ABSTRACT
Several viruses have been proven to inhibit the formation of RNA processing bodies (P-bodies); however, knowledge regarding whether enterovirus blocks P-body formation remains unclear, and the detailed molecular mechanisms and functions of picornavirus regulation of P-bodies are limited. Here, we show the crucial role of 2A protease in inhibiting P-bodies to promote viral replication during enterovirus 71 infection. Moreover, we found that the activity of 2A protease is essential to inhibit P-body formation, which was proven by the result that infection with EV71-2AC110S, a 2A protease activity-inactivated recombinant virus, failed to block the formation of P-bodies. Furthermore, we show that DDX6, a scaffolding protein of P-bodies, interacted with viral RNA to facilitate viral replication rather than viral translation, by using a Renilla luciferase mRNA reporter system and nascent RNA capture assay. Altogether, our data first demonstrate that the 2A protease of enterovirus inhibits P-body formation to facilitate viral RNA synthesis by recruiting the P-body components to viral RNA.
IMPORTANCE Processing bodies (P-bodies) are constitutively present in eukaryotic cells and play an important role in the mRNA cycle, including regulation of gene expression and mRNA degradation. The P-body is the structure that viruses manipulate to facilitate their survival. Here, we show that the 2A protease alone was efficient to block P-body formation during enterovirus 71 infection, and its activity is essential. When the assembly of P-bodies was blocked by 2A protease, DDX6 and 4E-T, which were required for P-body formation, bound to viral RNA to facilitate viral RNA synthesis. We propose a model revealing that EV71 manipulates P-body formation to generate an environment that is conducive to viral replication by facilitating viral RNA synthesis: 2A protease blocked P-body assembly to make it possible for virus to take advantage of P-body components.
KEYWORDS: processing body, 2A protease, enterovirus, DDX6/4E-T complex
INTRODUCTION
Processing bodies (P-bodies), one type of cytoplasmic ribonucleoprotein (RNP) granules, are present in eukaryotic cells under normal growth conditions. During P-body formation, the translation of messenger RNAs (mRNAs) within P-bodies is inhibited by the repression machinery, but the translation initiation factors and ribosomes are excluded from P-bodies (2, 3). Generally, a P-body is thought to be a highly dynamic and reversible structure without cytomembrane and to assemble by liquid-liquid phase separation (LLPS). The size and number of P-bodies increase in response to a variety of stresses, such as glucose deprivation, osmotic stress, and UV light (3). Hundreds of proteins contained in the P-body form a huge, complicated network, and the components have been identified as being critical for the formation of P-bodies, including the decapping enzyme complex DCP1/DCP2, the deadenylase complex Ccr4/Pop2/Not, and the mRNA translational repression complex (4E-T, Rap55, and DDX6) (4–6). More recently, microRNA (miRNA)-induced silencing complex (RISC) and nonsense-mediated mRNA decay (NMD) have been identified as being components of P-bodies (1, 7). Hubstenberger and colleagues proved that up to two-thirds of P-body proteins that play different roles during P-body formation bind RNA (7). Some of these RNA binding proteins decide the fate of their bound mRNA within P-bodies to undergo degradation or return to the cytoplasmic translation pool, showing the essential role of protein-RNA interactions during P-body formation. The interactions in the P-body network can be divided into the specific interactions between protein and protein, protein and RNA, and RNA and RNA, which are mediated by intrinsically disordered regions (IDRs) , well-folded domain, and the transient interaction such as π-π and cation-π (8). Although the details of P-body formation are limited, many previous studies have shown that the silencing of specific RNPs such as PAT1, GW182, and LSM14 and the destruction of some specific interactions between proteins (for example, the breaking up of the complex between DDX6 and 4E-T) lead to the failure of P-body assembly (9). Furthermore, thousands of mRNAs are assembled into P-bodies, and mRNA self-assembly is considered to be necessary for P-body assembly (10).
Based on these multiple components of the P-body, the P-body has been considered to be the site of trapped mRNA degradation, which starts when the poly(A) tail of mRNA is shortened by the deadenylase complex, and then the 5′ cap structure is removed by the decapping complex: afterward comes exonucleolytic degradation by the 5′-to-3′ exoribonuclease XRN1 (4, 11–14). Moreover, several recent studies show that mRNA located in the P-body can also be stored to reenter translation (2, 15). In addition, Parker et al. have proposed a hypothesis about the mRNA cycle in eukaryotic cells, which indicates the important role of P-bodies in mitigating the abnormal surge of RNA in the cytoplasm and maintaining cytoplasmic homeostasis (2, 14, 17–19).
Enterovirus 71 (EV71) belongs to the genus Enterovirus in the family Picornaviridae and is a main pathogen of hand-foot-and-mouth disease (HFMD), a disease that is usually mild but may be associated with a severe form of brainstem encephalitis, along with pulmonary edema and high case fatality rates (20, 21). In addition, EV71 is a nonenveloped virus with a capsid with icosahedral symmetry containing a nonsegmented, single-stranded, positive-sense genomic RNA that is about 7,500 nucleotides long with a single open reading frame (ORF) flanked by untranslated regions (UTRs) at the 5′ and 3′ termini. The genomic RNA is not only the template for translation, but it is the template for the synthesis of RNA; however, the exact temporal mechanisms of translation and transcription remain unclear. Translation of the RNA genome is driven by the internal ribosome entry site (IRES) located in the 5′ UTR but is not dependent on the eukaryotic cap structure, and the polyprotein is processed by 2A protease and 3C protease to form 11 viral proteins. Genome replication depends on negative strands synthesized by the RNA-dependent RNA polymerase 3D (22, 23).
As the P-body is involved in gene expression and mRNA degradation and plays an important role in the mRNA cycle, it is possible for viruses to manipulate the formation of P-bodies to interfere with cellular gene expression to generate an environment that is conducive to viral replication. Previous reports have shown that the formation of P-bodies was blocked during positive-sense, single-stranded RNA virus infection by disrupting or co-opting P-body components (24–26). Although it has been reported that 3C protease cleaves DCP1A, which leads to P-body foci being invisible during poliovirus (PV) infection (24), there is doubt about whether 3C protease plays a decisive role in inhibiting P-bodies during picornavirus infection. On one hand, a previous article showed the decrease of DCP1A has no effect on P-body formation (27). On the other hand, Harendra et al. found that the expression levels of the proteins contained in the P-body remained unchanged when West Nile virus (WNV), another positive-sense, single-stranded RNA virus belonging to the family of Flaviviridae, infects the host cells (25). In addition, the specific mechanism of picornavirus regulation of P-body assembly remains unknown.
In this study, we demonstrate that the 2A protease but not 3C protease is essential for EV71 inhibition of P-body formation. Besides, the infection of the 2A protease activity-inactivated recombinant virus EV71-2AC110S fails to block the formation of P-bodies, indicating the importance of the 2A protease activity. Furthermore, we provide experimental evidence of viral RNA (vRNA) taking advantage of P-body components to facilitate genome RNA synthesis during enterovirus infection.
RESULTS
EV71 infection inhibits the formation of P-bodies.
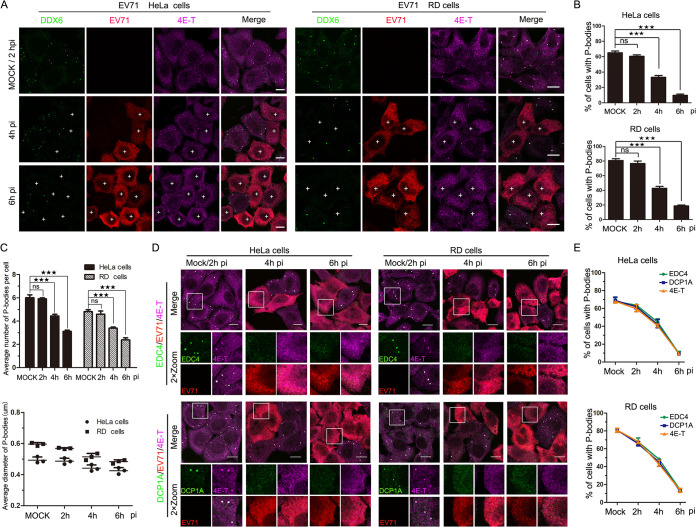
To investigate whether EV71 infection inhibited P-body formation, HeLa and RD (rhabdomyosarcoma) cells were continuously infected with EV71 for up to 6 h, and the distributions of P-body marker proteins DDX6 and 4E-T at different time points postinfection (p.i.) were analyzed (Fig. 1A). Antibody against EV71 was used to visualize EV71-infected cells. In mock-infected or EV71-infected cells for 2 h p.i. (hpi), when EV71 infection was undetectable by immunofluorescence (IF), the distributions of DDX6 and 4E-T had no significant changes, and they were colocalized and coassembled into foci that represent the P-bodies (Fig. 1A). As infection progressed, when 85% of cells were infected at 4 hpi and 95% at 6 hpi, DDX6 and 4E-T foci were undetected in EV71-infected cells, indicating the P-bodies failed to assembly in EV71-infected cells. To measure the change in formation of P-bodies, the percentage of cells containing P-bodies was analyzed, and the result showed that it had dropped to ∼35% in HeLa cells and ∼40% in RD cells at 4 hpi, and ∼10% in HeLa cells and ∼18% in RD cells at 6 hpi (Fig. 1B), and this ratio remained stable until 8 hpi (data not shown). To further quantitatively analyze the change in P-body formation, P-body size and the number of P-bodies per cell were analyzed, and the results showed the average number and size of P-bodies decreased as EV71 infection progressed (Fig. 1C). In addition, we also examined the distribution of DCP1A and EDC4, the other two typical P-body marker proteins, during EV71 infection in HeLa and RD cells and found that P-bodies failed to assembly in EV71-infected cells (Fig. 1D). The number of cells containing P-bodies decreased to about 40% at 4 hpi and to about 17% at 6 hpi (Fig. 1E). Taken together, the results indicated that the formation of P-bodies was blocked during EV71 infection and the blocking was not cell specific because similar results were observed in HeLa cells and RD cells.
FIG 1.
EV71 infection inhibits the formation of P-bodies. (A to C) HeLa and RD cells were mock infected or infected with EV71 (MOI of 10) under normal growth conditions for consecutive periods (0, 2, 4, and 6 h, with mock-infected cells converged at 2 h). (A) Cells were analyzed by IF after being immunostained for DDX6 (green), EV71 (red), and 4E-T (magenta). (B) Quantitative analysis of the percentage of cells containing P-bodies in panel A. (C) Quantitative analysis of the average number of P-bodies per cell and the average diameter of P-bodies in panel A. (D and E) HeLa and RD cells were mock infected or infected with EV71 (MOI of 10) under normal growth conditions for consecutive periods (0, 2, 4, and 6 h, with mock-infected cells converged at 2 h). (D) Cells were analyzed by IF after being immunostained for EV71 (red), 4E-T (magenta), and EDC4 (green) or DCP1A (green). (E) Quantitative analysis of the percentage of cells containing P-bodies in panel D. “+” indicates EV71-infected cells. Error bars represent the means ± SDs of values from three independent experiments (n = 3). A total of 100 cells and 50 P-bodies were counted each time. By Student’s test: ★★★, P < 0.001; ns, not significant. Scale bars, 10 μm.
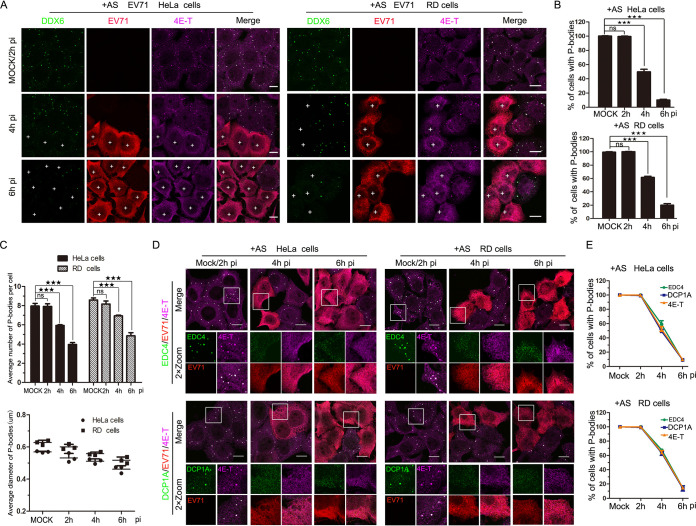
To further confirm the above results, HeLa and RD cells were treated with sodium arsenite (AS [200 μM]), which was an external stress inducing 100% of cells containing P-bodies when cells were mock infected; similar results were observed, which suggested that EV71 infection blocked the formation of AS-induced P-bodies (Fig. 2). Altogether, these data indicate that the infection with EV71 inhibits P-body formation.
FIG 2.
EV71 infection inhibits AS-induced P-bodies. (A to C) HeLa and RD cells were mock infected or infected with EV71 (MOI of 10) for consecutive periods (0, 2, 4, and 6 h, with mock-infected cells converged at 2 h) and treated with AS (200 μM) for 1 h prior to fixation. (A) Cells were analyzed by IF after being immunostained for DDX6 (green), EV71 (red), and 4E-T (magenta). (B) Quantitative analysis of the percentage of cells containing P-bodies in panel A. (C) Quantitative analysis of the average number of P-bodies per cell and the average diameter of P-bodies in panel A. (D and E) HeLa and RD cells were mock infected or infected with EV71 (MOI of 10) for consecutive periods (0, 2, 4, and 6 h, with mock-infected cells converged at 2 h) and treated with AS (200 μM) for 1 h prior to fixation. (D) Cells were analyzed by IF after being immunostained for EV71 (red), 4E-T (magenta), and EDC4 (green) or DCP1A (green). (E) Quantitative analysis of the percentage of cells containing P-bodies in panel D. “+” indicates EV71-infected cells. Error bars represent the means ± SDs of values from three independent experiments (n = 3). A total of 100 cells and 50 P-bodies were counted each time. By Student’s test: ★★★, P < 0.001; ns, not significant. Scale bars, 10 μm.
2A protease alone is efficient at inhibiting the formation of P-bodies.
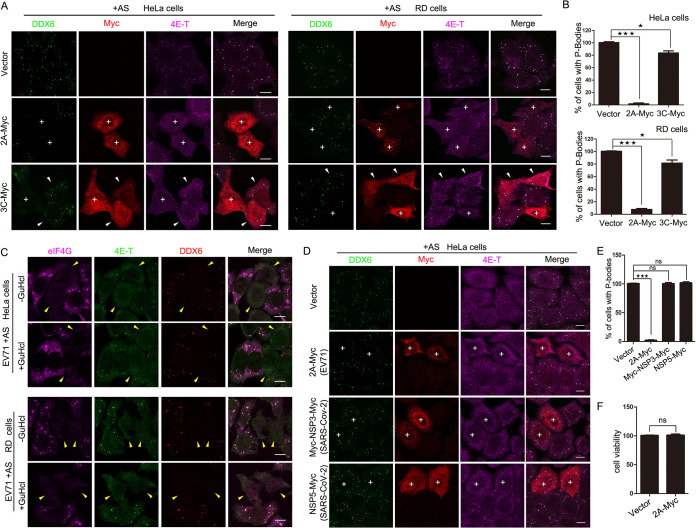
The single ORF of the EV71 genome can be translated into four structural proteins and seven nonstructural proteins; therefore, to explore which protein plays the key role in inhibiting the formation of P-bodies during EV71 infection, HeLa cells were transfected with plasmids expressing Myc-tagged viral proteins in the presence of AS (data not shown). We found that P-bodies marked by EDC4 and 4E-T disappeared in cells expressing 2A protease, whereas few foci vanished in cells expressing 3C protease. Moreover, we expressed Myc-tagged 2A and 3C in AS-treated HeLa and RD cells and counted the percentages of cells containing P-bodies marked by DDX6 and 4E-T (Fig. 3A and B). As the data show, P-bodies disappeared in almost all HeLa cells expressing 2A protease, in 92% of RD cells expressing 2A protease, and in only about 20% of cells expressing 3C protease, implying that both 2A protease and 3C protease can block the formation of P-bodies, but 2A protease plays a more important role.
FIG 3.
2A protease alone is sufficient to inhibit the P-body formation during EV71 infection. (A and B) HeLa cells or RD cells grown in 24-well plates were transfected with empty vector, 2A-Myc, or 3C-Myc for 23 h and then treated with AS for another 1 h to ensure every cell contained P-bodies. (A) Cells were analyzed by IF after being immunostained for DDX6 (green), Myc tag (red), and 4E-T (magenta). P-bodies were marked by DDX6 and 4E-T. “+” indicates cells expressing 2A-Myc or 3C-Myc without P-bodies. White arrowheads indicate 3C-expressing cells still containing P-bodies. (B) Quantitative analysis of the percentage of cells containing P-bodies in panel A. (C) HeLa cells or RD cells were infected with EV71 in the presence or absence of GuHCl and then immunostained for eIF4G (magenta), 4E-T (green), and DDX6 (red). eIF4G was detected to indicate the activity of 2A protease. Yellow arrowheads indicate EV71-infected cells, and 2A protease activity was almost unaffected by additional GuHCl treatment. (D and E) HeLa cells were transfected with empty vector, Myc-2A of EV71, Myc-NSP3-Myc of SARS-Cov-2, and NSP5-Myc of SARS-Cov-2 for 23 h and then treated with additional AS (200 μM) for another 1 h. (D) Cells were stained with antibodies against Myc tag (red), DDX6 (green), and 4E-T (magenta). P-bodies were marked by DDX6 and 4E-T. “+” indicates cells expressing 2A-Myc, Myc-NSP3-Myc, or NSP5-Myc. (E) Quantitative analysis of the percentage of cells containing P-bodies in panel D. (F) HeLa cells were transfected with vector and Myc-2A for 24 h, and the cell viability was analyzed using the Cell Counting kit-8 (ZOMANBIO). Error bars represent the means ± SDs of values from three independent experiments (n = 3). A total of 100 cells were counted each time. ★, P < 0.05; ★★★, P < 0.001. Scale bars, 10 μm.
To further verify whether 2A protease alone is sufficient to inhibit the formation of P-bodies, we treated HeLa cells with guanidine hydrochloride (GuHCl) during EV71 infection. GuHCl specifically inhibits picornavirus RNA synthesis but has no effect on the translation of viral RNA at a concentration that does not adversely affect host cells (28, 29). Therefore, an extremely low level of viral proteins was expressed in infected cells in the presence of GuHCl. More importantly, our previous article showed that low-level expression of 2A protease sufficiently resulted in the cleavage of eIF4GI in GuHcl-treated cells, whereas the function of 3C protease was suppressed by GuHcl (30). Therefore, we treated EV71-infected HeLa and RD cells with AS for 1 h and found that the P-bodies disappeared in infected cells in the presence of GuHcl (Fig. 3C), suggesting that 2A protease, but not 3C protease, was essential for inhibiting P-body formation in EV71-infected cells. Subsequently, to confirm it is specifically 2A protease that inhibits P-bodies during EV71 infection, HeLa cells were transfected with empty vector, 2A protease of EV71, and the NSP3 and NSP5 proteases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which belongs to the Coronaviridae family of the single-stranded, plus-stranded RNA viruses. The result showed that NSP3 and NSP5 of SARS-CoV-2 have no effect on P-body formation (Fig. 3D and E). In addition, to exclude the possibility that the expression of 2A protease made the cells sick overall rather than necessarily targeting the P-bodies, HeLa cells were transfected with empty vector and 2A-Myc for 24 h. Then, HeLa cell viability was analyzed by using Cell Counting Kit-8 (ZOMANBIO). The results showed that 2A protease has no significant effect on cell viability compared with cells transfected with empty vector (Fig. 3F). Taken together, these data demonstrate that 2A protease alone is efficient to block the formation of P-bodies.
The activity of 2A protease is critical for blocking P-body formation during EV71 infection.
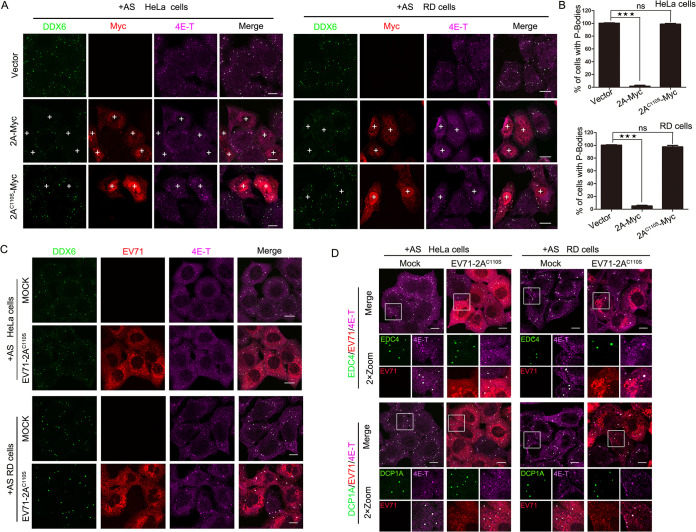
2A protease and 3C protease are required for the replication of EV71 in host cells, and they not only are essential for the translated polypeptide chain’s maturation into different viral proteins, but they also interact with host factors to create an environment conducive to viral survival (23, 31). Based on the result that 2A protease is essential for inhibiting the information of P-bodies, we hypothesized that the activity of 2A protease plays a critical role in blocking P-body formation. To test this hypothesis, we expressed a protease-inactivated mutant of 2A, 2AC110S, in HeLa and RD cells treated with AS and found that AS-induced P-bodies (marked by DDX6 and 4E-T) appeared in 100% of empty vector-transfected and 2AC110S-expressing cells, whereas 2A-expressing cells failed to form P-bodies, suggesting that 2AC110S failed to inhibit the formation of P-bodies and that the activity of 2A protease is critical to block P-body formation (Fig. 4A and B).
FIG 4.
The activity of 2A protease is critical to block P-body formation during EV71 infection. (A and B) HeLa cells or RD cells grown in 24-well plates were transfected with empty vector, 2A-Myc, or 2AC110S-Myc for 23 h and then treated with AS for another 1 h. (A) Cells were analyzed by IF after being immunostained for DDX6 (green), Myc tag (red), and 4E-T (magenta). P-bodies were marked by DDX6 and 4E-T. “+” indicates cells expressing 2A-Myc or 2AC110S-Myc. (B) Quantitative analysis of the percentage of cells containing P-bodies in panel A. Error bars represent the means ± SDs of values from three independent experiments (n = 3). A total of 100 cells were counted each time. ★★★, P < 0.001; ns, not significant. Scale bars, 10 μm. (C and D) HeLa or RD cells were mock infected or EV71-2AC110S infected for 23 h with AS treatment for another 1 h and then stained with antibodies as indicated. DDX6, EDC4, DCP1A, and 4E-T are all markers of P-bodies. The infected cells were revealed with the EV71 antibody. Scale bars, 10 μm.
To further confirm that the activity of 2A protease is critical to the EV71-induced blocking of P-bodies, we examined the formation of P-bodies marked by DDX6 and 4E-T via IF in mock-infected cells or in cells infected with EV71-2AC110S, the 2A protease activity-inactivated recombinant virus mentioned in our previous article (30). As expected, when AS-treated HeLa and RD cells were infected with EV71-2AC110S, the distribution of DDX6 and 4E-T had no difference compared to the mock-infected cells (Fig. 4C), indicating that EV71-2AC110S infection failed to block the formation of P-bodies. We also examined the distribution of EDC4 and DCP1A, other markers of P-bodies in EV71-2AC110S-infected HeLa and RD cells with AS treatment, and found the results were consistent with the above findings (Fig. 4D). Taken together, these data highlight the necessity of the 2A protease activity in blocking the formation of P-bodies in EV71-infected cells.
EV71 replication is impaired by blocking P-body formation.
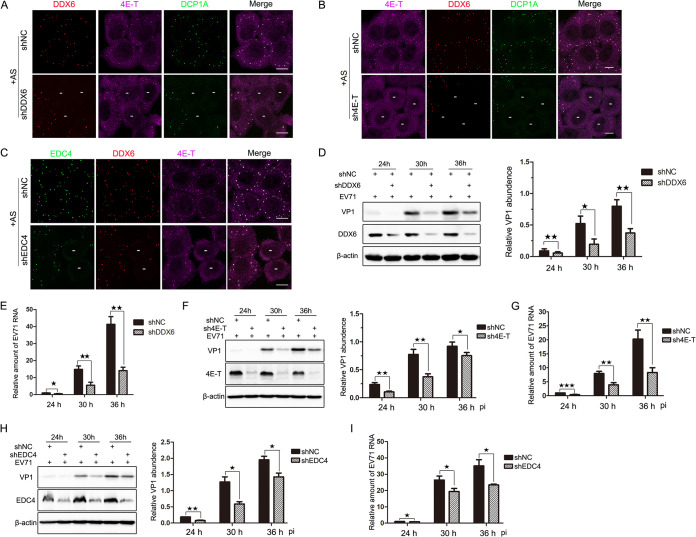
Despite the details of P-body formation remaining unclear, hundreds of proteins are known to be located in P-bodies, and based on their important roles in P-body assembly, some of them are considered to be scaffold proteins. DDX6, which can interact with half of the P-body proteins, is one of the scaffold proteins (7). By analogy, knockdown (KD) of the gene expression of these scaffold proteins leads to the failure of P-body assembly, and then we can detect EV71 replication under the condition of blocking P-body formation. As the data show, P-bodies (marked by DDX6, 4E-T, and DCP1A) failed to assemble in KD cells (Fig. 5A to C), and based on this, stable KD cell lines were constructed to determine the influence on EV71 replication when P-body formation was inhibited. Subsequently, KD cells were infected with EV71 for consecutive periods (24, 30, and 36 h), the expression of VP1 and DDX6 proteins was examined via Western blotting (WB), and the relative amount of viral RNA was detected via reverse transcription-quantitative PCR (RT-qRCR). As the infection processed, the levels of VP1 and viral RNA were both significantly reduced compared to those in the negative-control cells, although the overall trends of VP1 and viral RNA were to gradually increase in KD-DDX6 cells (Fig. 5D and E). Furthermore, we knocked down the expression of 4E-T and EDC4. Similar results were observed in the KD-4E-T and KD-EDC4 cell lines (Fig. 5F to I), which suggests that EV71 replication was impaired when P-body formation was blocked.
FIG 5.
EV71 replication is impaired by blocking P-body formation. (A to C) The formation of P-bodies was blocked by knocking down the components of P-bodies. HeLa cells were cultured on 6-well plates overnight, transfected with shRNAs, and treated with additional AS for 1 h before being harvested. (A and B) Cells were fixed and stained with antibodies against DDX6 (red), 4E-T (magenta), and DCP1A (green). (C) Cells were fixed and stained with antibodies against EDC4 (green), DDX6 (red), and 4E-T(magenta). “−” indicates cells in which the target gene was unable to express due to knockdown. Scale bars, 10 μm. (D and E) EV71 replication was impaired in KD-DDX6 cells, wherein P-bodies failed to assemble. Stable KD cells were infected with EV71 (MOI of 0.1) and harvested at various time points postinfection as indicated. (D) Cell lysates were analyzed via Western blotting using anti-VP1, anti-DDX6, and anti-β-actin antibodies. The relative abundances of VP1 were quantified from three independent experiments. (E) Total RNA was isolated for qPCR analysis to measure the amount of EV71 RNA that normalized to β-actin mRNA. (F and G) EV71 replication was impaired in KD-4E-T cells, where P-bodies failed to assemble. Stable KD cells were treated as described above. Cells were analyzed via Western blotting (F) and qPCR analysis (G). (H and I) EV71 replication was impaired in KD-EDC4 cells, where P-bodies failed to assemble. Stable KD cells were treated as described above. Cells were analyzed via Western blotting (H) and qPCR analysis (I). Error bars represent the means ± SDs of values from three independent experiments (n = 3). ★, P < 0.05; ★★, P < 0.01; ★★★, P < 0.001.
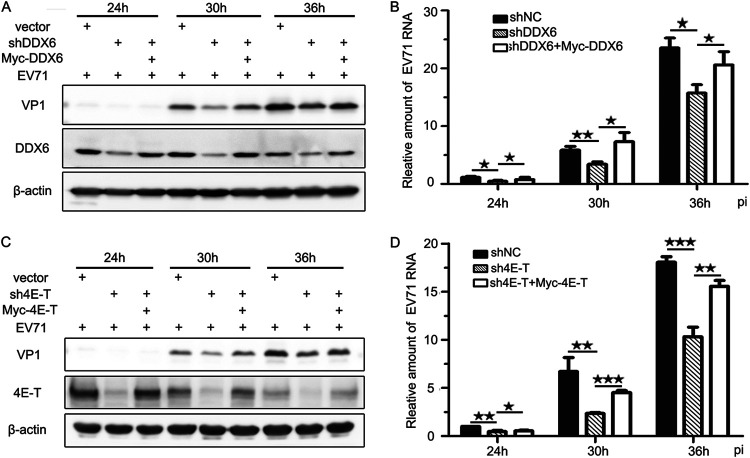
To further confirm this result, rescue assays were performed by expressing the synonymous mutants of DDX6 and 4E-T, each with a Myc tag in the knockdown cell lines. We found that not only the abundance of VP1 but also the amount of viral RNA increased compared to those in the KD cell lines (Fig. 6). Collectively, these data show that EV71 replication is impaired when the formation of P-bodies is inhibited, suggesting that the reason why EV71 infection blocks P-body formation is likely to be that it hijacks or rearranges the components of the P-body to facilitate replication.
FIG 6.
EV71 replication is affected by P-body formation. Six-well plates were seeded with stable cells expressing the synonymous mutants of DDX6 and 4E-T, each with a Myc tag, or the KD cells for 24 h, and then cells were infected by EV71 (MOI of 0.1) and harvested at various time points postinfection as indicated. (A) Cell lysates were analyzed via Western blotting using anti-VP1, anti-DDX6, and anti-β-actin antibodies. (B) Total RNA was isolated for qPCR analysis to measure the amount of EV71 RNA that normalized to β-actin mRNA. (C) Cell lysates were analyzed via Western blotting using anti-VP1, anti-4E-T, and anti-β-actin antibodies. (D) Total RNA was isolated for qPCR analysis to measure the amount of EV71 RNA that normalized to β-actin mRNA. Error bars represent the means ± SDs of values from three independent experiments (n = 3). ★, P < 0.05; ★★, P < 0.01; ★★★, P < 0.001.
EV71 infection inhibits P-body formation to facilitate viral RNA synthesis.
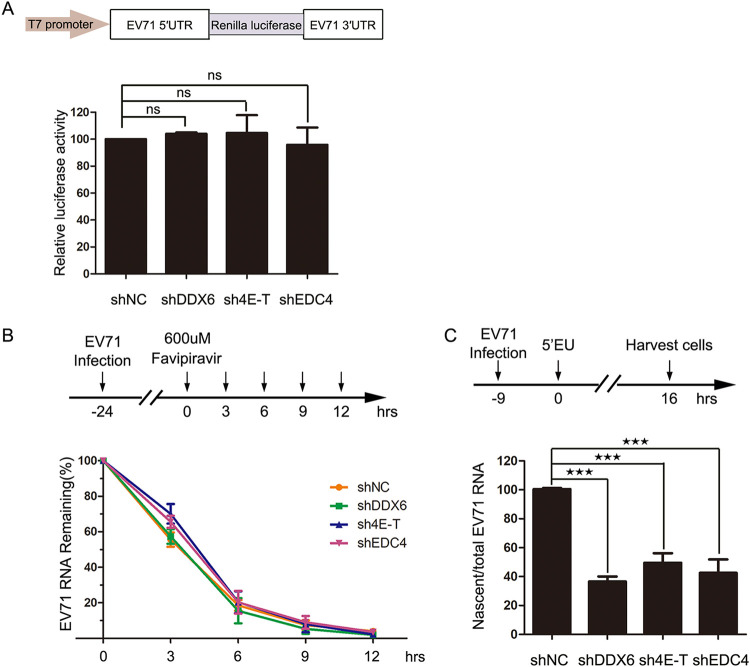
Given that mRNA translation repression, miRNA-mediated silence, and mRNA decay are involved in P-body assembly, we further wondered whether the translation and stability of viral RNA during EV71 replication were affected when P-body formation was inhibited. To verify the impact on IRES-mediated virus translation, stable KD cell lines were transfected with the RNA of the Renilla luciferase mRNA reporter UTREV71-Rluc, in which the Renilla luciferase sequence is flanked by the viral 5′ and 3′ UTRs, as shown in Fig. 6A, and the T7 promoter was used for in vitro transcription (30). We observed that there was no effect on the luciferase activity in KD-DDX6 cells, KD-4E-T cells, and KD-EDC4 cells (Fig. 7A), indicating the blocking of P-body formation did not affect IRES-mediated translation of viral RNA.
FIG 7.
The blocking of P-body formation affects viral RNA synthesis. (A) The blocking of P-body formation had no effect on viral translation. Shown is a graphic illustration of UTREV71-Rluc-derived Renilla luciferase reporter expression and reporter assays. Stable KD cells derived from 293T cells were cultured on 12-well plates overnight and transfected with UTREV71-Rlu reporter RNA (0.4 μg/well) for 12 h. Relative luciferase activity was measured according to the manufacturer’s instructions. Error bars represent the means ± SDs of values from three independent experiments (n = 3). ns, not significant. (B) The blocking of P-body formation had no effect on viral RNA stability. Stable KD cells derived from HeLa cells were infected with EV71 (MOI of 0.1) and then treated with favipiravir at 24 hpi, which was regarded as 0 h. Cells were harvested at the indicated time points after favipiravir treatment, and total RNA was isolated for qPCR analysis. The EV71 RNA remaining was measured and normalized to β-actin mRNA. Error bars represent the means ± SDs of values from three independent experiments (n = 3). (C) The blocking of P-body formation led to a decrease of nascent viral RNA. Stable KD cells derived from HeLa cells were cultured in 10-cm dishes overnight and infected by EV71 (MOI of 0.1) and then treated with 0.25 mM 5-EU at 9 hpi for another 16 h. Cells were harvested to isolate total RNA and 5-EU-labeled nascent RNA for qPCR analysis. The nascent EV71 RNA was measured and normalized to total EV71 RNA in KD cells. Error bars represent the means ± SDs of values from three independent experiments (n = 3). ★★, P < 0.01; ★★★, P < 0.001.
Subsequently, to determine whether the stability of viral RNA was affected, the stable KD-DDX6, KD-4E-T, and KD-EDC4 cells were infected with EV71 at a low multiplicity of infection (MOI of 0.1) for 24 h to accumulate viral RNA and then treated with favipiravir (a potent inhibitor of the 3D RNA-dependent RNA polymerase) (32) to inhibit viral RNA synthesis. Then, cells were harvested at intervals, and the remaining viral RNA was quantified by RT-qPCR (Fig. 7B). As the data show, there was no difference between KD cell lines and negative-control cells, suggesting the P-body does not affect viral RNA stability.
According to the data shown in Fig. 5 and discussed above, the decrease in viral protein levels is probably caused by the decrease in viral RNA. To further assess the effect of the P-body on viral RNA synthesis, the stably knocked-down cells were infected with EV71 (MOI of 0.1) and incubated with 5-ethynyl uridine (5-EU) for another 16 h at 9 hpi (1 h of viral adsorption is counted) to label nascent RNA, including newly synthesized viral RNA and cellular RNA, as described previously (33). Then, 5-EU-labeled RNAs were linked with azide-labeled biotin via click chemistry reaction before being harvested to homogenize the cell lysates, which made it possible to isolate nascent RNAs with streptavidin-conjugated magnetic beads. Nascent viral RNA, normalized to the total amount of EV71 RNA used to capture nascent RNA transcripts on streptavidin-conjugated magnetic beads, was significantly reduced to ∼40% in KD-DDX6 cells, ∼48% in KD-4E-T cells, and ∼43% in KD-EDC4 cells compared with negative-control cells (Fig. 7C). Taken together, these data suggest that EV71 regulates P-body formation to facilitate viral RNA synthesis, but has no effect on translation of viral RNA or viral RNA stability.
2A protease promotes the interaction of DDX6/4E-T complex and viral RNA during EV71 infection.
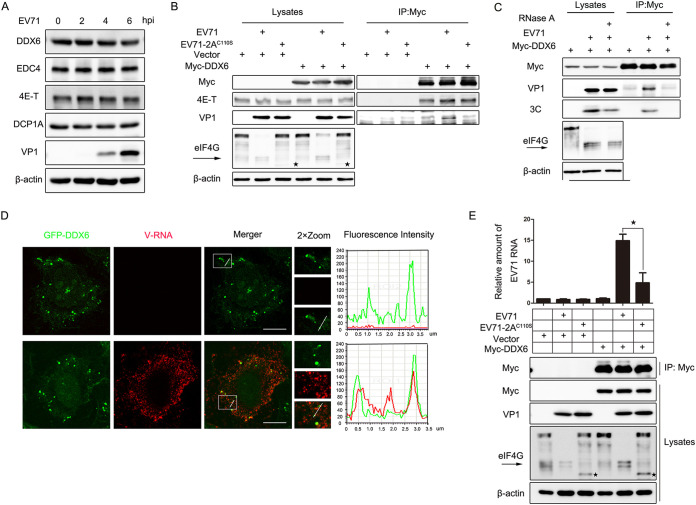
To explore how 2A protease during EV71 infection inhibits the formation of P-bodies, first, HeLa cells were infected with EV71 (MOI of 10) for up to 6 h, and then the protein levels of DDX6, 4E-T, and EDC4, which are necessary for P-body assembly, were examined via WB. As the data show, there were no significant differences between mock-infected and EV71-infected cells (Fig. 8A). Second, previous studies have shown that the interaction between DDX6 and 4E-T is required for P-body assembly (9, 34); therefore, to determine whether EV71 infection or 2A protease transfection breaks this interaction, coimmunoprecipitation (co-IP) experiments were carried out to detect the influence of 2A protease and 2AC110S on the interaction of DDX6 and 4E-T. We found that 2A and 2AC110S neither affect the interaction of DDX6 and 4E-T nor interact with DDX6 or 4E-T (data not shown). Furthermore, HeLa cells were transfected with empty vector or Myc-DDX6 for 24 h and infected with EV71 and EV71-2AC110S for another 24 h, and then cells were harvested for subsequent IP experiments. eIF4G was detected to indicate the activity of 2A protease during virus infection. IP bands were not detected in cells transfected with empty vector, and the interaction of DDX6 and 4E-T was still detectable during EV71-infected and EV71-2AC110S-infected cells compared to the mock-infected cells transfected with Myc-DDX6 (Fig. 8B), indicating that the way EV71 infection blocks P-body formation is not to destroy the DDX6/4E-T complex. However, we surprisingly observed that VP1 was immunoprecipitated with DDX6 in EV71-infected cells but not in those infected with EV71-2AC110S, indicating that 2A protease plays an essential role in the interaction of DDX6 and VP1.
FIG 8.
2A protease-regulated viral RNA recruiting DDX6-4E-T during EV71 infection. (A) HeLa cells were infected with EV71 (MOI of 10) for up to 6 h. Cell lysates were analyzed via Western blotting. VP1 indicated EV71 replication. β-Actin was the sample loading control. (B) HeLa cells were transfected with empty vector or Myc-DDX6 for 24 h and then mock infected or infected with EV71 (MOI of 0.1) or EV71-2AC110S (MOI of 0.6) for another 24 h. Cell lysates were subjected to IP and analyzed via Western blotting. VP1 indicated EV71 replication, and β-actin was the sample loading control. An arrow indicates eIF4G products cleaved by 2A, and ★ indicates eIF4G products cleaved by caspases (45, 46). (C) HeLa cells were transfected with Myc-DDX6 for 24 h and then infected with EV71 (MOI of 0.1) for another 24 h. Cell lysates were treated with or without RNase A before being subjected to IP and analyzed via Western blotting. VP1 and 3C indicated EV71 replication, and β-actin was the sample loading control. (D) HeLa cells expressing GFP-DDX6 were mock infected or infected with EV71 (MOI of 0.1) for 24 h. EV71 RNA (vRNA) was detected via RNA-FISH. The fluorescence intensity profiles of DDX6 (green) and vRNA (red) were measured along the line drawn on a 2×Zoom panel by Leica Application Suite Advanced Fluorescence Lite. Scale bars, 10 μm. (E) HeLa cells were transfected with empty vector or Myc-DDX6 for 24 h and then infected with EV71 or EV71-2AC110S. Subsequently, the RNA-IP assay was performed for analysis. IP and cell lysates were analyzed via Western blotting. An arrow indicates eIF4G products cleaved by 2A, and “★” indicates eIF4G products cleaved by caspases. The relative amount of EV71 was analyzed via RT-qPCR. Error bars represent means ± SDs of values from three experiments (n = 3). ★, P < 0.05.
DDX6 is a member of DEAD box protein family, containing the conserved motifs IV and V, RNA-binding motifs (35), and previous studies of positive-stranded RNA viruses showed that DDX6 interacted with the RNA of dengue virus (DENV) and hepatitis C virus (HCV) (36, 37). Therefore, the interaction of DDX6 and VP1 should be mediated by viral RNA. To confirm this, we first treated EV71-infected cells with or without RNase A before the co-IP assay. We found that VP1 was not detected in the presence of RNase A, while it was detected in cells without RNase A treatment, and the result was further confirmed by 3C, suggesting DDX6 was binding to the viral RNA during EV71 infection (Fig. 8C). Then, we constructed cells stably expressing green fluorescent protein (GFP)-tagged DDX6 and evaluated the localization of DDX6 by RNA fluorescence in situ hybridization (FISH) assays in mock-infected or EV71-infected cells. The fluorescence signal of EV71 viral RNA (vRNA) was not detected in mock-infected cells, which indicated that the pore of vRNA was specific, and GFP-DDX6 was colocalized to vRNA in EV71-infected cells, which indicated the colocation was specific (Fig. 8D). In addition, to demonstrate the important role of 2A protease during EV71 infection, HeLa cells were transfected with empty vector or Myc-DDX6 for 24 h and then infected with EV71 or EV71-2AC110S. Subsequently, an RNA immunoprecipitation assay was performed to detect whether there was a difference between cells infected with EV71 and those infected with EV71-2AC110S (Fig. 8E). IP and cell lysates were analyzed via WB, and IP bands were not detected in cells transfected with empty vector. The relative amount of EV71 was analyzed via RT-qPCR, and we found that relative amount of viral RNA bound by DDX6 was much less in EV71-2AC110S-infected cells than in EV71-infected cells. Taken together, these data not only suggest that 2A protease regulates the binding of DDX6 and viral RNA, but also highlight the essential role of 2A protease in blocking P-body formation during EV71 infection.
DISCUSSION
In summary, our data demonstrate that EV71 infection inhibits the formation of P-bodies to promote viral RNA synthesis (Fig. 1, 2, 7C, and 8C and D). Most strikingly, we show the essential role of 2A and its protease activity in blocking P-body formation, making it possible for viral RNA to take advantage of the P-body components by interacting with them (Fig. 3, 4, and 8B and E). These findings highlight the importance of 2A protease while EV71 manipulates the assembly of P-bodies for its replication. Furthermore, our data promote a deeper understanding of the mechanism by which picornaviruses modulate P-body formation and provide a novel mode whereby RNA viruses regulate host cellular structure to benefit their replication.
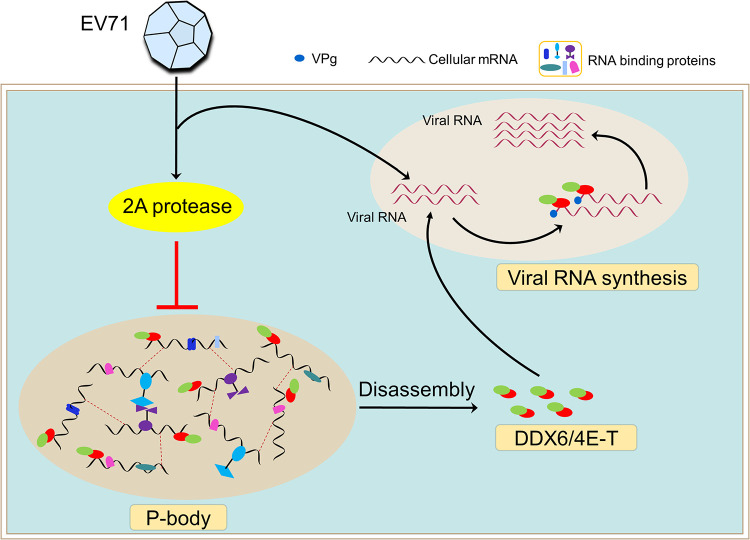
According to our findings, we propose the following model for the regulation of the P-body during EV71 infection, which is modulated by 2A protease to generate an optimal intracellular environment for viral RNA synthesis (Fig. 9). Upon entering the host cell, EV71 releases genomic RNA, which is immediately used as the template to be translated to express the 2A protease protein. Then, the formation of the P-body is blocked by 2A protease, and correspondingly, the P-body disassembles and the components are released. Therefore, the DDX6/4E-T complex can be recruited to viral RNA to facilitate viral RNA synthesis but not viral translation or viral RNA stability.
FIG 9.
Models of the regulation of P-bodies during EV71 infection. Once entering the host cell, EV71 releases genomic RNA, which is immediately used as a template to be translated into 2A protease. Then, the formation of P-bodies is blocked by 2A protease, and correspondingly, the P-body is disassembled and the components are released. As a result, DDX6/4E-T can be recruited to viral RNA rather than intracellular mRNA to facilitate viral RNA synthesis.
Similar to previous studies on other positive-stranded RNA viruses (25, 38, 39), here we found that EV71 infection blocks the formation of P-bodies and has no effect on the protein level of the components (Fig. 1, 2, and 8A). Furthermore, we also found that 2A protease alone is essential and specific to inhibition of P-body formation, and 3C protease is dispensable (Fig. 3). On one hand, we treated EV71-infected cells with additional GuHCl to inhibit 3C protease and found that the formation of P-bodies still was blocked by the low level of 2A protease expression (Fig. 3C). On the other hand, we found the EV71-2AC110S, a recombinant EV71 strain with 2A protease activity inactivated but with 3C protease activity, failed to suppress P-body formation (Fig. 4C and D), not only confirming the essential role of 2A protease activity during EV71 infection but also reflecting the true situation of the EV71-infected cell.
In principle, the virus manipulates P-body assembly during infection to generate a beneficial intracellular environment for the viral biological cycle. As expected, we found that the levels of viral protein and viral RNA were both decreased when P-body formation was blocked, indicating EV71 replication was impaired (Fig. 5D to I). Consistent with our finding, previous studies have shown that P-body components such as DDX6, Lsm1–7, Xrn1, and Ago2 are required during HCV replication (37–40). Given that P-bodies are constitutively present in cells and known to be involved in many aspects of the mRNA cycle in the cytoplasm, including mRNA translation repression and decay, we excluded the possibility that viral RNA translation and stability are affected when P-body formation is blocked (Fig. 7A and B). Similarly, a previous study showed that DDX6 was not required by internal ribosome entry site-directed translation (37). All of these data suggest the decrease in viral protein levels is probably caused by the decrease in viral RNA (Fig. 5D to I). Subsequently, we confirmed the hypothesis by using the nascent viral RNA synthesis capture assay under the condition of P-body assembly being blocked (Fig. 7C).
GW182, DDX3, and DDX6 have been proven to be recruited to the replication site by interacting with viral NS3 protein during WNV infection (25). DDX3 has been shown to be needed for the export and translation of unspliced HIV RNA from the nucleus (41). Lsm1–7, Xrn1, and Ago2 are required for HCV replication (37, 39, 40), and a recent study demonstrated Kaposi’s sarcoma-associated herpesvirus (KSHV) inhibited P-bodies by the interaction of viral ORF57 protein and GW182/Ago2 (27). Based on our findings and previous studies of P-bodies mentioned above, on one hand, in addition to the DDX6/4E-T complex, viruses can also make use of many P-body components to benefit viral replication. On the other hand, although viruses take advantage of P-body formation, it seems likely that different viruses co-opt different components of the P-body to promote viral replication at different stages of the viral life cycle, reflecting the complexity and adaptability of viruses. To our knowledge, we are the first to demonstrate that EV71 inhibits P-body formation to facilitate viral RNA synthesis, providing further understanding of enterovirus regulation of P-body formation to generate a beneficial environment for viral replication.
We found that EV71 infection has no effect on the interaction of DDX6 and 4E-T (Fig. 8B), although their relationship is required for P-body formation (9). Furthermore, we found that DDX6 was recruited to viral RNA (Fig. 8C and D), and correspondingly, 4E-T can be indirectly recruited to viral RNA as well. Consistent with our finding, DDX6 has been identified to interact with pseudoknots in the RNA 3′ UTR of DENV and HCV, other positive-stranded RNA viruses (36, 37). Although the molecular mechanism of how DDX6/4E-T promotes EV71 replication should be further explored, we should also pay enough attention to this. On one hand, both DDX6 and 4E-T are the scaffold proteins during P-body assembly: DDX6 can interact with half of the P-body proteins and is involved in mRNA translational repression, mRNA decay, and miRNA-mediated silencing. On the other hand, similarly, a previous study showed that the Lsm1-7-Pat1 complex promotes cellular mRNA decay by binding its 3' end; however, it promotes the translation and replication of genomic RNA of Brome mosaic virus (BMV), a plant positive-stranded RNA virus, by binding cis-acting regulatory sequences (42).
We further found that 2A protease plays a crucial role in the process of viral RNA interacting with DDX6 (Fig. 8B and E). As a member of the DEAD box family of helicases, DDX6 is conserved from unicellular eukaryotes to vertebrates and contains the highly conserved RNA-binding motifs IV and V. Therefore, there is no significant difference between DDX6 binding to mRNA and its binding to viral RNA, and consistently, previous studies have shown that DDX6 interacts with RNA of HCV, DENV, BMV, and the Ty3 retrotransposon in yeast (36, 37, 43), indicating the diversity and conservatism of DDX6 binding to RNA. Correspondingly, viral RNA and intracellular mRNA competitively bind to DDX6 during EV71 infection. However, as a result of the blocking of P-body formation by 2A protease, DDX6 tends to bind to viral RNA rather than mRNA, and this is why we still detected a small amount of EV71 RNA binding to DDX6 in EV7-2AC110S-infected cells (Fig. 8E). Further studies are needed to clarify the specific mechanisms of 2A-induced suppression of P-body formation, although 2A protease of PV has been proven to inhibit the nuclear export of cellular mRNAs (44), which itself is necessary for P-body assembly, and its self-assembly can also promote P-body formation (10).
In conclusion, this is the first report to reveal the essential role of 2A protease during EV71-induced blocking of P-body formation to promote viral RNA synthesis, which also provides a deeper understanding of the importance of picornavirus 2A protease. Furthermore, our research in these areas provides fascinating insights into the regulation of cytoplasmic RNP granules by picornavirus and may also offer new therapeutic strategies for diseases induced by picornavirus.
MATERIALS AND METHODS
Plasmids and shRNA oligonucleotides.
The coding regions of DDX6 and 4E-T were obtained from HeLa cells by RNA extraction and subsequently reverse transcription PCR (RT-PCR), and then each was cloned into a pCAGGS vector with an N-terminal Myc tag and into a pHAGE(puro) vector (replacing the fluorescent tag in pHAGE-CMV-MCS-IRES-ZsGreen [EvNO00061605] with puromycin) with an N-terminal Myc tag. All of the plasmids expressing EV71 proteins and pBS-T7-EV71-2AC110S plasmid have been described previously (30). Short hairpin RNA (shRNA) oligonucleotides corresponding to the target sequences were cloned into pLKO.1, and the target sequences were as follows: shNC, GCGCGATAGCGCTAATAATTT; shDDX6, GAGACCTATCTCCATCGTATT; sh4E-T, GGATCACCGTCTTAGCGATAA; and shEDC4, GCTTTGGTATCCAGGTTGTGA.
Cell culture.
HEK293T (human embryonic kidney 293 cells, obtained from the China Center for Type Culture Collection), HeLa cells (human cervical cancer epithelial cells, obtained from the China Center for Type Culture Collection), and human RD cells (rhabdomyosarcoma cells, obtained from the China Center for Type Culture Collection) were cultured in Dulbecco’s modified eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin (Gibco) at 37°C with 5% CO2. Other stable overexpression cells (Myc-DDX6-HeLa/293T, 293T-T7, and GFP-DDX6-HeLa) and stable knockdown (KD) cells (shRNA-HeLa) or negative-control cells (shNC-HeLa) were cultured in DMEM with 10% FBS, 1% penicillin–streptomycin (Gibco), and 1 μg/ml puromycin (Sigma-Aldrich) at 37°C with 5% CO2.
Transfection and infection.
For transfection, plasmids were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, and cells were harvested at 24 h posttransfection (hpt) or underwent further treatment as indicated. For infection, HeLa or RD cells were infected with DMEM containing viruses at various multiplicities of infection (MOI), as indicated in the figure legends. After 1 h of incubation, the medium was replaced with fresh DMEM with 4% FBS, and this time point was considered 0 h postinfection (hpi). Cells were then harvested for further analysis as indicated.
Stable knockdown and overexpression cell lines.
HEK293T cells were cotransfected with psPAX2 , pMD2.G, and shRNA with PLKO vector or target protein expression plasmids for 48 h to generate lentiviral particles. Then, supernatants were filtered with a 0.45-μm-pore filter and collected to infect HeLa cells. Fresh culture medium was added after 24 h postinfection, and then we conducted the next round for another 24 h. To ensure the infection’s efficiency, HeLa cells were infected with lentivirus again, then were continuously cultured in complete growth medium with the addition of puromycin, and then were harvested to detect the deletion or overexpression of target proteins. The stable knockdown or overexpression cell lines were used for subsequent experiments.
Antibodies and reagents.
Rabbit polyclonal anti-DCP1A was purchased from Abcam (catalogue no. ab183709) and ABclonal (catalogue no. A7376). Rabbit polyclonal anti-DDX6 was purchased from ABclonal (catalogue no. A16270), Bethyl (catalogue no. A300-460A-M), and Santa Cruz (catalogue no. sc-376433). Rabbit polyclonal anti-EDC4 (catalogue no. ab72408) and goat polyclonal anti-4E-T (catalogue no. ab6034) were purchased from Abcam. Mouse monoclonal anti-c-Myc (catalogue no. sc-40) and rabbit polyclonal anti-c-Myc (catalogue no. sc-789) were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-β-actin monoclonal was purchased from ABclonal (catalogue no. AC026). Mouse monoclonal anti-VP1 was purchased from Abmax (clone 22A14). Rabbit polyclonal anti-3C polyclonal was purchased from ABclonal (catalogue no. A10003). Mouse monoclonal anti-EV71 (catalogue no. MAB979) was purchased from Millipore. Alexa Fluor 647-conjugated donkey anti-goat immunoglobulin (IgG) H+L (catalogue no. A21447), Alexa Fluor 488-conjugated donkey anti-rabbit IgG H+L (catalogue no. A21206), and Alexa Fluor 594-conjugated donkey anti-mouse IgG H+L (catalogue no. A21203) were purchased from Life Technologies. Rabbit monoclonal anti-eIF4G (catalogue no. 2469S) was purchased from Cell Signaling Technology. AS was purchased from Sigma-Aldrich and was used at concentrations of 200 μM. Recombinant RNase inhibitor (RRI) was purchased from Takara. Biotin-azide and 5-ethyluridine (5-EU) were purchased from Ribobio. Favipiravir was purchased from Med Chem Express (MCE).
Western blotting and RNA immunoprecipitation.
For Western blotting, cells were harvested and lysed in lysis buffer (150 nM NaCl, 50 nM Tris-HCl [pH 7.4], 1% Triton X-100, 1 mM EDTA [pH 8.0], and 0.1% SDS with a protease inhibitor cocktail) for 30 min at 4°C and centrifuged at 4°C for 20 min at 12,000 × g. Then, the supernatants were boiled in 1× loading buffer (0.08 M Tris-HCl [pH 6.8], 2.0% SDS,10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue) at 100°C for 10 min, followed by separation by 8 to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto a nitrocellulose membrane (GE Healthcare). The membrane was incubated with the primary antibodies after blocking with 5% nonfat milk, followed by horseradish peroxidase-conjugated secondary antibodies (Thermo Fisher Scientific). The proteins were detected on a Fujifilm LAS-4000 imaging system.
For RNA immunoprecipitation, briefly, cells were harvested and lysed in RNase-free NP-40 buffer (50 nM Tris-HCl [pH 7.4], 50 mM NaCl, 50 mM NaF, and 0.5% NP-40 with a protease inhibitor cocktail) on ice for 30 min and then centrifuged at 4°C for 20 min at 12,000 × g. The supernatants were collected and divided: 8% of the supernatant was subjected to WB to detect the expression of plasmids, 12% was resuspended in 1 ml TRIzol (Ambion, Invitrogen) to extract the total cellular RNA, and the rest was subjected to IP for 6 h at 4°C. Anti-Myc agarose beads (MBL) were centrifuged at 4°C for 2 min at 5,000 rpm and washed 3 times with NP-40 buffer. The beads were finally resuspended in 1 ml NP-40 buffer and divided into two parts. One portion was subjected to WB to detect IP bands, and all of the other beads were resuspended in 1 ml TRIzol to extract the coimmunoprecipitated RNA. The amount of RNA was quantified by RT-qPCR.
Immunofluorescence assay.
HeLa or RD cells were cultured on coverslips in 24-well plates overnight before being transfected or infected as indicated and then fixed with 4% (wt/vol) paraformaldehyde–phosphate-buffered saline (PBS). Subsequently, cells were washed with PBS for 5 min at room temperature, and this step were repeated twice. Then, cells were permeated with 0.2% (wt/vol) Triton X-100–PBS solution for 20 min at room temperature and washed three times as before. Next, cells were blocked with 3% (wt/vol) bovine serum albumin (BSA) in PBS at room temperature for at least 30 min and incubated with primary antibodies for 2 h at room temperature. After washing, cells were incubated with secondary antibodies for 2 h at room temperature, followed by staining with 1 mg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 5 min to label nuclei. At last, cells were mounted with Prolong Diamond antifade mountant (Life Technology) and observed by using a Leica confocal microscope.
Nascent RNA synthesis assay.
The nascent RNA synthesis assay was performed as described earlier (33). HeLa cells were cultured in 10-cm dishes overnight and were infected by EV71. At 9 hpi, cells were pulsed with 0.25 mM 5-ethynyl uridine (5-EU) for another 16 h to incorporate 5-EU into newly synthesized RNA. Cells were washed three times with ice-cold PBS after discarding the culture medium and irradiated with 0.15 J/cm2 UV light at 254 nm. Then, cells were fixed with 90% ethanol for 30 min at 4°C, followed by permeabilization with 0.5% Triton X-100 in PBS for 30 min at 4°C. Next, to conjugate the biotin azide to the 5-EU-labeled RNA in a copper-catalyzed click reaction, the click reaction solution containing biotin was added to cells. Then, cells were harvested, homogenized, and centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was divided into two parts to quantify EV71 RNA. One portion was subjected to extract total RNA to quantify the total amount of EV71 RNA. The other was subjected to IP with prepared streptavidin-conjugated magnetic beads to extract biotin-conjugated, 5-EU-labeled EV71 RNA. Nascent EV71 RNA was normalized to the total amount of EV71 RNA used to capture nascent RNA transcripts on streptavidin-conjugated magnetic beads, and is expressed as nascent RNA/total EV71 RNA.
RNA stability assay.
Stable KD cells were cultured in 12-well plates overnight, infected with EV71 (MOI of 0.1) for 24 h, and then treated with 600 μM favipiravir. Cells were harvested at different times as indicated, and EV71 RNA was extracted and quantified by RT-qPCR and normalized to the amount of β-actin mRNA. The following primers were used: EV71 5′ UTR forward (5′-GAAACTTAGAAGCAGCAAAC-3′), EV71 5′ UTR reverse (5′-TCATCGACCTGATCTACACT-3′), human β-actin forward (5′-TGAGACCTTCAACACCCCAG-3′), and human β-actin reverse (5′-TGTCACGCACGATTTCCCGC-3′).
RNA-FISH assay.
The RNA-FISH assay was performed according to the manufacturer’s instructions for the ViewRNA ISH cell assay kit (Affymetrix). Briefly, HeLa cells were cultured on coverslips in 24-well plates overnight, followed by EV71 infection for 24 h, and fixed in 4% paraformaldehyde solution for 30 min at room temperature, followed by permeabilization with detergent solution for 5 min at room temperature. To expose viral RNA and to be identified by the specific probe, cells were incubated with protease solution at a suitable dilution (1:1,000 to 1:4,000) in PBS for 10 min at room temperature. Then, cells were incubated with the probe set for 3 h at 40°C, followed by individual incubation with the solution containing preamplifier, amplifier, and label probe at 40°C for 30 min. After being stained with DAPI, cells were mounted with Prolong Diamond antifade mountant (Life Technology) and observed by using a Leica confocal microscope. The probes for EV71 positive-stranded RNA (vRNA) were purchased from Affymetrix.
Luciferase assay.
The Renilla luciferase reporter mRNA was transcribed with the TranscriptAid T7 high-yield transcription kit (Thermo Fisher Scientific) and purified with the RNeasy minikit (Qiagen). Cells were cultured in 24-well plates overnight and transfected with the transcribed RNA for 12 h. Cells were harvested, and the Renilla luciferase activity was examined with a Renilla luciferase assay kit (Promega) according to the manufacturer’s instructions.
Statistical analysis.
Statistical significance (★, P < 0.05; ★★, P < 0.01; ★★★, P < 0.001; ns, not significant [P > 0.05]) was determined with an unpaired Student's t test by using GraphPad Prism 6.01. All results shown in the article are expressed as means ± standard deviation (SD) of results from three independent experiments (n = 3).
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China (2017YFA0505801), the National Natural Science Foundation of China (81825015, 81871650, and 31630086), and the Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory (2019ACCP-MS06).
Contributor Information
Yali Qin, Email: yqin@whu.edu.cn.
Mingzhou Chen, Email: chenmz@whu.edu.cn.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7:719–723. 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489. 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382. 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808. 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu JH, Yang W-H, Gulick T, Bloch KD, Bloch DB. 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11:1795–1802. 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717–727. 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, Daunesse M, Bertrand E, Pierron G, Mozziconacci J, Kress M, Weil D. 2017. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68:144–157.e5. 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Mittag T, Parker R. 2018. Multiple modes of protein-protein interactions promote RNP granule assembly. J Mol Biol 430:4636–4649. 10.1016/j.jmb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamenska A, Simpson C, Vindry C, Broomhead H, Benard M, Ernoult-Lange M, Lee BP, Harries LW, Weil D, Standart N. 2016. The DDX6-4E-T interaction mediates translational repression and P-body assembly. Nucleic Acids Res 44:6318–6334. 10.1093/nar/gkw565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Treeck B, Parker R. 2018. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174:791–802. 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22. 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 12.Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4:a012286. 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol Cell 32:605–615. 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Y, Na Z, Slavoff SA. 2018. P-bodies: composition, properties, and functions. Biochemistry 57:2424–2431. 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker R, Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25:635–646. 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Ivanov P, Kedersha N, Anderson P. 2019. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol 11:a032813. 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson P, Kedersha N. 2006. RNA granules. J Cell Biol 172:803–808. 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoecklin G, Kedersha N. 2013. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol 768:197–211. 10.1007/978-1-4614-5107-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev 26:91–107. 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 21.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 10:778–790. 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 22.Baggen J, Thibaut HJ, Strating J, van Kuppeveld FJM. 2018. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol 16:368–381. 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 23.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. 2014. Picornavirus morphogenesis. Microbiol Mol Biol Rev 78:418–437. 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dougherty JD, White JP, Lloyd RE. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol 85:64–75. 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chahar HS, Chen S, Manjunath N. 2013. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 436:1–7. 10.1016/j.virol.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85:6882–6892. 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma NR, Majerciak V, Kruhlak MJ, Yu L, Kang JG, Yang A, Gu S, Fritzler MJ, Zheng ZM. 2019. KSHV RNA-binding protein ORF57 inhibits P-body formation to promote viral multiplication by interaction with Ago2 and GW182. Nucleic Acids Res 47:9368–9385. 10.1093/nar/gkz683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baltera RF, Tershak DR. 1989. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J Virol 63:4441–4444. 10.1128/jvi.63.10.4441-4444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenneth E, Murray MLN. 2007. Guanidine hydrochloride inhibits mammalian orthoreovirus growth by reversibly blocking the synthesis of double-stranded RNA. J Virol 81:4572–4584. 10.1128/JVI.02106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Hu Z, Fan S, Zhang Q, Zhong Y, Guo D, Qin Y, Chen M. 2018. Picornavirus 2A protease regulates stress granule formation to facilitate viral translation. PLoS Pathog 14:e1006901. 10.1371/journal.ppat.1006901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. 2009. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci 16:103. 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Li G, Yuan S, Gao Q, Lan K, Altmeyer R, Zou G. 2016. In vitro assessment of combinations of enterovirus inhibitors against enterovirus 71. Antimicrob Agents Chemother 60:5357–5367. 10.1128/AAC.01073-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao X, Guo X, Yin M, Tariq M, Lai Y, Kanwal S, Zhou J, Li N, Lv Y, Pulido-Quetglas C, Wang X, Ji L, Khan MJ, Zhu X, Luo Z, Shao C, Lim DH, Liu X, Li N, Wang W, He M, Liu YL, Ward C, Wang T, Zhang G, Wang D, Yang J, Chen Y, Zhang C, Jauch R, Yang YG, Wang Y, Qin B, Anko ML, Hutchins AP, Sun H, Wang H, Fu XD, Zhang B, Esteban MA. 2018. Capturing the interactome of newly transcribed RNA. Nat Methods 15:213–220. 10.1038/nmeth.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozgur S, Basquin J, Kamenska A, Filipowicz W, Standart N, Conti E. 2015. Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep 13:703–711. 10.1016/j.celrep.2015.09.033. [DOI] [PubMed] [Google Scholar]
- 35.Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37. 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 36.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3' UTR structures. RNA Biol 8:1173–1186. 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jangra RK, Yi M, Lemon SM. 2010. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol 84:6810–6824. 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85:6882–6892. 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheller N, Mina LB, Galão RP, Chari A, Giménez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA 106:13517–113522. 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Wang L, Rivera-Serrano EE, Chen X, Lemon SM. 2019. TNRC6 proteins modulate hepatitis C virus replication by spatially regulating the binding of miR-122/Ago2 complexes to viral RNA. Nucleic Acids Res 47:6411–6424. 10.1093/nar/gkz278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381–392. 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 42.Jungfleisch J, Chowdhury A, Alves-Rodrigues I, Tharun S, Diez J. 2015. The Lsm1-7-Pat1 complex promotes viral RNA translation and replication by differential mechanisms. RNA 21:1469–1479. 10.1261/rna.052209.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilanchone V, Clemens K, Kaake R, Dawson AR, Matheos D, Nagashima K, Sitlani P, Patterson K, Chang I, Huang L, Sandmeyer S. 2015. Ty3 retrotransposon hijacks mating yeast RNA processing bodies to infect new genomes. PLoS Genet 11:e1005528. 10.1371/journal.pgen.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castello A, Izquierdo JM, Welnowska E, Carrasco L. 2009. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J Cell Sci 122:3799–3809. 10.1242/jcs.055988. [DOI] [PubMed] [Google Scholar]
- 45.Zamora M, Marissen WE, Lloyd RE. 2002. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. J Virol 76:165–177. 10.1128/JVI.76.1.165-177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu YY, Liu YN, Lu WW, Kung SH. 2009. Visualizing and quantifying the differential cleavages of the eukaryotic translation initiation factors eIF4GI and eIF4GII in the enterovirus-infected cell. Biotechnol Bioeng 104:1142–1152. 10.1002/bit.22495. [DOI] [PubMed] [Google Scholar]