ABSTRACT
The emergence of the CRISPR/Cas system as a technology has transformed our ability to modify nucleic acids, and the CRISPR/Cas13 system has been used to target RNA. CasRx is a small type VI-D effector (Cas13d) with RNA knockdown efficiency that may have an interference effect on RNA viruses. However, the RNA virus-targeting activity of CasRx still needs to be verified in vivo in vertebrates. In this study, we successfully engineered a highly effective CasRx system for fish virus interference. We designed synthetic mRNA coding for CasRx and used CRISPR RNAs to guide it to target the red-spotted grouper nervous necrosis virus (RGNNV). This technique resulted in significant interference with virus infections both in vitro and in vivo. These results indicate that CRISPR/CasRx can be used to engineer interference against RNA viruses in fish, which provides a potential novel mechanism for RNA-guided immunity against other RNA viruses in vertebrates.
IMPORTANCE RNA viruses are important viral pathogens infecting vertebrates and mammals. RNA virus populations are highly dynamic due to short generation times, large population sizes, and high mutation frequencies. Therefore, it is difficult to find widely effective ways to inhibit RNA viruses, and we urgently need to develop effective antiviral methods. CasRx is a small type VI-D effector (Cas13d) with RNA knockdown efficiency that can have an interference effect on RNA viruses. Nervous necrosis virus (NNV), a nonenveloped positive-strand RNA virus, is one of the most serious viral pathogens, infecting more than 40 cultured fish species and resulting in huge economic losses worldwide. Here, we establish a novel effective CasRx system for RNA virus interference using NNV and grouper (Epinephelus coioides) as a model. Our data showed that CasRx was most robust for RNA virus interference applications in fish, and we demonstrate its suitability for studying key questions related to virus biology.
KEYWORDS: CRISPR/CasRx, RNA virus, virus interference, virus resistance, in vivo
INTRODUCTION
In recent years, numerous genetic modification tools have been developed. The CRISPR/Cas system offers sequence-specific DNA-editing methods to correct mutant genes. CRISPR/Cas systems are divided into two main classes, i.e., class I, in which multieffector complexes mediate the interference, and class II, which employs single, multidomain effectors to mediate the interference (1). These classes are further subdivided into 6 types and 33 subtypes according to the genomic architecture of the CRISPR array and the signature interference effector. The class II CRISPR/Cas systems include types II, V, and VI. Types II and V consist of endonucleases that operate at the DNA level, whereas type VI systems exclusively target RNA molecules (1, 2).
Class II type VI CRISPR/Cas systems are RNA-guided and RNA-targeting machineries that provide prokaryotes with immunity against RNA (3). All type VI CRISPR/Cas systems have a single effector protein known as the Cas13 effector (formerly called the C2c2 CRISPR/Cas system) (3). CRISPR/LshCas13a from the bacterium Leptotrichia shahii was the first Cas13 orthologue to be harnessed for programmable RNA-targeting activities (3). Since then, only a few studies have shown that LshCas13a mediates specific RNA virus targeting in plants (4–6), and several studies have identified more variants of Cas13 proteins belonging to different Cas13 families, which have been classified into four type VI subtypes (subtypes A to D) (7–9). Additionally, researchers recently identified a new Cas13 subtype called CRISPR/Cas13d (CasRx), which shows minimal sequence identity with previous Cas13 effectors (9, 10).
Compared to other Cas13 effectors, CasRx is more efficient and has more robust activation during RNA-guided RNA cleavage in mammalian cells in vitro (9, 10). It also has the smallest size and exhibits high targeting specificity and efficiency, making it ideal for in vivo therapeutic applications (10). In plants, CasRx systems have been reported to have a remarkable ability to inactivate or silence RNA viruses (11). Thus, CasRx systems could be a new paradigm for treating viral infections, and the need is great, considering recent outbreaks of Nipah virus, Zika virus, Ebola virus, and coronavirus disease 2019 (COVID-19), which are all RNA viruses (12–14). Recently, RNA-targeting Cas13a and Cas13b were shown to have antiviral activity in cell lines infected with single-stranded RNA viruses (15). However, application of CasRx systems to vertebrate RNA viruses has not been studied to date.
Nervous necrosis virus (NNV) is a nonenveloped positive-strand RNA virus classified in the family Nodaviridae (16, 17). According to the International Committee on Taxonomy of Viruses, NNV is classified into four genotypes, namely, striped jack NNV, tiger puffer NNV, barfin flounder NNV, and red-spotted grouper NNV (RGNNV) (18). NNVs are about 25 to 30 nm in diameter, icosahedral, and nonenveloped and have a bipartite positive-sense RNA genome. RNA1 encodes the RNA-dependent RNA polymerase (RdRp) and nonstructural protein B. RNA2 encodes the capsid protein (CP) (19). NNV infection has a mortality rate of more than 90% in several cultured marine fish species in the larval and juvenile stages (20). Recently, a number of NNV isolates were identified from different species of fishes, including groupers, gilthead sea bream, Asian sea bass, and tilapia, and NNV as a causative agent of viral nervous necrosis disease has become one of the most important threats to cultured marine and freshwater fish worldwide (17, 21). RGNNV mainly destroys the brain tissue of grouper, and it has resulted in huge losses in the populations of groupers and other marine fish, as well as economic losses to aquaculture (22–24). Here, we chose grouper (Epinephelus spp.) and RGNNV as models to study CasRx interference with vertebrate RNA viruses, and we investigated the feasibility of using the CRISPR/CasRx system for this purpose.
RESULTS
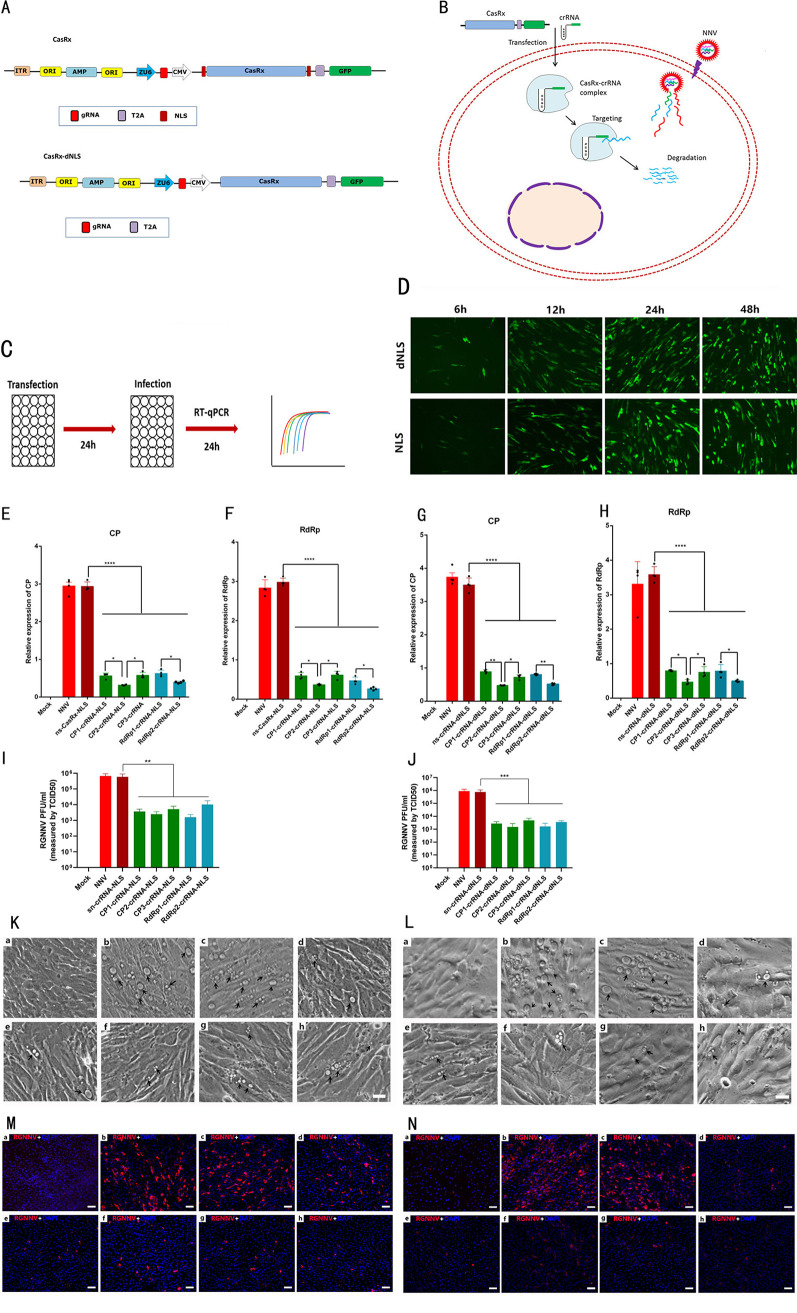
Construction of the CasRx/crRNA system and assessment of its effects on RGNNV infection.
For efficient RNA targeting in fish, we optimized the CasRx-CRISPR RNA (crRNA) system (Fig. 1A and B). First, we optimized codons encoding CasRx to improve its protein content. Second, we used the zebrafish U6 promoter to improve crRNA expression. Third, we engineered CasRx with or without the nuclear localization sequence (NLS) to exclude the cellular location discrepancies.
FIG 1.
In vitro RGNNV targeting via the CasRx system. (A) Schematic representation of different CasRx protein variants used in this study. ZU6, zebrafish U6 promoter. (B) Schematic representation of the mechanism of RGNNV RNA cleavage by CasRx. (C) Schematic representation of the experimental set-up to test RGNNV inhibition in GS cells in panels E to H. (D) Expression kinetics of CasRx in GS cells. GS cells were transfected with 1 μg of CasRx plasmid. Cells were fixed at 6, 12, 24, and 48 h. (E and F) The CasRx-NLS system prevented RGNNV infection when transfected with CasRx-NLS and crRNAs targeting the RGNNV CP gene (CP1, CP2, and CP3) (E) and RdRp gene (RdRp1 and RdRp2) (F), Data are expressed as mean ± SD. *, P < 0.05; ****, P < 0.0001. (G and H) The CasRx-dNLS system prevented RGNNV infection when transfected with CasRx-dNLS and crRNAs targeting the RGNNV CP gene (CP1, CP2, and CP3) (G) and RdRp gene (RdRp1 and RdRp2) (H). Data are expressed as mean ± SD. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. (I) RGNNV PFU per milliliter measured as TCID50 with transfection of targeting or nontargeting crRNAs and CasRx-NLS encoding plasmids in GS cells. (J) RGNNV PFU per milliliter measured as TCID50 with transfection of targeting or nontargeting crRNAs and CasRx-dNLS-encoding plasmids in GS cells (n = 3). Data are expressed as mean ± SD. **, P < 0.01; ***, P < 0.001. (K) Cell morphology and CPEs (vacuolation) after infection with RGNNV, with or without the CasRx-NLS system. a, Mock; b, Lipofectamine 3000 plus RGNNV; c, CasRx-NLS plus ns-crRNA plus RGNNV; d, CasRx-NLS plus CP1-crRNA plus RGNNV; e, CasRx-NLS plus CP2-crRNA plus RGNNV; f, CasRx-NLS plus CP3-crRNA plus RGNNV; g, CasRx-NLS plus RdRp1-crRNA plus RGNNV; h, CasRx-NLS plus RdRp1-crRNA plus RGNNV. Arrows indicate the vacuoles induced by RGNNV infection. Scale bar = 20 μm. (L) Cell morphology and CPEs (vacuolation) after infection with RGNNV, with or without the CasRx-dNLS system. a, Mock; b, Lipofectamine 3000 plus RGNNV; c, CasRx-dNLS plus ns-crRNA plus RGNNV; d, CasRx-dNLS plus CP1-crRNA plus RGNNV; e, CasRx-dNLS plus CP2-crRNA plus RGNNV; f, CasRx-dNLS plus CP3-crRNA plus RGNNV; g, CasRx-dNLS plus RdRp1-crRNA plus RGNNV; h, CasRx-dNLS plus RdRp1-crRNA plus RGNNV. Arrows indicate the vacuoles induced by RGNNV infection. Scale bar = 20 μm. (M) Immunofluorescence staining of RGNNV after infection with RGNNV, with or without the CasRx-NLS system. a, Mock; b, Lipofectamine 3000 plus RGNNV; c, CasRx-NLS plus ns-crRNA plus RGNNV; d, CasRx-NLS plus CP1-crRNA plus RGNNV; e, CasRx-NLS plus CP2-crRNA plus RGNNV; f, CasRx-NLS plus CP3-crRNA plus RGNNV; g, CasRx-NLS plus RdRp1-crRNA plus RGNNV; h, CasRx-NLS plus RdRp1-crRNA plus RGNNV. Red indicates the RGNNV CP protein. Scale bar = 50 μm. (N) The immunofluorescence staining of RGNNV after infection with RGNNV, with or without the CasRx-dNLS system. a, Mock; b, Lipofectamine 3000 plus RGNNV; c, CasRx-dNLS plus ns-crRNA plus RGNNV; d, CasRx-dNLS plus CP1-crRNA plus RGNNV; e, CasRx-dNLS plus CP2-crRNA plus RGNNV; f, CasRx-dNLS plus CP3-crRNA plus RGNNV; g, CasRx-dNLS plus RdRp1-crRNA plus RGNNV; h, CasRx-dNLS plus RdRp1-crRNA plus RGNNV. Red indicate the RGNNV CP protein. Scale bar = 50 μm.
RGNNV is a positive-sense, single-stranded RNA genome virus composed of CP and RdRp. Therefore, we tested whether CasRx could target the CP and RdRp of RGNNV to mediate efficient and robust RNA virus interference. We designed and prepared three crRNAs to target the coding sequences of CP mRNA (CP1, CP2, and CP3) and two crRNAs to target the coding sequence of RdRp mRNA (RdRp1 and RdRp2).
To assess the effects of the CasRx/crRNA system in cellular models of RGNNV infection (Fig. 1A and B), plasmids encoding CasRx and each crRNA were transfected into grouper spleen (GS) cells using the transfection agent Lipofectamine 3000 (Fig. 1C). CasRx expression was observed as early as 6 h posttransfection. Its expression increased at 12 and 24 h but did not increase further from 24 to 48 h (Fig. 1D). GS cells were simultaneously transfected with plasmids encoding CasRx-dNLS or CasRx-NLS and each of the five crRNAs followed by RGNNV infection (Fig. 1C). All 10 combinations reduced viral RNA copy numbers (Fig. 1E to H). Furthermore, the virus titer results showed that, when cells were transfected with either CasRx-dNLS or CasRx-NLS and each crRNA, viral titers were significantly reduced; RGNNV infectivity was reduced >300-fold for the CP2 and RdRp2 crRNAs tested and >200-fold for the CP1, CP3, and RdRp1 crRNAs tested (Fig. 1I and J).
Previous studies reported that cell vacuolation is a remarkable cytopathic effect (CPE) that occurs after RGNNV infection (25, 26). When GS cells were exposed to only RGNNV or to CasRx plus nonspecific (ns)-crRNA and RGNNV, extensive CPEs were observed. However, when cells were transfected with either CasRx-dNLS or CasRx-NLS and each crRNA, few CPEs were detected (Fig. 1K and L). In addition, the immunofluorescence assay results demonstrated that, when GS cells were exposed to only RGNNV or to CasRx plus ns-crRNA and RGNNV, extensive fluorescence signals of the RGNNV CP protein were observed. However, when cells were transfected with either CasRx-dNLS or CasRx-NLS and each crRNA (Fig. 1M and N), positive fluorescence signals were significantly decreased. These results indicated that the CasRx system was able to efficiently and functionally interfere with RGNNV infections in vitro.
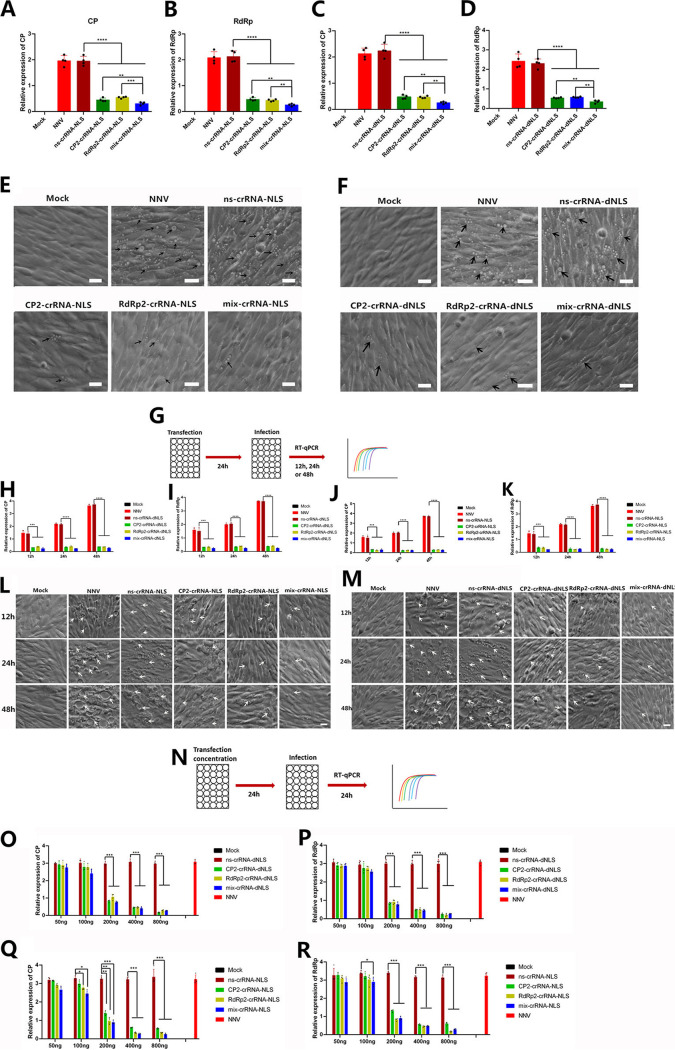
Effects of crRNA combinations, time, and concentrations on CasRx interference with RGNNV.
To characterize the performance of the CasRx/crRNA system, we tested the effects of combinations of different crRNAs, as well as exposure time and concentration, on RGNNV infection. In the previous experiment, we found that CP2-crRNA and RdRp2-crRNA were most efficient among the five crRNAs tested; therefore, we combined them (mix-crRNA). Targeting either CP2 or RdRp2 (i.e., CP2-crRNA or RdRp2-crRNA alone) reduced viral copies, but the mix-crRNA further reduced viral RNA copies for both the CasRx-dNLS and CasRx-NLS groups (Fig. 2A to D). Similarly, compared to transfection with CP2-crRNA or RdRp2-crRNA alone, few CPEs appeared when cells were transfected with mix-crRNA for both CasRx-dNLS and CasRx-NLS plasmids (Fig. 2E and F).
FIG 2.
CasRx preventing viral infection. (A) Expression of CP after transfection with CasRx-NLS and single-target CP2 (CP2-crRNA) or RdRp2 (RdRp2-crRNA) crRNAs or combined-target CP2 and RdRp2 crRNA (mix-crRNA). (B) Expression of RdRp after transfection with CasRx-NLS and CP2-crRNA, RdRp2-crRNA or mix-crRNA. (C) Expression of CP after transfection with CasRx-dNLS and CP2-crRNA, RdRp2-crRNA or mix-crRNA. (D) Expression of RdRp after transfection with CasRx-dNLS and CP2-crRNA, RdRp2-crRNA or mix-crRNA. (E and F) Cell morphology and CPEs (vacuolation) after infection with RGNNV, with or without CasRx-NLS (E) or CasRx-dNLS (F). Scale bar = 20 μm. (G) Schematic representation of the CasRx system preventing RGNNV infection at different time points. (H to K) The CasRx system prevented RGNNV infection at different time points when transfected with either CasRx-dNLS (H and I) or CasRx-NLS (J and K) and crRNAs targeting the RGNNV CP gene (CP2), RdRp gene (RdRp2), or both (mix). (H) Expression of CP after RGNNV infection 12 h, 24 h, 48 h at cells transfected with CasRx-dNLS and crRNAs targeting the RGNNV CP gene (CP2), RdRp gene (RdRp2), or both (mix). (I) Expression of RdRp after RGNNV infection 12 h, 24 h, 48 h at cells transfected with CasRx-dNLS and crRNAs targeting the CP2, RdRp2, or mix. (J) Expression of CP after RGNNV infection 12 h, 24 h, 48 h at cells transfected with CasRx-NLS and crRNAs targeting the CP2, RdRp2, or mix. (K) Expression of RdRp after RGNNV infection 12 h, 24 h, 48 h at cells transfected with CasRx-NLS and crRNAs targeting the CP2, RdRp2, or mix. (L and M) Cell morphology and CPEs (vacuolation) at different time points after RGNVV infection in cells treated with CasRx-NLS (L) or CasRx-dNLS (M). Arrows indicate the vacuoles induced by RGNNV infection. Scale bar = 20 μm. (N) Schematic representation of the CasRx system preventing RGNNV infection at different crRNA concentrations. (O) Expression of CP after transfection with CasRx-dNLS and different concentrations (50, 100, 200, 400, or 800 ng) of crRNAs (CP2, RdRp2, or mix). (P) Expression of RdRp after transfection with CasRx-dNLS and different concentrations (50, 100, 200, 400, or 800 ng) of crRNAs (CP2, RdRp2, or mix). (Q) Expression of CP after transfection with CasRx-NLS and different concentrations (50, 100, 200, 400, or 800 ng) of crRNAs (CP2, RdRp2, or mix). (R) Expression of RdRp after transfection with CasRx-NLS and different concentrations (50, 100, 200, 400, or 800 ng) of crRNAs (CP2, RdRp2, or mix). Data are expressed as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We detected time-dependent degradation of viral RNA during infection (Fig. 2G). Viral RNA copy numbers increased from 12 to 48 h in only the NNV- and ns-crRNA-infected groups, whereas targeting CP2, RdRp2, or both reduced viral copy numbers from 12 to 48 h for both the CasRx-dNLS and CasRx-NLS groups (Fig. 2H to K). Over the same time period, cell vacuolation increased and vacuoles grew larger in the NNV- and ns-crRNA-infected groups, whereas few vacuoles appeared in the crRNA-treated groups at any time point (Fig. 2L and M).
We also examined crRNA plasmid concentration-dependent degradation of viral RNA during infection (Fig. 2N). The viral RNA copy number was reduced significantly in cells transfected with 200, 400, and 800 ng of CP2-crRNA, RdRp2-crRNA, or mix-crRNA and CasRx-NLS (Fig. 2O and P). However, only 100 ng of mix-crRNA treatment obviously reduced viral copy numbers for cells transfected with CasRx-dNLS plasmid. Furthermore, 200, 400, and 800 ng CP2-crRNA, RdRp2-crRNA, or mix-crRNA transfected with CasRx-dNLS or CasRx-NLS significantly inhibited virus replication (Fig. 2Q and R). Thus, treatment with CasRx interrupted RGNNV replication and reduced the number of viral copies.
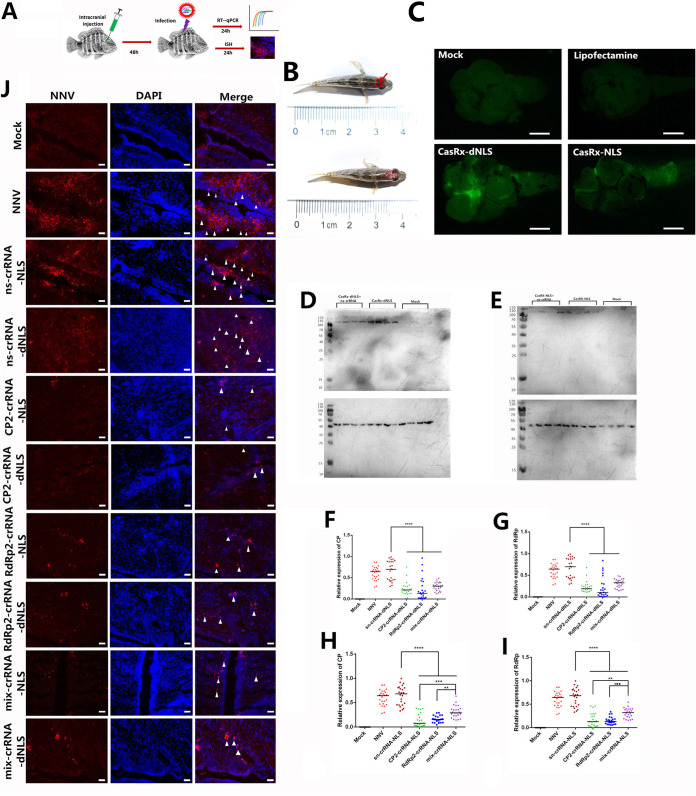
In vivo interference effects of the CasRx/crRNA system on RGNNV.
We next investigated the in vivo effects of CasRx/crRNA interference on RGNNV. RGNNV mainly destroys the brain tissue of grouper; therefore, we delivered the plasmids encoding CasRx and CP2-crRNA, RdRp2-crRNA, or mix-crRNA (with Lipofectamine 3000 as the transfection agent) to the grouper brain by intracranial injection (Fig. 3A and B). We then tested whether CasRx was expressed in the brain tissues. At 48 h postinjection, the green fluorescence of green fluorescent protein (GFP) was detected only in the CasRx-NLS and CasRx-dNLS injection groups but not in fish injected with Lipofectamine 3000 (Fig. 3C). Western blot analysis showed that samples injected with CasRx-NLS or CasRx-dNLS and ns-crRNA also expressed GFP (Fig. 3D and E). These results indicated successful vector delivery in vivo.
FIG 3.
In vivo RGNNV targeting via the CasRx system. (A) Schematic representation of intracranial injection of CasRx. (B) Diagram of intracranial injection location. a, the dotted red line indicates the site of the intracranial injection; b, the dotted red line indicates brain tissue. The red arrow indicates the injection area. (C) GFP expression in the brain of representative fish after injection with Lipofectamine 3000, CasRx-dNLS, or CasRx-NLS. (D and E) Western blot analysis of GFP expression in the brain after plasmid injection. The mouse anti-GFP antibody was used to detect the expression of the CasRx-dNLS (D) and CasRx-NLS (E) systems in the brain. (F to I) Intraperitoneal injection of CasRx prevented RGNNV infection. (F) Expression of CP after injection with CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (G) Expression of RdRp after injection with CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (H) Expression of CP after injection with CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (I) Expression of RdRp after injection with CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (J) Representative RGNNV FISH staining of brain sections from fish injected with the CasRx system (n = 3). Little fluorescence was observed after injection with CasRx-dNLS or CasRx-NLS targeting CP2-crRNA, RdRp2-crRNA, or mix-crRNA. Scale bar = 50 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Compared to the control (injected with the CasRxs and ns-crRNA), intracranial injection of CasRx-dNLS or CasRx-dNLS with CP2-crRNA or RdRp2-crRNA significantly decreased the CP and RdRp mRNA levels (Fig. 3F to I). Also, compared to mix-crRNA, the CP2-crRNA and RdRp2-crRNA had better interference of RGNNV in vivo (Fig. 3H and I). We further performed fluorescence in situ hybridization (FISH) for the CP genome. Cells transfected with either CasRx-NLS or CasRx-dNLS plasmid and CP2-crRNA, RdRp2-crRNA, or mix-crRNA showed reduced RGNNV infection (Fig. 3J). These results indicated that the CasRx/crRNA system was able to efficiently and functionally interfere with RGNNV infections in vivo.
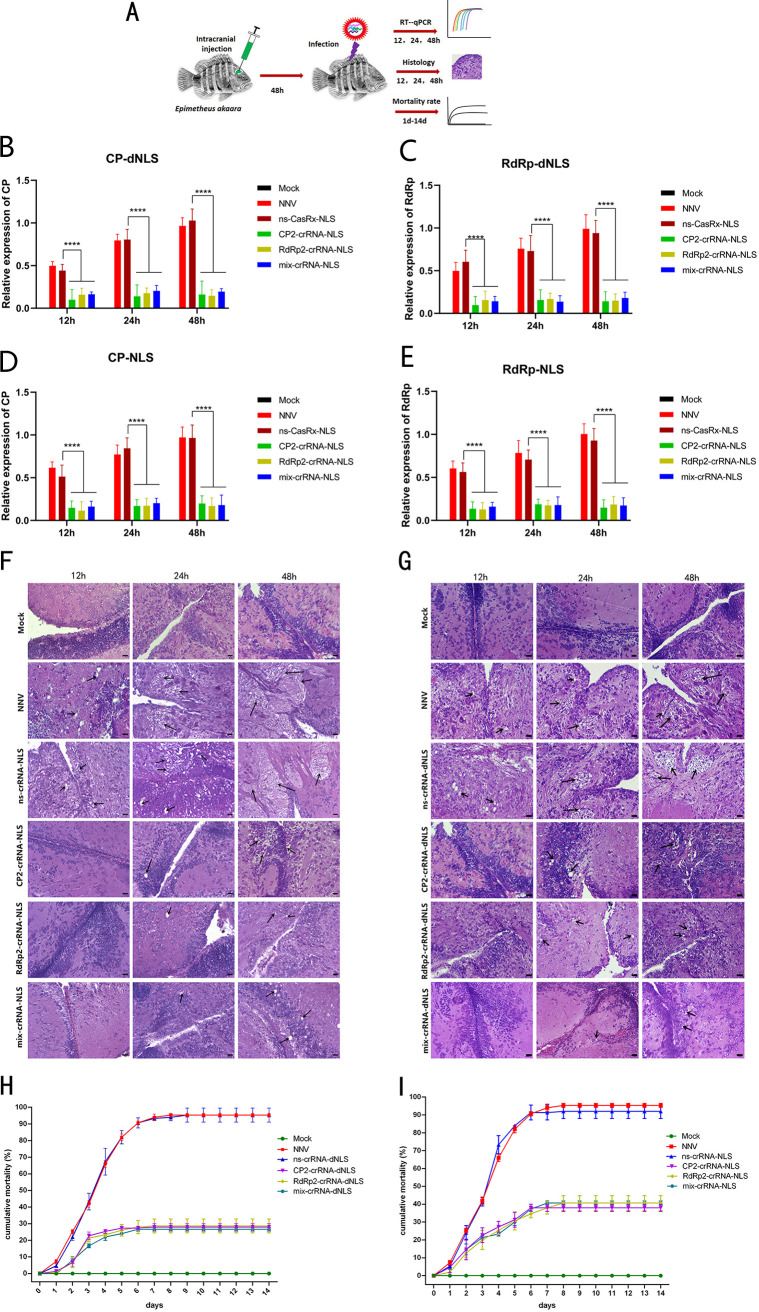
Lasting interference effect of the CasRx/crRNA system on RGNNV in vivo.
We detected time-dependent degradation of viral RNA during infection in vivo (Fig. 4A). Viral RNA copy numbers increased from 12 to 48 h only in the NNV- and ns-crRNA-infected groups, whereas viral RNA copy numbers decreased significantly in the CP2-, RdRp2-, or both-crRNA-treated groups at 12, 24, and 48 h after virus infection (Fig. 4B to E). Over the same time period, the brain histology analysis showed that vacuolation increased and vacuoles grew larger in the brain tissue of NNV- and ns-crRNA-infected groups, whereas few vacuoles appeared in the crRNA-treated groups at any time point (Fig. 4F and G).
FIG 4.
Lasting interference effect of the CasRx system. (A) Schematic representation of intracranial injection of the CasRx. (B) Expression of CP after RGNNV infection 12 h, 24 h and 48 h at different groups which were injected with CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (C) Expression of RdRp after RGNNV infection 12 h, 24 h and 48 h at different groups which were injected with CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (D) Expression of CP after RGNNV infection 12 h, 24 h and 48 h at different groups which were injected with CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (E) Expression of RdRp after RGNNV infection 12 h, 24 h and 48 h at different groups which were injected with CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (F and G) Brain histology of fish injected with or without the CasRx system. (F) Brain histology at different time points (12 h, 24 h, and 48 h) when injected with CasRx-dNLS and crRNAs targeting the RGNNV CP gene (CP2), RdRp gene (RdRp2), or both (mix). The black arrows indicate vacuoles. Scale bar = 10 μm. (G) Brain histology at different time points (12 h, 24 h, and 48 h) when injected with CasRx-NLS and crRNAs targeting the RGNNV CP gene (CP2), RdRp gene (RdRp2), or both (mix). The black arrows indicate vacuoles. Scale bar = 10 μm. (H) Values indicate the cumulative mortality rate for each group of orange-spotted groupers during the 14-day experimental period after different injection treatments (mock, Lipofectamine 3000, CasRx-dNLS plus ns-crRNA, CasRx-dNLS plus CP2-crRNA, CasRx-dNLS plus RdRp2-crRNA, or CasRx-dNLS plus mix-crRNA). (I) Values indicate the cumulative mortality rate for each group of orange-spotted groupers during the 14-day experimental period after different injection treatments (mock, Lipofectamine 3000, CasRx-NLS plus ns-crRNA, CasRx-NLS plus CP2-crRNA, CasRx-NLS plus RdRp2-crRNA, or CasRx-NLS plus mix-crRNA). ***, P < 0.001; ****, P < 0.0001.
Fourteen days after injection, no fish had died in the mock group. In the RGNNV only and CasRx-dNLS plus ns-crRNA groups, almost 90% of the groupers had died by 8 days postinfection. However, the cumulative mortality rate of groupers injected with CasRx-dNLS and CR2-crRNA, RdRp2-crRNA, or mix-crRNA was just 20 to 30% (Fig. 4H). Compared with a mortality rate of almost 90% in the CasRx-NLS plus ns-crRNA group, the cumulative mortality rate was just 30 to 40% in fish injected with CasRx-NLS and CR2-crRNA, RdRp2-crRNA, or mix-crRNA (Fig. 4I). These results indicate that the CasRx/crRNA system inhibits RGNNV infection in the orange-spotted grouper.
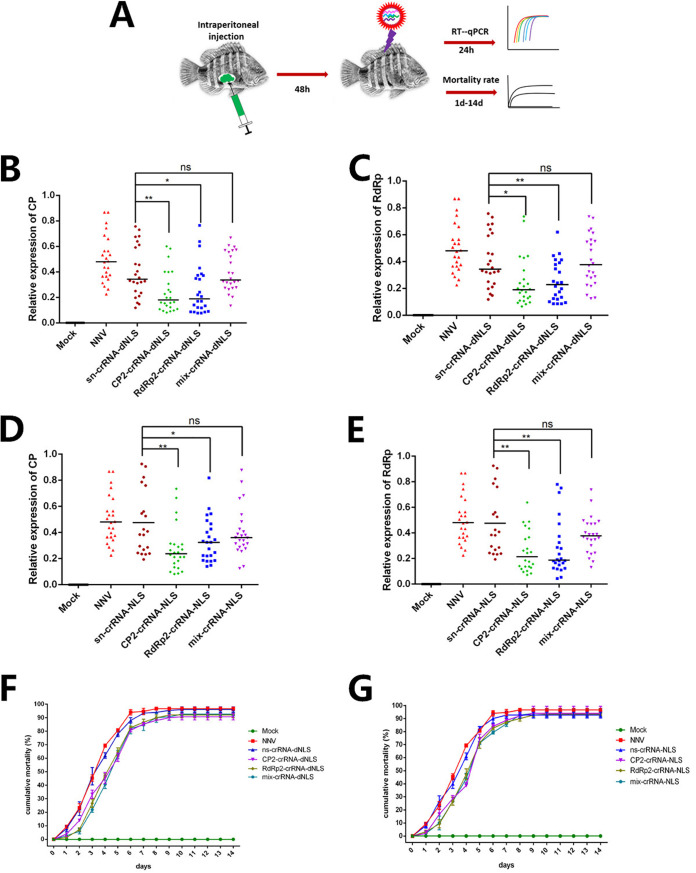
Effect of intraperitoneal injection of the CasRx/crRNA system on RGNNV.
We then investigated whether intraperitoneal injection of CasRx/crRNA could interfere with RGNNV (Fig. 5A). Fish injected with CasRx-NLS or CasRx-dNLS plasmid and CP2-crRNA or RdRp2-crRNA, but not the mix-crRNA, showed decreased CP and RdRp mRNA levels (Fig. 5B and E). The final cumulative mortality rate was almost 90% in all treatment groups, but the temporal trend differed among groups (Fig. 5F and G). The mortality rate increased rapidly on day 2 and the cumulative mortality rate reached 90% on day 7 in the RGNNV-only and CasRx plus ns-crRNA groups, whereas the mortality rate increased rapidly on day 3 and the cumulative mortality rate reached 90% on day 9 in the CasRx plus CP2-crRNA, RdRp2-crRNA, or mix-crRNA groups.
FIG 5.
Intraperitoneal delivery of CasRx. (A) Schematic representation of intraperitoneal injection of the CasRx system in vivo. (B) The mRNA levels of CP decreased after intraperitoneal injection of CasRx-dNLS targeting CP2 and RdRp2, but injection of mix-crRNA (CP2-crRNA and RdRp2-crRNA together) did not decrease the mRNA level. (C) The mRNA levels of RdRp decreased after intraperitoneal injection of CasRx-dNLS targeting CP2 and RdRp2, but injection of mix-crRNA (CP2-crRNA and RdRp2-crRNA together) did not decrease the mRNA level. (C) The mRNA levels of CP decreased after intraperitoneal injection of CasRx-NLS targeting CP2 and RdRp2, but injection of mix-crRNA (CP2-crRNA and RdRp2-crRNA together) did not decrease the mRNA level. (D) The mRNA levels of RdRp decreased after intraperitoneal injection of CasRx-NLS targeting CP2 and RdRp2, but injection of mix-crRNA (CP2-crRNA and RdRp2-crRNA together) did not decrease the mRNA level. (E) Values indicate the cumulative mortality rates for each group of groupers during the 14-day experimental period after intraperitoneal injection of CasRx-dNLS and CP2-crRNA, RdRp2-crRNA, or mix-crRNA. (F) Values indicate the cumulative mortality rates for each group of groupers during the 14-day experimental period after intraperitoneal injection of CasRx-NLS and CP2-crRNA, RdRp2-crRNA, or mix-crRNA. *, P < 0.05; **, P < 0.01; ns, not significant.
Effect of the CasRx/crRNA system on RGNNV in different grouper varieties.
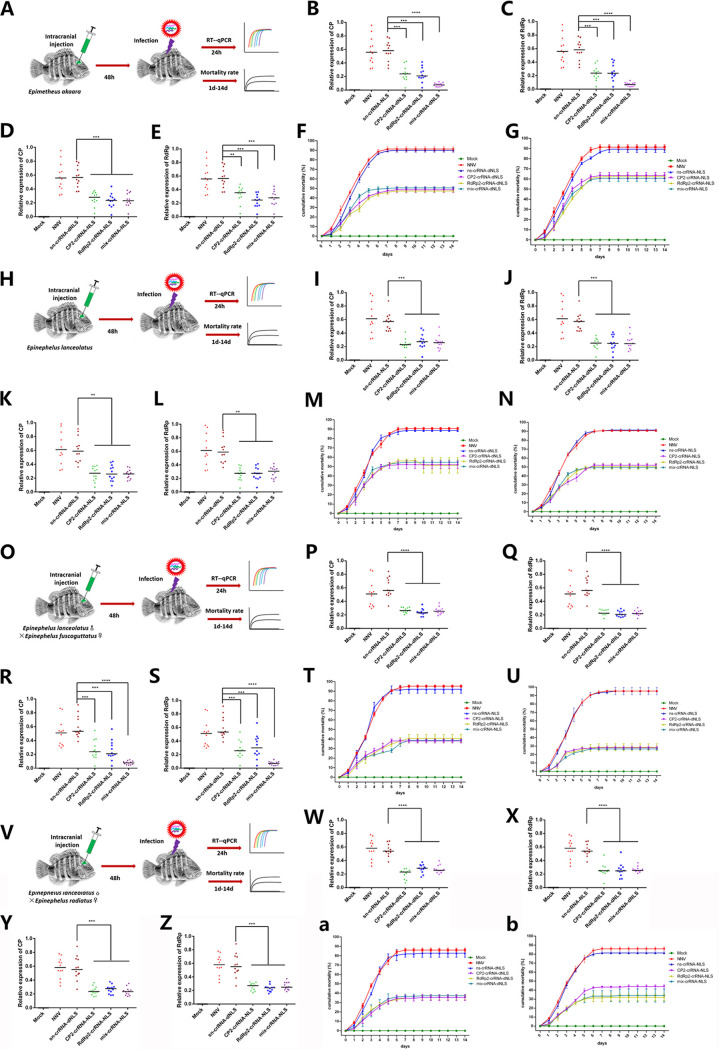
To investigate whether the CasRx/crRNA system has effects in other grouper species, we tested our system in the red-spotted grouper (Epinephelus akaara), giant grouper (Epinephelus lanceolatus), and two hybrid groupers (E. lanceolatus♂ × Epinephelus fuscoguttatus♀ and E. lanceolatus♂ × Epinephelus radiatus♀). In E. akaara, intracranial injection of CasRx-NLS or CasRx-dNLS and CP2-crRNA, RdRp2-crRNA, or mix-crRNA significantly reduced CP and RdRp mRNA levels and CasRx-dNLS plus mix-crRNA had the best effect (Fig. 6A to E). The final cumulative mortality rates for fish in the CasRx-NLS plus three crRNAs groups were 50 to 60%, whereas the rate was almost 90% in the CasRx-NLS plus ns-crRNA control group (Fig. 6F). The cumulative mortality rates in the CasRx-dNLS plus three crRNAs groups were 40 to 50%, lower than the mortality rate of almost 90% in the CasRx-dNLS plus ns-crRNA control group (Fig. 6G). The cumulative mortality rates did not differ significantly among the CP2-crRNA, RdRp2-crRNA, and mix-crRNA with CasRx-NLS or CasRx-dNLS cotreatments.
FIG 6.
RGNNV targeting via CasRx in different grouper species. (A) Schematic representation of intracranial injection of the CasRx system in E. akaara. (B) The mRNA level of CP after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (C) The mRNA level of RdRp after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (D) The mRNA level of CP after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (E) The mRNA level of RdRp after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (F and G) Values indicate the cumulative mortality rate for each group of E. akaara during the 14-day experimental period after intracranial injection of CasRx-dNLS (F) or CasRx-NLS (G) with crRNAs. (H) Schematic representation of intracranial injection of the CasRx system in E. lanceolatus. (I) The mRNA level of CP after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (J) The mRNA level of RdRp after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (K) The mRNA level of CP after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (L) The mRNA level of RdRp after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (M and N) Values indicate the cumulative mortality rate for each group of E. lanceolatus during the 14-day experimental period after intracranial injection of CasRx-dNLS (M) or CasRx-NLS (N) with crRNAs. (O) Schematic representation of intracranial injection of the CasRx system in E. lanceolatus♂ × E. fuscoguttatus♀. (P) The mRNA level of CP after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (Q) The mRNA level of RdRp after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (R) The mRNA level of CP after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (S) The mRNA level of RdRp after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (T and U) Values indicate the cumulative mortality rate for each group of E. lanceolatus♂ × E. fuscoguttatus♀ during the 14-day experimental period after intracranial injection of CasRx-dNLS (T) or CasRx-NLS (U) and crRNAs. (V) Schematic representation of intracranial injection of the CasRx system in E. lanceolatus♂ × E. radiatus♀. (W) The mRNA level of CP after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (X) The mRNA level of RdRp after intracranial injection of CasRx-dNLS and different crRNAs (CP2, RdRp2, or mix). (Y) The mRNA level of CP after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (Z) The mRNA level of RdRp after intracranial injection of CasRx-NLS and different crRNAs (CP2, RdRp2, or mix). (a and b) Values indicate the cumulative mortality rate for each group of E. lanceolatus♂ × E. radiatus♀ during the 14-day experimental period after intracranial injection of CasRx-dNLS (a) or CasRx-NLS (b) with crRNAs. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In E. lanceolatus, intracranial injection of CasRx-NLS or CasRx-dNLS with CP2-crRNA, RdRp2-crRNA, or mix-crRNA resulted in significant reductions in CP and RdRp mRNA levels, compared to the CasRxs plus ns-crRNA control groups, and the interference effects were similar among the crRNA treatments (Fig. 6H to L). The final cumulative mortality rates for groups treated with CasRx-NLS or CasRx-dNLS and CP2-crRNA, RdRp2-crRNA, or mix-crRNA were about 50%, whereas the CasRx-NLS or dNLS plus ns-crRNA groups had cumulative mortality rates of almost 90% (Fig. 6M and N).
In the hybrid grouper E. lanceolatus♂ × E. fuscoguttatus♀, intracranial injection of CasRx-NLS or CasRx-dNLS and CP2-crRNA, RdRp2-crRNA, or mix-crRNA significantly reduced CP and RdRp mRNA levels, compared to the CasRxs plus ns-crRNA control treatments, and CasRx-NLS plus mix-crRNA had the best effect (Fig. 6O to S). The final cumulative mortality rate for fish with CasRx-NLS treatments targeting CP2, RdRp2, or both were 30 to 40%, lower than that for the CasRx-NLS plus ns-crRNA control group (almost 90%) (Fig. 6T). With the CasRx-dNLS treatments targeting CP2, RdRp2, or both, cumulative mortality rates were 20 to 30%, lower than that of the CasRx-dNLS plus ns-crRNA control group (Fig. 6U).
In the hybrid grouper E. lanceolatus♂ × E. radiatus♀, treatment with CasRx-NLS or CasRx-dNLS and CP2-crRNA, RdRp2-crRNA, or mix-crRNA significantly reduced CP and RdRp mRNA levels, compared to the CasRxs-ns-crRNA control treatments (Fig. 6V to Z). The final cumulative mortality rates for fish with the CasRx-NLS or CasRx-dNLS treatments targeting CP2, RdRp2, or both were 30 to 40%, whereas the rates were almost 90% in the CasRx-NLS or dNLS plus ns-crRNA control groups (Fig. 6a and b).
These results showed that the CasRx system was able to efficiently and functionally interfere with the RNA of RGNNV in different grouper species, with best effects in the hybrid groupers. Moreover, CasRx-dNLS had better effects than CasRx-NLS when targeting RGNNV.
DISCUSSION
CRISPR/Cas systems provide bacteria and archaea with immunity to fend off invading nucleic acids. They also provide synthetic bioengineers with rich resources to build systems to edit and regulate the genome and epigenome and to modulate, modify, and monitor the transcriptome. Therefore, CRISPR/Cas systems can be harnessed for functional biology, biotechnology, and genetic medicine applications. In this study, we used CRISPR/CasRx to selectively interfere with the RGNNV. Our data demonstrate that CRISPR/CasRx can mediate molecular interference against a vertebrate RNA virus. Targeting different viral genomic regions for degradation resulted in reduced levels of CP and RdRp mRNA, cell vacuolation in vitro, and cumulative death in vivo. These findings indicated that the treatment attenuated RGNNV replication and spread, thereby illustrating the effectiveness of CRISPR/CasRx in targeting and interfering with an RNA virus in a vertebrate.
In this study, we constructed CasRx and five corresponding crRNAs. The different crRNAs exhibited different interference efficiencies. A previous study of LshCas13a showed that the most effective crRNAs were clustered in regions of strong interference, suggesting that target RNA accessibility impacts the CasRx activity (3). Similar conclusions were drawn for other Cas13 variants, including the LwaCas13a (7) and BzCas13b (27) CRISPR systems. These findings suggest that careful selection of RNA target sites as well as exploration of the efficiencies of various crRNAs might be essential to maximize CasRx activity and to achieve successful CasRx-mediated RNA interference.
To test whether different crRNAs have synergetic effects in virus RNA interference, we codelivered CP2-crRNA and RdRp2-crRNA with CasRx and found that the pooled crRNAs provided the most robust interference against RGNNV in vitro. Similarly, Freije et al. (15) reported that pooled crRNAs were more efficient than a single crRNA. Additionally, He et al. (28) performed gene knockdown and found that CasRx combined with single guide RNAs (sgRNAs) was more efficient in reducing target mRNA levels than was the sgRNA. However, our in vivo results indicated that injection with mix-crRNA (CP2-crRNA and RdRp2-crRNA) was less effective in suppressing the virus than treatment with either crRNA alone, especially in conjunction with CasRx-NLS in the orange-spotted grouper (Fig. 3H and I), whereas injection with mix-crRNA was more effective than treatment with either single crRNA in the hybrid grouper E. lanceolatus♂ × E. fuscoguttatus♀ (Fig. 5R and S). We propose that this difference may be related to the different transfection efficiencies of codelivering two or more crRNAs in different species. Moreover, other mechanisms may be involved in controlling the targeting accessibility and efficiencies of CasRx systems. Moreover, cytoplasmic-localized Cas13 (Cas13-dNLS) provided better interference than nuclear-localized variants (Cas13-NLS), which could be explained by the simultaneous localization of CasRx proteins and virus in the cytoplasm, where the RGNNV replicates.
Our new findings are striking in light of the challenges associated with targeting replicating viruses. We found that the CasRx system performed with high interfering efficiency with RGNNV in GS cells. In an in vivo context, direct intracranial injection stringently inhibited viral RNA, indicating that the CasRx system has strong effects on viral infectivity. However, intraperitoneal injection had lower interference efficiency, as it reduced the viral RNA level but did not reduce the final mortality rate. Unlike mRNA targeting via Cas13 (7, 8, 28, 29), viral RNA is actively replicating and evolving; therefore, CasRx must act at high efficiency. The mode of CasRx delivery is critical. We found that intraperitoneal delivery of CasRx resulted in reduced virus RNA expression levels and delayed virus outbreak, whereas direct intracranial delivery was more effective at virus suppression and mortality rate reduction. Our results represent an essential first step in the repurposing of CasRx for targeting vertebrate RNA viruses, and they provide the foundation for testing applications of the system in vivo.
We also showed that the CasRx system could be used for virus interference in different grouper species, and the effect was best in the hybrid groupers. The CasRx RNA interference discrepancy among grouper species may be due to differences in their autoimmunity, as studies have shown that hybrid groupers inherit good traits from their parents with high disease resistance (30–32). The CasRx system does not kill all invasive viruses and may just inhibit virus replication, and degraded parts of the virus need to be cleaned up by the host’s own immune system. This may explain why the CasRx system works better in hybrids with stronger immunity. These results suggest that the CasRx system may work with the host immune system to combat virus infection.
Overall, our results indicate that engineered CasRx systems are highly effective in RNA virus targeting in fish both in vitro and in vivo. This platform will allow researchers to target other RNA viruses in other species, which may lead to development of therapeutics for RNA virus diseases.
MATERIALS AND METHODS
Generation of plasmids.
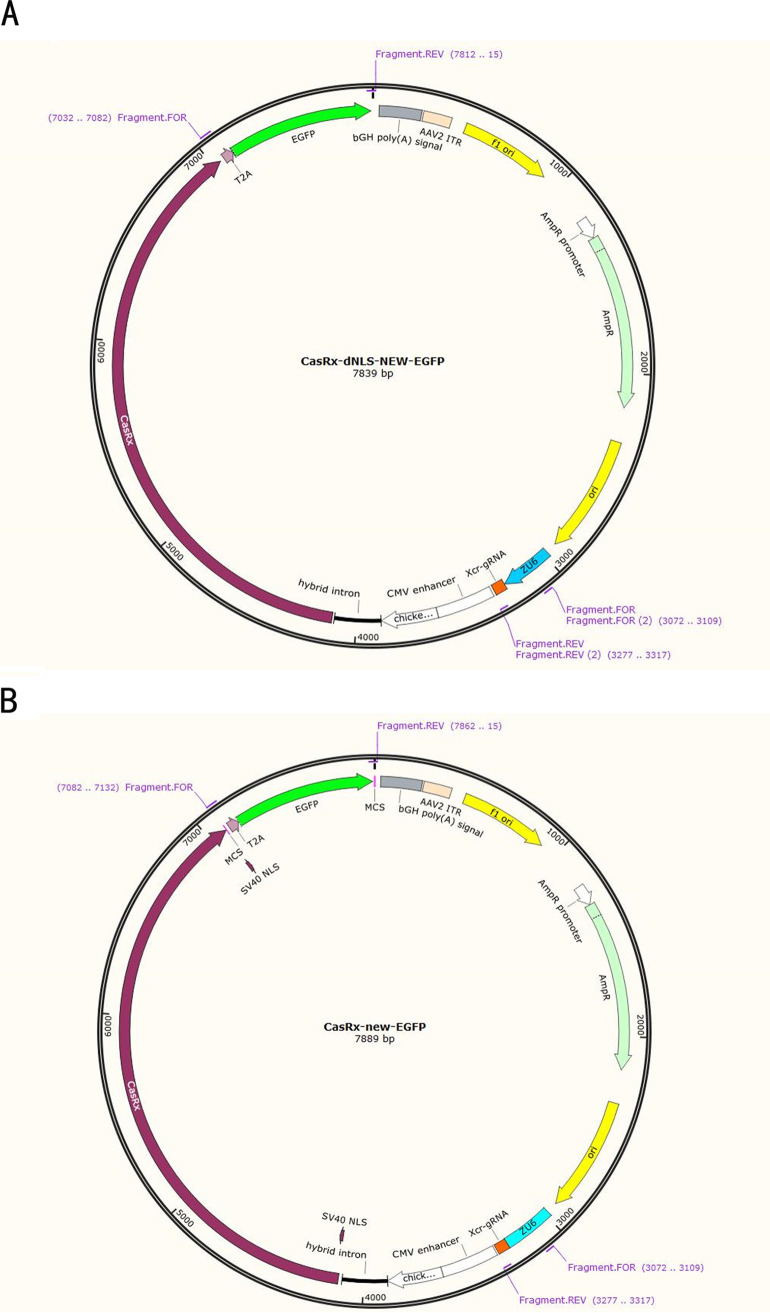
The recombinant px458-CasRx-enhanced GFP (EGFP) plasmid was designed mainly based on the px458 plasmid. We first synthesized the Cas13d protein sequence and then subcloned Cas13d into px458 using XbaI and EcoRI enzymes to generate a single clone. We further synthesized the zebrafish U6 promoter along with guide RNA (gRNA) (Rfx) to replace the original U6 and gRNA in px458 using PciI and XbaI enzymes to generate the px458-CasRx plasmid (10). Based on the restriction enzyme site of XbaI, a double-stranded DNA fragment of the cytomegalovirus (CMV) promoter was also synthesized using PCR and inserted at the front of the Cas13d protein sequence by the homologous recombination method using a one-step cloning kit (Vazyme Biotech). Finally, to facilitate observation at the cellular level, the EGFP coding sequence was added behind px458-CasRx, with the 2A peptide from the Thosea asigna virus CP (T2A) sequence ahead of it, using EcoRI enzyme and the homologous recombination method, thereby constructing the px458-CasRx-dNLS-EGFP plasmid (CasRx-dNLS) (Fig. 7A). We also synthesized the Cas13d protein sequence, including the NLS of the SV40 large T antigen, and further constructed the px458-CasRx-NLS-EGFP plasmid (CasRx-NLS) as described previously (Fig. 7B). All constructs were confirmed by sequencing. The amino acid and nucleotide sequences of CasRx were used in accordance with a previous study (10).
FIG 7.
Plasmid profile of the CasRx system. (A) Plasmid profile of CasRx-dNLS-EGFP. (B) Plasmid profile of CasRx-NLS-EGFP.
Design of RGNNV crRNAs.
The crRNA backbone plasmid sequences were used according to a previous study (10). The genomes of four NNVs (striped jack NNV, tiger puffer NNV, barfin flounder NNV, and RGNNV) were downloaded from the NCBI database and simply aligned to identify highly conserved windows that could be CasRx target sites. Five target sites (CP1, CP2, CP3, RdRp1, and RdRp2) were designed to target regions of the RGNNV genome with high sequence conservation. The length of the targeting site sequences ranged from 24 to 32 nucleotides. The targeting site sequences were cloned into the CasRx crRNA backbone plasmid using overlapped PCR and the primers listed in the Table 1.
TABLE 1.
Primers and sequence used in the study
| Primer or sequence | Sequence (5′ to 3′) |
|---|---|
| Primers for plasmid construction | |
| Targeting-CP1-F | AAACCAGGTGCGTCAGTGCGATTACTAC |
| Targeting-CP1-R | CTTGGTAGTAATCGCACTGACGCACCTG |
| Targeting-CP2-F | AAACCCTGAGAGATGGACGTCATTGGTC |
| Targeting-CP2-R | CTTGGACCAATGACGTCCATCTCTCAGG |
| Targeting-CP3-F | AAACTCCAGTGTTTCAACAGCGTATCGCTGGAAG |
| Targeting-CP3-R | CTTGCTTCCAGCGATACGCTGTTGAAACACTGGA |
| Targeting-RdRp1-F | AAACCACTGACGACTCCGTTCACTACCGTGTTGC |
| Targeting-RdRp1-R | CTTGGCAACACGGTAGTGAACGGAGTCGTCAGTG |
| Targeting-RdRp2-F | AAACCGATGACTGGAATGACGTTGTAGCCAACGAGC |
| Targeting-RdRp2-R | CTTGGCTCGTTGGCTACAACGTCATTCCAGTCATCG |
| Primers for RT-qPCR | |
| RGNNV CP-RT-F | CAACTGACAACGATCACACCTTC |
| RGNNV CP-RT-R | CAATCGAACACTCCAGCGACA |
| RGNNV RdRp-RT-F | GTGTCCGGAGAGGTTAAGGATG |
| RGNNV RdRp-RT-R | CTTGAATTGATCAACGGTGAACA |
| Primers for riboprobe synthesis | |
| DIG-CP-F | CCCAAGCTTCAGAACAGTCCGACCTCAGTAC |
| DIG-CP-R | CGGGGTACCGTCAACCCTAGTGCAGACAGTG |
| Sequence for probe synthesis | CAGAACAGTCCGACCTCAGTACACCCGCACGCTCCTCTGGACCTCGTCGGGAAAGGAGCAGCGTCTCACGTCACCTGGTCGGCTGATACTCCTGTGTGTCGGCAACAACACTGATGTGGTCAACGTGTCAGTGCTGTGTCGCTGGAGTGTTCGACTGAGCGTTCCATCTCTTGAGACACCTGAAGAGACCACCGCTCCCATCATGACACAAGGTTCCCTGTACAACGATTCCCTTTCCACAAATGACTTCAAGTCCATCCTCCTAGGATCCACACCACTGGACATTGCCCCTGATGGAGCAGTCTTCCAGCTGGACCGTCCGCTGTCCATTGACTACAGCCTTGGAACTGGAGATGTTGACCGTGCTGTTTATTGGCACCTCAAGAAGTTTGCTGGAAATGCTGGCACACCTGCAGGCTGGTTTCGCTGGGGCATCTGGGACAACTTCAACAAGACGTTCGCAGATGGCGTTGCCTACTACTCTGATGAGCAGCCTCGTCAAATCCTGCTGCCTGTTGGCACTGTCTGCACTAGGGTTGAC |
Cell lines and viruses.
GS cells were established from the spleen of the grouper Epinephelus akaara. GS cells were propagated at 25°C in Leibovitz L15 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA) as described previously (33). Propagation of RGNNV was performed as described previously (33). For viral stock propagation, GS cells at 80% confluence were inoculated with virus for 2 h. After 2 h, the cells were washed three times with Leibovitz L15 medium supplemented with 2% FBS to remove excess RGNNV, and Leibovitz L15 medium supplemented with 10% FBS was added. Virus-containing cellular supernatant was harvested 48 to 60 h postinfection, centrifuged to remove cell debris, aliquoted, and stored at −80°C until use. The viral titers of RGNNV were 107 times the 50% tissue culture infective dose (TCID50)/ml.
Virus titers.
The titers of the RGNNV viral stocks were determined by the TCID50 as described previously (34). In short, GS cells were seeded in 96-well plates 1 to 2 days prior to inoculation to allow a full monolayer to form. RGNNV viral stocks were thawed on ice, viral stocks were diluted via 10-fold dilutions, and inoculation was performed. After 2 h, the inoculation medium was washed off and replaced with Leibovitz L15 medium supplemented with 10% FBS. Plaques were counted 1 day postinfection, and titers were calculated using the Reed-Muench method.
Detection of plasmid transfection.
Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) transfection methods were used to transfect CasRx plasmid into GS cells. Cells were seeded on coverslips (12 mm) overnight and transfected the next day with 1 μg of CasRx plasmid according to the manufacturer’s protocol. Cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 1 h. Finally, cells were mounted with 50% glycerol and observed under a fluorescence microscope (Leica, Wetzler, Germany).
RNA extraction and RT-qPCR.
After the cells were washed three times with PBS, the total RNA was extracted by adding 500 μl of TRIzol (Invitrogen) and 200 μl of chloroform. The total RNA from brain tissue was extracted by adding 1,000 μl of TRIzol and 200 μl of chloroform. After centrifugation of the cells at 12,000 rpm for 15 min at 4°C, the supernatant was transferred to a 1.5-ml RNase-free tube. Isopropanol and 75% alcohol were used to precipitate and purify the RNA. Total RNA was reverse transcribed to synthesize first-strand cDNA using the ReverTra Ace kit (TOYOBO, Osaka, Japan) according to the manufacturer's instructions. RT-qPCR amplification was performed on an ABI QuantStudio 5 system (Applied Biosystems, Waltham, MA, USA) using the 2 × SYBR green real-time PCR mix (TOYOBO) as described previously (35). β-Actin was used as the internal control to verify the successful transcription and to calibrate the cDNA template for corresponding samples. Each assay was carried out in triplicate with the following cycling conditions: 1 min for activation at 95°C followed by 40 cycles of 15 s at 95°C, 15 s at 60°C, and 45 s at 72°C. Triplicate CT values were calculated using the comparative CT (ΔΔCT) method. The data are presented as mean ± standard deviation (SD). All reagents were precooled in advance. The results of RT-qPCR were calculated based on the expression levels of targeted genes normalized to that of β-actin at the indicated time. All primers used for qPCRs are listed in Table 1.
FISH and Western blotting.
Sense and antisense digoxigenin (DIG)-labeled riboprobes were synthesized from the open reading frame sequence of the RGNNV CP gene (Table 1) using the DIG RNA labeling kit (Roche Diagnostics, Mannheim, Germany). The procedures for RNA FISH followed those described by Ragoczy et al. (36), Ho et al. (37), and Beliveau et al. (38) with modifications. Briefly, brain tissues from grouper were fixed in buffered 4% paraformaldehyde for 24 h. The brain samples were then dehydrated with a series of graded ethanol solutions (70 to 100%), cleared in xylene, and embedded in paraffin; 10-μm sections were cut for FISH. Prior to hybridization, the slides were washed with PBS, sequentially dehydrated in 70%, 90%, and 100% ethanol solutions, and equilibrated in 10% formamide/2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (pH 7.0). The mixture of the primary sense and antisense probes and the secondary probes were hybridized to the cells in 10% formamide-10% dextran sulfate-2× SSC-5 mmol/liter ribonucleotide vanadate complex-0.05% bovine serum albumin-1 μg/μl Escherichia coli tRNA and hybridized overnight at 55°C in a humidified chamber. Slides were sequentially washed at 37°C in 10% formamide-2× SSC (pH 7) followed by 2× SSC and then were mounted with Fluoroshield with 6-diamidino-2-pheny-lindole (DAPI) (Sigma, St. Louis, MO, USA). Fluorescent signals from FISH were imaged with a confocal microscope (Zeiss, Oberkochen, Germany).
Total proteins were extracted from brain tissue using extraction buffer (100 mM Tris-HCl [pH 8], 150 mM NaCl, 0.6% IGEPAL, 1 mM EDTA, and 3 mM dithiothreitol [DTT]) with protease inhibitors (phenylmethylsulfonyl fluoride [PMSF], leupeptin, aprotinin, pepstatin, antipain, chymostatin, Na2VO3, NaF, MG132, and MG115). Equal amounts of protein were subjected to SDS-PAGE and then transferred to polyvinylidene difluoride membranes. After blocking with 5% nonfat dry milk, immunoblot analysis was conducted using mouse anti-GFP antibody (1:3,000; Sigma) for detection of CasRx proteins; anti-actin (1:5,000) was used for detection of β-actin. The antigens were detected by chemiluminescence using an enhanced chemiluminescence (ECL) detection reagent (Thermo Fisher Scientific, USA).
Immunofluorescence assay.
The immunofluorescence assay was performed as described previously (39). In brief, GS cells were seeded onto coverslips in 6-well plates and infected with RGNNV for 24 h after transfection with or without the CasRx-NLS or CasRx-dNLS system. RGNNV-infected GS cells were fixed and then incubated with anti-CP antibody (1:200); the specificity of the antibody was reported in our previous report (40). After washing with PBS, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated secondary antibody. Finally, cells were stained with 1 mg/ml DAPI and observed under fluorescence microscopy (Zeiss).
In vitro antiviral assay.
CasRx-dNLS and CasRx-NLS with different crRNA systems against RGNNV were assessed to determine their efficacy. In a 24-well plate, GS cells that had been grown overnight were transfected with CasRx (400 ng) and targeting crRNAs (400 ng) or ns-crRNA (400 ng) using Lipofectamine 3000. After 24 h, RGNNV was added to the cells. After 2 h of infection, cells were washed three times with Leibovitz L15 medium supplemented with 2% FBS to remove excess RGNNV, and new Leibovitz L15 medium supplemented with 10% FBS was added to enable measurement of newly produced viral RNA at subsequent postinfection time points. Finally, 24 h after infection, the cells were washed three times with PBS, and total RNA was extracted.
For different time points, in a 24-well plate, GS cells that had been grown overnight were transfected with CasRx (400 ng) and crRNA (400 ng) or ns-crRNA (400 ng) using Lipofectamine 3000. After 24 h, RGNNV was added to the cells. After 2 h, the virus was washed off and new Leibovitz L15 medium supplemented with 10% FBS was added. Total RNA was extracted 12, 24, and 48 h after infection. The cells were washed three times with PBS before extraction of the RNA.
To evaluate the effects of different concentrations, GS cells that had been grown overnight were added to a 24-well plate and transfected with CasRx (400 ng) and crRNAs (50, 100, 200, 400, or 800 ng) or nontarget crRNA (50, 100, 200, 400, or 800 ng) using Lipofectamine 3000. RGNNV at a multiplicity of infection of 0.5 was added to the cells 24 h after transfection. After 2 h, the virus was washed off and new Leibovitz L15 medium supplemented with 10% FBS was added. Total RNA was extracted 24 h after infection. The cells were washed three times with PBS before extraction of the RNA. cDNA synthesis and RT-qPCR were performed as described above.
In vivo antiviral assay.
Four groups (mock, Lipofectamine 3000, CasRx-NLS, and CasRx-dNLS) of orange-spotted grouper (Epinephelus coioides) fry were cultured for 1 week before the experiment began. The weights and lengths of the fish are shown in Table 2. Healthy fry were starved for 24 h before being injected intracranially with 400 ng of CasRx (NLS or dNLS) and 400 ng of ns-crRNA mixed with Lipofectamine 3000, following the manufacturer’s instructions. The fish were sacrificed at 48 h postinjection, and the whole brain was separated from the body and examined under a Nikon inverted microscope.
TABLE 2.
Body weight and length data for different grouper species
| Species | Body weight (g) | Body length (cm) |
|---|---|---|
| Orange-spotted grouper (Epinephelus coioides) | 2.36 ± 0.24 | 2.62 ± 0.45 |
| Red-spotted grouper (Epinephelus akaara) | 2.61 ± 0.25 | 2.47 ± 0.38 |
| Giant grouper (Epinephelus lanceolatus) | 2.98 ± 0.26 | 3.25 ± 0.41 |
| Hybrid grouper Epinephelus lanceolatus♂ × Epinephelus fuscoguttatus♀ | 2.56 ± 0.30 | 2.76 ± 0.37 |
| Hybrid grouper Epinephelus lanceolatus♂ × Epinephelus radiatus♀ | 2.42 ± 0.26 | 2.96 ± 0.33 |
For the intracranial injection experiment, the following 10 groups were created: mock, Lipofectamine 3000, CasRx-NLS plus ns-crRNA, CasRx-dNLS plus ns-crRNA, CasRx-NLS plus CP2-crRNA, CasRx-dNLS plus CP2-crRNA, CasRx-NLS plus RdRp2-crRNA, CasRx-dNLS plus RdRp2-crRNA, CasRx-NLS plus mix-crRNA, and CasRx-dNLS plus mix-crRNA. Each group had four parallel groups (n = 50 fish), one of which was used for RT-qPCR analysis; the other three were used to calculate mortality rates. Fish were injected intraperitoneally with 40 μl of 107 TCID50/ml RGNNV at 48 h after intracranial injection with the plasmid (400 ng CasRx and 400 ng crRNAs). After 12, 24, and 48 h, 13 fish were randomly selected from each parallel group, and brain tissue was extracted. Brain tissues from eight fish were used for virus gene expression analysis conducted using RT-qPCR, and tissues from the other five fish were fixed in Bouin’s fluid for histological analysis. Deaths were recorded daily for 14 days. The same strategy was used for intracranial injection of different species of grouper, i.e., red-spotted grouper (Epimetheus akaara), giant grouper (Epinephelus lanceolatus), hybrid grouper E. lanceolatus♂ × E. fuscoguttatus♀, and hybrid grouper E. lanceolatus♂ × E. radiatus♀. The weights and lengths of the fish are shown in Table 2. This work received research ethics approval from the Animal Research and Ethics Committees of South China Agriculture University (approval SYXK-2019-0136).
For the intraperitoneal injection experiment, the same 10 groups were established but the injected plasmid amounts were 800 ng of CasRx and 1 μg of crRNAs; the fish were injected intraperitoneally with 40 μl of 107 TCID50/ml RGNNV at 48 h postinjection with the plasmid. After 24 h, 24 fish were randomly selected from each parallel group, brain tissue was extracted, and virus gene expression analysis was conducted using RT-qPCR. Deaths were recorded daily for 14 days.
Brain histology.
Brains were dissected from fish in the mock, Lipofectamine 3000, CasRx-NLS plus ns-crRNA, CasRx-dNLS plus ns-crRNA, CasRx-NLS plus CP2-crRNA, CasRx-dNLS plus CP2-crRNA, CasRx-NLS plus RdRp2-crRNA, CasRx-dNLS plus RdRp2-crRNA, CasRx-NLS plus mix-crRNA, and CasRx-dNLS plus mix-crRNA groups, fixed in Bouin’s fluid for 24 h at room temperature, dehydrated, and embedded in paraffin wax. All tissue blocks were sectioned at 5 μm and stained with hematoxylin and eosin for analysis.
Statistical analysis.
All data are expressed as mean ± SD. Statistics analyses were performed using the software SPSS v17.0. One-way analysis of variance was used, and statistical significance is indicated. GraphPad Prism 6 was used for all statistical analysis.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grants U20A20102, 31930115, 41806151, and 31972768), the Science and Technology Planning Project of Guangzhou (grant 202002030206), the China Agriculture Research System of MOF and MARA(CARS-47-G16), the Provincial Projects with Special Funds for Promoting Economic Development of Marine and Fisheries Department of Guangdong (grant SDYY-2018-05), the Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (grant 311021006), and the Guangdong Provincial Special Fund For Modern Agriculture Industry Technology Innovation Teams(2019KJ143).
Contributor Information
Huihong Zhao, Email: zhaohh@scau.edu.cn.
Yong Zhang, Email: lsszy@mail.sysu.edu.cn.
Qiwei Qin, Email: qinqw@scau.edu.cn.
Julie K. Pfeiffer, University of Texas Southwestern Medical Center
REFERENCES
- 1.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. 2015. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60:385–397. 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M. 2018. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19:1. 10.1186/s13059-017-1381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan X, Zhang F, Zhong Z, Chen R, Wang Y, Chang L, Bock R, Nie B, Zhang J. 2019. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol J 17:1814–1822. 10.1111/pbi.13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, Zhao Y, Ye J, Cao X, Xu C, Chen B, An H, Jiao Y, Zhang F, Yang X, Zhou G. 2019. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol J 17:1185–1187. 10.1111/pbi.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. 2017. RNA targeting with CRISPR-Cas13. Nature 550:280–284. 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. 2017. RNA editing with CRISPR-Cas13. Science 358:1019–1027. 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA. 2018. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell 70:327–339. 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. 2018. Transcriptome engineering with RNA-targeting type VID CRISPR effectors. Cell 173:665–676. 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahas A, Aman R, Mahfouz M. 2019. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol 20:263. 10.1186/s13059-019-1881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. 2017. An update on Zika virus infection. Lancet 390:2099–2109. 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- 14.Rizzardini G, Saporito T, Visconti A. 2018. What is new in infectious diseases: Nipah virus, MERS-CoV and the Blueprint List of the World Health Organization. Infez Med 26:195–198. [PubMed] [Google Scholar]
- 15.Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, Yozwiak NL, Zhang F, Sabeti PC. 2019. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell 76:826–837. 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishizawa T, Furuhashi M, Nagai T, Nakai T, Muroga K. 1997. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl Environ Microbiol 63:1633–1636. 10.1128/aem.63.4.1633-1636.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandín I, Souto S. 2020. Betanodavirus and VER disease: a 30-year research review. Pathogens 9:106. 10.3390/pathogens9020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shetty M, Maiti B, Shivakumar Santhosh K, Venugopal MN, Karunasagar I. 2012. Betanodavirus of marine and freshwater fish: distribution, genomic organization, diagnosis and control measures. Indian J Virol 23:114–123. 10.1007/s13337-012-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto T, Okinaka Y, Mise K, Mori K, Arimoto M, Okuno T, Nakai T. 2004. Identification of host-specificity determinants in betanodaviruses by using reassortants between striped jack nervous necrosis virus and sevenband grouper nervous necrosis virus. J Virol 78:1256–1262. 10.1128/jvi.78.3.1256-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parameswaran V, Kumar Rajesh S, Ishaq Ahmed VP, Sahul Hameed AS. 2008. A fish nodavirus associated with mass mortality in hatchery-reared Asian Sea bass, Lates calcarifer. Aquaculture 275:366–369. 10.1016/j.aquaculture.2008.01.023. [DOI] [Google Scholar]
- 21.Crane M, Hyatt A. 2011. Viruses of fish: an overview of significant pathogens. Viruses 3:2025–2046. 10.3390/v3112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi S, Lo C, Kou G, Chang P, Peng S, Chen S. 1997. Mass mortalities associated with viral nervous necrosis (VNN) disease in two species of hatchery‐reared grouper, Epinephelus fuscogutatus and Epinephelus akaara (Temminck & Schlegel). J Fish Dis 20:185–193. 10.1046/j.1365-2761.1997.00291.x. [DOI] [Google Scholar]
- 23.Chi S, Lo B, Lin S. 2001. Characterization of grouper nervous necrosis virus GNNV. J Fish Dis 24:3–13. 10.1046/j.1365-2761.2001.00256.x. [DOI] [Google Scholar]
- 24.Zhou L, Wang S, Yu Q, Wei S, Liu M, Wei J, Huang Y, Huang X, Li P, Qin Q. 2020. Characterization of novel aptamers specifically directed to red-spotted grouper nervous necrosis virus (RGNNV)-infected cells for mediating targeted siRNA delivery. Front Microbiol 11:660. 10.3389/fmicb.2020.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Zhang J, Liu J, Hu Y, Ni S, Yang Y, Yu Y, Huang X, Qin Q. 2017. Fish TRIM35 negatively regulates the interferon signaling pathway in response to grouper nodavirus infection. Fish Shellfish Immunol 69:142–152. 10.1016/j.fsi.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Wang L, Huang X, Wang S, Huang Y, Qin Q. 2019. Fish cholesterol 25-hydroxylase inhibits virus replication via regulating interferon immune response or affecting virus entry. Front Immunol 10:322. 10.3389/fimmu.2019.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, Koonin EV, Zhang F. 2017. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell 65:618–630. 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He B, Peng W, Huang J, Zhang H, Zhou Y, Yang X, Liu J, Li Z, Xu C, Xue M, Yang H, Huang P. 2020. Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell 11:518–524. 10.1007/s13238-020-00700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, Liu L, Lang J, Cheng K, Wang Y, Li X, Shi J, Wang Y, Nie G. 2018. A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment. Cancer Lett 431:171–181. 10.1016/j.canlet.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Tseng WY, Poon CT. 1983. Hybridization of Epinephelus species. Aquaculture 34:177–182. 10.1016/0044-8486(83)90302-2. [DOI] [Google Scholar]
- 31.Othman AR, Kawamura G, Senoo S, Fui CF. 2015. Effects of different salinities on growth, feeding performance and plasma cortisol level in hybrid TGGG (tiger grouper, Epinephelus fuscoguttatus × giant grouper, Epinephelus lanceolatus) juveniles. Int J Biol Sci 4:15–20. [Google Scholar]
- 32.Sun Y, Guo CY, Wang DD, Li XF, Xiao L, Zhang X, You X, Shi Q, Hu GJ, Fang C, Lin HR, Zhang Y. 2016. Transcriptome analysis reveals the molecular mechanisms underlying growth superiority in a novel grouper hybrid (Epinephelus fuscogutatus ♀ × Epinephelus lanceolatus ♂). BMC Genet 17:24. 10.1186/s12863-016-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang XH, Huang YH, Sun JJ, Han X, Qin QW. 2009. Characterization of two grouper Epinephelus akaara cell lines: application to studies of Singapore grouper iridovirus (SGIV) propagation and virus-host interaction. Aquaculture 292:172–179. 10.1016/j.aquaculture.2009.04.019. [DOI] [Google Scholar]
- 34.Szretter KJ, Balish AL, Katz JM. 2006. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol 29:15G.1.1–15G.1–24. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Liu J, Zhang X, Wei S, Huang X, Huang Y, Wei J, Qin Q. 2019. Fish autophagy protein 5 exerts negative regulation on antiviral immune response against iridovirus and nodavirus. Front Immunol 10:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. 2006. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20:1447–1457. 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho Y, Shewchuk BM, Liebhaber SA, Cooke NE. 2013. Distinct chromatin configurations regulate the initiation and the maintenance of hGH gene expression. Mol Cell Biol 33:1723–1734. 10.1128/MCB.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beliveau BJ, Boettiger AN, Avendaño MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, Hannan MA, Hoang HG, Colognori D, Lee JT, Shih WM, Yin P, Zhuang X, Wu CT. 2015. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun 6:7147. 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Huang Y, Huang X, Li C, Ni SW, Yu Y, Qin Q. 2019. Grouper DDX41 exerts antiviral activity against fish iridovirus and nodavirus infection. Fish Shellfish Immunol 91:40–49. 10.1016/j.fsi.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Wang L, Zheng J, Huang L, Wang S, Huang X, Qin Q, Huang Y. 2021. Grouper interferon-induced transmembrane protein 1 inhibits iridovirus and nodavirus replication by regulating virus entry and host lipid metabolism. Front Immunol 12:636806. 10.3389/fimmu.2021.636806. [DOI] [PMC free article] [PubMed] [Google Scholar]