Abstract
Expression of C/EBPα is required for differentiation of 3T3-L1 preadipocytes into adipocytes. Previous investigations indicated that transcription of the C/EBPα gene is sequentially activated during differentiation, initially by C/EBPβ and C/EBPδ and later by C/EBPα (autoactivation). These events are mediated by a C/EBP regulatory element in the promoter of the C/EBPα gene. This article presents evidence that members of the Sp family, notably Sp1, act repressively on the C/EBPα promoter prior to the induction of differentiation. Sp1 was shown to bind to a GC box at the 5′ end of the C/EBP regulatory element in the C/EBPα promoter and, in so doing, to competitively prevent binding to and transactivation of the promoter by the C/EBPs. One of the differentiation inducers methylisobutylxanthine (a cAMP phosphodiesterase inhibitor) or Forskolin, both of which increase the cellular cAMP level, causes down-regulation of Sp1. This decrease in Sp1 level early in the differentiation program appears to facilitate access of C/EBPβ and/or C/EBPδ to the C/EBP regulatory element and, thereby, derepression of the C/EBPα gene.
C/EBPα plays a pivotal role in adipocyte differentiation (7, 10, 23). C/EBPα not only is required for differentiation (18, 24) but is sufficient to initiate the differentiation program without the hormonal inducers normally required (19). When differentiation is triggered by hormonal inducers, growth-arrested preadipocytes reenter the cell cycle and undergo two to three rounds of mitotic clonal expansion (1, 7). As mitosis ceases, C/EBPα is expressed and then serves as a pleiotropic transcriptional activator of numerous adipocyte genes which, when coordinately expressed, contribute to acquisition of the adipocyte phenotype (3–5, 9, 11, 14).
When it became evident that C/EBPα is an important regulator of adipogenesis, our efforts were redirected toward identifying the cis-elements and their cognate trans-acting factors which control transcription of the C/EBPα gene (4, 13, 20, 27, 29). DNaseI footprinting of the proximal 5′ flanking region of the mouse C/EBPα gene revealed several binding sites for nuclear factors that are differentially expressed during adipocyte differentiation (4, 17). Dual repressor binding sites (26) and a C/EBP binding site (4) were identified in this region of the promoter. CUP (C/EBP undifferentiated protein), a nuclear protein that binds to these repressor binding sites, is expressed by undifferentiated preadipocytes but not by differentiated adipocytes (26, 28). Purification and characterization of CUP showed it to be an isoform of the transcription factor AP-2α, which appears to function as a repressor of the C/EBPα gene. Consistent with its apparent repressive role in adipocyte differentiation, expression of CUP/AP-2α is down-regulated concurrently with the transcriptional activation of the C/EBPα gene (13). Moreover, CUP/AP-2α was found to transinhibit reporter gene expression mediated by the C/EBPα promoter (13). With respect to the C/EBP binding site, it was shown that C/EBPα, C/EBPβ, and C/EBPδ can bind to and trans-activate reporter gene expression mediated by the C/EBPα gene promoter (20–22). Evidence suggests that C/EBPβ and C/EBPδ, which are expressed early in the differentiation program (2, 32), initially activate transcription of the gene (23) and that C/EBPα, which is expressed later in the program, autoactivates transcription of the gene as the cells undergo terminal differentiation (20, 23).
Many adipocyte gene promoters possess functional C/EBP binding sites that mediate transcriptional activation by C/EBPα (7, 10, 23). It has been shown that these sites are DNaseI footprinted by nuclear extract from adipocytes, which express C/EBPα, but not by nuclear extracts from preadipocytes, which do not (5, 11, 14). An exception to this pattern is seen with the C/EBPα promoter, which, as indicated above, also possesses a functional C/EBP binding site (5, 20). Unexpectedly, this promoter was found to be footprinted by nuclear extracts from both preadipocytes and adipocytes (5). However, the footprint by preadipocyte nuclear extract extended further 5′ than that by adipocyte nuclear extract. Closer examination of the nucleotide sequence of this binding site revealed a consensus Sp binding site at the 5′ end of the C/EBP site that is not found in the C/EBP binding sites of other adipocyte genes (5, 11, 14). In the present article, we show that Sp1 does in fact bind at this site and in so doing obscures the C/EBP binding site, thereby preventing C/EBPβ or C/EBPα from binding. Evidence which suggests that early in the differentiation program Sp1 is down-regulated, thereby allowing C/EBPβ to bind and to transcriptionally activate the C/EBPα gene, is presented. This and other evidence suggests that Sp1 may serve to maintain the C/EBPα gene in a repressed state prior to differentiation and thereby prevent premature expression of the gene.
MATERIALS AND METHODS
Cell culture and induction of differentiation.
3T3-L1 preadipocytes were propagated and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (vol/vol) calf serum as previously described (26). To induce differentiation, 2-day postconfluent (designated day 0) cells were treated with DMEM containing 10% (vol/vol) fetal bovine serum (FBS), 1 μg of insulin (INS) per ml, 1 μM dexamethasone (DEX), and 0.5 mM 3-isobutyl-1-methyl-xanthine (MIX) until day 2. Cells were then fed DMEM supplemented with 10% FBS and 1 μg of INS per ml for 2 days, after which they were fed every other day with DMEM containing 10% FBS. Adipocyte gene expression and acquisition of the adipocyte phenotype began on day 3 and was maximal by day 8.
EMSAs and DNaseI footprinting.
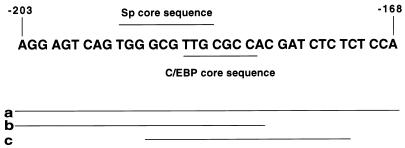
Nuclei were isolated, and nuclear extracts were prepared by using 1× NUN buffer (15) containing 0.3 M NaCl, 1 M urea, 1% Nonidet P-40, 25 mM HEPES (pH 7.9), and 1 mM dithiothreitol. Protein concentration was determined by the Bradford method (Bio-Rad). An electrophoretic mobility shift assay (EMSA) was performed essentially as described previously (21, 22), with the following modifications. Reaction mixtures containing ∼0.25 ng of 32P-labeled oligonucleotide, 2 μg of poly(dI-dC), and 10 μg of nuclear extract protein in 30 μl of buffer (10 mM HEPES, 0.1 mM EDTA, 5% glycerol, 100 mM NaCl, 0.3 M urea, 0.3% Nonidet P-40) were incubated on ice for 15 min and at room temperature for 15 min and were then separated electrophoretically on 5% polyacrylamide gels–0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA [pH 8.3]). For competition experiments, a 100-fold excess of unlabeled competitor oligonucleotide was added to reaction mixtures prior to the addition of labeled probe. For supershift experiments, 1 μl of antiserum (∼5 μg of immunoglobulin G protein) was added to the reaction mixture prior to the addition of labeled probe. For the experiments with limiting probe, the amount of labeled oligonucleotide probe was reduced to ∼1/30th of that used above. Recombinant Sp1 and C/EBPβ were obtained from Promega Corporation and S. McKnight (University of Texas Southwestern Medical Center), respectively. The labeled double-stranded oligonucleotide probes (derived from the 5′ flanking sequence of the C/EBPα gene) included a, b, c, and a-mut (probe a with a mutated Sp site) and had the following sequences: probe a, (−203) AGGAG T CAG TGGGCG T TGCGCCACGATC TC TCTCCA (−168 ) ; probe b, (−203)AGGAGTCAGTGGGCGTTGCGCCAC(−180); probe c, (−191)GCGTTGCGCCACGATCTCTC(−172); and probe a-mut, (−203)AGGAGTCAGTAGATCTTGCGCCACGATCTCTCTCCA(−168) (mutated nucleotides are underlined).
DNaseI footprinting.
Mouse 3T3-L1 preadipocytes were maintained and differentiated as described above. A 204-bp SmaI-StyI fragment of the C/EBPα gene (nucleotides [nt] −348 to −144) was 5′ end labeled on the noncoding strand. Nuclear extracts were prepared by the NUN method (described above) and desalted by passage through a PD-10 column (Pharmacia) previously equilibrated with buffer containing Sp1 storage buffer (Promega). DNaseI footprinting was performed in accordance with a protocol obtained from Promega (23a).
Gene constructs, mutations, and transfection.
The p468 C/EBPα gene promoter-reporter construct was prepared as previously described (27). Briefly, a 468-bp segment of the 5′ region of the C/EBPα gene (from nt −343 to +125) containing 343 bp of 5′ flanking sequence and the entire 5′ untranslated region was excised from pC/EBP9.7 (4) with SmaI and NcoI and inserted into the same sites of the pGL3-BA luciferase expression vector. The Sp site mutant in which the core Sp binding site sequence (GGGCG) was mutated to AGATC (mutated bases are underlined) was generated by PCR as previously described (6). The C/EBP site mutant, in which the core C/EBP binding site sequence (TTGCGC) was mutated to AGATCT, was generated by PCR as described above. The authenticity of the mutations was verified by sequencing. The C/EBPα expression vector (pMSV) was provided by S. McKnight (University of Texas Southwestern Medical Center). The cDNAs for C/EBPβ and Sp1 (provided by D. Nathans, Johns Hopkins University School of Medicine) were inserted into the pMT2 expression vector. The TK promoter-luciferase construct (pRL-TK) was from Promega Corp. The obese gene promoter (700 bp of 5′ flanking sequence)-luciferase construct was as previously described (11).
Transient transfections were performed on proliferating preconfluent (at 60 to 70% of confluent cell density) 3T3-L1 preadipocytes with 2 μg of the C/EBPα promoter-reporter construct (wild type or mutant) without or with 2 μg of the C/EBPβ or C/EBPα expression vector and without or with different amounts of the Sp1 expression vector. Transfections were performed by the calcium phosphate coprecipitation method (6). After 48 h, at which time the cells had reached confluence, cell extracts were prepared and assayed for luciferase activity.
Immunoblotting and alkaline phosphatase treatment.
To follow changes in the level of Sp1 protein following various treatments 2-day postconfluent (day 0) 3T3-L1 preadipocytes, cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting. Treatments with various agents (alone or in combination), including 0.5 mM MIX, 1 μM DEX, 1 μg of INS/ml, or 100 μM Forskolin alone or in combination, were performed. At various times thereafter, cell monolayers (6-cm dishes) were washed once with cold phosphate-buffered saline (pH 7.4) and then scraped into lysis buffer containing 1% SDS and 60 mM Tris-Cl (pH 6.8). Lysates were heated at 100°C for 10 min, clarified by centrifugation, and then subjected to immunoblotting with Sp1 antibody (Santa Cruz). To ascertain whether Sp1 undergoes phosphorylation or dephosphorylation when 3T3-L1 preadipocytes are induced to differentiate, cell lysates from day 0 (undifferentiated) cells or day 8 (fully differentiated) were subjected to alkaline phosphatase treatment. Cell lysates (∼200 μg of protein) were incubated for 30 min at 37°C without or with 100 U of calf intestinal alkaline phosphatase (Boehringer Mannheim) in a final volume of 60 μl, followed by the addition of 100 U of phosphatase and incubation for another 30 min under the same conditions. The reaction mixtures were then subjected to immunoblotting with anti-Sp1 antibody.
RESULTS
DNaseI footprinting of the C/EBP binding site in the C/EBPα promoter.
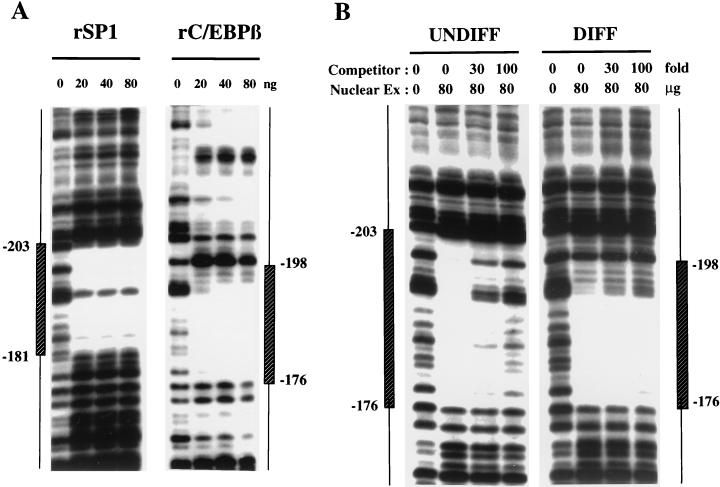
Many adipocyte gene promoters possess C/EBP binding sites that are DNaseI footprinted by nuclear extracts from adipocytes but not by those from preadipocytes (5, 9, 11, 14). This is consistent with the fact that C/EBPα is expressed by adipocytes but not preadipocytes and functions as a transactivator of these gene promoters during the adipocyte differentiation program (7, 23). In contrast, the C/EBPα gene promoter, which also possesses a functional C/EBP binding site, is footprinted by both preadipocyte and adipocyte nuclear extracts (Fig. 1 and reference 4). However, the footprint with preadipocyte nuclear extract (nt −203 to −176) differs from that (nt −198 to −176) with adipocyte nuclear extract in that it extends ∼5 bp further 5′ of the C/EBP binding site (Fig. 1). These findings suggested that another nuclear factor present in preadipocyte nuclear extract might at least in part be responsible for this footprinting pattern.
FIG. 1.
Nucleotide sequences in the C/EBPα gene promoter protected from DNaseI digestion (footprinted) by nuclear proteins from 3T3-L1 preadipocytes and adipocytes. Nuclear extracts were prepared from 3T3-L1 cells either maintained in the undifferentiated state (UNDIFF) as preadipocytes or induced to differentiate into adipocytes (DIFF). A 204-bp SmaI-StyI fragment (nt −348 to −144) of the C/EBPα gene promoter was incubated with increasing amounts (20 to 80 μg of protein) of nuclear extract and then subjected to digestion with DNaseI. The footprinted regions are indicated by vertical hatched boxes indicating the number of nucleotides from the transcriptional start site (determined by a sequencing gel run in parallel).
EMSA with oligonucleotides that contain the overlapping C/EBP and Sp binding sites present in the C/EBPα promoter.
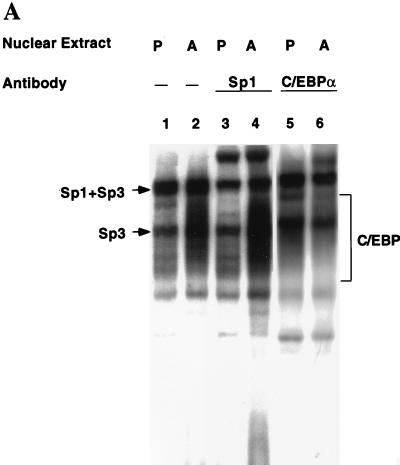
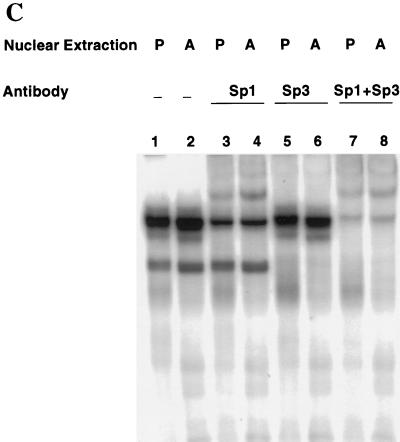
Unlike the C/EBP binding sites in other adipocyte gene promoters examined to date, the C/EBP binding site in the C/EBPα gene promoter possesses an overlapping Sp consensus sequence near its 5′ end (see Fig. 2). The possibility was considered, therefore, that the unique footprinting pattern with preadipocyte nuclear extract might be due to Sp1. To determine whether the factor present in preadipocyte nuclear extract that footprints the C/EBP binding site is Sp1, a 36-bp oligonucleotide (with excess oligonucleotide probe a; Fig. 2) encompassing this region was subjected to an EMSA with preadipocyte and adipocyte nuclear extracts. An array of nuclear protein-oligonucleotide complexes was produced (Fig. 3A, lanes 1 and 2), most of which are due to members of the C/EBP and Sp families of transcription factors. Consistent with previous findings (4, 21), a rather diffuse group of complexes (labeled C/EBP in Fig. 3A), attributable largely to homo- and heterodimers of the two C/EBPα isoforms (p42 and p30 [20]), is produced with adipocyte, but not preadipocyte, nuclear extract. This is indicated by the fact that most of these complexes were supershifted by anti-C/EBPα antibody (lanes 5 and 6). This region of the gel also contains small amounts of C/EBPα heterodimers with the two isoforms (LAP and LIP) of C/EBPβ (21). The slowest-moving complex (lanes 1 and 2 in Fig. 3A) is formed with both preadipocyte and adipocyte nuclear extract and is due primarily (70 to 80%) to Sp1, as most of this complex is supershifted by anti-Sp1 antibody (lanes 3 and 4). A smaller fraction of this complex and all of two other complexes contain Sp3, as judged by the fact that the equivalent complexes (with a probe containing the Sp site but lacking the C/EBP binding site) are partially or totally supershifted, respectively, by antibody against Sp3 (Fig. 3C).
FIG. 2.
Nucleotide sequence encompassing the C/EBP and Sp binding sites in the C/EBP α gene promoter. The consensus Sp core binding site and the consensus C/EBP core sequence for members of the C/EBP family are indicated. Lines labeled a, b, and c indicate the lengths of the labeled oligonucleotide probes used in the EMSAs.
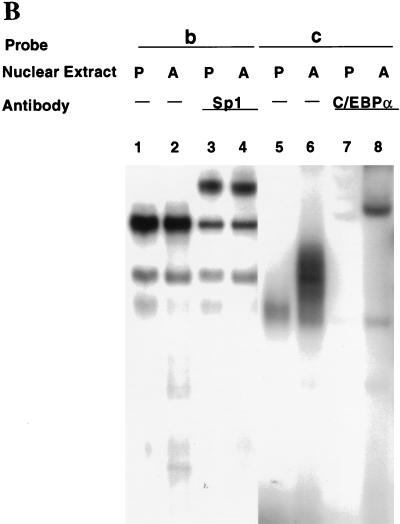
FIG. 3.
EMSA of oligonucleotides corresponding to the Sp and/or C/EBP binding sites in the C/EBPα gene promoter. (A) EMSA of preadipocyte (P) and adipocyte (A) nuclear extract and 32P-labeled oligonucleotide a (nt −203 to −168; see Fig. 2) encompassing both the Sp and C/EBP binding sites. In lanes 3 and 4, nuclear extracts were supershifted with antibody directed against Sp1 and in lanes 5 and 6 extracts were supershifted with antibody directed against C/EBPα. (B) EMSA of preadipocyte (P) and adipocyte (A) nuclear extract and 32P-labeled oligonucleotide b (nt −203 to −180, which encompass the Sp core binding site; Fig. 2) or oligonucleotide c (nt −191 to −172, which encompass the C/EBP binding site; Fig. 2). In lanes 3 and 4, nuclear extracts were supershifted with antibody directed against Sp1 and in lanes 7 and 8 the extracts were supershifted with antibody directed against C/EBPα. (C) EMSA of preadipocyte (P) and adipocyte (A) nuclear extract and 32P-labeled oligonucleotide b (nt −203 to −180, which encompass the Sp core binding site; Fig. 2). In lanes 3 and 4, nuclear extracts were supershifted with antibody directed against Sp1, in lanes 5 and 6, extracts were supershifted with antibody directed against Sp3, and in lanes 7 and 8, extracts were supershi with antibody directed against both Sp1 and Sp3. In all cases, a 100-fold excess of unlabeled oligonucleotide probes a, b, and c effectively competed away virtually all detectable protein–32P-labeled oligonucleotide complexes on the gels shown above (results not shown).
To verify these assignments, an EMSA was also performed with oligonucleotide probes for each of the overlapping Sp and C/EBP binding sites in the C/EBPα promoter, i.e., probes b and c (Fig. 2), respectively. As shown in Fig. 3B (lanes 1 and 2 versus lanes 3 and 4) most of the major (i.e., lowest-mobility) protein complex with the Sp site probe (probe b) was supershifted by anti-Sp1 antibody, the remainder being supershifted by anti-Sp3 antibody (Fig. 3C). The amounts of the Sp1- and Sp3-containing protein complexes did not differ significantly in preadipocyte and adipocyte nuclear extracts. In contrast, the amount of protein-oligonucleotide complex formed with the C/EBP-site probe (probe c) increased dramatically in terminally differentiated adipocytes (Fig. 3B; compare lanes 5 and 6), almost all of which was supershifted with anti-C/EBPα antibody (Fig. 3B; compare lanes 6 and 8). It can be concluded that Sp1, and to a lesser extent Sp3, can account for the binding observed at the Sp site (nt −203 to −180) which overlaps the 5′ end of the C/EBP binding site in the C/EBPα promoter.
Sp1 competes with members of the C/EBP family for binding to the C/EBPα gene promoter.
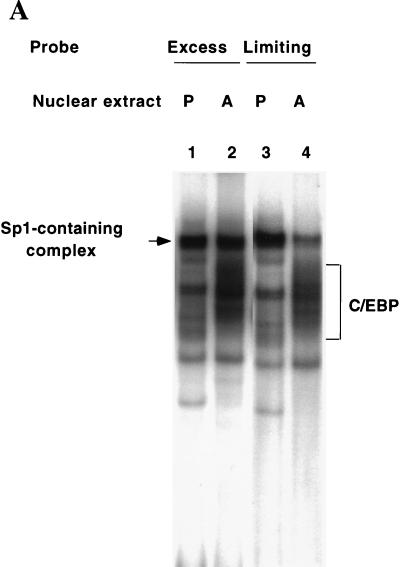
Since the Sp and C/EBP binding sites in the C/EBPα gene promoter overlap (Fig. 2), it seemed likely that binding of Sp1 and members of the C/EBP family would be mutually exclusive. Experiments in which the level of probe a (containing both binding sites) was reduced to a limiting concentration were conducted. Preadipocytes express Sp1 but not C/EBPα, while adipocytes express both Sp1 (as well as a smaller amount of Sp3) and C/EBPα. Therefore, the relative amount of Sp-containing protein-oligonucleotide complex should be lower with adipocyte than with preadipocyte nuclear extract, since the former contains “competitor” C/EBPα. Lowering the concentration of probe a by 30-fold while maintaining the level of preadipocyte and adipocyte nuclear extract constant markedly reduced the relative amount of Sp-containing complex (compare lane 1 relative to lane 2 and lane 3 relative to lane 4 in Fig. 4A). These findings suggested that C/EBPα competes with Sp1 and possibly Sp3 for binding to probe a and is dominant in nuclear extracts from the adipocyte.
FIG. 4.
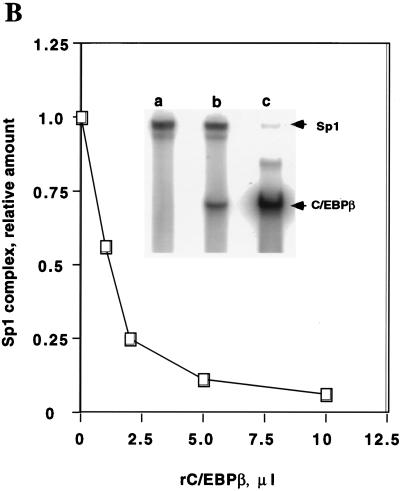
Bindings of C/EBPα, C/EBPβ, and Sp1 at the C/EBP and Sp sites in the C/EBPα gene promoter are mutually exclusive. (A) EMSA of preadipocyte (P) and adipocyte (A) nuclear extract and differing amounts (the standard level, 0.25 ng or the “limiting” level, 0.008 ng) of 32P-labeled oligonucleotide a (nt −203 to −168; see Fig. 2) which encompasses both the Sp and C/EBP binding sites. Exposure of film for lanes 1 and 2 was 12 h and for lanes 3 and 4 exposure was 96 h. (B) Effect of rC/EBPβ on the binding of rSp1 to oligonucleotide a. EMSA was conducted with a limiting amount (see above) of probe a, the amount of rSp1 was held constant (10 ng), and the amount of C/EBPβ (8.25 ng per μl) was increased as shown. The inset shows the autoradiograms for gels, corresponding to the 0, 1, and 10 μl levels of rC/EBPβ. (C) Effect of rSp1 on the binding of rC/EBPβ to oligonucleotide a. EMSA was conducted with a limiting amount (see above) of probe a, the amount of C/EBPβ was held constant (8.25 ng) and the amount of rSp1 (10 ng per μl) was increased as shown. The inset shows the autoradiograms for gels corresponding to the 0-, 1-, and 10-μl levels of rSp1 added.
To directly determine whether Sp1 interferes with binding of C/EBPβ to the C/EBPα gene promoter, the effect of rC/EBPβ on the binding of rSp1 to an oligonucleotide containing both the C/EBP and Sp binding sites, i.e., probe a, was tested by an EMSA. The effect of rSp1 on the binding of rC/EBPβ to the same oligonucleotide was also tested. As shown in Fig. 4B and C, increasing the concentration of either transcription factor caused decreased binding of the other. Similar results were obtained with rC/EBPδ (results not shown). (It should be noted that C/EBPα, -β, and -δ all bind to the C/EBP binding sites in the 422/aP2 and C/EBPα gene promoters [21, 22].) In other experiments, a 4-base mutation in the Sp regulatory element of probe a eliminated its activity as a competitor and prevented the formation of a protein-oligonucleotide complex with Sp1 or Sp3 (see below). Together, these findings demonstrate that members of the C/EBP family and Sp1 compete with one another for binding to probe a.
DNaseI footprinting experiments were conducted with recombinant Sp1 to verify that Sp1 per se binds to the Sp binding site at the 5′ end of the C/EBP regulatory element in the C/EBPα promoter. A segment of the proximal 5′ flanking region of the gene containing the overlapping Sp and C/EBP binding sites was subjected to DNaseI footprinting with rSp1 and with rC/EBPβ, which served as a representative of the C/EBP family. It should be noted that C/EBPα, C/EBPβ, and C/EBPδ are known to bind to C/EBP binding sites and to transactivate promoters containing these sites (7, 23). As illustrated in Fig. 5A, the regions footprinted by rSp1 and rC/EBPβ overlap. Moreover, the regions footprinted by both rSp1 (Fig. 5A) and preadipocyte nuclear extract (Fig. 1) are similar in that they both extend further (and to the same extent) 5′ than the regions footprinted by adipocyte nuclear extract (Fig. 1) or rC/EBPβ (Fig. 5A), which are virtually identical. The footprint of rSp1 does not, however, extend as far 3′ as that of preadipocyte nuclear extract (compare Fig. 1 with Fig. 5A). This difference in footprinting patterns could be due to covalent modification of Sp1 derived from preadipocytes which alters its conformation. Alternatively, it could be due to the interaction of Sp1 with another nuclear factor present in preadipocytes, but not present in the cells from which rSp1 is derived, which extends the footprint further 3′. Additional evidence that an Sp family member is primarily responsible for the footprinting pattern of preadipocyte nuclear extract is provided in Fig. 5B. Thus, preadipocyte nuclear extract but not adipocyte nuclear extract (whose footprint is due primarily to C/EBPα) is competed away by oligonucleotide b (Fig. 2), which contains the Sp binding site. These findings, together with the results of the gel shift experiments (Fig. 3 and 4), suggest that Sp1 (or Sp3) is primarily responsible for the footprinting pattern of the C/EBPα promoter and that in the preadipocyte, Sp1 or Sp3 might interfere with transactivation by members of the C/EBP family.
FIG. 5.
DNaseI footprint analysis of the C/EBPα proximal promoter. (A) DNaseI footprint of a segment (nt −348 to −144; cut with SmaI and StyI) of the 5′ flanking region of the C/EBPα gene was subjected to digestion with DNaseI in the presence of increasing amounts (20 to 80 ng) of rSp1 or rC/EBPβ. The footprinted regions are indicated by vertical boxes indicating the number of nucleotides from the transcriptional start site (determined by a sequencing gel run in parallel). (B) DNaseI footprint of a segment (nt −348 to −144) of the 5′ flanking region of the C/EBPα gene was subjected to digestion with DNaseI in the presence or absence of nuclear extract (80 μg of protein) from undifferentiated (UNDIFF) or differentiated (DIFF) 3T3-L1 cells and increasing amounts of unlabeled oligonucleotide corresponding to the Sp binding site (nt −203 to −180) in the C/EBPα promoter.
Sp1 blocks transactivation by C/EBPβ and C/EBPα of the C/EBPα gene promoter.
To determine whether the competition between Sp1 and C/EBPβ in binding to the overlapping Sp and C/EBP sites in the C/EBPα promoter observed in vitro occurs ex vivo (in preadipocytes), the effect of Sp1 on transactivation of the C/EBPα promoter by C/EBPβ or C/EBPα was investigated. A C/EBPα promoter-luciferase reporter gene construct containing the overlapping Sp and C/EBP binding sites was cotransfected into 3T3-L1 preadipocytes with either a C/EBPβ or C/EBPα expression vector along with increasing levels of an Sp1 expression vector. In the absence of Sp1, C/EBPβ or C/EBPα activated reporter gene expression by 12- to 15-fold (not shown). As shown in Fig. 6A, increasing levels of a Sp1 expression vector produced corresponding decreases of reporter gene expression. At the highest level of Sp1 expression vector, inhibition was ∼95%. To assess the specificity of the inhibitory effect of Sp1, another adipocyte gene promoter, i.e., the obese gene promoter, which contains a functional C/EBP binding site (11) but lacks an internal or adjacent Sp binding site, was tested. Cotransfection of a C/EBPα expression vector along with increasing levels of the Sp1 expression vector did not inhibit reporter gene expression. Rather, Sp1 activated reporter gene expression to a small extent (Fig. 6B). Similarly, Sp1 activated rather than inhibited reporter gene expression driven by the C/EBPα promoter in which the Sp site was mutated (see below). A positive control, i.e., a minimal TK promoter-luciferase construct (pTK-Luc; Promega), containing two proximal Sp sites, transfected into 3T3-L1 cells, produced “dose-dependent” transactivation by the Sp1 expression vector (2 and 4 μg of the expression vector produced ∼3- and ∼8-fold increases in reporter gene expression, respectively).
FIG. 6.
Effect of Sp1 on transactivation by C/EBPβ or -α mediated by the C/EBPα or obese gene promoters. (A) A C/EBPα promoter-luciferase reporter gene construct (2 μg) containing 343 bp of 5′ flanking sequence and 125 bp of 5′ untranslated sequence of the promoter was cotransfected into 3T3-L1 preadipocytes with a C/EBPβ or C/EBPα expression vector (2 μg) and increasing amounts of an Sp1 expression vector. After 48 h, luciferase assays with cell lysates were conducted. (B) An obese gene promoter-luciferase reporter gene construct (2 μg) containing 700 bp of 5′ flanking sequence of the promoter (containing a C/EBP regulatory element at nt −55) was cotransfected into 3T3-L1 preadipocytes with a C/EBPα expression vector (2 μg) and increasing amounts of an Sp1 expression vector. After 48 h, luciferase assays with cell lysates were conducted.
To verify that “transinhibition” by Sp1 occurs through the Sp site, which overlaps the C/EBP binding site in the C/EBPα promoter, this site was mutated. Initially, the effect of the mutation on Sp1 binding was determined by EMSA. As shown in Fig. 7A (lanes 1 and 2 versus lanes 5 and 6), mutation of the Sp site in probe a completely eliminated the complexes produced by preadipocyte or adipocyte nuclear extract attributable to Sp1 and Sp3 (Fig. 3A to C). Moreover, excess unlabeled mutant probe a-mut had no effect on binding at the Sp site (Fig. 7A; compare lanes 1 and 2 and lanes 3 and 4), but effectively competed away the complexes attributable to members of the C/EBP family, primarily C/EBPα (compare lanes 1 and 2 and lanes 3 and 4 in Fig. 7A; see also Fig. 3A and B). Probe a-mut, which has a mutation in the Sp site, had no effect on binding at the C/EBPα site as indicated by the persistence of the protein-oligonucleotide complexes ascribed to C/EBPα (compare lanes 2 and 6, Fig. 7A) and verified by supershifting with anti-C/EBPα antibody (compare lanes 6 and 8, Fig. 7A).
FIG. 7.
Effect of mutation of the Sp core element in the C/EBPα gene promoter on binding and inhibition of transactivation by Sp1. (A) An EMSA was performed with oligonucleotide a and a-mut (mutated in the Sp binding site; see Materials and Methods) with preadipocyte (P) or adipocyte (A) nuclear extract. A 100-fold excess of unlabeled a-mut was added during the binding reaction in lanes 3 and 4. Antibody against C/EBPα was used for the supershift experiments (lanes 7 and 8). (B) C/EBPα promoter-luciferase reporter gene constructs (2 μg) containing 343 bp of 5′ flanking sequence and 125 bp of 5′ untranslated sequence either mutated in the Sp binding site (■) or the C/EBP binding site (●) (see Materials and Methods) were cotransfected into 3T3-L1 preadipocytes with a C/EBPα expression vector (2 μg) and increasing amounts of an Sp1 expression vector. After 48 h, luciferase assays with cell lysates were conducted. (C) C/EBPα promoter-reported constructs (wild types or constructs mutated in the Sp site as in panel B) were cotransfected into 3T3-L1 preadipocytes with an Sp1 expression vector (1 μg) without or with different amounts of a C/EBPα expression vector. After 48 h, luciferase assays with cell lysates were conducted. (D) Effect of mutating the C/EBP and Sp binding sites in the C/EBPα promoter on reporter gene expression following induction of differentiation. Wild-type (wt) promoter-luciferase, C/EBP site-mutated promoter-luciferase (C/EBP Mut), and Sp site mutated promoter-luciferase (Sp Mut) constructs were transfected into D0 postconfluent 3T3-L1 preadipocytes. Twenty-four hours later, the cells were induced to differentiate by using the standard protocol, and after an additional 24 h, the cells were lysed and luciferase assays with the cell lysates were performed.
Mutation of the same 4 nt in the C/EBPα promoter-reporter gene construct completely prevented Sp1-mediated inhibition of transactivation by C/EBPα (compare Fig. 7B with Fig. 6A). Rather, Sp1 had a small additive activating effect on transactivation by C/EBPα, a pattern similar to that of another adipose-specific promoter (the obese gene promoter; Fig. 6B) that possesses a C/EBP binding site but lacks a Sp binding site. Furthermore, as shown in Fig. 7C, the magnitude of transactivation of the C/EBPα promoter by C/EBPα was increased by a factor greater than twofold by mutating the Sp site in 3T3-L1 preadipocytes, presumably as a result of release from inhibition by Sp1 with the mutant construct. Mutation of the C/EBP binding site in the promoter markedly decreased transactivation by members of the C/EBP family (results not shown). While as might be expected, Sp1 had no inhibitory effect as the C/EBPs (with which Sp1 competes with the wild-type promoter construct) could not bind and transactivate, Sp1 did have an small activating effect (Fig. 7B).
The effect of the Sp1 and C/EBP site mutations was also assessed during the early stage of the differentiation program. Thus, the wild-type promoter-luciferase, C/EBP site-mutated promoter-luciferase, and Sp site-mutated promoter-luciferase constructs were transfected into postconfluent (D0) 3T3-L1 preadipocytes. Twenty-four hours after transfection, the cells were induced to differentiate and after an additional 24 h, cell extracts were prepared and luciferase assays were performed. It should be noted that it is during this period that Sp1 levels and activity decline and C/EBPβ and -δ increase (though not yet maximally). As shown in Fig. 7D, reporter gene activity was increased approximately twofold 24 h after induction of differentiation, and mutation of the C/EBP binding site prevented this increase. Consistent with a repressive role for Sp1 during this time window, mutation of the Sp site gave rise to an approximately fivefold increase in reporter gene activity. These results support our proposal that Sp1 prevents transactivation of the C/EBPα promoter by C/EBPβ and/or -δ. Taken together, these results indicate that Sp1 acts as a competitor with the C/EBPs which interact with the C/EBP/Sp binding site in the C/EBPα promoter and suggest that Sp1 (and possibly Sp3) contributes to repression of the C/EBPα gene in the preadipocyte prior to induction of differentiation.
Effect of differentiation inducers on the phosphorylation, level, and binding activity of Sp1.
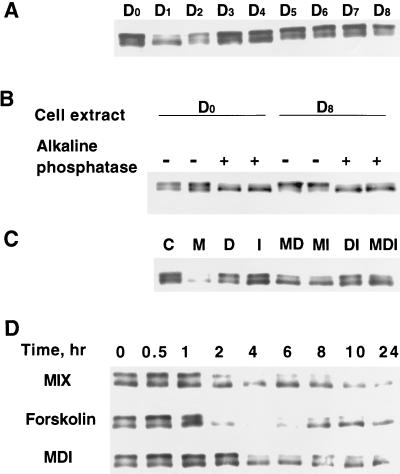
Given that Sp1 can repress the C/EBPα gene promoter, the question is raised as to whether the level and/or binding activity of Sp1 changes during the time window of the differentiation program during which transcription of the C/EBPα gene is initiated. To address this issue, the cellular level of Sp1 was monitored by immunoblotting during the course of the differentiation. Sp1 gives rise to two bands of ∼95 and 105 kDa by SDS-PAGE (Fig. 8A). Previous studies (12) have shown that the doublet pattern of Sp1 on SDS gels is due to differences in phosphorylation state, the more highly phosphorylated form exhibiting slower mobility. To verify that the doublet of Sp1 observed in Fig. 8A is due to a difference in phosphorylation state, cell extracts from day 0 and day 8 3T3-L1 preadipocytes and adipocytes were subjected to alkaline phosphatase treatment prior to SDS-PAGE. As shown in Fig. 8B phosphatase treatment caused both doublets to collapse into the higher-mobility form. This finding indicates that both phosphorylated and dephosphorylated forms of Sp1 were present.
FIG. 8.
Effect of adipocyte differentiation inducers on the level and phosphorylation state of Sp1. (A) Two-day postconfluent 3T3-L1 preadipocytes were induced to differentiate into adipocytes by using the standard differentiation protocol described in Materials and Methods. At different times (subscripts indicate to the number of days following induction of differentiation) after the induction of differentiation, cell lysates were subjected to SDS-PAGE and then Western blotted with anti-Sp1 antibody. (B) Cell lysates on day 0 and Day 8 were subjected to dephosphorylation by alkaline phosphatase treatment (see Materials and Methods). Following dephosphorylation, cell lysates were subjected to SDS-PAGE and Western blotting with anti-Sp1 antibody. (C) Two-day postconfluent 3T3-L1 preadipocytes were exposed to individual components of the differentiation inducer mixture either alone or in combination. After 24 h, cell lysates were analyzed as described for panel A. M, MIX; D, DEX; I, INS. (D) Two-day postconfluent 3T3-L1 preadipocytes were exposed to MIX, Forskolin, or MDI for the times indicated, after which cell lysates were analyzed as described for panel in A.
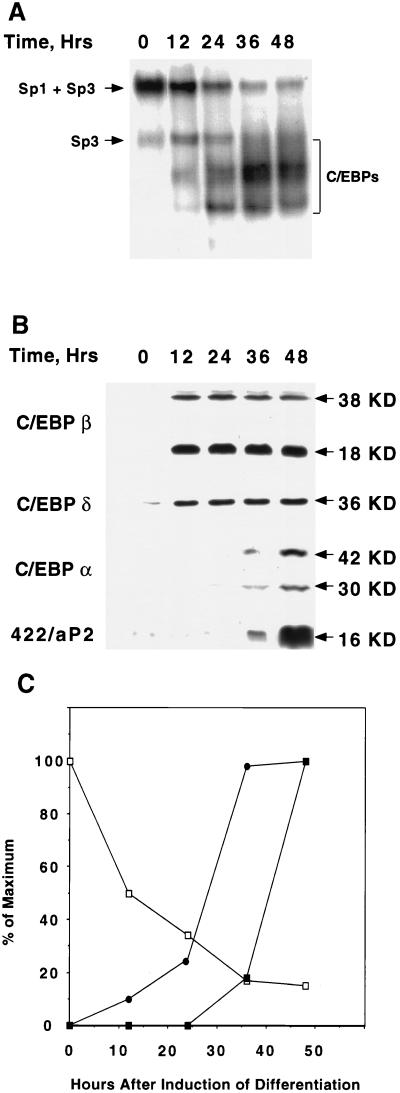
Of particular interest is the fact that the level of Sp1 (most notably the slower migrating and apparently more phosphorylated form) falls abruptly, i.e., on days 1 and 2, following exposure of preadipocytes to the differentiation inducers (MIX, INS, and DEX) (Fig. 8A). This decrease in Sp1 level is reflected in a decrease in binding activity (measured by EMSA with a limiting level of an oligonucleotide, i.e., probe a, which contains both the Sp and C/EBP binding sites found in the C/EBPα promoter) by nuclear extracts from preadipocytes following exposure to the differentiation inducers (Fig. 9). It is evident (Fig. 9A and C) that the amount of the low-mobility protein-oligonucleotide complex (due primarily to Sp1; see above) decreases and the complexes containing C/EBPβ and -δ followed by C/EBPα increase. The increases in the complexes due to members of the C/EBP family correspond well to the levels of the C/EBPs assessed by Western blotting (Fig. 9B and C). It should be noted that during differentiation, the level of Sp3 does not change significantly (results not shown); hence, the decrease in the amount of the low-mobility complex (Fig. 9A and C) appears to reflect the fall in the level and/or state of phosphorylation of Sp1 (Fig. 8A and D). By day 3, however, Sp1 returns to the preinduction level (Fig. 8A) while the level of C/EBPα remains high (5). It is not known at present why the return of Sp1 later in the differentiation program is not accompanied by a decrease in the expression of C/EBPα. Conceivably, C/EBPα becomes covalently modified late in the program, which increases its binding affinity. Further studies will be required to address this issue.
FIG. 9.
Changes in Sp1/Sp3 binding activity and expression of C/EBPβ, -δ, and -α following induction of differentiation. (A) An EMSA was performed with a limiting amount (0.004 ng) of an oligonucleotide (probe a) containing the Sp and C/EBP binding sites in the proximal C/EBPα gene promoter. An EMSA was performed with nuclear extracts from 3T3-L1 cells at 12, 24, 36, and 48 h after induction of differentiation. (B) Western blot analyses for C/EBPβ, -δ, and -α, and 422/aP2 using the cell lysates as described for panel A. (C) Changes in the binding activity of Sp1 + Sp3 (□) and C/EBPβ and -δ (●) and the expression of C/EBPα (■) at 12, 24, 36, and 48 h after induction of differentiation. Data are from panels A and B above.
To determine which of the differentiation inducers causes the changes in Sp1 level and phosphorylation state, each component was tested alone and in combination. As shown in Fig. 8C, MIX was found to be the inducer responsible for lowering the level of Sp1 after exposure for 24 h. MIX is an inhibitor of cAMP phosphodiesterase and is capable of increasing the cellular cAMP concentration of 3T3-L1 preadipocytes (12a). To determine whether cAMP per se has an effect on Sp1 level, the effect of Forskolin, a specific activator of adenyl cyclase (25), was compared with the effects of MIX and the complete mixture of the differentiation inducers MIX, DEX, and INS (MDI). As shown in Fig. 8D, Forskolin, like MIX and MDI, caused a rapid (within 2 to 4 h) decrease in both forms of Sp1. It can be concluded that an increase in cAMP following exposure of growth-arrested preadipocytes to the differentiation inducers, notably MIX, causes the decrease in the cellular level of Sp1.
DISCUSSION
The proximal promoter of the C/EBPα gene possesses overlapping Sp and C/EBP binding sites (Fig. 2). Previous studies (4, 20–22) have shown that members of the C/EBP family of transcription factors bind to and transactivate the C/EBPα promoter through the C/EBP element. As shown in the present article, the ubiquitous transcription factor Sp1 binds to the Sp site located at the 5′ end of the C/EBP binding site in the promoter (Fig. 2). In vitro binding experiments revealed that binding of Sp1 to the Sp site prevents binding to the C/EBP binding site by members of the C/EBP family, e.g., C/EBPα and C/EBPβ (Fig. 4; see also Fig. 9) as well as C/EBPδ (results not shown). Competition among members of the Sp and C/EBP families also occurs ex vivo in the 3T3-L1 adipocytes (Fig. 4A). Thus, transactivation of a C/EBPα promoter-reporter gene by C/EBPβ or C/EBPα was blocked by overexpression of Sp1 (Fig. 6A). Several lines of evidence show that the effect of Sp1 is mediated through the Sp site per se, rather than through an indirect effect on members of the C/EBP family. Thus, mutation of the Sp site prevents both binding and trans-inhibition by Sp1 (Fig. 7A and B) and actually increased the response to C/EBPα, presumably by eliminating competition by Sp1 (Fig. 7C). Since Sp1 is constitutively expressed at relatively high levels by preadipocytes and binds to the Sp site in the C/EBPα promoter with high affinity, it seems likely that this site would be occupied by Sp1 in the preadipocyte before induction of differentiation. These findings suggest that Sp1 acts as a repressor of the C/EBPα gene, and thereby of adipocyte differentiation. It should be noted that in other contexts Sp1 and specific C/EBPs can cooperate positively to activate transcription of a promoter. For example, C/EBPβ, but not C/EBPα, can act synergistically (in HepG2 cells) with Sp1 to activate transcription mediated by the CYP2D5 gene promoter, a promoter that contains both C/EBP and Sp binding sites (16).
Evidence from several laboratories suggests that several members of the C/EBP family of transcription factors participate in the adipocyte differentiation program through a cascade mechanism (2, 30–32). C/EBPβ and C/EBPδ are expressed early in the differentiation program preceding the expression of C/EBPα (2, 6a, 32). Moreover, all three of these C/EBPs have the capacity to bind and to transactivate the C/EBPα gene through a C/EBP binding site in the proximal promoter (20–22, 32). Therefore, it would be necessary for Sp1 to vacate the Sp site in the promoter at the appropriate point in the differentiation program. C/EBPβ and C/EBPδ are expressed within 8 h after the exposure of 3T3-L1 preadipocytes to differentiation inducers and are maximal by 14 to 16 h (Fig. 9). Expression of C/EBPα begins about 36 h after induction, when the levels of C/EBPβ and C/EBPδ are at their maximum (Fig. 9). However, as mentioned above, in the preadipocyte, the C/EBP binding site is obscured by Sp1 and must be vacated before C/EBPβ and C/EBPδ can bind. The present investigation suggests a mechanism by which the C/EBP binding site (with an Sp site at its 5′ end) may be vacated to allow transcriptional activation of the gene by C/EBPβ and C/EBPδ. We found that one of the differentiation inducers, notably MIX (a cAMP phosphodiesterase inhibitor) or Forskolin (or dibutyryl-cAMP; results not shown), triggers the decrease in the level of Sp1. This decrease in Sp1 level is accompanied by a decrease in Sp1 binding capacity as indicated by EMSA (Fig. 9A). Compelling evidence obtained with other cell systems indicates that cAMP increases the rate of turnover of Sp1 (8). Importantly, a decrease in the cellular level of Sp1 would allow C/EBPβ and C/EBPδ access to the C/EBP binding site in the C/EBPα gene promoter and thus, transactivation of the gene.
It is important that C/EBPα not be expressed prematurely, i.e., before preadipocytes undergo mitotic clonal expansion, since C/EBPα is antimitotic (28) and would prevent this critical mitosis. A delay in expression of the gene would guarantee that mitosis occurs before expression of C/EBPα. Considerable indirect evidence indicates that the “critical mitosis” is required for terminal differentiation (7, 23), presumably because changes in chromatin structure that accompany DNA replication allow access of cis-elements to trans-acting factors which activate (or derepress) transcription of the genes (e.g., the C/EBPα gene) that are necessary for differentiation.
We have shown that, in addition to negative regulation of the C/EBPα gene in the preadipocyte by Sp1, this gene is also controlled through another repression mechanism (13, 27). Previously, we identified dual repressive elements in the C/EBPα gene promoter and a transacting factor (CUP) expressed by preadipocytes but not adipocytes that binds to these elements (27). CUP was purified from nuclear extracts of 3T3-L1 preadipocytes. Amino acid sequencing and mass spectral analysis of the protein showed it to be an isoform of the transcription factor AP-2α (13). CUP/AP-2α is expressed by preadipocytes and during differentiation its expression decreases concomitantly with the transcriptional activation of the C/EBPα gene. Consistent with a repressive role of AP-2α/CUP, an AP-2α1 expression vector cotransfected with a C/EBPα promoter-reporter construct into 3T3-L1 adipocytes inhibits reporter gene transcription. Thus, two mechanisms guarantee that expression of the C/EBPα gene is repressed in the preadipocyte, repressed by Sp1 at the C/EBP binding site as reported in this article, and repressed by CUP/AP-2α at dual CUP repressor binding sites in the promoter and 5′ untranslated region of the gene (13, 27).
ACKNOWLEDGMENTS
This work was supported by a research grant from the National Institutes of Health (NIDDK).
REFERENCES
- 1.Bernlohr D A, Bolanowski M A, Kelly T J, Lane M D. Evidence for an increase in transcription of specific mRNAs during differentiation of 3T3-L1 preadipocytes. J Biol Chem. 1985;260:5563–5567. [PubMed] [Google Scholar]
- 2.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 3.Cheneval D, Christy R J, Geiman D, Cornelius P, Lane M D. Cell-free transcription directed by the 422 adipose P2 gene promoter: activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1991;88:8465–8469. doi: 10.1073/pnas.88.19.8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christy R J, Kaestner K H, Geiman D E, Lane M D. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 6.Cooke D W, Lane M D. A sequence element in the GLUT4 gene that mediates repression by insulin. J Biol Chem. 1998;273:6210–6217. doi: 10.1074/jbc.273.11.6210. [DOI] [PubMed] [Google Scholar]
- 6a.Cornelius, P., and M. D. Lane. Unpublished results.
- 7.Cornelius P, MacDougald O A, Lane M D. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- 8.Han I, Kudlow J E. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrera R, Ro H S, Robinson G S, Xanthopoulos K G, Spiegelman B M. A direct role for C/EBP and the AP-I binding site in gene expression linked to adipocyte differentiation. Mol Cell Biol. 1989;9:5331–5339. doi: 10.1128/mcb.9.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang C-S, Loftus T, Mandrup S, Lane M. Adipocyte differentiation and leptin expression. Annu Rev Cell Biol Dev. 1997;13:231–259. doi: 10.1146/annurev.cellbio.13.1.231. [DOI] [PubMed] [Google Scholar]
- 11.Hwang C-S, Mandrup S, MacDougald O M, Geiman D E, Lane M D. Transcriptional activation of the mouse obese gene by CCAAT/enhancer binding protein α. Proc Natl Acad Sci USA. 1996;93:873–877. doi: 10.1073/pnas.93.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 12a.Janicot, M., and M. D. Lane. Unpublished results.
- 13.Jiang M-S, Tang Q-Q, McLenithan J, Geiman D, Shillinglaw W, Henzel W J, Lane M D. Derepression of the C/EBPα gene during adipogenesis: identification of AP-2α as a repressor. Proc Natl Acad Sci USA. 1998;95:3467–3471. doi: 10.1073/pnas.95.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestner K H, Christy R J, Lane M D. Mouse insulin-responsive glucose transporter gene: characterization of the gene and trans-activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavery D J, Schibler U. Circadian transcription of the cholesterol 7α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y-H, Williams S C, Baer M, Sternbeck E, Gonzalez F J, Johnson P F. The ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legraverend C, Antonson P, Flodby P, Xanthopoulos K G. High level activity of the mouse CCAAT/enhancer binding protein (C/EBPα) gene promoter involves autoregulation and several ubiquitous transcription factors. Nucleic Acids Res. 1993;21:1735–1742. doi: 10.1093/nar/21.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin F-T, Lane M D. Antisense CCAAT/enhancer-binding protein RNA suppresses coordinate gene expression and triglyceride accumulation during differentiation of 3T3-L1 preadipocytes. Genes Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 19.Lin F-T, Lane M D. CCAAT/enhancer binding protein α is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin F-T, MacDougald O A, Diehl A M, Lane M D. A 30 kilodalton alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci USA. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDougald O A, Cornelius P, Lin F-T, Chen S S, Lane M D. Glucocorticoids reciprocally regulate expression of the CCAAT/enhancer-binding protein α and δ genes in 3T3-L1 adipocytes and white adipose tissue. J Biol Chem. 1994;269:19041–19047. [PubMed] [Google Scholar]
- 22.MacDougald O A, Cornelius P, Liu R, Lane M D. Insulin regulates transcription of the CCAAT/enhancer binding protein (C/EBP) α, β, and δ genes in fully-differentiated 3T3-L1 adipocytes. J Biol Chem. 1995;270:647–654. doi: 10.1074/jbc.270.2.647. [DOI] [PubMed] [Google Scholar]
- 23.MacDougald O A, Lane M D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 23a.Promega Corporation. Promega Technical Bulletin no. 137. Madison, Wis: Promega Corporation; 1997. [Google Scholar]
- 24.Samuelsson L, Stromberg K, Vikman K, Bjursell G, Enerback S. The CCAAT/enhancer binding protein and its role in adipocyte differentiation: evidence for direct involvement in terminal adipocyte development. EMBO J. 1991;10:3787–3793. doi: 10.1002/j.1460-2075.1991.tb04948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seamon K B, Daly J W. Forskolin: a unique diterpene activator of cyclic AMP-generating systems. J Cyclic Nucleotide Res. 1981;7:201–224. [PubMed] [Google Scholar]
- 26.Student A K, Hsu R Y, Lane M D. Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J Biol Chem. 1980;255:4745–4750. [PubMed] [Google Scholar]
- 27.Tang Q-Q, Jiang M-S, Lane M D. Repression of transcription mediated by dual elements in the CCAAT/enhancer binding protein α gene. Proc Natl Acad Sci USA. 1997;94:13571–13575. doi: 10.1073/pnas.94.25.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umek R M, Friedman A D, McKnight S L. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 29.Vasseur-Cognet M, Lane M D. CCAAT/enhancer binding protein α (C/EBPα) undifferentiated protein: a developmentally regulated nuclear protein that binds to the C/EBPα gene promoter. Proc Natl Acad Sci USA. 1993;90:7312–7316. doi: 10.1073/pnas.90.15.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Z, Bucher N L R, Farmer S R. Induction of peroxisome proliferator-activated receptor γ during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol Cell Biol. 1996;16:4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Xie Y, Bucher N L, Farmer S R. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–63. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 32.Yeh W-C, Cao Z, Classon M, McKnight S L. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]