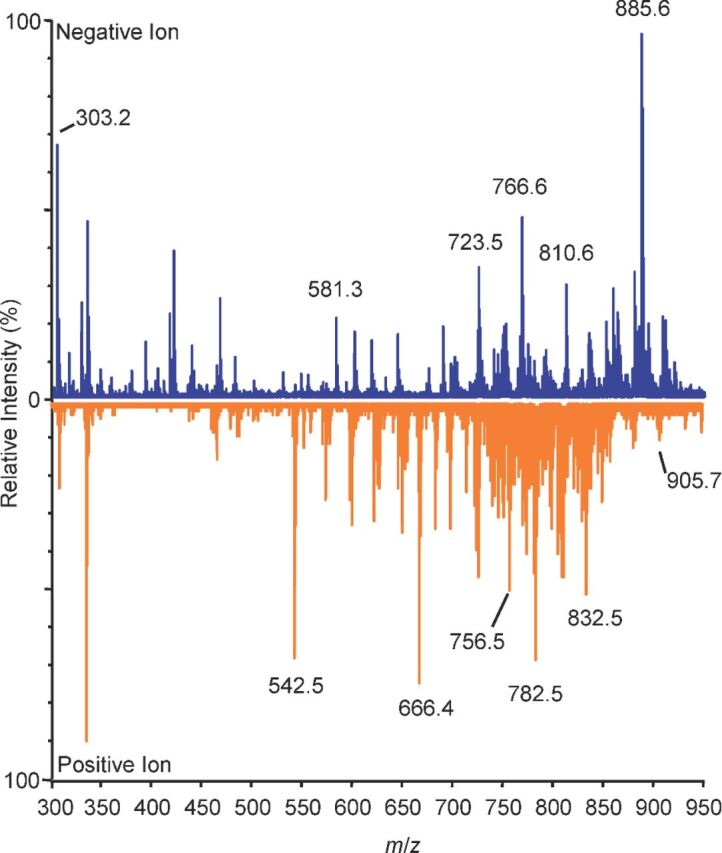
Differential tissue fingerprints of spleen in both positive- and negative-ion modes using NRM. Spot profiling of lipids,m/z 500-1000. Blue: negative-ion mode TIC, orange: positive-ion mode, NRM matrix, 12 µm sections, data collected in both red and white pulp arbitrarily for summation, TIC, relative intensity. Ions indicated bym/z values are for reference purposes.
Profiling the extent of incorporation of PUFAs in the phospholipid repertoire may have interesting implications for host lipid-based inflammatory response, including our understanding of the overall potential for a tissue to absorb damage from reactive oxygen species. Additionally, release of specific PUFAs from the parent phospholipid is a critical initiation point for production of lipid signaling molecules that can be potent modulators in the context of inflammation and immunity. Prostaglandins, produced from liberated AA, have an important, yet poorly defined role inFrancisella infections. Woolardet al. (2007) demonstrated that the T-cell blocking mechanism observed inFrancisella tularensis LVS-infected macrophages was due to production of prostaglandin E2. Mapping the AA-containing phospholipids upstream of these immunomodulatory effects will be the focus of further study.
Simultaneous mapping of unique bacterial and host lipids
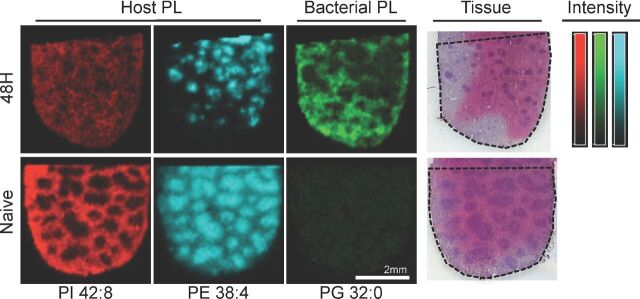
In an effort to harness the improved performance of our lipid detection and mapping experiments, we sought to use NRM to further describe the host-pathogen interaction within infected tissue.Fn is a Gram-negative species with two membranes: the inner membrane consisting of phospholipids and the asymmetrical outer membrane. Comprising the outer membrane are phospholipids on the inner leaflet of the outer membrane and LPS (the lipid A anchor component) on the outer leaflet of the outer membrane. Bacterial membranes contain a dominant fraction of PG and PE, though generalizations about individual bacterial backgrounds are difficult since the unique lipid composition is linked to taxonomy (Ratledge and Wilkinson1988). We have previously characterized thein vitro phospholipid and lipid A populations ofFn using standard lipid extraction methods and determined that PG 32:0 was a major component of the bacterial membrane (Zhang and Rock2008; Li, Wang and Ernst2011). Using MSI, we sought to map the distribution of PG 32:0 inFn-infected tissue. Figure 4 demonstrates the distribution of PG 32:0 (m/z 721.5) withinFn-infected spleen versus naïve spleen, with high relative abundances observed in the red pulp following infection. At 48 h post-infection, organisms are present in the spleen, especially the red pulp (Conlanet al.2003; Elkins, Cowley and Bosio2007; Kanistanonet al.2008; Ojedaet al.2008; Rasmussenet al.2012). Although PG 32:0 is not exclusively a bacterial phospholipid, the relative abundance in the naive spleen profile (Fig. 3) is low and near the detection threshold. PG levels in mouse tissues (liver, <5%) are modest compared to other phospholipid classes (White1973). It is worth noting that PG 32:0 was not reported present in human plasma samples analyzed by the Lipid Maps Consortium; however, absent complete comparative descriptions of the mouse and human splenic lipidomes, it is impossible to directly compare PG content (Quehenbergeret al.2010). For the purposes of this work, PG was considered an abundant bacterial marker, though the possibility exists that the presence of bacteria or simply of an activated immune response may be sufficient to stimulate production of host-borne PG 32:0.
Figure 4.
Simultaneous host and pathogen lipid mapping. Top:Fn-infected spleens at 48 h post-infection. Bottom: Naïve spleens. Red: PI 42:8 (m/z 933.5) is moderately distributed in the infected spleen, decreasing in intensity from naive. Cyan: PE 38:4 (m/z 766.5) condenses in the white pulp following infection. Green: PG 32:0 (m/z 721.5) is a highly abundant bacterial phospholipid that appears in the red pulp following infection. Post-MSI H&E reference stain of 48 hFn-infected spleen, capture area outlined for reference. Bars: False color scale for intensity (arbitrary) with limits as given (low-high) for PI 42:8 (red, 0-33), PE 38:4 (cyan, 0-102), PG 32:0 (green, 0-32). Negative polarity ion image of mouse spleen infected withFn for 48 h, 1/3 of whole spleen tissue, NRM matrix, MALDI-MSI, 75 µm resolution.
In contrast, several classes of lipid are not made byFn including the PIs; thus, they are exclusively host lipids in this infection (Li, Wang and Ernst2011). To demonstrate the simultaneous host-pathogen lipid monitoring made, we mapped PI 42:8 (m/z 933.5), an extensively polyunsaturated PI with 42 total acyl carbons (Table S1, Supporting Information), throughout the infected spleen with some bias toward a red pulp distribution. Similarly, PE 38:4 (m/z 766.5) is another exclusively host lipid as it is not found in lipid extracts ofFn. Curiously, PE 38:4 was found in the white pulp with high relative intensity organized puncta (Fig. 4) suggesting that this may be a marker for a specific cell type or a highly localized immune process. By colocalizing components of the immune response toFn in tandem with the host and pathogen lipid distributions, we aim to further describe the basic pathogenic mechanisms of this infection. Achieving higher sensitivity for a wide variety of pathogen lipids is crucial to the success of this approach and the studies herein implicate its feasibility for numerous infection models. Further studies will be necessary to define imaging parameters for exclusive bacterial lipids, such as lipid A; however, highly expressed bacterial lipids can serve as proxy markers to map bacterial infection from a new perspective while simultaneously mapping the host response. This is a valuable combination for future host-pathogen interaction studies.
CONCLUSIONS
When used in MALDI-coupled techniques for bacterial, vector and host lipid analysis, NRM is a powerful and versatile matrix allowing picogram-level detection of MPLA and enabling analysis of lipid A from primary extracts ofin vitro andin vivo infection model systems. Beyond MALDI spot analysis, the use of NRM in alternative MALDI-coupled techniques resulted in the detection of a wide range of lipids and facilitated analysis of both lipid A and phospholipids directly from 2D-TLC-MALDI plates making it possible to survey many lipid components of bacterial membranes in a single scan. It also increased the presence of higher mass lipids, extending the useable information range from a single experiment. The overall utility of NRM is underlined by improved LOD of lipid A, robust performance in positive- and negative-ion modes, and versatility across multiple MALDI applications. Finally, we demonstrated the capability of coupling bacterial lipid mapping to host lipid mapping using this simultaneous monitoring approach. Lipids exclusive to the host were found in the same regions as specific bacterial lipids as well as in unique tissue structures (organized splenic white pulp) involved in the immune response toFn infection. Future studies will focus on the direct mapping of lipid A within infected tissues, including optimization of the on-tissue LPS hydrolysis steps that will be necessary for robust MSI of lipid A signal from bacterial infections bearing smooth LPS. Together, our results establish a path to describe novel lipid-based mechanisms of microbial pathogenesis that will find wide utility within the infection and immunity fields.
Supplementary Material
SUPPLEMENTARY DATA
FUNDING
This work was supported by the National Institute for Allergy and Infectious Diseases at the National Institutes of Health [ R21 AI101691 to RKE] with training support for AJS [T32 AI007540 and T32 AI095190].
Conflict of interest. None declared.
REFERENCES
- Bligh EG, Dyer WJ A rapid method of total lipid extraction and purification. Can J Biochem Phys. 1959;37:1–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brown T, Clipston NL, Simjee N, et al. Matrix-assisted laser desorption/ionization of amphiphilic fullerene derivatives. Int J Mass Spectrom. 2001;210/211:249–63. [Google Scholar]
- Caprioli RM, Farmer TB, Gile J Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Cerruti CD, Benabdellah F, Laprévote O, et al. MALDI imaging and structural analysis of rat brain lipid negative ions with 9-aminoacridine matrix. Anal Chem. 2012;84:2164–71. doi: 10.1021/ac2025317. [DOI] [PubMed] [Google Scholar]
- Chaurand P. Imaging mass spectrometry of thin tissue sections: a decade of collective efforts. J Proteomics. 2012;75:4883–92. doi: 10.1016/j.jprot.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Conlan JW, Chen W, Shen H, et al. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb Pathogenesis. 2003;34:239–48. doi: 10.1016/s0882-4010(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Cornett DS, Reyzer ML, Chaurand P, et al. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–33. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- d'Hauteville H, Khan S, Maskell DJ, et al. Two msbB genes encoding maximal acylation of lipid A are required for invasive Shigella flexneri to mediate inflammatory rupture and destruction of the intestinal epithelium. J Immunol. 2002;168:5240–51. doi: 10.4049/jimmunol.168.10.5240. [DOI] [PubMed] [Google Scholar]
- Elkins KL, Cowley S, Bosio CM Innate and adaptive immunity to Francisella. Ann N Y Acad Sci. 2007;1105:284–324. doi: 10.1196/annals.1409.014. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fuchs B, Bischoff A, Süss R, et al. Phosphatidylcholines and -ethanolamines can be easily mistaken in phospholipid mixtures: a negative ion MALDI-TOF MS study with 9-aminoacridine as matrix and egg yolk as selected example. Anal Bioanal Chem. 2009;395:2479–87. doi: 10.1007/s00216-009-3032-1. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Schiller J, Süss R, et al. A direct and simple method of coupling matrix-assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF MS) to thin-layer chromatography (TLC) for the analysis of phospholipids from egg yolk. Anal Bioanal Chem. 2007;389:827–34. doi: 10.1007/s00216-007-1488-4. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Süss R, Schiller J An update of MALDI-TOF mass spectrometry in lipid research. Prog Lipid Res. 2010;49:450–75. doi: 10.1016/j.plipres.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- Gunn JS, Ernst RK The structure and function of Francisella lipopolysaccharide. Ann NY Acad Sci. 2007;1105:202–18. doi: 10.1196/annals.1409.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev AI, Proctor A, Rabinovich YI, et al. Thin-layer chromatography combined with matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 1995;67:1805–14. [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, et al. Cytoplasmic LPS activates Caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–3. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar AM, Ernst RK, Tsai JH, et al. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3:354–9. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- Hamidi El A, Tirsoaga A, Novikov A, et al. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res. 2005;46:1773–8. doi: 10.1194/jlr.D500014-JLR200. [DOI] [PubMed] [Google Scholar]
- Heeren RMA. Getting the picture: the coming of age of imaging MS. Int J Mass Spectrom. 2015;377:672–80. [Google Scholar]
- Kanistanon D, Kanistanon D, Hajjar AM, et al. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 2008;4:e24. doi: 10.1371/journal.ppat.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JH, Fausone MK, Du H, et al. Impact of laboratory-reported urine culture colony counts on the diagnosis and treatment of urinary tract infection for hospitalized patients. Am J Clin Pathol. 2012;137:778–84. doi: 10.1309/AJCP4KVGQZEG1YDM. [DOI] [PubMed] [Google Scholar]
- Li Y, Powell DA, Shaffer SA, et al. LPS remodeling is an evolved survival strategy for bacteria. P Natl Acad Sci USA. 2012;109:8716–21. doi: 10.1073/pnas.1202908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang X, Ernst RK A rapid one-step method for the characterization of membrane lipid remodeling in Francisella using matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometry. Rapid Commun Mass Sp. 2011;25:2641–8. doi: 10.1002/rcm.5168. [DOI] [PubMed] [Google Scholar]
- Luxembourg SL, Mcdonnell LA, Duursma MC, et al. Effect of local matrix crystal variations in matrix-assisted ionization techniques for mass spectrometry. Anal Chem. 2003;75:2333–41. doi: 10.1021/ac026434p. [DOI] [PubMed] [Google Scholar]
- Murphy RC, Hankin JA, Barkley RM Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2008;50:S317–22. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE The genus Francisella. Prokaryotes. 2006;6:1119–32. [Google Scholar]
- Nchoutmboube JA, Grüner BM, Spector AA, et al. Increased long chain acyl-coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog. 2013;9:e1003401. doi: 10.1371/journal.ppat.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BD, Trent MS Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–81. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AJ, Gusev AI, Hercules DM Direct quantitative analysis from thin-layer chromatography plates using matrix-assisted laser desorption/ionization mass spectrometry. Soc Appl Spectrosc. 1996:1479–82. [Google Scholar]
- O'Hara JA, Ambe LA, Casella LG, et al. Activities of vancomycin-containing regimens against colistin-resistant Acinetobacter baumannii clinical strains. Antimicrob Agents Chemoth. 2013;57:2103–8. doi: 10.1128/AAC.02501-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda SS, Doyle MR, Wang ZJ, et al. Rapid dissemination of Francisella tularensis and the effect of route of infection. BMC Microbiol. 2008;8:215. doi: 10.1186/1471-2180-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier MR, Casella LG, Jones JW, et al. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Ch. 2013;57:4831–40. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JW, Tam JW, Okan NA, et al. Phenotypic, morphological, and functional heterogeneity of splenic immature myeloid cells in the host response to Tularemia. Infect Immun. 2012;80:2371–81. doi: 10.1128/IAI.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C, Wilkinson SG An overview of microbial lipids. Microb Lipids. 1988;1:3–22. [Google Scholar]
- Schmeltz I, Hoffmann D Nitrogen-containing compounds in tobacco and tobacco smoke. Chem Rev. 1977;77:295–311. [Google Scholar]
- Schmiemann G, Kniehl E, Gebhardt K, et al. The diagnosis of urinary tract infection. Dtech Arztebl Int. 2010;107:361–7. doi: 10.3238/arztebl.2010.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn K, Caprioli RM Molecular imaging by mass spectrometry — looking beyond classical histology. Nat Pub Group. 2010;10:639–46. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- Scott AJ, Jones JW, Orschell CM, et al. Mass spectrometry imaging enriches biomarker discovery approaches with candidate mapping. Health Phys. 2014;106:120–8. doi: 10.1097/HP.0b013e3182a4ec2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer SA, Harvey MD, Goodlett DR, et al. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J Am Soc Mass Spectr. 2007;18:1080–92. doi: 10.1016/j.jasms.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey KA, Lai W, Scott AJ, et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013;497:498–502. doi: 10.1038/nature12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltwisch J, Kettling H, Vens-Cappell S, et al. Mass spectrometry imaging with laser-induced postionization. Science. 2015;348:211–5. doi: 10.1126/science.aaa1051. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Staab D, Staufenbiel M, et al. Molecular imaging of amyloid ² peptides in mouse brain sections using mass spectrometry. Anal Biochem. 2002;311:33–9. doi: 10.1016/s0003-2697(02)00386-x. [DOI] [PubMed] [Google Scholar]
- van Hove ERA, Smith DF, Heeren RMA A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217:3946–54. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Watson SW, Novitsky TJ, Quinby HL, et al. Determination of bacterial number and biomass in the marine environment. Appl Environ Microb. 1977;33:940–6. doi: 10.1128/aem.33.4.940-946.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DA. The phospholipid composition of mammalian tissues. In: Ansell GG Dawson RMC Hawthorne JN , editor. Form and Function of Phospholipids. 3rd edn. NY: Elsevier; 1973. [Google Scholar]
- Wojtowicz E, Zawirska-Wojtasiak R, Przygoński K, et al. Bioactive β-carbolines norharman and harman in traditional and novel raw materials for chicory coffee. Food Chem. 2015;175:280–3. doi: 10.1016/j.foodchem.2014.11.143. [DOI] [PubMed] [Google Scholar]
- Woolard MD, Wilson JE, Hensley LL, et al. Francisella tularensis-infected macrophages release prostaglandin E2 that blocks T cell proliferation and promotes a Th2-like response. J Immunol. 2007;178:2065–74. doi: 10.4049/jimmunol.178.4.2065. [DOI] [PubMed] [Google Scholar]
- Yamagaki T, Nakanishi H A new technique distinguishing α2-3 sialyl linkage from α2-6 linkage in sialyllactoses and sialyl-N-acetyllactosamines by post-source decay fragmentation method of MALDI-TOF mass spectrometry - Springer. Glycoconjugate J. 1999;16:385–9. doi: 10.1023/a:1007050909293. [DOI] [PubMed] [Google Scholar]
- Zemiski Berry KA, Li B, Reynolds SD, et al. MALDI imaging MS of phospholipids in the mouse lung. J Lipid Res. 2011;52:1551–60. doi: 10.1194/jlr.M015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-M, Rock CO Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–33. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.