SUMMARY
Hematopoietic ontogeny consists of two broad programs: an initial hematopoietic stem cell (HSC)-independent program followed by HSC-dependent hematopoiesis that sequentially seed the fetal liver and generate blood cells. However, the transition from HSC-independent to HSC-derived hematopoiesis remains poorly characterized. To help resolve this question, we developed Mds1CreERT2 mice, which inducibly express Cre-recombinase in emerging HSCs in the aorta and label long-term adult HSCs, but not HSC-independent yolk-sac-derived primitive or definitive erythromyeloid (EMP) hematopoiesis. Our lineage-tracing studies indicate that HSC-derived erythroid, myeloid, and lymphoid progeny significantly expand in the liver and blood stream between E14.5 and E16.5. Additionally, we find that HSCs contribute the majority of F4/80+ macrophages in adult spleen and marrow, in contrast to their limited contribution to macrophage populations in brain, liver, and lungs. The Mds1CreERT2 mouse model will be useful to deconvolute the complexity of hematopoiesis as it unfolds in the embryo and functions postnatally.
In brief
Zhang et al. develop Mds1CreERT2 mice, which inducibly express Cre-recombinase in emerging fetal hematopoietic stem cells (HSCs). Lineage tracing reveals increasing HSC contributions to hematopoietic cells in E14.5–E16.5 embryos, as well as the majority of macrophages in adult spleen and marrow. Mds1CreERT2 mice will help deconvolute the complexity of hematopoietic ontogeny.
Graphical Abstract
INTRODUCTION
The emergence of the hematopoietic system during embryonic and fetal development has been the focus of extensive study, both for its paradigmatic value but also vis-à-vis its relevance to human diseases. Studies primarily in the murine model system indicate that the ontogeny of the hematopoietic system consists of two broad programs, an initial HSC-independent program followed by the emergence and establishment of an HSC-dependent program that arises from arterial vessels within the body of the embryo. HSC-independent hematopoiesis is complex, consisting of overlapping waves of primitive hematopoietic progenitors, erythro-myeloid progenitors (EMPs), and small numbers of lymphoid progenitors (Böiers et al., 2013; McGrath et al., 2015; Palis et al., 1999; Yoshimoto et al., 2011, 2012). Primitive hematopoiesis is derived from progenitors that emerge in the yolk sac beginning at embryonic day 7.25 (E7.25), producing circulating maturing primitive erythroid, megakaryocyte, and macrophage cells by E10 (Palis et al., 1999). Lineage-tracing experiments indicate that these first macrophages persist as adult self-renewing tissue-resident microglia (Hoeffel et al., 2015; Palis et al., 1999; Stremmel et al., 2018). Multilineage EMPs emerge from hemogenic endothelium in the yolk sac from E8.25 through E10.5 and seed the fetal liver as early as E10 to generate definitive erythroid, megakaryocyte, granulocyte, and monocyte lineage cells (Frame et al., 2016; Gomez Perdiguero et al., 2015; Lux et al., 2008; McGrath et al., 2015; Palis and Yoder, 2001). EMPs also provide tissue-resident macrophages that persist into adulthood in multiple organs including the liver, lungs, and skin (Gomez Perdiguero et al., 2015). Rare B cell and T cell progenitors arise independently of HSCs in the yolk sac and embryo proper by E9.5 (Yoshimoto et al., 2011, 2012), and lymphomyeloid-restricted progenitors (LMP) are already present in the fetal liver by E10.5, before HSC activity is detectable there (Boiers et al., 2013; Luis et al., 2016)
Small numbers of HSCs capable of long-term reconstitution of adult mouse recipients begin to emerge at E10.5 from the ventral aspect of the dorsal aorta (Yokomizo and Dzierzak, 2010). At E11.5–12.0, HSCs are found in the fetal liver where they subsequently expand in numbers (Ema and Nakauchi, 2000; Kumaravelu et al., 2002). However, the transition from HSC-independent to HSC-derived hematopoiesis in the murine fetus remains poorly characterized, particularly since the maturing erythroid, megakaryocytic, and myeloid progenies of EMPs and HSCs are not currently easily distinguishable. Several different inducible lineage-tracing approaches have been used to distinguish HSC-independent versus HSC-derived contributions based on promoters active both in HSCs and in HSC-independent progenitors, including (Cdh5 (Zovein et al., 2008), CSF1R-Cre (Qian et al., 2011), Kit-Cre (Stremmel et al., 2018), Runx1-Cre (Samokhvalov et al., 2007), Tie2-Cre (Gomez Perdiguero et al., 2015), and TLR2-Cre (Balounová et al., 2019). The efficacy of these CreER alleles relies on the timing of induction during the staggered emergence of HSC-independent progenitors and of HSCs. However, the significant temporal overlap in the emergence of HSC-independent progenitors and of HSCs, coupled with the broad cellular expression of these promoters, and the imprecision of in vivo CreER induction often results both in incomplete and in mixed labeling (Gentek et al., 2018; Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Soares-da-Silva et al., 2021). An alternative approach is to specifically label emerging HSCs. Studies of emerging HSCs have identified several genes preferentially or specifically expressed in HSCs, including Hlf (Yokomizo et al., 2019), Ly6A (Sca-1; (Ma et al., 2002), Procr (Iwasaki et al., 2010), and Mecom (Yuasa et al., 2005).
The Mecom or Mds1-Evi1 complex locus was first identified as a site of proviral insertion in murine myeloid leukemias (Morishita et al., 1988; Mucenski et al., 1988) and was also found to be activated via chromosomal rearrangements in human AML (Bitter et al., 1985; Rubin et al., 1990). Importantly, the Mds1-Evi1 gene harbors two widely separated transcription start sites (TSSs) (Fears et al., 1996) that encode a series of different isoforms, with the upstream TSS encoding MDS1-EVI1, a 160 kDa isoform harboring an N-terminal PR domain, thus placing this isoform in the PR/SET family of transcriptional regulators and chromatin modifiers (Hohenauer and Moore, 2012). The downstream TSS encodes at least three zinc finger protein isoforms of EVI1 (135/145, 123, and 103 kDa). Roles for Mecom in HSCs during embryogenesis have been documented by several groups. In the mouse, homozygous loss of all but the 103 kDa isoform results in loss of competitive repopulating ability within the para-aortic splanchnopleura and lethality by E10.5 (Hoyt et al., 1997; Yuasa et al., 2005). Knockout of all isoforms is associated with HSCs in the fetal liver that lack competitive repopulation ability and with lethality at E13.5 (Goyama et al., 2008). In contrast, the elimination of the 160 kDa MDS1-EVI1 isoform results in viable mice with diminished numbers of adult HSCs (Zhang et al., 2011) and with skeletal defects (Juneja et al., 2014). Using a zebrafish model, Konantz et al. (2016) documented expression of Evi1 in the ventral aspect of the dorsal aorta, and downregulation of Evi1 led to impaired HSC emergence from hemogenic endothelium.
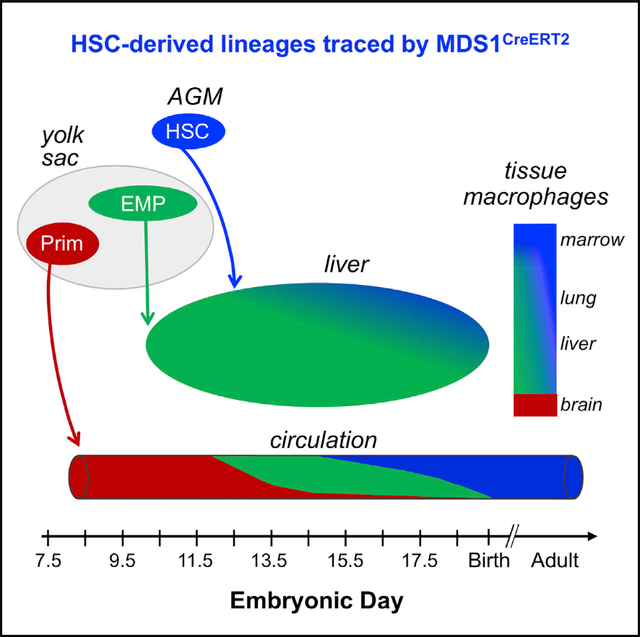
To more clearly define the role of the different Mecom isoforms in the mammalian hematopoietic system, we introduced a lacZ reporter into the upstream TSS of murine Mecom (Mds1lacZ) facilitating the examination of its expression histologically within the murine embryo. Mds1lacZ is expressed in Runx1+ clusters in the ventral aspect of the dorsal aorta of the mouse but is not expressed in the fetal liver (Zhang et al., 2011). To probe this further, we developed a Cre-ER knockin into the Mds1 locus (Mds1CreERT2). Lineage-tracing studies using this mouse model revealed that the progeny of Mds1-expressing cells are subsequently found in the fetal liver as immunophenotypic and functional LT-HSCs. In contrast, neither EMPs themselves nor their progeny were labeled, nor were the progeny of primitive hematopoietic progenitors. Lineage tracing with Mds1CreERT2 demonstrated that HSC-derived hematopoietic progeny are found in increasing numbers in the livers of E14.5 to E16.5 murine embryos and contribute significantly to circulating erythroid, myeloid, and lymphoid cells by E16.5. In addition, while varying degrees of tissue-resident macrophages in the brain, liver, and lung of adult mice originate from HSC-independent sources, the large majority of F4/80+ macrophages in the spleen and bone marrow of adult mice are lineage labeled, consistent with their derivation from HSCs. The Mds1CreERT2 model will be useful to address multiple questions regarding the complexity of the hematopoietic system as it unfolds in the murine embryo and continues to function postnatally.
RESULTS
Embryonic expression of Mecom
We reported previously the expression pattern of Mecom transcripts during embryogenesis using in situ hybridization probes that hybridized to both Evi1 and Mds1 transcripts. These studies revealed Mds1-Evi1 transcripts in the developing limb bud, lung epithelium, and developing renal system among other locations, with no discernable expression in the fetal liver (Perkins et al., 1991). In an effort to understand more precisely where Mds1 is expressed in the embryonic hematopoietic system, we created a nuclear localized NLS-lacZ knockin allele that puts the lacZ gene under transcriptional control of the Mecom upstream promoter (termed Mds1) (Fears et al., 1996; Zhang et al., 2011). Using this allele, termed Mds1lacZ (referred to as Mds1m1 in Zhang et al., 2011), we conducted whole-mount labeling of E8.5 to E14.5 embryos with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal). β-galactosidase staining was evident in the limb buds, mesonephros, and vertebral bodies (Figures 1A–1E), overlapping considerably with the expression of Mds1-Evi1 described previously by in situ hybridization (Perkins et al., 1991).
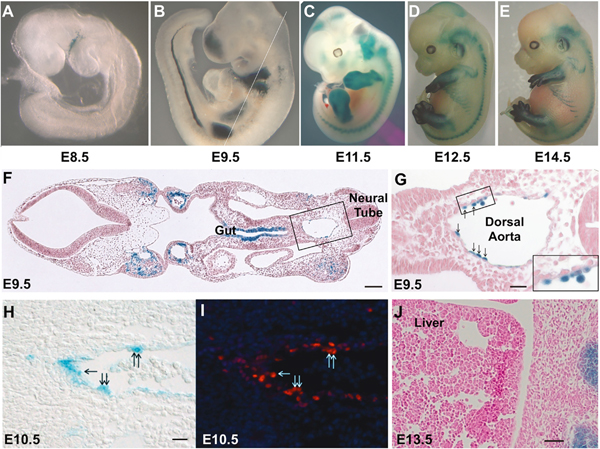
Figure 1. Mds1 is expressed in aortic hemogenic endothelium.
Embryo-fetal expression of Mds1 documented using the Mds1lacZ allele (Zhang et al., 2011).
(A–E) Whole-mount β-galactosidase staining of heterozygous Mds1lacZ/wt embryos at various stages of development as indicated. Mean crown-rump length for C57 E8.5 = 1.88, E9.5 = 3.47, E11.5 = 6.83, E12.5 = 8.60, E14.5 = 12.29 mm (Mu et al., 2008).
(F) Photomicrograph of a X-gal-stained section of an E9.5 embryo with plane of section indicated by the white line in (B). Scale bar, 100 μM.
(G) Close-up of boxed region in (F), showing the dorsal aorta, left being ventral, right being dorsal, with β-galactosidase-positive cells seen budding from the endothelium (arrows). Scale bar, 25 μM.
(H and I) Photomicrographs of E10.5 aorta (left is ventral) β-galactosidase-positive cells (H) that also express Runx1 detected by immunocytochemistry (I). Scale bar, 20 μM.
(J) X-gal staining reveals no β-galactosidase-positive cells in the liver of the E13.5 mouse embryo. Scale bar, 25 μM.
Coronal sections of E9.5, E10.5, and E11.5 embryos (Figures 1F–1H; Figures S1A and S1C) revealed LacZ staining of endothelial cells on the ventral aspect of the dorsal aorta, as well as in intra-aortic hematopoietic clusters (IAHC). Importantly, these LacZ-positive aortic endothelial cells were found to co-express Runx1, consistent with a hemogenic endothelial identity (Figure 1I; Figures S1B and S1D) (Yzaguirre et al., 2017). Given that HSCs are known to seed and expand within the fetal liver, we expected to detect β-galactosidase-staining cells in the fetal liver. Paradoxically, we did not find evidence of Mds1-Evi1 expression in the fetal liver (Figure 1J), consistent with previous findings (Perkins et al., 1991). These data suggest that the expression of Mds1-Evi1 in the fetal hematopoietic system is remarkably transient, in contrast to the expression of Mds1-Evi1 in adult LT- and ST-HSCs (Zhang et al., 2011).
Generation of a mouse model of TAM-inducible Cre recombinase knocked into the Mds1 locus
To assess whether the β-gal-positive cells identified at E9.5 in the dorsal aorta of Mds1lacZ mice represent functional hematopoietic cells, we performed lineage-tracing experiments. We constructed an Mds1CreERT2 allele in which a tamoxifen (TAM)-inducible Cre recombinase was inserted into the Mecom locus so it would be expressed by the upstream TSS (Figures 2A and 2B). We first performed lineage tracing with the Mds1CreERT2 allele in adult mice by combining it with the ubiquitously transcribed, Cre-dependent Rosa26loxP-STOP-loxP-YFP (Rosa26LSL-YFP) reporter. Based on the restricted expression of Mds1-Evi1 to adult LT- and ST-HSCs, as well as a small percentage of MPPs (Christodoulou et al., 2020; Zhang et al., 2011), we expected that activation of Mds1CreERT2 via TAM treatment would result in increasing and broad YFP positivity in hematopoietic cells over time. Mds1CreERT2 Rosa26LSL-YFP mice received five treatments of TAM over 5 days (Figure 2C). Analysis of bone marrow at 4, 10, and 15 days (relative to the first treatment) revealed a time-dependent increase in YFP+ cells (Figure 2D). By 15 days, up to 40% of marrow cells were YFP+ and they consisted of myeloid, erythroid, and lymphoid cells (data not shown). These labeled cells in the marrow contributed to YFP+ progeny detected in the peripheral blood at 30 days (Figure 2E).
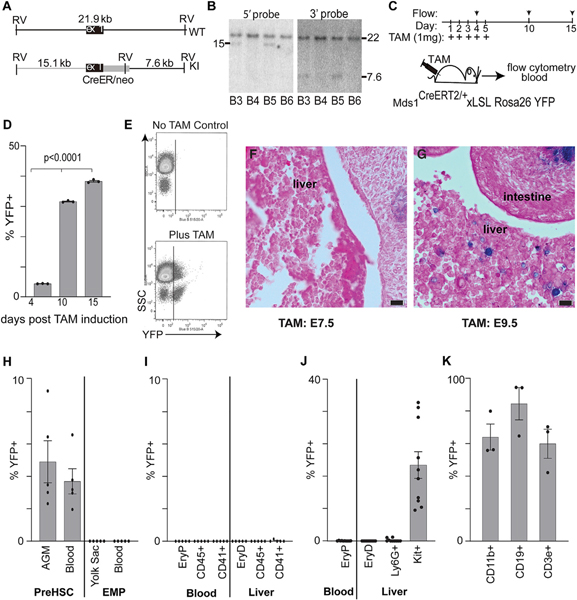
Figure 2. Tamoxifen-inducible Mds1CreERT2 allele construction and expression in adult hematopoiesis and fetal HSC but not EMP- or primitive-derived hematopoiesis.
(A) A DNA cassette encoding the TAM-inducible Cre-ER fusion protein was introduced into the Mecom locus in embryonic stem (ES) cells via homologous recombination.
(B) ES cell DNA was screened by Southern blotting, cutting the genomic DNA with EcoRV and blotting with 5′ and 3′ probes situated outside of the recombination cassette. Clones B3 and B5 show the expected sizes of 15.1 Kb and 7.6 kb, with 5′ and 3′ probes, respectively. Mice were created from ES cell clone B5, which were backcrossed onto a C57Bl6 background and crossed with mice bearing the Rosa26 LSL-YFP allele.
(C–E) These mice were treated with TAM, and the peripheral blood was analyzed by flow cytometry at 30 days and bone marrow at days 4, 10, and 15 for YFP+ cells, which progressively increased. (D) Quantitation of percentage of bone-marrow YFP-positive cells following the first TAM treatment on the days indicated. Average ± SEM, n = 3. (E) Representative scatterplot (day 30) showing the appearance of YFP+ blood cells following TAM treatment.
(F and G) X-gal staining of fetal liver in E13.5 mouse embryos, after a single treatment with TAM at E7.5 (F) or E9.5 (G). Eosin counter-stain; bar denotes 10 μm.
(H) Flow cytometric analysis of E11.5 AGM, yolk sac, and blood for pre-HSC (kit+CD31+VECadherin+) versus EMP (kit+CD41+CD16/32+) as indicated. For each analysis, the percentage of total cells that were YFP+ is charted as average ± SEM, n = 5. See Figures S2A and S2B, for the gating strategy.
(I) Flow cytometric analysis of E11.5 blood and liver for lineage-positive cell subsets (EryP Ter119+ blood, EryD Ter119+FSChi to distinguish from circulating EryP in the liver) as indicated. For each analysis, the percentage of total cells that were YFP+ is charted as average ± SEM, n = 5. See Figures S2D and S2E for the gating strategy.
(J) Flow cytometric analysis of E12.5 fetal liver and blood for cell subsets as indicated gated as in (I). For each analysis, the percentage of total cells that were YFP+ is charted as average ± SEM, n = 10.
(K) Analysis of mice competitively transplanted with 106 E12.5 liver cells from TAM-treated (at E9.5) Mds1CreERT2/WT, Rosa26LSL-YFP/LSL-YFP mice and 105 unlabeled adult bone-marrow cells. Flow cytometric analysis of circulating YFP+ and YFP-negative granulocyte/monocyte cells (CD11b+), B cells (CD19+), and T cells (CD3e+) at 12 weeks post-transplant. See Figure S2F for gating strategy. Average ± SEM, n = 3.
Lineage-tracing preHSCs and fetal liver cells with Mds1CreERT2
To test whether the Mds1lacZ-expressing cells present in the dorsal aorta might contain precursors of fetal liver hematopoiesis, we conducted lineage-tracing studies using the Mds1CreERT2 allele crossed with a Rosa26LSL-lacZ reporter. Timed pregnant mice were treated with a single dose of TAM either at E7.5 or at E9.5, and embryos were harvested and stained for β-gal at E13.5. Labeling at E9.5 with TAM lineage traced small numbers of β-gal-positive cells in the fetal liver at E13.5, which were not seen when TAM was delivered at E7.5 (Figures 2F and 2G). While fetal liver cells themselves do not express Mds1, the fetal liver contains a population of cells derived from earlier Mds1-expressing cells.
We next investigated the specificity of the Mds1CreERT2 lineage-tracing mouse model by analyzing E11.5 fetuses. Pregnant Mds1CreERT2/wt Rosa26LSL-YFP/LSL-YFP mice were treated with TAM at E9.5 and cells from aorta-gonad-mesonephros (AGM), yolk sac, blood, and liver in Mds1CreERT2/wt Rosa26LSL-YFP/wt(referred to as Mds1CreERT2 Rosa26LSL-YFP) fetuses were analyzed by flow cytometry. Cells from littermate negative control Mds1WT/WT Rosa26LSL-YFP/wt mice were used to determine bone fide YFP+ expression in all experiments (data not shown). Consistent with the staining seen with Mds1lacz, we found YFP+CD31+kit+VE-cadherin+ pre-HSCs ((Azzoni et al., 2018; Gao et al., 2020; Yokomizo and Dzierzak, 2010) in the AGM and in the circulation. In contrast, we found no labeling of CD41+kit+CD16/32+ EMPs in the yolk sac, Ter119+ primitive erythroid cells (EryP) in the bloodstream(Kingsley et al., 2004), or EMP-derived definitive erythroid cells (EryD) in the liver (McGrath et al., 2015; Gomez Perdiguero et al., 2015) (Figures 2H and 2I; Figures S2A and S2B). Other HSC-independent CD45+ early myeloid cells and CD41+ megakaryocytes in the bloodstream and in the liver were also not labeled (Figure 2I). The Mds1CreERT2 promoter did not label HSC-independent hematopoietic cells in E11.5 fetuses even when activated at E8.5 (Figure S2C). TAM treatment at E8.5 also did not label pre-HSCs, despite labeling a subset of cells in the paw, consistent with Mds1 expression in limb buds (Figure S2C; Figure 1).
We next asked whether the Mds1CreERT2 can label functional HSC in the murine embryo. As E12.5 is the first time that a significant number of transplantable HSCs are found in the liver (Ema and Nakauchi, 2000; Kumaravelu et al., 2002), pregnant Mds1CreERT2 Rosa26LSL-YFP mice were treated with TAM at E9.5, and blood and liver were analyzed at E12.5. As seen at E11.5, primitive erythroid cells in the blood stream and definitive erythroid and granulocyte progeny of EMPs in the liver were not labeled (Figure 2J; Figures S2D and S2E). In contrast, YFP+ cells in the E12.5 liver were found to constitute a subset of Kit+ cells, suggesting that Mds1CreERT2 might be lineage-labeling HSCs. To directly test this hypothesis, we transplanted 2 × 106 E12.5 Mds1CreERT2 Rosa26LSL-YFP liver cells (induced with TAM at E9.5) into irradiated syngeneic recipient mice along with 2 × 105 wild-type bone-marrow cells. At 12 weeks post-transplant, peripheral blood from these recipients revealed a high percentage of YFP+ T cells, B cells, and myeloid cells (Figure 2K; Figure S2F). Taken together, these data demonstrate that Mds1CreERT2, combined with TAM induction at E9.5, lineage-traces fetal HSCs but not primitive erythropoiesis or EMP-derived hematopoiesis.
Mds1CreERT2 lineage tracing of later fetal hematopoiesis
We utilized the Mds1CreERT2 lineage-tracing mouse model to examine the transition from HSC-independent to HSC-derived hematopoiesis in livers of E14.5 and E16.5 mouse embryos (Figure 3). The level of labeling of HSC-enriched LSK (lineage−, Sca1+, Kit+) cells was used to normalize for different efficiencies of excision. In contrast to E12.5 livers, YFP+ Ter119+ (definitive erythroid), Ly6G+ (granulocyte), Ly6C+ (monocyte), and CD19+ (lymphoid) cells were readily evident at E14.5. This suggests that HSCs have begun to contribute to multiple hematopoietic lineages by E14.5 (Figure 3A; Figures S3A and S3B). Furthermore, at E14.5, the higher contribution of Mds1CreERT2-labeled cells to the hematopoietic progenitor (LK, Lineage−, Sca1−, Kit+) compartment compared to the downstream lineage-positive cells suggests that HSC contribution to fetal hematopoiesis is expanding (Figure 3A). Indeed, this is evident at E16.5, with the increased contribution of HSCs to the LK hematopoietic progenitor pool, as well as downstream to maturing erythroid-, granulocyte-, and monocyte-lineage cells (Figure 3B, left panel). Consistent with the progressive contribution of HSCs to fetal hematopoiesis, labeled myeloid cells and red blood cells are now also found in the bloodstream (Figure 3B, right panel, Figure S3C). Since the Rosa26 locus is progressively downregulated in maturing erythroid cells (Asari et al., 2004; Zambrowicz et al., 1997), the YFP+ fraction of circulating erythrocytes may underrepresent the full contribution from HSCs. Further analysis of E16.5 hematopoietic stem and progenitor cells indicated that fetal HSCs contribute equally to all lineage-biased MPP subsets, and to a lesser, but uniform degree to lineage-committed lymphoid (CD127+), myeloid (CD16/32+), and erythroid/megakaryocyte (CD16/32−) progenitors (Figure 3C; Figure S3D; Pronk et al., 2007). Taken together, these data indicate that HSCs provide an increasing contribution to hematopoiesis in the fetal liver between E14.5 and E16.5, the time points associated with significant expansion of HSC numbers and differentiating hematopoietic progenitors and precursors in the fetal liver (Ema and Nakauchi, 2000; Kumaravelu et al., 2002)
Figure 3. Mds1CreERT2Rosa26LSL-YFP/E9.5 TAM labeling demonstrates increasing presence of HSC-dependent hematopoiesis through E16.5.
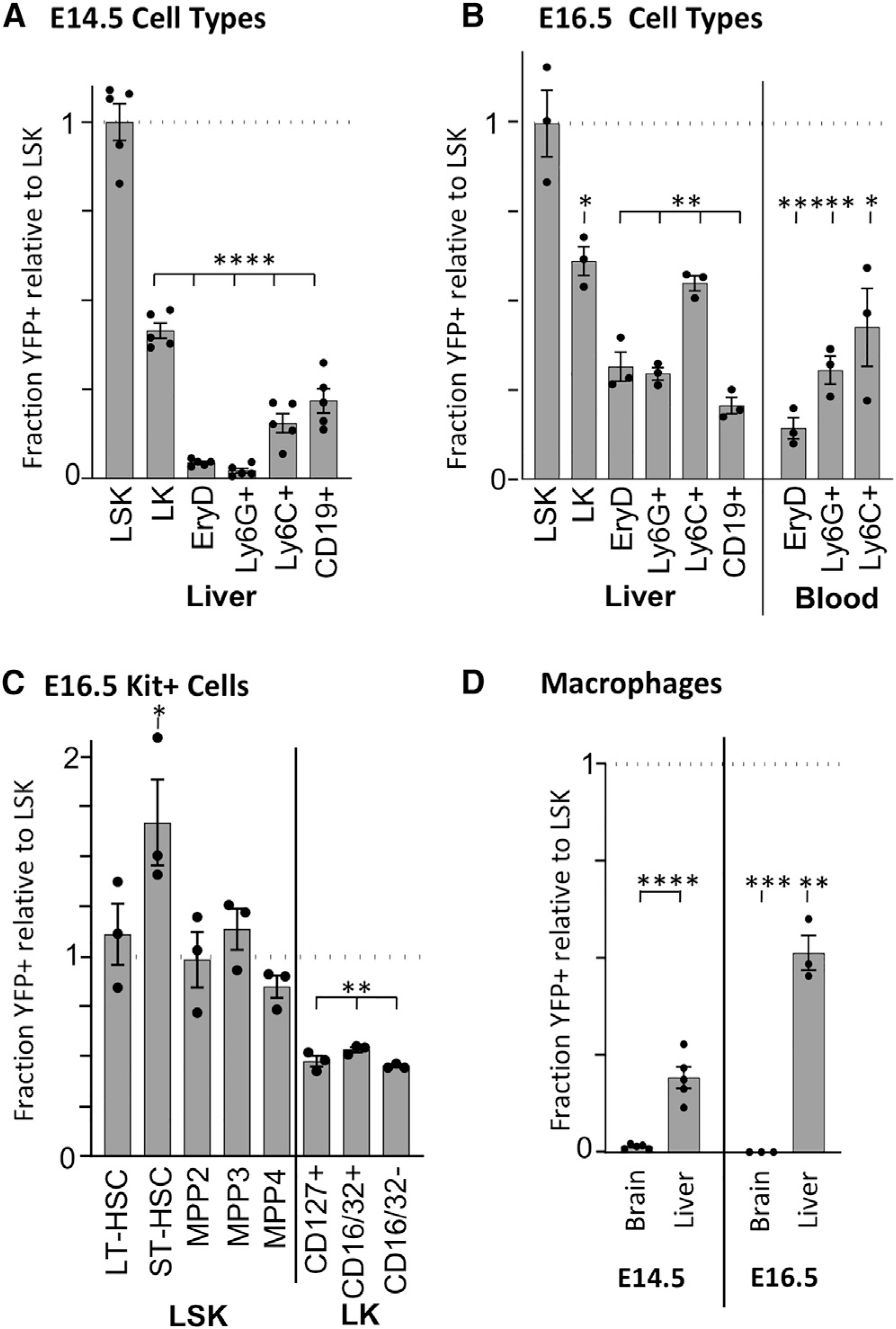
(A) Flow cytometric analysis of E14.5 liver cells. Levels of YFP positivity were normalized to the level found in LSK (Lin-Kit+Sca1+, average 23.3% YFP+) to control for excision rates. Averages ± SEM of 5 individual embryos are plotted for LK (Lin-Kit+Sca1−), erythroid (Ter119+FSChi), granulocyte (Ly6G+), monocyte (Ly6C+), and B (CD19+) cells. See Figures S3A and S3B for gating strategy.
(B) Flow cytometric analysis of circulating blood cells and liver cells at E16.5. Averages ± SEM of 3 individuals are plotted. Gating is as in (A) with the addition of circulating definitive erythroid (EryD, Ter119+) cells gated as in Figure S3C. Levels of YFP positivity were normalized to the level found in LSK average 8.1% YFP+) to control for excision rates.
(C) Flow cytometric analysis of E16.5 progenitors in the liver. LSK was further refined into LT-HSC, ST-HSC, MPP2, MPP3, and MPP4 based on Flt3, CD150, and CD48 positivity (see Figures S3A and S3D). LK were further refined into lymphoid (CD127+), myeloid (CD16/32+), and erythroid/megakaryocyte (CD16/32−) progenitors. Values normalized to YFP levels in LSK (B). Averages ± SEM, n = 3.
(D) Analysis by flow cytometry of YFP+ F4/80+ macrophages from E14.5 and E16.5 brain and liver normalized to YFP positivity of LSK (panels A and B). Details of gating are found in Figure S3E. Average ± SEM, n = 3. Unpaired two tail Student’s t test was performed comparing YFP labeling in cell populations to LSK. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Several tissue-resident macrophage populations, established from HSC-independent sources, persist through embryogenesis as self-renewing populations (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Liu et al., 2019). This is particularly evident for microglia, which are seeded early in development and not normally replaced by later fetal or adult myeloid cells (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015). Consistent with these findings, no YFP labeling of F4/80+ macrophage cells in the E14.5 or 16.5 brain was evident in Mds1CreERT2 fetuses (Figure 3D; Figure S3E). In contrast, a high proportion of macrophages in the fetal liver are HSC-derived by E16.5 consistent with contribution of HSCs to the monocyte lineage (Figures 3B and 3D).
Lineage tracing of hematopoiesis in adult mice
TAM administration at E9.5 clearly activated Cre-ER in fetal cells capable of long-term engraftment when transplanted into adult recipients (Figure 2K). However, differences in the regulation of Flt3 in subsets of fetal HSCs has indicated that there is heterogeneity in fetal transplantable HSCs, including subsets that may not normally contribute to adult HSCs (Beaudin et al., 2016). To directly test whether precursors of adult HSCs were being labeled in the Mds1CreERT2 mouse model, E9.5 TAM-treated pregnancies containing Mds1CreERT2 RosaLSL-YFP fetuses were allowed to come to term. After 1 year, these mice were analysed for YFP+ cells both in blood and in the bone marrow. YFP+ cells constituted nearly a third of the leukocytes in the blood, and similar proportions of bone-marrow immunophenotypic LT-HSCs, MPPs, more restricted hematopoietic progenitors, as well as kit-CD45+ cells (Figures 4A and 4B; Figure S4A). Treatment with TAM at E8.5 did not lead to the labeling of hematopoietic cells in the adult, consistent with lack of preHSC labeling at E11.5 (Figure S2E).
Figure 4. TAM treatment at E9.5 of Mds1CreERT2 Rosa26LSL-YFP efficiently labels HSCs.
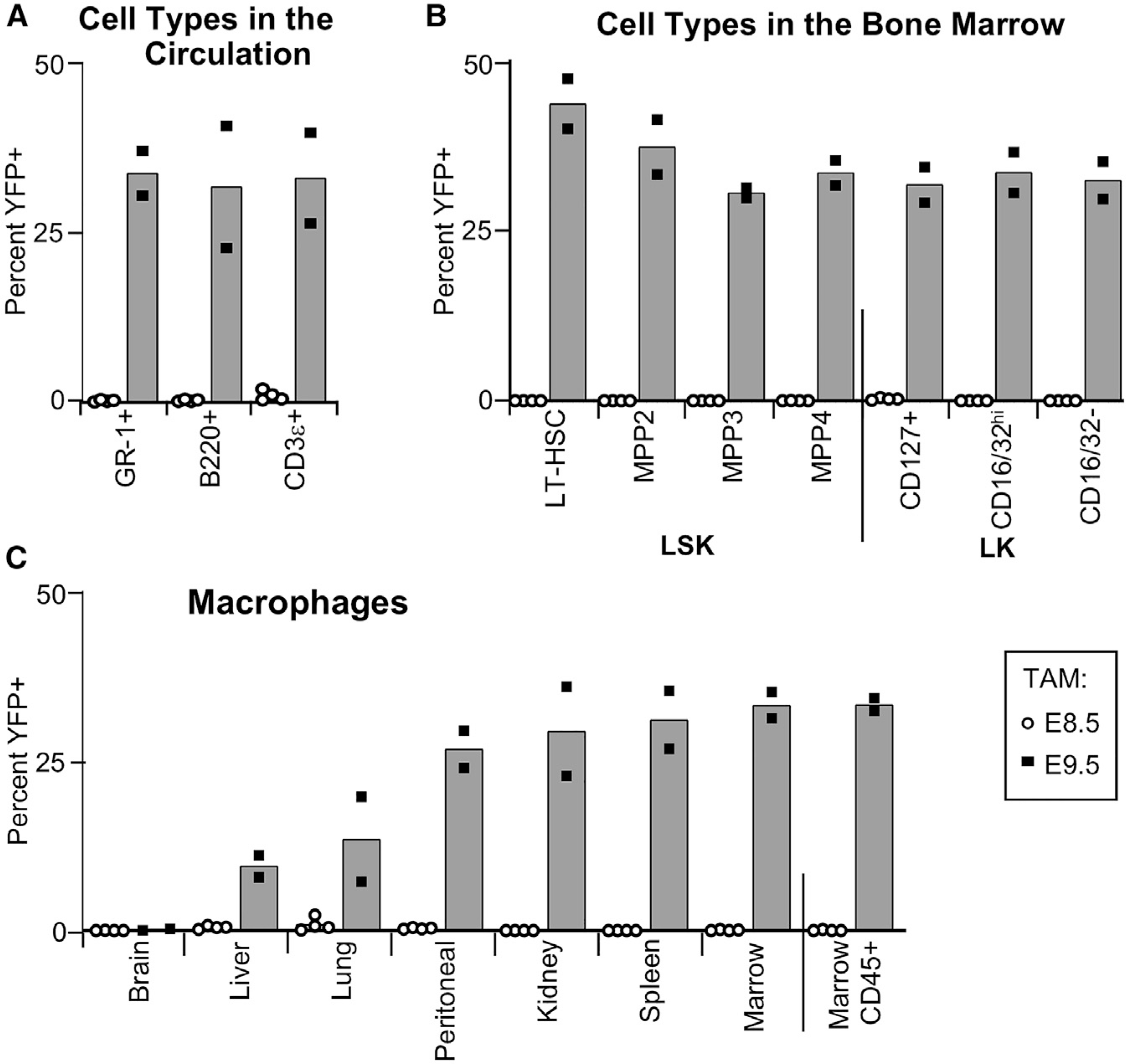
(A) Flow cytometric analysis of the proportion of YFP+ granulocyte/monocyte cells (GR-1+), B cells (B220+), and T cells (CD3ε+) in the circulation of 1-year-old Mds1CreERT2 Rosa26LSL-YFP mice after TAM treatment at E8.5 (empty circles, n = 4) or at E9.5 (solid circles, n = 2).
(B) Flow cytometric analysis of bone marrow of the same mice as in (A) demonstrating a high percentage of YFP+ progenitor cells in LSK subsets, as well as of lineage-restricted lymphoid (CD127+), myeloid (CD16/32+), and erythroid/megakaryocyte (CD16/32−) LK subsets. See Figure S4A for gating strategy. Total marrow CD45+ YFP+ cells shown in (C).
(C) Percentage of YFP+ F4/80+ tissue-resident macrophages in these mice. See Figure S4B for detailed gating strategy. In all panels, the average is plotted.
We also examined the contribution of HSCs to F4/80+ tissue-resident macrophage populations in the adult. As expected, there was no HSC contribution to microglia, and lower but increasing contribution to macrophages in liver, lung, and kidney (Figure 4C; Figure S4B). Surprisingly, a high proportion of F4/80+ macrophages in the spleen and bone marrow were labeled, indicating that they are predominately HSC-derived. Mds1CreERT2 Rosa26LSL-YFP activated by TAM at E8.5 did not lead to the labeling of any of these macrophage populations indicating that HSC-independent macrophages are not labeled by the Mds1creERT2 allele (Figure 4C).
DISCUSSION
HSCs, which transiently emerge in the embryo from hemogenic endothelial cells in large arterial vessels, serve as the foundational source of all circulating blood cells in the postnatal organism. A number of genes, including Mecom, Hlf, Procr, and HoxA family members, have been specifically associated with HSC emergence in mice (Iwasaki et al., 2010; Ng et al., 2016; Yokomizo et al., 2019). Mecom is one of the most differentially expressed genes present in hemogenic endothelium of the aorta compared both to hemogenic endothelium in the yolk sac and to non-hemogenic endothelial cells (Baron et al., 2018; Gao et al., 2018; Solaimani Kartalaei et al., 2015; Yokomizo et al., 2019). Recently, Mecom expression has been identified in primary murine aortic arterial-identity cells, as well as in their subsequent transition in pseudotime to hemogenic endothelium and preHSC identity cells (Hou et al., 2020; Zhu et al., 2020). The MECOM locus is complex, with multiple isoforms expressed from two TSS. Here, we report the development of an Mds1CreERT2 mouse model that contains a TAM-inducible Cre cassette within the first TSS of the MECOM locus. Utilizing an Mds1lacZ mouse (Zhang et al., 2011), we found expression of the LacZ reporter in a subset of cells of the E9.5 aorta that also express Runx1 (Figures 1G–1I), consistent with the reported expression of MECOM in aortic endothelial cells as they transition to hemogenic endothelium (Konantz et al., 2016; Yokomizo et al., 2019; Zhu et al., 2020, Hou, 2020 #42). Our data suggest that the MECOM locus is transiently expressed during HSC emergence and subsequently downregulated in the fetal liver, unlike its expression in LSK cells in adult mice (Zhang et al., 2011). Our RNA expression studies in adult tissues revealed that the two TSSs are coordinately regulated (Zhang et al., 2011), and analysis of publicly available transcription factor binding data reveals coordinate binding of transcription factors to the two TSSs, as well as similar histone markings and DNaseI hypersensitive sites (unpublished findings). However, the factors regulating transcription from the Mds1 and Evi1 promoters in adults or during development are poorly characterized and are an avenue for future research.
Subsets of myeloid and specialized lymphoid cells are formed in the fetus and self-renew postnatally with little input from adult HSCs (Gentek et al., 2018; Ghosn and Yang, 2015; Hashimoto et al., 2013). The overlapping emergence and lineage output of HSC-independent and HSC-derived hematopoiesis raise important questions regarding the identity and potential functional differences of their mature progeny. Exploring these questions will require ways to distinguish and isolate cells derived from different developmental sources. Current lineage-tracing approaches rely largely on the temporal induction of promoters, including Cdh5 (VECadherin), Runx1, Csf1r, and Kit, that are expressed both in HSC-independent progenitors and also in HSCs (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Soares-daSilva et al., 2021; Stremmel et al., 2018; Zovein et al., 2008). An alternative approach would be to use promoters for genes such as Hlf, Procr, or Mecom that are expressed more specifically in fetal HSCs (Iwasaki et al., 2010; Yokomizo et al., 2019; Yuasa et al., 2005). Although a caveat common to all of these systems, including Mds1CreERT2, is that it remains unclear to what degree rare lymphoid or lympho-myeloid progenitors might also be labeled. Our studies reveal the usefulness of the Mds1CreERT2 Rosa26LSLYFP mouse model that labels HSCs as they first emerge from hemogenic endothelium, but does not label the major HSC-independent waves of primitive hematopoietic progenitors and EMPs (Figure S4C).
EMPs begin to colonize the fetal liver between E9.5 and E10.5 and rapidly provide differentiating progeny, particularly the first circulating definitive erythrocytes (McGrath et al., 2015; Palis et al., 1999). Transplantable HSCs subsequently start to seed the liver between E11.5 and E12.5; however, the transition between these two sources of blood cells has only recently begun to be scrutinized (Ema and Nakauchi, 2000; Kumaravelu et al., 2002; Soaresda-Silva et al., 2021). Our analysis of TAM-treated Mds1CreERT2Rosa26LSL-YFP embryos indicates that HSCs have begun to generate small numbers of differentiating erythroid, myeloid, and lymphoid progeny between E12.5 and E14.5. By E16.5, a significant proportion of differentiating erythroid, myeloid, and B cell lineage cells in the liver are HSC-derived, and HSCs have now begun to contribute erythroid and myeloid cells to the rapidly expanding pool of circulating blood cells. A similar result was reported in a recent paper that found increased HSC contribution to hematopoiesis from E14.5 to E16.5 (Soaresda-Silva et al., 2021). These authors also observed a strong lineage bias, concluding that erythropoiesis is HSC independent until birth, which we did not observe. This could possibly be due to the challenges of using different lineage-tracing mouse models. Our data support the conclusion that HSCs contribute increasing proportions of erythroid, myeloid, and lymphoid cells to the rapidly developing late gestation murine fetus, supplementing the robust blood cell output of EMPs prior to birth.
Many tissue-resident macrophage populations in adult organs contain contributions of fetal cells that persist by self-renewal. These self-renewing macrophage populations were thought to derive primarily from HSC-independent sources. Here, we used the Mds1CreERT2Rosa26LSL-YFP model to examine the contribution of HSCs to F4/80+ macrophage populations in several organs over developmental time. As expected, we found no evidence of HSC contribution to microglia, but increasing contributions of HSCs to macrophages in liver, lung, and kidney (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Liu et al., 2019). Interestingly, the majority of F4/80+ cells in the adult spleen and bone marrow, though also self-maintaining (Hashimoto et al., 2013), appear to be predominantly derived from fetal HSCs (Figure 4C). Macrophage cells in the marrow perform multiple functions, including regulation of the HSC niche, support of erythroid precursor maturation in erythroblastic islands, and clearance of pyrenocytes. The surprisingly high proportion of HSC-derived F4/80+ cells in the fetal compared to the adult liver may reflect the need for those macrophage cells to also fulfill these specific functions in the fetus.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to Archibald Perkins (archibald_perkins@urmc.rochester.edu).
Material availability
Mds1lacZ mice (also referred to as Mds1m1 mice) have been submitted to MMRRC (Mds1-lacZ, Stock No. MMRRC:68126); please contact the MMRRC regarding availability. Mds1CreERT2 mice (also referred to as Mds1m2 mice) have been submitted to Jackson Labs for distribution (Mds1-CreERT2, Stock No. 032863). Please contact Jen Merriman regarding availability (Jen.Merriam@jax.org). There are no restrictions on the distribution of these lines.
Data and code availability
Any data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code.
Any additional information requires to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mds1lacZ mice have been described previously (Zhang et al., 2011). Mds1CreERT2 (also referred to as Mds1m2) mice were generated as described below. Rosa26-YFP reporter mice (Srinivas et al., 2001) Gt(ROSA)26Sortm1(EYFP)Cos/J, Stock No: 006148, abbreviated Rosa26YFP) and C57BL/6 were obtained from Jackson Labs. Both female and male embryos and adults were analyzed in these experiments. For timed pregnancies, dams were aged 6–16 weeks; sires were aged 7–60 weeks. For bone marrow transplantation, recipient mice were aged 8–12 weeks. All animals were housed and cared for in facilities operated by the Division of Animal Care at the University of Rochester Medical Center and used in accordance with the guidelines of the University of Rochester School of Medicine and Dentistry Institutional Animal Care and Use Committee.
METHOD DETAILS
Construction of Mds1CreERT2 allele
Plasmid construction was carried out using BAC recombineering (Liu et al., 2003). Bacterial artificial chromosome for Mecom (clone RP24–120B18) was introduced via electroporation into bacterial strain DY380 to create GS1650. The ApSacB cassette was then inserted into exon 1 of Mecom destroying the exon’s splice donor to create GS1656. The CreERT2 cassette (Metzger et al., 1995), derived from plasmid pXL119 (Zhao et al., 2007), together with a FRT-neo-FRT cassette, was inserted into GS1656, replacing the ApSacB cassette, to create GS1690. The PciI site of plasmid pL253 (Liu et al., 2003) was changed to an NsiI site, creating p1342. The NotI site of p1342 was altered to a FseI site, creating p1343. The NsiI fragment of GS1690 was then transferred into p1343 to create the final plasmid, p1347, which was linearized with FseI and submitted to the Genome Editing Facility at the University of Rochester, which created embryonic stem cell clones that were screened by Southern blotting, cutting DNAs with EcoRV for both the 5° and 3° hybridization probes. Ten positives were identified from 192 clones, one of which was used to create mice. These were then crossed with a FLPE deleter strain (Gt(ROSA)26Sortm1(FLP1)Dym/RainJ) to delete the Frt-neo-Frt cassette; the resultant allele was designated Mds1CreERT2.
Time pregnancy and competitive BM transplantation
Time pregnancy experiments were setup using male Mds1+/CreERT2 Rosa26LSL-YFP/LSL-YFP crossed with Rosa26LSL-YFP or C57BL/6 females. Mice were mated overnight and the morning of vaginal plug detection was considered embryonic day (E) 0.3. Pregnant mice were treated with tamoxifen (TAM) freshly dissolved in corn oil at 10mg/ml and 1mg/mouse delivered by gavage at E7.5, E8.5 or E9.5, and harvested at E11.5, E12.5, E14.5 or E16.5 for tracing embryonic hematopoiesis, or harvested 12 months after birth for long term tracing experiments. For competitive repopulation assays, C57BL/6 recipient mice were given 10 Gy of radiation, split in two 5-Gy doses separated by 24 hours and injected i.v. with 2 × 106 E12.5 liver cells mixed with 100,000 C57BL/6 control bone marrow cells
Tissue collection and processing
For complete blood count (CBC) values, blood was collected by capillary action by holding a heparinized 147 mm glass blood collection tube to a razor-pricked mouse tail. CBC measurements on fresh blood samples were taken using a scil Vet ABC Plus+ hematology analyzer.
Marrow cells were collected from mouse tibias and femurs as follows. Mice were euthanized with carbon dioxide and cervical dislocation, and tibias and femurs were dissected free of muscle, tendons, and connective tissue (Amend et al., 2016) and placed into room temperature sterile phosphate-buffered saline (PBS). Marrow contents were flushed from femurs with 1 mL of PBS by cutting the bones at the distal ends of the central marrow cavity. Pooled marrow contents in PBS were washed once with PBS and used in subsequent hematopoietic analyses (see section below: Flow Cytometry).
Embryonic tissues were isolated from staged embryos were dissected from dams as follows. Dams were euthanized with carbon dioxide and cervical dislocation. Uteri were removed from the peritoneum and washed with several changes of phosphate-buffered saline (PBS). Embryos were dissected free of decidual tissues and Reichert’s membrane in IMDM/PBS with 0.2% delipidated BSA using #5 watchmakers forceps (Dumoxal; Electron Microscopy Science, Ft. Washington, PA). Vestiges of maternal blood cells were removed by multiple transfers of tissues into fresh medium during the dissection process. Somite stage embryos (E8.25-E11.5) were grouped according to somite number. Most of the blood was derived from the larger vessels of the embryo proper. At E > 11.5, the liver was isolated separately from the remainder of the embryo proper. Embryonic livers were dissociated with Type I Collagenase (Stem Cell Technologies, diluted 1:3 with Dulbecco’s Phosphate Buffered Saline (DPBS or PB2), with 0.1 g/L CaCl2, 0.3% BSA (Gemini Bio-Products) and 0.1% glucose with gentle pipetting (McGrath et al., 2011). Cells were counted and used for Flow Cytometry (see below) or transplantation into irradiated recipient mice (see Competitive BM transplantation section above).
Histochemical and β-galactosidase staining
Timed pregnancies between dams and sires of appropriate genotypes. Embryos at different stages were harvested and dissected clear of yolk sac, amniotic tissue, and placenta. β-galactosidase staining (Loughna and Henderson, 2007; Nagy et al., 2007; Sanes et al., 1986) of whole-mount embryos was performed as follows: Embryos were fixed in 0.4% paraformaldehyde (in PBS, pH 7.4) on ice for 15 minutes for embryos up to E12.5 dpc and for 30 min for E13.5-E14.5. Following fixation, embryos were rinsed 3 times in PBS. Embryos were then stained in 2 mM MgCl2, 0.02% IGEPAL, 0.1 M phosphate buffer pH 7.2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.01% sodium deoxycholate, and 1 mg/ml X-gal in a 24-well tissue culture plate, incubating at 37°C in a humidified tissue culture incubator until color develops (2–8 hr). After the staining is complete, embryos were rinsed 4–5 times in PBS, then post-fixed in 2% paraformaldehyde/PBS overnight at 4°C. Embryos were then stored in 70% ethanol. Whole-mount photography was performed on a Zeiss 9901 Stereo Zoom microscope. Embryos were then dehydrated, embedded in paraffin, sectioned, and counterstained with eosin. Slides were then dehydrated and coverslipped.
Runx1 Immunohistochemistry
β-galactosidase-stained sections were hydrated, treated with 3% hydrogen peroxide, and subjected to HIER with 10mM Citric acid pH6, 0.05% Tween. Sections were blocked in PBST with 5% BSA and 5% goat serum, incubated overnight at 4°C with Rabbit antiRunx1 (Abcam Ab92336), washed and incubated with an AF647 secondary antibody (Thermofisher A-21244). Sections were counterstained with DAPI and mounted in antifade Prolong Gold.
Flow cytometric analysis
All antibody used in this study are listed in Key Resources Table and combinations and gating are shown in Supplemental Figures. Flow cytometry was performed on a LSRii or LSR Fortessa X20 (BD Biosciences) and analyzed with FlowJo v10.3 software (TreeStar).
KEY RESOURCES TABLE.
| Reagent or resource | Source | Identifier |
|---|---|---|
|
| ||
| Antibodies | ||
| Hámster Anti-Mouse CD3e PE-CF594 Clone 145–2C11 | BD | (BD Biosciences Cat# 562286, RRID:AB_11153307) |
| Anti-Mouse CD3e PE Clone 145–2C11 | eBioscience | Cat# 17–0031-82 |
| Anti-Mouse CD3e APC-AF780 Clone 17A2 | eBioscience | Cat# 47–0032-82 |
| Rat Anti Mouse -CD11b APC Clone M1/70 | BD | (BD Biosciences Cat# 553312, RRID:AB_398535) |
| Rat Anti Mouse -CD11b APC-EF780 Clone M1/70 | eBioscience | Cat# 27–0112-82 |
| Rat Anti Mouse CD11b PEcy7 Clone M1/70 | BD | (BD Biosciences Cat# 561098, RRID:AB_2033994) |
| Anti-Mouse CD16/32 PE Clone 93 | eBioscience | Cat# 12–0161-82 |
| Anti-Mouse CD16/32 PEcy7 Clone 93 | eBioscience | Cat# 25–0161-81 |
| Rat Anti-Mouse CD16/CD32 APC-Cy7 Clone 2.4G2 | BD | (BD Biosciences Cat# 560541, RRID:AB_1645229) |
| Rat Anti-mouse CD19 APCeFluor780 Clone ID3 | eBioscience | Cat# 47–0193-82 |
| Rat Anti-mouse CD19 PE Clone ID3 | eBioscience | Cat# 12–0193-82 |
| Rat Anti-mouse CD19 PerCP-Cy5.5 Clone ID3 | eBioscience | Cat #25–0193-82 |
| Rat Anti-Mouse CD34 AF700 Clone RAM34 | BD | (BD Biosciences Cat# 560518, RRID:AB_1727471) |
| Rat Anti-Mouse CD45 PE Clone 30-F11 | eBioscience | Cat #12–0451-82 |
| Rat Anti-Mouse CD45R/B220 PE-Cy7 Clone RA3–6B2 | BD | (BD Biosciences Cat# 552772, RRID:AB_394458) |
| Armenian Hamster anti-Mouse CD48 PE, Clone: HM48–1, | eBioscience | Cat# 12–0481-81 |
| Armenian Hamster anti-Mouse CD48 APC-eFluor780, Clone: HM48–1, | eBioscience | Cat# 50–112-4034 |
| Rat Anti-Mouse CD115 (CSF1R) PE clone AFS98 | eBioscience | Cat# 12–1152-82 |
| Rat Anti-Mouse CD117 (Kit) PE-cy5, Clone: 2B8 | eBioscience | Cat# 50–140-92 |
| Rat Anti-Mouse CD117 (Kit) PE Clone: 2B8 | eBioscience | Cat #12–1171-82 |
| Rat Anti-Mouse CD117 (Kit) PECF594 Clone: 2B8 | BD | (BD Biosciences Cat# 562417, RRID:AB_11154233) |
| Rat Anti-Mouse CD117 (Kit) PE-cy7, Clone: 2B8 | eBioscience | Cat #25–1171-82 |
| Rat Anti-Mouse CD127 BUV737 Clone SB/199 | BD | (BD Biosciences Cat# 612841, RRID:AB_2870163) |
| Rat Anti-Mouse CD127 APC clone A7R34 | eBioscience | Cat#17–1271-82 |
| Rat anti-Mouse CD135 (Flt3) PE, Clone: A2F10 | eBioscience | Cat# 50–106-20 |
| Rat anti-Mouse CD135 (Flt3) PE-CF594, Clone: A2F10 | BD | (BD Biosciences Cat# 562537, RRID:AB_2737639) |
| Rat anti-Mouse CD144 (VECadherin) EF660 Clone BV13 | eBioscience | Cat#50–106-20 |
| Rat anti-Mouse CD150 APC Clone: 9D1 | eBioscience | Cat# 50–150-87 |
| Rat Anti-mouse F4/80 APC Clone BM8 | eBioscience | Cat #17–4801-82 |
| Rat Anti-Mouse GR1 (Ly-6G and Ly6C) APC-Cy7 Clone RB6–8C5 | BD | (BD Biosciences Cat# 557661, RRID:AB_396775) |
| Rat Anti-mouse Ly6C APC Clone HK1.4 | eBioscience | Cat #17–5932-82 |
| Rat Anti-mouse Ly6C PEcy7 Clone HK1.4 | eBioscience | Cat #25–5932-82 |
| Rat antimouse Sca-1 (Ly-6A/E) PerCP-Cy5.5, Clone: D7 | eBioscience | Cat# 50–158-66 |
| Rat Anti-Mouse Ter119 Percp cy5.5 Clone TER-119 | BD | (BD Biosciences Cat# 560512, RRID:AB_10561844) |
| Rat Anti-Mouse Ter119 APC-eFluor780 Clone TER-119 | eBioscience | Cat #47–5921–80 |
| Rabbit anti-Runx | Abcam | (Abcam Cat# ab92336, RRID:AB_2049267) |
| Goat anti-rabbit IgG AF647 | ThermoFisher | (Thermo Fisher Scientific Cat# A-21244, RRID:AB_2535812) |
| ProLong Gold antifade reagent | ThermoFisher | Cat # P36934 |
|
Bacterial and Virus Strains | ||
| bacterial line DY380 | Neal Copeland (Liu et al., 2003) | n/a |
|
Chemicals, Peptides, and Recombinant Proteins | ||
| PE-CF594 Streptavidin | BD Biosciences | Cat# 562318 |
| Tamoxifen (CAS 10540–29–1) | Santa Cruz Biotechnology | Cat# sc-208414 |
| DAPI | Thermofisher | Cat# EN62248 |
|
Critical Commercial Assays | ||
| SuperScript IV One-Step RT-PCR System | Thermofisher | Cat# 12594025 |
| GoTaq | Promega | Cat# M7422 |
|
Experimental Models: Organisms/Strains | ||
| B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | Jackson Lab | Stock No: 006148 |
| B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ | Jackson Lab | Stock No: 009086 |
| Mds1 CreEr | This paper | n/a |
| Mds1 LacZ | This paper | n/a |
| C57BL/6 wildtype Inbred mice | Charles River | C57BL/6NCrl |
|
Oligonucleotides | ||
| M2 wtfw GGTGTCCAAACTGACAATGC | This paper | n/a |
| M2 fw ACTCACCCTGAAGTTCTCAGG | This paper | n/a |
| M2 rv CGGAGTTGCCACAGCTGG | This paper | n/a |
| YFP fw AAAGTCGCTCTGAGTTGTTAT | This paper | n/a |
| YFP rv AAGACCGCGAAGAGTTTGTC | This paper | n/a |
|
Recombinant DNA | ||
| BAC RP24–120B18 | BACPAC Resources Center (BPRC) | RP24–120B18 |
| GS1650 | This paper | n/a |
| GS1656 | This paper | n/a |
| GS1690 | This paper | n/a |
|
Software and Algorithms | ||
| FlowJo 8.5.3 software (TreeStar) | BD | FJ8.5.3 |
QUANTITATION AND STATISTICAL ANALYSIS
Differences between groups were compared with unpaired two-tailed Student’s t test using Prism 7.0 (GraphPad Software, San Diego, CA). A two-tailed paired Student’s t test and a Two-Way ANOVA analyses with Bonferroni correction made when appropriate and p values indicated on figures. All experiments are representative of at least three independent experiments, unless otherwise noted.
Supplementary Material
Highlights.
HSCs express Mds1 as they emerge in the aorta of the murine fetus
Inducible Mds1Cre-ERT2 mice facilitate the lineage tracing of HSC-derived hematopoiesis
HSC increasingly contribute to hematopoiesis in the fetal liver after E12.5
The majority of macrophages in adult spleen and marrow are derived from HSCs
ACKNOWLEDGMENTS
Funding: NIH NCI R01CA175761 and R01CA120313 (to A.S.P.), NIH R01 DK119285 (to J.P.), NYSTEM Grant C029547 (to Y.Z.), and the Department of Pathology and Laboratory Medicine, University of Rochester Medical Center (to A.S.P. and Y.Z.). We thank Stacey Duemmel, Leah Vit, Emma R. Guilfoyle, and Anne Koniski for technical assistance and the URMC Flow Cytometry Core Facility for technical support.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109562.
REFERENCES
- Amend SR, Valkenburg KC, and Pienta KJ (2016). Murine Hind Limb Long Bone Dissection and Bone Marrow Isolation. J. Vis. Exp. Published online April 14, 2021. 10.3791/53936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asari S, Okada S, Ohkubo Y, Sakamoto A, Arima M, Hatano M, Kuroda Y, and Tokuhisa T (2004). Beta-galactosidase of ROSA26 mice is a useful marker for detecting the definitive erythropoiesis after stem cell transplantation. Transplantation 78, 516–523. [DOI] [PubMed] [Google Scholar]
- Azzoni E, Frontera V, McGrath KE, Harman J, Carrelha J, Nerlov C, Palis J, Jacobsen SEW, and de Bruijn MF (2018). Kit ligand has a critical role in mouse yolk sac and aorta-gonad-mesonephros hematopoiesis. EMBO Rep. 19, e45477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balounová J, Šplíchalová I, Dobešová M, Kolář M, Fišer K, Procházka J, Sedlacek R, Jurisicova A, Sung HK, Kořínek V, et al. (2019). Toll-like receptor 2 expression on c-kit+ cells tracks the emergence of embryonic definitive hematopoietic progenitors. Nat. Commun. 10, 5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron CS, Kester L, Klaus A, Boisset J-C, Thambyrajah R, Yvernogeau L, Kouskoff V, Lacaud G, van Oudenaarden A, and Robin C (2018). Single-cell transcriptomics reveal the dynamic of haematopoietic stem cell production in the aorta. Nat. Commun. 9, 2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin AE, Boyer SW, Perez-Cunningham J, Hernandez GE, Derderian SC, Jujjavarapu C, Aaserude E, MacKenzie T, and Forsberg EC (2016). A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell Stem Cell 19, 768–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter MA, Neilly ME, Le Beau MM, Pearson MG, and Rowley JD (1985). Rearrangements of chromosome 3 involving bands 3q21 and 3q26 are associated with normal or elevated platelet counts in acute nonlymphocytic leukemia. Blood 66, 1362–1370. [PubMed] [Google Scholar]
- Böiers C, Carrelha J, Lutteropp M, Luc S, Green JC, Azzoni E, Woll PS, Mead AJ, Hultquist A, Swiers G, et al. (2013). Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 13, 535–548. [DOI] [PubMed] [Google Scholar]
- Christodoulou C, Spencer JA, Yeh SA, Turcotte R, Kokkaliaris KD, Panero R, Ramos A, Guo G, Seyedhassantehrani N, Esipova TV, et al. (2020). Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H, and Nakauchi H (2000). Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95, 2284–2288. [PubMed] [Google Scholar]
- Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley JD, and Nucifora G (1996). Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl. Acad. Sci. USA 93, 1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame JM, Fegan KH, Conway SJ, McGrath KE, and Palis J (2016). Definitive Hematopoiesis in the Yolk Sac Emerges from Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. Stem Cells 34, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Tober J, Gao P, Chen C, Tan K, and Speck NA (2018). RUNX1 and the endothelial origin of blood. Exp. Hematol. 68, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Chen C, Howell ED, Li Y, Tober J, Uzun Y, He B, Gao L, Zhu Q, Siekmann AF, et al. (2020). Transcriptional regulatory network controlling the ontogeny of hematopoietic stem cells. Genes Dev. 34, 950–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, Launay P, Chen J, Ginhoux F, and Bajénoff M (2018). Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 48, 1160–1171. [DOI] [PubMed] [Google Scholar]
- Ghosn EE, and Yang Y (2015). Hematopoietic stem cell-independent B-1a lineage. Ann. N Y Acad. Sci. 1362, 23–38. [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, and Rodewald HR (2015). Tissue-resident macrophages originate from yolk-sac-derived erythromyeloid progenitors. Nature 518, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, Chiba S, and Kurokawa M (2008). Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell 3, 207–220. [DOI] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. (2013). Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenauer T, and Moore AW (2012). The Prdm family: expanding roles in stem cells and development. Development 139, 2267–2282. [DOI] [PubMed] [Google Scholar]
- Hou S, Li Z, Zheng X, Gao Y, Dong J, Ni Y, Wang X, Li Y, Ding X, Chang Z, et al. (2020). Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 30, 376–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt PR, Bartholomew C, Davis AJ, Yutzey K, Gamer LW, Potter SS, Ihle JN, and Mucenski ML (1997). The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech. Dev. 65, 55–70. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Arai F, Kubota Y, Dahl M, and Suda T (2010). Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood 116, 544–553. [DOI] [PubMed] [Google Scholar]
- Juneja SC, Vonica A, Zeiss C, Lezon-Geyda K, Yatsula B, Sell DR, Monnier VM, Lin S, Ardito T, Eyre D, et al. (2014). Deletion of Mecom in mouse results in early-onset spinal deformity and osteopenia. Bone 60, 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley PD, Malik J, Fantauzzo KA, and Palis J (2004). Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 104, 19–25. [DOI] [PubMed] [Google Scholar]
- Konantz M, Alghisi E, Müller JS, Lenard A, Esain V, Carroll KJ, Kanz L, North TE, and Lengerke C (2016). Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. 35, 2315–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaravelu P, Hook L, Morrison AM, Ure J, Zhao S, Zuyev S, Ansell J, and Medvinsky A (2002). Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 129, 4891–4899. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, and Copeland NG (2003). A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gu Y, Chakarov S, Bleriot C, Kwok I, Chen X, Shin A, Huang W, Dress RJ, Dutertre CA, et al. (2019). Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell 178, 1509–1525. [DOI] [PubMed] [Google Scholar]
- Loughna S, and Henderson D (2007). Methodologies for staining and visualisation of beta-galactosidase in mouse embryos and tissues. Methods Mol. Biol. 411, 1–11. [DOI] [PubMed] [Google Scholar]
- Luis TC, Luc S, Mizukami T, Boukarabila H, Thongjuea S, Woll PS, Azzoni E, Giustacchini A, Lutteropp M, Bouriez-Jones T, et al. (2016). Initial seeding of the embryonic thymus by immune-restricted lympho-myeloid progenitors. Nat. Immunol. 17, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, and Yoder MC (2008). All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood 111, 3435–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Robin C, Ottersbach K, and Dzierzak E (2002). The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem Cells 20, 514–521. [DOI] [PubMed] [Google Scholar]
- McGrath KE, Frame JM, Fromm GJ, Koniski AD, Kingsley PD, Little J, Bulger M, and Palis J (2011). A transient definitive erythroid lineage with unique regulation of the β-globin locus in the mammalian embryo. Blood 117, 4600–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, and Palis J (2015). Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 11, 1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, and Chambon P (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA 92, 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, and Ihle JN (1988). Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell 54, 831–840. [DOI] [PubMed] [Google Scholar]
- Mu J, Slevin JC, McCormick S, and Adamson SL (2008). In vivo quantification of embryonic and placental growth during gestation in mice using micro-ultrasound. Reprod. Biol. Endocrinol. 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski ML, Taylor BA, Ihle JN, Hartley JW, Morse HC 3rd, Jenkins NA, and Copeland NG (1988). Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol. Cell. Biol. 8, 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, and Behringer R (2007). Staining Whole Mouse Embryos for beta-Galactosidase (lacZ) Activity. CSH Protoc. 2007, pdb.prot4725. [DOI] [PubMed] [Google Scholar]
- Ng ES, Azzola L, Bruveris FF, Calvanese V, Phipson B, Vlahos K, Hirst C, Jokubaitis VJ, Yu QC, Maksimovic J, et al. (2016). Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 34, 1168–1179. [DOI] [PubMed] [Google Scholar]
- Palis J, and Yoder MC (2001). Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp. Hematol. 29, 927–936. [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, and Keller G (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073–5084. [DOI] [PubMed] [Google Scholar]
- Perkins AS, Mercer JA, Jenkins NA, and Copeland NG (1991). Patterns of Evi-1 expression in embryonic and adult tissues suggest that Evi-1 plays an important regulatory role in mouse development. Development 111, 479–487. [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, and Bryder D (2007). Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1, 428–442. [DOI] [PubMed] [Google Scholar]
- Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, and Pollard JW (2011). CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CM, Larson RA, Anastasi J, Winter JN, Thangavelu M, Vardiman JW, Rowley JD, and Le Beau MM (1990). t(3;21)(q26;q22): a recurring chromosomal abnormality in therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood 76, 2594–2598. [PubMed] [Google Scholar]
- Samokhvalov IM, Samokhvalova NI, and Nishikawa S (2007). Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 446, 1056–1061. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Rubenstein JL, and Nicolas JF (1986). Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 5, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-da-Silva F, Freyer L, Elsaid R, Burlen-Defranoux O, Iturri L, Sismeiro O, Pinto-do-Ó P, Gomez-Perdiguero E, and Cumano A (2021). Yolk sac, but not hematopoietic stem cell-derived progenitors, sustain erythropoiesis throughout murine embryonic life. J. Exp. Med. 218, e20201729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaimani Kartalaei P, Yamada-Inagawa T, Vink CS, de Pater E, van der Linden R, Marks-Bluth J, van der Sloot A, van den Hout M, Yokomizo T, van Schaick-Solernó ML, et al. (2015). Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med. 212, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremmel C, Schuchert R, Wagner F, Thaler R, Weinberger T, Pick R, Mass E, Ishikawa-Ankerhold HC, Margraf A, Hutter S, et al. (2018). Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat. Commun. 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, and Dzierzak E (2010). Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development 137, 3651–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, Watanabe N, Umemoto T, Matsuo J, Harai R, Kihara Y, Nakamura E, Tada N, Sato T, Takaku T, et al. (2019). Hlf marks the developmental pathway for hematopoietic stem cells but not for erythro-myeloid progenitors. J. Exp. Med. 216, 1599–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, Dorshkind K, and Yoder MC (2011). Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc. Natl. Acad. Sci. USA 108, 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Porayette P, Glosson NL, Conway SJ, Carlesso N, Cardoso AA, Kaplan MH, and Yoder MC (2012). Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood 119, 5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, Mucenski ML, Suda T, and Morishita K (2005). Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 24, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yzaguirre AD, de Bruijn MF, and Speck NA (2017). The Role of Runx1 in Embryonic Blood Cell Formation. Adv. Exp. Med. Biol. 962, 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, and Soriano P (1997). Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Stehling-Sun S, Lezon-Geyda K, Juneja SC, Coillard L, Chatterjee G, Wuertzer CA, Camargo F, and Perkins AS (2011). PR-domain-containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood 118, 3853–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhou X, Szabó N, Leitges M, and Alvarez-Bolado G (2007). Foxb1-driven Cre expression in somites and the neuroepithelium of diencephalon, brainstem, and spinal cord. Genesis 45, 781–787. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Gao P, Tober J, Bennett L, Chen C, Uzun Y, Li Y, Howell ED, Mumau M, Yu W, et al. (2020). Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 136, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. (2008). Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code.
Any additional information requires to reanalyze the data reported in this paper is available from the lead contact upon request.