Abstract
The role of circadian dysregulation is increasingly acknowledged in the background of depressive symptoms, and is also a promising treatment target. Similarly, stress shows a complex relationship with the circadian system. The CLOCK gene, encoding a key element in circadian regulation has been implicated in previous candidate variant studies in depression with contradictory findings, and only a few such studies considered the interacting effects of stress. We investigated the effect of CLOCK variation with a linkage-disequilibrium-based clumping method, in interaction with childhood adversities and recent negative life events, on two phenotypes of depression, lifetime depression and current depressive symptoms in a general population sample.
Methods: Participants in NewMood study completed questionnaires assessing childhood adversities and recent negative life events, the Brief Symptom Inventory to assess current depressive symptoms, provided data on lifetime depression, and were genotyped for 1054 SNPs in the CLOCK gene, 370 of which survived quality control and were entered into linear and logistic regression models with current depressive symptoms and lifetime depression as the outcome variable, and childhood adversities or recent life events as interaction variables followed by a linkage disequilibrium-based clumping process to identify clumps of SNPs with a significant main or interaction effect.
Results: No significant clumps with a main effect were found. In interaction with recent life events a significant clump containing 94 SNPs with top SNP rs6825994 for dominant and rs6850524 for additive models on current depression was identified, while in interaction with childhood adversities on current depressive symptoms, two clumps, both containing 9 SNPs were found with top SNPs rs6828454 and rs711533.
Conclusion: Our findings suggest that CLOCK contributes to depressive symptoms, but via mediating the effects of early adversities and recent stressors. Given the increasing burden on circadian rhythmicity in the modern lifestyle and our expanding insight into the contribution of circadian disruption in depression especially as a possible mediator of stress, our results may pave the way for identifying those who would be at an increased risk for depressogenic effects of circadian dysregulation in association with stress as well as new molecular targets for intervention in stress-related psychopathologies in mood disorders.
Keywords: clock gene, depression, stress, childhood adversities, negative life events, gene-environment interactions, circadian rhythm
Introduction
In order to successfully adapt to environmental changes including day-night cycles signaling rhythmic alterations in the availability of resources and presence of dangers, regulation of rhythmic metabolic, cognitive, and behavioral functions via optimization of physiological and biological processes to 24-h cycles is necessary (1). Circadian disruption may compromise survival and lead to the emergence of several somatic and mental disorders including depression (2, 3). Self-sustained biological rhythms are controlled by the circadian system composed of tissues expressing endogenous 24-h timekeeping activity (4). The nucleus suprachiasmaticus acts as central pacemaker, the activity of which is based on a transcriptional/posttranslational feedback loop with rhythmic expression of circadian clock genes (5), including the CLOCK gene (circadian locomotor output cycles kaput gene) which possesses a transcriptional activator role in the circadian clock mechanism.
Genetic mutations in clock genes may lead to disruptions in the period, phase and amplitude of circadian rhythms (6). In animal models, manipulation and mutation of clock genes causes alterations in behavioral and affective phenotypes, suggesting a direct connection between clock genes and brain functions relevant to psychiatric illness (7). Depression has been linked to circadian abnormalities for decades as suggested by somatic rhythms showing disruption in depressed patients, as well as a typical circadian rhythmicity of various symptoms (8). Prominent circadian disturbances in mood disorders include sleep problems, morning worsening and evening improvement of symptoms, changes in appetite, social interactions, as well as alteration in the circadian rhythmicity of blood pressure, body temperature or hormone levels (9–11). These have implicated a rhythm disruption of central origin involving the core molecular machinery underlying circadian rhythm generation, thus suggesting that disruption of circadian regulation is a factor possibly underlying the development and maintenance of the disorder (12–14).
There is a strong genetic background of mood disorders (15, 16) and some studies point to an association between CLOCK gene variation and bipolar disorder (17, 18) but weak association with major depressive disorder (MDD) (12, 17, 19). However, in case of depression, the heterogeneity of the disorder and the underlying neurobiological etiological processes raised the possibility that inaccurate or misinterpreted phenotypes (19) and lack of differentiation between depression subtypes may have obscured existing associations. Furthermore, the circadian clock and stress response systems are closely related (7), and stress/increased vulnerability to stress are risk factors for multiple psychiatric disorders. Modulation of stress response is a common mechanism by which circadian clock genes affect such illnesses including depression (7). It has been suggested that effects of SNPs in candidate gene studies and GWAS-s may be masked by lack of consideration of the interacting effects of various types of environmental events in spite of our increasing understanding that the majority of genes and variants contributing to the emergence of depression act via modulating sensitivity toward stress (16, 20, 21).
In previous studies, only a few candidate polymorphisms in the CLOCK gene, and most frequently rs1801260 (also known as 3111T/C) were investigated in association with depression with inconsistent findings. GWAS-s have not confirmed the role of this variant or the CLOCK gene, although suggested a role for circadian system genes (19, 22, 23). However, GWAS-s may overlook existing association due to strict p-value criteria to compensate for multiple testing with even true positive SNPs not achieving genome-wide significance (19, 24). One way of reducing multiple testing burden yet overcoming the hit-and-miss approach of candidate variant studies is employing a gene-wide approach focusing on variations along the CLOCK gene.
The high prevalence of depression coupled with the remarkable lack of efficacy of currently available antidepressive medications leaving ~30–35% of patients treatment resistant (25) on the one hand reflects the lack of in-depth understanding of the etiological processes in the background of depression, and on the other hand highlights the need for understanding novel processes and identifying novel molecular targets for intervention. Furthermore, given the well-known heterogeneity of depressive disorders not only on the clinical-symptomatic level, but also in the neurobiological and genetic background of such divergent clinical manifestations, understanding genetic background of various processes, such as circadian disruption, playing a role in the development of different depressive symptoms and syndromes may also help subtypization and, in the end, precision and personalized treatments of depression.
The aim of the present study was to investigate the association between variation in the CLOCK gene, lifetime depression, and current depressive symptoms in interaction with childhood adversities and recent negative life events in a general European population.
Materials and Methods
Study Sample
The present study was part of the NewMood study funded by the European Union (New Molecules in Mood Disorders, Sixth Framework Program of the EU, LSHM-CT-2004-503474). Seven hundred sixty-seven non-related participants (238 males, 529 females) of self-reported European white ethnic origin aged between 18 and 60 years were recruited from the general population through advertisements, a website and general practices; provided self-reported data on sociodemographic factors including age and gender, as well as lifetime and current depression, and early childhood adversities and recent negative life events occurring in the past year; and provided genetic data by a saliva sampling kit.
NewMood aimed to recruit participants with a diverse exposure to different types on environmental influences and adversities and with diverse socioeconomical background to allow for generalisability of results to real-life settings. Furthermore, as NewMood focused on a general population with a continuum approach to affective symptoms and disorders, our sample included previously depressed, currently depressed and never depressed participants as well. A detailed description of the study population is provided in previously published reports (26–28). The study was carried out in accordance with the Declaration of Helsinki, and it was approved by the Scientific and Research Ethics Committee of the Medical Research Council, Budapest, Hungary. All participants provided written informed consent prior to participating in the study.
Phenotypes
The present study focused on measuring two aspects of depression and two types of stressors. Lifetime depression (DEP) was ascertained based on self-report using a background questionnaire. This measure to capture the lifetime presence of major depression has been validated previously with face-to-face structured diagnostic interviews (SCID-I) within a subpopulation of our sample, yielding a 91.7% sensitivity and 89.8% specificity (28).
Current level of depression (BSI-Depression) was measured using the Brief Symptom Inventory (29), a questionnaire measuring psychopathological symptoms in several scales with items scored between 0 and 4 depending on the distress caused. For the present study, only the depression subscale score was used to reflect the actual levels of depression, calculated as the sum of depression and additional item scores divided by the number of completed items. Use of BSI-Depression to capture actual depressive symptom severity has been validated in a previous study in a subsample of our population using the Montgomery-Asberg Depression Rating Scale (MADRS) administered by trained interviewers (28).
The two types of stressors measured in our study included early childhood adversities (CHA) as distal and etiological stressors and recent negative life events (RLE) occurring in the past year as proximal, trigger stressors. The early childhood adversity measure was derived from the Childhood Trauma Questionnaire (CTQ) (30), and included four items referring to emotional and physical abuse and emotional and physical neglect, and two items about the loss of parents. This short childhood adversity measure has previously been validated with the 28-item CTQ within a subpopulation of our sample, yielding a high correlation between the original and derived measures (28). The sum of item scores was used in the analyses. Recent stressful life events (RLE) occurring in the past year related to financial difficulties, illnesses/injuries, personal problems, and intimate relationship or social network difficulties were measured using the List of Threatening Experiences (31, 32). The number of recent negative life events (RLEs) was used in the statistical analyses.
Genotyping and Imputation
Participants provided buccal mucosa cells collected by a cytology brush (Cytobrush plus C0012, Durbin PLC). Genomic DNA was extracted according to the protocol of Freeman et al. (33). Genotyping was performed by Illumina's CoreExom PsychChip. All laboratory work was performed under the ISO 9001:2000 quality management requirements and was blinded with regard to phenotype. Variants were positioned on the genome based on GRCh37/hg19. SHAPEIT was used to determine haplotype information, then missing genotypes were imputed using IMPUTE2 on the CLOCK gene with boundaries extended by 10 kilobase pairs on both sides. Imputation and subsequent filtering were carried out in line with multiple quality control steps (34), except that missingness rate (MR), Hardy-Weinberg equilibrium (HWE) and minor allele frequency (MAF) steps were limited to SNPs within the CLOCK gene. Variants with an imputation score certainty <0.7 or info <0.5 were excluded.
Statistical Analyses
Plink v1.90 was used to calculate MR (<0.05), HWE (>1 × 10−5) and MAF (>0.01) as part of quality control steps prior to the analyses; for clumping; and for building linear and logistic regression models to test for main and interaction effects of genetic variation in the CLOCK gene. Analyses were supported by scripts individually written in R 3.0.2 (35). R was also used to illustrate the effects of significant findings (version 4.0.3 with the ggplot2 package). Descriptive statistics were run using IBM SPSS Statistics 25.
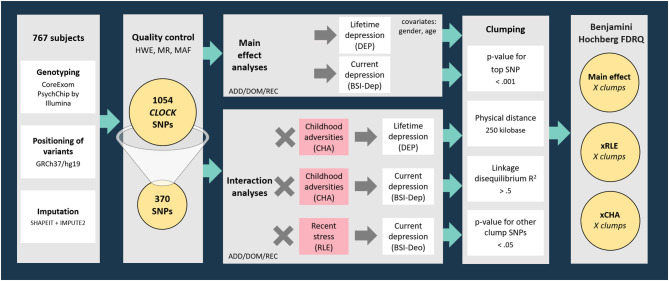
Genotyping provided a dataset incorporating 1054 SNPs in the region of CLOCK gene (with boundaries extended by 10 kb) available in the NewMood database. Three hundred and seventy SNPs survived quality control steps were analyzed with linear (for current depression with BSI-dep as the outcome variable) and logistic (for lifetime depression with DEP as the outcome variable) regression models to test for main effects of CLOCK variation on lifetime and current depression. To test for gene-environment correlation (rGE) effects, the main effects of CLOCK variants surviving quality control on childhood adversities (CHA) and recent life events (RLE) were also analyzed in linear regression models. After tests for main effects of genetic variants on the outcome variables, gene × environment interaction models with early childhood adversities (CHA) and recent negative life events (RLE) were also run. Regression data on all CLOCK SNPs surviving the quality control in all models are shown in Supplementary Tables 1–4). Regression models were in the next step followed by a clumping procedure both for main effect and for GxE interaction effects based on linkage disequilibrium (LD) estimates between the SNPs using the CLUMP function in Plink. The four parameters used for clumping included: (1) maximum p-value of the clump's top SNP was set at 0.001; (2) physical distance with top SNP was 250 kilobase; (3) minimum linkage disequilibrium R2 with top SNP was 0.5; and (4) maximum p-value for the clump's other SNPs was 0.05. Results of top SNPs, that is, the most significant SNP representing correlated SNPs in individual clumps are reported (Figure 1).
Figure 1.
Methods of investigating the effects of variation in CLOCK in interaction with childhood adversities and recent life events on lifetime and current depression: study population, quality control steps, and statistical analyses. RLE, recent life events; CHA, Childhood adversities; BSI-Dep, Brief Symptom Inventory depression score; ADD, additive model; DOM, dominant model; REC, recessive model; HWE, Hardy-Weinberg Equilibrium; MR, missingness rate; MAF, minor allele frequency; DEP, lifetime depression; FDRQ, False Discovery Rate Q value.
All analyses, including main effect and interaction effect models, were run according to additive, dominant, and recessive models. Age and gender were covariates in all Plink regression models. When testing an SNP × CHA/RLE interaction effect, main effects of both the SNP and CHA/RLE were also included as covariates in the model. Nominal significance threshold was p < 0.05. To correct for multiple comparisons in analyses for each of the above outcome variables, Benjamini-Hochberg False Discovery Rate (FDR) Q-values were calculated; results with a Q ≤ 0.05 were considered significant.
The data presented in this study are openly available in FigShare at https://doi.org/10.6084/m9.figshare.14258567.v1 (36).
Results
Descriptive Statistics
Descriptive statistics of our study sample are provided in Table 1.
Table 1.
Descriptive statistics of the study sample.
| Gender | n | % | ||
| Male | 238 | 21.76% | ||
| Female | 529 | 48.36% | ||
| Minimum | Maximum | Mean | SEM | |
| Age | 18 | 60 | 30.724 | 0.375 |
| Clinical data | n | % | ||
| Subjects reporting lifetime depression | 166 | 21.64% | ||
| Subjects reporting previous suicide attempts or self-harm | 35 | 4.56% | ||
| Minimum | Maximum | Mean | SEM | |
| BSI-depression score | 0 | 4 | 0.546 | 0.024 |
| Environmental influences | Minimum | Maximum | Mean | SEM |
| Childhood adversity | 0 | 15 | 2.761 | 0.104 |
| Recent negative life events | 0 | 8 | 1.091 | 0.042 |
| Sociodemographic descriptors | n | % | ||
| Employment status | ||||
| Working full-time Working part-time Student Retired Housewife/househusband Unemployed |
405 27 295 13 12 15 |
52.80% 3.52% 38.46% 1.69% 1.56% 1.95% |
||
| Marital status | ||||
| Single Married Cohabiting Divorced Separated Widowed Data missing |
360 251 98 35 11 7 5 |
46.94% 32.72% 12.78% 4.56% 1.43% 0.91% 0.65% |
||
| Financial situation | ||||
| Living comfortably Just getting by Not able to get along Data missing |
495 259 5 8 |
64.54% 33.76% 0.65% 0.91% |
||
BSI-depression score, current depressive symptom scores as measured by Brief Symptom Inventory; SEM, standard error of the mean.
Main Effects of Variation in CLOCK on Current Depressive Symptoms and Lifetime Depression
Linear and logistic regression models on BSI-Dep (for current depression) and DEP (for lifetime depression), respectively, identified a few SNPs with a nominally significant main effect, but significant clumps could not be formed, furthermore, all p-values exceeded the maximum threshold for top SNP (p = 0.001) [Supplementary Tables 1, 2 for lifetime depression (DEP) and current depressive symptoms (BSI-Dep, respectively), but significant clumps for main genetic effects could not be identified]. Nevertheless, top SNPs of significant clumps emerging in the interaction models were tested in main effect models for lifetime and current depression as well, the results of which are shown in Table 2.
Table 2.
Main effects of CLOCK variants emerging as top SNPs in the interaction models on lifetime depression and on current depression (BSI-dep) severity.
| Additive | Dominant | Recessive | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | 95% C.I. | p-value | FDRQ | β | 95% C.I. | p-value | FDRQ | β | 95% C.I. | p-value | FDRQ | ||||
| rs6828454 | Lifetime depression | 1.122 | 0.883 | 1.426 | 0.3482 | 0.497 | 1.034 | 0.710 | 1.506 | 0.8605 | 0.906 | 1.342 | 0.898 | 2.005 | 0.1514 | 0.293 |
| BSI depression | 0.015 | −0.050 | 0.079 | 0.6571 | 0.773 | 0.031 | −0.07 | 0.133 | 0.5456 | 0.668 | 0.006 | −0.106 | 0.118 | 0.9189 | 0.951 | |
| rs711533 | Lifetime depression | 1.524 | 1.114 | 2.084 | 0.0084 | 0.042 | 1.497 | 1.045 | 2.144 | 0.0279 | 0.084 | 2.945 | 1.152 | 7.530 | 0.0241 | 0.080 |
| BSI depression | 0.070 | −0.019 | 0.159 | 0.1233 | 0.264 | 0.074 | −0.024 | 0.174 | 0.1463 | 0.293 | 0.129 | −0.171 | 0.431 | 0.3988 | 0.532 | |
| rs6825994 | Lifetime depression | 1.465 | 1.149 | 1.868 | 0.0021 | 0.015 | 1.648 | 1.145 | 2.371 | 0.0071 | 0.039 | 1.730 | 1.102 | 2.715 | 0.0171 | 0.064 |
| BSI depression | 0.058 | −0.009 | 0.124 | 0.0913 | 0.211 | 0.086 | −0.010 | 0.181 | 0.0757 | 0.182 | 0.059 | −0.074 | 0.192 | 0.3822 | 0.533 | |
| rs6850524 | Lifetime depression | 1.351 | 1.061 | 1.721 | 0.0149 | 0.059 | 1.535 | 1.048 | 2.248 | 0.0279 | 0.088 | 1.470 | 0.963 | 2.242 | 0.0741 | 0.185 |
| BSI depression | 0.063 | −0.003 | 0.129 | 0.0608 | 0.159 | 0.106 | 0.007 | 0.205 | 0.0358 | 0.102 | 0.054 | −0.067 | 0.175 | 0.3832 | 0.523 | |
BSI-current depressive symptom scores as measured by Brief Symptom Inventory. DEP-self-reported lifetime depression. ADD, DOM, REC, additive, dominant and recessive heritability models. FDR, false discovery rate; Italic type shows nominally significant p-values, bold type denotes significant Q-values surviving correction for multiple testing (P < 0.05, FDR Q < 0.05).
Gene-Environment Correlation (rGE): Main Effects of Variation in CLOCK on Recent Life Events (RLE) and Childhood Adversities (CHA)
Main effect linear regression models on recent life events (RLE) and childhood adversities (CHA) did not identify any nominally significant SNPs (Supplementary Tables 5, 6 for RLE and CHA, respectively) suggesting no gene-environment correlation (rGE) effects in case of either early or recent adversities. Linear regression results for potential rGE effects on RLE or CHA in case of top SNPs of significant clumps emerging in the interaction models are also shown in Table 3.
Table 3.
Main effects of CLOCK variants emerging as top SNPs in the interaction models on recent stressful life events and childhood adversities (rGE models).
| Additive | Dominant | Recessive | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | 95% C.I. | p-value | β | 95% C.I. | p-value | β | 95% C.I. | p-value | ||||
| rs6828454 | RLE | 0.020 | −0.094 | 0.133 | 0.7341 | 0.061 | −0.117 | 0.239 | 0.5019 | −0.015 | −0.212 | 0.181 | 0.8788 |
| CHA | −0.146 | −0.423 | 0.131 | 0.3012 | −0.269 | −0.704 | 0.166 | 0.2251 | −0.111 | −0.592 | 0.369 | 0.6504 | |
| rs711533 | RLE | 0.037 | −0.119 | 0.193 | 0.6441 | 0.0505 | −0.125 | 0.226 | 0.5722 | −0.037 | −0.566 | 0.492 | 0.8910 |
| CHA | −0.224 | −0.604 | 0.157 | 0.2494 | −0.263 | −0.691 | 0.164 | 0.2279 | −0.177 | −1.470 | 1.115 | 0.7879 | |
| rs6825994 | RLE | 0.005 | −0.113 | 0.123 | 0.9337 | 0.073 | −0.095 | 0.241 | 0.3967 | −0.122 | 0.120 | −0.356 | 0.113 |
| CHA | 0.011 | −0.276 | 0.298 | 0.9417 | 0.113 | −0.295 | 0.522 | 0.5874 | −0.122 | −0.356 | 0.113 | 0.3085 | |
| rs6850524 | RLE | 0.046 | −0.071 | 0.164 | 0.4404 | 0.110 | −0.067 | 0.287 | 0.223 | −0.008 | 0.110 | −0.223 | 0.207 |
| CHA | −0.014 | −0.302 | 0.275 | 0.9245 | 0.039 | −0.395 | 0.473 | 0.8597 | −0.008 | −0.223 | 0.207 | 0.9439 | |
RLE, recent life stress; CHA, Childhood Adversity. ADD, DOM, REC, additive, dominant, and recessive heritability models.
Gene x Environment Effects of Variation in CLOCK on Current Depressive Symptoms: Interaction With Recent Life Events (RLE)
Our analyses for interaction with recent stressful life events (RLE) on current depressive symptoms (BSI-Dep) yielded one significant clump containing 94 SNPs (Supplementary Table 3) and with top SNPs rs6825994 for dominant and rs6850524 for additive models, both surviving correction for multiple testing (Table 4).
Table 4.
Interactions of CLOCK top SNPs rs6828454, rs711533, rs6850524, and rs6825994 with recent negative life events (RLE) and childhood adversity (CHA) on current depression (BSI-Dep) and lifetime depression (DEP) (GxE models).
| Additive | Dominant | Recessive | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | 95% C.I. | p | FDRQ | β | 95% C.I. | p-value | FDRQ | β | 95% C.I. | p | FDRQ | ||||
| rs6828454 | xRLE on BSI | 0.017 | −0.036 | 0.071 | 0.5240 | 0.655 | 0.046 | −0.044 | 0.136 | 0.3172 | 0.476 | 0.003 | −0.083 | 0.089 | 0.9469 | 0.947 |
| xCHA on BSI | 0.034 | 0.013 | 0.055 | 0.0015 | 0.0145 | 0.032 | −0.000 | 0.065 | 0.0510 | 0.1390 | 0.066 | 0.028 | 0.104 | 0.0006 | 0.0118 | |
| xCHA on DEP | 0.992 | 0.920 | 1.070 | 0.8424 | 0.903 | 0.974 | 0.865 | 1.096 | 0.6606 | 0.762 | 1.016 | 0.887 | 1.164 | 0.8143 | 0.905 | |
| rs711533 | xRLE on BSI | 0.088 | 0.018 | 0.158 | 0.0141 | 0.060 | 0.095 | 0.016 | 0.174 | 0.0188 | 0.066 | 0.139 | −0.085 | 0.364 | 0.2244 | 0.396 |
| xCHA on BSI | 0.053 | 0.022 | 0.084 | 0.0008 | 0.0092 | 0.058 | 0.024 | 0.092 | 0.0008 | 0.0114 | 0.066 | −0.054 | 0.186 | 0.2815 | 0.4692 | |
| xCHA on DEP | 1.045 | 0.934 | 1.168 | 0.4458 | 0.581 | 1.070 | 0.944 | 1.213 | 0.2878 | 0.467 | 0.892 | 0.605 | 1.315 | 0.5621 | 0.675 | |
| rs6850524 | xRLE on BSI | 0.098 | 0.044 | 0.152 | 0.0004 | 0.013 | 0.127 | 0.044 | 0.210 | 0.0028 | 0.019 | 0.132 | 0.037 | 0.228 | 0.0066 | 0.040 |
| xCHA on BSI | 0.014 | −0.008 | 0.036 | 0.2160 | 0.3927 | 0.017 | −0.016 | 0.050 | 0.3099 | 0.4768 | 0.020 | −0.020 | 0.061 | 0.3192 | 0.4671 | |
| xCHA on DEP | 1.057 | 0.973 | 1.148 | 0.1866 | 0.350 | 0.986 | 0.872 | 1.114 | 0.8173 | 0.892 | 1.230 | 1.047 | 1.445 | 0.0117 | 0.054 | |
| rs6825994 | xRLE on BSI | 0.091 | 0.034 | 0.147 | 0.0017 | 0.015 | 0.141 | 0.064 | 0.219 | 0.0004 | 0.023 | 0.061 | −0.055 | 0.176 | 0.3046 | 0.481 |
| xCHA on BSI | 0.013 | −0.010 | 0.036 | 0.2622 | 0.4495 | 0.024 | −0.008 | 0.056 | 0.1408 | 0.2913 | 0.002 | −0.043 | 0.047 | 0.9237 | 0.9394 | |
| xCHA on DEP | 1.028 | 0.946 | 1.117 | 0.5139 | 0.656 | 0.982 | 0.873 | 1.104 | 0.7614 | 0.862 | 1.145 | 0.967 | 1.357 | 0.1171 | 0.260 | |
BSI–current depressive symptom scores as measured by Brief Symptom Inventory. DEP–self–reported lifetime depression. ADD, DOM, REC, additive, dominant and recessive heritability models; CHA, Childhood Adversity; RLE, recent life stress; FDR, false discovery rate. Italic type shows nominally significant p-values, bold type denotes significant Q-values surviving correction for multiple testing (P < 0.05, FDR Q < 0.05). In case of all top SNPs statistical parameters are shown for all analyses, not only for those models where they emerged as top SNPs, thus gray shading indicates results for the models where the given SNP was top SNP.
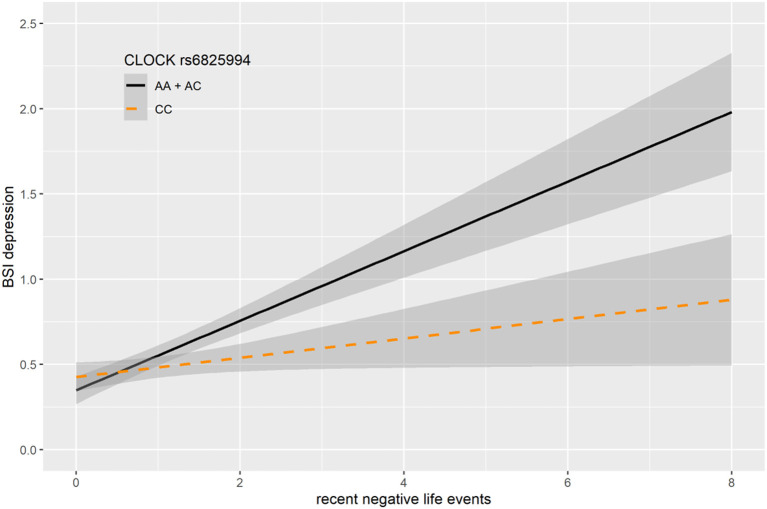
In case of rs6825994, we found a nominally significant interaction effect with recent negative life events (RLE) on BSI-depression in dominant (p = 0.0004) model as the lead SNP, and its effect in the additive model was also nominally significant (p = 0.0017), both of which remained significant after correction for multiple testing (FDRQadd = 0.0149 and FDRQdom = 0.0232, respectively) (Table 4). Subjects carrying minor A allele of rs6825994 had significantly higher current depression scores when exposed to recent stressful life events suggesting the minor allele to be a risk allele (Figure 2).
Figure 2.
Linear regression indicated a significant interaction between top SNPs rs6825994 of a clump containing 94 SNPs in the CLOCK gene and exposure to recent life events (RLE) on current depressive symptoms (BSI-Dep) in the dominant model, with the minor A allele as a risk allele. Linear regression indicated a significant interaction between CLOCK rs6825994 genotype and recent stressful life events (RLE) on current depression scores according to the dominant (p = 0.0004, FDR Q = 0.023, as top SNP) and additive (p = 0.0017, FDR Q = 0.015) models. Presence of the minor A allele was associated with higher depression scores in subjects exposed to more severe recent life events conveying a risk effect. On the vertical axis weighted depression (BSI-Dep) scores are shown. The horizontal axis shows recent life events (RLE) occurring within the past year as measured by the List of Threatening Experiences (31). BSI-Brief symptom inventory. Gray shading denotes 95%CI.
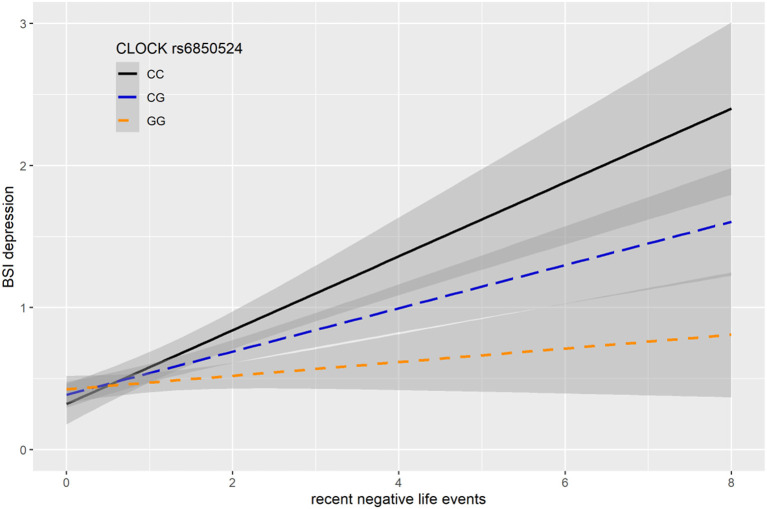
In case of rs6850524 we found nominally significant interaction effects with recent life events (RLE) on BSI-depression as top SNP in the additive model (p = 0.0004) which survived correction for multiple testing (FDRQadd = 0.0127). This SNP, although not as a top SNP, was also nominally significant in interaction with recent life events on BSI-depression in dominant (p = 0.0028) and recessive (p = 0.0066) models which all remained significant after correction for multiple testing (FDRQdom = 0.0187, FDRQrec = 0.0398) (Table 4). In these models, presence of the minor C allele was associated with a lower BSI-depression score if the subject was exposed to recent negative life events indicating a risk effect (Figure 3).
Figure 3.
Linear regression indicated a significant interaction between top SNP rs6850524 of a clump containing 94 SNPs in the CLOCK gene and exposure to recent life events (RLE) on current depression symptoms (BSI-dep) in the additive model, with the minor C allele as a risk allele. Linear regression indicated a significant interaction between CLOCK rs6850524 genotype and recent stressful life events (RLE) on current depression scores according to the additive model as top SNP (p = 0.0004, FDR Q = 0.013). Presence of the minor C allele was associated with higher depression scores in subjects exposed to more severe recent life events conveying a risk effect. On the vertical axis weighted depression (BSI-Dep) scores are shown. The horizontal axis shows recent life events (RLE) occurring within the past year as measured by the List of Threatening Experiences (31). BSI-Brief symptom inventory. Gray shading denotes 95%CI.
Gene × Environment Effects of Variation in CLOCK on Current Depressive Symptoms and Lifetime Depression: Interaction With Childhood Adversities (CHA)
In case of gene x environment interaction models with childhood adversity (CHA) on current depression (BSI-Dep), we identified two clumps with top SNPs rs6828454 and rs711533 (Table 4), both of them containing 9 SNPs (Supplementary Table 4).
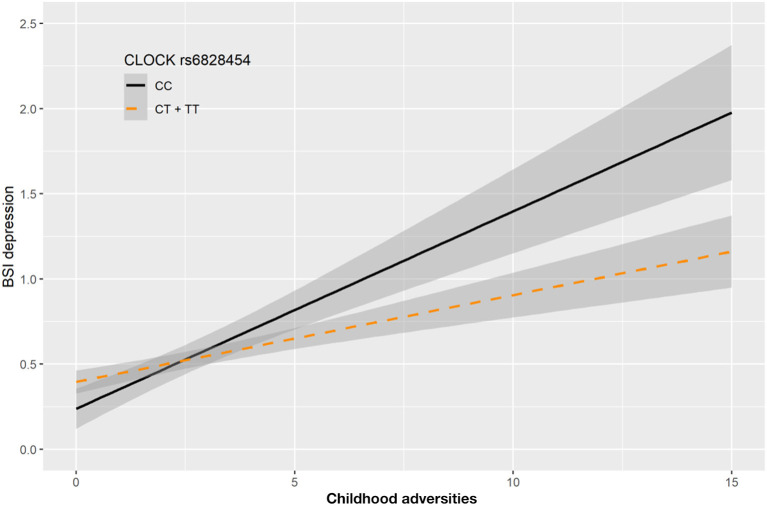
In case of rs6828454 we found a significant interaction effect with childhood adversities (CHA) on current depression (BSI-Dep) in the recessive model as top SNP (p = 0.0006, FDRQ = 0.0118). Minor C allele carriers reported significantly higher current depression levels when exposed to moderate or severe childhood adversity indicating the minor allele to be a risk allele (Figure 4).
Figure 4.
Linear regression indicated a significant interaction between top SNP rs6828454 of a clump containing 9 SNPs in the CLOCK gene and exposure to childhood adversities on current depression symptoms (BSI-dep) in the recessive model, with the minor C allele being a risk allele. Linear regression indicated a significant interaction between CLOCK rs6828454 genotype and childhood adverse life events (CHA) on current depression scores according to the recessive model as top SNP (p = 0.0006, FDR Q = 0.0118). Homozygous presence of the minor C allele was associated with higher depression scores in subjects exposed to more severe childhood adverse events conveying a risk effect. On the vertical axis weighted depression (BSI-Dep) scores are shown. The horizontal axis shows childhood adverse life events (CHA) as measured by an instrument derived from the CTQ (30). BSI-Brief symptom inventory. Gray shading denotes 95%CI.
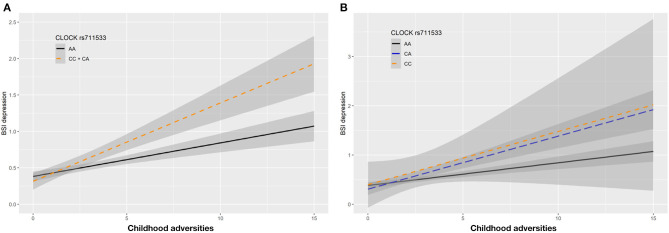
In case of rs711533 we found significant interaction effects with CHA on current depression (BSI-Dep) as top SNP in both additive (p = 0.0008, FDRQ = 0.0092) and dominant (p = 0.0008, FDRQ = 0.0114) models remaining significant after correcting for multiple testing (Table 4). Subjects carrying the minor C allele scored significantly higher on BSI-depression scale when exposed to childhood maltreatment reflecting a risk effect for the minor allele (Figure 5).
Figure 5.
Linear regression indicated a significant interaction between top SNP rs711533 of a clump containing 9 SNPs in the CLOCK gene and exposure to childhood adversity (CHA) on current depression symptoms (BSI-Dep) in the dominant (A) and additive (B) models, with the minor C allele being a risk allele. Linear regression indicated a significant interaction between CLOCK rs711533 genotype and childhood adverse life events (CHA) on current depression scores according to the dominant and additive models as top SNP (p = 0.0006, FDR Q = 0.0118). Presence of the minor C allele was associated with higher depression scores in subjects exposed to more severe childhood adverse events conveying a risk effect. On the vertical axis weighted depression (BSI-Dep) scores are shown. The horizontal axis shows childhood adverse life events (CHA) as measured by an instrument derived from the CTQ (30). BSI-Brief symptom inventory. Gray shading denotes 95%CI.
Interaction with childhood adversity did not have a significant effect on lifetime depression in case of any SNPs, therefore significant clumps could not be calculated (Table 4).
In silico Characterization and Functional Prediction of Identified Top SNPs rs6828454, rs711533, rs6825994, and rs6850524 as Well as SNPs in the Clumps Showing a Significant Effect on Depression
Genomic location of significant SNPs and top SNPs identified in the clumping procedure are shown in Figure 6. To detect the functional effect of the top significant SNPs, we utilized FuncPred tool (https://snpinfo.niehs.nih.gov). Two SNPs (rs28448438, rs28463765) are located in the transcriptional-factor-binding site of CLOCK gene, furthermore, rs726967 is located in the microRNA-binding site. Among the significant polymorphisms, several SNP showed high regulatory potential and conservation score.
Figure 6.
Genomic location of significant SNPs identified in the clumping procedure, in the analysis of variation in CLOCK in interaction with recent life events and early childhood adversities on current depressive symptoms. In interaction with recent life events (RLE), our analyses identified one clump containing 94 SNPs with top SNPs rs6825994 and rs6850524 (for dominant and additive models, respectively) on current depressive symptoms (BSI-Dep), marked in green. In interaction with childhood adversities (CHA), two clumps, each with 9 SNPs and top SNPs rs6828454 and rs711533 were identified on current depression symptoms which are shown in yellow and purple, respectively. UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) was used to visualize the location of polymorphisms on CLOCK gene.
In the literature exploratory analyzes on SNPedia (https://www.snpedia.com/), ClinVar (https://figshare.com/articles/dataset/CLOCK_stress_depression/14258567), GWAS Catalog (https://www.ebi.ac.uk/gwas/), we found some relevant sources related to the following SNPs: rs6853192, rs11726609, rs11133399, rs4865010, rs1000254, rs2412646, rs3736544, rs6811520, rs11931061, rs3817444, rs6832769, rs726967, and rs11735267. Results are shown in Supplementary Table 7 and discussed in the Discussion part.
Discussion
In our study investigating the effect of variation in the CLOCK gene with a linkage disequilibrium-based clumping method, we found no clumps of SNPs with a significant main effect either on lifetime depression or current depressive symptoms. However, we have identified significant clumps of SNPs in interaction with both early childhood adversities and recent negative life events on current depressive symptoms, but not in case of lifetime depression. While our results confirm the role of CLOCK variation in emergence of depressive symptoms, they also indicate that this effect is observable only in case of exposure to stress. Notably, we found that clumps of SNPs in the CLOCK gene interacted with both distal childhood traumas and adversities, which contribute to the emergence of a diathesis promoting susceptibility for affective disorders, and recent, proximal stressors, which have a role in triggering onset of the actual symptoms or illness episodes based on a diatheses. Finally, we detected no gene-environment correlation effects suggesting that CLOCK variation does not influence risk of exposure to early or recent adversities.
The Involvement of Circadian Disruption in Depression
Circadian rhythms regulate a multitude of physiological processes coordinating them both with each other and with the external environment, integrating outer sensory information, and environmental cues with internal physiological and psychological states, thus also influencing human cognition, affect, and behavior, which suggests that dysregulated or disturbed circadian rhythms are involved in the etiopathology of mental and mood disorders as well (6, 19). The presence of circadian abnormalities in depression have been evidenced for a long time with alterations in depressed patients in the circadian rhythmicity of somatic functions including body temperature, blood pressure, or urine metabolite excretion; altered hormone rhythms including prolactin, cortisol, GH, thyrotropin, and melatonin; and in the diurnal fluctuation of symptoms of depression including alteration in sleep-wake cycles, timing and structure of sleep, appetite, or social rhythms (8–14, 37). While in clinical studies risk and severity of depression correlated with the degree of circadian rhythm misalignment (19, 38), there is no definitive evidence whether circadian dysregulation precedes and plays a causative role in, or follows and results from, or merely coincides with depression, however, the complex relationship between the circadian system and mood disorders appears to be bidirectional (7, 39). Nevertheless, conditions leading to circadian rhythm disruption such as shift work may precipitate mood symptoms in those susceptible (40, 41), and in post-mortem brain studies in depressed patients severe disruption and desynchronization of daily rhythmic gene expression patterns were reported (42). One possible linking factor between the circadian system and mood disorders is that neural systems playing a key role in affective illness including the HPA-axis, limbic regions, and monoamine neurotransmitter household are under circadian regulation, and alterations in the biological clock could lead to neurobiological changes in neurotransmitter systems triggering depressive states (12, 14, 42–44). While variation in clock genes encoding elements of circadian rhythm mechanisms is associated with smaller alterations of circadian behaviors in healthy subjects, it appears to have a more pronounced impact on psychopathological features in mood disorder patients affecting key clinical and course features such as timing of disease onset and recurrence and treatment response (45–47).
Variation in the CLOCK Gene Does Not Directly Impact Lifetime Depression or Current Depressive Symptoms
Our findings indicate that variation in the CLOCK gene exerts no significant main effect either on lifetime depression, or current depressive symptoms. The CLOCK gene, located at chr4q12, is one of the chief genes in the endogenous master clock system playing a key role in the formation of circadian rhythms. Genomic variation in CLOCK gene in common polymorphisms with a MAF of ≥1% in 1000 Genomes Project (48) comprises 406 SNPs for African, 271 for European, 278 for Asian, and 277 for American continental populations (49). The CLOCK gene, as part of CLOCK(NPAS2)/ARNTL complex regulates rhythmic transcription of clock-controlled genes (CCG) in several tissues, including at least 15% of mammalian transcripts many of which gene expression patterns are disrupted in MDD (11, 19, 50, 51). The CLOCK machinery could provide mechanisms for control of circadian gene expression and responsivity to stimuli on cellular levels influencing activity of brain structures which control emotions and behavior (47). Furthermore, the function of CLOCK protein as a transcription factor and histone acetyltransferase also implies that genetic and epigenetic variations could contribute to physiological changes possibly leading to altered susceptibility of psychiatric disorders including depression (49). CLOCK in the mammalian brain is expressed in several brain structures beyond the master clock, the nucleus suprachiasmaticus, including the cortex (52), and molecular and behavioral studies support that CLOCK gene plays an important role in neuronal function involved in the regulation of several pathways implicated in psychiatric disorders and its genetic manipulation leads to marked changes in neurotransmitter activity and behavior. For example, CLOCK regulates expression of neurogenic transcription factors influencing the differentiation of adult neural stem cells in mice (53), controls transcription of tyrosine hydroxylase and cholecystokinin and other regulators of monoaminerg transmission (54), and disruption of CLOCK gene function leads to alterations in glutamatergic and GABAergic signaling (49, 55). Animal experiments on CLOCK gene in psychiatric condition-related behaviors have shown biological plausibility and promising findings (3), while in humans CLOCK variations have been implicated in susceptibility to phenotypes of common psychiatric disorders including autism spectrum disorders, schizophrenia, attention deficit/hyperactivity disorder, substance use disorder, major depressive disorder, bipolar disorder, and anxiety (49).
Several studies suggested an association between CLOCK and mood disorders, however, results are inconsistent in case of unipolar depression (56, 57). Previous studies employing a candidate variant approach focused only on a few variants and mainly on rs1801260, speculated to affect mRNA, which has been found to be associated with evening preference and a substantial, 10–44-min delay in preferred timing for activity and sleep in healthy C carriers (58). In some studies rs1801260 has been shown to be associated with clinical features mostly in case of bipolar disorder, especially with sleep problems including increased occurrence of lifetime and episode-related insomnia (59), higher recurrence of initial, middle and late insomnia in both MDD and BD patients (60), number and recurrence of manic episodes (17), and appetite disturbances in women (49), but a meta-analysis could not confirm an association between rs1801260 and either unipolar or bipolar mood disorder (61). These findings suggest that this variant is not associated with mood symptoms itself but only with certain, mainly sleep-related symptoms accompanying mood disorders. Other, less numerous studies focused on other CLOCK variants, with a nominally significant relationship between rs11932595 and SSRI efficacy, rs534654 and weight loss and rs12504300 and rs3825148 and SSRI side effects including nausea, constipation and vomiting in a Han Chinese population (11).
In light of the above contradictory findings and the general lack of robust and replicable positive associations between candidate CLOCK SNPs and depression, our present findings, investigating variation along the CLOCK gene corroborate previous reports suggesting that CLOCK does not have a significant direct main effect on lifetime depression or current depressive symptoms (61, 62).
CLOCK Variation Mediates the Effect of Childhood Adversities and Recent Negative Life Events on Current Depressive Symptoms
More importantly, in spite of a lack of a main effect of CLOCK variation on depressive phenotypes, we did detect a robust effect of several clumps of SNPs on current depressive symptoms in interaction with both distal, early childhood, and proximal, recent stressors, which is in line with our paradigm postulating that the majority of genes impact depression by increasing susceptibility toward the negative effects of stress rather than exerting a direct effect (16, 21).
According to the social Zeitgeber theory, mood disorders are triggered by life stressors disrupting normal routines and circadian rhythms thus altering mood and biological rhythms (63). Several depression-relevant brain functions are controlled simultaneously by the circadian clock and stress response systems which are themselves closely connected (7). The relationship between stress and the circadian rhythm regulation is bidirectional, with both circadian rhythms impacting stress response and the effects of stress impacting regulation of circadian rhythms. Diurnal cycles of glucocorticoids, regulating various stress response-related physiological and behavioral responses, are one of the most prominent endocrine manifestations of circadian rhythms and themselves play a role in synchronizing peripheral and circadian oscillators with promoters of several clock genes containing glucocorticoid responsive elements (GREs) (7, 64, 65). Furthermore, many stress-inducible genes are also under control of the circadian clock (7). The circadian clock and stress response systems show an extensive overlap and regulate multiple systems which control cognition, affect, reward processing, and other systems and functions implicated in depression (7), suggesting that modulation of glucocorticoid-mediated stress response may constitute a common mechanism by which circadian clock affects mood disorders (19). Animal studies have shown that chronic mild stress led to anhedonic behavior associated with disturbed diurnal oscillation in circadian gene expression and higher Clock expression in the basolateral amygdala (66) and reduced CLOCK protein levels in the prefrontal cortex (67). As variants of CLOCK gene show differential transcriptional response to glucocorticoids (19), it is possible that variation in the CLOCK gene may mediate the effects of stress on the development of depressive symptoms.
Although only a few human studies took stressors into consideration when investigating the effects of CLOCK variants, some both in healthy and depressed subjects reported that, similar to our findings, the impact of the investigated variants was detectable only in those exposed to some type of stress. In healthy subjects, presence of the C allele of rs1801260 has been associated with a greater disruption of sleep patterns following stressful life events, and a few studies in mood disorder patients found that this variant was associated with sleep change only in case of prior stressful experiences (10) concluding that environmental stress may increase vulnerability to circadian rhythm disruption (10) and suggesting an interaction between CLOCK variants in shaping individual risk for deleterious effects of environmental stress including depression (47). In another study in bipolar disorder patients, a significant interaction between CLOCK variants and early stress exposure on hopelessness and suicide in BP patients was reported (47). In a non-clinical Chinese population, G allele of rs11932595 was associated with altered sleep duration only in those exposed to high job stress, but showed no effect in case of low stress, while AA carriers showed less daytime dysfunction under low stress and more in high stress conditions compared to G carriers suggesting not only a gene-environment interaction effect, but also the role of the CLOCK as both a vulnerability and resilience gene with both positive and negative effects depending on stress (68).
Our present findings showing that CLOCK variation increases depressive symptoms only in those exposed to either early childhood adversities or recent stressful life events extend research suggesting that CLOCK gene is involved in the development of depressive symptoms by increasing vulnerability toward the disruptive effects of stress. More importantly, we found that both distal, early stressors, with a diathesis-forming etiological role, and recent negative life events, proximal stressors with a triggering role, interacted with CLOCK variation on severity of actual depressive symptoms. The exact mechanisms and phenotypes through which CLOCK in interaction with stress influences risk or severity of depression needs further study, but it has been suggested that CLOCK could hypothetically directly influence neural activity in brain structures playing a key role in generation and control of emotions and affect thus biasing depressive cognition in depression and resilience to detrimental effects of exposure to early stress (10, 69) and the interplay between CLOCK variation could influence the effects of stress on the central nervous system determining both vulnerability to or expression of phenotypes related to depression (7).
In silico Characterization and Functional Prediction of Top SNPs and SNPs in the Significant Clumps
In the final step of our analysis we carried out in silico characterization and functional prediction for the SNPs in the three clumps significantly impacting current depressive symptoms in interaction with either childhood adversities or current stressors. The search yielded several relevant findings (Supplementary Table 7). Notably, two SNPs, rs28448438 and rs28463765 are located in the transcriptional binding site of the CLOCK gene, while a third SNP, rs726967 is located in the microRNA-binding site possibly influencing transcriptional activity.
Of the four top SNPs, rs6850524, which significantly interacted with recent life events on current depressive symptoms has previously been associated with depression-relevant phenotypes including susceptibility to bipolar disorder (70), sleep quality (71), and risk of sleep disorders (72), which indicates its involvement in affective disorders and their symptomatology. This variant has also been found to be associated with somatic conditions including risk of obesity (73, 74), and non-alcoholic fatty liver disease (75) which may be clinically relevant considering the frequent comorbidity of the above illnesses with depression (76–78).
Among SNPs in the clumps significantly interacting with recent life events symptoms, rs6832769 has been reported to be related to emotional prosociality and agreeableness (79, 80), with response and remission with fluvoxamine in MDD but was not found to be associated with affective disorders (81–84), whereas rs2412646 has been associated with depression-comorbid alcohol use disorders and susceptibility to restless leg syndrome in schizophrenic patients (85–87). Several SNPs in the clumps significantly interacting with recent life events including rs11931061, rs3817444, and rs726967 have been reported to be associated with ADHD (88) or sleep problems, including rs11735267 showing a nominal association with late insomnia (70), and rs6853192 with longer sleep duration in general community sample of African Americans (89). In addition, several SNPs in the clumps interacting with recent life events were associated with risk of somatic conditions, including rs10002541 with abdominal obesity and diabetes (74), rs11726609 with BMI in African Americans (89), rs11133399 with gastric cancer overall survival and recurrence free survival and also a primary risk factor contributing to prognosis (90), rs4865010 with essential hypertension and coronary artery disease (91) and in men with hypertonia with testosterone levels corresponding to an androgenic deficient state itself a risk factor for cardiovascular complications (92), and rs6811520 with myocardial infarction (93), multiple sclerosis (94), male infertility (95).
In case of SNPs in clumps significantly interacting with childhood adversities on current depression, rs3749473 was associated with cognitive aging (96), rs6858749 with habitual sleep duration in interaction with protein intake (97, 98), and rs534654 with ADHD (88), disrupted sleep-wake cycles in BD (70), and development of depression (11).
Possible Clinical Implications of CLOCK Variation in Interaction With Early and Recent Stress in Depression
The association of stress and circadian vulnerability especially in case of mood disorders has significant clinical relevance in case of stress chronotherapy. As circadian disruptions and frequent stress exposure—especially in interaction—amplify the risks of development of a wide range of health-related disorders including mood disorders, simultaneous reduction of circadian and classical stressors including strategies stabilizing endogenous rhythms to counteract circadian perturbations would be beneficial. While the majority of such practices are hardly feasible in the modern lifestyle, there are several effective therapies for depression which act via modulating circadian parameters, including bright light, sleep deprivation, and phase resetting paradigms (12, 19, 99, 100) which improve symptoms within hours (101, 102). Scheduled meals or activity, or specific interventions such as social rhythm therapy to enhance circadian alignment of peripheral and central clocks also offer means of reducing circadian stress to prevent or improve mood disorder symptoms (103). Our study identifying an interaction between CLOCK variation and both childhood and recent stressors in the development of depressive symptoms could in the future help predict those at a higher risk for development of depression in case of circadian disruption, and also those who would benefit from chronotherapies for depression.
Furthermore, antidepressants including SSRIs, SNRIs and agomelatine also directly impact circadian rhythms as part of their effects improving depressive symptoms (6, 104–106). For example, fluoxetine normalizes disrupted light-induced entrainment, fragmented ultradian rhythms and altered hippocampal CLOCK expression in animal models of depression (107), while agomelatine, a melatonergic antidepressant is hypothesized to resynchronize disrupted circadian rhythms (108, 109). It has also been suggested that CLOCK expression may predict efficacy of antidepressants exerting their effects via influencing circadian mechanisms (68). Thus, understanding the role of CLOCK variation in depression may help identify new targets for pharmacological treatment possibly via modulating the impact of stress on brain function (7) as well as prediction of efficacy especially in subtypes of depression aiding precision therapy.
Limitations
When interpreting the impact of our findings, several limitations of our study must also be taken into account. First, both early childhood adversities and recent life events occurring in the past year were assessed retrospectively, and based on self-report of the subjects but not ascertained by other informants, and are thus subject to recall and reporting biases. Second, both lifetime depression and current depression severity was similarly based on a self-reported measure. Third, our population sample was relatively small, consisting approximately two thirds of female subjects, and limited to European white participants. Fourth, scoring childhood adversity and counting the number of recent negative life events does not take into consideration the differing severity and subjective impact of individual life events. Nevertheless, our study also has several strengths, including considering several hundred variants along the CLOCK gene with a clumping method rather than individual, hypothesis-based candidate SNPs, employing a dimensional approach capture current depression symptom severity, and using a GxE paradigm with two etiologically different types of stressors.
Conclusion
In conclusion, the results of our study investigating variation along the CLOCK gene with a linkage disequilibrium-based clumping of SNPs support the role of CLOCK in the background of depressive symptoms but only in association with recent stress and early adversities, underlining not only the depressogenic importance of circadian disruption but also that it acts in interaction with both early, etiological, and recent, trigger-like stressors. Thus, our findings strengthen the rationale of looking at the circadian system in search of new molecular targets for pharmacological interventions, and by specifically reporting that effects in clock variation are only observable in interaction with stress also helps to explain previous lack of consistent findings.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://figshare.com/articles/dataset/CLOCK_stress_depression/14258567.
Ethics Statement
The studies involving human participants were reviewed and approved by Scientific and Research Ethics Committee of the Medical Research Council, Budapest, Hungary. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XG, DG, DB, GJ, and GB: conceptualization. DB, NE, SS, and PP: methodology. XG, NE, DB, DT, and ZG: data collection. DG, ZK, SS, PP, DB, NE, DT, and ZG: data analysis. XG, DG, SS, ZK, ZG, DT, and DB: writing—original draft preparation. XG, DG, NE, ZG, DT, DB, ZK, SS, PP, GJ, and GB: writing—review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding. The study was supported by the Sixth Framework Program of the European Union (NewMood, LSHM-CT-2004-503474); by the Hungarian Brain Research Program (Grants KTIA_13_NAPA-II/14 and 2017-1.2.1-NKP-2017-00002), and the National Development Agency (Grant KTIA_NAP_13-1-2013-0001); by the Hungarian Academy of Sciences, Hungarian National Development Agency, Semmelweis University and the Hungarian Brain Research Program (Grant KTIA_NAP_13-2-2015-0001, MTA-SE-NAP B Genetic Brain Imaging Migraine Research Group); by the Hungarian Academy of Sciences (MTA-SE Neuropsychopharmacology and Neurochemistry Research Group); by OTKA 131629; by TAMOP-4.2.1.B-09/1/KMR-2010-0001; by the New National Excellence Program of The Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund (ÚNKP-20-3-II-SE-51, ÚNKP-20-4-II-SE-9); by the National Research, Development and Innovation Office, Hungary (2019-2.1.7-ERA-NET-2020-00005), under the frame of ERA PerMed (ERA-PERMED2019-108); and by the Thematic Excellence Programme (Tématerületi Kiválósági Program, 2020-4.1.1.-TKP2020) of the Ministry for Innovation and Technology in Hungary, within the framework of the Neurology and Translational Biotechnology thematic Programmes of the Semmelweis University and the European Union's Horizon 2020 Research and Innovation Programme under the Marie Skłodowska Curie grant agreement No. 766124. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2021.687487/full#supplementary-material
Main effect of CLOCK SNPs in the NewMood database surviving quality control on lifetime depression (DEP). Results of logistic regression models and quality control steps are shown. While there were nominally significant SNPs, significant clumps could not be identified.
Main effect of CLOCK SNPs in the NewMood database surviving quality control on current depressive symptoms (BSI-DEP). Results of linear regression models and quality control steps are shown. While there were nominally significant SNPs, significant clumps could not be identified.
CLOCK SNPs in the NewMood database surviving quality control, in interaction with recent life events (RLE) on current depressive symptoms (BSI-DEP). Results of linear regression models and quality control steps are shown. SNPs in significant clumps are marked in red, top SNP of the clump is marked in bold red. Results for additive, dominant, and recessive models are shown.
CLOCK SNPs in the NewMood database surviving quality control, in interaction with childhood adversities (CHA) on current depressive symptoms (BSI-DEP). Results of linear regression models and quality control steps are shown. SNPs in significant clumps are marked in red, top SNP of the clump is marked in bold red. Results for additive, dominant, and recessive models are shown.
Main effect of CLOCK SNPs in the NewMood database surviving quality control on recent negative life events (RLE). Results of linear regression models are shown. There were no nominally significant SNPs, significant clumps could not be identified.
Main effect of CLOCK SNPs in the NewMood database surviving quality control on childhood adversities (CHA). Results of linear regression models are shown. There were no nominally significant SNPs, significant clumps could not be identified.
In silico characterization and functional prediction of SNPs in the clumps significantly interacting with recent life events (RLE) and childhood adversities (CHA). Top SNP of each clump is marked in bold red.
References
- 1.Roenneberg T, Wirz-Justice A, Merrow M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. (2003) 18:80–90. 10.1177/0748730402239679 [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, Mcdearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. (2008) 9:764–75. 10.1038/nrg2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendoza J, Vanotti G. Circadian neurogenetics of mood disorders. Cell Tissue Res. (2019) 377:81–94. 10.1007/s00441-019-03033-7 [DOI] [PubMed] [Google Scholar]
- 4.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. (2010) 72:517–49. 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- 5.Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. (2011) 3:10. 10.1186/gm224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitsillou E, Liang J, Hung A, Karagiannis TC. The circadian machinery links metabolic disorders and depression: a review of pathways, proteins and potential pharmacological interventions. Life Sci. (2021) 265:118809. 10.1016/j.lfs.2020.118809 [DOI] [PubMed] [Google Scholar]
- 7.Landgraf D, Mccarthy MJ, Welsh DK. Circadian clock and stress interactions in the molecular biology of psychiatric disorders. Curr Psychiatry Rep. (2014) 16:483. 10.1007/s11920-014-0483-7 [DOI] [PubMed] [Google Scholar]
- 8.Van Cauter E, Turek FW. Depression: a disorder of timekeeping? Perspect Biol Med. (1986) 29:510–519. 10.1353/pbm.1986.0033 [DOI] [PubMed] [Google Scholar]
- 9.Reinink E, Bouhuys AL, Gordijn MC, Van Den Hoofdakker RH. Prediction of the antidepressant response to total sleep deprivation of depressed patients: longitudinal versus single day assessment of diurnal mood variation. Biol Psychiatry. (1993) 34:471–81. 10.1016/0006-3223(93)90238-9 [DOI] [PubMed] [Google Scholar]
- 10.Antypa N, Mandelli L, Nearchou FA, Vaiopoulos C, Stefanis CN, Serretti A, et al. The 3111T/C polymorphism interacts with stressful life events to influence patterns of sleep in females. Chronobiol Int. (2012) 29:891–7. 10.3109/07420528.2012.699380 [DOI] [PubMed] [Google Scholar]
- 11.Ma HY, Liu ZF, Xu YF, Hu XD, Sun N, Li XR, et al. The association study of CLOCK gene polymorphisms with antidepressant effect in Chinese with major depressive disorder. Per Med. (2019) 16:115–22. 10.2217/pme-2018-0123 [DOI] [PubMed] [Google Scholar]
- 12.Mcclung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. (2007) 114:222–32. 10.1016/j.pharmthera.2007.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vadnie CA, Mcclung CA. Circadian rhythm disturbances in mood disorders: insights into the role of the suprachiasmatic nucleus. Neural Plast. (2017) 2017:1504507. 10.1155/2017/1504507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker WH, III, Walton JC, Devries AC, Nelson RJ. Circadian rhythm disruption and mental health. Transl Psychiatry. (2020) 10:28. 10.1038/s41398-020-0694-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. (2000) 157:1552–62. 10.1176/appi.ajp.157.10.1552 [DOI] [PubMed] [Google Scholar]
- 16.Gonda X, Petschner P, Eszlari N, Baksa D, Edes A, Antal P, et al. Genetic variants in major depressive disorder: from pathophysiology to therapy. Pharmacol Therap. (2019) 194:22–43. 10.1016/j.pharmthera.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 17.Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, et al. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet B Neuropsychiatr Genet. (2003) 123B:23–6. 10.1002/ajmg.b.20038 [DOI] [PubMed] [Google Scholar]
- 18.Mccarthy MJ, Nievergelt CM, Kelsoe JR, Welsh DK. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS ONE. (2012) 7:e32091. 10.1371/journal.pone.0032091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi SQ, White MJ, Borsetti HM, Pendergast JS, Hida A, Ciarleglio CM, et al. Molecular analyses of circadian gene variants reveal sex-dependent links between depression and clocks. Transl Psychiatry. (2016) 6:e748. 10.1038/tp.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagdy G, Juhasz G, Gonda X. A new clinical evidence-based gene-environment interaction model of depression. Neuropsychopharmacol Hung. (2012) 14:213–20. 10.5706/nph201212001 [DOI] [PubMed] [Google Scholar]
- 21.Gonda X, Hullam G, Antal P, Eszlari N, Petschner P, Hokfelt TG, et al. Significance of risk polymorphisms for depression depends on stress exposure. Sci Rep. (2018) 8:3946. 10.1038/s41598-018-22221-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, et al. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. (2010) 67:133–8. 10.1016/j.biopsych.2009.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. (2010) 68:811–7. 10.1016/j.biopsych.2010.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. (2009) 460:748–52. 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. (2006) 163:28–40. 10.1176/appi.ajp.163.1.28 [DOI] [PubMed] [Google Scholar]
- 26.Lazary J, Lazary A, Gonda X, Benko A, Molnar E, Juhasz G, et al. New evidence for the association of the serotonin transporter gene (SLC6A4) haplotypes, threatening life events, depressive phenotype. Biol Psychiatry. (2008) 64:498–504. 10.1016/j.biopsych.2008.03.030 [DOI] [PubMed] [Google Scholar]
- 27.Juhasz G, Chase D, Pegg E, Downey D, Toth ZG, Stones K, et al. CNR1 gene is associated with high neuroticism and low agreeableness and interacts with recent negative life events to predict current depressive symptoms. Neuropsychopharmacology. (2009) 34:2019–27. 10.1038/npp.2009.19 [DOI] [PubMed] [Google Scholar]
- 28.Juhasz G, Dunham JS, Mckie S, Thomas E, Downey D, Chase D, et al. The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. Biol Psychiatry. (2011) 69:762–71. 10.1016/j.biopsych.2010.11.019 [DOI] [PubMed] [Google Scholar]
- 29.Derogatis LR. BSI: Brief Symptom Inventory: Administration, Scoring, and Procedures Manual Minneapolis: National Computer Systems, Minneapolis, MN, (1993). [Google Scholar]
- 30.Bernstein DP, Fink L, Handelsman L, Foote J, Lovejoy M, Wenzel K, et al. Initial reliability and validity of a new retrospective measure of child abuse and neglect. Am J Psychiatry. (1994) 151:1132–6. 10.1176/ajp.151.8.1132 [DOI] [PubMed] [Google Scholar]
- 31.Brugha T, Bebbington P, Tennant C, Hurry J. The list of threatening experiences: a subset of 12 life event categories with considerable long-term contextual threat. Psychol Med. (1985) 15:189–94. 10.1017/S003329170002105X [DOI] [PubMed] [Google Scholar]
- 32.Brugha TS, Cragg D. The list of threatening experiences: the reliability and validity of a brief life events questionnaire. Acta Psychiatr Scand. (1990) 82:77–81. 10.1111/j.1600-0447.1990.tb01360.x [DOI] [PubMed] [Google Scholar]
- 33.Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig IW. DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav Genet. (2003) 33:67–72. 10.1023/A:1021055617738 [DOI] [PubMed] [Google Scholar]
- 34.Eszlari N, Millinghoffer A, Petschner P, Gonda X, Baksa D, Pulay AJ, et al. Genome-wide association analysis reveals KCTD12 and miR-383-binding genes in the background of rumination. Transl Psychiatry. (2019) 9:119. 10.1038/s41398-019-0454-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/ [Google Scholar]
- 36.Gonda X, Gyorik D, Eszlari N, Gal Z, Torok D, Baksa D, et al. “CLOCK, Stress, Depression”. (FigShare) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rusting CL, Larsen RJ. Diurnal patterns of unpleasant mood: associations with neuroticism, depression, and anxiety. J Pers. (1998) 66:85–103. 10.1111/1467-6494.00004 [DOI] [PubMed] [Google Scholar]
- 38.Emens J, Lewy A, Kinzie JM, Arntz D, Rough J. Circadian misalignment in major depressive disorder. Psychiatry Res. (2009) 168:259–61. 10.1016/j.psychres.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 39.Lamont EW, Legault-Coutu D, Cermakian N, Boivin DB. The role of circadian clock genes in mental disorders. Dialogues Clin Neurosci. (2007) 9:333–42. 10.31887/DCNS.2007.9.3/elamont [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedrosian TA, Nelson RJ. Timing of light exposure affects mood and brain circuits. Transl Psychiatry. (2017) 7:e1017. 10.1038/tp.2016.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee A, Myung SK, Cho JJ, Jung YJ, Yoon JL, Kim MY. Night shift work and risk of depression: meta-analysis of observational studies. J Korean Med Sci. (2017) 32:1091–96. 10.3346/jkms.2017.32.7.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci USA. (2013) 110:9950–5. 10.1073/pnas.1305814110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. (2006) 4:163–73. 10.1016/j.cmet.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 44.Harbour VL, Robinson B, Amir S. Variations in daily expression of the circadian clock protein, PER2, in the rat limbic forebrain during stable entrainment to a long light cycle. J Mol Neurosci. (2011) 45:154–61. 10.1007/s12031-010-9469-z [DOI] [PubMed] [Google Scholar]
- 45.Mccarthy MJ, Welsh DK. Cellular circadian clocks in mood disorders. J Biol Rhythms. (2012) 27:339–52. 10.1177/0748730412456367 [DOI] [PubMed] [Google Scholar]
- 46.Benedetti F, Terman M. Much ado about.a moody clock. Biol Psychiatry. (2013) 74:236–7. 10.1016/j.biopsych.2013.05.037 [DOI] [PubMed] [Google Scholar]
- 47.Benedetti F, Riccaboni R, Dallaspezia S, Locatelli C, Smeraldi E, Colombo C. Effects of CLOCK gene variants and early stress on hopelessness and suicide in bipolar depression. Chronobiol Int. (2015) 32:1156–61. 10.3109/07420528.2015.1060603 [DOI] [PubMed] [Google Scholar]
- 48.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature. (2015) 526:75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuch JB, Genro JP, Bastos CR, Ghisleni G, Tovo-Rodrigues L. The role of CLOCK gene in psychiatric disorders: evidence from human and animal research. Am J Med Genet B Neuropsychiatr Genet. (2018) 177:181–98. 10.1002/ajmg.b.32599 [DOI] [PubMed] [Google Scholar]
- 50.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. (2006) 125:497–508. 10.1016/j.cell.2006.03.033 [DOI] [PubMed] [Google Scholar]
- 51.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. (2007) 450:1086–90. 10.1038/nature06394 [DOI] [PubMed] [Google Scholar]
- 52.Abe H, Honma S, Namihira M, Masubuchi S, Ikeda M, Ebihara S, et al. Clock gene expressions in the suprachiasmatic nucleus and other areas of the brain during rhythm splitting in CS mice. Brain Res Mol Brain Res. (2001) 87:92–9. 10.1016/S0169-328X(00)00295-3 [DOI] [PubMed] [Google Scholar]
- 53.Kimiwada T, Sakurai M, Ohashi H, Aoki S, Tominaga T, Wada K. Clock genes regulate neurogenic transcription factors, including NeuroD1, and the neuronal differentiation of adult neural stem/progenitor cells. Neurochem Int. (2009) 54:277–85. 10.1016/j.neuint.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 54.Sidor MM, Spencer SM, Dzirasa K, Parekh PK, Tye KM, Warden MR, et al. Daytime spikes in dopaminergic activity drive rapid mood-cycling in mice. Mol Psychiatry. (2015) 20:1479–80. 10.1038/mp.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mcclung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, et al. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci USA. (2005) 102:9377–81. 10.1073/pnas.0503584102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kishi T, Kitajima T, Ikeda M, Yamanouchi Y, Kinoshita Y, Kawashima K, et al. Association study of clock gene (CLOCK) and schizophrenia and mood disorders in the Japanese population. Eur Arch Psychiatry Clin Neurosci. (2009) 259:293–7. 10.1007/s00406-009-0869-4 [DOI] [PubMed] [Google Scholar]
- 57.Dmitrzak-Weglarz MP, Pawlak JM, Maciukiewicz M, Moczko J, Wilkosc M, Leszczynska-Rodziewicz A, et al. Clock gene variants differentiate mood disorders. Mol Biol Rep. (2015) 42:277–88. 10.1007/s11033-014-3770-9 [DOI] [PubMed] [Google Scholar]
- 58.Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. (1998) 21:569–76. 10.1093/sleep/21.6.569 [DOI] [PubMed] [Google Scholar]
- 59.Benedetti F, Dallaspezia S, Fulgosi MC, Lorenzi C, Serretti A, Barbini B, et al. Actimetric evidence that CLOCK 3111 T/C SNP influences sleep and activity patterns in patients affected by bipolar depression. Am J Med Genet B Neuropsychiatr Genet. (2007) 144B:631–5. 10.1002/ajmg.b.30475 [DOI] [PubMed] [Google Scholar]
- 60.Serretti A, Benedetti F, Mandelli L, Lorenzi C, Pirovano A, Colombo C, et al. Genetic dissection of psychopathological symptoms: insomnia in mood disorders and CLOCK gene polymorphism. Am J Med Genet B Neuropsychiatr Genet. (2003) 121B:35–8. 10.1002/ajmg.b.20053 [DOI] [PubMed] [Google Scholar]
- 61.Kishi T, Yoshimura R, Fukuo Y, Kitajima T, Okochi T, Matsunaga S, et al. The CLOCK gene and mood disorders: a case-control study and meta-analysis. Chronobiol Int. (2011) 28:825–33. 10.3109/07420528.2011.609951 [DOI] [PubMed] [Google Scholar]
- 62.Calati R, Gaspar-Barba E, Yukler A, Serretti A. T3111C clock single nucleotide polymorphism and mood disorders: a meta-analysis. Chronobiol Int. (2010) 27:706–21. 10.3109/07420521003681480 [DOI] [PubMed] [Google Scholar]
- 63.Grandin LD, Alloy LB, Abramson LY. The social zeitgeber theory, circadian rhythms, and mood disorders: review and evaluation. Clin Psychol Rev. (2006) 26:679–94. 10.1016/j.cpr.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 64.Nader N, Chrousos GP, Kino T. Interactions of the circadian CLOCK system and the HPA axis. Trends Endocrinol Metab. (2010) 21:277–86. 10.1016/j.tem.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalsbeek A, Van Der Spek R, Lei J, Endert E, Buijs RM, Fliers E. Circadian rhythms in the hypothalamo-pituitary-adrenal (HPA) axis. Mol Cell Endocrinol. (2012) 349:20–9. 10.1016/j.mce.2011.06.042 [DOI] [PubMed] [Google Scholar]
- 66.Savalli G, Diao W, Schulz S, Todtova K, Pollak DD. Diurnal oscillation of amygdala clock gene expression and loss of synchrony in a mouse model of depression. Int J Neuropsychopharmacol. (2014) 18:pyu095. 10.1093/ijnp/pyu095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calabrese F, Savino E, Papp M, Molteni R, Riva MA. Chronic mild stress-induced alterations of clock gene expression in rat prefrontal cortex: modulatory effects of prolonged lurasidone treatment. Pharmacol Res. (2016) 104:140–50. 10.1016/j.phrs.2015.12.023 [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Cao Z, Wu S, Wang C, He S, Dong Y, et al. Association of job stress, CLOCK gene polymorphism and their interaction with poor sleep quality. J Sleep Res. (2020) 30:e13133. 10.1111/jsr.13133 [DOI] [PubMed] [Google Scholar]
- 69.Benedetti F, Radaelli D, Bernasconi A, Dallaspezia S, Falini A, Scotti G, et al. Clock genes beyond the clock: CLOCK genotype biases neural correlates of moral valence decision in depressed patients. Genes Brain Behav. (2008) 7:20–5. 10.1111/j.1601-183X.2007.00312.x [DOI] [PubMed] [Google Scholar]
- 70.Shi J, Wittke-Thompson JK, Badner JA, Hattori E, Potash JB, Willour VL, et al. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. (2008) 147B:1047–55. 10.1002/ajmg.b.30714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi L, Liu Y, Jiang T, Yan P, Cao F, Chen Y, et al. Relationship between mental health, the CLOCK gene, and sleep quality in surgical nurses: a cross-sectional study. Biomed Res Int. (2020) 2020:4795763. 10.1155/2020/4795763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ning L, Shi L, Tao N, Li R, Jiang T, Liu J. Effects of occupational stress and circadian CLOCK gene polymorphism on sleep quality of oil workers in Xinjiang, China. Med Sci Monit. (2020) 26:e924202. 10.12659/MSM.924202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sookoian S, Gemma C, Gianotti TF, Burgueno A, Castano G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. (2008) 87:1606–15. 10.1093/ajcn/87.6.1606 [DOI] [PubMed] [Google Scholar]
- 74.Ye D, Cai S, Jiang X, Ding Y, Chen K, Fan C, et al. Associations of polymorphisms in circadian genes with abdominal obesity in Chinese adult population. Obes Res Clin Pract. (2016) 10(Suppl. 1):S133–41. 10.1016/j.orcp.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 75.Sookoian S, Castano G, Gemma C, Gianotti TF, Pirola CJ. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol. (2007) 13:4242–8. 10.3748/wjg.v13.i31.4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marx P, Antal P, Bolgar B, Bagdy G, Deakin B, Juhasz G. Comorbidities in the diseasome are more apparent than real: What Bayesian filtering reveals about the comorbidities of depression. PLoS Comput Biol. (2017) 13:e1005487. 10.1371/journal.pcbi.1005487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hullam G, Antal P, Petschner P, Gonda X, Bagdy G, Deakin B, et al. The UKB envirome of depression: from interactions to synergistic effects. Sci Rep. (2019) 9:9723. 10.1038/s41598-019-46001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soto-Angona O, Anmella G, Valdes-Florido MJ, De Uribe-Viloria N, Carvalho AF, Penninx B, et al. Non-alcoholic fatty liver disease (NAFLD) as a neglected metabolic companion of psychiatric disorders: common pathways and future approaches. BMC Med. (2020) 18:261. 10.1186/s12916-020-01713-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. (2010) 15:647–56. 10.1038/mp.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ci H, Wu N, Su Y. Clock gene modulates roles of OXTR and AVPR1b genes in prosociality. PLoS ONE. (2014) 9:e109086. 10.1371/journal.pone.0109086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kishi T, Kitajima T, Ikeda M, Yamanouchi Y, Kinoshita Y, Kawashima K, et al. CLOCK may predict the response to fluvoxamine treatment in Japanese major depressive disorder patients. Neuromolecular Med. (2009) 11:53–57. 10.1007/s12017-009-8060-7 [DOI] [PubMed] [Google Scholar]
- 82.Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms. (2009) 7:2. 10.1186/1740-3391-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mccarthy MJ, Nievergelt CM, Shekhtman T, Kripke DF, Welsh DK, Kelsoe JR. Functional genetic variation in the Rev-Erbalpha pathway and lithium response in the treatment of bipolar disorder. Genes Brain Behav. (2011) 10:852–61. 10.1111/j.1601-183X.2011.00725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Porcelli S, Drago A, Fabbri C, Gibiino S, Calati R, Serretti A. Pharmacogenetics of antidepressant response. J Psychiatry Neurosci. (2011) 36:87–113. 10.1503/jpn.100059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sjoholm LK, Kovanen L, Saarikoski ST, Schalling M, Lavebratt C, Partonen T. CLOCK is suggested to associate with comorbid alcohol use and depressive disorders. J Circadian Rhythms. (2010) 8:1. 10.1186/1740-3391-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jung JS, Lee HJ, Cho CH, Kang SG, Yoon HK, Park YM, et al. Association between restless legs syndrome and CLOCK and NPAS2 gene polymorphisms in schizophrenia. Chronobiol Int. (2014) 31:838–44. 10.3109/07420528.2014.914034 [DOI] [PubMed] [Google Scholar]
- 87.Partonen T. Clock genes in human alcohol abuse and comorbid conditions. Alcohol. (2015) 49:359–65. 10.1016/j.alcohol.2014.08.013 [DOI] [PubMed] [Google Scholar]
- 88.Carpena MX, Hutz MH, Salatino-Oliveira A, Polanczyk GV, Zeni C, Schmitz M, et al. CLOCK polymorphisms in Attention-Deficit/Hyperactivity Disorder (ADHD): further evidence linking sleep and circadian disturbances and ADHD. Genes. (2019) 10:88. 10.3390/genes10020088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Riestra P, Gebreab SY, Xu R, Khan RJ, Gaye A, Correa A, et al. Circadian CLOCK gene polymorphisms in relation to sleep patterns and obesity in African Americans: findings from the Jackson heart study. BMC Genet. (2017) 18:58. 10.1186/s12863-017-0522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Y, Wang D, Song Y, Zhang X, Jiao Z, Dong J, et al. Functional polymorphisms in circadian positive feedback loop genes predict postsurgical prognosis of gastric cancer. Cancer Med. (2019) 8:1919–29. 10.1002/cam4.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kolomeychuk S, Kurbatova I, Topchieva LV, Korneva VA, Poltorak AN, Chambers TC, et al. Association between CLOCK genetic variants and individual susceptibility to essential hypertension and coronary artery disease in Russian population. Exp Clin Cardiol. (2014) 20:1–17. [Google Scholar]
- 92.Kurbatova I, Topchieva LV, Korneva VA. The level of circulating testosterone and the blood-plasma lipid profile in men with essential Hypertension. carriers of GG genotype in relation to the−257T>G polymorphic marker of the clock gene. Proc Karelian Res Centre Russian Acad Sci. (2016) 6:39–48. 10.17076/eb291 [DOI] [Google Scholar]
- 93.Skrlec I, Milic J, Steiner R. The impact of the circadian genes CLOCK and ARNTL on myocardial infarction. J Clin Med. (2020) 9:484. 10.3390/jcm9020484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lavtar P, Rudolf G, Maver A, Hodzic A, Starcevic Cizmarevic N, Zivkovic M, et al. Association of circadian rhythm genes ARNTL/BMAL1 and CLOCK with multiple sclerosis. PLoS ONE. (2018) 13:e0190601. 10.1371/journal.pone.0190601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hodzic A, Ristanovic M, Zorn B, Tulic C, Maver A, Novakovic I, et al. Genetic variation in circadian rhythm genes CLOCK and ARNTL as risk factor for male infertility. PLoS ONE. (2013) 8:e59220. 10.1371/journal.pone.0059220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin CH, Lin E, Lane HY. Genetic biomarkers on age-related cognitive decline. Front Psychiatry. (2017) 8:247. 10.3389/fpsyt.2017.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dashti HS, Follis JL, Smith CE, Tanaka T, Cade BE, Gottlieb DJ, et al. Habitual sleep duration is associated with BMI and macronutrient intake and may be modified by CLOCK genetic variants. Am J Clin Nutr. (2015) 101:135–43. 10.3945/ajcn.114.095026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sridhar GR, Sanjana NS. Sleep, circadian dysrhythmia, obesity and diabetes. World J Diabetes. (2016) 7:515–22. 10.4239/wjd.v7.i19.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wehr TA, Wirz-Justice A, Goodwin FK, Duncan W, Gillin JC. Phase advance of the circadian sleep-wake cycle as an antidepressant. Science. (1979) 206:710–3. 10.1126/science.227056 [DOI] [PubMed] [Google Scholar]
- 100.Lewy AJ, Sack RL, Miller LS, Hoban TM. Antidepressant and circadian phase-shifting effects of light. Science. (1987) 235:352–4. 10.1126/science.3798117 [DOI] [PubMed] [Google Scholar]
- 101.Fritzsche M, Heller R, Hill H, Kick H. Sleep deprivation as a predictor of response to light therapy in major depression. J Affect Disord. (2001) 62:207–15. 10.1016/S0165-0327(00)00154-3 [DOI] [PubMed] [Google Scholar]
- 102.Dallaspezia S, Benedetti F. Chronobiological therapy for mood disorders. Expert Rev Neurother. (2011) 11:961–70. 10.1586/ern.11.61 [DOI] [PubMed] [Google Scholar]
- 103.Koch CE, Leinweber B, Drengberg BC, Blaum C, Oster H. Interaction between circadian rhythms and stress. Neurobiol Stress. (2017) 6:57–67. 10.1016/j.ynstr.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abe M, Herzog ED, Block GD. Lithium lengthens the circadian period of individual suprachiasmatic nucleus neurons. Neuroreport. (2000) 11:3261–3264. 10.1097/00001756-200009280-00042 [DOI] [PubMed] [Google Scholar]
- 105.Sprouse J, Braselton J, Reynolds L. Fluoxetine modulates the circadian biological clock via phase advances of suprachiasmatic nucleus neuronal firing. Biol Psychiatry. (2006) 60:896–9. 10.1016/j.biopsych.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 106.Germain A, Kupfer DJ. Circadian rhythm disturbances in depression. Hum Psychopharmacol. (2008) 23:571–85. 10.1002/hup.964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaufler J, Ronovsky M, Savalli G, Cabatic M, Sartori SB, Singewald N, et al. Fluoxetine normalizes disrupted light-induced entrainment, fragmented ultradian rhythms and altered hippocampal clock gene expression in an animal model of high trait anxiety- and depression-related behavior. Ann Med. (2016) 48:17–27. 10.3109/07853890.2015.1122216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kasper S, Hajak G, Wulff K, Hoogendijk WJ, Montejo AL, Smeraldi E, et al. Efficacy of the novel antidepressant agomelatine on the circadian rest-activity cycle and depressive and anxiety symptoms in patients with major depressive disorder: a randomized, double-blind comparison with sertraline. J Clin Psychiatry. (2010) 71:109–20. 10.4088/JCP.09m05347blu [DOI] [PubMed] [Google Scholar]
- 109.Zajecka J, Schatzberg A, Stahl S, Shah A, Caputo A, Post A. Efficacy and safety of agomelatine in the treatment of major depressive disorder: a multicenter, randomized, double-blind, placebo-controlled trial. J Clin Psychopharmacol. (2010) 30:135–44. 10.1097/JCP.0b013e3181d420a7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Main effect of CLOCK SNPs in the NewMood database surviving quality control on lifetime depression (DEP). Results of logistic regression models and quality control steps are shown. While there were nominally significant SNPs, significant clumps could not be identified.
Main effect of CLOCK SNPs in the NewMood database surviving quality control on current depressive symptoms (BSI-DEP). Results of linear regression models and quality control steps are shown. While there were nominally significant SNPs, significant clumps could not be identified.
CLOCK SNPs in the NewMood database surviving quality control, in interaction with recent life events (RLE) on current depressive symptoms (BSI-DEP). Results of linear regression models and quality control steps are shown. SNPs in significant clumps are marked in red, top SNP of the clump is marked in bold red. Results for additive, dominant, and recessive models are shown.
CLOCK SNPs in the NewMood database surviving quality control, in interaction with childhood adversities (CHA) on current depressive symptoms (BSI-DEP). Results of linear regression models and quality control steps are shown. SNPs in significant clumps are marked in red, top SNP of the clump is marked in bold red. Results for additive, dominant, and recessive models are shown.
Main effect of CLOCK SNPs in the NewMood database surviving quality control on recent negative life events (RLE). Results of linear regression models are shown. There were no nominally significant SNPs, significant clumps could not be identified.
Main effect of CLOCK SNPs in the NewMood database surviving quality control on childhood adversities (CHA). Results of linear regression models are shown. There were no nominally significant SNPs, significant clumps could not be identified.
In silico characterization and functional prediction of SNPs in the clumps significantly interacting with recent life events (RLE) and childhood adversities (CHA). Top SNP of each clump is marked in bold red.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://figshare.com/articles/dataset/CLOCK_stress_depression/14258567.