Abstract
Chromosome 11q13-14 amplification is a defining feature of high-risk hormone receptor-positive (HR+) breast cancer; however, the mechanism(s) by which this amplicon contributes to breast tumorigenesis remains unclear. In the current study, proteogenomic analyses of >3000 breast tumors from the TCGA, METABRIC and CPTAC studies demonstrated that carnitine palmitoyltransferase 1A (CPT1A), which is localized to this amplicon, is overexpressed at the mRNA and protein level in aggressive luminal tumors, strongly associated with indicators of tumor proliferation and a predictor of poor prognosis. In vitro genetic studies demonstrated that CPT1A is required for and can promote luminal breast cancer proliferation, survival, as well as colony and mammosphere formation. Since CPT1A is the rate-limiting enzyme during fatty acid oxidation (FAO), our data indicate that FAO may be essential for these tumors. Pharmacologic inhibition of FAO prevented in vitro and in vivo tumor growth and cell proliferation as well as promoted apoptosis in luminal breast cancer cells and orthotopic xenograft tumor models. Collectively, our data establish an oncogenic role for CPT1A and FAO in HR+ luminal tumors and provide preclinical evidence and rationale supporting further investigation of FAO as a potential therapeutic opportunity for the treatment of HR+ breast cancer.
INTRODUCTION
Luminal subtype or hormone receptor positive (HR+) breast cancers represent nearly 70% of the approximately 270 000 breast cancer cases diagnosed each year in the United States. The clinical implementation of endocrine-based therapies results in remission and improves prognosis in the majority of patients with HR+ tumors (1,2). However, approximately 30% of patients manifest primary resistance and about 50% of patients receiving endocrine therapy will acquire resistance leading to a high incidence of recurrence and progression (3–5). Phosphatidylinositol 3-kinase (PI3K)-family inhibitors as well as CDK4/6 inhibitors have been shown to be effective in preclinical and clinical studies in ER+ breast cancers; however, these treatments are limited to patients with specific genetic profiles and/or mutations (6–9). These clinical results highlight the need to identify additional genomic alterations and dysregulated signaling pathways that will be important for optimizing the identification and validation of novel therapeutic opportunities in HR+ breast cancer patients.
Cellular metabolism has been well established as a hallmark of cancer and recognized as a therapeutically exploitable vulnerability in cancer cells (10,11). While early studies established the role of glycolysis in tumor progression, more recent studies have demonstrated that tumor cells may use alternative pathways, including fatty acid β-oxidation (FAO), and/or multiple metabolic pathways throughout tumor development and progression (10–17). We recently reported the results of an integrative genomic analysis that identified a subset of loci and genes that were amplified and associated with poor clinical outcome in highly proliferative HR+ luminal breast tumors (18). This included amplification of 11q13-14 that was previously shown to be associated with high risk ER+ breast cancer (19). Further analysis of functional data from a genome-wide RNAi screen predicted that Carnitine palmitoyltransferase 1A (CPT1A), the rate limiting enzyme for FAO and localized on amplicon 11q13, is essential for cell viability in luminal/ER+ cell lines (18). These data thus suggest that CPT1A and FAO might play an essential role in aggressive luminal/HR+ breast tumors and suggest the therapeutic vulnerability of targeting FAO in these patients.
In the current study, we sought to establish a role for CPT1A in luminal or HR+ breast cancer tumorigenesis and examine the therapeutic potential of CPT1A and FAO in this subset of patients. Our data demonstrate that CPT1A is significantly overexpressed at both the mRNA and protein level in highly proliferative luminal tumors. CPT1A overexpression is associated with increased proliferation markers in luminal tumors and corresponds with poor overall survival. In vitro genetic studies establish a role for CPT1A and FAO in luminal tumor growth and survival. Importantly, we demonstrate through both in vitro cell line analyses and in vivo xenograft studies that pharmacological inhibition of CPT1A/FAO resulted in decreased cell proliferation, reduced beta oxidation, increased apoptosis and inhibited tumor growth and progression.
MATERIALS AND METHODS
Gene and protein expression analysis in human breast tumors and cell lines
RNA sequencing (RNAseq) data (n=1031) from human tumors was acquired from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/) and processed as previously described (18,20,21). Illumina HT-29 v3 expression data for the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) project (n=1992) was acquired from the European Genome-phenome Archive at the European Bioinformatics Institute (https://www.ebi.ac.uk/ega/) and data were median centered (19). PAM50 classification, as well as calculation of the 11-gene PAM50 proliferation signature score, was performed as previously described (22,23). To examine the relationship between CPT1A expression, ER status and proliferation score, samples were segregated into three groups: basal-like or non-basal (including Luminal A, Luminal B and HER2E subtype) with the non-basal/luminal tumors being further divided into high and low proliferation groups based on median PAM50 proliferation signature score. An ANOVA test followed by Tukey’s test for pairwise comparison was used to calculate differences between subgroups. Proteome-wide mass spectrometry data for 77 tumors was acquired from the CPTAC study (24). Samples were classified into the above three groups and were similarly analyzed to assess differences in Cpt1a protein expression. To analyze CPT1A isoform expression, RNAseq data from 817 tumors (20) was used to calculate the expression of CPT1A variant 1 (NM_001876.3) and variant 2 (NM_001031847.2) across all tumors; an unpaired t-test was used to calculate differences in isoform expression. Expression data for a panel of 51 breast cancer cell lines were acquired from GEO (GSE12777) (25). Affymetrix U133+2 data were MAS5.0 normalized using Affymetrix Expression Console (ver1.2.1.20) and log2 transformed. Expression probes were collapsed using the median gene value with the GenePattern (26) module CollapseProbes.
RPPA analyses
Replication Based Normalized Reverse Phase Protein Array (RPPA) data for 733 tumors from the TCGA cohort was acquired from The Cancer Proteome Atlas data portal (http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html). To examine the relationship between CPT1A mRNA expression and protein markers of proliferation, 606 luminal tumors were dichotomized into CPT1A high (top quartile) and CPT1A low (all other tumors) based on mRNA expression and an unpaired t-test was used to examine differences in expression of specific proliferation and cell cycle marker proteins.
Clinical analyses
To investigate the relationship between CPT1A mRNA expression and overall survival, clinical data for the 1992 patients in the METABRIC study were obtained. We extracted patients classified as LumA, LumB or HER2E for which survival data were reported (n = 1333). For survival analysis of the TCGA dataset, we extracted patients classified as LumA, LumB or HER2E for which clinical data were available (n = 807). Overall survival was calculated comparing patients with high CPT1A versus low CPT1A using the median for the dataset as the threshold. For each analysis, significance was calculated by a log-rank test and the hazard ratio (HR) is reported.
Cell culture and molecular cloning
Human breast cancer cell lines MCF7, T47D, ZR751, MCF10A, HCC1143, HCC1954, BT549, BT20, MDAMB231 and MDAMB468 were purchased from the American Tissue Culture Collection (Manassas, VA, USA) and cultured according to the suggested guidelines. MCF7 or ZR751 cell expressing one of two unique doxycycline (dox) inducible shRNA against CPT1A or non-silencing negative control (shNS) were created using the pTRIPZ Inducible Lentiviral shRNA system (GE Dharmacon). The catalogue number for shRNA is: V3THS_359756, for shRNA is: V3THS_359760 and for shNS is: RHS4743. The shRNA expression was induced using 2.0 μg/ml of doxycycline for either cell line. Silencing of CPT1A was verified by western blot and qRT-PCR analyses. CPT1A was overexpressed in MCF10A cells by lentivirus p7056-pHAGE-P-CMVt-N-HA-GAW-CPT1A (Addgene #100146) (27) using an MOI of 3. Sterile suspensions of etomoxir, ranolazine, perhexiline and doxycycline were prepared as per manufacturer’s protocol.
Preparation of protein lysates and western blot analyses
Protein lysates were prepared from cell lines by mechanical homogenization in 1.5% dodecyl maltoside (DDM, Sigma) under ice-cold conditions. Protein concentration was measured using the BCA protein assay kit. About 35–50 μg of protein was loaded on 10–12% mini-protean TGX gradient gel (BioRad) at 100 V for 2 h at room temperature and transferred on to nitrocellulose membrane at 35 V overnight at 4°C. The membranes were blocked using AdvanBlock-chemi blocking solution (Advansta) for 1 h at room temperature, incubated with primary antibody overnight at 4°C followed by incubation with HRP-conjugated secondary antibodies for 1 h at room temperature. The signal was developed using SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific) and digitally imaged using the ChemiDoc Touch Imaging System (BioRad). The primary antibodies for CPT1A (Proteintech #15184-1-AP), PARP (Cell Signaling #9542), cleaved PARP (Cell Signaling #5625) and β-actin (Cell Signaling #4970) were purchased from the noted manufacturer. HRP-conjugated anti-mouse and anti-rabbit secondary antibodies were purchased from Cell Signaling.
Oxygen consumption rate (OCR) assay
Mitochondrial respiration and ATP production were assessed by measuring OCR using a Seahorse Biosciences extracellular flux analyzer (XFe24) as per the manufacturer’s protocol. Briefly, MCF7 cells were seeded at 5 × 104 cells per well in XF24 plates in DMEM (10% FBS, 1% Pen-Strep) or HBSS and incubated for 20–24 h at 37°C and 5% CO2. Cells were treated with vehicle or doxycycline (2 μg/ml) in DMEM or HBSS for 24 h prior to the assay (4 wells/condition). Subsequently, cells were incubated with seahorse XF media with 10 mM glucose, 2 mM glutamine and 1 mM pyruvate, and basal OCR was measured. The results were analyzed using Wave software.
Oil Red O staining
For Oil Red O staining, parental MCF7 or ZR751 cells and tet-inducible shRNA expressing MCF7shNS, MCF7sh1, MCF7sh2, ZR751shNS, ZR751sh1 or ZR751sh2 cells were seeded onto Millicell EZ slide (EMD Millipore #EZGS0416) at a density of 50 000 cells per well. Parental cells were mock-treated or treated with etomoxir (75 μg/ml), perhexiline (6 μg/ml) or ranolazine (50 μg/ml) for 24 h and tet-inducible shRNA expressing MCF7shNS, MCF7sh1, MCF7sh2, ZR751shNS, ZR751sh1 or ZR751sh2 cells were mock-treated or treated with doxycycline (2 μg/ml) for 72 h and Oil Red O staining was performed. Briefly, cells were fixed with 4% PFA for 30 min, washed with 60% isopropanol and stained with Oil Red O (Sigma-Aldrich #O1391,) solution for 10 min. Nuclei were counterstained with hematoxylin (Fisher Scientific #3535-16) for 2 min, mounted with cytoseal mounting medium (VWR #48212-154), and images were captured using Nikon Eclipse 80i microscope at 100× using an oil immersion lens.
Cell viability and proliferation assays
MCF7 or ZR751 cells were seeded on to 96-well tissue culture plates at a density of 2000 cells per well. Cells were mock-treated or treated with etomoxir (25, 50 and 75 μg/ml), perhexiline (2, 4 and 6 μg/ml) or ranolazine (15, 25 and 50 μg/ml) and viability was assessed at 24, 48 and 72 h. To measure the effect of genetic inhibition of CPT1A on cell viability and proliferation MCF7shNS, MCF7sh1, MCF7sh2, ZR751shNS, ZR751sh1 or ZR751sh2 cells were seeded on to 96-well tissue culture plates at a density of ,000 cells per well. Cells were mock-treated or treated with doxycycline (2 μg/ml) for 72 h. To demonstrate the effect of CPT1A overexpression on short-term proliferation, MCF10A cells were seeded on a 6-well plate. When cells reached a density of ∼70–80% confluence, they were transduced with HA-CPT1A lentivirus or empty vector at an MOI of 3. After 48 h, 2000 cells were transplanted on to a 96-well plate for 72 h. For each experiment, at each time point, cells were incubated in 10% CellTiter 96 AQueous One Solution (Promega) according to the manufacturer’s protocol for 2 h at 37°C and colorimetric absorbance was measured at 490 nm by a Tecan F50 Microplate Reader.
Colony formation assay
For colony formation assay, tet-inducible shRNA expressing MCF7 or ZR751 cells were plated at a density of 2000 (MCF7) or 5000 (ZR751) cells, respectively, per 6-well plate. Cells were mock treated or treated with 2 μg/ml doxycycline to induce CPT1A silencing for up to 21 days. To demonstrate the effect of CPT1A overexpression on colony formation, MCF10A cells were transduced with either HA-CPT1A lentivirus or empty vector at an MOI of 3. The cells were allowed to recover for 48 h and then seeded on to a 6-well plate at a density of 2000 cells per well for up to 14 days. Colonies were fixed with 3.7% formaldehyde for 30 min at 37°C and stained with 0.01% crystal violet. To quantify these data, independent colonies were counted from three replicate experiments and an unpaired t-test used to assess statistical significance.
Mammosphere formation assay
For mammosphere formation assay, parental or shRNA expressing MCF7 or ZR751 cells were plated as a single cell suspension in ultra-low attachment plates (500 cells per well in a 24-well plate) and cultured in serum-free DMEM/F12 supplemented with B27 (Thermofisher #17504044), bFGF (10 ng/ml; Corning #354060) and hEGF (20 ng/ml; Sigma #E9644). After 24 h, cells were treated with doxycycline (2 μg/ml) that was replenished every 48 h. Alternatively, to demonstrate the effect of CPT1A overexpression on mammosphere formation, MCF10A cells which inherently do not form mammospheres were transduced with HA-CPT1A expressing or empty vector control lentivirus with an MOI of 3 for 48 h. Transduced cells were then seeded as a single cell suspension at a density of 1000 cells per well in a 6-well ultra-low attachment plate and cultured in serum-free DMEM/F12 supplemented with B27, bFGF and hEGF as described above. For all experiments, mammosphere formation was followed for up to 10 days, quantified and expressed as relative count to either the no dox treatment for shRNA expressing cell lines or empty vector control for CPT1A overexpressing cell line.
Xenograft tumor growth
All animal experiments were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the US National Institutes of Health and in accordance with an approved Rutgers University Institutional Animal Care and Use Committee protocol specific to this project (#PROTO999900087). For these studies, 12-week old virgin nod scid gamma (NSG) female mice were acquired from Jackson Laboratories. Forty-eight hours prior to xenograft transplant, a 0.36 mg 17-β estradiol pellet (Innovative Research of America) was subcutaneously implanted in the lateral side of the neck. For transplant studies, 3.5 × 106 MCF7 cells per animal were grown under normal in vitro culturing conditions. Immediately prior to transplant, cells were removed from culturing media, trypsinized, washed three times in sterile PBS and resuspended in Matrigel (100 µl total volume). Cells were injected into the fourth mammary fat pad of 12-week old NSG virgin female mouse (n = 24). Tumor nodules were observed approximately 6 weeks after transplant. When tumors reached an approximate volume of 250–350 mm3, mice were randomized into one of the three treatment cohorts (n = 4–8 mice per group): saline control, 20 mg/kg ranolazine, or 40 mg/kg ranolazine. Mice were treated by intraperitoneal (i.p) injection twice a week for 21 days. Tumor size was measured twice a week by electronic caliper and tumor volume was calculated by using the formula 0.5 × (width)2 × (length). Tumors were harvested when the average tumor volume of the control cohort exceeded 1500 mm3; animals were euthanized, tumors harvested and snap frozen in liquid nitrogen for immunohistochemistry (IHC) analyses.
Immunohistochemistry
All staining and IHC were performed using automated and standardized protocols by the Rutgers Cancer Institute Biorepository and Histopathology Core Facility. Xenograft tumors were fresh frozen and paraffin embedded (FFPE) using the Tissue Tek VIP 5 Tissue Processor and then sectioned at 4μm. To deparaffinize tumors for hematoxylin and eosin (H&E) or IHC staining, sections were baked for 1 h at 65°C and then cooled. H&E staining was performed with standardized protocols using an automated Sakura Tissue-Tek Routine Stainer. Briefly, the sample was hydrated with subsequent washes of xylene, 100% ethanol, 95% ethanol, 70% ethanol and water. Sections were incubated in hematoxylin for 8 min and then subsequently washed in water, acid alcohol and water again. Samples were incubated in bluing agent for 2 min, washed in water and then 95% ethanol prior to eosin staining (2 min). Slides were subsequently washed in 100% ethanol followed by xylene and ethanol dehydration and then mounted. For single antibody IHC, the Ventana Discovery XT Immunostainer and Ventana (Roche) proprietary solutions were used. As before, the slide was deparaffinized and rehydrated. Antibodies against Ki67 (Abcam SP6 #16667 at 1:200), cleaved PARP (Cell Signaling #5625 at 1:200) and perilipin-1 (Cell Signaling #9349 at 1:200) or secondary antibody (Jackson Immunochemical donkey anti-rabbit at 1:500) were prepared in DAKO diluent background reducing solution. Antigen retrieval was completed using the Ventana, heat antigen retrieval solution (CC1). Samples were then treated with peroxidase inhibitor solution (3% H2O2), incubated in primary and secondary antibody solutions, and developed using DAB peroxidase. Slides were then treated with Copper D to enhance DAB signal and counterstained using hematoxylin. As before bluing agent was used to enhance the hematoxylin signal. Samples were washed (PBS), dehydrated in 100% ethanol followed by xylene washes and then mounted for visualization. Images were captured using EVOS M500 microscope at 40× magnification. DAB intensity was quantified using FIJI (28), as described previously (29). Briefly, each image was processed using the color deconvolution tool in FIJI, maximum threshold value in all samples were tested and then an average maximum threshold value was calculated and set for all DAB quantification in all images testing the same antigen. Mean signal intensity was measured in at least three different fields/sections of each tumor sample. A student t-test was used to determine the significance of the difference between treated and control samples.
RESULTS
CPT1A is overexpressed in aggressive HR+ breast tumors
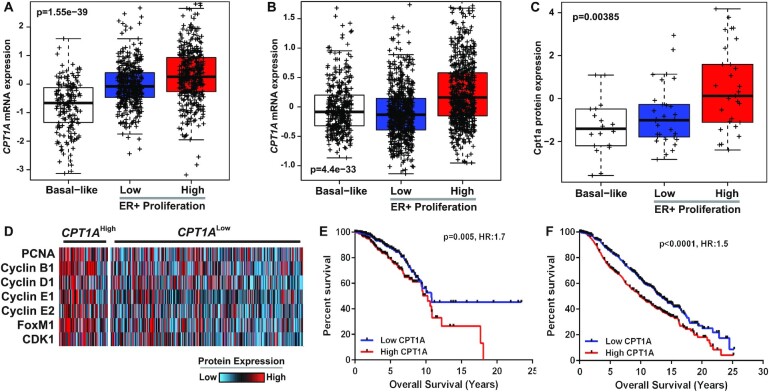
In order to examine the functional implications of CPT1A in HR+ breast cancer, we assessed CPT1A mRNA expression in both The Cancer Genome Atlas (TCGA, n = 1032) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC, n = 1998) datasets relative to the 11-gene PAM50 proliferation gene expression signature (22,23). Owing to the noted observation that breast tumors, when viewed across the pan-cancer spectrum, classify into basal-like and HR+/luminal (including HER2E, Luminal A and Luminal B) subtypes (30), we examined the association between CPT1A expression and proliferation as a function of these subgroups. CPT1A mRNA expression was found to be significantly upregulated in highly proliferative HR+ breast tumors compared to less aggressive HR+ tumors or basal-like tumors in both the TCGA (Figure 1A, P = 1.6 × 10–39) and METABRIC (Figure 1B, P = 4.4 × 10–33) datasets (two-way ANOVA, Tukey’s post-test). This association was further validated by demonstrating increased Cpt1a protein expression (P = 0.004, ANOVA) in highly proliferative HR+ human breast tumors using mass spectrometry data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) project (Figure 1C).
Figure 1.
CPT1A is overexpressed in highly proliferative ER+ breast tumors and is associated with poor overall survival. Tumor samples from the (A) TCGA (n = 1031) or (B) METABRIC (n = 1992) cohorts were dichotomized into basal-like or ER+ tumors with ER+ tumors further divided into high- and low-proliferation groups based on the PAM50 proliferation signature. Highly proliferative ER+ tumors show increased CPT1A expression (P < 0.0001, ANOVA, Tukey’s test). (C) Proteomic data from the CPTAC project (n = 77) were used to demonstrate increased Cpt1a protein expression in highly proliferative ER+ tumors (P < 0.0001, ANOVA, Tukey’s test).(D) RPPA data from ER+ tumors from the TCGA project (n = 606) demonstrate increased expression of proliferation markers (P<0.0001, unpaired t-test). High CPT1A mRNA expression was shown to correlate with poor overall survival in the (E) TCGA (P = 0.005, HR: 1.5) and (F) METABRIC (P < 0.0001, HR: 1.7) cohorts.
Given that CPT1A mRNA and protein expression are associated with increased proliferation, we next sought to verify the association between CPT1A mRNA expression and protein markers of proliferation using Reverse Phase Protein Array (RPPA) data from luminal TCGA tumors (n = 606). We determined that common makers of proliferation including PCNA, Cyclin D1, Cyclin E1, Cyclin E2, Cyclin B1, CDK1 and FOXM1 were significantly (P<0.001, t-test) upregulated in tumors with high CPT1A (top quartile) expression (Figure 1D). Importantly, analyses of clinical data demonstrated that high CPT1A mRNA levels correlated with a worse overall prognosis in luminal tumors and this relationship was apparent in both the TCGA (Figure 1E; HR: 1.7, P = 0.005) and METABRIC (Figure 1F; HR: 1.5, P<0.0001) datasets.
CPT1A is essential for FAO in ER+ cell lines
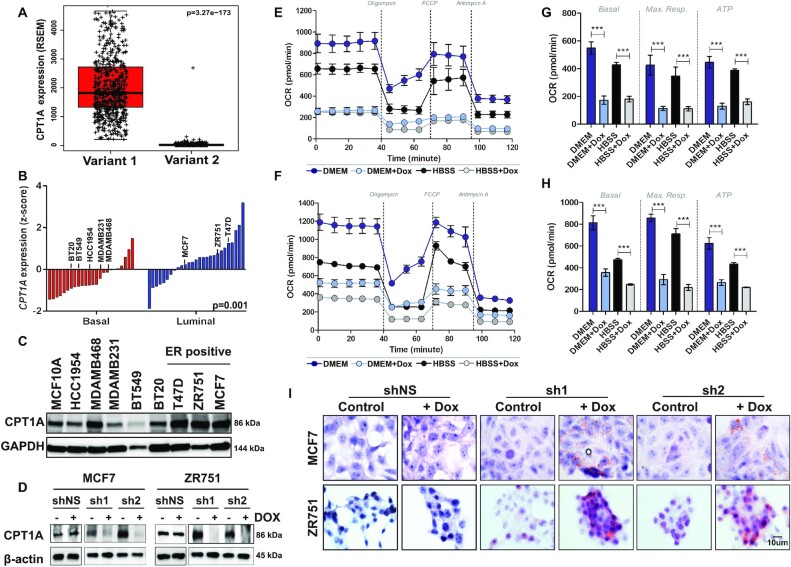
Previous studies have reported multiple isoforms of CPT1A: cytoplasmic variant 1 is the rate limiting enzyme required for transport of long chain fatty acids into the mitochondria during FAO while variant 2 is localized to the nucleus where it interacts with histone deacetylases to mediate epigenetic regulation of transcription (31). Analysis of mRNA isoform expression using RNA sequencing data from 817 human breast tumors from the TCGA cohort (20) demonstrated that variant 1 is the predominant isoform expressed in human tumors (Figure 2A) with approximately 99.2% of sequenced transcripts in a given tumor being classified as variant 1 irrespective of histological or PAM50 subtype.
Figure 2.
CPT1A mediates FAO in ER+ breast cancer. (A) Isoform analysis of RNAseq data from 817 tumors from the TCGA demonstrated increased expression of variant 1 with >99.2% of sequenced RNA called as variant 1 on a per tumor basis (P < 0.0001). (B) Gene expression analyses of 51 breast cancer cell lines (GSE12777) demonstrates that CPT1A is more highly expressed (z-score) in ER+/luminal subtype cell lines compared to basal-like cell lines (P = 0.001). (C) Western blot analyses confirm increased Cpt1a expression in ER+ cell lines. (D) MCF7 (MCF7sh1 or MCF7sh2) or ZR751 (ZR751sh1 or ZR751sh2) cells expressing one of two independent tet-inducible shRNA show a significant decreased in Cpt1a expression by western blot following dox treatment (2 μg/ml, 96 h); no effect on MCF7shNS or ZR751shNS cells was observed upon dox treatment. (E) MCF7sh1 and (F) MCF7sh2 cells show decreased OCR levels following shRNA-mediated silencing of CPT1A under normal (DMEM; blue scale) or starvation (HBSS; gray scale) growth conditions. (G) MCF7sh1 and (H) MCF7sh2 cells show decreased basal OCR, maximum respiration and ATP (P < 0.0001) following shRNA-mediated silencing of CPT1A; the mean of four replicates with standard error are shown for each sample (Student’s t-test, ***p<0.0005). (I) MCF7 or ZR751 CPT1A shRNA expressing cells show increased lipid droplet accumulation after dox (2 μg/ml, 96 h) treatment; no effect was observed in shNS expressing cells.
We next assessed CPT1A mRNA expression in a panel of 51 breast cancer cell lines (Figure 2B, GSE12777) (25) in order to identify model systems to examine the role of CPT1A in luminal breast tumorigenesis. Consistent with human tumors, CPT1A expression was uniformly higher in luminal cell lines when compared to basal-like cell lines (P = 0.001, t-test). Western blot analyses of a subset of cell lines confirmed these findings with luminal MCF7, T47D and ZR751 cells having consistently higher Cpt1a expression compared to basal-like breast cancer cell lines or MCF10A immortalized breast epithelial cells (Figure 2C). Based on these data, we engineered MCF7 or ZR751 cells to express one of two doxycycline (dox)-inducible shRNA against CPT1A or a non-silencing (NS) negative control; hereafter referred to as MCF7sh1, MCF7sh2, MCF7shNS, ZR751sh1, ZR751sh2 or ZR751shNS. As expected, dox treatment (2 μg/ml, 96 h) significantly reduced Cpt1a protein expression in each cell line expressing shRNA against CPT1A while no change in expression was evident in MCF7shNS or ZR751shNS cell lines (Figure 2D). Consistent with its role in regulating FAO, genetic inhibition of CPT1A resulted in reduced mitochondrial respiration as evident by reduced oxygen consumption rates (OCR) when assessed by Seahorse XF Bioanalyzer in both MCF7sh1 (Figure 2E) and MCF7sh2 (Figure 2F) cells. As illustrated in Figure 2G (MCF7sh1) and Figure 2H (MCF7sh2), a significant reduction (P<0.0001) in basal OCR, maximum respiration and ATP production was observed following CPT1A inhibition. Notably, a similar effect was observed upon dox treatment under normal (DMEM; blue scale) or starvation growth conditions (HBSS; gray scale). Finally, Oil Red O staining was used to demonstrate the accumulation of lipid droplets following dox treatment in CPT1A shRNA expressing MCF7 or ZR751 cell lines indicating that down-regulation of CPT1A expression resulted in reduced β-oxidation (Figure 2I); no effect on Oil Red O staining was noted in shNS expressing cells (+/-dox).
CPT1A is required for HR+ breast cancer cell line clonogenicity and mammosphere formation
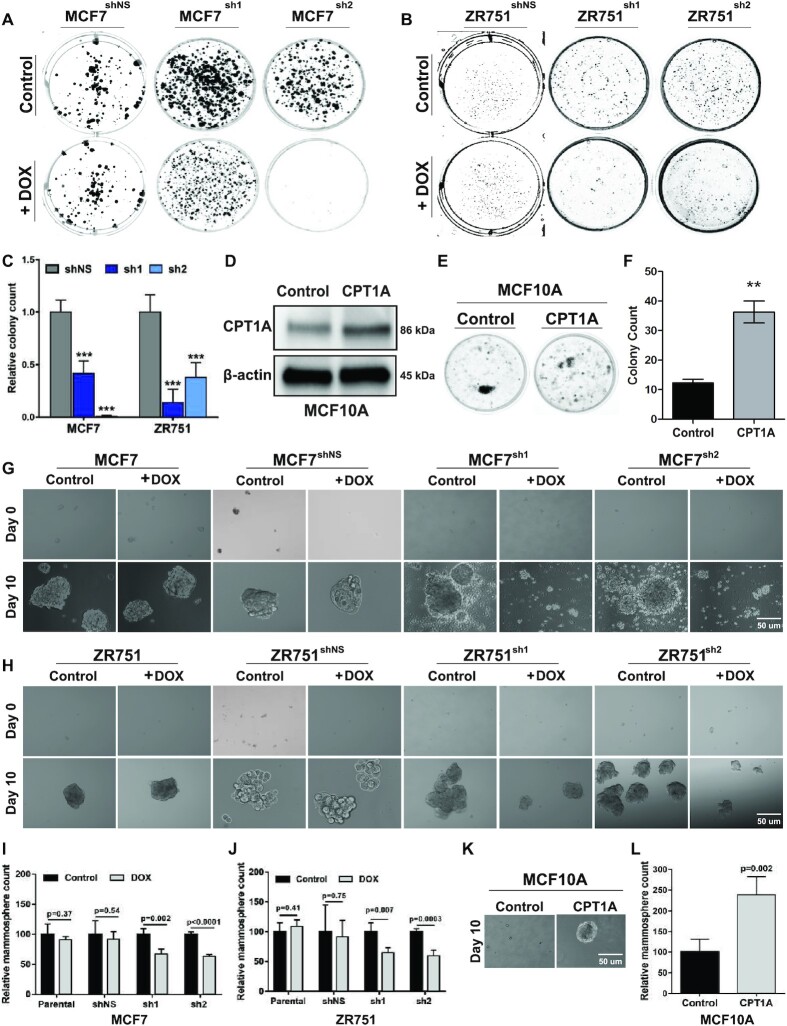
Given the observed association between CPT1A mRNA or protein expression and proliferation status in human breast tumors, we hypothesized that CPT1A and FAO are essential for HR+ breast tumor maintenance and growth. To test this hypothesis, we examined the effects of CPT1A knockdown on colony formation capacity. We determined that shRNA-mediated silencing of CPT1A in either MCF7 (Figure 3A) or ZR751 (Figure 3B) cells (2 μg/ml dox) significantly reduced colony formation over 21 days; no effect was seen in shNS expressing cell lines. Quantitation of these data demonstrate that MCF7sh1 (63.3%, P = 0.0005), MCF7sh2 (100%, P = 0.0003), ZR751sh1 (66.1%, P = 0.0006) and ZR751sh2 (79.9%, P = 0.002) cells showed significant reductions in colony formation upon dox treatment. As expected, colony forming capacity of MCF7shNS (10.9%, P = 0.31) and ZR751shNS (4.4%, P = 0.69) were not affected by dox treatment (Figure 3C). In order to confirm that the observed effects on colony formation were specifically due to CPT1A, MCF10A cells were transduced to overexpress either CPT1A or an empty vector control (MOI: 3, 72 h). As shown in Figure 3D, lentiviral overexpression resulted in a ∼68.5% increase in Cpt1a protein levels and this was associated with a 3.4-fold increase in MCF10A colony formation capacity (P < 0.0001) over a 14-day time course relative to empty vector transduced cells (Figure 3E and F).
Figure 3.
CPT1A is required for HR+ breast cancer cell line clonologenicity and mammosphere formation. (A) Silencing of CPT1A in MCF7 or (B) ZR751 dox-inducible shRNA expressing cells demonstrate reduced colony formation following dox treatment (2 μg/ml dox, 21 days); no change is seen in MCF7shNS or ZR751shNS cells +/-dox (C). Quantitation of colony formation assays (t-test) reported as the ratio of +/-dox. (D) Western blot analysis demonstrates relative Cpt1a overexpression in MCF10A cells following lentiviral transduction (MOI = 3, 72 h). (E) CPT1A overexpression in MCF10A cells result in increased colony formation capacity. (F) Quantitation of colony formation assay in (E). An unpaired-test was used for each analysis: ***p<0.0005; **p<0.005; mean and standard error are indicated. (G) MCF7 or (H) ZR751 cells expressing either sh1 or sh2 tet-inducible shRNA against CPT1A show reduced mammosphere formation following dox treatment (2 μg/ml; 21 days); no effect on mammosphere formation was apparent in parental or shNS expressing cells +/-dox. Quantification of mammosphere following shRNA-mediated silencing of CPT1A in (I) MCF7 or (J) ZR751 cells; data are presented relative to no dox treatment for each cell line. (K) CPT1A overexpression (MOI: 3, 10 days) induces mammosphere formation in MCF10A cells. (L) Quantification of (K) as relative mammosphere count. An unpaired t-test was performed for all assays.
Next we next sought to determine whether CPT1A overexpression could contribute to tumor initiation and three-dimensional growth capacity of luminal tumor cells. To test this hypothesis, we examined the effect of CPT1A on mammosphere formation. As shown in Figure 3G, control or shRNA expressing MCF7 cells readily form mammosphere within 10 days. However, induction of CPT1A-specific shRNA expression following dox (2 μg/ml) treatment resulted in 32.8% loss of mammosphere formation in MCF7sh1(P = 0.002) and 36.6% in MCF7sh2 (P < 0.0001) cells (Figure 3G and I). Similarly, CPT1A silencing in ZR751 cells resulted in a 35.4% decrease in mammosphere formation in ZR751sh1(P = 0.007) and 40.4% in ZR751sh2 (P = 0.0003) cells over a 10-day period (Figure 3H and J). As shown, mammosphere formation was not affected by dox treatment in parental or shNS expressing MCF7 or ZR751 cell lines. Finally, as illustrated in Figure 3K, MCF10A cells form a limited number of small mammospheres within the 10-day time course under normal growth conditions. However, lentivirus overexpression of CPT1A resulted in an approximately 2-fold increase in mammosphere formation relative to empty vector transduced cells (Figure 3K and L, P = 0.002). These data indicate that CPT1A is essential for mammosphere initiation and formation as well as for the three-dimensional growth of luminal breast cancer cell lines.
CPT1A is essential for luminal cell growth and survival
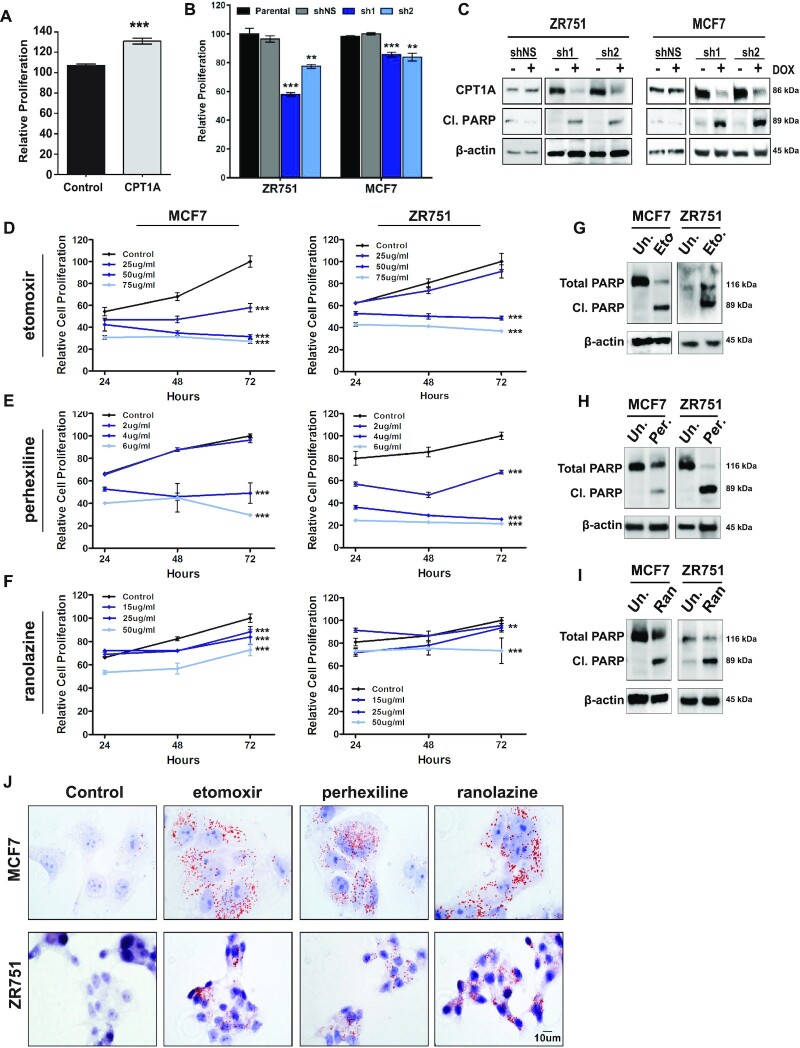
Given the notable impact of CPT1A on both colony and mammosphere formation, we next sought to determine whether modulation of CPT1A expression levels altered cell viability and growth. To do so, we first examined the effects of CPT1A overexpression on short-term cell proliferation (72 h). As shown in Figure 4A, CPT1A overexpression in MCF10A cells resulted in a ∼30.9% increase in MCF10A proliferation as measured by MTS assay (P < 0.0001, t-test). Similarly, shRNA-mediated silencing of CPT1A in ZR751 cells resulted in a 23.0% (sh1, P < 0.0001) and 42.1% (sh2, P = 0.0007) reduction in proliferation over this time course. Likewise, a more modest, yet statistically significant 15.5% (sh1, P = 0.0003) and 16.2% (sh2, P = 0.0009) reduction in short-term proliferation was observed in MCF7 cells expressing either dox-inducible shRNA. No effect on proliferation was noted in MCF7shNS or ZR751shNS cells treated with dox (2 μg/ml) relative to parental control cells (Figure 4B). Importantly, decrease proliferation is associated with increased apoptosis as demonstrated by increased cleaved PARP expression upon shRNA-mediated silencing of CPT1A in either MCF7 or ZR751 cells; no effect was observed in shNS expressing cells (Figure 4C).
Figure 4.
CPT1A is essential for luminal cell line growth and survival. (A) CPT1A overexpression results in an ∼30.9% increase in MCF10A cell proliferation when assessed by MTS assay. (B) shRNA mediated silencing of CPT1A in ZR751 or MCF7 cells expressing one of two tet-inducible shRNA (2 μg/ml dox, 72 h) results in reduced cell proliferation (MTS, unpaired t-test); shNS (+dox) had no effect on cell growth. (C) Western blot analyses demonstrate increased cleaved PARP expression following dox treatment of MCF7 and ZR751 CPT1A sh1 or sh2 expressing cells (2 μg/ml dox, 72 h); no effect was observed in shNS expressing cells. An unpaired-test was used for each analysis: ***P < 0.0005; **P < 0.005; mean and standard error are indicated. (D–F) MCF7 or ZR751 cells demonstrate a dose and time-dependent (24–96 h) response to (D) etomoxir (0, 25, 50 or 75 μg/ml), (E) perhexiline (0, 2, 4 or 6 μg/ml) or (F) ranolazine (0, 15, 25 or 50 μg/ml). For each analysis, values were normalized to untreated (72 h) and are plotted as a percentage; each time point represents the mean of at last three independent replicates and the standard error is indicated. A Student’s t-test was used at each time point to calculate statistical significance. (G–I) Increased cleaved-PARP was observed in each cell line in response to (G) etomoxir (H) perhexiline or (I) ranolazine. (J) MCF7 or ZR751 cells show increased accumulation of lipid droplets indicative of reduced lipid oxidation in response to drug treatment.
FAO is a drug-able cellular process with a number of compounds that target CPT1A (etomoxir), CPT-family members (perhexiline) or 3-ketoacyl CoA thiolase (ranolazine) commercially available (11,15,17). The latter compounds have been approved internationally (perhexiline) or by the United States Food and Drug Administration (FDA) (ranolazine) for treatment of chest pain due to chronic angina pectoris and/or ischemia. Given that shRNA-mediated silencing of CPT1A results in significant apoptosis of HR+ breast cancer cell lines, our data suggest that pharmacological inhibition of CPT1A/FAO may represent a therapeutic opportunity with available compounds that could be repurposed to treat breast cancer. To test this hypothesis, we examined the impact of FAO inhibitors on luminal cell line activity to investigate the clinical applicability of CPT1A/FAO inhibition. MCF7 and ZR751 cells were treated with increasing doses of perhexiline (0, 2, 4 or 6 μg/ml), etomoxir (0, 25, 50 or 75 μg/ml) or ranolazine (0, 15, 25 or 50 μg/ml) and the effect on proliferation measured at 24, 48 and 72 ho by MTS assay. Focusing first on the CPT1A-specific inhibitor etomoxir, we determined that MCF7 cells showed a 29.9–61.1% (P < 0.0001) decrease in cell proliferation in a time- and dose-dependent manner (Figure 4D). Similarly, ZR751 cells showed up to a 51.7% reduction (P < 0.0001) in proliferative capacity at the highest doses and time points (50–75 μg/ml, 48–72 h; Figure 4D). Extending these studies to currently approved reagents showed similar results. Treatment of MCF7 or ZR751 cells with the internationally approved pan-CPT family inhibitor perhexiline resulted in a 70.4% (MCF7; P < 0.0001) and 78.4% (ZR751; P < 0.0001) decrease in proliferation at the highest dose (6 μg/ml, 72 h) (Figure 4E). Similarly, ranolazine treatment of MCF7 (27.3%, P < 0.0001) or ZR751 (26.7%, P < 0.0001) cells resulted in a significant decrease in viability (Figure 4F). Importantly, western blot analyses indicate that decreased cell growth corresponds with increased apoptosis as illustrated by increased cleaved PARP expression in each cell line in response to etomoxir (Figure 4G), perhexiline (Figure 4H) or ranolazine (Figure 4I). Oil Red O staining following treatment with each compound illustrated increased lipid droplet accumulation (Figure 4J), which is attributed to inhibition of FAO.
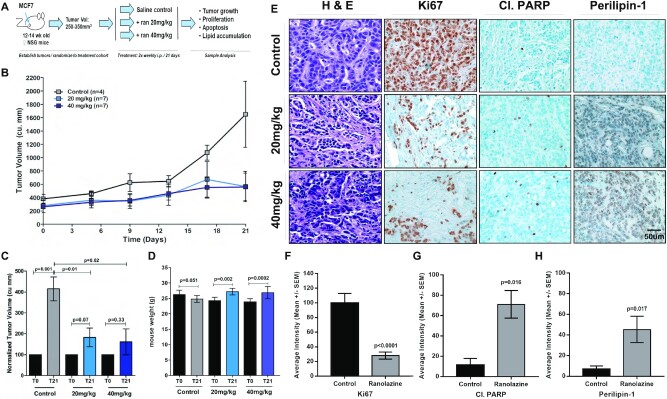
Ranolazine inhibits MCF7 xenograft tumor growth
Given in vitro effects of genetic or pharmacological inhibition of CPT1A or FAO, we next assessed the potential impact of FDA-approved ranolazine on tumor growth in vivo. MCF7 cells (3.5 × 106) were transplanted into the inguinal mammary gland of 12-week-old virgin nod scid gamma (NSG) mice. Each animal had a 17-β estradiol tablet (0.36 mg) implanted 48 h prior to transplant to facilitate estrogen-dependent tumor growth. Once the tumor reached an initial volume of 250–350 mm3, the mice were randomized to three treatment cohorts: saline control, 20 mg/kg ranolazine or 40 mg/kg ranolazine. Mice were treated twice weekly by intraperitoneal (i.p.) injection and tumor volume assessed for 21 days (Figure 5A). No differences in initial tumor volume between control (381.7 mm3) and 20 mg/kg (241.8 mm3, P = 0.24) or 40 mg/kg (262.4 mm3, P = 0.09) ranolazine-treated cohorts was noted (t-test) (Figure 5B). As expected, tumors in the control group increased 4.3-fold to 1650 mm3 by Day 21 (P = 0.001) (Figure 5B and C). Conversely, treatment with either the 20 mg/kg (P = 0.07) or 40 mg/kg dose (P = 0.33) of ranolazine showed no significant increase in tumor volume over this time course relative to Day 0 measurements (Figure 5B and C). Animals treated with the 20 mg/kg dose had a final tumor volume of 530.7 mm3 which corresponds to a 2.3-fold decrease relative to tumor volume of control saline treated animals (Figure 5B and C; P = 0.01). Likewise, animals treated with the 40 mg/kg ranolazine dose showed a 2.6-fold decrease in tumor volume relative to control animals with a final volume of 558.9 mm3 (Figure 5B and C; P = 0.02). Importantly, we determined that mice treated with either dose of ranolazine were generally healthy and showed no apparent side effects, including loss of body weight (Figure 5D); ranolazine treated mice showed a modest, yet statically significant (P < 0.002) increase in body weight during treatment while no change was observed in control mice. Finally, tumors were stained with hematoxylin and eosin (H&E) to assess tumor architecture while Ki67 and cleaved PARP IHC was used to examine differences in proliferation and apoptosis in a subset of tumors from each cohort. As illustrated in representative images in Figure 5E, control tumors were highly proliferative and demonstrated low levels of apoptosis as indicated by strong Ki67 and weak cleaved PARP staining whereas ranolazine-treated tumors showed high cleaved PARP and low Ki67 expression. Importantly, ranolazine treatment resulted in a significant increase in perilipin-1 staining which is indicative of increased lipid droplet accumulation and decreased β-oxidation compared to the control animals (Figure 5E). Quantification of the data demonstrate a 72% decrease (P < 0.0001, t-test) in Ki67 staining intensity (Figure 5F), as well as a 6.3-fold increase in cleaved PARP (Figure 5G; P = 0.016, t-test) and a 6.6-fold increase in perilipin-1 (Figure 5H; P = 0.017, t-test) staining intensity, respectively. Consistent with in vitro shRNA and pharmacological data, in vivo data indicate that ranolazine treatment reduced FAO, tumor growth and cell proliferation and promoted apoptosis.
Figure 5.
Ranolazine inhibits MCF7 xenograft tumor growth. (A) Schematic outlining experimental approach. (B) Treatment of MCF7 xenograft tumors twice weekly with either 20 or 40 mg/kg ranolazine resulted in a significant reduction in tumor volume after 21 days of treatment (lower dose: P = 0.01; higher dose: P = 0.02); mean tumor volume and standard error are indicated at each time point. (C) Tumor volume at Day 21 normalized to Day 0 for each sample demonstrates a significant (P = 0.001) increase in tumor volume in untreated samples (n = 4) and no significant change in tumor volume in mice treated with 20 mg/kg (P = 0.07; n = 7) or 40 mg/kg (P = 0.33, n = 7) ranolazine; both doses resulted in decreased tumor volume relative to untreated samples. (D) Increased body weight was observed after 21 days in mice treated with 20 mg/kg (P = 0.002) or 40 mg/kg (P = 0.0002) ranolazine; no change was observed in control animals (P = 0.051). (E) H&E staining of control or ranolazine treated tumors showed normal tumor architecture. Ki67 and cleaved PARP IHC staining demonstrate decreased cell proliferation and increased cell death while perilipin-1 staining indicates increased lipid droplet accumulation in drug treated tumors, respectively. (F–H) Quantification of IHC analyses indicates a significant reduction in (F) Ki67 staining, (G) increase cleaved PARP, (H) and increase in perilipin-1 staining following ranolazine treatment.
DISCUSSION
During the course of cellular transformation and tumorigenesis, tumor cells and other cells within the tumor microenvironment undergo metabolic reprogramming (11). These changes enable tumor cells to develop and maintain their malignant phenotype in both the primary and metastatic niche. While reprogramming of energy metabolism has been considered as one of the hallmarks of cancer, much of the focus has been on the Warburg effect (11,32,33). However, an increasing number of studies have begun to establish FAO as an essential process that contributes to transformation and tumorigenesis (12,14,15,34). CPT1A is the rate-limiting enzyme in FAO and mediates the import of long chain fatty acids through the outer mitochondrial membrane. DNA amplification and/or activation of CPT1A as a result of altered oncogenic signaling, including Myc-activation or K-Ras mutations, has been reported to promote tumorigenesis by promoting proliferation, survival, stemness, drug resistance and/or metastasis. Importantly, increased CPT1A expression or FAO is associated with poor clinical outcome and has been shown to be a potential therapeutic opportunity in several forms of cancer (13,35–49).
Amplification of chromosome 11q13-14 has been identified as a defining feature of high-risk ER+ breast cancers and has been shown to be strongly associated with increased proliferation in luminal breast cancer (18,19). However, the mechanism(s) by which this amplicon contributes to breast tumor development and/or progression remains unclear. In the current study, we demonstrate that CPT1A, which is localized to chromosome 11q13, is significantly overexpressed in highly proliferative luminal breast tumors compared to less proliferative luminal or basal-like tumors at both the mRNA and protein level. Further supporting these data, it was recently reported that lipid metabolism-related proteins, including CPT1A, are highly expressed in HER2+ and luminal B tumors when assessed by IHC (50). In the current study, within the context of luminal breast cancer, CPT1A mRNA expression corresponds with protein markers of proliferation and is associated with a worse clinical prognosis. Importantly, in vitro genetic manipulation of CPT1A resulted in altered mitochondrial respiration, cell proliferation, cell survival, colony formation and mammosphere formation. Given that CPT1A overexpression can promote and is required to maintain these phenotypes and that genetic silencing of CPT1A results in cell death, our data support the premise that aberrant CPT1A expression is oncogenic in this subset of tumors and that CPT1A and/or FAO may be essential for luminal breast cancer. Interestingly, while our previous study demonstrated that increased CPT1A mRNA expression is associated with copy number gains, our data also indicate that not all tumors that show increased mRNA expression are defined by CPT1A DNA amplification which may explain why CPT1A copy number status is not prognostic while increased CPT1A mRNA expression is associated with a worse outcome in ER+ tumors (18,19,51). These data further suggests that increased CPT1A expression may be due to various genomic or proteomic events, including ER activity which has been observed in ER+ cell lines (52) and/or altered oncogenic signaling, and that these events likely occur in a subtype-specific manner.
FAO inhibitors which have been developed for the treatment of chest pain due to chronic angina and/or ischemia are generally safe and well tolerated. Two predominant pharmacological targets have emerged to inhibit this pathway: CPT1A and 3-KAT which mediates the final step in FAO (10,15). Among the agents that have been approved and/or are in clinical trials are the CPT1 family inhibitor perhexiline, which has been approved by the European Medicine Administration (EMA) and the 3-KAT inhibitor ranolazine that has been approved by the EMA and FDA (14,15). In our studies, pharmacological-based experiments demonstrate that FAO inhibitors reduce cell proliferation, inhibit tumor growth, induce apoptosis and decrease lipid oxidation in luminal breast cancer cell lines and cell line-derived xenograft models. These results indicate that inhibition of CPT1A or FAO may represent a novel therapeutic target in specific subsets of ER+ breast tumor and that current drugs could be repurposed to address this need. It is important to note that our data (Figures 1A–C and Figure 2C) demonstrate that in addition to aggressive ER+ tumors, CPT1A is variably expressed in basal-like breast tumors and cell lines suggesting that some tumors or cell lines may be sensitive to FAO inhibition irrespective of molecular or clinical subtype. Consistent with this premise, studies by Camarda et al. have demonstrated that etomoxir mediated inhibition of FAO decreased energy metabolism and blocked tumor growth in a MYC-driven transgenic TNBC model (49). As such, we believe that the effects of FAO inhibitors may not be restricted to the ER+ breast cancer; however additional studies will be required to address this question and to identify potential predictive biomarkers. Collectively, the potential therapeutic implications of our work is consistent with previous studies that have demonstrated that inhibition of CPT1A or FAO in prostate, nasopharyngeal, breast or lung adenocarcinoma cancer models can sensitize cells to androgen blockade chemotherapy or radiotherapy, respectively (13,39,42–44).
Endocrine-based therapies are the standard-of-care treatment for patients with HR+ breast cancer (7). While advances in these therapies have improved clinical outcome and decreased adverse side effects, a significant percentage of patients will still acquire resistance leading to disease progression and death (3–5). More recent therapeutic strategies incorporating the mTOR inhibitor everolimus; CDK4/6 inhibitors palbociclib, ribociclib and abemaciclib; or the PI3K inhibitor alpelisib have led to improved clinical response and quality of life; however, these regimens are limited to patients with specific genomic alterations, and not all patients will respond equally (6,7). As such, there remains a critical need to incorporate novel therapies that can be used in parallel or sequentially with current treatments to improve the long-term prognosis and quality of life. Consistent with this argument, a number of studies have shown that etomoxir, perhexiline or ranolazine can sensitize prostate cancer cells, including enzalutamide resistant cells, to hormone-based therapies (43,44). Recent reports have indicated that invasive lobular breast cancer cell lines that have developed resistance to long-term estrogen depletion show increased expression of SREBP1 (a key regulator of fatty acid synthase) or CPT1A (53). These studies, in conjunction with our work, collectively raise the possibility that CPT1A/FAO may contribute to development of endocrine resistance and/or that FAO inhibitors could similarly enhance the response to endocrine therapies in ER+ breast carcinomas.
In addition to the primary role of CPT1A in mediating FAO, Pucci et al. have recently reported a potential epigenetic role for CPT1A in breast cancer oncogenesis. The authors demonstrated that CPT1A variant 2 which differs in 11 amino acids from CPT1A variant 1 at the C-terminus is specifically localized to the nucleus in breast cancer cells and regulates the expression of genes involved in apoptosis and invasion through interactions with HDAC1 (31). These results indicate that CPT1A contributes to cell survival by not only regulating FAO but by also stimulating histone acetylase activity in the nucleus. However, our analysis of CPT1A mRNA isoform expression from the TCGA data demonstrated that CPT1A variant 1 is the predominant isoform expressed in human breast tumors suggesting that FAO is the primary mechanism by which CPT1A contributes to breast tumorigenesis. Nonetheless, additional studies are required to delineate the isoform specific contribution of CPT1A in the regulation of breast oncogenesis.
In summary, our data establish an oncogenic role for CPT1A in luminal/ER+ breast cancer and provide a mechanism, albeit incomplete, by which amplification of 11q13-14 promotes tumor development and progression in these tumors. While additional studies will be required to elucidate the mechanisms by which CPT1A contributes to tumor development and progression, identify mechanism(s) that regulate CPT1A expression or determine the impact of CPT1A/FAO on development of resistance, our studies do indicate that CPT1A is an essential gene in highly aggressive luminal breast tumors and is required to promote cell proliferation, tumor growth and survival. Importantly, in vitro and in vivo genetic and pharmacological based studies provide preclinical evidence and rationale to support the further investigation of current and next generation FAO inhibitors for treatment of these patients.
ACKNOWLEDGEMENTS
We would like to thank the members of our laboratory and colleagues for their comments and suggests on this project and for critical review of the manuscript.
Authors Contributions: N.J., G.A.M. and M.L.G. conceived and designed the study. N.J., J.S.P. and M.L.G. performed computational analyses. N.J., G.A.M., V.B., S.H., K.A.P., N.Y. and J.G. contributed to experimental design, execution and analyses. N.J., G.A.M. and M.L.G. wrote the manuscript. All authors have reviewed and approved the final manuscript.
Contributor Information
Nidhi Jariwala, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Radiation Oncology, Robert Wood Johnson Medical School, New Brunswick, NJ, 08903, USA.
Gaurav A Mehta, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Radiation Oncology, Robert Wood Johnson Medical School, New Brunswick, NJ, 08903, USA.
Vrushank Bhatt, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Medicine, Robert Wood Johnson Medical School, New Brunswick, NJ, 08901, USA.
Shaimaa Hussein, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Radiation Oncology, Robert Wood Johnson Medical School, New Brunswick, NJ, 08903, USA.
Kimberly A Parker, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Radiation Oncology, Robert Wood Johnson Medical School, New Brunswick, NJ, 08903, USA.
Neha Yunus, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Radiation Oncology, Robert Wood Johnson Medical School, New Brunswick, NJ, 08903, USA.
Joel S Parker, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill NC, 27599, USA.
Jessie Yanxiang Guo, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Medicine, Robert Wood Johnson Medical School, New Brunswick, NJ, 08901, USA; Department of Chemical Biology, Rutgers Ernest Mario School of Pharmacy, Piscataway, NJ, 08901, USA.
Michael L Gatza, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, 08903, USA; Department of Radiation Oncology, Robert Wood Johnson Medical School, New Brunswick, NJ, 08903, USA.
FUNDING
National Cancer Institute, National Institutes of Health [R00-CA166228]; V Foundation for Cancer Research [V2016-013]; American Cancer Society [RSG-19-160-01-TBE]; New Jersey Health Foundation [PC-52-16 to M.L.G.]; New Jersey Commission for Cancer Research [DHFS-19PPC-019 to N.J.]; Cox Foundation for Cancer Research (to G.A.M.); National Institute of Health [R01 CA237347-01A1]; American Cancer Society [134036-RSG-19-165-01-TBG]; GO2 Foundation for Lung Cancer (to J.Y.G.); NJCCR [DCHS19PPC013]; Mistletoe Research Fellowship (to V.B.).; National Cancer Institute [P30-CA072720].
Conflict of interest statement. None declared.
REFERENCES
- 1. Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M., Jemal A., Kramer J.L., Siegel R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019; 69:363–385. [DOI] [PubMed] [Google Scholar]
- 2. Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. 2019; 321:288–300. [DOI] [PubMed] [Google Scholar]
- 3. Musgrove E.A., Sutherland Robert L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009; 9:631. [DOI] [PubMed] [Google Scholar]
- 4. Jeselsohn R., Buchwalter G., De Angelis C., Brown M., Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015; 12:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lumachi F., Brunello A., Maruzzo M., Basso U., Basso S.M. Treatment of estrogen receptor-positive breast cancer. Curr. Med. Chem. 2013; 20:596–604. [DOI] [PubMed] [Google Scholar]
- 6. Presti D., Quaquarini E. The PI3K/AKT/mTOR and CDK4/6 Pathways in Endocrine Resistant HR+/HER2- Metastatic Breast Cancer: Biological Mechanisms and New Treatments. Cancers. 2019; 11:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cardoso F., Senkus E., Costa A., Papadopoulos E., Aapro M., André F., Harbeck N., Aguilar Lopez B., Barrios C.H., Bergh J. et al. 4th ESO-ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 4)†. Ann. Oncol. 2018; 29:1634–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Narayan P., Prowell T.M., Gao J.J., Fernandes L.L., Li E., Jiang X., Qiu J., Fan J., Song P., Yu J. et al. FDA Approval Summary: Alpelisib plus fulvestrant for patients with HR-positive, HER2-negative, PIK3CA-mutated, advanced or metastatic breast cancer. Clin. Cancer Res. 2020; 27:1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yardley D.A., Noguchi S., Pritchard K.I., Burris H.A. 3rd, Baselga J., Gnant M., Hortobagyi G.N., Campone M., Pistilli B., Piccart M. et al. Everolimus plus exemestane in postmenopausal patients with HR(+) breast cancer: BOLERO-2 final progression-free survival analysis. Adv. Ther. 2013; 30:870–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carracedo A., Cantley L.C., Pandolfi P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer. 2013; 13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016; 23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butler L.M., Perone Y., Dehairs J., Lupien L.E., de Laat V., Talebi A., Loda M., Kinlaw W.B., Swinnen J.V. Lipids and cancer: emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug. Deliv. Rev. 2020; 159:245–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han S., Wei R., Zhang X., Jiang N., Fan M., Huang J.H., Xie B., Zhang L., Miao W., Butler A.C.-P. et al. CPT1A/2-mediated FAO enhancement—a metabolic target in radioresistant breast cancer. Front. Oncol. 2019; 9:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlaepfer I.R., Joshi M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology. 2020; 161:bqz046. [DOI] [PubMed] [Google Scholar]
- 15. Ma Y., Temkin S.M., Hawkridge A.M., Guo C., Wang W., Wang X.Y., Fang X. Fatty acid oxidation: an emerging facet of metabolic transformation in cancer. Cancer Lett. 2018; 435:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y.Y., Attané C., Milhas D., Dirat B., Dauvillier S., Guerard A., Gilhodes J., Lazar I., Alet N., Laurent V. et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017; 2:e87489–e87489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qu Q., Zeng F., Liu X., Wang Q.J., Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis. 2016; 7:e2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gatza M.L., Silva G.O., Parker J.S., Fan C., Perou C.M. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat. Genet. 2014; 46:1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012; 486:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., McLellan M., Yau C., Kandoth C. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015; 163:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mehta G.A., Parker J.S., Silva G.O., Hoadley K.A., Perou C.M., Gatza M.L. Amplification of SOX4 promotes PI3K/Akt signaling in human breast cancer. Breast Cancer Res. Treat. 2017; 162:439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009; 27:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nielsen T.O., Parker J.S., Leung S., Voduc D., Ebbert M., Vickery T., Davies S.R., Snider J., Stijleman I.J., Reed J. et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 2010; 16:5222–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P., Wang X., Qiao J.W., Cao S., Petralia F. et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016; 534:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoeflich K.P., O’Brien C., Boyd Z., Cavet G., Guerrero S., Jung K., Januario T., Savage H., Punnoose E., Truong T. et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. 2009; 15:4649–4664. [DOI] [PubMed] [Google Scholar]
- 26. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006; 38:500–501. [DOI] [PubMed] [Google Scholar]
- 27. Galligan J.T., Martinez-Noël G., Arndt V., Hayes S., Chittenden T.W., Harper J.W., Howley P.M. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J. Proteome Res. 2015; 14:953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crowe A.R., Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc. 2019; 9:e3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoadley K.A., Yau C., Wolf D.M., Cherniack A.D., Tamborero D., Ng S., Leiserson M.D., Niu B., McLellan M.D., Uzunangelov V. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014; 158:929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pucci S., Zonetti M.J., Fisco T., Polidoro C., Bocchinfuso G., Palleschi A., Novelli G., Spagnoli L.G., Mazzarelli P. Carnitine palmitoyl transferase-1A (CPT1A): a new tumor specific target in human breast cancer. Oncotarget. 2016; 7:19982–19996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zu X.L., Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem. Biophys. Res. Commun. 2004; 313:459–465. [DOI] [PubMed] [Google Scholar]
- 33. Warburg O. On respiratory impairment in cancer cells. Science. 1956; 124:269–270. [PubMed] [Google Scholar]
- 34. Currie E., Schulze A., Zechner R., Walther T.C., Farese R.V. Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013; 18:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi J., Fu H., Jia Z., He K., Fu L., Wang W. High expression of CPT1A predicts adverse outcomes: a potential therapeutic target for acute myeloid leukemia. EBioMedicine. 2016; 14:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cirillo A., Di Salle A., Petillo O., Melone M.A., Grimaldi G., Bellotti A., Torelli G., De' Santi M.S., Cantatore G., Marinelli A. et al. High grade glioblastoma is associated with aberrant expression of ZFP57, a protein involved in gene imprinting, and of CPT1A and CPT1C that regulate fatty acid metabolism. Cancer Biol. Ther. 2014; 15:735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du Q., Tan Z., Shi F., Tang M., Xie L., Zhao L., Li Y., Hu J., Zhou M., Bode A. et al. PGC1α/CEBPB/CPT1A axis promotes radiation resistance of nasopharyngeal carcinoma through activating fatty acid oxidation. Cancer Sci. 2019; 110:2050–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu J., Wu G., Song L., Cao L., Tan Z., Tang M., Li Z., Shi D., Zhang S., Li J. NKX2-8 deletion-induced reprogramming of fatty acid metabolism confers chemoresistance in epithelial ovarian cancer. EBioMedicine. 2019; 43:238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan Z., Xiao L., Tang M., Bai F., Li J., Li L., Shi F., Li N., Li Y., Du Q. et al. Targeting CPT1A-mediated fatty acid oxidation sensitizes nasopharyngeal carcinoma to radiation therapy. Theranostics. 2018; 8:2329–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flaig T.W., Salzmann-Sullivan M., Su L.-J., Zhang Z., Joshi M., Gijón M.A., Kim J., Arcaroli J.J., Van Bokhoven A., Lucia M.S. et al. Lipid catabolism inhibition sensitizes prostate cancer cells to antiandrogen blockade. Oncotarget. 2017; 8:56051–56065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sung G.J., Choi H.K., Kwak S., Song J.H., Ko H., Yoon H.G., Kang H.B., Choi K.C. Targeting CPT1A enhances metabolic therapy in human melanoma cells with the BRAF V600E mutation. Int. J. Biochem. Cell Biol. 2016; 81:76–81. [DOI] [PubMed] [Google Scholar]
- 42. Dheeraj A., Agarwal C., Schlaepfer I.R., Raben D., Singh R., Agarwal R., Deep G. A novel approach to target hypoxic cancer cells via combining β-oxidation inhibitor etomoxir with radiation. Hypoxia (Auckland, N.Z.). 2018; 6:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Flaig T.W., Salzmann-Sullivan M., Su L.J., Zhang Z., Joshi M., Gijon M.A., Kim J., Arcaroli J.J., Van Bokhoven A., Lucia M.S. et al. Lipid catabolism inhibition sensitizes prostate cancer cells to antiandrogen blockade. Oncotarget. 2017; 8:56051–56065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlaepfer I.R., Rider L., Rodrigues L.U., Gijón M.A., Pac C.T., Romero L., Cimic A., Sirintrapun S.J., Glodé L.M., Eckel R.H. et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol. Cancer Ther. 2014; 13:2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shi Z.Z., Liang J.W., Zhan T., Wang B.S., Lin D.C., Liu S.G., Hao J.J., Yang H., Zhang Y., Zhan Q.M. et al. Genomic alterations with impact on survival in esophageal squamous cell carcinoma identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2011; 50:518–526. [DOI] [PubMed] [Google Scholar]
- 46. Ricciardi M.R., Mirabilii S., Allegretti M., Licchetta R., Calarco A., Torrisi M.R., Foa R., Nicolai R., Peluso G., Tafuri A. Targeting the leukemia cell metabolism by the CPT1a inhibition: functional preclinical effects in leukemias. Blood. 2015; 126:1925–1929. [DOI] [PubMed] [Google Scholar]
- 47. Shao H., Mohamed E.M., Xu G.G., Waters M., Jing K., Ma Y., Zhang Y., Spiegel S., Idowu M.O., Fang X. Carnitine palmitoyltransferase 1A functions to repress FoxO transcription factors to allow cell cycle progression in ovarian cancer. Oncotarget. 2016; 7:3832–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Casciano J.C., Perry C., Cohen-Nowak A.J., Miller K.D., Vande Voorde J., Zhang Q., Chalmers S., Sandison M.E., Liu Q., Hedley A. et al. MYC regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br. J. Cancer. 2020; 122:868–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Camarda R., Zhou A.Y., Kohnz R.A., Balakrishnan S., Mahieu C., Anderton B., Eyob H., Kajimura S., Tward A., Krings G. et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 2016; 22:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim S., Lee Y., Koo J.S. Differential expression of lipid metabolism-related proteins in different breast cancer subtypes. PLoS One. 2015; 10:e0119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cancer Genome Atlas, N Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Monaco M.E. Fatty acid metabolism in breast cancer subtypes. Oncotarget. 2017; 8:29487–29500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Du T., Sikora M.J., Levine K.M., Tasdemir N., Riggins R.B., Wendell S.G., Van Houten B., Oesterreich S. Key regulators of lipid metabolism drive endocrine resistance in invasive lobular breast cancer. Breast Cancer Res. 2018; 20:106. [DOI] [PMC free article] [PubMed] [Google Scholar]