Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is a chronic, progressive liver disease with an increasing incidence rate. This study investigated the protective effects of live combined Bacillus subtilis and Enterococcus faecium (LCBE) on NAFLD, and its possible mechanisms.
Material/Methods
Five-week-old C57BL/6 mice were randomly divided into 3 groups: chow, HFD, and HFD+LCBE groups. The levels of serum biochemical markers, glucose tolerance, insulin, the inflammatory cytokines IL-1β, IL-6, and TNF-α, LPS, and histological staining were measured using commercial kits. qPCR was used to examine the mRNA expression levels of inflammatory cytokines in the liver. Western blotting was used to determine the protein levels of TLR4, NF-κB p65, PPAR-α, and CPT-1 in the liver, and occludin and Claudin1 in the intestine. The intestinal flora of the mice was analyzed by high-throughput sequencing of the V3–V4 region of 16S rDNA.
Results
LCBE significantly lowered the body weight, liver/body weight ratio, and serum glucose level, and increased the serum insulin level in NAFLD mice. In addition, LCBE treatment improved the liver function and lipid profile, decreased the levels of LPS and inflammatory cytokines, and downregulated the expression of TLR4 and NF-κB p65. Moreover, LCBE enhanced the intestinal barrier function by increasing the expression of occludin and Claudin1. Furthermore, LCBE modulated the composition of the gut microbiota by reducing the Firmicutes to Bacteroidetes ratio, and the proportion of inflammation-related and LPS-producing bacteria, thus re-arranging the structure of the gut microbiota.
Conclusions
LCBE protects against NAFLD by alleviating inflammation, restoring the intestinal barrier, and modulating gut microbiota composition.
Keywords: Bacillus subtilis, Enterococcus faecium, Inflammation, Liver Diseases, Microbiota
Background
Owing to the significant increase in obesity rates worldwide, the incidence of non-alcoholic fatty liver disease (NAFLD), a metabolic syndrome, has also increased, and it is not restricted to developed countries. The disease spectrum of NAFLD is wide, ranging from simple steatosis to more serious forms, including non-alcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and hepatic cellular carcinoma [1]. This disorder affects the health of both adults and children, leading to a worldwide disease burden [2,3]. Despite the in-depth research on NAFLD, its pathogenesis remains poorly understood. A series of studies has focused on the role of inflammation in the pathogenesis of NAFLD [4]. Excess LPS flux to the liver from the gut triggers an inflammatory response in the liver [5]. Additionally, obesity induces the recruitment of inflammatory cells, such as macrophages, and results in an increase in the release of inflammatory cytokines, including TNF-α and IL-1β, aggravating the pathogenesis of NAFLD [6]. It is widely accepted that the “multiple hits” hypothesis is involved in the pathogenesis of NAFLD. Recent studies have provided evidence that gut microbiota, one of the key factors in the “multiple hits,” play an important role in the progression of NAFLD [7–10].
The number of human intestinal microbial cells, ranging from 100 trillion to 1000 trillion, is approximately 10 times the number of human cells in the body [11]. An imbalance of the gut microbiota is related to chronic diseases, including NAFLD. Firmicutes and Bacteroidetes are the major components of gut microbiota. One study showed that NAFLD patients had fewer Bacteroidetes and more Firmicutes than healthy individuals [10]. Another study reported that sterile mice transplanted with microbiota from high-fat diet (HFD)-fed mice were prone to NAFLD [12]. Moreover, germ-free mice transplanted with cold-exposed microbiota had higher insulin sensitivity and lower fat levels [13]. The long-term consumption of a diet with high sugar and fat contents results in gut dysbiosis, which causes the production of LPS. Specifically, the gut-liver axis is under intense scrutiny to determine whether it is involved in the incidence and progression of NAFLD. Studies have shown that LPS produced by gram-negative bacteria in the intestine impairs the integrity of the intestinal barrier, and activates inflammation by binding to TLR4 on the cell surface, leading to the progression of NAFLD [14–17]. Recently, increasing attention has been focused on the effects of probiotics on host health, as well as NAFLD. Some studies have indicated that probiotic supplementation with Lactobacillus species can alleviate inflammation, enhance intestinal barrier function, and thus ameliorate NAFLD [18]. Xue et al found that probiotic administration could improve NAFLD patient health by alleviating the inflammatory response [19].
Live combined Bacillus subtilis and Enterococcus faecium (LCBE) has been reported to reduce the mortality rate by modulating the inflammatory response and intestinal barrier function in CLP-induced sepsis [20]. However, the effect of LCBE on NAFLD remains unclear. Therefore, we hypothesized that LCBE may amend NAFLD by alleviating inflammation, restoring the intestinal barrier, and modulating gut microbiota. This study demonstrates that LCBE administration is a potential candidate for treating metabolic diseases, including NAFLD.
Material and Methods
Animals and Experimental Design
All experimental procedures were conducted in accordance with the guidelines of the National Research Council Guide for Care and Use of Laboratory Animals and were approved by the Committee on Care and Use of Laboratory Animals of the Medical College of Tongji University, China (TJHBLAC-2020-72).
Male C57BL/6J mice (5 weeks old) were procured from Shanghai Silaike Experimental Animal Co. Ltd. (Shanghai, China). All the mice were acclimatized in a controlled environment (temperature 20–24°C; relative humidity 60–70%, 12-h daylight cycle) with free access to food and water. After 1 week of acclimation, the mice were randomly allocated into 3 groups: (1) chow group (the mice received a chow diet, n=5); (2) HFD group (the mice received an HFD, n=5); and (3) LCBE group (the mice received an HFD and LCBE by gavage once daily, n=6). The chow group was fed a chow diet, and the HFD and LCBE groups were fed with an HFD providing 60% kcal fat from TROPHIC Animal Feed High-Tech Co. Ltd. (Jiangsu, China), for 16 weeks. The LCBE group mice received 0.5 mL LCBE containing 1.4×109 CFU of B. subtilis and 1.55×1010 CFU of E. faecium by gavage per day for 16 weeks. Mice in the chow and HFD groups received the same volume of distilled water by gavage. The LCBE used in the experiments was obtained from Beijing Hanmi Pharmaceutical Co., Ltd. (China). After a 16-h fast, the mice were anesthetized, and blood, liver, intestine, and colonic contents were collected.
Biochemical Analysis
Serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), triglyceride (TG), total cholesterol (Cho), low-density lipoprotein cholesterol (LDL), and high-density lipoprotein cholesterol (HDL) were measured using commercial kits (Jiancheng Biomedical Company, Nanjing, China) according to the manufacturer’s instructions. The serum levels of inflammatory cytokines, such as TNF-α, IL-β, and IL-6, were detected using ELISA kits (Jingmei, Jiangsu, China). After a 16-h fast, the mice were administered glucose (2.0 g/kg, intraperitoneally). Blood was obtained from the tail vein at 0, 15, 30, 60, and 120 min to measure blood glucose levels using a glucometer (ACCU-CHEK, Roche Diabetes, Germany). ELISA was performed using commercial kits to measure the serum insulin levels (Cusabio, Wuhan, China) and serum LPS levels (Bioendo Technology Co., Xiamen, China). The method for representing insulin sensitivity was based on the homeostatic model assessment for insulin resistance (HOMA-IR), which was calculated as follows:
Histological Analysis
Livers and intestines were fixed in 4% paraformaldehyde for 24 h at 4°C, embedded in paraffin, and sliced into 4-μm slices for hematoxylin and eosin (H&E) staining. To detect lipid deposition in the liver, liver samples were frozen, cut into 8-μm-thick sections, and stained with Oil Red O. The sections were visualized and imaged using a light microscope (Nikon, Tokyo, Japan).
RNA Isolation and Quantitative Real-time PCR (qPCR)
Total RNA was isolated from liver tissues using TRIzol reagent (Takara, Dalian, China) and reverse-transcribed into cDNA using a cDNA synthesis kit (RR036A, Takara, Dalian, China).
Specific gene expression was detected by qPCR using SYBR Green (Qiagen, Hilden, Germany) with an Applied Biosystems QuantStudio 5 instrument. Relative gene expression was calculated using the 2−ΔΔCt method. The primers used in this study are shown in Table 1.
Table 1.
List of primer sequences used for qRT-PCR.
| Genes | Forward primer | Reverse primer |
|---|---|---|
| IL-1β | TTCACCATGGAATCCGTGTC | GTCTTGGCCGAGGACTAAGG |
| TNF-α | GGCCTCTCTACCTTGTTGCC | CAGCCTGGTCACCAAATCAG |
| IL-6 | TTGCCTTCTTGGGACTGATG | CCACGATTTCCCAGAGAACA |
| GAPDH | TGTGTCCGTCGTGGATCTGA | TTGCTGTTGAAGTCGCAGGAG |
Western Blotting
Protein extracts were prepared using a protein lysis kit (Beyotime, China) with a protease inhibitor (Beyotime), and protein lysates were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Next, the proteins in the gel were transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore). The membranes were washed and blocked with 5% BSA in Tris-buffered saline and Tween-20 (TBST) for 1 h at room temperature. The membranes were then incubated with primary antibodies, including anti-NF-κB (1: 2000, Proteintech, 66535-1-lg), anti-TLR4 (3.5 μg/mL, Abcam, ab13867), anti-CPT-1 (1: 1000, Proteintech,15184-1-AP), anti-PPAR-α (1 μg/mL, Abcam, ab126285), anti-Claudin1 (1: 5000, Abcam, ab180158), anti-occludin (1: 1000, Abcam, ab167161), and anti-GAPDH (1: 25000, Proteintech, 60004-1-lg) for 16 h at 4°C. After washing with TBST, the membranes were incubated with IgG conjugated with horseradish peroxidase (HRP; Thermo Fisher, USA) for 1 h at room temperature. Finally, enhanced chemiluminescence (ECL; Beyotime) was employed to detect protein bands using the ChemiDoc XRS system (Bio-Rad Laboratories, CA, United States). Densitometry analysis of each band was conducted using ImageJ software.
Bacterial DNA Extraction and 16S rDNA Bioinformatics Analysis
Total DNA from colonic contents was extracted using the TIANamp Stool DNA Kit (Tiangen Biotech, Beijing, China). After confirming the DNA integrity, the V3–V4 region of the bacterial 16S rDNA was amplified according to the following cycling conditions: initial denaturation at 95°C for 3 min, and 30 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s. The 16S rDNA PCR primers used were as follows: 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). After extraction based on quantitative analysis, and purification, the amplicons were merged at a ratio of 1: 1. A library was constructed using an Illumina HiSeq 2500 platform. Operational taxonomic units (OTUs) were clustered with 97% sequence identity based on the 16S raw data sequence, which was analyzed using QIIME (Quantitative Insights Into Microbial Ecology).
Statistical Analysis
All data are expressed as means±standard deviations (SD) for each group, and data analysis was performed using SPSS software version 20.0. The differences among groups were determined by one-way analysis of variance (ANOVA), and P<0.05 was considered to indicate a significant difference.
Results
LCBE Prevents Abnormal Gain of Body Weight and Liver Index And Improves Glucose Tolerance and Insulin Resistance
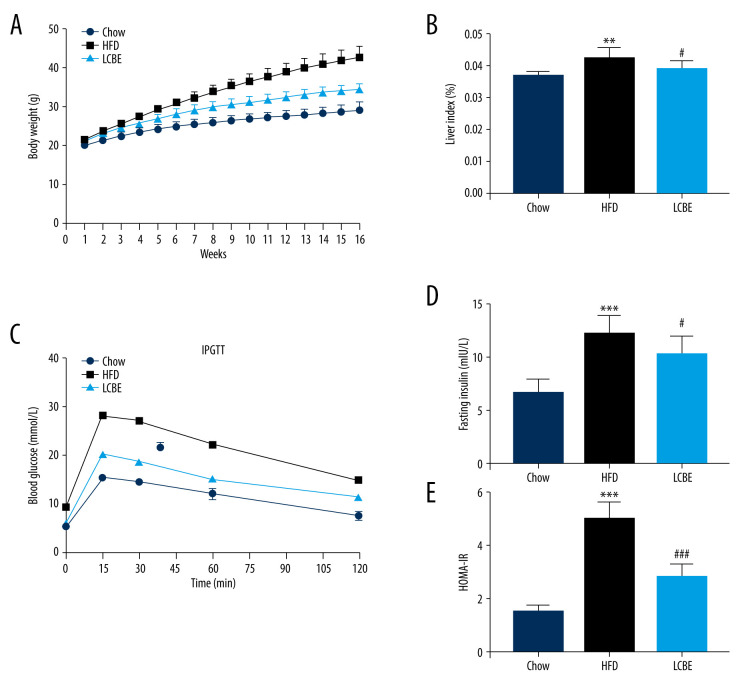
To examine the effects of LCBE on NAFLD in mice, we determined their body weights, and found that the body weight in the HFD group was higher than that in the chow group for 16 weeks (Figure 1A). LCBE treatment significantly prevented abnormal body weight gain in HFD-fed mice (Figure 1A). Furthermore, the HFD increased the liver index compared with that in the chow group, and LCBE intervention reversed this (Figure 1B). Additionally, we assessed the glucose levels and insulin sensitivity, and the results showed that HFD-fed mice had higher blood glucose, serum insulin, and HOMA-IR levels than mice in the chow group (Figure 1C–1E). LCBE administration improved the blood glucose, serum insulin, and HOMA-IR levels, indicating that LCBE improved glucose tolerance and IR in the HFD-fed mice.
Figure 1.
LCBE prevents abnormal weight, liver weight, and liver index levels, and improves glucose tolerance and insulin resistance in mice fed a high-fat diet (HFD). (A) Body weight; (B) liver index; and (C) blood glucose concentration in the intraperitoneal glucose tolerance test (IPGTT); (D) levels of serum insulin; and (E) determination of HOMA-IR. The data are expressed as means±SD. # P<0.05, ## P<0.01, ### P<0.001 vs Chow, * P<0.05, ** P<0.01, *** P<0.001 vs HFD.
LCBE Alleviate Hepatic Steatosis
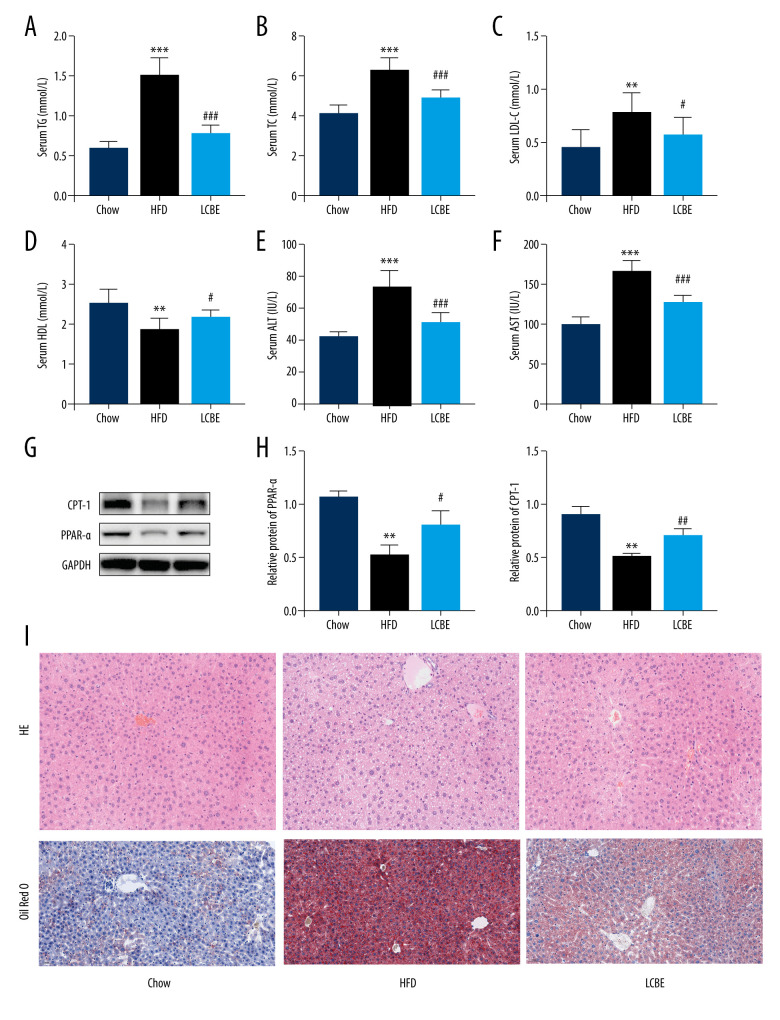
We assessed the serum lipid parameters in all the groups and found that HFD remarkably elevated the levels of serum TG, Cho, and LDL, and decreased the level of serum HDL (Figure 2A–2D). LCBE intervention markedly decreased the serum TG, Cho, and LDL levels, and upregulated the serum HDL levels compared with those in the HFD group (Figure 2A–2D). In addition, LCBE significantly reduced the high serum levels of ALT and AST induced by the HFD (Figure 2E, 2F). Since LCBE affected serum lipid homeostasis and liver function in NAFLD mice, we detected the expression of PPAR-α and CPT-1, and conducted histological analyses of the liver, including H&E and Oil Red O staining, to determine the role of LCBE in hepatic steatosis. We found that LCBE treatment elevated the protein expression levels of PPAR-α and CPT-1 in the livers of HFD-fed mice (Figure 2G, 2H). As shown in Figure 2I, there were more lipid droplets in the livers of NAFLD mice than in the chow group, whereas LCBE treatment distinctly alleviated abnormal lipid accumulation in the liver.
Figure 2.
Effect of LCBE on serum lipid metabolism, ALT, and AST levels in mice fed a high-fat diet. (A) Serum triglyceride; (B) serum total cholesterol; (C) serum low-density lipoprotein cholesterol; (D) serum high-density lipoprotein cholesterol; (E) activity of serum ALT; (F) activity of serum AST; (G) histopathology images of liver tissues ((I) H&E, 200× magnification; Oil Red O, 200× magnification); and (H) protein expression levels of PPAR-α and CPT-1 in the liver. GAPDH was used in the loading chow. The data are expressed as means±SD. * P<0.05, ** P<0.01, *** P<0.001 vs Chow, # P<0.05, ## P<0.01, ### P<0.001 vs HFD.
LCBE Reduces Inflammatory Response
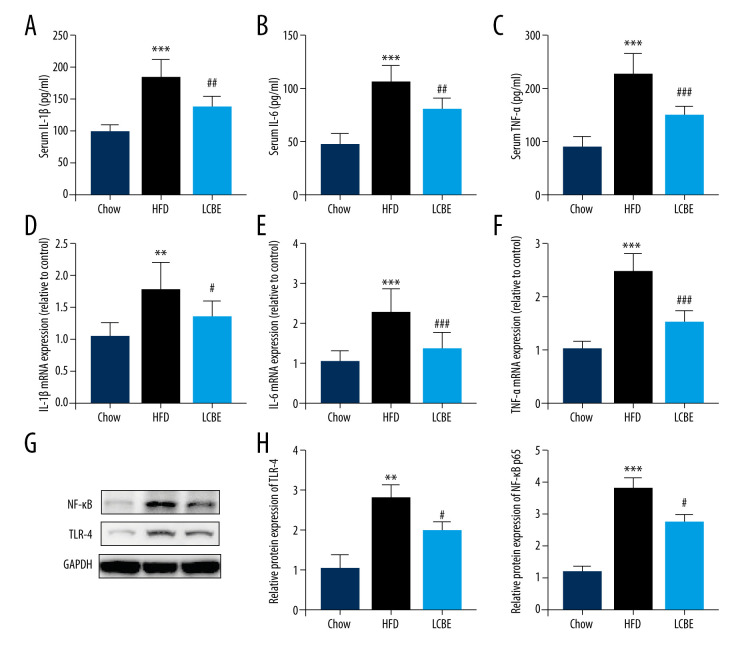
Based on the important role of the inflammatory response in the development and progression of NAFLD, the expression levels of genes associated with inflammation were determined. The expression levels of serum IL-1β, IL-6, and TNF-α were higher in the HFD group than in the chow group (Figure 3A–3C). Interestingly, treatment with LCBE remarkably reduced the IL-1β, IL-6, and TNF-α levels (Figure 3A–3C). We measured the expression of inflammatory cytokines in the liver. Consistent with the above results, the expression of IL-1β, IL-6, and TNF-α was significantly higher in the HFD group than in the chow group, but the expression of these inflammatory cytokines was markedly decreased by LCBE (Figure 3D–3F). Furthermore, we performed western blotting to determine the expression of genes related to inflammatory signaling pathways, such as those of TLR4 and NF-κB p65. Compared with the control group, the HFD group showed higher expression of TLR4 and NF-κB, whereas LCBE intervention prevented the increase in TLR4 and NF-κB p65 levels in the liver (Figure 3G, 3H).
Figure 3.
LCBE mitigates systemic inflammation in mice fed a high-fat diet (HFD). The levels of (A, D) interleukin 1β (IL-1β), (B, E) interleukin 6 (IL-6), and (C, F) tumor necrosis factor α (TNF-α) in the serum and liver. (G–H) Protein expression levels of Toll-like receptor 4 (TLR4) and nuclear factor κB (NF-κB) in the liver. The data are expressed as means±SD. * P<0.05, ** P<0.01, *** P<0.001 vs Chow, # P<0.05, ## P<0.01, ### P<0.001 vs HFD.
LCBE Reduces Serum LPS Levels and Improves Intestinal Barrier Integrity
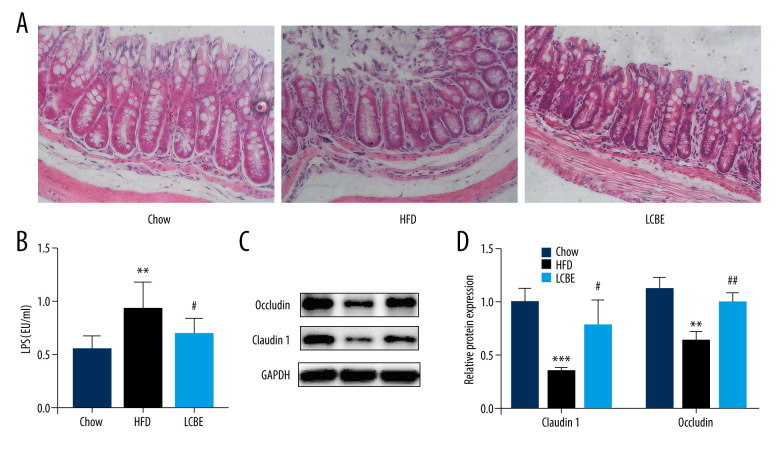
Intestinal barrier integrity not only affects systemic inflammation but is also involved in the development of NAFLD. H&E staining of the intestine revealed that the intestinal mucosa villi in chow group were slim, neatly arranged, and integral. However, the intestinal mucosa villi in the HFD group showed edema and the villus height was decreased; this was repaired to a certain degree by LCBE intervention (Figure 4A). Moreover, LPS, translocating into the blood recycle from a leaky gut, reflects the integrity of the intestinal barrier. We found that the expression of serum LPS was higher in the HFD group than in the chow group, and LCBE intervention decreased LPS expression (Figure 4B). Epithelial tight junction molecules play a critical role in intestinal barrier integrity. As expected, the protein levels of occludin and Claudin1 were lower in the HFD group than in the chow group (Figure 4C, 4D). In contrast, LCBE treatment reversed the downregulation of these proteins in NAFLD mice (Figure 4C, 4D).
Figure 4.
LCBE consolidates the intestinal barrier in mice fed a high-fat diet. (A) Histopathology images of intestine tissues (H&E, 200× magnification). (B) LPS levels in the serum. (C, D) Protein expression levels of occludin and Claudin1 in the intestine. The data are expressed as means±SD. * P<0.05, ** P<0.01, *** P<0.001 vs Chow, # P<0.05, ## P<0.01, ### P<0.001 vs HFD.
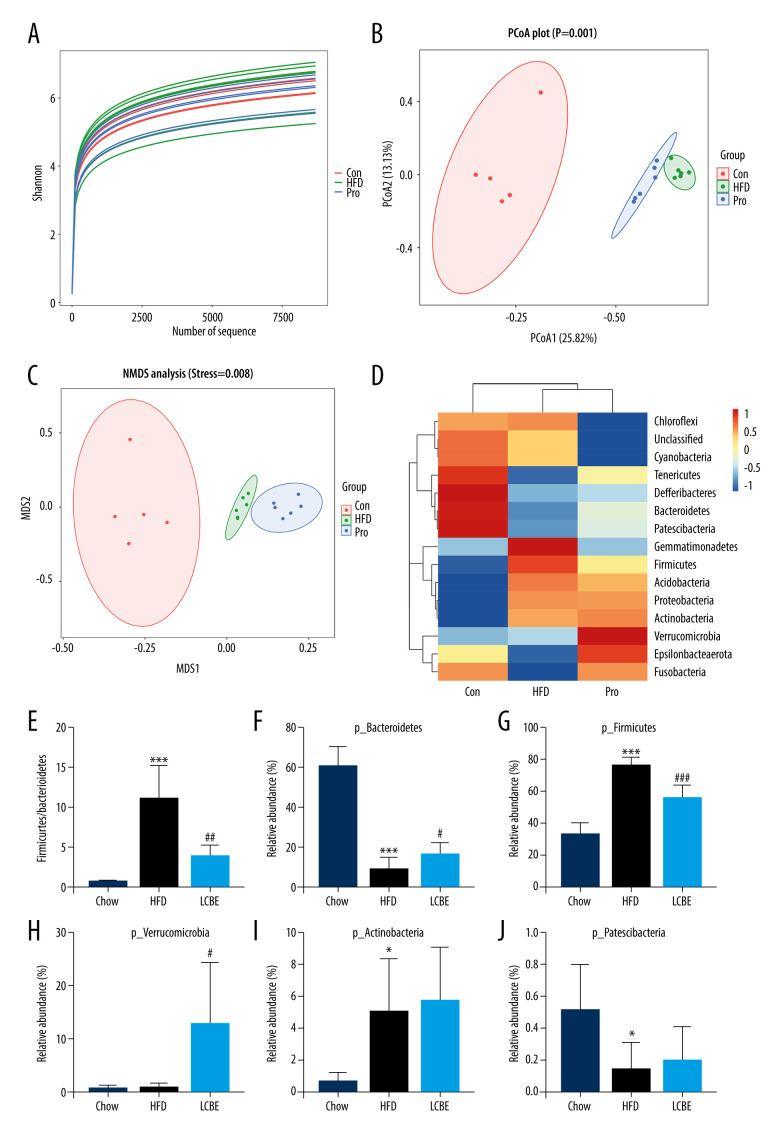
LCBE Regulates Intestinal Flora Composition of Gut Microbiota
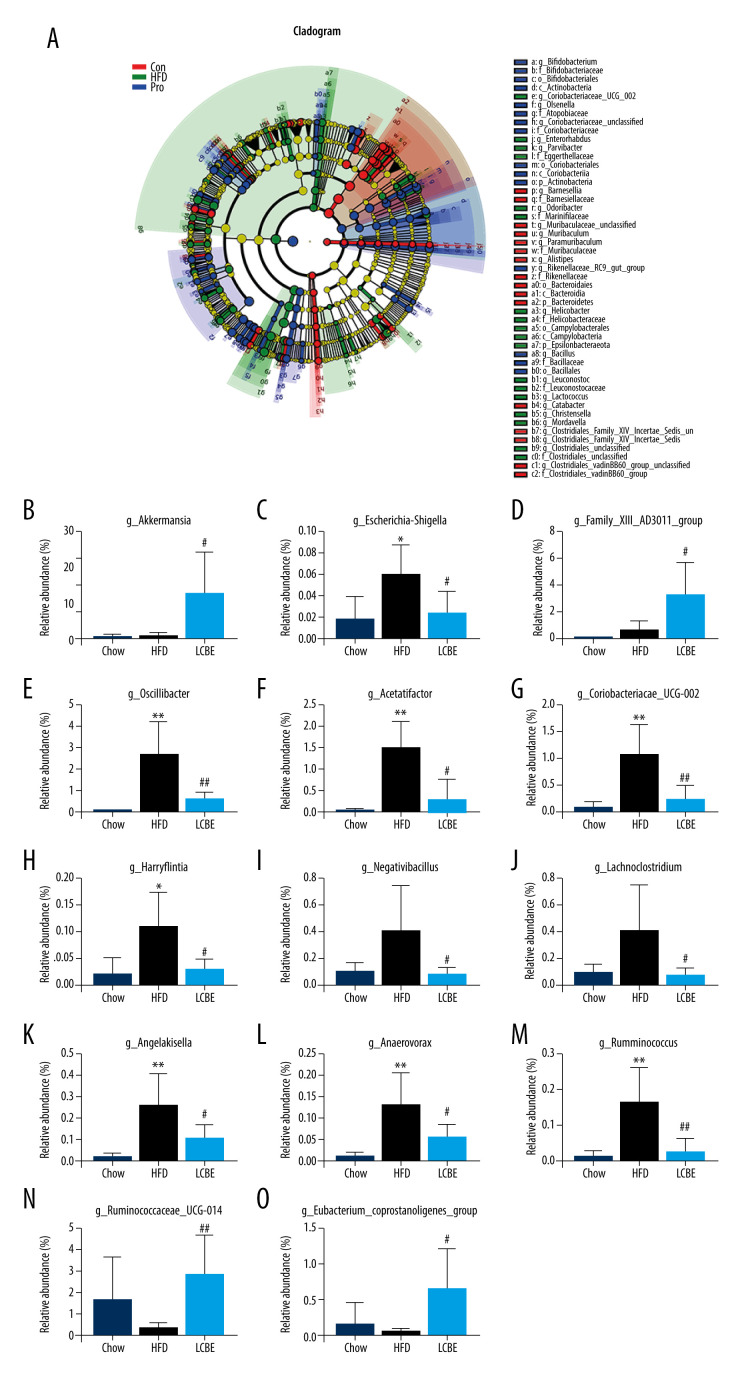
The V3–V4 region of 16S rDNA in mice fecal samples was used to evaluate the effects of LCBE on the microbiota. The Shannon dilution curve tended to be smooth for all the samples, indicating that the sequencing depth was sufficient to reveal the biodiversity in the samples (Figure 5A). Next, we conducted PCoA and NMDS analyses to investigate the effects of LCBE on the gut microbiota structure. The PCoA and NMDS of the unweighted UniFrac distance showed a remarkable separation among the compositions of gut microbiota in the chow, HFD, and LCBE groups (Figure 5B, 5C). In addition, the microbes in the gut of the mice could be classified into 15 phyla, and Firmicutes and Bacteroidetes were the dominant bacterial phyla in all the groups. As shown in Figure 5D, the microbial composition in the HFD group was remarkably different from that in the chow group. At the phylum level, the HFD increased the abundance of Firmicutes and decreased that of Bacteroidetes compared with that in the chow group (Figure 5F, 5G). Therefore, the HFD significantly increased the ratio of Firmicutes to Bacteroidetes (F/B ratio). LCBE treatment reversed the F/B ratio by downregulating the abundance of Firmicutes and upregulating the abundance of Bacteroidetes (Figure 5E). In addition, the relative abundance of Verrucomicrobia, Actinobacteria, and Patescibacteria changed at the phylum level (Figure 5H–5J). Furthermore, the LDA effect size (LEfSe) method was used to determine the different bacterial taxa. The results from LEfSe analysis demonstrated that some genera (Figure 6A), such as Akkermansia, Ruminococcus_UCG-014, Family_XIII_AD3011_group, Oscillibacter, Acetatifactor, Coriobacterium_UCG-002, Eubacterium_coprostanoligenes_group, Anaerovorax, Negativibacillus, Lachnoclostridium, Angelakisella, Ruminococcus, Harryflintia, and Escherichia-Shigella, were notably different. Compared with that in the chow group, the HFD promoted a remarkable increase in the relative abundance of Akkermansia, Lachnoclostridium, Oscillibacter, Family_XIII_AD3011_group, Anaerovorax, Acetatifactor, Coriobacterium_UCG-002, Negativibacillus, Angelakisella, Ruminococcus, Harryflintia, and Escherichia-Shigella (Figure 6B–6M), while decreasing the relative abundance of Ruminococcus_UCG-014 and Eubacterium_coprostanoligenes_group (Figure 6N, 6O). The administration of LCBE dramatically elevated the relative abundance of Akkermansia, Ruminococcus_UCG-014, and Eubacterium_coprostanoligenes_group, and reduced that of Lachnoclostridium, Oscillibacter, Family_XIII_AD3011_group, Anaerovorax, Acetatifactor, Coriobacterium_UCG-002, Negativibacillus, Angelakisella, Ruminococcus, Harryflintia, and Escherichia-Shigella. These results indicated that LCBE modulated gut microbiota dysbiosis by promoting beneficial bacterial proliferation and inhibiting harmful flora expansion.
Figure 5.
LCBE modulates the gut microbiota community in mice fed a high-fat diet. (A) Shannon curve of each sample. (B) Principal coordinates analysis (PCoA) of microbial communities based on the unweighted Unifrac cluster for each sample at the phylum level. (C) Non-metric multidimensional scaling (NMDS) of microbial communities based on the unweighted Unifrac cluster for each sample at the phylum level. (D) Relative abundance analysis of microbiota at the phylum level between groups. (E–J) Ratio of Firmicutes to Bacteroidetes and relative abundance of bacteria among groups. * P<0.05, ** P<0.01, *** P<0.001 vs Chow, # P<0.05, ## P<0.01, ### P<0.001 vs HFD.
Figure 6.
LCBE amends the gut microbiota at the genus level in mice fed a high-fat diet. (A) Taxonomic cladogram depicting enriched taxa that are differentially abundant in all groups with a LDA score >3.0. (B–O) Relative abundance of different bacteria in all groups. * P<0.05, ** P<0.01, *** P<0.001 vs Chow, # P<0.05, ## P<0.01, ### P<0.001 vs HFD.
Discussion
NAFLD is a worldwide health issue that is associated with obesity. Although lifestyle modifications and exercise have been suggested for NAFLD treatment, both are difficult to execute. In addition, there is a lack of effective drugs targeting NAFLD. Therefore, it is of great importance to identify drugs that can treat NAFLD. Previous studies have indicated that probiotics significantly prevent HFD-induced NAFLD in mice [21,22]. Hence, LCBE is believed to alleviate NAFLD. In this study, we found that treatment with LCBE protected NAFLD mice fed with an HFD by partially enhancing insulin sensitivity, alleviating the inflammatory response, improving the function of the intestinal barrier, and regulating the composition of gut microbiota.
Abnormal blood glucose levels and the accumulation of excessive lipids in the liver are common in NAFLD mice. The results of our research showed that the HFD increased the body weight and liver index, and caused dyslipidemia and glucose intolerance, which were reversed by the LCBE intervention. Triglyceride accumulation in the liver is affected by many factors, including fatty acid uptake, synthesis, oxidation, and transporters [23]. Insulin resistance is the “first hit” in NAFLD [24]. Insulin resistance decreases lipid oxidation in the liver, and the PPAR-α and CPT-1 genes have been identified as the key fatty acid oxidation genes that can alleviate NAFLD by promoting lipid oxidation [25]. The results of this study showed that the HFD enhanced insulin resistance and HOMA-IR, and this was prevented by LCBE. In addition, we found that LCBE treatment significantly upregulated the expression levels of hepatic PPAR-α and CPT-1, indicating that fatty acid β-oxidation plays a critical role in alleviating dyslipidemia and hepatic steatosis upon the administration of LCBE. Therefore, an increase in insulin sensitivity plays an important role in alleviating hepatic steatosis upon LCBE intervention.
Since the inflammatory response participates in the development of NAFLD, we assessed the effect of LCBE on inflammation in NAFLD mice. In addition, a previous study verified that E. faecium and B. subtilis, the 2 components of LCBE, could protect mice from CLP-induced sepsis by ameliorating inflammation [20]. Inflammatory cytokines, such as IL-1β, IL-6, and TNF-α, were detected. LCBE prevented an increase in the levels of inflammatory mediators in NAFLD mice. LPS, produced by gram-negative bacteria, is one of the main factors involved in inflammation in NAFLD [26]. TLR4 plays a critical role in the production of pro-inflammatory cytokines in various diseases, including NAFLD [27]. LPS can translocate from a leaky gut into the blood recycle, and then bind to TLR4 in the liver, causing the release of inflammatory mediators [28]. Therefore, the levels of serum LPS and the expression of TLR4 and NF-κB in the liver were assessed; LCBE reduced the serum LPS levels and the protein expression of TLR4 and NF-κB in HFD-fed mice. Therefore, LCBE inhibited the inflammatory response via the LPS-TLR4/NF-κB pathway.
Previous studies have shown that the intestinal barrier plays a key role in NAFLD induced by a high-fat or high-fructose diet [29–31]. The physical intestinal barrier is an important component of the intestinal barrier [32]. Recently, the physical intestinal barrier was widely studied in relation to HFD-obesity gut permeability, which enhances the translocation of LPS and harmful bacteria from the gut to the liver via blood circulation [28,33]. Since LPS reflects the leaky gut and LCBE can reduce the serum LPS levels, the intestinal integrity in NAFLD mice was affected by LCBE treatment. LCBE promoted the upregulation of tight junction proteins such as occludin and Claudin1. In addition, unlike the chow group, the HFD group exhibited damaged intestinal structure integrity, which was restored by LCBE. These data indicated that LCBE maintained the strength of the intestinal physical barrier in NAFLD mice.
An increasing number of studies are showing that gut microbiota play a vital role in the development of NAFLD by harvesting energy and modulating energy metabolism. Firmicutes and Bacteroidetes are the major phyla involved in energy metabolism [34]. There was a decrease in the relative abundance of Bacteroidetes and an increase in the relative abundance of Firmicutes and the F/B ratio in HFD-fed mice [35,36]. Based on the above discussion, we speculated that LCBE might prevent liver inflammation and protect the intestinal barrier by modulating the gut microbiota structure in NAFLD mice. Interestingly, the results of our study showed that the HFD increased the abundance of Firmicutes and the F/B ratio, and reduced the relative abundance of Bacteroidetes; these changes were reversed by LCBE intervention, suggesting that LCBE maintained the intestinal flora balance. In addition to the abundance at the phylum level, our results indicated that the abundance of some bacterial genera changed. Interestingly, LCBE reduced the abundance of bacterial genera associated with obesity and inflammation, such as Akkermansia, Lachnoclostridium, and Oscillibacter. In this study, we found that LCBE upregulated the abundance of Akkermansia in the gut of HFD-fed mice. Previous studies have shown that Akkermansia, an important genus in the microbial community, negatively correlates with body weight and obesity in mice and humans [37]. Zhao et al showed that Lactobacillus fermentum CQPC06 alleviated NAFLD by modulating gut microbiota; for example, by increasing the abundance of Akkermansia [38]. Moreover, an HFD increased the levels of Lachnoclostridium and Oscillibacter, and LCBE reversed this. A study showed that Lachnoclostridium was related to obesity and inflammation induced by an HFD [39]. In addition to causing obesity and inflammation, bacteria in the genus Oscillibacter produce LPS [40]. The abundance of another harmful bacterial genus, Escherichia-Shigella, which produces LPS, significantly decreased after treatment with LCBE. Similarly, Zhuang et al found that eicosapentaenoic acid and docosahexaenoic acid reversed the levels of LPS in the serum and the abundance of Escherichia-Shigella in HFD-fed mice [41]. In addition, we found that an HFD changed the abundance of some genera, including Ruminococcus_UCG-014, Family_XIII_AD3011_group, Eubacterium_coprostanoligenes_group, Anaerovorax, Acetatifactor, Coriobacterium_UCG-002, Negativibacillus, Angelakisella, Ruminococcus, and Harryflintia, and this could be prevented by treatment with LCBE. These bacteria may act as new targets for the treatment of NAFLD.
Our study has one limitation. Additional methods, such as FITC dextran tests, should be used to determine the intestinal permeability.
Conclusions
In summary, our study showed that LCBE administration significantly alleviated HFD-induced NAFLD. Mechanistically, LCBE modulated gut microbiota dysbiosis by increasing species richness and diversity. Moreover, LCBE alleviated liver inflammation, reduced serum LPS levels, and improved the intestinal barrier function. These data suggest that LCBE may be a potential candidate for the treatment of NAFLD.
Abbreviations
- NAFLD
non-alcoholic fatty liver disease
- LCBE
live combined Bacillus subtilis and Enterococcus faecium
- IL-1β
interleukin 1β
- IL-6
interleukin 6
- TNF-α
tumor necrosis factor α
- LPS
lipopolysaccharide
- TLR4
toll-like receptor 4
- NF-κB p65
nuclear factor kappa-B p65
- PPAR-α
peroxisome proliferator-activated receptor alpha
- CPT-1
carnitine palmitoyl transferase 1
- CLP
cecal ligation and puncture
- AST
aspartate aminotransferase
- ALT
alanine aminotransferase
- TG
triglyceride
- Cho
total cholesterol
- LDL
low-density lipoprotein cholesterol
- HDL
high-density lipoprotein cholesterol
- HFD
high-fat diet
Footnotes
Conflict of Interest
None.
Declaration of Figures Authenticity
All figures submitted have been created by the authors who confirm that the images are original with no duplication and have not been previously published in whole or in part.
Financial support: This study was supported by funds from the National Natural Science Foundation of China (81700509)
References
- 1.Karlas T, Wiegand J, Berg T. Gastrointestinal complications of obesity: Non-alcoholic fatty liver disease (NAFLD) and its sequelae. Best Pract Res Clin Endocrinol Metab. 2013;27(2):195–208. doi: 10.1016/j.beem.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Fan J-G, Farrell GC. Epidemiology of non-alcoholic fatty liver disease in China. J Hepatol. 2009;50(1):204–10. doi: 10.1016/j.jhep.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Wang F-S, Fan J-G, Zhang Z, et al. The global burden of liver disease: The major impact of China. Hepatology (Baltimore, Md) 2014;60(6):2099–108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne CD. Fatty liver: Role of inflammation and fatty acid nutrition. Prostaglandins Leukot Essent Fatty Acids. 2010;82(4–6):265–71. doi: 10.1016/j.plefa.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Carpino G, Del Ben M, Pastori D, et al. Increased liver localization of lipopolysaccharides in human and experimental NAFLD. Hepatology (Baltimore, Md) 2020;72(2):470–85. doi: 10.1002/hep.31056. [DOI] [PubMed] [Google Scholar]
- 6.Oh DY, Morinaga H, Talukdar S, et al. Increased macrophage migration into adipose tissue in obese mice. Diabetes. 2012;61(2):346–54. doi: 10.2337/db11-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang S, Demir M, Martin A, et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology. 2020;159(5):1839–52. doi: 10.1053/j.gastro.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven L, Rahman A, Nair Parvathy S, et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: A randomized control trial. Am J Gastroenterol. 2020;115(7):1055–65. doi: 10.14309/ajg.0000000000000661. [DOI] [PubMed] [Google Scholar]
- 9.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–77. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Aron-Wisnewsky J, Vigliotti C, Witjes J, et al. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17(5):279–97. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 11.Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science (New York, NY) 2005;307(5717):1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 12.Le Roy T, Llopis M, Lepage P, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62(12):1787–94. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 13.Chevalier C, Stojanović O, Colin DJ, et al. Gut microbiota orchestrates energy homeostasis during cold. Cell. 2015;163(6):1360–74. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Compare D, Coccoli P, Rocco A, et al. Gut–liver axis: The impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22(6):471–76. doi: 10.1016/j.numecd.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Paolella G, Mandato C, Pierri L, et al. Gut-liver axis and probiotics: Their role in non-alcoholic fatty liver disease. World J Gastroenterol. 2014;20(42):15518–31. doi: 10.3748/wjg.v20.i42.15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10(4):311–23. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miele L, Marrone G, Lauritano C, et al. Gut-liver axis and microbiota in NAFLD: Insight pathophysiology for novel therapeutic target. Curr Pharm Des. 2013;19(29):5314–24. [PubMed] [Google Scholar]
- 18.Zhao Z, Chen L, Zhao Y, et al. Lactobacillus plantarum NA136 ameliorates nonalcoholic fatty liver disease by modulating gut microbiota, improving intestinal barrier integrity, and attenuating inflammation. Appl Microbiol Biotechnol. 2020;104(12):5273–82. doi: 10.1007/s00253-020-10633-9. [DOI] [PubMed] [Google Scholar]
- 19.Xue L, He J, Gao N, et al. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci Rep. 2017;7:45176. doi: 10.1038/srep45176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Meng M, Wei Y, et al. Protective effects of live combined and in polymicrobial sepsis through modulating activation and transformation of macrophages and mast cells. Front Pharmacol. 2018;9:1506. doi: 10.3389/fphar.2018.01506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan A, Ding Z, Ishaq M, et al. Understanding the effects of gut microbiota dysbiosis on non-alcoholic fatty liver disease and the possible probiotics role: Recent updates. Int J Biol Sci. 2021;17(3):818–33. doi: 10.7150/ijbs.56214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitetta L, Henson JD. Probiotics and synbiotics targeting the intestinal microbiome attenuate non-alcoholic fatty liver disease. Hepatobiliary Surg Nutr. 2020;9(4):526–29. doi: 10.21037/hbsn.2019.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masarone M, Rosato V, Dallio M, et al. Role of oxidative stress in pathophysiology of nonalcoholic fatty liver disease. Oxid Med Cell Longev. 2018;2018:9547613. doi: 10.1155/2018/9547613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Guo F, Yu M, et al. Reduced Nogo expression inhibits diet-induced metabolic disorders by regulating ChREBP and insulin activity. J Hepatol. 2020;73(6):1482–95. doi: 10.1016/j.jhep.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Fu Y, Hu F, et al. PIK3R3 regulates PPARα expression to stimulate fatty acid β-oxidation and decrease hepatosteatosis. Exp Mol Med. 2018;50(1):e431. doi: 10.1038/emm.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita M, Kuraji R, Ito H, et al. Histological effects and pharmacokinetics of lipopolysaccharide derived from Porphyromonas gingivalis on rat maxilla and liver concerning with progression into non-alcoholic steatohepatitis. J Periodontol. 2018;89(9):1101–11. doi: 10.1002/JPER.17-0678. [DOI] [PubMed] [Google Scholar]
- 27.Zhao G-N, Zhang P, Gong J, et al. Tmbim1 is a multivesicular body regulator that protects against non-alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat Med. 2017;23(6):742–52. doi: 10.1038/nm.4334. [DOI] [PubMed] [Google Scholar]
- 28.Fei N, Bruneau A, Zhang X, et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio. 2020;11(1):e03263–19. doi: 10.1128/mBio.03263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambertz J, Weiskirchen S, Landert S, et al. Fructose: A dietary sugar in crosstalk with microbiota contributing to the development and progression of non-alcoholic liver disease. Front Immunol. 2017;8:1159. doi: 10.3389/fimmu.2017.01159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volynets V, Louis S, Pretz D, et al. Intestinal barrier function and the gut microbiome are differentially affected in mice fed a western-style diet or drinking water supplemented with fructose. J Nutr. 2017;147(5):770–80. doi: 10.3945/jn.116.242859. [DOI] [PubMed] [Google Scholar]
- 31.Trigueros L, Peña S, Ugidos AV, et al. Food ingredients as anti-obesity agents: A review. Crit Rev Food Sci Nutr. 2013;53(9):929–42. doi: 10.1080/10408398.2011.574215. [DOI] [PubMed] [Google Scholar]
- 32.Cui Y, Wang Q, Chang R, et al. Intestinal barrier function-non-alcoholic fatty liver disease interactions and possible role of gut microbiota. J Agric Food Chem. 2019;67(10):2754–62. doi: 10.1021/acs.jafc.9b00080. [DOI] [PubMed] [Google Scholar]
- 33.Feng D, Zou J, Su D, et al. Curcumin prevents high-fat diet-induced hepatic steatosis in ApoE mice by improving intestinal barrier function and reducing endotoxin and liver TLR4/NF-κB inflammation. Nutr Metab (Lond) 2019;16:79. doi: 10.1186/s12986-019-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Chierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology (Baltimore, Md) 2017;65(2):451–64. doi: 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]
- 35.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 37.Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110(22):9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu J, Tan F, Zhou X, Zhao X. Lactobacillus fermentum CQPC06 in naturally fermented pickles prevents non-alcoholic fatty liver disease by stabilizing the gut-liver axis in mice. Food Funct. 2020;11(10):8707–23. doi: 10.1039/d0fo01823f. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Chen L, Hu M, et al. Dietary type 2 resistant starch improves systemic inflammation and intestinal permeability by modulating microbiota and metabolites in aged mice on high-fat diet. Aging. 2020;12(10):9173–87. doi: 10.18632/aging.103187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song JJ, Tian WJ, Kwok L-Y, et al. Effects of microencapsulated Lactobacillus plantarum LIP-1 on the gut microbiota of hyperlipidaemic rats. Br J Nutr. 2017;118(7):481–92. doi: 10.1017/S0007114517002380. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang P, Zhang Y, Shou Q, et al. Eicosapentaenoic and docosahexaenoic acids differentially alter gut microbiome and reverse high-fat diet-induced insulin resistance. Mol Nutr Food Res. 2020;64(10):e1900946. doi: 10.1002/mnfr.201900946. [DOI] [PubMed] [Google Scholar]