Abstract
The spread of coronavirus disease 2019 (COVID-19) throughout the world has resulted in stressful healthcare burdens and global health crises. Developing an effective measure to protect people from infection is an urgent need. The blockage of interaction between angiotensin-converting enzyme 2 (ACE2) and S protein is considered an essential target for anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) drugs. A full-length ACE2 protein could be a potential drug to block early entry of SARS-CoV-2 into host cells. In this study, a therapeutic strategy was developed by using extracellular vesicles (EVs) with decoy receptor ACE2 for neutralization of SARS-CoV-2. The EVs embedded with engineered ACE2 (EVs-ACE2) were prepared; the EVs-ACE2 were derived from an engineered cell line with stable ACE2 expression. The potential effect of the EVs-ACE2 on anti-SARS-CoV-2 was demonstrated by both in vitro and in vivo neutralization experiments using the pseudovirus with the S protein (S-pseudovirus). EVs-ACE2 can inhibit the infection of S-pseudovirus in various cells, and importantly, the mice treated with intranasal administration of EVs-ACE2 can suppress the entry of S-pseudovirus into the mucosal epithelium. Therefore, the intranasal EVs-ACE2 could be a preventive medicine to protect from SARS-CoV-2 infection. This EVs-based strategy offers a potential route to COVID-19 drug development.
KEY WORDS: SARS-CoV-2, COVID-19, Spike protein, Pseudovirus, Extracellular vesicles, ACE2, Intranasal administration, Neutralization
Abbreviations: ACE2, angiotensin-converting enzyme 2; BSA, bovine albumin; EVs, extracellular vesicles; FBS, fetal bovine serum; NTA, nanoparticle tracking analysis; PAGE, polyacrylamide gel electrophoresis; RIPA, radio immunoprecipitation assay; RLU, relative luminescence units; S protein, spike protein; SDS, sodium dodecyl sulfate; TEM, transmission electron microscope; WB, western blot
Graphical abstract
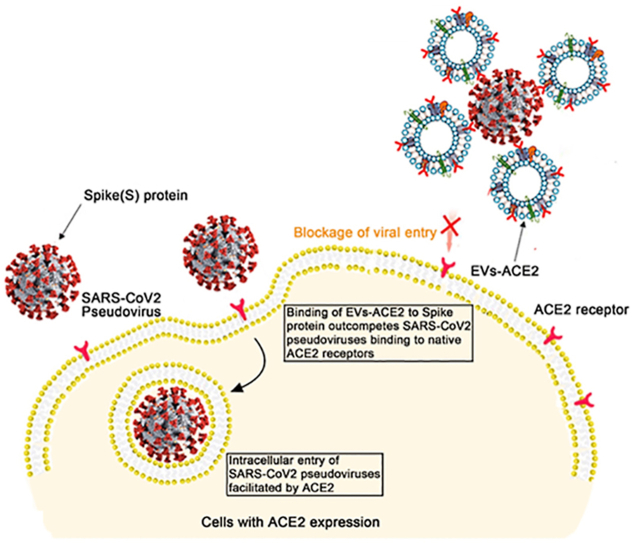
Extracellular vesicles (EVs) embedded with engineered ACE2 can inhibit the transfection of S-pseudovirus in the host cells by serving as decoy receptors and competitively binding with the virus.
1. Introduction
The outbreak of coronavirus disease 2019 (COVID-19) infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has posed significant threats to international health and the economy. As of July 30, 2021, there were 197,295,441 cases and 4,212,931 deaths from the COVID-19 pandemic1. In addition, there are increasing reports of various sequelae from the SARS-CoV-2 infection, and the sequela rate is high in the infected patients discharged after hospitalization2,3. Considering the limited availability and accessibility of COVID-19 vaccination, as well as the increasing incidence of immune escape after vaccination, the development of anti-SARS-CoV-2 therapeutics is still a pressing need for the world. However, there are currently no specific antiviral drugs for SARS-CoV-2 despite great effort and input from the scientific community. For example, an initial randomized, double-blinded trial of Remdesivir in China showed no substantial benefit to patients4. Several repurposed drugs (e.g., chloroquine and anti-IL 6) have been evaluated in clinical trials, ending with little therapeutic benefit5,6.
The viral structural S proteins in SARS-CoV-2, which form a characteristic crown on the virion surface, govern the entry of coronavirus into host cells7. The interaction between angiotensin-converting enzyme 2 (ACE2) in the host cells and S proteins of SARS-CoV-2 is the essential mechanism for infection, and ACE2 serves as a major entry receptor for mediating the entry of SARS-CoV-2 into host cells8,9. Further down, ACE2 was seen to be upregulated in COVID-19 patients, and therefore, the blockage of ACE2 and the S protein is considered a target for drug design10,11. For instance, a recent study demonstrated that human recombinant soluble ACE2 (hrsACE2) protein can inhibit SARS-CoV-2 entry into host cells12. However, recombinant ACE2 exhibits a fast clearance rate, with a dose-independent terminal half-life of only 10 h reported in clinical pharmacokinetic studies13. The short half-life could be a huge barrier to practical use.
Extracellular vesicles (EVs) are defined as cell-derived vesicles averaging 100 nm in diameter14,15, and also referred to as the natural “Trojan horses” for drug delivery and therapy16,17. Clinical trials based on EVs have shown positive results in various diseases18. Because of the great promise of EVs in serving as a carrier for biomacromolecules, big pharma companies have invested heavily in the R&D of EVs19. In this study, EVs with genetically engineered embedded human ACE2 (termed EVs-ACE2) were used as antagonists against the S proteins, thereby neutralizing the S-pseudovirus and inhibiting its entry into the host cells (Fig. 1A).
Figure 1.
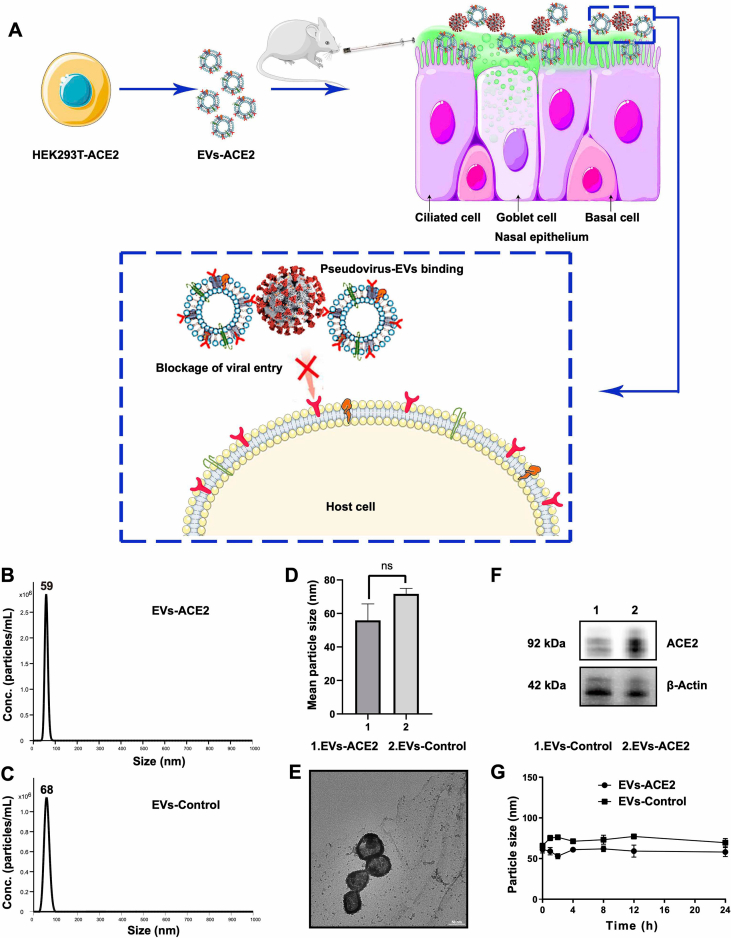
(A) Schematic mechanism of EVs-ACE2 inhibiting SARS-CoV-2 infection. The EVs-ACE2 were derived from the engineered HEK293T cells with stable ACE2 expression. EVs-ACE2 can competitively bind with the viruses via ACE2/S–protein interaction, thus blocking the virus to enter the host cells. (B) Size distributions of EVs-ACE2 measured by NTA. (C) Size distributions of EVs-control. (D) The median diameters of the EVs. (E) TEM images of EVs-ACE2. Scale bar = 50 nm. (F) ACE2 expression in EVs-ACE2 and EVs-Control. (G) The colloidal stability of the EVs. Data are presented as mean ± SD (n = 3); ns, no significance.
2. Materials and methods
2.1. Animals
The BALB/c mice (female, 6–8 weeks) were procured from Shanghai Laboratory Animal Center (SLAC) Co., Ltd. (Shanghai, China). The mice had free access to water and food during the experimental period. All animal experiment procedures, complying with the animal experiment guidelines, have been approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Institute of Materia Medica, Chinese Academy of Sciences, China.
2.2. Materials
Anti-ACE2 antibody and anti-SARS-CoV-2 spike glycoprotein antibody (Abcam, Cambridge, UK); Plasmid Mini, Midi and Maxi Kits (Qiagen, Hilden, Germany); Firefly Luciferase Reporter Assay Kit (Meilunbio, Dalian, China); Human recombinant ACE2 protein (Sino Biological, Beijing, China); Hoechst 33342 (Meilunbio, Dalian, China).
2.3. Preparation of the EVs-ACE2
Prior to cell culture, FBS (fetal bovine serum) was centrifuged at 100,000×g (CP100NX ultracentrifuge, Hitachi, Tokyo, Japan) for 2 h to deplete the serum-derived extracellular vesicles (EVs-free FBS). HEK293T and HEK293T-ACE2 (stable transfection) cells, utilized for extracellular vesicle production, were cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% EVs-free FBS for 48 h. Briefly, the EVs were isolated from the cell culture medium according to the method from a previous report20. After three centrifugations (300×g 10 min, 2000×g 10 min, and 10,000×g 30 min) using the Heraeus Multifuge X1R (ThermoFisher, Osterode am Harz, Germany), the pellets, including cells, dead cells, cell debris, and large vesicles, were discarded. The supernatant was ultracentrifuged at 100,000×g for 70 min at 4 °C using the CP100NX ultracentrifuge (Hitachi). The EVs thus obtained were then resuspended using PBS and ultracentrifuged again at 100,000×g for 70 min (Hitachi). The purified EVs were collected for the subsequent experiments.
2.4. Characterization of the EVs
A nanoparticle tracking analysis (NTA) was performed using a Nanosight NS300 instrument (Malvern Instruments, Worcestershire, UK). For transmission electron microscopy (TEM), the EVs were dropped onto the grids for 1 min, contrasted with 1% uranyl acetate, and dried. The micrographs were captured under a Talos L120C TEM (Thermo Scientific, Waltham, MA, USA) at 120 kV. For the WB analysis, the EVs were lysed with a radioimmunoprecipitation assay buffer (RIPA) containing a protease inhibitor cocktail (100:1, v/v, Sigma–Aldrich, St. Louis, MO, USA). The samples of EVs were analyzed using SDS-PAGE (Bio-Rad, Hercules, CA, USA) following a standard procedure. The blots were probed with antibodies specific to ACE2 (Rabbit, Abcam, Cambridge, UK). The bands were visualized on the ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA). The freshly isolated EVs were stored at 4 °C in an EVs-free culture medium and the particle size changes in the test time frame were measured using NTA. The ACE2 expression in EVs-ACE2 was determined by SDS-PAGE and Coomassie brilliant blue method using human ACE2 protein (Sino Biological, Beijing, China) as a standard. During the quantification procedure, a standard protein human ACE2 was separately loaded in 0.025, 0.5, 0.75, 1.0, and 1.5 μg to five lanes. The standard protein was used to give a standard curve of known concentration. The whole EVs-ACE2 lysate (2 and 5 μg of total protein amount) was also separately loaded to two lanes on the same gel. The content of ACE2 in each sample lane was then quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.5. Production of the SARS-CoV-2 S-pseudovirus
To generate the SARS-CoV-2 S-pseudovirus, the HEK293T cells were co-transfected with pNL4-3.Luc.R-E, nCOV.his-SPIKE-FL, and a Golgi location pTagRFP plasmid using the transfection reagent PEI25k. The HEK293T cells were seeded in the 12-well plates at a density of 5 × 105 cells per well and cultured for 20 h. The cells per well were treated with the PEI25k/plasmids complex (1.3:1, w/w), including 1 μg pNL4-3.Luc.R-E, 0.5 μg pTagRFP, and a varying dose of nCOV.his-SPIKE-FL (0.25, 0.33, 0.5, or 0.75 μg) in a fresh DMEM medium without FBS for 4 h at 37 °C, and then replaced with a fresh medium with 10% FBS21, 22, 23. Two days post-transfection, the supernatant containing the SARS-CoV-2 S-pseudovirus were harvested and filtered through a membrane with 0.45-μm pore size. Subsequently, to investigate the best proportion of the three plasmids for the pseudovirus preparation, the HEK293T-ACE2 cells with a density of 5 × 105 cells/well were seeded in 12-well plates. After 24 h, the cells were incubated for 12 h with the pseudovirus produced by the different ratios of pNL4-3.Luc.R-E, nCOV.his-SPIKE-FL, and pTagRFP. The cells were then washed with PBS three times and then used for the fluorescent imaging (CARL ZEISS, Oberkochen, Germany).
2.6. Characterization of the S-pseudovirus
The level of the S protein was analyzed using a WB analysis with Anti-SARS-CoV-2 spike glycoprotein antibody (Abcam), according to a standard procedure. The total protein concentration was measured using a standard BCA method. The median size and size distribution of the S-pseudovirus were analyzed using NTA.
2.7. WB analysis of ACE2 expression in various cell lines
The cells were seeded in 12-well plates at a density of 5 × 105 cells per well and incubated for 24 h. The cells were then collected and the levels of ACE2 analyzed by WB with anti-ACE2 antibody (Abcam), according to a standard procedure. The total protein concentration was measured using a standard BCA method.
2.8. Inhibition of viral attachment by EVs-ACE2
To investigate the inhibition of pseudovirus attachment by EVs-ACE2, the cells were seeded in the 24-well plates at a density of 2.5 × 105 cells per well. After 24 h culture, the S-pseudovirus (5 × 107 particles per well) were incubated with EVs-ACE2 or EVs-Control at 37 °C for 4 h (the ratio of pseudovirus and EVs was 1:5, based on the NTA particle quantity), followed by culture with different cells for 12 h. Cell nuclei were then stained with Hoechst 33342 for 5 min, and thoroughly washed with PBS three times to perform the fluorescent imaging (CARL ZEISS). For the quantitative measurements, the cells were digested, collected, and then analyzed using a flow cytometer (NovoCyte, Agilent, Santa Clara, USA). The S-pseudovirus without EV-pretreatment was used as a control.
2.9. Inhibition of viral infection by EVs-ACE2
To investigate the inhibition effect on pseudovirus infection by EVs-ACE2, the S-pseudovirus were also pretreated with the EVs-ACE2 4 h prior to adding to the cells. After 48 h of co-incubation, the cells were harvested and treated with 200 μL of the RIPA lysis buffer. After centrifugation at 12,000×g for 20 min (Heraeus Multifuge X1R, ThermoFisher), the luciferase activity of the supernatants was detected using the Luciferase Assay Kit (Meilunbio, Dalian, China), and the luminescence was measured using the EnSpire Multimode Plate Reader (PerkinElmer, Waltham, USA). The luminescence was normalized to the protein concentration of each sample, which was measured using a BCA Microplate Protein Assay Kit (Beyotime, Shanghai, China). Additionally, the quantification of luciferase expression in the transfected cells was determined by quantitative real-time polymerase chain reaction PCR (qRT-PCR). Total RNA was isolated from cells with Trizol (Tiangen, Beijing, China). The reverse transcription was finished with the iScriptTM gDNA Clear cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). And, the real-time PCR was finished by using multiple kits (SYBR Premix Ex TaqTM, RR036A, Takara Bio, Kusatsu, Japan). Furthermore, qRT-PCR reactions were finished in an ABI 7500FAST Sequence Detector System (ABI, Foster City, CA, USA). The luciferase forward and reverse primers were 5′-AATGTCCGTTCGGTTGGCAG-3′ and 5′-GGCTGCGAAATGCCCATACT-3′, respectively. And the actin (the loading control) forward and reverse primers were 5′-GGTCATCACTATTGGCAACG-3′ and 5′- ACGGATGTCAACGTCACACT-3′, respectively.
2.10. In vivo inhibition test
The BALB/c mice were randomly divided into three groups. At the beginning of the experiment, the mice were placed in an animal anesthesia machine (E-ZSystem, Palmer, PA, USA) and 1% isoflurane was used as an anesthetic. The mice received PBS (the blank control), the DiO-labeled EVs-ACE2 (60 μg, calculated by the total protein content), and an equal amount of the DiO-labeled EVs-Control (the negative control) via intranasal administration. Thirty minutes later, all of the mice were given 20 μL (12 μg, calculated by the total protein content) of the S-pseudovirus. Another 30 min later, they were sacrificed using a high dose of isoflurane. The nasal mucosa tissues of the mice were dissected and fixed with 4% paraformaldehyde for preparation of the cryosection slices with a thickness of 10 μm (CM1950, Leica, Solms, Germany). The tissue slices were imaged using a fluorescence microscope (Carl Zeiss, Dublin, CA, USA). The overlap proportion was determined using ImageJ (NIH) by calculating Pearson's value. The nasal mucosa tissues were also used for preparing the single-cell suspension for flow cytometry assay to detect the RFP fluorescence signal. Additionally, the quantification of luciferase expression in the nasal mucosa tissues was determined by qRT-PCR. Total RNA was isolated from the nasal mucosa tissues with Trizol (Tiangen). The reverse transcription was performed using the iScriptTM gDNA Clear cDNA Synthesis Kit (Bio-Rad).
2.11. WB analysis of the nasal mucosa tissue
The dissected nasal mucosa tissues were used to verify whether the native ACE2 protein was expressed. After they were cut into pieces, two mL of the cell lysate was added, and they were placed on a shaker at 37 °C for digestion for 1 h. The lysate was then filtered using nylon mesh to remove the residual tissue. The total protein was determined using a BCA kit, and the samples were processed by SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with Anti-ACE2 and were visualized using a ChemiDoc MP Imaging System (Bio-Rad).
2.12. In vivo preliminary safety studies
The EVs-ACE2 or EVs-Control (60 μg in 20 μL PBS) were administered into the nasal cavity of the BALB/c mice, and the blood was collected via the orbital vein three days later. The serum was collected and analyzed using an automated hematology analyzer (XT-2000i, Sysmex, Kobe, Japan) for the blood chemistry test, including alanine aminotransferase (ALT), total protein (TP), albumin (ALB), urea nitrogen (Urea), creatinine (CRE), calcium (Ca), phosphorus (P), potassium (K), and sodium (Na).
For the hematoxylin-eosin staining, the BALB/c mice were randomly divided into three groups. The mice received PBS (the blank control), EVs-ACE2 (60 μg), and the EVs-Control (60 μg) via nasal administration. The animals were then humanely sacrificed 72 h post-administration, and the major organs (heart, liver, spleen, lung, and kidney) were collected and fixed using 4% paraformaldehyde for histopathological examination.
2.13. Data analysis
Statistical analysis was performed using t-tests and one-way analysis of variance (ANOVA). Data were expressed as mean ± standard deviation (SD). Statistically, significant difference was defined as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3. Results
3.1. Characterization of EVs-ACE2
The engineered 293T cells with stable expression of full-length human ACE2 (HEK293T-ACE2) were constructed using a lentivirus. The 293T cells are an experimentally amenable, homogenous cell line that is routinely used in cell engineering24 and as exosome donors and packaging cells25,26. Moreover, the EVs derived from 293T cells have been demonstrated to be immunologically inert27,28. The EVs-ACE2 were purified from the cell culture supernatants using differential ultracentrifugation. Meanwhile, the EVs derived from the HEK293T control cells without ACE2 transfection (termed EVs-Control) were also prepared. The median particle diameter of the EVs-ACE2 was 58.5 nm, and that of the EVs-Control was 68.2 nm (Fig. 1B–D), determined by NTA. Transmission electronic microscopy showed the morphology of the EVs-ACE2 with a double-layer membrane (Fig. 1E).
The ACE2 expression on the EVs-ACE2 was confirmed by Western blot (WB) analysis (Fig. 1F). The quantitative analysis of ACE2 expression in EVs-ACE2 was determined by SDS-PAGE with Coomassie Brilliant Blue staining, using human ACE2 as the standard protein (Fig. 3B), and the ACE2 expression in EVs-ACE2 is 35 μg/mg total proteins (Fig. 3B). Furthermore, the stability of the resuspended EVs after ultracentrifugation was evaluated by NTA, and the EVs remained stable in the test time frame (Fig. 1G).
Figure 3.
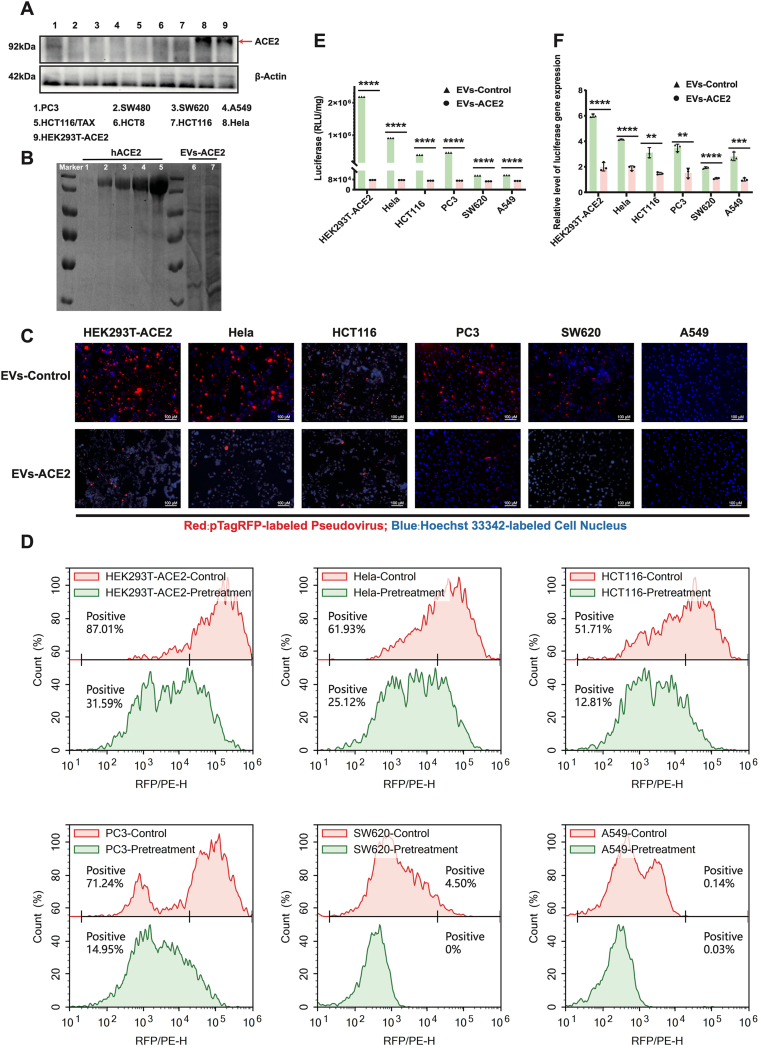
The EVs-ACE2 inhibited the pseudoviral infections in vivo. (A) The ACE2 levels in various cell lines. (B) Coomassie brilliant blue method: a standard protein human ACE2 was separately loaded in 0.025, 0.5, 0.75, 1.0, 1.5 μg to five lanes (lane 1–5); the whole EVs-ACE2 lysate was also separately loaded in amounts of 2.0 or 5.0 μg (total proteins) to two lanes (lane 6&7). (C) EVs-ACE2 inhibited the cell entry of S-pseudovirus. Scale bars = 100 μm. (D) Flow cytometry assay results of (C). (E) EVs-ACE2 inhibited the infection of S-pseudovirus; RLU detected at 48 h after pseudoviral inoculation; scale bar = 100 μm. (F) The luciferase expression levels were qualified by qRT-PCR assay. Data are presented as mean ± SD (n = 3). ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001; ns, no significance.
3.2. Characterization of SARS-CoV-2 S-pseudovirus
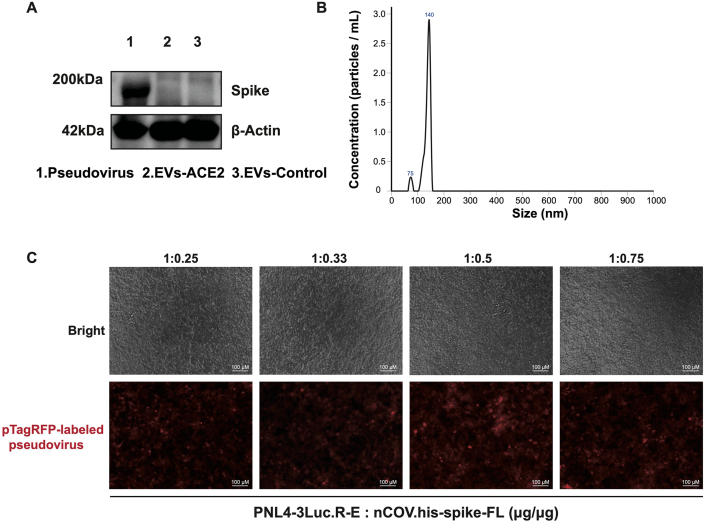
S-protein pseudovirus was generated by using a lentivirus-based pseudoviral system co-packing pNL4-3.Luc.R-E, nCOV.his-SPIKE-FL, and pTagt RFP. The transfection efficiency of the pseudovirus was reflected by the S-protein expression, which showed a high level by the WB analysis (Fig. 2A). The particle size of the S-pseudovirus is shown in Fig. 2B. Because pseudoviral particles could bind together, they showed different peaks. For the pTagRFP-labeled S-protein pseudovirus, the fluorescence intensity served as an indicator of the ability of the pseudovirus to enter the cells. According to the results (Fig. 2C), a packing ratio of 1:0.5 (w/w) between pNL4-3.Luc.R-E, nCOV.his-SPIKE-FL was optimal for preparing the pseudovirus.
Figure 2.
Characterization of the S-pseudovirus. (A) S-protein expression in the S-pseudovirus and EVs. (B) Size distributions of S-pseudovirus determined by NTA. (C) The S-pseudovirus prepared with a packing ratio of pNL4-3.Luc.RE and nCOV.his-spike-FL of 1:0.5 (w/w) had the highest efficiency of cell entry. Scale bar = 100 μm.
S-pseudovirus provides a useful model to safely study SARS-CoV-2 with benefits of non-replicability and tractability21, 22, 23. The one created in our study is characterized by three major functional components, namely, the viral envelope full-length S protein, a tracer protein red fluorescence protein (RFP), and a reporter luciferase gene. Thus, the infection process of the S-pseudovirus can be indicated by the RFP signal and luciferase activity.
3.3. Blockage of pseudovirus attachment onto the cells
The first step for a virus to infect a host cell is attachment onto the cell membrane via the viral receptors. In SARS-CoV-2, the S2 subunit of the S protein is highly conserved, and the S protein binds to the ACE2 receptor29. In order to investigate the cell entry of S-pseudovirus, several ACE2+ cell lines were applied. WB analysis showed that HeLa and HEK293T-ACE2 cells had a high level of ACE2 expression. HCT116 and PC3 cells had a moderate expression level, and SW620 and A549 cells had a minimal level of ACE2 expression (Fig. 3A). It was revealed that the cell entry efficiency was closely associated with the ACE2 expression level; for example, the cell entry efficiency of the S-pseudovirus was high in the HeLa and HEK293T-ACE2 cells but low in the SW620 and A549 cells. Importantly, the treatment with EVs-ACE2 resulted in the inhibition of the S-pseudovirus to enter the cells, which was reflected by the significant reduction of the pseudovirus-labeled fluorescence (Fig. 3C and D). These results demonstrated that EVs-ACE2 efficiently blocked the entry of S-pseudovirus into the cells.
3.4. Blockage of pseudovirus infection
The infection efficiency was performed by titrating the S-pseudovirus. After transfection, the co-packed pNL4-3.Luc.RE expressed luciferase. Therefore, luminescence intensity was a transfection indicator. The results showed that the HeLa, HCT116, and PC3 cells were more effectively transduced by the S-pseudovirus than the SW620 and A549 cells, and the highest transduction efficiency (approximately 2 × 106 RLU) was observed in the HeLa cells with the highest ACE2 expression, 20-fold higher than the SW620 or A549 cells with the low ACE2 expression (Fig. 3E). However, the infection efficiency in the cells was decreased by EVs-ACE2 treatment, and Fig. 3E shows that EVs-ACE2 treatment reduces 96% luciferase activity compared to the control group. It was demonstrated that the EVs-ACE2 significantly blocked the S-pseudovirus infection in the HeLa, HT116, and PC3 cells with high ACE2 expression. The results suggest that the entry and infection of SARS-CoV-2 were blocked by administering EVs-ACE2 to neutralize the virus. This was also verified by qRT-PCR (Fig. 3F).
3.5. Blockage against S-pseudovirus in the nasal epithelium
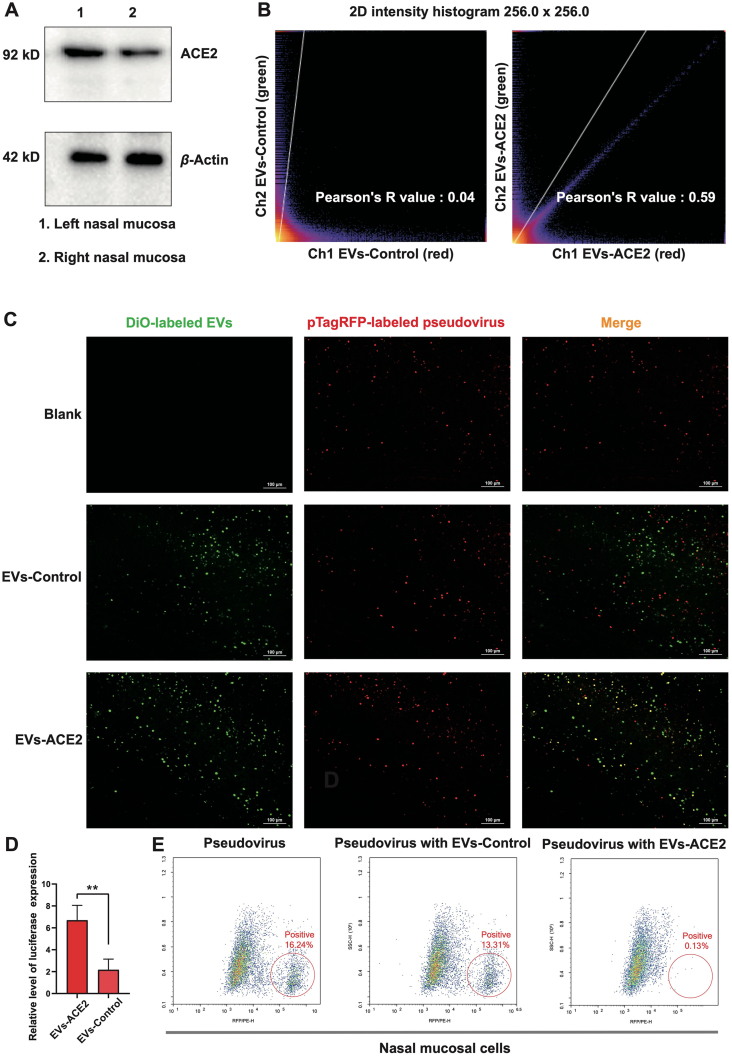
In symptomatic and asymptomatic patients, nasal swabs typically have been found to have a higher viral load of SARS-CoV-2 than throat swabs do30. Notably, nasal epithelial cells, containing goblet cells and ciliated cells, show high expression of ACE2. This indicates that the nasal cavity is the primary portal and incubator for SARS-CoV-2 to enter the human body31. The ACE2 protein was highly expressed in the nasal epithelium of the mice (Fig. 4A). The S-pseudovirus (red) was successfully captured by the EVs-ACE2 (green) that were intranasally pre-administered to the mice, reflected by the major overlap of the fluorescence of red and green. By contrast, in the EVs-Control group, there was much less overlap of the fluorescence (Fig. 4C). The overlap proportion was reflected in the Pearson's value: 0.59 (EVs-ACE2 group) vs. 0.04 (EVs-Control group) (Fig. 4B). In the qRT-PCR assay, we obtained the same conclusions (Fig. 4D).
Figure 4.
In vivo neutralization test. (A) ACE2 expression in the murine nasal mucosa. (B) The quantitative analysis of the overlap proportion in the images of (C) using ImageJ. (C) Fluorescence images of the nasal mucosa cryosection slices from the mice challenged by the S-pseudovirus with the DiO-labeled EVs-ACE2/EVs-Control pretreatment. Scale bars = 100 μm. (D) The luciferase expression levels of nasal mucosa cryosection tissues were qualified by qRT-PCR assay (n = 3). (E) Flow cytometry assay of nasal mucosal tissues after S-pseudovirus challenge. Data are presented as mean ± SD (n = 3). ∗∗P < 0.01, ns, no significance.
Furthermore, the epithelium tissues were processed for flow cytometry analysis, which showed that the S-pseudovirus positive rates in the non-treatment and EVs-Control groups were more than 100 times higher than the EVs-ACE2 group (16.2% and 13.1% vs. 0.13%, Fig. 4E).
There are olfactory epithelium and respiratory epithelium in the nasal cavity. Although they have a different biological function, both of them overexpress ACE232, 33, 34. As a whole, the results show an effective blockage against S-pseudovirus by treatment with ACE2-expressing EVs.
3.6. Preliminary safety evaluation
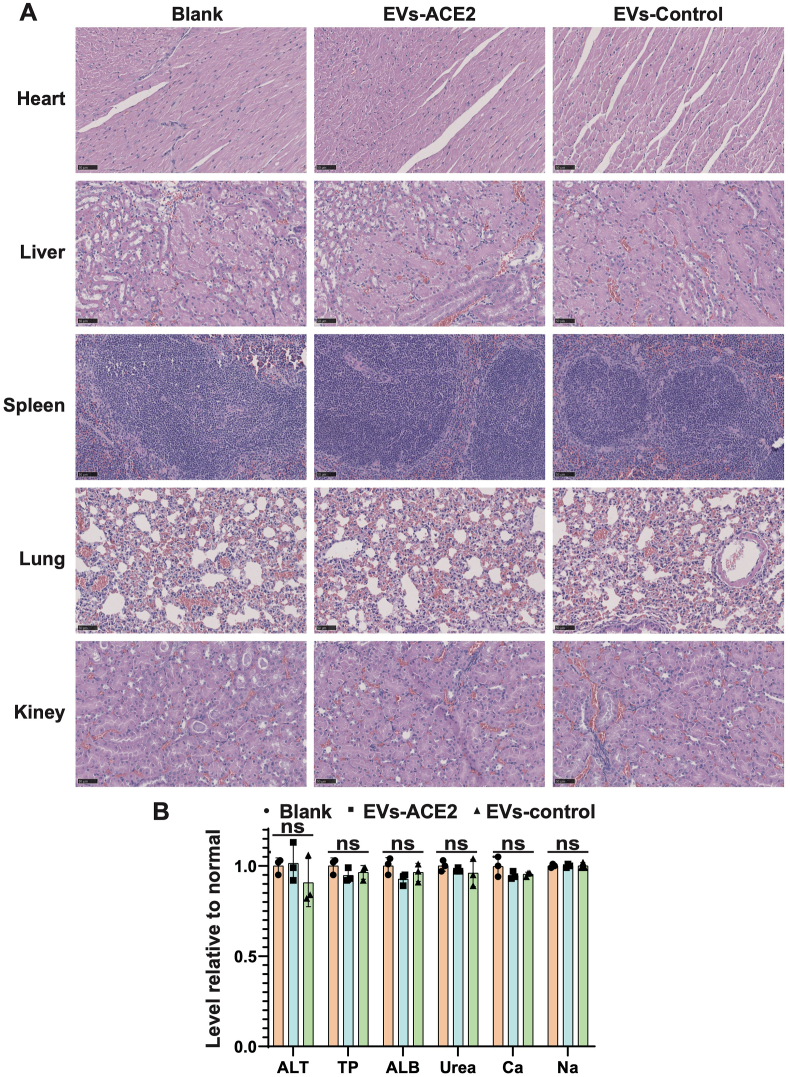
After intranasal administration of EVs-ACE2, a blood chemistry test was conducted three days later. The results showed no obvious changes, and all remained at the baseline levels (Fig. 5B). The histopathological analysis also reveals that there was no evidence of lesion or tissue damage (Fig. 5A).
Figure 5.
(A) Hematoxylin and eosin staining of the sections from the major organs taken 3 days after nasal administration of the EVs. Scale bars = 50 μm. (B) Serum chemistry test. TP, total protein; ALB, albumin; ALT, alanine aminotransferase; Urea, urea nitrogen; CRE, creatinine; Ca, calcium; P, phosphorus; K, potassium; and Na, sodium. Data are presented as mean ± SD (n = 3); ns, no significance.
4. Discussion
Neutralization strategy plays an important role in antivirus therapy. For example, monoclonal antibodies35,36, antisera37, and recombinant human ACE2 protein38 have also been investigated for neutralization treatment. Among them, the recombinant human ACE2 has been considered a promising therapeutic agent against COVID-1939. However, due to its short half-life, the therapeutic success has been limited. To address this issue, the use of nanocarriers to deliver ACE2 protein was developed to neutralize SARS-CoV-240. In this work, we proposed ACE2-expressing EVs as a means for preventing healthy individuals.
Engineered EVs have been actively explored as potent therapeutic candidates. Via cell engineering technology, it is feasible to prepare EVs bearing various types of functional proteins, with extra advantages of non-toxicity and good biocompatibility and stability in biofluids (e.g., plasma)41. Till July 2021, more than 200 clinical trials involving EVs-related treatments and diagnoses of different diseases have been registered at https://clinicaltrials.gov/.
EVs bearing decoy receptors that competitively bind with the target receptors as a potential treatment has been proposed in skeletal muscle pathophysiology42. It was proposed that ACE2-expressing EVs that bind with SARS-Cov-2 could be a possible therapy43, and subsequently, it was demonstrated by in vitro tests44. In this study, EVs with decoy ACE2 were used as an antagonist to neutralize SARS-CoV-2 pseudovirus via the ACE2/S–protein interaction. The EVs-ACE2 demonstrated the potent ability to neutralize pseudovirus in both in vitro and in vivo experiments. Specifically, we first showed that the intranasal pretreatment with EVs-ACE2 can block the viruses to enter the nasal epithelium, which typically serves as the primary portal for SARS-CoV-2. Of note, 41% of the people with COVID-19 had reported experiencing a loss of smell45. As nasal mucosa is a front-line defense against respiratory infection in the human body, the intranasal administration of therapeutics could be promising in limiting the spread of COVID-19. Importantly, intranasal dosing can be self-administrated and the formulation is easy to be given intranasally. It should be mentioned that physical protective equipment might not provide complete protection in the virus-rich indoor environments, as evidenced by the frequently reported cases in medical centers and hospitals. Therefore, to develop an intranasal medicine (e.g., EVs-ACE2) that could provide additional protection is clinically meaningful.
5. Conclusions
We reported a novel antivirus strategy where engineered EVs with decoy receptors as a nanoplatform can act as a safe and effective therapeutic avenue.
Acknowledgments
We are thankful for the support of National Special Project for Significant Drugs Development (2018ZX09711002-010-002, China), National Natural Science Foundation of China (81925035 and 81521005, China), Shanghai Sci-Tech Innovation Initiative (19431903100, 18430740800, China), the Shanghai Collaborative Innovation Group of Early Diagnosis and Precise Treatment of Hemangiomas and Vascular Malformations (SSMU-ZDCX20180701, China), the Sanofi-SIBS Yong Faculty Award, and The Youth Innovation Promotion Association. We thank the Molecular Imaging Center and TEM Facility at SIMM for the technical support.
Author contributions
Yongzhuo Huang, Hong Qiu and Huiyuan Wang designed the research. Canhao Wu and Bin Tu carried out the experiments and performed data analysis. Jiaxin Zeng, Pengfei Zhao and Mingjie Shi participated part of the experiments. Qin Xu and Hong Qiu provided experimental drugs and quality control. Huiyuan Wang wrote the manuscript. Yongzhuo Huang revised the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.09.004.
Contributor Information
Huiyuan Wang, Email: wanghuiyuan@simm.ac.cn.
Hong Qiu, Email: hongqiu@simm.ac.cn.
Yongzhuo Huang, Email: yzhuang@simm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.COVID-19 coronavirus pandemic. https://www.worldometers.info/coronavirus/ Available from:
- 2.Weng J., Li Y., Li J., Shen L., Zhu L., Liang Y., et al. Gastrointestinal sequelae 90 days after discharge for COVID-19. Lancet Gastroenterol Hepatol. 2021;6:344–346. doi: 10.1016/S2468-1253(21)00076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Huang L., Wang Y., Li X., Ren L., Gu X., et al. 6-Month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh B., Ryan H., Kredo T., Chaplin M., Fletcher T. Chloroquine or hydroxychloroquine for prevention and treatment of COVID-19. Cochrane Database Syst Rev. 2021;2 doi: 10.1002/14651858.CD013587.pub2. CD013587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosas I.O., Brau N., Waters M., Go R.C., Hunter B.D., Bhagani S., et al. Tocilizumab in hospitalized patients with severe COVID-19 pneumonia. N Engl J Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zamorano Cuervo N., Grandvaux N. ACE2: evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife. 2020;9:e61390. doi: 10.7554/eLife.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G., Yang M., Duan Z., Liu F., Jin, Long C., et al. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y., Li J., Pang Z. Recent insights for the emerging COVID-19: drug discovery, therapeutic options and vaccine development. Asian J Pharm Sci. 2021;16:4–23. doi: 10.1016/j.ajps.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.004. 905-13.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haschke M., Schuster M., Poglitsch M., Loibner H., Salzberg M., Bruggisser M., et al. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52:783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu J., Ren L., Li S., Li W., Zheng X., Yang Y., et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021 doi: 10.1016/j.apsb.2021.01.001. https://www.sciencedirect.com/science/article/pii/S2211383521000058 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samanta S., Rajasingh S., Drosos N., Zhou Z., Dawn B., Rajasingh J. Exosomes: new molecular targets of diseases. Acta Pharmacol Sin. 2018;39:501–513. doi: 10.1038/aps.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syn N.L., Wang L., Chow E.K., Lim C.T., Goh B.C. Exosomes in cancer nanomedicine and immunotherapy: prospects and challenges. Trends Biotechnol. 2017;35:665–676. doi: 10.1016/j.tibtech.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Wang Y., Lv Q., Li X. Exosomes: from garbage bins to translational medicine. Int J Pharm. 2020;583:119333. doi: 10.1016/j.ijpharm.2020.119333. [DOI] [PubMed] [Google Scholar]
- 18.Wiklander O.P.B., Brennan M.A., Lotvall J., Breakefield X.O., El Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aav8521. eaav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zipkin M. Big pharma buys into exosomes for drug delivery. Nat Biotechnol. 2020;38:1226–1228. doi: 10.1038/s41587-020-0725-7. [DOI] [PubMed] [Google Scholar]
- 20.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30 doi: 10.1002/0471143030.cb0322s30. 3.22.1-3.9. [DOI] [PubMed] [Google Scholar]
- 21.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7:551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S., Tai C., Hsu Y., Cheng D., Hung S., Chai K.M., et al. Assessing the application of a pseudovirus system for emerging SARS-CoV-2 and re-emerging avian influenza virus H5 subtypes in vaccine development. Biomed J. 2020;43:375–387. doi: 10.1016/j.bj.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas P., Smart T.G. HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods. 2005;51:187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Wang Y., Bai M., Wang J., Zhu K., Liu R., et al. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor siRNA. Cancer Sci. 2018;109:629–641. doi: 10.1111/cas.13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Q., Shi X., Han M., Smbatyan G., Lenz H.J., Zhang Y. Reprogramming exosomes as nanoscale controllers of cellular immunity. J Am Chem Soc. 2018;140:16413–16417. doi: 10.1021/jacs.8b10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X., Badawi M., Pomeroy S., Sutaria D.S., Xie Z., Baek A., et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J Extracell Vesicles. 2017;6:1324730. doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Virgiliis F., Di Giovanni S. Lung innervation in the eye of a cytokine storm: neuroimmune interactions and COVID-19. Nat Rev Neurol. 2020;16:645–652. doi: 10.1038/s41582-020-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DosSantos M.F., Devalle S., Aran V., Capra D., Roque N.R., Coelho-Aguiar J.M., et al. Neuromechanisms of SARS-CoV-2: a review. Front Neuroanat. 2020;14:37. doi: 10.3389/fnana.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soltani Zangbar H., Gorji A., Ghadiri T. A Review on the neurological manifestations of COVID-19 infection: a mechanistic view. Mol Neurobiol. 2021;58:536–549. doi: 10.1007/s12035-020-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cathcart A.L., Havenar-Daughton C., Lempp F.A., Ma D., Schmid M.A., Agostini M.L., et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2021 https://www.biorxiv.org/content/10.1101/2021.03.09.434607v6.full Available from: [Google Scholar]
- 36.Ledford H. COVID antibody treatments show promise for preventing severe disease. Nature. 2021;591:513–514. doi: 10.1038/d41586-021-00650-7. [DOI] [PubMed] [Google Scholar]
- 37.Hueso T., Pouderoux C., Pere H., Beaumont A., Raillon L., Ader F., et al. Convalescent plasma therapy for B-cell-depleted patients with protracted COVID-19. Blood. 2020;136:2290–2295. doi: 10.1182/blood.2020008423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoufaly A., Poglitsch M., Aberle J.H., Hoepler W., Seitz T., Traugott M., et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnamurthy S., Lockey R.F., Kolliputi N. Soluble ACE2 as a potential therapy for COVID-19. Am J Physiol Cell Physiol. 2021;320:C279–C281. doi: 10.1152/ajpcell.00478.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q., Honko A., Zhou J., Gong H., Downs S.N., Vasquez J.H., et al. Cellular nanosponges inhibit SARS-CoV-2 infectivity. Nano Lett. 2020;20:5570–5574. doi: 10.1021/acs.nanolett.0c02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu W., Hurley J., Roberts D., Chakrabortty S.K., Enderle D., Noerholm M., et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466–477. doi: 10.1016/j.annonc.2021.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conceicao M., Forcina L., Wiklander O.P.B., Gupta D., Nordin J.Z., Vrellaku B., et al. Engineered extracellular vesicle decoy receptor-mediated modulation of the IL6 trans-signalling pathway in muscle. Biomaterials. 2021;266:120435. doi: 10.1016/j.biomaterials.2020.120435. [DOI] [PubMed] [Google Scholar]
- 43.Inal J.M. Decoy ACE2-expressing extracellular vesicles that competitively bind SARS-CoV-2 as a possible COVID-19 therapy. Clin Sci. 2020;134:1301–1304. doi: 10.1042/CS20200623. Lond. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cocozza F., Nevo N., Piovesana E., Lahaye X., Buchrieser J., Schwartz O., et al. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 spike protein-containing virus. J Extracell Vesicles. 2020;10:e12050. doi: 10.1002/jev2.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agyeman A.A., Chin K.L., Landersdorfer C.B., Liew D., Ofori-Asenso R. Smell and taste dysfunction in patients with COVID-19: a systematic review and meta-analysis. Mayo Clin Proc. 2020;95:1621–1631. doi: 10.1016/j.mayocp.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.