Abstract
Simple Summary
In this multicentre study of 1117 patients with stage I–IV non-squamous non-small cell lung carcinoma (NSCLC), we investigated associations between KRAS and clinical characteristics and survival. We investigated survival among the following groups of patients: those with no KRAS mutations (wild type) versus those with mutated tumours, those with KRAS wild type versus KRAS G12C versus KRAS non-G12C mutated tumours and among patients with different KRAS mutation subtypes. We also grouped KRAS mutated patients according to mutation preference for the Raf, PI3K/Akt and RalGDS/Ral intracellular signalling pathways and investigated whether there were differences in survival according to their preference for these pathways. We found that the proportion of KRAS mutated patients and frequency of KRAS mutation subtypes in our study is consistent with other studies of non-Asian patients with NSCLC. In multivariable analyses, we found no significant differences in the time to disease progression or overall survival within any of the analysed groups.
Abstract
Background: due to emerging therapeutics targeting KRAS G12C and previous reports with conflicting results regarding the prognostic impact of KRAS and KRAS G12C in non-small cell lung cancer (NSCLC), we aimed to investigate the frequency of KRAS mutations and their associations with clinical characteristics and outcome. Since mutation subtypes have different preferences for downstream pathways, we also aimed to investigate whether there were differences in outcome according to mutation preference for the Raf, PI3K/Akt, or RalGDS/Ral pathways. Methods: retrospectively, clinicopathological data from 1233 stage I–IV non-squamous NSCLC patients with known KRAS status were reviewed. KRAS’ associations with clinical characteristics were analysed. Progression free survival (PFS) and overall survival (OS) were assessed for the following groups: KRAS wild type (wt) versus mutated, KRAS wt versus KRAS G12C versus KRAS non-G12C, among KRAS mutation subtypes and among mutation subtypes grouped according to preference for downstream pathways. Results: a total of 1117 patients were included; 38% had KRAS mutated tumours, 17% had G12C. Among KRAS mutated, G12C was the most frequent mutation in former/current smokers (45%) and G12D in never smokers (46%). There were no significant differences in survival according to KRAS status, G12C status, among KRAS mutation subtypes or mutation preference for downstream pathways. Conclusion: KRAS status or KRAS mutation subtype did not have any significant influence on PFS or OS.
Keywords: non-small cell lung cancer, cohort study, survival, signalling pathway, KRAS, KRAS G12C
1. Introduction
Mutations of the v-Ki-ras2 Kirsten rat sarcoma viral oncogene homology gene (KRAS) gene are the most common oncogenic drivers of non-squamous non-small cell lung carcinoma (NSCLC) and occur in approximately 25–38% of non-Asian and 8–10% of Asian lung adenocarcinoma patients [1,2,3]. Although associated with smoking, KRAS mutations also occur in approximately 5–15% of never-smoking patients [1,4]. KRAS’ role as a prognostic factor has been investigated in numerous studies, but with conflicting results [5,6,7,8,9,10,11]. This may be attributed to heterogeneity of the study populations regarding sample size, disease stage, ethnicity, histological subtypes, study end points, and therapeutic history. Importantly, evaluation of the prognostic value is further complicated by the diverse and complex biological effects of mutated KRAS in signal transduction. Co-occurring genetic alterations in other genes have also been shown to have an impact on survival, exemplified by co-mutations in STK11 or KEAP1, which are associated with inferior survival compared to KRAS mutation only [2,12,13].
The KRAS gene encodes a small, cell-membrane bound guanosine triphosphate (GTP)ase, which is central in signal transduction through receptor tyrosine kinases via the Raf/Mek/Erk, PI3K/Akt, RalGDS-RalA/B and other signalling pathways. The Ras protein switches between an inactive guanosine diphosphate (GDP) bound and active GTP bound state [14]. The active state is promoted by a Ras guanine exchange factor, which enhances dissociation of GDP from Ras and Ras binding to GTP. GTPase activating protein (GAP) and the Ras protein’s intrinsic GTPase activity facilitate hydrolysis of GTP, returning Ras to its inactive GDP-bound state.
KRAS hot spot mutations are clustered on codon 12 and 13 in exon 2 and codon 61 in exon 3 [15]. The most common mutation subtypes in KRAS mutated lung adenocarcinoma are the codon 12 transversion mutations (substitution of a purine with a pyrimidine nucleotide, or opposite) G12C (39%) and G12V (18–21%), followed by the transition mutations (substitution of a purine by a purine, or a pyrimidine with a pyrimidine) G12D (14–18%) and G12A (10–11%) [1,2,16]. While KRAS transversion mutations are associated with a history of smoking, KRAS transition mutations are more common in never-smokers [1]. The oncogenic Ras proteins interfere with the GDP/GTP exchange and GTP hydrolysis, leaving the mutated Ras protein in a constitutively active GTP-bound state [17] with subsequent continuous activation of downstream pathways. The Ras oncoproteins may have some differences in affinity for downstream effector proteins. G12A, G12C, G13D, Q61L, and Q61H have been shown to have higher preference for Raf interaction [18]. G12C also has a high preference for RalGDS-RalA/B while G12D has been associated with preference for interaction with PI3K/Akt [19,20].
With the prospect of targeted treatment of patients with KRAS G12C mutated NSCLC, we retrospectively aimed to explore the frequency of KRAS mutations, clinical characteristics and the prognostic effects of KRAS in a cohort of patients diagnosed with non-squamous NSCLC disease stage I–IV in three university hospitals in Norway. We aimed to investigate potential differences in progression free survival (PFS) and overall survival (OS) in patients with KRAS wild type (wt) compared to those with KRAS mutated (mut) tumours, in patients with KRAS G12C compared to patients with KRAS wt and KRAS non-G12C mutations, and among KRAS mutation subtypes. Due to heterogeneity of biological effects of mutated Ras proteins, we also investigated whether survival was associated with KRAS mutation preference for interaction with either the PI3K/Akt, Raf- or RalGDS/Ral pathways.
2. Material and Methods
2.1. Ethics
This study was approved by the Regional Committees for Medical and Health Research Ethics (REC) in Eastern, Central, and Northern Norway (identification number 82144). The study also included patients enrolled in the regional research biobanks of Eastern, Central and Northern Norway. These research biobanks are approved by REC in Eastern, Central and Northern Norway, the Ministry of Health and Care Services and the Norwegian Data Protection Authority. Patients enrolled in the biobanks are over 18 years old and have given written informed consent.
2.2. Patient Inclusion and Tumour Specimens
Patients diagnosed with non-squamous non-small cell lung carcinoma (NSCLC) stage I–IV at St. Olav’s Hospital (STO, n = 676), the University Hospital of North Norway (UNN, n = 293) and Oslo University Hospital (OUH, n = 264) between 2003 and 2020 were evaluated for inclusion. Of these were 594 patients included in the regional biobanks (STO n = 266, UNN n = 64, OUH n = 264). Patients fulfilling all of the following criteria were included: (a) non-squamous (non-neuroendocrine), histology or immunophenotype; (b) known KRAS mutation status; (c) known mutational, rearrangement status of the Epidermal Growth Factor Receptor (EGFR)/Anaplastic Lymphoma Receptor Tyrosine Kinase (ALK)/ROS Proto-Oncogene 1, Receptor Tyrosine Kinase (ROS1) genes, or if status was unknown, treatment naïve to tyrosine kinase inhibitors (TKI); and (d) no non-pulmonary synchronous malignancy. Patients treated with curative intent for other malignancies and who were recurrence free ≥ 5 years before the lung cancer were also evaluated for the study. Patients with ≥2 pulmonary nodules at the time of diagnosis were excluded, unless the tumours were in the same lobe and of the same histology and same EGFR, KRAS, ALK, or ROS1 status. The following information was retrieved from the hospital medical records and pathology reports: age, sex, smoking history (current smoker, former smoker > 1 year prior to diagnosis or never smoker), Eastern Cooperative Oncology Group performance status (ECOG PS), pathological disease stage if surgical treatment, clinical disease stage if no surgery, extent of disease and metastatic sites at the time of diagnosis, results of molecular analyses, first line tumour treatment, history of second and later treatment lines, history of treatment with TKI or immune checkpoint inhibitors (ICI), date of first relapse, and date of death. All tumour specimens were reviewed and classified according to the fourth edition of the WHO Classification of Lung Tumours [21] by experienced lung pathologists (authors S.G.F.W., E.R., M.L.I.) in the respective pathology departments of the three hospitals. Clinical or pathologic restaging was performed according to the eighth edition of The New American Joint Committee on Cancer/International Union Against Cancer TNM stage classification for lung cancer [22].
Tumour specimens were analysed for KRAS mutations either by mutation specific real time PCR targeting codon 12 and 13 of exon 2 and codon 61 of exon 3 (OUH only) according to protocols implemented for routine diagnostics in the respective pathology departments at STO and OUH, or by next generation sequencing (NGS). NGS was performed using Illumina TruSight® Tumour 15 and TruSight® Tumour 26 (Illumina®, San Diego, CA, USA; used at UNN and STO, respectively) or QIAseqTM Comprehensive Targeted DNA Panel (Qiagen, Hilden, Germany; used at STO).
2.3. Statistics
The chi-square test for independence was used for comparison of categorical variables. PFS was defined as the time from the first diagnostic tissue specimen (biopsy or cytology) until objective progression or death by any cause. OS was defined as the time from the first diagnostic tissue specimen until death by any cause. Median follow-up time for PFS and OS was estimated using the reversed Kaplan–Meier method. Survival was estimated using the Kaplan–Meier method and compared using the log-rank test. The Cox proportional hazards model was used for univariable and multivariable analyses. The significance level was defined as a two-sided p < 0.05. All analyses were performed using the IBM SPSS Statistics for Windows version 27.0. (Armonk, NY, USA: IBM Corp.).
3. Results
3.1. Patient Characteristics
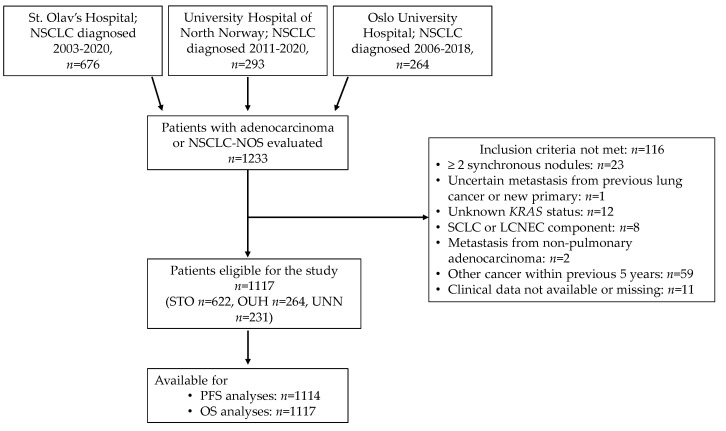
Of the 1233 patients evaluated for inclusion, 1117 were eligible for this study (Figure 1). Of these, 622 (55.7%) were diagnosed with NSCLC at STO, 264 (23.6%) at OUH and 231 (20.7%) at UNN.
Figure 1.
Outline of patient selection. Abbreviations: NSCLC, non-small cell lung carcinoma; NOS, not otherwise specified; STO, St. Olav’s Hospital; UNN, University Hospital of North Norway; OUH, Oslo University Hospital; SCLC, small cell lung carcinoma; LCNEC, large cell neuroendocrine carcinoma; PFS, progression free survival; OS, overall survival.
Patients’ characteristics according to KRAS mutation status are presented in Table 1. Median age was 69 (range 32–90) years, 592 (53%) were women, 950 (89%) were former or current smokers, 1063 (95%) were diagnosed with adenocarcinoma; ECOG PS was 0–1 in 1037 (93%) patients. The distribution of patients according to disease stage I-IV was 359 (32%), 148 (13%), 230 (21%), and 380 (34%), respectively.
Table 1.
Baseline patient characteristics according to KRAS status.
| Characteristic | Total | KRAS wt n = 697 |
KRAS mut n = 420 |
|||
|---|---|---|---|---|---|---|
| Age (years) | ||||||
| <50 | 46 | (4.1) | 29 | (4.2) | 17 | (4.0) |
| 50–60 | 169 | (15.1) | 104 | (14.9) | 65 | (15.5) |
| >60 | 902 | (80.8) | 564 | (80.9) | 338 | (80.5) |
| Hospital | ||||||
| STO | 622 | (55.7) | 391 | (56.1) | 231 | (55.0) |
| UNN | 231 | (20.7) | 144 | (20.7) | 87 | (20.7) |
| OUH | 264 | (23.6) | 162 | (23.2) | 102 | (24.3) |
| Sex | ||||||
| Female | 592 | (53.0) | 353 | (50.6) | 239 | (56.9) |
| Male | 525 | (47.0) | 344 | (49.4) | 181 | (43.1) |
| Smoking history | ||||||
| Never smoker | 126 | (11.3) | 113 | (16.2) | 13 | (3.1) |
| Former/current smoker | 991 | (88.7) | 584 | (83.8) | 407 | (96.9) |
| Ethnicity | ||||||
| Scandinavian/European | 1106 | (99.0) | 686 | (98.4) | 420 | (100.0) |
| African | 4 | (0.4) | 4 | (0.6) | 0 | (0.0) |
| Asian | 7 | (0.6) | 7 | (1.0) | 0 | (0.0) |
| ECOG PS | ||||||
| 0–1 | 1037 | (92.8) | 650 | (93.3) | 387 | (92.1) |
| 2 | 59 | (5.3) | 35 | (5.0) | 24 | (5.7) |
| 3–4 | 21 | (1.9) | 12 | (1.7) | 9 | (2.1) |
| Histology | ||||||
| Adenocarcinoma | 1063 | (95.2) | 650 | (93.3) | 413 | (98.3) |
| Adenosquamous carcinoma | 6 | (0.5) | 6 | (0.9) | 0 | (0.0) |
| Large cell carcinoma | 3 | (0.3) | 3 | (0.4) | 0 | (0.0) |
| NSCLC-NOS | 36 | (3.2) | 31 | (4.4) | 5 | (1.2) |
| MIA | 2 | (0.2) | 2 | (0.3) | 0 | (0.0) |
| Other non-squamous NSCLC | 7 | (0.6) | 5 | (0.7) | 2 | (0.5) |
| EGFR status | ||||||
| No | 948 | (84.9) | 528 | (75.8) | 420 | (100.0) |
| Yes | 142 | (12.7) | 142 | (20.4) | 0 | (0.0) |
| Not assessed | 27 | (2.4) | 27 | (3.9) | 0 | (0.0) |
| ALK rearrangement | ||||||
| No | 1011 | (90.5) | 620 | (89.0) | 391 | (93.1) |
| Yes | 12 | (1.1) | 12 | (1.7) | 0 | (0.0) |
| Not assessed | 94 | (8.4) | 65 | (9.3) | 29 | (6.9) |
| ROS1 rearrangement | ||||||
| No | 552 | (49.4) | 342 | (49.1) | 210 | (50.0) |
| Yes | 3 | (.3) | 3 | (0.4) | 0 | (0.0) |
| Not assessed | 562 | (50.3) | 352 | (50.5) | 210 | (50.0) |
| Disease stage | ||||||
| IA | 227 | (20.3) | 141 | (20.2) | 86 | (20.5) |
| IB | 132 | (11.8) | 77 | (11.0) | 55 | (13.1) |
| IIA | 40 | (3.6) | 19 | (2.7) | 21 | (5.0) |
| IIB | 108 | (9.7) | 75 | (10.8) | 33 | (7.9) |
| IIIA | 138 | (12.4) | 83 | (11.9) | 55 | (13.1) |
| IIIB | 67 | (6.0) | 42 | (6.0) | 25 | (6.0) |
| IIIC | 25 | (2.2) | 16 | (2.3) | 9 | (2.1) |
| IVA | 206 | (18.4) | 140 | (20.1) | 66 | (15.7) |
| IVB | 174 | (15.6) | 104 | (14.9) | 70 | (16.7) |
| Treatment intention | ||||||
| Curative | 671 | (60.1) | 414 | (59.4) | 257 | (61.2) |
| Palliative | 400 | (35.8) | 251 | (36.0) | 149 | (35.5) |
| No treatment | 46 | (4.1) | 32 | (4.6) | 14 | (3.3) |
Abbreviations: wt, wild type; mut, mutated; STO, St. Olav’s Hospital; UNN, University Hospital of North Norway; OUH, Oslo University Hospital; ECOG PS, Eastern Cooperative Oncology Group performance status; NSCLC-NOS, non-small cell lung carcinoma, not otherwise specified; MIA, minimal invasive adenocarcinoma.
Of the 1117 patients, 46 (4%) had no treatment due to comorbidities, 671 (60%) had potentially curative treatment and of these had 572 (85%) complete surgical resection. Of the 400 (36%) patients with advanced disease treated with palliative intention, the dominant first line treatments were platinum-doublet chemotherapy in 137 (34%) and radiochemotherapy in 91 (23%). Detailed overviews of curative and palliative treatments are presented in Supplementary Table S1. Of the 1117 patients included, 420 (38%) had KRAS mut tumours, 142 (13%) had EGFR mut tumours, 12 (1%) had ALK rearranged tumours, and 3 (0.3%) patients had ROS1 rearranged tumours. EGFR, ALK, and ROS1 analyses were not performed in 27, 94, and 562 of the patients, respectively, since these analyses were not routinely performed in Norway at the time of diagnosis. None of the patients with unknown EGFR/ALK/ROS1 status was treated with TKIs.
3.2. KRAS Mutation Status and Correlations with Clinical Characteristics
An overview of associations between patient characteristics and KRAS status is presented in Supplementary Table S2. Among patients with KRAS mutated tumours, 407 (97%) were current or former smokers and 13 (3%) were never smokers (p < 0.001). The proportion of women with KRAS mutated tumours was higher compared to men (57% versus 43%, respectively, p = 0.042). There were no associations between the presence of KRAS mutation and age, ECOG PS, disease stage, treatment history of surgery and number of metastatic sites at the time of diagnosis. At the time of diagnosis, the proportion of patients with pleural metastases, as either the only metastatic site or concurrent with other metastatic sites, was higher for patients with KRAS wt tumours (13.1%) than patients with KRAS mut tumours (6.5%, p < 0.001). No associations between KRAS status and metastases in other sites (adrenal gland, liver, skeleton and brain) were found. We found no associations between KRAS G12C, G12V or G12D and clinical characteristic.
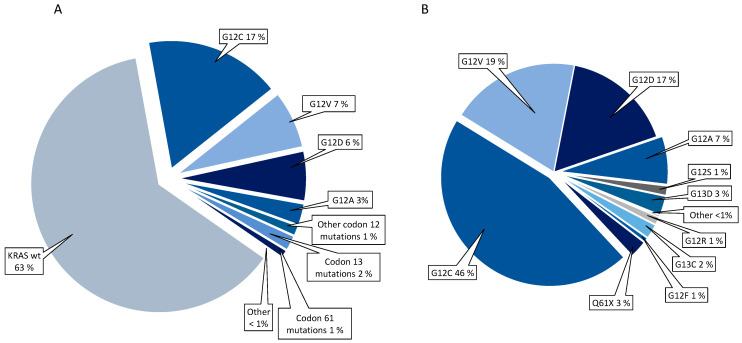
In the whole cohort of 1117 patients, 192 (17%) had KRAS G12C, 81 (7%) G12V, 70 (6%) G12D, and 30 (3%) G12A. The frequencies of KRAS mutation subtypes are presented in Figure 2.
Figure 2.
Frequencies of KRAS mutations in the whole cohort (A) and within the group of KRAS mutated patients (B).
Within the group of patients with KRAS mut tumours, G12C was the most frequent mutation in former/current smokers (45%), while G12D was more frequent in KRAS mut never smokers (46%; p = 0.016; Supplementary Table S2). Among patients with the three most common KRAS mutation subtypes, KRAS G12C, G12V, and G12D, there were no significant differences in distribution of mutation subtype according to sex, age, disease stages, surgical history, and the number of metastatic sites or metastatic site at time of diagnosis (Supplementary Table S2).
3.3. Mutation Status and Survival
Median follow-up for PFS was 52.7 (95%CI 44.3–61.2) months and for OS 52.7 (95%CI 45.7–59.6) months; 419 patients were progression-free, and 547 patients were alive at the time of data completion (April 2020). In the whole cohort, estimated median PFS was 17.2 (95%CI 13.6–20.7) months and estimated median OS 38.1 (95%CI 30.1–46.0) months.
3.3.1. Whole Cohort (Stage I–IV)
In the univariable analyses, neither KRAS status, G12C status (G12C versus KRAS wt versus KRAS non-G12C), nor KRAS mutation subtypes grouped according to signalling pathway preference, had effect on PFS or OS. Among KRAS mutation subtypes, G12C was associated with an effect on PFS (HR 0.62; 95%CI 0.38–1.00, p = 0.050) and OS (HR 0.59; 95%CI 0.035–0.99, p = 0.044) compared to G12A, but not to G12V or G12D. An overview of univariable analyses of relations between all covariates and survival is presented in Table 2.
Table 2.
Univariable analyses (Cox proportional hazards model) of patient and clinical characteristics and their associations with PFS and OS in the whole cohort.
| Variable | PFS | OS | ||||||
|---|---|---|---|---|---|---|---|---|
| n | HR | 95% CI | p | n | HR | 95% CI | p | |
| Age (years) | ||||||||
| <50 | 46 | 1 (ref) | 46 | 1 (ref) | ||||
| 50–60 | 166 | 1.07 | 0.71–1.60) | 0.762 | 169 | 1.49 | (0.90–2.45) | 0.118 |
| >60 | 902 | 1.04 | (0.72–1.51) | 0.818 | 902 | 1.68 | (1.06–2.67) | 0.027 |
| Sex | ||||||||
| Women | 590 | 1 (ref) | 592 | 1 (ref) | ||||
| Men | 524 | 1.10 | (0.94–1.27) | 0.233 | 525 | 1.13 | (0.96–1.34) | 0.137 |
| Smoking history | ||||||||
| Never | 126 | 1 (ref) | 126 | 1 (ref) | ||||
| Former/current | 988 | 1.48 | (1.15–1.91) | 0.002 | 991 | 1.59 | (1.19–2.12) | 0.002 |
| ECOG PS | ||||||||
| 0–1 | 1034 | 1 (ref) | 1037 | 1 (ref) | ||||
| 2 | 59 | 4.30 | (3.24–5.71) | <0.001 | 59 | 6.41 | (4.80–8.56) | <0.001 |
| 3–4 | 21 | 4.55 | (2.76–7.48) | <0.001 | 21 | 6.91 | (4.18–11.41) | <0.001 |
| Stage | ||||||||
| I | 359 | 1 (ref) | 359 | 1 (ref) | ||||
| II | 148 | 2.15 | (1.61–2.86) | <0.001 | 148 | 2.11 | (1.51–2.95) | <0.001 |
| III | 230 | 4.37 | (3.43–5.57) | <0.001 | 230 | 3.89 | (2.93–5.15) | <0.001 |
| IV | 377 | 12.74 | (10.01–16.09) | <0.001 | 380 | 12.46 | (9.61–16.14) | <0.001 |
| Surgery | ||||||||
| No | 534 | 1 (ref) | 534 | 1 (ref) | ||||
| Yes | 580 | 0.15 | (0.12–0.17) | <0.001 | 583 | 0.13 | (0.11–0.16) | <0.001 |
| Curative RT +/− CT first line | ||||||||
| No | 1023 | 1 (ref) | 1023 | 1 (ref) | ||||
| Yes | 91 | 1.28 | (0.99–1.64) | 0.060 | 91 | 1.05 | (0.78–1.43) | 0.740 |
| Palliative CT and/or RT first line | ||||||||
| No | 843 | 1 (ref) | 846 | 1 (ref) | ||||
| Yes | 271 | 5.75 | (4.85–6.81) | <0.001 | 271 | 5.54 | ((4.64–6.63)) | <0.001 |
| History of TKI (any line) | ||||||||
| No | 1025 | 1 (ref) | 1028 | 1 (ref) | ||||
| Yes | 89 | 1.56 | (1.22–1.99) | <0.001 | 89 | 1.32 | (0.86–2.03) | 0.198 |
| History of ICI (any line) | ||||||||
| No | 939 | 1 (ref) | 941 | 1 (ref) | ||||
| Yes | 175 | 2.19 | (1.81–2.64) | <0.001 | 176 | 1.39 | ((1.11–1.74)) | 0.006 |
| KRAS status | ||||||||
| Wild type | 695 | (1 (ref) | 697 | 1 (ref) | ||||
| Mutated | 419 | 1.04 | (0.89–1.21) | 0.664 | 420 | 1.01 | (0.85–1.19) | 0.940 |
| KRAS G12C status | ||||||||
| Wild type | 695 | 1 (ref) | 697 | 1 (ref) | ||||
| G12C | 192 | 0.91 | (0.74–1.12) | 0.379 | 193 | 0.91 | (0.72–1.14) | 0.414 |
| KRAS non-G12C | 227 | 1.15 | (0.96–1.39) | 0.132 | 227 | 1.10 | (0.89–1.35) | 0.383 |
| Raf vs. PI3K preference | ||||||||
| Raf | 245 | 1 (ref) | 245 | 1 (ref) | ||||
| PI3K | 69 | 1.11 | (0.79–1.55) | 0.554 | 70 | 1.09 | (0.75–1.58) | 0.649 |
| Ral A/B vs. PI3K preference | ||||||||
| RalA/B | 192 | 1 (ref) | 192 | 1 (ref) | ||||
| PI3K | 69 | 1.20 | (0.85–1.70) | 0.312 | 70 | 1.19 | (0.81–1.74) | 0.385 |
| Type KRAS mutation | ||||||||
| G12A | 30 | 1 (ref) | 30 | 1 (ref) | ||||
| G12C | 192 | 0.62 | (0.38–1.00) | 0.050 | 192 | 0.59 | (0.35–0.99) | 0.044 |
| G12V | 81 | 0.72 | (0.43–1.21) | 0.220 | 81 | 0.60 | (0.34–1.06) | 0.080 |
| G12D | 69 | 0.74 | (0.44–1.26) | 0.274 | 70 | 0.69 | (0.39–1.24) | 0.216 |
Abbreviations: PFS, progression free survival; OS, overall survival; HR, hazard ratio; ref reference; ECOG PS, Eastern Cooperative Oncology Group performance status; CT, chemotherapy; RT, radiotherapy; TKI, tyrosine kinase inhibitor; ICI, immune checkpoint inhibitor.
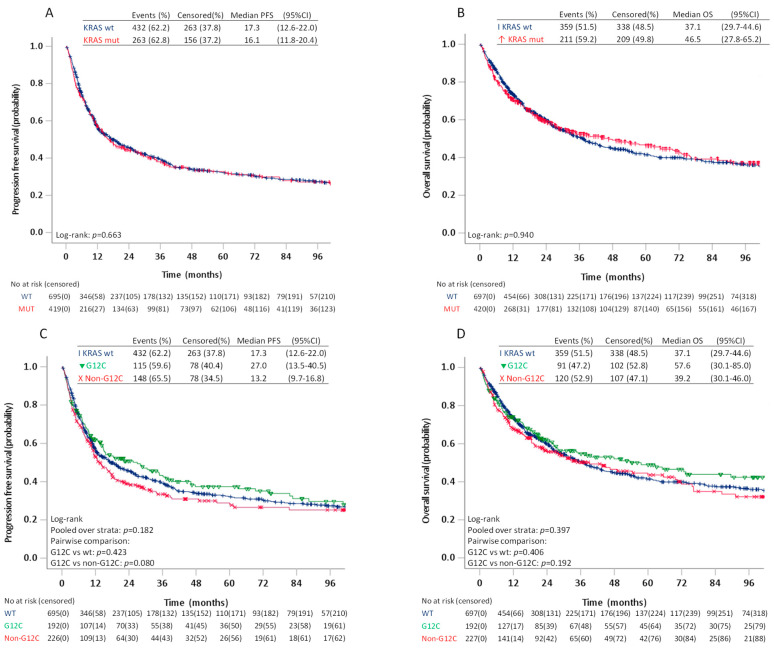
There were no differences in estimated median PFS or OS between patients with KRAS wt/KRAS mut and between patients with KRAS wt, KRAS G12C, or KRAS non-G12C, neither in the log-rank tests (Figure 3) or multivariable analyses adjusting for age, sex, smoking history, ECOG PS, disease stage and treatment type (Table 3).
Figure 3.
Progression free survival and overall survival in patients stage I–IV with KRAS wild type and KRAS mutated tumours (A,B) and patients with KRAS wild type, KRAS G12C, and KRAS non-G12C (C,D).
Table 3.
Multivariable analyses of progression free survival and overall survival in patients st. I–IV with KRAS wild type, KRAS mutated, KRAS G12C, and KRAS non-G12C mutated tumours.
| Variable | PFS | OS | PFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | p | HR | 95%CI | p | HR | 95%CI | p | HR | 95%CI | p | |
| Age (years) | 1.01 | (1.00–1.01) | 0.207 | 1.02 | (1.01–1.03) | <0.001 | 1.01 | (1.00–1.01) | 0.214 | 1.02 | (1.01–1.03) | <0.001 |
| Sex | ||||||||||||
| Women | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Men | 1.09 | (0.93–1.27) | 0.280 | 1.05 | (0.88–1.24) | 0.603 | 1.09 | (0.93–1.27) | 0.283 | 1.05 | (0.89–1.24) | 0.596 |
| Smoking history | ||||||||||||
| Never | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Former/current | 1.56 | (1.19–2.05) | <0.001 | 1.71 | (1.24–2.33) | 0.001 | 1.56 | (1.19–2.05) | 0.001 | 1.70 | (1.25–2.33) | 0.001 |
| ECOG PS | ||||||||||||
| 0–1 | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| 2 | 2.02 | (1.51–2.71) | <0.001 | 2.90 | (2.15–3.92) | <0.001 | 2.01 | (1.50–2.70) | <0.001 | 2.91 | (2.16–3.94) | <0.001 |
| 3–4 | 4.27 | (2.56–7.14) | <0.001 | 6.51 | (3.83–11.05) | <0.001 | 4.25 | (2.54–7.11) | <0.001 | 6.56 | (3.86–11.14) | <0.001 |
| Stage | ||||||||||||
| I | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| II | 2.11 | (1.58–2.81) | <0.001 | 2.09 | (1.49–2.92) | <0.001 | 2.11 | (1.58–2.82) | <0.001 | 2.08 | (1.49–2.92) | <0.001 |
| III | 3.02 | (2.28–3.99) | <0.001 | 2.67 | (1.92–3.71) | <0.001 | 3.02 | (2.28–3.99) | <0.001 | 2.67 | (1.92–3.72) | <0.001 |
| IV | 4.61 | (3.18–6.67) | <0.001 | 4.34 | (2.91–6.47) | <0.001 | 4.61 | (3.19–6.68) | <0.001 | 4.34 | (2.91–6.47) | <0.001 |
| Surgery | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 0.36 | (0.25–0.52) | <0.001 | 0.25 | (0.11–0.37) | <0.001 | 0.36 | (0.25–0.52) | <0.001 | 0.25 | (0.17–0.37) | <0.001 |
| Curative RT +/− CT first line | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 0.74 | (0.51–1.09) | 0.127 | 0.47 | (0.31–0.73) | <0.001 | 0.74 | (0.51–1.09) | 0.130 | 0.47 | (0.31–0.73) | 0.001 |
| Palliative CT and/or RT first line | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 1.49 | (1.19–1.88) | <0.001 | 1.16 | (0.90–1.48) | 0.250 | 1.49 | (1.18–1.87) | 0.001 | 1.16 | (0.91–1.48) | 0.236 |
| History of TKI (any line) | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 0.78 | (0.58–1.03) | 0.081 | 0.65 | (0.47–0.90) | 0.009 | 0.78 | (0.58–1.03) | 0.081 | 0.65 | (0.47–0.90) | 0.009 |
| KRAS status | ||||||||||||
| Wild type | 1 (ref) | 1 (ref) | - | - | - | - | - | - | ||||
| Mutated | 0.98 | (0.83–1.15) | 0.801 | 0.96 | (0.80–1.15) | 0.678 | - | - | - | - | - | - |
| KRAS G12C status | ||||||||||||
| Wild type | - | - | - | - | - | - | 1 (ref) | 1 (ref) | ||||
| G12C | - | - | - | - | - | - | 0.96 | (0.77–1.19) | 0.691 | 1.00 | (0.79–1.28) | 0.972 |
| KRAS non-G12C | - | - | - | - | - | - | 1.00 | (0.82–1.21) | 0.977 | 0.93 | (0.75–1.16) | 0.530 |
Abbreviations: PFS, progression free survival; OS, overall survival; ref reference; ECOG PS, Eastern Cooperative Oncology Group performance status; CT, chemotherapy; RT, radiotherapy; TKI, tyrosine kinase inhibitor; ICI, immune checkpoint inhibitor.
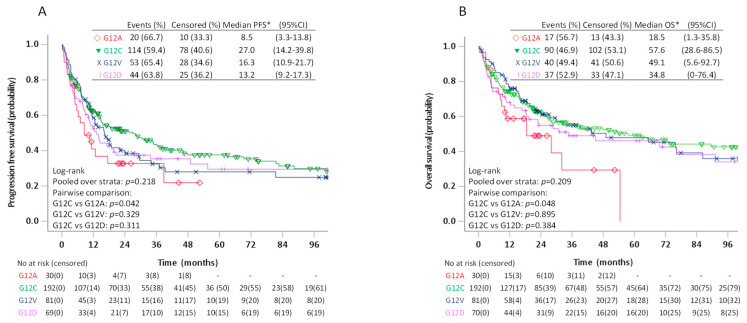
Due to a trend towards better PFS for patients with G12C compared to patients with non-G12C KRAS mutations on pairwise log-rank test (p = 0.080), we further compared survival among patients with the four most frequent KRAS mutations (Figure 4). In these analyses, the estimated median PFS for G12C was 27.0 (95%CI 14.2–39.8) months compared to 16.3 (95%CI 10.9–21.7) months for G12V, 13.2 (95%CI 9.2–17.3) for G12D and 8.5 (95%CI 3.3–13.8) months for G12A (p = 0.218). The pairwise log-rank test showed significantly better PFS for patients with G12C compared to G12A (p = 0.042), but not G12V (p = 0.329) or G12D (p = 0.311). The estimated median OS was 57.6 (95%CI 28.6–86.5) months for G12C, 49.1 (95%CI 5.6–92.7) months for G12V, 34.8 (95%CI 0–76.4) months for G12D and 18.5 (95%CI 1.3–35.8) months for G12A. On pairwise comparison, patients with G12C also had better OS compared to patients with G12A (p = 0.048), but not G12V (p = 0.895) or G12D (p = 0.384). The differences in PFS or OS between patients with G12C and G12A, however, did not remain statistically significant in multivariable analyses adjusting for age, sex, smoking history, ECOG PS, and treatment type (Table 4).
Figure 4.
Progression free survival (A) and overall survival (B) in patients stage I–IV with KRAS G12C, G12V, G12D, and G12A mutated tumours.
Table 4.
Multivariable analyses of progression free survival and overall survival in patients stage I–IV with KRAS G12C, G12V, G12D, and G12A.
| Variable | PFS | OS | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | p | HR | 95%CI | p | |
| Age at time of diagnosis | 1.00 | (0.99–1.02) | 0.820 | 1.01 | (0.99–1.03) | 0.142 |
| Sex | ||||||
| Women | 1 (ref) | 1/ref) | ||||
| Men | 1.16 | (0.89–1.52) | 0.283 | 1.01 | (0.75–1.37) | 0.940 |
| Smoking history | ||||||
| Never smoker | 1 (ref) | 1 (ref) | ||||
| Former/current smoker | 0.93 | (0.40–2.14) | 0.858 | 0.77 | (0.31–1.94) | 0.582 |
| ECOG PS | ||||||
| 0–1 | 1 (ref) | 1 (ref) | ||||
| 2 | 1.99 | (1.16–3.40) | 0.012 | 2.66 | (1.53–4.61) | 0.001 |
| 3–4 | 3.82 | (1.64–8.88) | 0.002 | 6.76 | (2.86–16.00) | 0.000 |
| Stage | ||||||
| I | 1 (ref) | 1 (ref) | ||||
| II | 1.66 | (1.01–2.74) | 0.047 | 1.45 | (0.80–2.60) | 0.218 |
| III | 2.69 | (1.66–4.35) | 0.000 | 2.02 | (1.13–3.60) | 0.018 |
| IV | 2.49 | (1.29–4.82) | 0.007 | 2.06 | (0.99–4.29) | 0.054 |
| Surgery | ||||||
| No | 1 (ref) | 1 (ref) | ||||
| Yes | 0.24 | (0.13–0.46) | 0.000 | 0.17 | (0.08–0.34) | 0.000 |
| Curative RT +/− CT first line | ||||||
| No | 1 (ref) | 1 (ref) | ||||
| Yes | 0.53 | (0.27–1.03) | 0.062 | 0.26 | (0.11–0.59) | 0.001 |
| Palliative CT and/or RT first line | ||||||
| No | 1 (ref) | 1 (ref) | ||||
| Yes | 1.53 | (1.01–2.33) | 0.046 | 1.26 | (0.80–1.99) | 0.314 |
| KRAS mutation | ||||||
| G12A | 1 (ref) | 1 (ref) | ||||
| G12C | 0.76 | (0.45–1.26) | 0.281 | 0.97 | (0.54–1.73) | 0.907 |
| G12V | 0.79 | (0.46–1.37) | 0.399 | 0.83 | (0.44–1.56) | 0.561 |
| G12D | 0.76 | (0.44–1.33) | 0.333 | 0.87 | (0.46–1.64) | 0.659 |
Abbreviations: PFS, progression free survival; OS, overall survival; ref, reference; ECOG PS, Eastern Cooperative Oncology Group performance status; CT, chemotherapy; RT, radiotherapy.
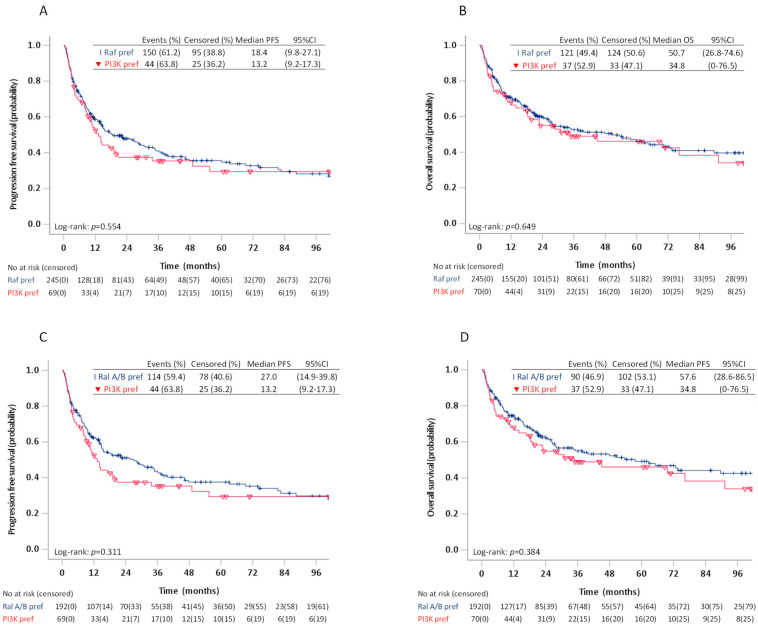
We then investigated PFS and OS in patients with mutation preference for the Raf pathway (G12C, G12A, G13D, Q61L, Q61H) versus patients with G12D favouring PI3K/Akt, and for patients with G12D favouring PI3K/Akt versus G12C favouring the Ral A/B pathway. We found no differences in estimated median PFS or OS between any of these groups, neither in the log-rank tests (Figure 5) or multivariable analyses (Table 5).
Figure 5.
Progression free survival and overall survival in patients stage I–IV with KRAS mutants favouring the Raf pathway (G12C, G12A, G13D, Q61L/H) versus patients with KRAS G12D favouring PI3K/Akt (A,B) and patients with KRAS G12C favouring the RalA/B pathway versus patients with KRAS G12D favouring PI3K/Akt (C,D).
Table 5.
Multivariable analyses of progression free survival and overall survival in stage I–IV patients with KRAS mutations favouring PI3K/Akt (G12D), Raf (G12C, G12A, G13D, Q61L/H) and Ral (G12C).
| Variable | PFS | OS | PFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | p | HR | 95%CI | p | HR | 95%CI | p | HR | 95%CI | p | |
| Age (years) | 1.02 | (1.00–1.03) | 0.070 | 1.02 | (1.00–1.04) | 0.027 | 1.02 | (1.00–1.04) | 0.085 | 1.01 | (0.99–1.03) | 0.200 |
| Sex | ||||||||||||
| Women | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Men | 1.00 | (0.75–1.35) | 0.981 | 0.88 | (0.63–1.22) | 0.431 | 0.90 | (0.64–1.25) | 0.529 | 0.69 | (0.47–1.01) | 0.054 |
| Smoking history | ||||||||||||
| Never | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Former/current | 1.02 | (0.36–2.88) | 0.975 | 0.93 | (0.28–3.09) | 0.911 | 1.31 | (0.40–4.33) | 0.653 | 1.34 | (0.31–5.67) | 0.695 |
| ECOG PS | ||||||||||||
| 0–1 | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| 2 | 2.23 | (1.32–3.76) | 0.003 | 3.03 | (1.79–5.14) | <0.001 | 2.77 | (1.46–5.23) | 0.002 | 4.89 | (2.49–9.58) | <0.001 |
| 3–4 | 6.68 | (2.89–15.5) | <0.001 | 9.21 | (3.85–22.02) | <0.001 | 6.02 | (2.41–14.99) | <0.001 | 10.95 | (4.18–28.68) | <0.001 |
| Stage | ||||||||||||
| I | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| II | 1.55 | (0.88–2.71) | 0.128 | 1.52 | (0.79–2.94) | 0.212 | 1.35 | (0.71–2.58) | 0.363 | 1.03 | (0.47–2.26) | 0.946 |
| III | 2.61 | (1.49–4.58) | 0.001 | 2.71 | (1.45–5.06) | 0.002 | 3.21 | (1.77–5.85) | <0.001 | 2.61 | (1.30–5.25) | 0.007 |
| IV | 2.74 | (1.36–5.52) | 0.005 | 3.08 | (1.50–6.32) | 0.002 | 3.24 | (1.46–7.19) | 0.004 | 1.94 | (0.80–4.71) | 0.143 |
| Surgery | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 0.234 | (0.11–0.48) | <0.001 | 0.22 | (0.1–0.48) | <0.001 | 0.28 | (0.13–0.62) | 0.002 | 0.16 | (0.07–0.39) | <0.001 |
| Curative RT +/− CT first line | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 0.617 | (0.30–1.27) | 0.189 | 0.35 | (0.14–0.83) | 0.017 | 0.57 | (0.26–1.28) | 0.173 | 0.22 | (0.08–0.58) | 0.002 |
| Palliative CT and/or RT first line | ||||||||||||
| No | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) | ||||||||
| Yes | 1.56 | (1.00–2.44) | 0.052 | 1.27 | (0.77–2.09) | 0.350 | 1.15 | (0.94–2.48) | 0.089 | 1.32 | (0.78–2.24) | 0.302 |
| PI3K vs. Raf | ||||||||||||
| Raf | 1 (ref) | 1 (ref) | - | - | - | - | - | - | ||||
| PI3K | 0.903 | (0.64–1.28) | 0.570 | 0.89 | (0.60–1.30) | 0.530 | - | - | - | - | - | - |
| PI3K vs. RalA/B | ||||||||||||
| RalA/B | - | - | - | - | - | - | 1 (ref) | 1 (ref) | ||||
| PI3K | - | - | - | - | - | - | 0.94 | (0.65–1.35) | 0.720 | 0.86 | (0.58–1.29) | 0.474 |
Abbreviations: PFS, progression free survival; OS, overall survival; ref, reference; ECOG PS, Eastern Cooperative Oncology Group performance status; CT, chemotherapy; RT, radiotherapy.
3.3.2. Curative Surgery
There were no differences in PFS or OS between patients with KRAS mut/KRAS wt or KRAS wt/KRAS G12C/ KRAS non-G12C (Supplementary Figures S1 and S2 and Supplementary Table S3). We then compared survival among patients with G12C, G12V, G12D and G12A (Supplementary Figure S3). Patients with G12C had longer estimated median PFS than G12V (88.4 versus 29.7 months, respectively), but was not estimated for G12D and G12A since the estimated survival probability did not reach 50% for these groups. The difference in estimated median PFS was significant for G12C compared to G12V in pairwise log-rank test (p = 0.037) but did not remain significant in multivariable analyses adjusting for sex, age, smoking history, treatment and disease stage (p = 0.180, Supplementary Table S4). There were no significant differences in OS between patients with G12C, G12V, and G12D, but median OS was only estimated for G12V (Supplementary Figure S3 and Supplementary Table S4).
3.3.3. Non-Curative Treatment
No differences in the estimated median PFS or OS were observed between patients with KRAS wt/KRAS mut (Supplementary Figure S4). In the KRAS wt/KRAS G12C/KRAS non-G12C subgroups (Supplementary Figure S5), KRAS wt patients had better OS compared to KRAS G12C (9.6 versus 6.7 months) in the pairwise log-rank test (p = 0.047), but this association did not remain significant in multivariable analysis adjusting for sex, age, smoking history, ECOG PS, history of chemotherapy, history of TKI, history of ICI, and disease stage (Supplementary Table S5). There were no differences in PFS or OS between patients with G12C, G12V, G12D, and G12A; neither in the overall or pairwise log-rank tests or in multivariable analyses (Supplementary Figure S6 and Supplementary Table S5).
4. Discussion
In this retrospective multicentre study of 1117 patients with non-squamous NSCLC, we describe associations between KRAS status and various clinicopathological characteristics and survival. The presence of KRAS mutation was significantly associated with a history of smoking, with G12C being the most frequent mutation in former and current smokers and G12D the most common mutation in never smokers. We also found a significantly higher proportion of women with KRAS mut tumours compared to men. The associations with survival were investigated according to KRAS status (KRAS wt versus KRAS mut), KRAS G12 status (KRAS wt versus KRAS G12C versus KRAS non-G12C mutations) and KRAS mutation type (G12C, G12V, G12D, and G12A). We found no associations with survival for any of the compared groups in the multivariable analyses, in the analyses of the whole cohort, separately for resected patients with curative disease, or for patients with advanced disease. Furthermore, we found no associations with survival in subgroup analyses of KRAS mut patients grouped, according to mutation preference for interaction with the PI3K/Akt, Raf- or Ral pathways.
Our study is one of the largest studies on the prognostic effect of KRAS in non-squamous NSCLC in all disease stages. Since many studies on the prognostic value of KRAS in NSCLC have focused on patients with either local or advanced disease, we also performed isolated subgroup analyses of patients with resected curative disease and advanced disease, in addition to the analyses of the whole cohort. Furthermore, to our knowledge we present the first study of patients with NSCLC where specific KRAS mutations and their preference for signalling pathways have been taken into consideration in survival analyses.
Regarding survival in patients with KRAS wt and KRAS mut tumours, our results are consistent with other studies [5,6,7,8,9,23]. Moreover, we found no significant differences in PFS or OS between patients with KRAS wt, KRAS G12C, and KRAS non-G12C mutated tumours or among patients with G12C, G12V, G12D, and G12A mutated tumours, which agrees with other studies [2,13,16,24,25,26,27]. However, worse survival in patients with KRAS mutated tumours (as one group) compared to KRAS wt, as well as in patients with KRAS G12C compared to patients with KRAS non-G12C mutations, have also been reported [2,10,11,28].
The conflicting results on KRAS as a prognostic factor may be attributed to several factors, including differences of the study populations as mentioned in the introduction. Cross-study comparison is also challenging due to differences in follow-up, definitions of endpoints and variability in covariates adjusted for in multivariable analyses.
Investigations of the prognostic impact of KRAS mutations may further be complicated by the diverse biological effects of the mutated Ras proteins. In addition to different preferences for signalling pathways, studies on cell lines have also shown that different K-Ras oncoprotein subtypes also have phenotypical biochemical differences in terms of GTP affinity, the ability to speed up the GDP to GTP exchange and the ability to reduce the speed of intrinsic and GAP mediated hydrolysis [18,29]. Hence, grouping patients according to pathway preference for survival analyses is a simplified approach.
Evaluation of the prognostic value of KRAS is also complicated by co-occurring mutations in other genes. The presence of concurrent mutations in STK11 and KEAP1 have been reported at frequencies of 12–29% and 8–27%, respectively, and have been associated with worse recurrence free -or overall survival compared to KRAS mutation only [2,12,13]. Concurrent genomic alterations of KEAP1 and CDKN2A are also associated with reduced T-cell inflammation and low levels of PD-L1 expression, predictive of reduced response to immune checkpoint inhibitors [12,30,31,32,33]. KRAS mutated tumours with co-occurring mutations in the TP53 gene (reported frequency of 39–42%), on the other hand, are associated with active inflammation, high expression of PD-L1 and increased response to ICI [12,13,30,33,34]. The study by Scheffler et al. [33] also indicates that co-occurring mutations in specific genes may be associated with specific KRAS mutation subtypes.
Other less studied mechanisms, including mutant allele specific imbalance (MASI) and expression patterns, may also have effects on survival. Villaruz et al. showed that high levels of KRAS mutated alleles compared to KRAS wild type alleles was associated with significant worse PFS [24]. Nagy et al. combined gene expression data in patients with KRAS mut adenocarcinomas and generated a gene expression signature based on the five strongest genes expressed secondary to KRAS mutation [35]. Patients with high gene signature expressions had significantly shorter OS compared to the KRAS mutated patients with low expression.
Taken together, the traditional approaches to evaluate the prognostic value of mutated KRAS in mixed groups comprising patients with different mutation subtypes with different biological properties, may be too narrow. It is an increasing understanding that KRAS mutated NSCLCs are genetically heterogenous diseases. Hence, the complex biological diversity of KRAS mutated NSCLC should be taken into consideration when exploring associations with clinicopathological characteristics and outcome
There are some limitations to our study. These include the retrospective nature of the study. In our subgroup analyses of mutation preference for signalling pathways, we included G12C, G12A, G13D, and Q61L/H in the group “favouring Raf”, since these mutations were found to have high affinity for Raf in the study by Hunter et al. [18]. However, when considering the lower intrinsic hydrolysis rates for KRAS Q61L and G12A compared to G12C and G13D in this study, KRAS Q61L and G12A were predicted to be stronger activators of Raf. Due to the low number of patients with KRAS G12A and Q61L mutations in our study, we also included G12C and G12D in the group “favouring Raf”.
Another limitation is that we did not perform any additional molecular analyses to explore differences in expression of the main targets of the mutant Ras proteins. In a recent study of patients with KRAS G12C mutated colon cancer, it was shown that comprehensive analyses of gene expression profiles, co-occurring alterations of other genes and protein expression might shed light on the involvement of signalling pathways [36].
We also wanted to explore whether KRAS was of predictive value in patients with local disease treated with stereotactic body radiation therapy or conventional radiotherapy 60–66 Gy, but the number of these patients in our cohort was too small for analyses. Comprehensive next generation sequencing was only performed for a small subset of the patients. Hence, we did not have sufficient molecular data on co-occurring mutations in other genes, including STK11, KEAP1, CDKN2A, and TP53, for exploration of clinicopathological associations, associations with specific KRAS mutation subtypes, the prognostic value of concurrent genetic alterations or the predictive value with respect to ICI therapy. It would also be of interest to explore associations between KRAS mutation subtypes and tumour expression of programmed death ligand 1 (PD-L1), but we did not collect information on PD-L1 expression.
Although KRAS was the first oncogene to be associated with NSCLC [37], designing targeted therapies targeting the mutated K-Ras proteins has proven to be challenging due to the complex biology of the oncogenic Ras proteins and their high affinity for GTP. However, the development of G12C inhibitors which irreversibly bind to cysteine in the mutant G12C, locking the protein in an inactive GDP-bound state, have shown promising results in recent clinical phase I and II trials [38,39]. G12C inhibitors may improve the treatment options for a substantial proportion of patients with non-squamous NSCLC, with subsequent increased interest in the prognostic value of KRAS G12C. However, recent reports suggest a diversity of molecular alterations and mechanisms conferring adaption and resistance to G12C inhibitors [40,41]. Comprehensive molecular testing beyond KRAS mutation subtype may therefore be warranted before and during treatment with G12C inhibitors to identify possible alterations conferring resistance.
5. Conclusions
In this multicentre study of patients with non-squamous NSCLC, we found no differences in PFS or OS between patients with KRAS mutated and KRAS wild type NSCLC, between patients with KRAS wild type, G12C and KRAS non-G12 mutations, or among KRAS mutation subtypes. Furthermore, we found no differences in survival among patients grouped according to their mutation’s preference for either Raf, PI3K/Akt, or Ral pathways.
Acknowledgments
The authors thank all patients who agreed to be included in the biobanks, the Norwegian Cancer Society, and Northern Norway Health Region Authority.
Supplementary Materials
Supplementary Materials are available online at https://www.mdpi.com/article/10.3390/cancers13174294/s1.
Author Contributions
S.G.F.W.: conceptualisation, methodology, formal analysis, investigation, resources, writing—original draft, visualisation, project administration. H.Y.D.: investigation, resources, writing—review and editing. E.F.E.: investigation, resources, writing—review and editing. T.B.: investigation, resources, writing–review and editing. T.O.H.: investigation, resources, writing—review and editing. A.L.O.: resources, writing—review and editing. M.L.-I.: investigation, resources, writing—review and editing O.T.B.: conceptualisation, investigation, resources, writing—review and editing. D.F.: investigation, resources, writing—review and editing. E.-E.P.: conceptualisation, investigation, resources, writing—review and editing. T.D.: conceptualisation, investigation, resources, writing—review and editing. S.A.: conceptualisation, investigation, resources, writing—review and editing. B.H.G.: conceptualisation, resources, methodology, writing—review and editing, funding acquisition. E.R.: conceptualisation, methodology, formal analysis, investigation, resources, writing—original draft, visualisation, supervision, project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Liaison Committee for Education, Research and Innovation in Central Norway (reference number 2016/29014 and the Cancer Fund at St. Olav’s Hospital (reference number 2020/14-301/2020).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Regional Committees for Medical and Health Research Ethics (REC) in Eastern, Central, and Northern Norway (REC identification number 82144).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available within the manuscript and the supplementary material.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dogan S., Shen R., Ang D.C., Johnson M.L., D’Angelo S.P., Paik P.K., Brzostowski E.B., Riely G.J., Kris M.G., Zakowski M.F., et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: Higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin. Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El Osta B., Behera M., Kim S., Berry L.D., Sica G., Pillai R.N., Owonikoko T.K., Kris M.G., Johnson B.E., Kwiatkowski D.J., et al. Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2019;14:876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohno T., Nakaoku T., Tsuta K., Tsuchihara K., Matsumoto S., Yoh K., Goto K. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl. Lung Cancer Res. 2015;4:156–164. doi: 10.3978/j.issn.2218-6751.2014.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finn S.P., Addeo A., Dafni U., Thunnissen E., Bubendorf L., Madsen L.B., Biernat W., Verbeken E., Hernandez-Losa J., Marchetti A., et al. Prognostic Impact of KRAS G12C Mutation in Patients With NSCLC: Results From the European Thoracic Oncology Platform Lungscape Project. J. Thorac. Oncol. 2021;16:990–1002. doi: 10.1016/j.jtho.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 5.D’Angelo S.P., Janjigian Y.Y., Ahye N., Riely G.J., Chaft J.E., Sima C.S., Shen R., Zheng J., Dycoco J., Kris M.G., et al. Distinct Clinical Course of EGFR-Mutant Resected Lung Cancers: Results of Testing of 1118 Surgical Specimens and Effects of Adjuvant Gefitinib and Erlotinib. J. Thorac. Oncol. 2012;7:1815–1822. doi: 10.1097/JTO.0b013e31826bb7b2. [DOI] [PubMed] [Google Scholar]
- 6.Shepherd F.A., Domerg C., Hainaut P., Jänne P.A., Pignon J.-P., Graziano S., Douillard J.-Y., Brambilla E., Le Chevalier T., Seymour L., et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rulli E., Marabese M., Torri V., Farina G., Veronese S., Bettini A., Longo F., Moscetti L., Ganzinelli M., Lauricella C., et al. Value of KRAS as prognostic or predictive marker in NSCLC: Results from the TAILOR trial. Ann. Oncol. 2015;26:2079–2084. doi: 10.1093/annonc/mdv318. [DOI] [PubMed] [Google Scholar]
- 8.Zer A., Ding K., Lee S.M., Goss G.D., Seymour L., Ellis P.M., Hackshaw A., Bradbury P.A., Han L., O’Callaghan C.J., et al. Pooled Analysis of the Prognostic and Predictive Value of KRAS Mutation Status and Mutation Subtype in Patients with Non Small Cell Lung Cancer Treated with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2016;11:312–323. doi: 10.1016/j.jtho.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Shepherd F.A., Lacas B., Le Teuff G., Hainaut P., Jänne P.A., Pignon J.-P., Le Chevalier T., Seymour L., Douillard J.-Y., Graziano S., et al. Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J. Clin. Oncol. 2017;35:2018–2027. doi: 10.1200/JCO.2016.71.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izar B., Zhou H., Heist R.S., Azzoli C.G., Muzikansky A., Scribner E.E.F., Bernardo L.A., Dias-Santagata D., Iafrate A.J., Lanuti M. The Prognostic Impact of KRAS, Its Codon and Amino Acid Specific Mutations, on Survival in Resected Stage I Lung Adenocarcinoma. J. Thorac. Oncol. 2014;9:1363–1369. doi: 10.1097/JTO.0000000000000266. [DOI] [PubMed] [Google Scholar]
- 11.Kadota K., Sima C.S., Arcila M.E., Hedvat C., Kris M.G., Jones D.R., Adusumilli P.S., Travis W.D. KRAS Mutation Is a Significant Prognostic Factor in Early-stage Lung Adenocarcinoma. Am. J. Surg. Pathol. 2016;40:1579–1590. doi: 10.1097/PAS.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbour K.C., Jordan E., Kim H.R., Dienstag J., Yu H.A., Sanchez-Vega F., Lito P., Berger M., Solit D.B., Hellmann M., et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018;24:334. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aredo J.V., Padda S.K., Kunder C.A., Han S.S., Neal J.W., Shrager J.B., Wakelee H.A. Impact of KRAS mutation subtype and concurrent pathogenic mutations on non-small cell lung cancer outcomes. Lung Cancer. 2019;133:144–150. doi: 10.1016/j.lungcan.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vetter I.R., Wittinghofer A. The Guanine Nucleotide-Binding Switch in Three Dimensions. Science. 2001;294:1299. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 15.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2018;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebastian M., Eberhardt W.E.E., Hoffknecht P., Metzenmacher M., Wehler T., Kokowski K., Alt J., Schütte W., Büttner R., Heukamp L.C., et al. KRAS G12C-mutated advanced non-small cell lung cancer: A real-world cohort from the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315) Lung Cancer. 2021;154:51–61. doi: 10.1016/j.lungcan.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Scheffzek K., Ahmadian M.R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 18.Hunter J.C., Manandhar A., Carrasco M.A., Gurbani D., Gondi S., Westover K.D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015;13:1325. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 19.Céspedes M.V., Sancho F.J., Guerrero S., Parreño M., Casanova I., Pavón M.A., Marcuello E., Trias M., Cascante M., Capellà G., et al. K-ras Asp12 mutant neither interacts with Raf, nor signals through Erk and is less tumorigenic than K-ras Val12. Carcinogenesis. 2006;27:2190–2200. doi: 10.1093/carcin/bgl063. [DOI] [PubMed] [Google Scholar]
- 20.Ihle N.T., Byers L.A., Kim E.S., Saintigny P., Lee J.J., Blumenschein G.R., Tsao A., Liu S., Larsen J.E., Wang J., et al. Effect of KRAS Oncogene Substitutions on Protein Behavior: Implications for Signaling and Clinical Outcome. JNCI J. Natl. Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travis W.D., Brambilla E., Burke A.P., Marx A., Nicholson A.G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. WHO Classification of Tumours. 4th ed. Volume 7 International Agency for Research on Cancer; Lyon, France: 2015. [Google Scholar]
- 22.Detterbeck F.C., Boffa D.J., Kim A.W., Tanoue L.T. The Eighth Edition Lung Cancer Stage Classification. Chest. 2017;151:193–203. doi: 10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 23.La Fleur L., Falk-Sörqvist E., Smeds P., Berglund A., Sundström M., Mattsson J.S.M., Brandén E., Koyi H., Isaksson J., Brunnström H., et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50–58. doi: 10.1016/j.lungcan.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Villaruz L.C., Socinski M.A., Cunningham D.E., Chiosea S.I., Burns T.F., Siegfried J.M., Dacic S. The prognostic and predictive value of KRAS oncogene substitutions in lung adenocarcinoma. Cancer. 2013;119:2268–2274. doi: 10.1002/cncr.28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H.A., Sima C.S., Shen R., Kass S., Gainor J., Shaw A., Hames M., Iams W., Aston J., Lovly C.M., et al. Prognostic Impact of KRAS Mutation Subtypes in 677 Patients with Metastatic Lung Adenocarcinomas. J. Thorac. Oncol. 2015;10:431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui W., Franchini F., Alexander M., Officer A., Wong H.-L., Ijzerman M., Desai J., Solomon B.J. Real world outcomes in KRAS G12C mutation positive non-small cell lung cancer. Lung Cancer. 2020;146:310–317. doi: 10.1016/j.lungcan.2020.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Ruppert A.-M., Beau-Faller M., Debieuvre D., Ouafik L.H., Westeel V., Rouquette I., Mazières J., Bringuier P.-P., Monnet I., Escande F., et al. Outcomes of advanced Non-Small Cell Lung Cancer patients From the IFCT Biomarkers France study by KRAS Mutation Subtypes. JTO Clin. Res. Rep. 2020:100052. doi: 10.1016/j.jtocrr.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadal E., Chen G., Prensner J.R., Shiratsuchi H., Sam C., Zhao L., Kalemkerian G.P., Brenner D., Lin J., Reddy R.M., et al. KRAS-G12C Mutation Is Associated with Poor Outcome in Surgically Resected Lung Adenocarcinoma. J. Thorac. Oncol. 2014;9:1513–1522. doi: 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 29.Stolze B., Reinhart S., Bulllinger L., Fröhling S., Scholl C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci. Rep. 2015;5:8535. doi: 10.1038/srep08535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoulidis F., Byers L.A., Diao L., Papadimitrakopoulou V.A., Tong P., Izzo J., Behrens C., Kadara H., Parra E.R., Canales J.R., et al. Co-occurring Genomic Alterations Define Major Subsets of KRAS Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discov. 2015;5:860. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama S., Akbay E.A., Li Y.Y., Aref A.R., Skoulidis F., Herter-Sprie G.S., Buczkowski K.A., Liu Y., Awad M.M., Denning W.L., et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Res. 2016;76:999. doi: 10.1158/0008-5472.CAN-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skoulidis F., Goldberg M.E., Greenawalt D.M., Hellmann M.D., Awad M.M., Gainor J.F., Schrock A.B., Hartmaier R.J., Trabucco S.E., Gay L., et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheffler M., Ihle M.A., Hein R., Merkelbach-Bruse S., Scheel A.H., Siemanowski J., Brägelmann J., Kron A., Abedpour N., Ueckeroth F., et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019;14:606–616. doi: 10.1016/j.jtho.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y., Tu H.Y., Chen H.J., Sun Y.L., Zhou Q., et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 35.Nagy Á., Pongor L.S., Szabó A., Santarpia M., Győrffy B. KRAS driven expression signature has prognostic power superior to mutation status in non-small cell lung cancer. Int. J. Cancer. 2017;140:930–937. doi: 10.1002/ijc.30509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry J.T., Coker O., Chowdhury S., Shen J.P., Morris V.K., Dasari A., Raghav K., Nusrat M., Kee B., Parseghian C., et al. Comprehensive Clinical and Molecular Characterization of KRASG12C-Mutant Colorectal Cancer. JCO Precis. Oncol. 2021:613–621. doi: 10.1200/PO.20.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos E., Martin-Zanca D., Reddy E.P., Pierotti M.A., Della Porta G., Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 38.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skoulidis F., Li B.T., Dy G.K., Price T.J., Falchook G.S., Wolf J., Italiano A., Schuler M., Borghaei H., Barlesi F., et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue J.Y., Zhao Y., Aronowitz J., Mai T.T., Vides A., Qeriqi B., Kim D., Li C., de Stanchina E., Mazutis L., et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awad M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W., Johnson M.L., Heist R.S., Patil T., Riely G.J., et al. Acquired Resistance to KRASG12C Inhibition in Cancer. N. Engl. J. Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available within the manuscript and the supplementary material.